ABSTRACT
The environmental transport of Cryptosporidium spp. through combined sewer overflow (CSO) and the occurrence of several emerging human-pathogenic Cryptosporidium species in developing countries remain unclear. In this study, we collected 40 CSO samples and 40 raw wastewater samples from Shanghai, China, and examined them by PCR and DNA sequencing for Cryptosporidium species (targeting the small subunit rRNA gene) and Giardia duodenalis (targeting the triosephosphate isomerase, β-giardin, and glutamate dehydrogenase genes) and Enterocytozoon bieneusi (targeting the ribosomal internal transcribed spacer) genotypes. Human-pathogenic Cryptosporidium species were further subtyped by sequence analysis of the 60-kDa glycoprotein gene, with additional multilocus sequence typing on the emerging zoonotic pathogen Cryptosporidium ubiquitum. Cryptosporidium spp., G. duodenalis, and E. bieneusi were detected in 12 and 15, 33 and 32, and 37 and 40 CSO and wastewater samples, respectively, including 10 Cryptosporidium species, 3 G. duodenalis assemblages, and 8 E. bieneusi genotypes. In addition to Cryptosporidium hominis and Cryptosporidium parvum, two new pathogens identified in industrialized nations, C. ubiquitum and Cryptosporidium viatorum, were frequently detected. The two novel C. ubiquitum subtype families identified appeared to be genetic recombinants of known subtype families. Similarly, the dominant group 1 E. bieneusi genotypes and G. duodenalis subassemblage AII are known human pathogens. The similar distribution of human-pathogenic Cryptosporidium species and E. bieneusi and G. duodenalis genotypes between wastewater and CSO samples reaffirms that storm overflow is potentially a significant contamination source of pathogens in surface water. The frequent identification of C. ubiquitum and C. viatorum in urban wastewater suggests that these newly identified human pathogens may be endemic in China.
IMPORTANCE Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi are major waterborne pathogens. Their transport into surface water through combined sewer overflow, which remains largely untreated in developing countries, has not been examined. In addition, the identification of these pathogens to genotypes and subtypes in urban storm overflow and wastewater is necessary for rapid and accurate assessment of pathogen transmission in humans and transport in the environment. Data from this study suggest that, like untreated urban wastewater, combined sewer overflow is commonly contaminated with human-pathogenic Cryptosporidium, G. duodenalis, and E. bieneusi genotypes and subtypes, and urban storm overflow potentially plays a significant role in the contamination of drinking source water and recreational water with human pathogens. They also indicate that Cryptosporidium ubiquitum and Cryptosporidium viatorum, two newly identified human pathogens, may be common in China, and genetic recombination can lead to the emergence of novel C. ubiquitum subtype families.
KEYWORDS: Cryptosporidium, Giardia duodenalis, Enterocytozoon bieneusi, wastewater, combined sewer overflow, environmental transport, microbial source tracking
INTRODUCTION
Waterborne pathogens, such as Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi, are significant causes of diarrhea in children worldwide, especially in developing countries. Cryptosporidium spp. and E. bieneusi further cause high morbidity and mortality in immunocompromised persons, such as HIV patients, cancer patients, and organ transplant recipients (1, 2). The presence of these pathogens in the environment is a major threat to public health. Humans can be infected with these parasites via contaminated food, water, and fomites or physical contact with infected persons or animals. Nearly 90% of documented outbreaks of these pathogens are associated with water (3).
The environmental ecology of these waterborne parasites remains unclear, especially in developing countries. In industrialized nations, urban wastewater discharge appears to play an important role in their environmental transport, as is well demonstrated in the investigation of the massive cryptosporidiosis outbreak in Milwaukee, WI, in 1993, which was attributed to contamination of drinking source water by untreated urban wastewater due to an extreme weather event (4). The role of wastewater and storm water in environmental transport of waterborne pathogens, especially emerging Cryptosporidium species in developing countries, however, has not been thoroughly examined. There is also a need to characterize these pathogens using molecular tools for rapid and accurate assessment of their environmental transport.
Shanghai, one of the most populous cities in China, has numerous pump stations to collect raw wastewater and storm water to wastewater treatment plants. In some districts, a combined sewer overflow (CSO) system is used in wastewater disposal, in which runoff from rains is collected using the same network for sewage and other types of wastewater. After heavy rains, the wastewater mixed with storm runoff is discharged directly to a network of streams and creeks, as its volume exceeds the capacity of the reservoirs in pump stations. These streams and creeks are connected to the Huangpu River, which is one of the drinking water sources in Shanghai. Although the city is located near the outlet of the river, storm overflow at the upstream suburbs can potentially contaminate drinking source water and recreational water.
There are many reports of Cryptosporidium spp. and G. duodenalis in wastewater in recent years, but characterizations of pathogens using molecular methods are scarce. In China, one study characterized Cryptosporidium spp., G. duodenalis, and E. bieneusi in raw wastewater samples from four cities using PCR-sequencing tools (5), whereas another genotyped and subtyped Cryptosporidium spp. and G. duodenalis in wastewater samples in another city using the same approach (6). The results of the two studies suggest that wastewater may contribute to environmental contamination of waterborne pathogens. The role of CSO in the environmental transport of these major waterborne pathogens in China and elsewhere has not been examined using molecular tools.
This study was conducted to evaluate the transport of human-pathogenic Cryptosporidium spp., G. duodenalis, and E. bieneusi in Shanghai by genotyping and subtyping pathogens in both untreated wastewater and CSO. The identification of pathogens in wastewater and CSO at the genotype and subtype levels provides much needed data on the genetic diversity and environmental transport of major waterborne pathogens as well as the occurrence of some newly identified human-pathogenic Cryptosporidium species, such as Cryptosporidium ubiquitum and Cryptosporidium viatorum, in developing countries. Results of multilocus sequencing typing (MLST) indicate that genetic recombination may be responsible for the emergence of novel C. ubiquitum subtype families.
RESULTS
Occurrence of Cryptosporidium spp.
PCR analysis revealed that 12 (30.0%) CSO samples and 15 (37.5%) wastewater samples were positive for Cryptosporidium spp. (P = 0.637 for the difference). Based on sequence analysis of the partial small-subunit (SSU) rRNA gene, seven species were identified in both CSO and wastewater samples, including Cryptosporidium hominis (5/40 and 4/40, respectively), C. ubiquitum (4/40 and 5/40), C. viatorum (4/40 and 3/40), Cryptosporidium parvum (2/40 and 4/40), Cryptosporidium muris (2/40 and 3/40), Cryptosporidium meleagridis (3/40 and 1/40), and Cryptosporidium baileyi (1/40 and 3/40). Three other Cryptosporidium species or genotypes, including Cryptosporidium felis (1/40), rat genotype I (1/40), and rat genotype IV (1/40), were each found in one wastewater sample (Table 1).
TABLE 1.
Occurrence of Cryptosporidium spp. in 40 combined sewer overflow samples and 40 wastewater samples from Shanghai, China
| Species/genotype or subtype | No. of positive samples (no. of positive PCR products) |
|
|---|---|---|
| CSO | Wastewater | |
| Species in humans | ||
| C. hominis | 5 (8) | 4 (8) |
| IaA18R4 | 1 (5) | 1 (1) |
| IbA19G2 | 1 (2) | 1 (5) |
| C. viatorum | 4 (8) | 3 (3) |
| XVaA6 | 1 (1) | |
| Species in domestic animals | ||
| C. parvum | 2 (4) | 4 (4) |
| IIdA19G1 | 1 (1) | |
| C. felis | 1 (1) | |
| Species in birds | ||
| C. baileyi | 1 (1) | 3 (5) |
| C. meleagridis | 3 (3) | 1 (1) |
| IIIbA22G1R1c | 1 (1) | |
| Species in rodents | ||
| C. ubiquitum | 4 (4) | 5 (7) |
| XIIg | 3 (4) | |
| XIIh | 1 (1) | |
| C. muris | 2 (2) | 3 (3) |
| Rat genotype I | 1 (1) | |
| Rat genotype IV | 1 (1) | |
| Totala (%) | 12/40 (30.0)b | 15/40 (37.5)b |
A sample containing multiple genotypes was counted as one positive case.
P = 0.637 for the difference between CSO and wastewater samples.
Subtype distribution of Cryptosporidium spp.
Two CSO samples and two wastewater samples containing C. hominis and one wastewater sample containing C. parvum were successfully amplified at the gp60 locus. Two distinct C. hominis subtypes (IaA18R4 and IbA19G2) and one C. parvum subtype (IIdA19G1) were identified (Table 1).
Four C. ubiquitum-positive wastewater samples produced the expected gp60 PCR product, leading to the identification of two new subtype families that were named as XIIg and XIIh according to the established nomenclature (7). XIIg had 88% to 91% nucleotide sequence similarity to known subtype families XIIa to XIId, whereas XIIh appeared to be a genetic recombinant between XIId and XIIc, with sequence mostly identical to XIId except for the 150-bp sequence at the 3′ end, which was identical to XIIc. In phylogenetic analysis, XIIh clustered with XIId, whereas XIIg was placed outside the clade containing XIId and XIIc (Fig. 1).
FIG 1.
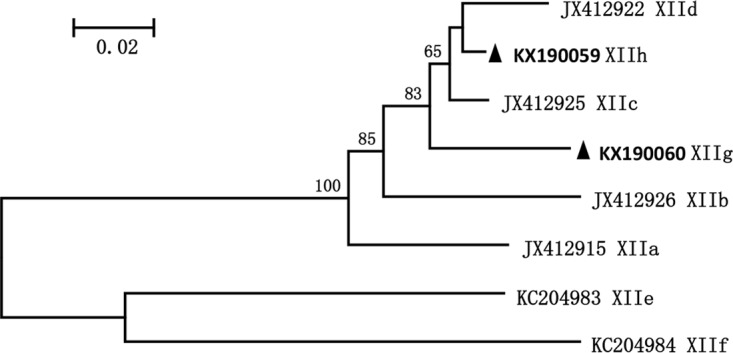
Phylogenetic relationship of novel Cryptosporidium ubiquitum subtype families detected in wastewater and combined sewer overflow samples as determined by a neighbor-joining analysis of gp60 nucleotide sequences based on Kimura 2-parameter distances. Genotypes with ▲ are novel genotypes found in this study.
One wastewater sample positive for C. meleagridis was successfully amplified and subtyped as IIIbA22G1R1c (GenBank accession number KJ210617) based on DNA sequence analysis. Similarly, one C. viatorum-positive wastewater sample was identified as a new subtype, XVaA6, according to the established nomenclature (8).
Cryptosporidium ubiquitum MLSTs.
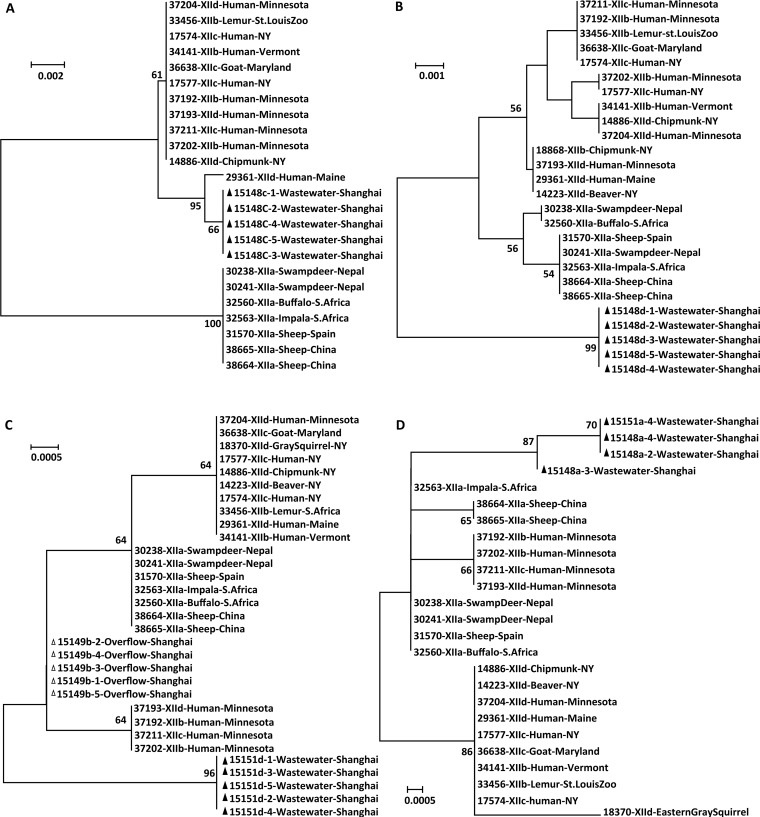
Two wastewater samples and one CSO sample were positive for C. ubiquitum as MLST analysis indicated. As they belonged to two new subtype families, these C. ubiquitum samples were further compared with data from known subtype families at four genetic loci. (i) At the cgd6_60 locus, all five sequences obtained from one wastewater sample were similar to the sequence KX28286380 obtained from a human patient infected with the C. ubiquitum XIId subtype family in Maine, with two nucleotide substitutions. In phylogenetic analysis, they formed a cluster that is related to other XIIb, XIIc, and XIId subtype families but without clear separation of cgd6_60 sequences from these subtype families (Fig. 2A). (ii) At the cgd6_1590 locus, all five sequences obtained from one wastewater sample were identical to each other and had 93% to 98% nucleotide sequence similarity to those from XIIa, XIIb, XIIc, and XIId. In phylogenetic analysis, they formed a cluster that was divergent from clusters formed by cgd6_1590 sequences from other subtype families. There was no consistent separation of cgd6_1590 sequences from XIIb, XIIc, and XIId subtype families (Fig. 2B). (iii) At the cgd2_3690 locus, five sequences obtained from one CSO sample were identical to each other and had only one nucleotide difference from cgd2_3690 sequences from XIIa, where the other five sequences from a wastewater sample had 5 to 6 nucleotide differences from XIIa and other subtype families. In phylogenetic analysis, both formed their own clusters. There was no consistent separation of cgd2_3690 sequences from XIIb, XIIc, and XIId subtype families (Fig. 2C). (iv) At the cgd4_370 locus, the four sequences obtained from two wastewater samples were almost identical to each other, with only 2 to 3 nucleotide differences from the cgd4_370 sequences previously obtained from XIIa. In phylogenetic analysis, they formed a cluster that was divergent from clusters formed by other subtype families. There was no consistent clustering of cgd4_370 sequences from subtype families XIIb, XIIc, and XIId (Fig. 2D).
FIG 2.
Phylogenetic relationship of Cryptosporidium ubiquitum in wastewater and combined sewer overflow samples at the cgd6_60 (A), cgd6_1590 (B), cgd2_3690 (C), and cgd4_370 (D) loci as determined by neighbor-joining analyses of nucleotide sequences based on Kimura 2-parameter distances. Sequences with △ are from combined sewer overflow samples, and those with ▲ are from wastewater samples.
Occurrence of Giardia duodenalis assemblages and subtypes.
Among the 40 wastewater samples, 32, 30, and 29 were PCR positive at the tpi, gdh, and bg loci, respectively. Similarly, of the 40 CSO samples, 33, 30, and 31 were PCR positive at these loci, respectively. The most commonly detected subtype in wastewater and CSO was A2 at all three genetic loci, indicating that it belonged to subassemblage AII. In addition, assemblage B, assemblage G, and three new assemblage A subtypes related to A2 were seen in a few wastewater and CSO samples (Table 2).
TABLE 2.
Assemblages and subtypes of Giardia duodenalis in 40 combined sewer overflow samples and 40 wastewater samples from Shanghai, China, as indicated by sequence analysis of three genetic loci (tpi, gdh, and bg)
| Source | Genotype or subtype (accession no.) | No. of positive samples (no. of positive PCR products) |
||
|---|---|---|---|---|
| tpi | gdh | bg | ||
| CSO | A2 (tpi, U57897; gdh, AY178737; bg, AY072723) | 28 (47) | 29 (51) | 27 (43) |
| A3 (FJ971415) | 4 (5) | |||
| A2-like (1 SNP compared to KJ888992) | 2 (3) | |||
| B (1 SNP compared to KP687778) | 1 (1) | |||
| G (2 SNPs compared to EU781013) | 1 (1) | |||
| A (AB516350) | 2 (2) | |||
| Subtotal | 33 | 30 | 31 | |
| Wastewater | A2 (tpi, U57897; gdh, AY178737; bg, AY072723) | 30 (44) | 30 (48) | 23 (37) |
| A3 (FJ971415) | 5 (5) | |||
| B (KP687778) | 1 (1) | |||
| A (AB516350) | 2 (2) | |||
| Subtotal | 32 | 30 | 29 | |
Occurrence of Enterocytozoon bieneusi genotypes.
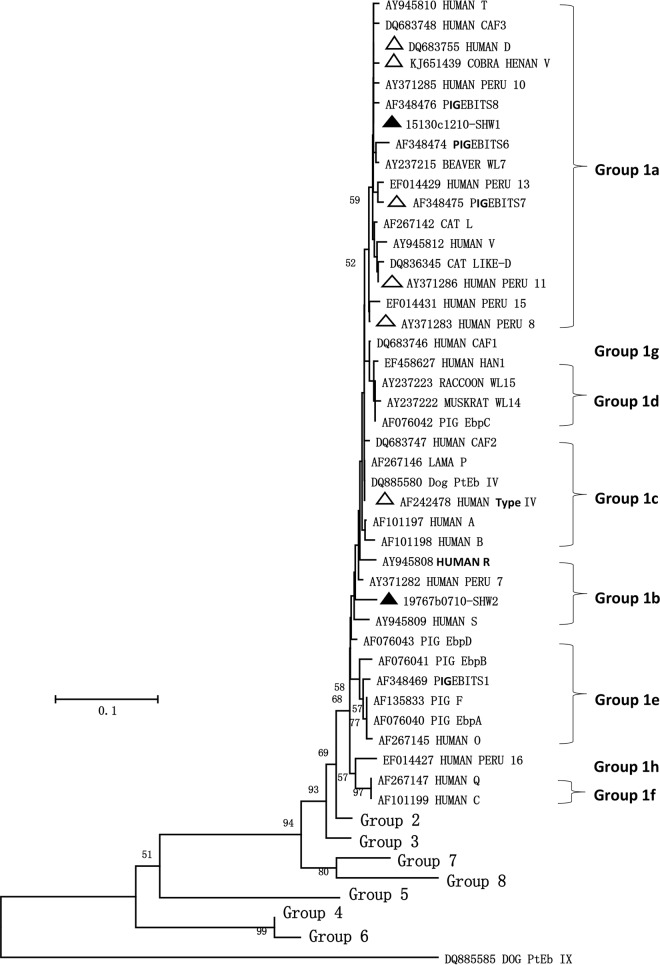
E. bieneusi was detected in all 40 wastewater samples and 37 (92.5%) of the 40 CSO samples. Eight E. bieneusi genotypes were detected in wastewater samples, including D, PigEBITS7, Henan V, type IV, Peru 8, Peru 11, and two new genotypes SHW1 and SHW2. Among them, 17 samples had two or more E. bieneusi genotypes. These genotypes were also found in CSO samples, except SHW1 (Table 3). Among genotypes detected in this study, D, PigEBITS7, Henan V, Peru 8, Peru 11, and SHW1 belonged to subgroup 1a; SHW2 belonged to subgroup 1b; and type IV belonged to subgroup 1c (Fig. 3).
TABLE 3.
Genotype of Enterocytozoon bieneusi in 40 combined sewer overflow samples and 40 wastewater samples from Shanghai, China
| Genotypea | GenBank accession no.b | No. of detections in CSO | No. of detections in wastewater |
|---|---|---|---|
| D | DQ683755 | 17 | 20 |
| PigEBITS7 | AF348475 | 0 | 1 |
| Peru 11 | AY371286 | 1 | 0 |
| Type IV | AF242478 | 0 | 1 |
| SHW2 | KX190063 | 0 | 1 |
| D, PigEBITS7 | 7 | 5 | |
| D, SHW1 | 3 | 1 | |
| D, SHW2 | 0 | 1 | |
| D, Peru 8 | 0 | 1 | |
| D, Peru 11 | 0 | 1 | |
| D, Henan V | 7 | 7 | |
| D, PigEBITS7, SHW1 | 0 | 1 | |
| D, Peru 8, type IV | 1 | 0 | |
| D, Peru 8, Peru 11, Henan V | 1 | 0 | |
| Total | 37 | 40 |
FIG 3.
Phylogenetic analysis of Enterocytozoon bieneusi genotypes detected in wastewater and combined sewer overflow as determined by a neighbor-joining analysis of ribosomal ITS sequences based on Kimura 2-parameter distances. Genotypes with △ are known genotypes detected in wastewater and combined sewer overflow samples, and those with ▲ are novel genotypes found in this study.
DISCUSSION
In this study, the transport of waterborne pathogens Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi through CSO and the occurrence of several emerging Cryptosporidium species in China were examined using molecular diagnostic tools. The genetic characteristics of C. ubiquitum and C. viatorum were further assessed using newly established subtyping and MLST tools. Data from the study suggest that the same waterborne pathogens presented in urban wastewater are frequently detected in CSO. The most frequently identified Cryptosporidium species and G. duodenalis and E. bieneusi genotypes included C. hominis, C. parvum, C. meleagridis, G. duodenalis assemblages A, and E. bieneusi group 1 genotypes, which are all recognized human pathogens (9–12). This is similar to the observations of previous studies of wastewater samples from Shanghai and other cities in China (5, 6, 13) but different from other studies conducted in cities with large slaughterhouses, where some Cryptosporidium species from farm animals, such as Cryptosporidium andersoni, are frequently detected in wastewater samples (14, 15). The human origin of the pathogens in the present study is apparently responsible for the similar distribution of Cryptosporidium, G. duodenalis, and E. bieneusi genotypes between wastewater and CSO. The discharge of CSO into the river system is obviously a public health issue that remains to be addressed fully in urban areas of developing countries.
Subtyping Cryptosporidium spp. and G. duodenalis supports the human-origin nature of dominant pathogens in CSO. The C. hominis (IaA18R4 and IbA19G2), C. parvum (IIdA19G1), and G. duodenalis (AII) subtypes found in the present study are dominant subtypes of these pathogens in humans in Shanghai (16, 17). In contrast, G. duodenalis subassemblage AI, which commonly infects animals, was not detected in both wastewater and CSO.
Phylogenetic analysis of E. bieneusi sequences lends further support to the human-origin nature of common pathogens in wastewater and CSO. Six known genotypes of E. bieneusi (i.e., D, PigEBITS7, Henan V, Peru 8, Peru 11, and type IV) were found in this study. Among them, Peru 8 and Peru 11 have been reported only in humans (18), whereas others infect humans as well as animals. Within group 1, except for type IV, which is zoonotic and belongs to subgroup 1c, all of the known genotypes in this study belong to subgroup 1a. One of the two novel genotypes, SHW1, also belonged to subgroup 1a, whereas the other one, SHW2, belonged to subgroup 1b. Genotypes in subgroups 1a, 1b, and 1c are well-known E. bieneusi genotypes that infect humans, even though they are also found in some animals (19). In contrast, some host-adapted group 1 genotypes, such as those in subgroup 1e, which are common pathogens in pigs, were not found in this study. The dominant E. bieneusi genotype in wastewater and CSO in this study, genotype D, thus far has not been identified in humans in China because of the lack of studies. However, it was identified as the dominant E. bieneusi genotype in raw wastewater in Shanghai and other cities and in nonhuman primates in China, indicating that it may also be a major human pathogen in China (5, 20).
Two emerging human-pathogenic Cryptosporidium species, including C. ubiquitum and C. viatorum, were found in both wastewater and CSO samples at a higher frequency than other species. C. ubiquitum is a common parasite of small ruminants and rodents. However, it is becoming an important Cryptosporidium species in humans in industrialized nations (21, 22), although it has been reported in only a few human cases in developing countries (21, 23, 24). In contrast, C. viatorum is known as a human pathogen in developing countries, as human infections in the United Kingdom and Sweden have been linked to travels to India, Pakistan, Bangladesh, Kenya, Barbados, and Guatemala (8). However, its presence in developing countries has been confirmed only in Ethiopia and Nigeria (25, 26). In the present study, C. viatorum was detected at a frequency similar to those of C. hominis, C. parvum, and C. ubiquitum. The detection of C. ubiquitum and C. viatorum in wastewater and CSO suggests that these two Cryptosporidium species are probably circulating in humans in China, and waterborne transmission plays a potential role in their epidemiology. Previously, they were not detected in raw wastewater in Shanghai and three other major Chinese cities (5). As there have been increased population exchanges between China and African nations in recent years, further studies are needed to monitor their environmental transport and human infection in China.
Subtype analysis of C. ubiquitum and C. viatorum has shown the genetic uniqueness of these two Cryptosporidium species in China. The two C. ubiquitum subtypes found in this study belong to two novel subtype families (21), and the C. viatorum subtype (XVaA6) found in this study has not been reported elsewhere (8). In phylogenetic analysis of gp60 sequences, the new C. ubiquitum subtype families formed a cluster with the rodent subtype families XIIb, XIIc, and XIId rather than the ruminant subtype family XIIa. The genetic uniqueness of the new C. ubiquitum subtype families has been confirmed by data from a newly developed MLST analysis, with sequences from China clustered together at each of the four polymorphic loci in addition to gp60. Both gp60 sequence and MLST analyses suggest that the two new C. ubiquitum subtype families are probably genetic recombinants. Previously, phylogenetic analysis of sequences from these MLST loci also showed no consistent cluster formations among XIIb, XIIc, and XIId isolates (27). Thus, unlike the ruminant-adapted XIIa subtype family, XIIb, XIIc, and XIId from rodents and the two novel subtype families identified in China are all genetic recombinants and different subtype families identified in rodents at the gp60 locus are in fact members of the same host-adapted group. The MLST data further suggest that human C. ubiquitum infection in China may be caused by pathogens of rodent rather than ruminant origin as seen previously in the United States (21).
In summary, data from this study suggest that molecular diagnostic tools can be used effectively in characterizing the transport of human-pathogenic Cryptosporidium spp., G. duodenalis, and E. bieneusi through CSO, and the distribution of species and genotypes of these pathogens in CSO from Shanghai, China, is similar to that in wastewater. They confirm CSO as a threat to contamination of drinking source water in developing countries and suggest that it may be a vehicle for environmental transport of emerging human-pathogenic Cryptosporidium species, such as C. viatorum and C. ubiquitum. Genetic recombination appears to be a driving force for the emergence of novel C. ubiquitum gp60 subtype families. Efforts should be made to confirm the occurrence of these pathogens in humans in China and elucidate their environmental ecology.
MATERIALS AND METHODS
Study sites.
Two wastewater pump stations located in the Yangpu and Hongkou districts in Shanghai were studied. The population densities in the two districts are 21,747 people/km2 and 36,014 people/km2, respectively. The areas studied adopt a CSO system in wastewater disposal, in which storm water and wastewater are not separated as in other districts of the inner city. In days with heavy rains, storm water mixed with sewage is discharged into a network of streams, which flow into the Huangpu River.
Sample collection and processing.
Grab samples of untreated wastewater and CSO were used in this study. A total of 80 samples were collected from the two pump stations from June through September in 2012 and 2014. Among the 80 samples, 40 were CSO samples collected in days with heavy rains and 40 samples were wastewater samples collected from the same sites 1 day after rain events, with one sample at each site each time. Each sample was collected using a 3-liter plastic container. The 3-liter samples were transported to the laboratory immediately, and pathogens in them were concentrated by centrifugation at 1,500 × g for 20 min. After the process, 600 to 1,000 μl of pellet was generated. Half (300 to 500 μl) of the pellet from each sample was used in DNA extraction immediately or stored at −80°C for less than 2 weeks.
DNA extraction and pathogen detection by PCR.
DNA was extracted from the sample concentrates using a FastDNA SPIN kit for soil (BIO 101, Carlsbad, CA) and eluted into 100 μl of reagent-grade water. The extracted DNA was kept at −20°C before being used in PCR analysis. Each sample was analyzed in quintuplicate using 2 μl of DNA per PCR, with the inclusion of two positive PCR controls and one negative PCR control. No measurement of the DNA concentration was made prior to PCR analysis.
To detect and identify Cryptosporidium spp., an ∼580-bp fragment of the small-subunit (SSU) rRNA gene was amplified by nested PCR (28). Specimens that were positive for C. hominis or C. parvum were subtyped by nested PCR analysis of the 60-kDa glycoprotein (gp60) gene (7). Those containing C. meleagridis, C. ubiquitum, and C. viatorum were subtyped at the gp60 locus using PCR methods developed specifically for these human pathogens (8, 21, 29). C. ubiquitum was further characterized by a newly established MLST method (27). Only four polymorphic loci, including the cgd2_3690, cgd4_370, cgd6_60, and cgd6_1590 genes, were targeted.
The detection of G. duodenalis was conducted using nested PCR analysis of the triosephosphate isomerase (tpi), β-giardin (bg), and glutamate dehydrogenase (gdh) genes as described previously (30). Nested PCR analysis of an ∼500-bp fragment of the rRNA gene containing the entire internal transcribed spacer (ITS) was used in the detection of E. bieneusi (5).
Genotyping and subtyping of pathogens by sequence analysis.
To identify genotypes and subtypes of pathogens, all positive secondary PCR products from the genetic loci described above were sequenced directly using the BigDye Terminator v3.1 cycle sequencing kits (Applied Biosystems, Foster City, CA). The sequence accuracy was confirmed by bidirectional sequencing. The sequences were read on an ABI3170 genetic analyzer (Applied Biosystems) and assembled using ChromasPro (v1.33) software (http://technelysium.com.au). Nucleotide sequences obtained were aligned with references of each gene downloaded from GenBank using ClustalX (http://www.clustal.org) to determine the genotype or subtype of these pathogens. The accepted terminology was used in naming Cryptosporidium subtypes and E. bieneusi genotypes (31, 32). Neighbor-joining analysis implemented in Mega 6.0 (http://www.megasoftware.net) was used to assess the phylogenetic positions of new C. ubiquitum subtype families and E. bieneusi genotypes found in this study based on genetic distances calculated using the Kimura 2-parameter model.
Statistical analysis.
The frequency of Cryptosporidium occurrence between wastewater and overflow was compared using the χ2 test implemented in SPSS 19.0 for Windows (SPSS, Inc., Chicago, IL, USA). Differences with a P value of <0.05 were considered significant.
Accession number(s).
Representative nucleotide sequences generated from the study were submitted to GenBank under accession numbers KX190059 to KX190063 and KY676886 to KY676890.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (31630078, 31425025, 31502055, 31602042), the China Postdoctoral Science Foundation (grant 2016M601531), and the Water Special Project (2014ZX07104006).
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the U.S. Centers for Disease Control and Prevention.
REFERENCES
- 1.Didier ES. 2005. Microsporidiosis: an emerging and opportunistic infection in humans and animals. Acta Trop 94:61–76. doi: 10.1016/j.actatropica.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 2.Chalmers RM, Davies AP. 2010. Minireview: clinical cryptosporidiosis. Exp Parasitol 124:138–146. doi: 10.1016/j.exppara.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 3.Baldursson S, Karanis P. 2011. Waterborne transmission of protozoan parasites: review of worldwide outbreaks—an update 2004–2010. Water Res 45:6603–6614. doi: 10.1016/j.watres.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 4.Mac Kenzie WR, Hoxie NJ, Proctor ME, Gradus MS, Blair KA, Peterson DE, Kazmierczak JJ, Addiss DG, Fox KR, Rose JB, Davis JP. 1994. A massive outbreak in Milwaukee of Cryptosporidium infection transmitted through the public water supply. N Engl J Med 331:161–167. doi: 10.1056/NEJM199407213310304. [DOI] [PubMed] [Google Scholar]
- 5.Li N, Xiao L, Wang L, Zhao S, Zhao X, Duan L, Guo M, Liu L, Feng Y. 2012. Molecular surveillance of Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi by genotyping and subtyping parasites in wastewater. PLoS Negl Trop Dis 6:e1809. doi: 10.1371/journal.pntd.0001809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu A, Ji H, Wang E, Liu J, Xiao L, Shen Y, Li Y, Zhang W, Ling H. 2011. Molecular identification and distribution of Cryptosporidium and Giardia duodenalis in raw urban wastewater in Harbin, China. Parasitol Res 109:913–918. doi: 10.1007/s00436-011-2333-4. [DOI] [PubMed] [Google Scholar]
- 7.Feng Y, Lal AA, Li N, Xiao L. 2011. Subtypes of Cryptosporidium spp. in mice and other small mammals. Exp Parasitol 127:238–242. doi: 10.1016/j.exppara.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 8.Stensvold CR, Elwin K, Winiecka-Krusnell J, Chalmers RM, Xiao L, Lebbad M. 2015. Development and application of a gp60-based typing assay for Cryptosporidium viatorum. J Clin Microbiol 53:1891–1897. doi: 10.1128/JCM.00313-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiao L, Feng Y. 2008. Zoonotic cryptosporidiosis. FEMS Immunol Med Microbiol 52:309–323. doi: 10.1111/j.1574-695X.2008.00377.x. [DOI] [PubMed] [Google Scholar]
- 10.Xiao L. 2010. Molecular epidemiology of cryptosporidiosis: an update. Exp Parasitol 124:80–89. doi: 10.1016/j.exppara.2009.03.018. [DOI] [PubMed] [Google Scholar]
- 11.Feng Y, Xiao L. 2011. Zoonotic potential and molecular epidemiology of Giardia species and giardiasis. Clin Microbiol Rev 24:110–140. doi: 10.1128/CMR.00033-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matos O, Lobo ML, Xiao L. 2012. Epidemiology of Enterocytozoon bieneusi infection in humans. J Parasitol Res 2012:981424. doi: 10.1155/2012/981424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng Y, Li N, Duan L, Xiao L. 2009. Cryptosporidium genotype and subtype distribution in raw wastewater in Shanghai, China: evidence for possible unique Cryptosporidium hominis transmission. J Clin Microbiol 47:153–157. doi: 10.1128/JCM.01777-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou L, Singh A, Jiang J, Xiao L. 2003. Molecular surveillance of Cryptosporidium spp. in raw wastewater in Milwaukee: implications for understanding outbreak occurrence and transmission dynamics. J Clin Microbiol 41:5254–5257. doi: 10.1128/JCM.41.11.5254-5257.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ben Ayed L, Yang W, Widmer G, Cama V, Ortega Y, Xiao L. 2012. Survey and genetic characterization of wastewater in Tunisia for Cryptosporidium spp., Giardia duodenalis, Enterocytozoon bieneusi, Cyclospora cayetanensis and Eimeria spp. J Water Health 10:431–444. doi: 10.2166/wh.2012.204. [DOI] [PubMed] [Google Scholar]
- 16.Feng Y, Wang L, Duan L, Gomez-Puerta LA, Zhang L, Zhao X, Hu J, Zhang N, Xiao L. 2012. Extended outbreak of cryptosporidiosis in a pediatric hospital, China. Emerg Infect Dis 18:312–314. doi: 10.3201/eid1802.110666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang L, Zhang H, Zhao X, Zhang L, Zhang G, Guo M, Liu L, Feng Y, Xiao L. 2013. Zoonotic Cryptosporidium species and Enterocytozoon bieneusi genotypes in HIV-positive patients on antiretroviral therapy. J Clin Microbiol 51:557–563. doi: 10.1128/JCM.02758-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Santín M, Fayer R. 2011. Microsporidiosis: Enterocytozoon bieneusi in domesticated and wild animals. Res Vet Sci 90:363–371. doi: 10.1016/j.rvsc.2010.07.014. [DOI] [PubMed] [Google Scholar]
- 19.Thellier M, Breton J. 2008. Enterocytozoon bieneusi in human and animals, focus on laboratory identification and molecular epidemiology. Parasite 15:349–358. doi: 10.1051/parasite/2008153349. [DOI] [PubMed] [Google Scholar]
- 20.Karim MR, Zhang S, Jian F, Li J, Zhou C, Zhang L, Sun M, Yang G, Zou F, Dong H, Rume FI, Qi M, Wang R, Ning C, Xiao L. 2014. Multilocus typing of Cryptosporidium spp. and Giardia duodenalis from non-human primates in China. Int J Parasitol 44:1039–1047. doi: 10.1016/j.ijpara.2014.07.006. [DOI] [PubMed] [Google Scholar]
- 21.Li N, Xiao L, Alderisio K, Elwin K, Cebelinski E, Chalmers R, Santin M, Fayer R, Kvac M, Ryan U, Sak B, Stanko M, Guo Y, Wang L, Zhang L, Cai J, Roellig D, Feng Y. 2014. Subtyping Cryptosporidium ubiquitum, a zoonotic pathogen emerging in humans. Emerg Infect Dis 20:217–224. doi: 10.3201/eid2002.121797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elwin K, Hadfield SJ, Robinson G, Crouch ND, Chalmers RM. 2012. Cryptosporidium viatorum n. sp. (Apicomplexa: Cryptosporidiidae) among travellers returning to Great Britain from the Indian subcontinent, 2007–2011. Int J Parasitol 42:675–682. doi: 10.1016/j.ijpara.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 23.Moore CE, Elwin K, Phot N, Seng C, Mao S, Suy K, Kumar V, Nader J, Bousfield R, Perera S, Bailey JW, Beeching NJ, Day NPJ, Parry CM, Chalmers RM. 2016. Molecular characterization of Cryptosporidium species and Giardia duodenalis from symptomatic Cambodian children. PLoS Negl Trop Dis 10:e0004822. doi: 10.1371/journal.pntd.0004822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Molloy SF, Smith HV, Kirwan P, Nichols RAB, Asaolu SO, Connelly L, Holland CV. 2010. Identification of a high diversity of Cryptosporidium species genotypes and subtypes in a pediatric population in Nigeria. Am J Trop Med Hyg 82:608–613. doi: 10.4269/ajtmh.2010.09-0624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ayinmode AB, Zhang H, Dada-Adegbola HO, Xiao L. 2014. Cryptosporidium hominis subtypes and Enterocytozoon bieneusi genotypes in HIV-infected persons in Ibadan, Nigeria. Zoonoses Public Health 61:297–303. doi: 10.1111/zph.12072. [DOI] [PubMed] [Google Scholar]
- 26.Adamu H, Petros B, Zhang G, Kassa H, Amer S, Ye J, Feng Y, Xiao L. 2014. Distribution and clinical manifestations of Cryptosporidium species and subtypes in HIV/AIDS patients in Ethiopia. PLoS Negl Trop Dis 8:e2831. doi: 10.1371/journal.pntd.0002831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang Y, Li N, Song M, Roellig DM, Feng Y, Xiao L. 2016. Development of a multilocus sequence typing tool for high-resolution subtyping and genetic structure characterization of Cryptosporidium ubiquitum. Infect Genet Evol 45:256–261. doi: 10.1016/j.meegid.2016.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu Y, Feng Y, Huang C, Xiao L. 2014. Occurrence, source, and human infection potential of Cryptosporidium and Enterocytozoon bieneusi in drinking source water in Shanghai, China, during a pig carcass disposal incident. Environ Sci Technol 48:14219–14227. doi: 10.1021/es504464t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stensvold CR, Beser J, Axén C, Lebbad M. 2014. High applicability of a novel method for gp60-based subtyping of Cryptosporidium meleagridis. J Clin Microbiol 52:2311–2319. doi: 10.1128/JCM.00598-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ye J, Xiao L, Wang Y, Guo Y, Roellig DM, Feng Y. 2015. Dominance of Giardia duodenalis assemblage A and Enterocytozoon bieneusi genotype BEB6 in sheep in Inner Mongolia, China. Vet Parasitol 210:235–239. doi: 10.1016/j.vetpar.2015.04.011. [DOI] [PubMed] [Google Scholar]
- 31.Santin M, Fayer R. 2009. Enterocytozoon bieneusi genotype nomenclature based on the internal transcribed spacer sequence: a consensus. J Eukaryot Microbiol 56:34–38. doi: 10.1111/j.1550-7408.2008.00380.x. [DOI] [PubMed] [Google Scholar]
- 32.Ryan U, Fayer R, Xiao L. 2014. Cryptosporidium species in humans and animals: current understanding and research needs. Parasitology 141:1667–1685. doi: 10.1017/S0031182014001085. [DOI] [PubMed] [Google Scholar]