ABSTRACT
The Duluth Complex in northeastern Minnesota hosts economically significant deposits of copper, nickel, and platinum group elements (PGEs). The primary sulfide mineralogy of these deposits includes the minerals pyrrhotite, chalcopyrite, pentlandite, and cubanite, and weathering experiments show that most sulfide-bearing rock from the Duluth Complex generates moderately acidic leachate (pH 4 to 6). Microorganisms are important catalysts for metal sulfide oxidation and could influence the quality of water from mines in the Duluth Complex. Nevertheless, compared with that of extremely acidic environments, much less is known about the microbial ecology of moderately acidic sulfide-bearing mine waste, and so existing information may have little relevance to those microorganisms catalyzing oxidation reactions in the Duluth Complex. Here, we characterized the microbial communities in decade-long weathering experiments (kinetic tests) conducted on crushed rock and tailings from the Duluth Complex. Analyses of 16S rRNA genes and transcripts showed that differences among microbial communities correspond to pH, rock type, and experimental treatment. Moreover, microbial communities from the weathered Duluth Complex rock were dominated by taxa that are not typically associated with acidic mine waste. The most abundant operational taxonomic units (OTUs) were from the genera Meiothermus and Sulfuriferula, as well as from diverse clades of uncultivated Chloroflexi, Acidobacteria, and Betaproteobacteria. Specific taxa, including putative sulfur-oxidizing Sulfuriferula spp., appeared to be primarily associated with Duluth Complex rock, but not pyrite-bearing rocks subjected to the same experimental treatment. We discuss the implications of these results for the microbial ecology of moderately acidic mine waste with low sulfide content, as well as for kinetic testing of mine waste.
IMPORTANCE Economic sulfide mineral deposits in the Duluth Complex may represent the largest undeveloped source of copper and nickel on Earth. Microorganisms are important catalysts for sulfide mineral oxidation, and research on extreme acidophiles has improved our ability to manage and remediate mine wastes. We found that the microbial assemblages associated with weathered rock from the Duluth Complex are dominated by organisms not widely associated with mine waste or mining-impacted environments, and we describe geochemical and experimental influences on community composition. This report will be a useful foundation for understanding the microbial biogeochemistry of moderately acidic mine waste from these and similar deposits.
KEYWORDS: 16S rRNA, Duluth Complex, Sulfuriferula, amplicon, humidity cell, microbial communities, mine waste, mining, pyrrhotite, sulfide mineral
INTRODUCTION
The Duluth Complex in northeastern Minnesota contains magmatic sulfide mineral deposits that are collectively estimated to represent one of the largest undeveloped sources of copper, nickel, and platinum group elements (PGEs) on Earth (1). Some of these economically significant deposits have been targeted for exploration and production. The most abundant sulfides in the Duluth Complex are pyrrhotite (Fe1-xS, where 0 ≤ x ≤ 0.2), chalcopyrite (CuFeS2), cubanite (CuFe2S3), and pentlandite [(Fe,Ni)9S8], with much smaller amounts of other Cu-, Ni-, and PGE-bearing sulfides (2, 3). The potential for sulfide minerals to generate sulfuric acid and release metals when exposed to air and water has raised concerns over degrading water quality in the region if these prospects were mined.
Microorganisms are important catalysts for sulfide mineral oxidation. Under extremely acidic conditions (pH <3), microbial activity can substantially accelerate sulfide mineral oxidation and intensify the generation of acidic drainage (see reference 4 for a recent review). Decades of research on the microbiology of metal sulfides have improved our management of sulfidic mine wastes (5, 6) and led to new strategies for mineral extraction (7, 8). Although important new microorganisms are still being discovered in low pH environments (e.g., see references 9 and 10), many of the taxa common to extremely acidic mine drainage are now well known (11–13), as are some of the environmental factors that control their distribution (e.g., see references 14–21).
By contrast, the Duluth Complex has a relatively low sulfide content, and its silicate mineralogy provides acid-neutralizing capacity. This type of mineral system is less documented than extremely acidic environments and presents a challenge with respect to predicting which microorganisms and microbial processes will occur in waste rock and tailings that could be produced from mining the Duluth Complex. Field and laboratory rock weathering experiments showed that leachate from Duluth Complex rock with sulfur contents below 1% is only moderately acidic (pH 4 to 6) and rarely reaches pH values less than 4 (22). Compared with extremely acidic environments, much less is known about the microbial ecology of moderately acidic sulfide-bearing mine waste. Microbial assemblages associated with circumneutral to moderately acidic waste rock and mine tailings are often characterized by the presence of sulfur-oxidizing microorganisms such as Thiobacillus denitrificans and Thiobacillus thioparus (23–26) but also often contain uncultivated taxa (23, 27, 28). The extent to which many of these populations impact the rate and products of sulfide mineral oxidation under moderately acidic to circumneutral conditions is not well studied. Although microorganisms are not thought to significantly accelerate the oxidation of the acid-insoluble sulfide pyrite under these conditions (26, 29–31, but see also reference 32), it is not clear if the same applies to pyrrhotite and other acid-soluble metal sulfides. To better predict the effect that microbial processes may have on water quality, there is a need to better understand the ecology and function of microbial communities in circumneutral and moderately acidic systems dominated by acid-soluble sulfide minerals.
The Minnesota Department of Natural Resources (MN DNR) is actively operating multiple laboratory and field rock weathering experiments to assess the potential for acid generation and release of metals and sulfate from existing and potential mine wastes. These experiments, some of which have been in operation for more than 35 years, provide an opportunity to study the microorganisms and microbial processes that are likely to occur during the weathering of sulfide-bearing waste rock and tailings generated from the Duluth Complex. In this study, we surveyed the microbial communities that developed during these weathering experiments to address the following questions. (i) What microbial taxa populate weathered Duluth Complex rock? (ii) How does the composition and diversity of the microbial assemblages relate to leachate geochemistry and other environmental factors? (iii) Do different laboratory and field weathering experiments (kinetic testing procedures) impact microbial community composition? (iv) Are certain microorganisms unique to the sulfide mineral assemblage of the Duluth Complex? We initially hypothesized that microbial communities in the weathering experiments would differ by pH and rock type and that more sulfur-oxidizing microorganisms and different sulfur-oxidizing taxa would occur in the Duluth Complex samples than in pyrite-bearing rock subjected to the same experimental treatments.
RESULTS
16S rRNA gene libraries.
We created 113 16S rRNA gene amplicon libraries from three different types of weathering experiments, including 12 humidity cell experiments, 16 reactors, and 3 field piles (Table 1). Humidity cells and reactors are laboratory kinetic tests in which crushed rock or tailings are weathered in short cylindrical columns (see Fig. S1 in the supplemental material for examples) (33, 34). Six of the humidity cells and two of the field rock piles contained Ely Greenstone (a pyrite-bearing greenschist from Northern Minnesota), and the rest of the experiments contained rock from the Duluth Complex. Libraries from the laboratory experiments created with the “full service” method (see Materials and Methods) had 10,559 to 478,165 sequences per sample, with an average library size of 116,899 sequences (standard deviation, 115,057). Libraries created with the “in-house” method were more even in size, with 18,320 to 134,401 sequences per sample and an average size of 39,882 sequences (standard deviation, 21,696). Despite the different amplification methods and slightly different primers, there was good agreement between the libraries produced by the two methods (Fig. 1; see also Fig. S2).
TABLE 1.
Sample and library summary
| Rock source | Expt type | Template | No. of expts | No. of libraries | Leachate pH range |
|---|---|---|---|---|---|
| Duluth Complex | Humidity cell | DNA | 6 | 20 | 4.5–6.9 |
| RNA | 6 | 7 | 4.5–6.9 | ||
| Reactor | DNA | 13 | 17 | 5.3–7.2 | |
| Covered reactor | DNA | 3 | 4 | 6.6–6.7 | |
| Field rock pilea | DNA | 1 | 29 | 4.7 | |
| Humidity cell, subsampledb | DNA | 3 | 6 | 4.5–6.9 | |
| Ely Greenstone | Humidity cell | DNA | 6 | 15 | 3.7–6.4 |
| RNA | 6 | 6 | 3.7–6.2 | ||
| Field rock pilea | DNA | 2 | 18 | 7.2–7.3 | |
| Humidity cell, subsampledb | DNA | 2 | 4 | 3.7–4.5 |
Field rock piles were sampled at multiple different locations and at two depths (2.5 and 15 cm). Details in Data Set S1 in the supplemental material.
Material was subsampled from the top and middle of certain humidity cells.
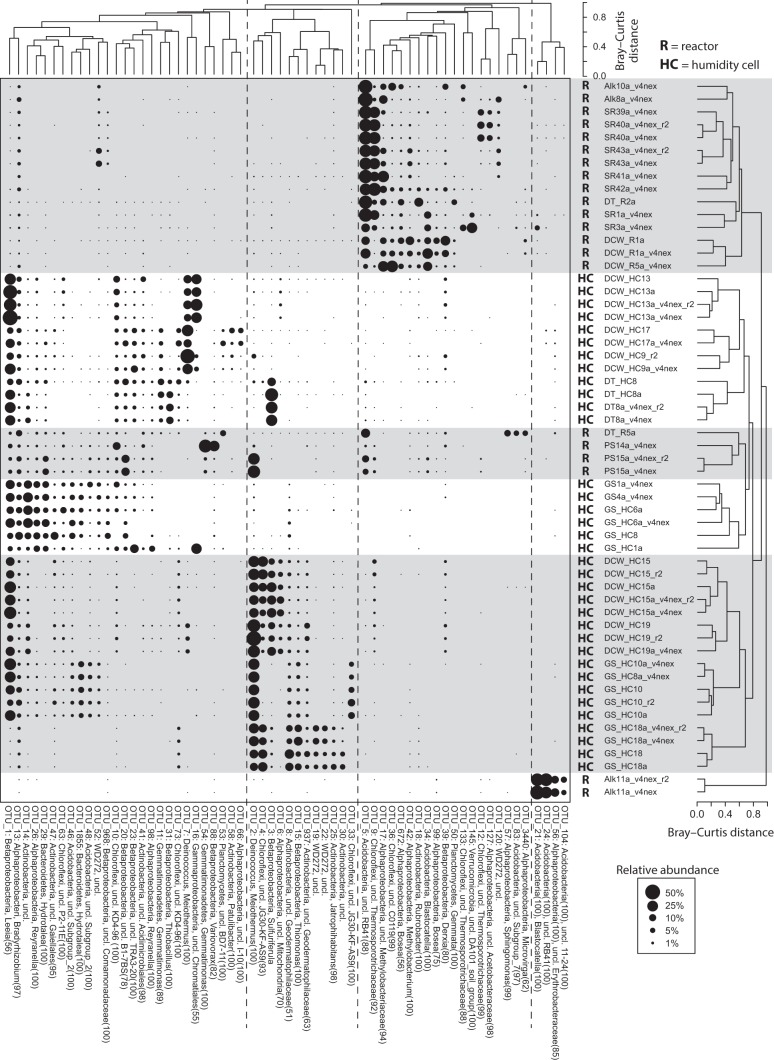
FIG 1.
Two-way cluster analysis of laboratory samples. The sizes of the points scale with the relative abundance of the OTUs. The Q-mode cluster analysis was calculated with all OTUs, while R-mode clustering only included those OTUs that were present at >5% in at least one sample. Multiple libraries from the same sample are included to show the variability among replicate libraries. The taxonomic affiliation of each OTU includes its phylum- and genus-level classifications, if available, and confidence scores >50 are provided in parentheses. OTUs that are unclassified at the genus level are indicated as such, and the highest available taxonomic classifications are provided.
Replicate libraries from humidity cell samples preserved in RNAlater were generally more consistent than those from frozen aliquots, likely because the addition of the RNAlater solution made it easier to homogenize the crushed rock in the collection tube after sampling. After realizing this potential bias, we either used the RNAlater-preserved samples or, in the case of the frozen samples from the field weathering experiments, performed 2 to 3 separate DNA extractions from each sample and combined extracts prior to analyses.
Several operational taxonomic units (OTUs) were abundant in the rRNA gene libraries. The most abundant OTU in the bulk humidity cell samples (OTU_1) was initially classified with low confidence (confidence score, 56) as Leeia spp. in the family Neisseriaceae, but a phylogenetic analysis showed that it actually belonged to a separate group of uncultivated Betaproteobacteria (see Fig. S3). Other abundant OTUs in the humidity cell samples included Meiothermus spp. (OTU_2 and OTU_7) (see Fig. S4) and Sulfuriferula spp. (OTU_3) (Fig. S3). With respect to the humidity cell subsamples with depth, sulfur-oxidizing populations (Sulfuriferula spp. and Thiomonas spp.) were less abundant near the surface, while some of the other abundant populations, such as Meiothermus spp., either showed the reverse trend or, as with OTU_1, exhibited little difference with depth (Table 1; see also Fig. S5). The most abundant OTUs in the reactors included an unclassified Acidobacteria (OTU_5) in the RB41 family (35) and an unclassified Chloroflexi in Thermosporotrichaceae (OTU_9) (see Fig. S6). OTUs from the classes Ktedonobacteria and Thermomicrobia in the phylum Chloroflexi were abundant in the field experiments (see Fig. S6), as were Sulfuriferula spp. (OTU_3). Certain sulfur-oxidizing betaproteobacteria were abundant in the field and laboratory experiments, notably Thiobacillus, Thiomonas, and Sulfuricella, in addition to Sulfuriferula. A phylogenetic analysis confirmed the classification of the Sulfuriferula, Thiobacillus, and Thiomonas OTUs but indicated that the “Sulfuricella” OTUs (OTU_74 and OTU_936) were also closely related to Sulfurirhabdus and may have originated from a related group of uncultivated organisms (Fig. S3).
Relationships of microbial community composition with geochemical variables and experimental treatments.
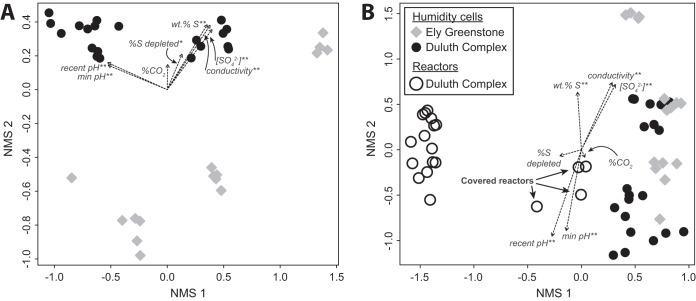
Overall, the microbial community composition from laboratory experiments was related to weathering conditions, rock type, and the geochemical characteristics of the rock and leachate. When only humidity cells were compared, libraries separated by rock type along the second nonmetric multidimensional scaling (NMS) ordination axis, while the first ordination axis correlated with leachate pH (Fig. 2A). The weight percentage of sulfur (a measure of sulfide mineral content in the rock at the start of the experiment), leachate conductivity, leachate sulfate concentration, and, to a lesser degree, percentage of sulfur depleted (an estimate of the sulfide mineral oxidation during the experiment) were also all significantly correlated with the ordination axes (Fig. 2A). When libraries from the humidity cells and reactors were compared (Fig. 2B), the libraries separated by experimental treatment along the first ordination axis, while the aforementioned geochemical parameters correlated with the second NMS axis. A hierarchical agglomerative cluster analysis was consistent with the ordination analyses (Fig. 1). Reactor libraries clustered separately from humidity cells, and humidity cell libraries formed three clusters, namely, libraries from higher pH Ely Greenstone cells (pH 5.0 to 6.4), libraries from higher pH Duluth Complex cells (pH 6.1 to 7.0), and libraries from the lowest pH humidity cells (pH 3.7 to 5.3). Libraries from the three covered reactors, which remained moist, clustered with the humidity cells (Fig. 1).
FIG 2.
NMS ordinations of rRNA gene libraries from humidity cells (A) and all laboratory samples (B). Fitted vectors of variables indicated by double asterisks (**) are statistically significant (in A, P < 0.001, r2 > 0.8; in B, P < 0.001, r2 > 0.4), and the variable indicated by a single asterisk is only statistically significant in panel A (P = 0.04, r2 0.18).
Amplicon libraries from the field experiments differed from those generated from laboratory experiments (both humidity cells and reactors) (Fig. 3), with laboratory and field experiments from the same rock type sharing few abundant OTUs. Many of the abundant OTUs from the laboratory experiments did not occur above 1% abundance in the field experiments, and vice versa. Only OTU_3 (Sulfuriferula spp.) was >1% in abundance in the Duluth Complex field experiment and some of the humidity cells.
FIG 3.
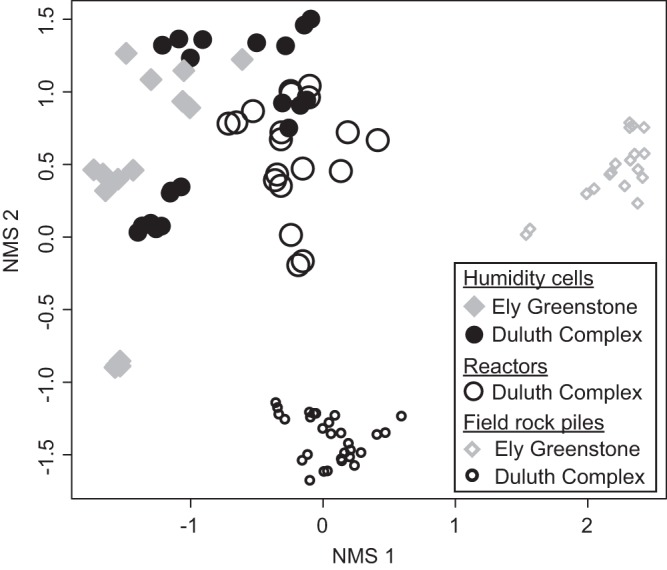
NMS ordination of rRNA gene libraries from all experiments.
Microbial diversity and cell counts.
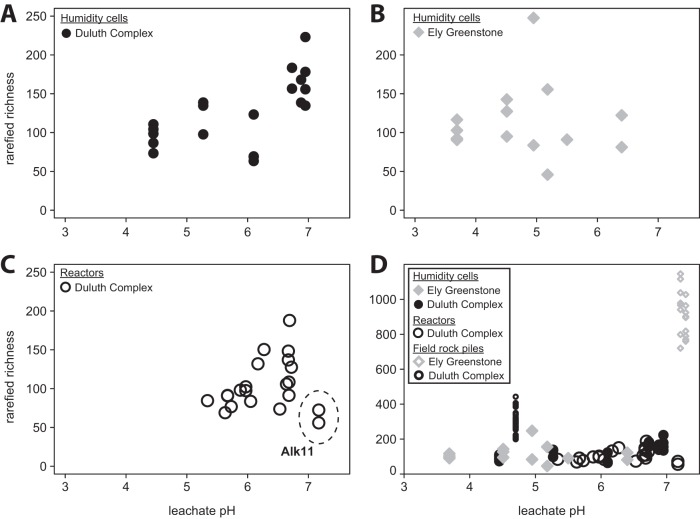
Microbial diversity, measured as expected richness by rarefaction, increased with pH in the Duluth Complex samples and was similar between reactors and humidity cells (Fig. 4A and C) (humidity cells, r2 = 0.38, P = 0.004; reactors, r2 = 0.34, P = 0.01). By contrast, the richness among the Ely Greenstone humidity cell communities did not trend with pH (Fig. 4B) (r2 < 0.01, P = 0.997). The field rock piles had a much greater richness than the laboratory experimental samples (Fig. 4D).
FIG 4.
Relationships between expected richness and pH. Correlations are statistically significant for Duluth Complex humidity cells (r2 = 0.38, P = 0.004) (A) and reactors (r2 = 0.34, P = 0.01, excluding Alk11 outliers [see Fig. 1]) (C), but not for Ely Greenstone humidity cells (r2 <0.01, P = 0.997) (B). (D) The field rock piles had a much greater richness than the laboratory experimental samples.
Total microbial cell counts in humidity cells ranged between 1.1 × 107 and 1.9 × 108 cells · g−1 rock (see Fig. S7A). Microbial cell counts in reactors were up to an order of magnitude greater, from 2.0 × 108 to 3.0 × 109 cells · g−1 rock (Fig. S7B). The counts were generally higher in the lower pH experiments, but the correlation between pH and microbial cell counts was only statistically significant among reactor samples (r2 = 0.32, P = 0.02). The majority of microbial cells in the laboratory experiments were attached to mineral surfaces (Fig. S7C). Sonication was necessary to detach the paraformaldehyde-fixed cells and achieve a homogenous distribution for counting, and no significant difference in counts was observed with sonication times from 15 s to 5 min.
rRNA transcript libraries.
Thirteen rRNA transcript libraries were created from 12 humidity cell samples (Table 1). Although the relative abundance of most OTUs differed between the corresponding 16S rRNA gene and transcript libraries, many of the same OTUs from the 16S rRNA gene libraries were present in the transcript libraries, likely indicating that those populations were alive and active at the time of sampling (Fig. 5). A notable exception was OTUs representing Meiothermus spp. (OTU_2 and OTU_7) (Fig. S4), which had little or no presence in the transcript libraries and may have been largely inactive or dead.
FIG 5.
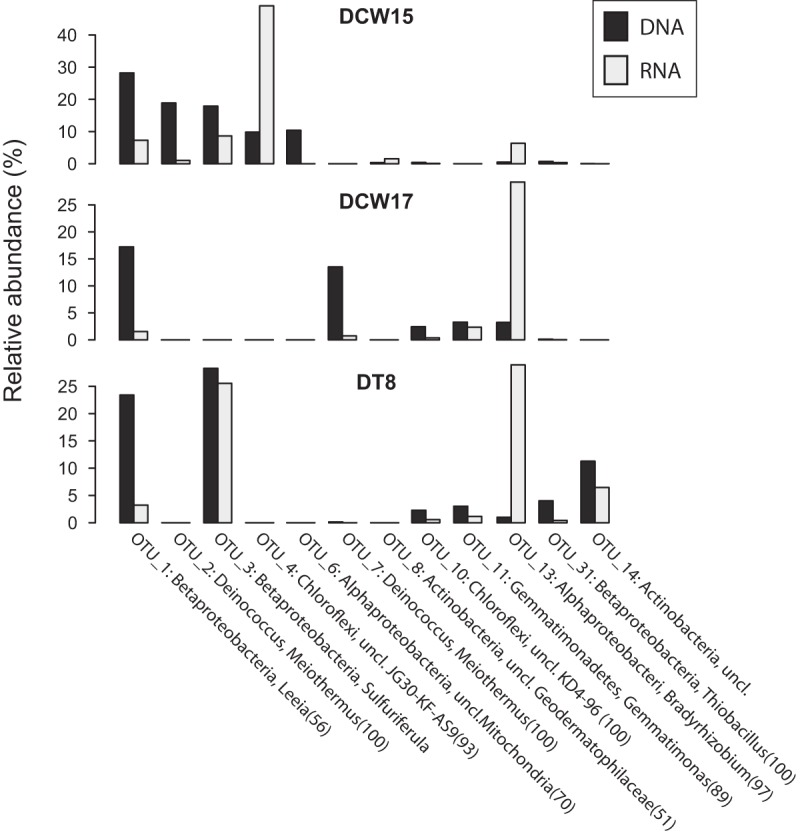
Relative abundances of select OTUs in rRNA gene and transcript libraries for three Duluth Complex humidity cell samples.
As with the 16S rRNA gene data sets, an NMS analysis showed that variance among the RNA libraries was correlated with leachate chemistry (see Fig. S8). Nevertheless, rRNA transcript libraries did not clearly differentiate by rock type (Duluth Complex versus Ely Greenstone) as the rRNA gene libraries had.
DISCUSSION
Duluth Complex weathering communities and novel diversity.
Under extremely acidic conditions (e.g., pH <3), sulfide minerals in ores and mine waste (waste rock and tailings) are oxidized by microbial consortia that include iron-oxidizing, sulfur-oxidizing, and heterotrophic populations that, together, regenerate ferric iron, maintain low pH, and remove toxic organic acids (7, 13, 36, 37). By contrast, the rock-associated microbial assemblages sampled here were primarily composed of taxa associated with sulfur-oxidizing or organoheterotrophic lifestyles. Conspicuously absent from the Duluth Complex communities were any abundant known iron-oxidizers, even in the most acidic experiments (pH 4.5 to 5.3). Many of the abundant OTUs, however, were classified as members of groups that are only known from environmental samples, such as the OTUs from unnamed clades in Chloroflexi that dominated the field rock pile (see Fig. S2 and S6 in the supplemental material) and the unclassified betaproteobacterial OTU (OTU_1) that was abundant in most humidity cells (Fig. 1; see also Fig. S3). Additional information on the function and metabolism of these organisms will await future culture- or meta-omics-based analyses, with some of the abundant unclassified populations possibly representing novel iron-oxidizers. Iron-oxidizing acidophiles in the Firmicutes are often found in acidic mine waste (see reference 9 and references therein), and could have been missed by the amplicon-based approach used in this study, especially if they were present as spores.
The most abundant populations identified here are not widely known from other mine environments. This may be a result of the moderately acidic conditions under which the Duluth Complex weathers. The microbial ecology of moderately acidic mine wastes has not been as well studied as that of extremely acidic mine-impacted environments (e.g., see reference 27). Moderately acidic waters draining mine waste are better characterized, but to our knowledge, the abundant Duluth Complex populations are also not common to those environments (e.g., see references 9, 17, 38–40). Because many mines produce waste with low sulfide content and moderately acidic drainage (4, 41), more research is needed to understand the microbial communities and microbe-mineral interactions in these less acidic mining-impacted environments, where the role of microbial processes in exacerbating or mitigating the release of metals and other harmful contaminants is of great interest and importance.
Geochemical and experimental influences on microbial community composition.
Although the weathering experiments sampled in this study were not initiated with microbial analyses in mind, microbial communities became established over the course of the experiments. Because the geochemical and mineralogical conditions in the experiments evolved over more than 10 years (Fig. S1F), the extant communities at the time of sampling likely included populations adapted to the contemporary geochemistry as well as “relict” populations that may have been more prominent earlier in the weathering experiments.
Inactive populations (Fig. 5) could represent these relicts, or they could be recent colonists from external sources that failed to thrive in the weathering experiments. Regardless, at the time of sampling, community composition was not random. Microbial assemblages trended with pH and other parameters, and so turnover was apparently rapid enough for microbial communities in geochemically similar cells to converge (Fig. 1 to 3; see also Fig. S8). Contemporary geochemical parameters in the experiments evidently impacted communities, and this selection was imprinted on the total microbial assemblages, despite the presence of inactive populations and a legacy of evolving geochemical conditions.
Among samples subjected to the same experimental treatment, trends in community composition were correlated with leachate geochemistry, including pH (Fig. 2). Our findings are consistent with those from other studies that have identified pH as an important explanatory variable in mine drainage (e.g., see references 11, 16, 17, 21, and 42). Leachate pH, sulfate, and conductivity were correlated with weight percentage of sulfur in the ordinations (Fig. 2), consistent with numerous Duluth Complex weathering experiments that have shown similar correlations with sulfide mineral content of the rock (34, 43, 44). From the available data, we were not able to discern whether the observed trends in microbial community structure and diversity were related to pH itself, to sulfide mineral content, or to a combination of these and other factors.
Humidity cells, reactors, and field rock piles supported different microbial communities. These differences might reflect reduced colonization opportunities in the laboratory compared with those the field, as well as specific treatment conditions, such as moisture content. Most of the reactors dried out completely after 4 to 5 days, and so organisms in those experiments would have had to repeatedly persist in a dry environment. Libraries from the covered reactors that remained moist, however, were intermediate to the humidity cell and uncovered reactor samples in ordinations (Fig. 2B) and grouped with humidity cell libraries in cluster analyses (Fig. 1). Furthermore, a phylum-level comparison of communities from a “normal” (uncovered) reactor (R2a), a covered reactor (R5a), and a humidity cell (DT8a) that were all initiated with the same tailings material showed that the covered reactor community was intermediate to that of the reactor and the humidity cell (see Fig. S9), pointing to the importance of moisture in community evolution. Paradoxically, cell counts were higher in the reactors than in the humidity cells (Fig. S7). It could be that the smaller particle size used in the reactors, and consequent high specific surface area, either helped sorb and retain cells or provided more chemical energy from an increased reactive mineral surface area.
The Ely Greenstone humidity cells contained distinct microbial communities compared with those generated from Duluth Complex rock (Fig. 2A). These two formations have different sulfide mineral assemblages, with pyrite in the Ely Greenstone and pyrrhotite and other acid-soluble sulfides in the Duluth Complex. These differences in sulfide mineralogy may be responsible for the differences in the dominant sulfur-oxidizing populations, as discussed in the next section.
Sulfur-oxidizing microorganisms.
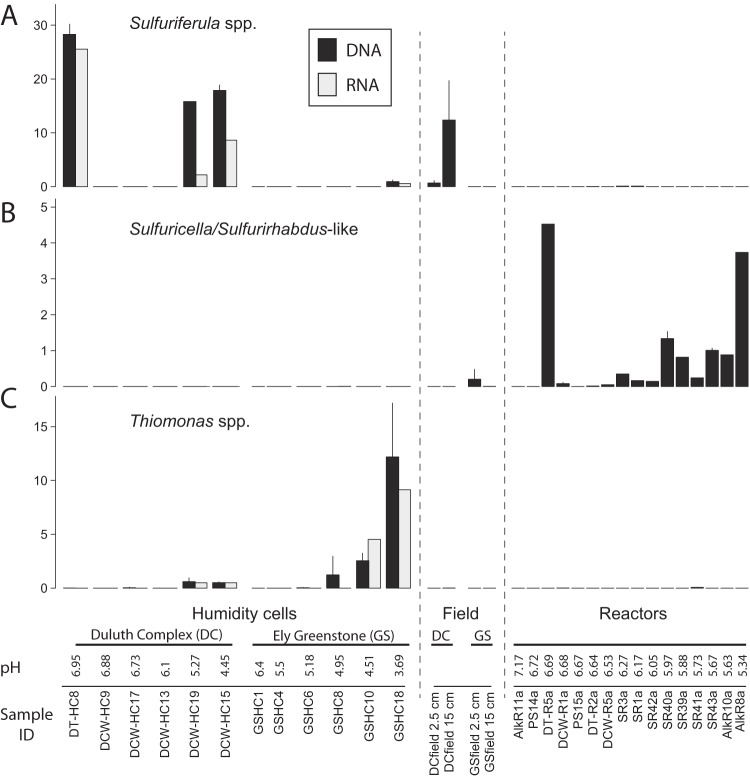
The abundance of OTUs from genera of known sulfur-oxidizing bacteria indicates that sulfur-oxidizers were an important part of the Duluth Complex weathering consortia and that the oxidation of reduced inorganic sulfur compounds is an important biogeochemical process in moderately acidic mine waste. Furthermore, different rock types and experimental treatments apparently selected for specific groups of sulfur-oxidizers. Sulfuriferula spp. were abundant in some Duluth Complex humidity cells (in both the rRNA gene and transcript libraries) and the Duluth Complex field rock pile (Fig. 6A). By contrast, Thiomonas spp. were abundant in the acidic Ely Greenstone humidity cells (Fig. 6C), whereas Duluth Complex reactors contained OTUs related to Sulfuricella and Sulfurirhabdus (Fig. 6B). Thiobacillus spp. (OTU_31) (Fig. S3), which are commonly observed in circumneutral mine waste (23, 25, 26, 45), were only present in the tailings humidity cell DT8 (Fig. 1).
FIG 6.
Distribution of the three most abundant sulfur-oxidizing genera from the leaching experiments, namely, Sulfuriferula spp. (OTU_3) (A), Sulfuricella/Sulfurirhabdus-like OTUs (OTU_74 and OTU_936) (B), and Thiomonas spp. (OTU_15, OTU_1848, OTU_3813, and OTU_5411) (C) (see also Fig. S3 in the supplemental material). Samples are arranged in order of decreasing pH and separated by rock type and experimental treatment. rRNA transcript libraries were only generated for the humidity cell samples. Where replicate libraries from the same sample are available, values represent the means and error bars are standard deviations. All samples from 2.5- and 15-cm depths were averaged for the field experiments, and only data from custom Nextera libraries are shown.
The Sulfuriferula spp. are of particular interest because they were abundant in the Duluth Complex humidity cell and field weathering experiments but not in pyrite-bearing Ely Greenstone subjected to the same experimental treatments. Further, we hypothesize that their uneven distribution among the Duluth Complex humidity cells (Fig. 6A) may be related to sulfide mineral content. Sulfuriferula spp. were present in the two most acidic cells (DCW15 and DCW19, pH 4.5 to 5.3) that also had the highest initial sulfide mineral content (0.61 to 1.03% sulfur) (Fig. 6A) but not the less acidic cells with lower sulfide content (DCW9, DCW13, and DCW17, pH 6.1 to 6.9, 0.13 to 0.55% sulfur). Even though the calculated “percentage of sulfur remaining” values indicate that all humidity cells still contained sulfides (see Data Set S1), some of the remaining sulfides were likely encased within larger particles with little or no exposed surface area. Therefore, the smaller amount of sulfides in the higher pH samples would have offered less area for microbial colonization than in the greater sulfur content samples. In further support of this hypothesis, Sulfuriferula spp. were also abundant in DT8, a high pH cell (pH 6.95) that contained tailings rather than crushed waste rock. DT8 still contained fine-grained and presumably reactive pyrrhotite (see Fig. S10), perhaps because the much smaller grain sizes in this experiment either increased the abundance of sulfide mineral surfaces available for reaction or caused it to retain more water and slowed oxygen penetration at depth (as proposed in reference 46).
If our hypothesis that the distribution of Sulfuriferula is related to reactive sulfide mineral surfaces is correct, then Sulfuriferula may be important sulfur-cycling organisms in moderately acidic mine waste that contains pyrrhotite or other acid-soluble sulfides. While this genus has not been widely associated with waste rock, tailings, or other mining environments, S. plumbophila (formerly “Thiobacillus plumbophilus”) was isolated from a uranium mine (47–49), and one of the few mining environments where Sulfuriferula-like clones have been previously reported was also moderately acidic pyrrhotite-bearing mine waste (27). Sulfur-oxidizing autotrophs are frequently observed in circumneutral mine wastes (24–26), but different populations may be important in the Duluth Complex because of its mineral assemblage and moderately acidic weathering conditions. Sulfuriferula's abundance in the Duluth Complex humidity cell and field experiments here suggest that it may be more widely important in sulfidic mine waste than previously recognized.
Implications for kinetic testing of mine waste.
Field and laboratory rock weathering experiments are widely used to assess acid generation and metal release from mine waste (http://www.gardguide.com [33, 50]), but few humidity cell studies have included microbial analysis (e.g., see references 9, 41, and 51). Microbial community analysis could be useful because microorganisms are a sensitive indicator of geochemical conditions. For example, the presence of Sulfuriferula spp. in both the humidity cell and field experiments (Fig. 6A) seems to indicate that humidity cells are a good reflection of the field when considering sulfur-cycling taxa. Microbial communities in these kinetic tests are also an important consideration because microorganisms can impact rock weathering rates (51). For example, under extremely acidic conditions, microorganisms can dramatically accelerate sulfide mineral oxidation, and rates can change based on microbial biomass (52) and the composition of the microbial community (37). Because of the paucity of research on microbial sulfide mineral oxidation under moderately acidic conditions, these poorly understood communities may have important yet unrecognized impacts on water quality as well.
MATERIALS AND METHODS
Samples.
Weathered rock samples were collected from kinetic tests, including both humidity cells and reactors (see Data Set S1 in the supplemental material). The kinetic test samples consisted of crushed rock representing a range of sulfur and metal contents and a Duluth Complex tailings sample from a pilot ore-processing test. The humidity cell apparatus and procedures are described by Lapakko (33) according to an earlier version of American Society for Testing and Materials (ASTM) method, D5744-13e1 (53). The table in Data Set S1 includes references to MN DNR reports or other publications detailing the specific laboratory and field experiments sampled here. Briefly, the humidity cells consisted of short cylindrical columns filled with 1 kg of crushed rock (<0.64 cm diameter) or tailings. Each week, the cells were rinsed with 500 ml of deionized H2O (dH2O) and the leachate was collected for geochemical analysis. Reactors are a smaller apparatus filled with 75 g of finely crushed rock (0.053 to 0.149 mm) (34, 45). Each week, the reactors were rinsed with 200 ml of dH2O. Reactors without covers dried out completely after approximately 4 to 5 days, whereas reactors that were covered (samples DT-R5, PS14, and PS15) retained moisture between rinse intervals. One covered reactor contained Duluth Complex tailings (DT-R5), and the other two contained Duluth Complex rock (PS14 and PS15). All the other reactors and six of the humidity cells contained Duluth Complex rock or tailings, and six of the humidity cells contained pyrite-bearing greenschist from the Ely Greenstone formation of northern Minnesota (Data Set S1). Details of the kinetic tests and leachate analyses can be found in reports referenced in the table in Data Set S1.
Samples for microbiological analysis were collected at the termination of some laboratory experiments, on 12 May and 3 June 2014. Material was collected aseptically by taking a vertical section from the center of the humidity cell or reactor with sterile metal spatulas. For some samples, subsamples were collected from the middle and the top 2 cm of the cell. Samples for DNA and RNA extraction were homogenized by shaking the collection tube and were either immediately frozen on dry ice or first preserved in RNAlater (Applied Biosystems, Foster City, CA, USA) and then frozen. Samples were subsequently stored at −80°C until analysis. Samples for cell counting were immediately preserved in 3% paraformaldehyde (PFA) in 1× phosphate-buffered saline (PBS). Samples were incubated for 24 h, and the PFA was removed by gently washing once with 1× PBS and transferring samples to a 1:1 solution of 1× PBS and 100% ethanol for storage at 4°C.
Samples from three field rock weathering experiments (one Duluth Complex and two Ely Greenstone) were collected on 24 April 2015 (Table 1; see also Fig. S1). Samples for microbiological analysis were collected from the surface (0- to 2.5-cm depth) and at a 15-cm depth by digging a small hole with a shovel and sampling material from the side of the hole. To representatively sample the shallow surface of the large field rock piles, multiple samples were collected from the top and the sides of each pile (Data Set S1). Samples were immediately frozen on dry ice in the field and stored at −80°C until analysis.
Cell counts.
To detach cells from mineral surfaces, PFA-fixed samples were diluted and sonicated for 45 s (setting 6 [usually 5 to 8 W] on a model 60 Sonic Dismembrator; Fisher Scientific), similar to the procedure described by Ravenschlag et al. (54). Aliquots (25 to 100 μl) of the sonicated samples were diluted in 1× PBS, stained with SYBR Gold (Fisher Scientific) for 10 min, and filtered through a 0.2-μm-pore-size black polycarbonate filter (Whatman, GE Healthcare Bio-Sciences, Marlborough, MA, USA). Counts were based on ≥10 randomly selected fields. The sonication procedure was optimized by testing sonication times from 15 s to 5 min. The standard error among replicates (sonication and counting of separate aliquots) was 25%. Cells in unsonicated PFA-fixed samples were imaged by applying samples to multiwell Teflon-coated glass slides, dehydrating them with ethanol, and staining them with SYBR Gold for 10 min. Fluorescently stained samples were mounted with Vectashield H-1000 (Vector Laboratories, Burlingame, CA, USA) and viewed with either an Olympus BX61 compound microscope with an Olympus XM10 camera or an Olympus IX61 inverted compound microscope with an Olympus DP73 camera, both with CellSens Dimensions software (Olympus, Japan).
Nucleic acid extraction and amplicon library preparation.
Total DNA was extracted using the Mo Bio PowerSoil DNA isolation kit (Mo Bio Laboratories, Inc., Carlsbad, CA, USA). To reduce DNA extraction bias, aliquots of each sample were bead beaten using a Vortex-Genie 2 (Scientific Industries, Inc., Bohemia, NY, USA) for 5, 10, and 15 min, and then recombined. For RNAlater-preserved samples, the RNAlater was removed prior to extraction by diluting samples 1:1 in either 1× PBS or nucleic acid-free water and removing the supernatant after centrifugation. Total RNA was extracted from RNAlater-preserved samples using the Mo Bio PowerBiofilm RNA isolation kit (Mo Bio) after RNAlater removal as described above. DNA was removed from the extracted RNA with two separate DNase treatments, first with an on-column DNase 1 treatment included in the PowerBiofilm kit (Mo Bio) and subsequently with the Turbo DNase enzyme (Applied Biosystems, Foster City, CA, USA). The absence of DNA was verified by attempting to amplify 16S rRNA genes with 35 PCR cycles using the primers 27f and 1492r (55), and RNA quality was checked by fragment analysis (Agilent Bioanalyzer; Agilent Technologies, Santa Clara, CA, USA) at the University of Minnesota Genomics Center (UMGC).
16S rRNA gene amplicon libraries were produced by sequencing the V4 hypervariable region of the 16S rRNA gene in two ways. Initially, V4 libraries were created with the full-service amplicon sequencing service at the UMGC (56) (referred to as “full-service” libraries) as described in reference 57 but with primers 515f (GTG CCA GCM GCC GCG GTA A) and 806r (GGA CTA CHV GGG TWT CTA AT). Briefly, the full-service amplicon sequencing service used the Kapa HiFidelity hot start polymerase (Kapa Biosystems, USA) to amplify the V4 region, and barcodes and Illumina sequencing adaptors were attached with a second amplification step. The first amplification step was performed as a quantitative PCR to optimize the template concentration for each sample and reduce the number of PCR cycles (56).
Because of low DNA yields from the humidity cell and reactor samples, the libraries had variable sizes (uneven sample pool balance), and some samples were unsuccessful. Therefore, amplicon libraries were subsequently created by first amplifying the V4 region in-house with custom Nextera primers (“in-house” libraries). The custom Nextera primers were the “improved” V4 amplification primers of Walters et al. (58) (515f modified, GTG YCA GCM GCC GCG GTA A; 806r modified, GGA CTA CNV GGG TWT CTA AT), with tail sequences to allow subsequent barcoding (forward primer tail, TCG TCG GCA GCG TCA GAT GTG TAT AAG AGA CAG; and reverse primer tail, GTC TCG TGG GCT CGG AGA TGT GTA TAA GAG ACA G). The HotStarTaq Plus polymerase (Qiagen, Hilden, Germany) was used because it had reduced adapter dimer formation with the Nextera primers compared with that with the Kapa Taq (see reference 56 for a discussion of this phenomenon). PCR was performed with 5 min for initial denaturation at 94°C, 30 cycles of 45 s for denaturation at 94°C, 60 s for annealing at 50°C, and 90 s for elongation at 72°C, with 10 min for final elongation at 72°C. Products were subsequently barcoded by the UMGC. Blank controls, consisting of only kit reagents, were included with all DNA extractions, and samples were reextracted if any product was visible in the blanks after 35 PCR cycles. Multiple DNA extraction and water blanks were included with all sequencing runs and had negligible amplification during library preparation and barcoding (see Table S1). Libraries were pooled and sequenced on an Illumina MiSeq (Illumina, San Diego, CA, USA) using 2 × 250 cycles.
The rRNA transcript libraries were created by amplifying RNA extracts by reverse transcription PCR (RT-PCR) with the SuperScript OneStep RT-PCR system (Thermo Fisher Scientific, Waltham, MA, USA) using the custom Nextera primers as described above. RT-PCR reaction mixtures were incubated for 30 min at 50°C (first strand synthesis), followed by 2 min for initial denaturation at 94°C, 30 cycles of 15 s for denaturation at 94°C, 30 s for annealing at 50°C, and 90 s for elongation at 68°C, with 5 min for final elongation at 68°C (second strand synthesis).
Quality trimming and filtering, assembly, and OTU calling.
Raw sequences were quality filtered and trimmed using Sickle (https://github.com/najoshi/sickle) to an average quality of ≥28 (5′ trimming only) and a minimum length of 100 bp. Residual adapters were trimmed using cutadapt (59), forward and reverse reads were assembled with PEAR (60), and primers were removed by trimming the assembled reads with prinseq v.0.20.4 (61). OTU calling (97% similarity) and chimera removal were performed with the UPARSE pipeline (USEARCH v.8.0; [62]), except that the “derep_fulllength” script from VSEARCH v.1.9.5 (63) was used to circumvent the memory limitations of the 32-bit version of USEARCH. The taxonomic affiliation of each OTU was determined by classifying representative sequences with mother v.1.3.2 using the Silva database v.123 (64) and a confidence cutoff of 50.
Statistical and phylogenetic analyses.
Libraries with less than 10,000 sequence reads (after quality filtering) were excluded from subsequent analyses. Multivariate analyses were performed with libraries generated from the bulk humidity cell samples, and unless otherwise noted, subsamples from the top and middle of the cells were excluded. To account for uneven library sizes, data were first converted to proportional values by dividing by the number of sequences in each library. OTUs that occurred at less than 0.01% were removed from the data set. Data were subsequently transformed using an arcsine square root transformation, bij = (2/π)arcsine[(xij)0.5], where xij is an element in the original data matrix and bij is an element in the transformed data matrix (65). The impacts of transformation on the data structure are depicted in Fig. S11. The use of transformed data lowered the stress in NMS analyses and improved the correlations with fitted environmental variables.
Diversity was compared by calculating rarefied richness at a sample size of 10,000 sequences using the rarefy() function in Vegan package v.2.4-0 (66) in R v.3.2.4 (67). NMS was performed using either 2 or 3 dimensions, with rotation to principal components, using the metaMDS() function in the vegan package. Environmental overlays of the NMS ordinations were created with the envfit() function in vegan, which computes a multiple alignment of environmental variables against the ordination axes. Significance was assessed with 999 permutations. Hierarchical agglomerative cluster analyses were performed with Bray-Curtis dissimilarity and unweighted pair-group method using arithmetic averages (UPGMA) clustering (65). Q-mode cluster analyses (clustering of samples) were calculated, including all OTUs in the transformed data set, while R-mode cluster analysis (clustering of OTUs) only included those OTUs that were present at >5% in at least one sample. The table of OTU occurrence across samples that was used for the statistical analyses is included as Data Set S2.
The phylogenetic analyses of amplicon sequences used to confirm the taxonomic assignments for certain OTUs were performed by first computing a maximum likelihood phylogeny of full-length sequences in RAxML v.8.0.24 (68). Trees were generated using the general time reversible model of nucleotide substitution, gamma distributed rates, and the proportion of invariant sites and base frequencies estimated from the data. Bootstrap support was calculated with 100 rapid bootstrap replicates. Short amplicon sequences were then placed into the trees with the evolutionary placement algorithm (EPA) using default parameters (69).
Accession number(s).
Amplicon libraries (including blank controls) have been deposited in NCBI's Sequence Read Archive under project SRP103471.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Minnesota Department of Natural Resources Division of Lands and Minerals for facilitating sampling of the weathered rock samples used in this study. Special thanks to S. Koski and P. Geiselman for advice and assistance with sample collection, to D. Gohl, A. Hague, A. MacLean, and K. Beckman for insightful discussion and assistance with amplicon sequencing at the UMGC, to T. Lösekann and D. Bond for advice and the use of laboratory equipment for cell counting, and to N. Seaton and K. Hobart for assistance with scanning electron microscope analyses. C. Nguyen, L. Thomas, the University of Minnesota College of Science and Engineering systems staff, and the Minnesota Supercomputing Institute provided computing support and the use of facilities.
This work was supported by University of Minnesota MnDRIVE initiative and the University of Minnesota BioTechnology Institute.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00909-17.
REFERENCES
- 1.Schulz KJ, Woodruff LG, Nicholson SW, Seal RR II, Piatak NM, Chandler VW, Mars JL. 2014. Occurrence model for magmatic sulfide-rich nickel-copper-(platinum-group element) deposits related to mafic and ultramafic dike-sill complexes. Scientific Investigations Report 2010–5070–I U.S. Geological Survey, Denver, CO. doi: 10.3133/sir20105070I. [DOI] [Google Scholar]
- 2.Ripley EM, Alawi JA. 1986. Sulfide mineralogy and chemical evolution of the Babbitt Cu-Ni deposit, Duluth Complex, Minnesota. Can Mineral 24:347–368. [Google Scholar]
- 3.Severson MJ, Miller J, Peterson DM, Green JC, Hauck SA. 2002. Mineral potential of the Duluth Complex and related intrusions, p 164–200. In Miller JD Jr, Green JC, Severson MJ, Chandler VW, Hauck SA, Peterson DM, Wahl TE (ed), RI-58 Geology and mineral potential of the Duluth Complex and related rocks of northeastern Minnesota. Minnesota Geological Survey, St. Paul, MN: http://hdl.handle.net/11299/58804. [Google Scholar]
- 4.Nordstrom DK, Blowes DW, Ptacek CJ. 2015. Hydrogeochemistry and microbiology of mine drainage: an update. Appl Geochem 57:3–16. doi: 10.1016/j.apgeochem.2015.02.008. [DOI] [Google Scholar]
- 5.Johnson DB. 2014. Recent developments in microbiological approaches for securing mine wastes and for recovering metals from mine waters. Minerals (Basel) 4:279–292. doi: 10.3390/min4020279. [DOI] [Google Scholar]
- 6.Johnson DB, Hallberg KB. 2005. Acid mine drainage remediation options: a review. Sci Total Environ 338:3–14. doi: 10.1016/j.scitotenv.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 7.Johnson DB. 2014. Biomining—biotechnologies for extracting and recovering metals from ores and waste materials. Curr Opin Biotechnol 30:24–31. doi: 10.1016/j.copbio.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 8.Rawlings DE, Dew D, du Plessis C. 2003. Biomineralization of metal-containing ores and concentrates. Trends Biotechnol 21:38–44. doi: 10.1016/S0167-7799(02)00004-5. [DOI] [PubMed] [Google Scholar]
- 9.Holanda R, Hedrich S, Nǎncucheo I, Oliveira G, Grail BM, Johnson DB. 2016. Isolation and characterisation of mineral-oxidising “Acidibacillus” spp. from mine sites and geothermal environments in different global locations. Res Microbiol 167:613–623. doi: 10.1016/j.resmic.2016.04.008. [DOI] [PubMed] [Google Scholar]
- 10.Johnson DB, Hallberg KB, Hedrich S. 2014. Uncovering a microbial enigma: isolation and characterization of the streamer-generating, iron-oxidizing, acidophilic bacterium “Ferrovum myxofaciens.” Appl Environ Microbiol 80:672–680. doi: 10.1128/AEM.03230-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baker BJ, Banfield JF. 2003. Microbial communities in acid mine drainage. FEMS Microbiol Ecol 44:139–152. doi: 10.1016/S0168-6496(03)00028-X. [DOI] [PubMed] [Google Scholar]
- 12.Schippers A, Breuker A, Blazejak A, Bosecker K, Kock D, Wright T. 2010. The biogeochemistry and microbiology of sulfidic mine waste and bioleaching dumps and heaps, and novel Fe(II)-oxidizing bacteria. Hydrometallurgy 104:342–350. doi: 10.1016/j.hydromet.2010.01.012. [DOI] [Google Scholar]
- 13.Vera M, Schippers A, Sand W. 2013. Progress in bioleaching: fundamentals and mechanisms of bacterial metal sulfide oxidation–part A. Appl Microbiol Biotechnol 97:7529–7541. doi: 10.1007/s00253-013-4954-2. [DOI] [PubMed] [Google Scholar]
- 14.Edwards KJ, Gihring TM, Banfield JF. 1999. Seasonal variations in microbial populations and environmental conditions in an extreme acid mine drainage environment. Appl Environ Microbiol 65:3627–3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones DS, Kohl C, Grettenberger C, Larson LN, Burgos WD, Macalady JL. 2015. Geochemical niches of iron-oxidizing acidophiles in acidic coal mine drainage. Appl Environ Microbiol 81:1242–1250. doi: 10.1128/AEM.02919-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuang J-L, Huang L-N, Chen L-X, Hua Z-S, Li S-J, Hu M, Li J-T, Shu W-S. 2013. Contemporary environmental variation determines microbial diversity patterns in acid mine drainage. ISME J 7:1038–1050. doi: 10.1038/ismej.2012.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lear G, Niyogi D, Harding J, Dong Y, Lewis G. 2009. Biofilm bacterial community structure in streams affected by acid mine drainage. Appl Environ Microbiol 75:3455–3460. doi: 10.1128/AEM.00274-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palacios C, Zettler E, Amils R, Amaral-Zettler L. 2008. Contrasting microbial community assembly hypotheses: a reconciling tale from the Rio Tinto. PLoS One 3:e3853. doi: 10.1371/journal.pone.0003853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rawlings D, Tributsch H, Hansford G. 1999. Reasons why “Leptospirillum”-like species rather than Thiobacillus ferrooxidans are the dominant iron-oxidizing bacteria in many commercial processes for the biooxidation of pyrite and related ores. Microbiology 145:5–13. doi: 10.1099/13500872-145-1-5. [DOI] [PubMed] [Google Scholar]
- 20.Schrenk MO, Edwards KJ, Goodman RM, Hamers RJ, Banfield JF. 1998. Distribution of Thiobacillus ferrooxidans and Leptospirillum ferrooxidans: implications for generation of acid mine drainage. Science 279:1519. doi: 10.1126/science.279.5356.1519. [DOI] [PubMed] [Google Scholar]
- 21.Sheng Y, Bibby K, Grettenberger C, Kaley B, Macalady JL, Wang G, Burgos WD. 2016. Geochemical and temporal influences on the enrichment of acidophilic iron-oxidizing bacterial communities. Appl Environ Microbiol 82:3611–3621. doi: 10.1128/AEM.00917-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lapakko K, Antonson D. 2012. Duluth Complex rock dissolution and mitigation techniques: a summary of 35 years of DNR research. Minnesota Department of Natural Resources, Division of Lands and Minerals, St Paul, MN: http://files.dnr.state.mn.us/lands_minerals/northmet/permit_to_mine/03_mdnr_2012_duluth_complex_rock_dissolution_mitigation_techniques.pdf. [Google Scholar]
- 23.Chen LX, Li JT, Chen YT, Huang LN, Hua ZS, Hu M, Shu WS. 2013. Shifts in microbial community composition and function in the acidification of a lead/zinc mine tailings. Environ Microbiol 15:2431–2444. doi: 10.1111/1462-2920.12114. [DOI] [PubMed] [Google Scholar]
- 24.Lindsay MB, Condon PD, Jambor JL, Lear KG, Blowes DW, Ptacek CJ. 2009. Mineralogical, geochemical, and microbial investigation of a sulfide-rich tailings deposit characterized by neutral drainage. Appl Geochem 24:2212–2221. doi: 10.1016/j.apgeochem.2009.09.012. [DOI] [Google Scholar]
- 25.Schippers A, Hallmann R, Wentzien S, Sand W. 1995. Microbial diversity in uranium mine waste heaps. Appl Environ Microbiol 61:2930–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schippers A, Von Rège H, Sand W. 1996. Impact of microbial diversity and sulfur chemistry on safeguarding sulfudic mine waste. Miner Eng 9:1069–1079. doi: 10.1016/0892-6875(96)00099-4. [DOI] [Google Scholar]
- 27.Korehi H, Blöthe M, Schippers A. 2014. Microbial diversity at the moderate acidic stage in three different sulfidic mine tailings dumps generating acid mine drainage. Res Microbiol 165:713–718. doi: 10.1016/j.resmic.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 28.Mendez MO, Neilson JW, Maier RM. 2008. Characterization of a bacterial community in an abandoned semiarid lead-zinc mine tailing site. Appl Environ Microbiol 74:3899–3907. doi: 10.1128/AEM.02883-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arkesteyn GJ. 1980. Pyrite oxidation in acid sulphate soils: the role of microorganisms. Plant Soil 54:119–134. doi: 10.1007/BF02182004. [DOI] [Google Scholar]
- 30.Schippers A. 2004. Biogeochemistry of metal sulfide oxidation in mining environments, sediments, and soils. GSA Spec Pap 379:49–62. doi: 10.1130/0-8137-2379-5.49. [DOI] [Google Scholar]
- 31.Schippers A, Jørgensen BB. 2002. Biogeochemistry of pyrite and iron sulfide oxidation in marine sediments. Geochim Cosmochim Acta 66:85–92. doi: 10.1016/S0016-7037(01)00745-1. [DOI] [Google Scholar]
- 32.Percak-Dennett E, He S, Converse B, Konishi H, Xu H, Corcoran A, Noguera D, Chan C, Bhayyacharyya A, Borch T, Boyd E, Roden EE. 2017. Microbial acceleration of aerobic pyrite oxidation at circumneutral pH. Geobiology 2017:1–14. doi: 10.1111/gbi.12241. [DOI] [PubMed] [Google Scholar]
- 33.Lapakko KA. 2015. Preoperational assessment of solute release from waste rock at proposed mining operations. Appl Geochem 57:106–124. doi: 10.1016/j.apgeochem.2015.01.010. [DOI] [Google Scholar]
- 34.Lapakko KA, Antonson DA. 1994. Oxidation of sulfide minerals present in Duluth Complex rock: a laboratory study, p 593–607. In Alpers C, Blowes DW (ed), Environmental geochemistry of sulfide oxidation. American Chemical Society, Washington, DC. [Google Scholar]
- 35.Quaiser A, Ochsenreiter T, Lanz C, Schuster SC, Treusch AH, Eck J, Schleper C. 2003. Acidobacteria form a coherent but highly diverse group within the bacterial domain: evidence from environmental genomics. Mol Microbiol 50:563–575. doi: 10.1046/j.1365-2958.2003.03707.x. [DOI] [PubMed] [Google Scholar]
- 36.Brierley CL, Brierley JA. 2013. Progress in bioleaching: part B: applications of microbial processes by the minerals industries. Appl Microbiol Biotechnol 97:7543–7552. doi: 10.1007/s00253-013-5095-3. [DOI] [PubMed] [Google Scholar]
- 37.Rawlings DE, Johnson DB. 2007. The microbiology of biomining: development and optimization of mineral-oxidizing microbial consortia. Microbiology 153:315–324. doi: 10.1099/mic.0.2006/001206-0. [DOI] [PubMed] [Google Scholar]
- 38.Hallberg KB, Johnson DB. 2005. Microbiology of a wetland ecosystem constructed to remediate mine drainage from a heavy metal mine. Sci Total Environ 338:53–66. doi: 10.1016/j.scitotenv.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 39.Hallberg KB, Johnson DB. 2003. Novel acidophiles isolated from moderately acidic mine drainage waters. Hydrometallurgy 71:139–148. doi: 10.1016/S0304-386X(03)00150-6. [DOI] [Google Scholar]
- 40.Fabisch M, Beulig F, Akob DM, Küsel K. 2013. Surprising abundance of Gallionella-related iron oxidizers in creek sediments at pH 4.4 or at high heavy metal concentrations. Front Microbiol 4:390. doi: 10.3389/fmicb.2013.00390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Langman JB, Moore ML, Ptacek CJ, Smith L, Sego D, Blowes DW. 2014. Diavik waste rock project: evolution of mineral weathering, element release, and acid generation and neutralization during a five-year humidity cell experiment. Minerals (Basel) 4:257–278. doi: 10.3390/min4020257. [DOI] [Google Scholar]
- 42.Grettenberger CL, Pearce AR, Bibby KJ, Jones DS, Burgos WD, Macalady JL. 2017. Efficient low-pH iron removal by a microbial iron oxide mound ecosystem at Scalp Level Run. Appl Environ Microbiol 83:e00015-17. doi: 10.1128/AEM.00015-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lapakko K. 1988. Prediction of acid mine drainage from Duluth Complex mining wastes in northeastern Minnesota, p 180–190, vol 1 Proceedings of the 1988 Mine Drainage and Surface Mine Reclamation Conference. U.S. Bureau of Mines IC9183, Office of Surface Mining, Reclamation, and Enforcement, American Society for Surface Mining and Reclamation. doi: 10.21000/JASMR88010180. [DOI] [Google Scholar]
- 44.Lapakko K. 1994. Comparison of Duluth Complex rock dissolution in the laboratory and field, p 419–428. Proceedings of the International Land Reclamation and Mine Drainage Conference and the 3rd International Conference on the Abatement of Acidic Drainage, Pittsburgh, PA. [Google Scholar]
- 45.Lapakko KA. November 1988. Apparatus for environmental leaching testing. US Patent 4,783,318.
- 46.Lapakko K, Bernt M. 2009. Laboratory dissolution of tailings under three different test conditions, p 341–351. Proceedings of the 8th International Conference on Acid Rock Drainage (ICARD) and Securing the Future: Mining, Metals & the Environment in a Stable Society, Skelleftea, Sweden Curran Associates, Red Hook, NY. [Google Scholar]
- 47.Drobner E, Huber H, Rachel R, Stetter KO. 1992. Thiobacillus plumbophilus spec. nov., a novel galena and hydrogen oxidizer. Arch Microbiol 157:213–217. doi: 10.1007/BF00245152. [DOI] [PubMed] [Google Scholar]
- 48.Watanabe T, Kojima H, Fukui M. 2015. Sulfuriferula multivorans gen. nov., sp. nov., isolated from a freshwater lake, reclassification of “Thiobacillus plumbophilus” as Sulfuriferula plumbophilus sp. nov., and description of Sulfuricellaceae fam. nov. and Sulfuricellales ord. nov. Int J Syst Evol Microbiol 65:1504–1508. doi: 10.1099/ijs.0.000129. [DOI] [PubMed] [Google Scholar]
- 49.Watanabe T, Kojima H, Fukui M. 2016. Sulfuriferula thiophila sp. nov., a chemolithoautotrophic sulfur-oxidizing bacterium, and correction of the name Sulfuriferula plumbophilus Watanabe, Kojima and Fukui 2015 to Sulfuriferula plumbiphila corrig. Int J Syst Evol Microbiol 66:2041–2045. doi: 10.1099/ijsem.0.000988. [DOI] [PubMed] [Google Scholar]
- 50.Verburg R, Bezuidenhout N, Chatwin T, Ferguson K. 2009. The global acid rock drainage guide (GARD Guide). Mine Water Environ 28:305–310. doi: 10.1007/s10230-009-0078-4. [DOI] [Google Scholar]
- 51.Maest AS, Nordstrom DK. 2017. A geochemical examination of humidity cell tests. Appl Geochem 81:109–131. doi: 10.1016/j.apgeochem.2017.03.016. [DOI] [Google Scholar]
- 52.Edwards KJ, Bond PL, Druschel GK, McGuire MM, Hamers RJ, Banfield JF. 2000. Geochemical and biological aspects of sulfide mineral dissolution: lessons from Iron Mountain, California. Chem Geol 169:383–397. doi: 10.1016/S0009-2541(00)00216-3. [DOI] [Google Scholar]
- 53.ASTM International. 2013. Standard test method for laboratory weathering of solid materials using a humidity cell, p 23 D5744-13e1, Annual book of ASTM standards, vol 1104 American Society for Testing and Materials International, West Conshohocken, PA. [Google Scholar]
- 54.Ravenschlag K, Sahm K, Amann R. 2001. Quantitative molecular analysis of the microbial community in marine Arctic sediments (Svalbard). Appl Environ Microbiol 67:387–395. doi: 10.1128/AEM.67.1.387-395.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lane D. 1991. 16S/23S rRNA sequencing, p 115–175. In Stackebrandt E, Goodfellow M (ed), Nucleic acid techniques in bacterial systematics. Wiley, New York, NY. [Google Scholar]
- 56.Gohl DM, Vangay P, Garbe J, MacLean A, Hauge A, Becker A, Gould TJ, Clayton JB, Johnson TJ, Hunter R. 2016. Systematic improvement of amplicon marker gene methods for increased accuracy in microbiome studies. Nat Biotechnol 34:942–949. doi: 10.1038/nbt.3601. [DOI] [PubMed] [Google Scholar]
- 57.Jones DS, Flood BE, Bailey JV. 2015. Metatranscriptomic analysis of diminutive Thiomargarita-like bacteria (“Candidatus Thiopilula” spp.) from abyssal cold seeps of the Barbados accretionary prism. Appl Environ Microbiol 81:3142–3156. doi: 10.1128/AEM.00039-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Walters W, Hyde ER, Berg-Lyons D, Ackermann G, Humphrey G, Parada A, Gilbert JA, Jansson JK, Caporaso JG, Fuhrman JA. 2016. Improved bacterial 16S rRNA gene (V4 and V4-5) and fungal internal transcribed spacer marker gene primers for microbial community surveys. mSystems 1:e00009-15. doi: 10.1128/mSystems.00009-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 60.Zhang J, Kobert K, Flouri T, Stamatakis A. 2014. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 30:614–620. doi: 10.1093/bioinformatics/btt593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schmieder R, Edwards R. 2011. Quality control and preprocessing of metagenomic datasets. Bioinformatics 27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Edgar RC. 2013. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 63.Rognes T, Flouri T, Nichols B, Quince C, Mahé F. 2016. VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glöckner FO. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McCune B, Grace JB, Urban DL. 2002. Analysis of ecological communities. MjM Software Design, Gleneden Beach, OR. [Google Scholar]
- 66.Oksanen J, Kindt R, Legendre P, O'Hara R, Simpson GL, Stevens MHH. 2008. Vegan: community ecology package, R package version 1.11-0. http://cran.r-project.org/, http://vegan.r-forge.r-project.org/.
- 67.R Development Core Team 2016. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org. [Google Scholar]
- 68.Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 69.Berger SA, Krompass D, Stamatakis A. 2011. Performance, accuracy, and web server for evolutionary placement of short sequence reads under maximum likelihood. Syst Biol 60:291–302. doi: 10.1093/sysbio/syr010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.