Abstract
Forkhead box class O (FOXO) proteins are transcription factors that function downstream of the PTEN tumor suppressor and directly control the expression of genes involved in apoptosis, cell cycle progression, and stress responses. In the present study, we show that FOXO1 interacts with four and a half LIM 2 (FHL2) in prostate cancer cells. This interaction occurred in the nucleus and was enhanced by lysophosphatic acid. FHL2 decreased the transcriptional activity of FOXO1 and the expression of known FOXO target genes and inhibited FOXO1-induced apoptosis. Interestingly, SIRT1, a mammalian homolog of yeast Sir2, bound to and deacetylated FOXO1 and inhibited its transcriptional activity. FHL2 enhanced the interaction of FOXO1 and SIRT1 and the deacetylation of FOXO1 by Sirtuin-1 (SIRT1). Overall, our data show that FHL2 inhibits FOXO1 activity in prostate cancer cells by promoting the deacetylation of FOXO1 by SIRT1.
Keywords: acetylation, FKHR, forkhead, prostate cancer, SIR2
Introduction
The forkhead transcription factors are winged helical proteins first known for their role in development. Each of the more than 90 members of the forkhead family contains a conserved DNA binding domain with a butterfly-shaped structure of three α-helices and two characteristic large loops or ‘wings' (Kaufmann and Knöchel, 1996).
Members of the forkhead family, as classified by phylogenetic analysis of the amino-acid sequence of the DNA binding domain, include the forkhead box class O (FOXO) factors. There are four FOXO factors: FOXO1 (FKHR, Forkhead in rhabdomyosarcoma) (Galili et al, 1993; Kaestner et al, 2000), FOXO3a (FKHRL1, FKHR-like 1) (Anderson et al, 1998), FOXO4 (AFX, acute-lymphocytic-leukemia-1) (Parry et al, 1994), and the recently cloned FOXO6 (Jacobs et al, 2003).
As demonstrated by genetic and biochemical analysis of lower organisms and mammalian systems, the FOXO factors are critical regulators of stress responses, insulin sensitivity, longevity, and oncogenesis. For example, in Caenorhabditis elegans, the FOXO-related protein Daf-16 increases longevity and signals formation of the dauer larva, which is a stage of developmental arrest and reduced metabolic activity that enhances survival during periods of environmental stress (Kenyon et al, 1993). Mutation of the insulin/IGF1 receptor homolog Daf-2 or of Age-1, the catalytic subunit of phosphoinositide 3-kinase (PI3K), mimics the effects of Daf-16, thus suggesting that the insulin/IGF receptor–PI3K pathway negatively regulates Daf-16 function. Consistent with this premise, Daf-16 and its human homologs contain multiple conserved phosphorylation sites for members of the protein kinase B family (AKT-1, AKT-2, AKT3, and SGK-1), all of which function downstream of PI3K (Brunet et al, 1999). Phosphorylation of the FOXO factors by PKB results in their nuclear export, their interaction with the 14-3-3 proteins in the cytosol, and consequently in the abrogation of their transcriptional activity. Recent studies suggest that kinases in addition to PKB inhibit FOXO activity (Brunet et al, 2001; Woods et al, 2001; Hu et al, 2004), as do nuclear hormone receptors (Schuur et al, 2001; Zhao et al, 2001; Dowell et al, 2003; Li et al, 2003) and other forkhead factors (Seoane et al, 2004).
The PI3K–PKB pathway is active in and enhances the growth and survival of many types of human cancer cells. Conversely, the FOXO factors, which function downstream of the PI3K antagonist PTEN in cancer cells, inhibit cell cycle progression and promote cell death. These factors accomplish these actions by modulating the expression of genes encoding apoptosis and growth regulatory proteins by two methods: (1) direct binding to the insulin response sequence (IRS) in gene promoters and (2) ‘tethering' to other transcription factors (Ramaswamy et al, 2002). Examples of genes regulated by direct binding of the FOXO factors to DNA include those encoding apoptotic proteins such as Bim and Fas ligand, and examples of genes regulated by ‘tethering' include those encoding the cell cycle regulators such as cyclin G2 and cyclin D1. Both wild-type FOXO and FOXO mutants that cannot bind DNA modulate the expression of cyclin D1 and cyclin G2 (Ramaswamy et al, 2002).
FHL2 (four and a half LIM 2) was first identified as a protein differentially expressed in normal human myoblasts and their malignant counterparts, rhabdomyosarcoma cells; thus, it is also named DRAL (Downregulated in Rhabdomyosarcoma LIM Protein) (Genini et al, 1997). FHL2 is a LIM-only protein with five LIM domains whereby the LIM domain in the amino terminus consists only of the second half of the consensus motif. Although not required for normal heart development (Chu et al, 2000), FHL2 may control the hypertrophic response to stress in cardiomyocytes, where it is enriched (Kong et al, 2001, Purcell et al, 2004). The present study shows that FHL2 interacts with FOXO1 both in vitro and in prostate cancer cells. Binding of FHL2 to FOXO1 inhibits the transcriptional activity of FOXO1 by facilitating its deacetylation through Sirtuin-1 (SIRT1), the mammalian homolog of the yeast Sir2 deacetylase. Our data raise the interesting possibility that interaction of FHL2 with FOXO1 and SIRT1 may promote prostate tumorigenesis in response to increased stress during aging.
Results
FHL2 interacts with the amino terminus of FOXO1 and inhibits the transcriptional activity of FOXO1
The functions of the amino terminus of FOXO1, in contrast to that of the carboxyl-terminal activation domain and of the central DNA binding domain, are unclear. The amino terminus interacts with transcriptional coactivators (Puigserver et al, 2003) and thus may contain an activation function. To better define its function and to identify additional binding partners, we used the amino terminus (amino acids 1–150) of FOXO1 as bait in a two-hybrid screen of a cDNA library from human prostate cancer. β-Galactosidase (β-gal)-based screening identified several positive clones, and sequencing of cDNA identified one of the positive clones as full-length FHL2 fused in-frame to the VP16 activation domain.
To determine whether FHL2 and FOXO1 interact in mammalian cells, we ectopically expressed Myc-FHL2 and Flag-FOXO1 in DU145 cells, which are human prostate tumor cells. Cell extracts were immunoprecipitated and immunoblotted with antibodies to the tags. In cells expressing both proteins, antibody to Myc co-precipitated FOXO1, and antibody to Flag co-precipitated Myc-FHL2 (Figure 1A). Co-precipitation did not occur in cells expressing Myc-FHL2 or Flag-FOXO1 alone, and thus is not the result of antibody crossreactivity.
Figure 1.

FHL2 interacts with FOXO1 and inhibits the transcriptional activity of the amino terminus of FOXO1. (A) DU145 cells were transfected with Myc-FHL2 (1 μg) and Flag-FOXO1 (1 μg). Cell extracts were immunoprecipitated with rabbit anti-Flag or anti-Myc antibody. Immunoprecipitates were immunoblotted with anti-Myc, rabbit anti-Flag, M2 anti-Flag, or goat anti-FHL2 antibody as indicated. (B) DU145 cells in medium containing 5% sFBS were transfected with Myc-FHL2 (1 μg) and Flag-FOXO1 (1 μg). Cells were stained with DAPI and incubated with anti-FHL2 and M2 antibodies. Immunoreactivity was detected with IgG conjugated to Alexa Fluor 594 (red, detects FHL2) or FITC (green, detects FOXO1). Colocalization was visualized by high-resolution imaging with deconvolution microscopy (panel B6). (C) DU145 cells were transfected with 1 μg of His-FOXO1(1–150), and cell extracts were incubated with GST or with GST-FHL2 fusion proteins prebound to glutathione beads. Proteins bound to the beads and the input (50% of extract protein used for precipitations) were immunoblotted with goat anti-FOXO1 antibody. (D) DU145 cells were transfected with Gal4 reporter 3 × 17merLuc (firefly, 0.3 μg) and pRL-null (Renilla, 0.01 μg) and the indicated amounts of Gal4-FOXO1(1–150) and Myc-FHL2. Activity of the firefly luciferase was normalized to that of the Renilla luciferase and expressed as relative luciferase unit (RLU).
To substantiate the interaction data, the subcellular distribution of FHL2 and FOXO1 in DU145 cells was determined by immunofluorescence microscopy. As shown in Figure 1B, a significant amount of ectopic FOXO1 was nuclear (panel B2). This finding is consistent with the presence of functional PTEN, and thus of relatively low amounts of PKB activity, in DU145 cells (Ramaswamy et al, 1999). Some cytoplasmic staining was detected, presumably because the cells were in medium containing 5% serum, which may activate PKB. FHL2 exhibited a similar pattern of distribution as did FOXO1 (panel B3) and apparently colocalized with FOXO1 in the nucleus, as indicated by the yellow color in the merged image (panel B4). The colocalization is more obvious in the high-resolution deconvolution image (panel B6). FHL2 and FOXO1 did not colocalize in the cytosol, thus suggesting that FHL2 interacts preferentially with nuclear FOXO1.
To define the region of FHL2 that interacts with FOXO1, cell extracts prepared from DU145 cells expressing His-tagged FOXO1 (amino acids 1–150) were incubated with recombinant FHL2 proteins of various lengths fused to glutathione S-transferase (GST). As determined by ‘pull-down' assays with glutathione beads, full-length FHL2 interacted with FOXO1, as did FHL2 lacking the Lim3 and Lim4 domains (Figure 1C). Deletion of the Lim2 domain, however, ablated the interaction of FHL2 with FOXO1. Thus, binding of FHL2 to FOXO1 requires Lim2 and perhaps also the one and a half Lim domain in the amino terminus of FHL2.
When fused to the DNA binding domain of Gal4, the amino terminus of FOXO1 (amino acids 1–150) activated the Gal4 reporter gene (Figure 1D). This finding indicates that this region contains an intrinsic activation function. Cotransfection of increasing amounts of FHL2 progressively inhibited Gal4 activation by FOXO1, thus suggesting that FHL2 is a FOXO1 suppressor.
FHL2 inhibits gene expression induced by direct binding of FOXO1 to DNA and the apoptosis of cells by FOXO1
FOXO1 activates the transcription of the IGFBP1 gene by directly binding to the IRS in the IGFBP1 promoter (Figure 2A). To determine whether FHL2 affects the transcriptional activity of FOXO1, we transfected DU145 cells with a luciferase reporter construct containing three copies of the IRS (3 × IRSLuc) (Guo et al, 1999; Tang et al, 1999) and with plasmids encoding FHL2 and full-length FOXO1. Ectopic expression of FOXO1 increased luciferase activity (and thus the expression of luciferase), and coexpression of FHL2 substantially reduced its capacity to do so (Figure 2B). FHL2 did not affect FOXO1 abundance (Figure 2B, inset) and thus apparently decreases the specific activity of FOXO1.
Figure 2.
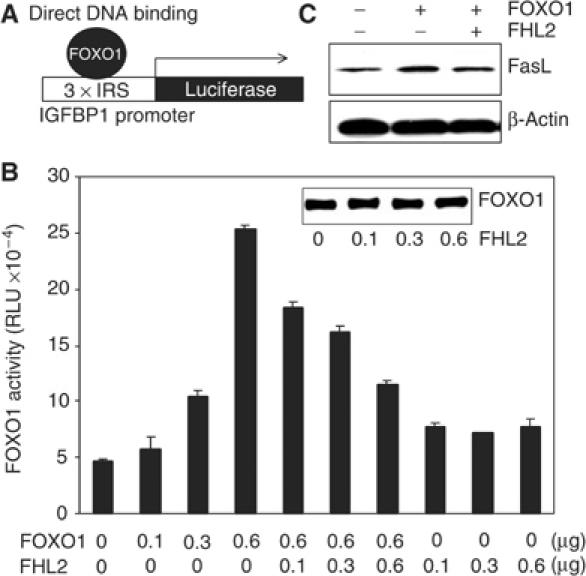
FHL2 suppresses FOXO1 action through the mode of direct DNA binding. (A) Diagram depicting FOXO1 action by direct DNA binding. (B) DU145 cells were transfected with 3 × IRSLuc (0.3 μg), pCMVβGal (0.3 μg), and the indicated amounts of FOXO1 and FHL2 vectors. FOXO1 activity is expressed as relative luciferase units (RLU) normalized to β-gal activity. (C) DU145 cells were transfected with Flag-FOXO1 (4 μg) and Myc-FHL2 (4 μg). Cell extracts were immunoblotted with anti-Fas ligand antibody 24 h post-transfection. A β-actin immunoblot is included as a loading control.
Another gene activated by FOXO1 by direct DNA binding is the gene encoding Fas ligand. To determine whether the inhibitory effects of FHL2 on FOXO1 activity translate into changes in the abundance of endogenous proteins encoded by FOXO1-responsive genes, we monitored amounts of Fas ligand in DU145 cells ectopically expressing FOXO1 or FOXO1 and FHL2 by immunoblotting. Expression of FOXO1 increased the abundance of Fas ligand, and coexpression of FHL2 reduced the extent of this increase (Figure 2C). These data support the results of the reporter assay in Figure 2B and importantly show that the inhibition of the mode of FOXO1 action through ‘direct DNA binding' by FHL2 is not limited to reporter genes. As estimated using green fluorescence protein (GFP) vectors, the transfection efficiency of DU145 cells in the current conditions is about 40% (data not shown). Therefore, the data shown underestimate the effects of FOXO1 on Fas ligand expression.
Our earlier studies show that FOXO1 induces the apoptosis of DU145 cells (Li et al, 2003). As shown here, FHL2 suppresses the transcriptional activity of FOXO1 in reporter assays and prevents the accumulation of Fas ligand (and presumably of other apoptotic proteins encoded by FOXO1-responsive genes). Thus, we predict that FHL2 will inhibit FOXO1-induced apoptosis. To test this prediction, DU145 cells were transfected with CD20 and either FOXO1 or FOXO1 and FHL2. After FACS-based sorting, the apoptotic index of CD20-positive cells was determined by the Annexin V method. FOXO1 increased the apoptotic index about two-fold in four independent experiments, and FHL2 reduced the extent of this increase (Figure 3A). We also found that caspase-3 was active in cells expressing FOXO1 but was not appreciably active in cells expressing both FOXO1 and FHL2 (Figure 3B). These findings show that FHL2 reverses the apoptotic actions of FOXO1 in DU145 cells. Such actions may or may not involve upregulation of the gene encoding Fas ligand. As shown below (Figure 8D), FOXO1 also increases the expression of the apoptotic protein Bim, and FHL2 antagonizes this action of FOXO1.
Figure 3.
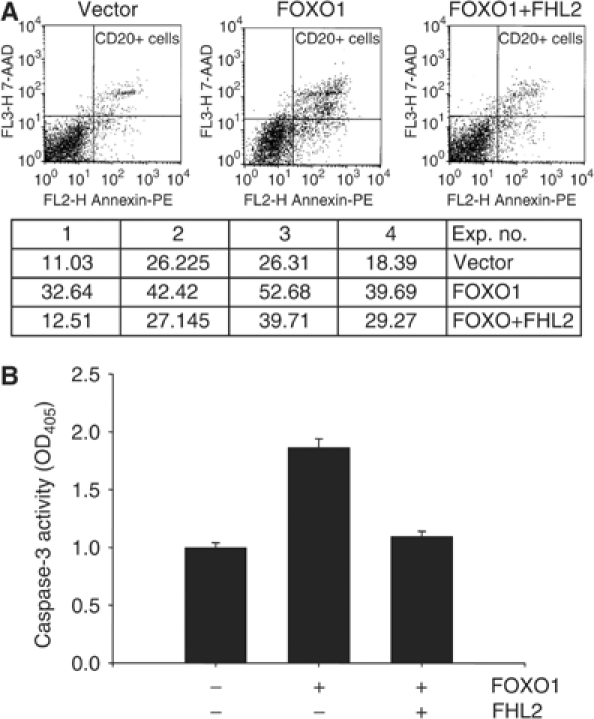
FHL2 suppresses FOXO1-induced apoptosis. (A) DU145 cells were transfected with pCMV-CD20 (3 μg), Flag-FOXO1 (2.5 μg), and Myc-FHL2 (2.5 μg) as indicated. At 16 h post-transfection, CD20-positive cells were separated from CD20-negative cells by FACS-based sorting. The apoptotic index of CD-20 positive cells was determined as described in Materials and Methods. Representative profiles are shown in the top graphs. Apoptotic indexes from four independent experiments are shown in the table below the graphs. (B) DU145 cells were transfected with FOXO1 (4 μg) and FHL2 (4 μg) as indicated. The activity of capase-3 was determined as described in Materials and methods.
Figure 8.
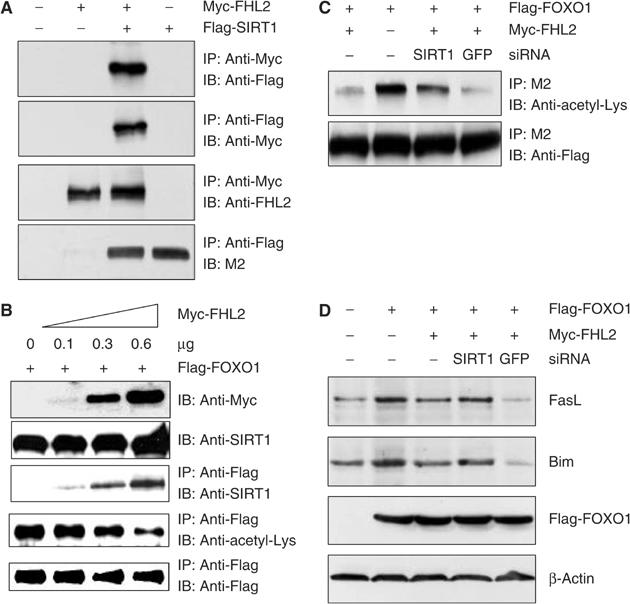
FHL2 decreases FOXO1 acetylation by promoting the interaction of FOXO1 with SIRT1. (A) DU145 cells were transfected with Myc-FHL2 (1 μg) and Flag-SIRT1 (1 μg) as indicated. Cell extracts were immunoprecipitated with anti-Myc or rabbit anti-Flag antibody, and immune complexes were immunoblotted with rabbit anti-Flag, anti-Myc, M2 anti-Flag, or anti-FHL2 antibody. (B) DU145 cells were transfected with Flag-FOXO1 (0.3 μg) and the indicated amounts of Myc-FHL2. Cell extracts were immunoprecipitated and immunoblotted with the indicated antibodies. (C) DU145 cells were transfected with Flag-FOXO1 (3 μg), Myc-FHL2 (3 μg), and 6 μg of siRNA vectors. Cell extracts were immunoprecipitated with the indicated antibodies. (D) DU145 cells were transfected with Flag-FOXO1 (3 μg), Myc-FHL2 (3 μg), and 6 μg of the siRNA vectors. Cells were harvested 24 h after transfection, and amounts of Fas ligand, Bim, and β-actin (loading control) were determined by immunoblotting.
Lysophosphatidic acid promotes the nuclear accumulation of FHL2 and the formation of FHL2-FOXO1 complexes and inhibits FOXO1 activity in prostate cancer cells
Prostate cancer cells express FHL2, and nuclear accumulation of FHL2 correlates with the progression of prostate cancers to higher Gleason scores (Muller et al, 2002). Thus, factors that promote the nuclear translocation of FHL2 may enhance prostatic tumorigenesis by inhibiting the expression of FOXO-responsive genes. Such factors include the serum factor lysophosphatidic acid (LPA), and the effects of LPA on the subcellular location of FHL2 and FOXO1 and on the activity of FOXO1 and its interaction with FHL2 were determined.
Serum-starved DU145 cells were treated with and without LPA for 10 h, and nuclear and cytosolic extracts were immunoblotted with antibodies to FOXO1 and FHL2. FOXO1 was predominantly nuclear in both serum-starved and LPA-treated cells (Figure 4A). In contrast, FHL2 resided in both the cytosol and nucleus of serum-starved cells and translocated to the nucleus in response to LPA, as has been reported previously (Muller et al, 2002). Similar to ectopically expressed proteins, endogenous FOXO1 interacted with FHL2, as demonstrated by co-precipitation of FHL2 with anti-FOXO1 antibody (Figure 4B). There were more FOXO1–FHL2 complexes in LPA-treated cells than in serum-starved cells; this presumably reflects the increased abundance of FHL2 in the nucleus and not an increase in binding affinity of the two proteins for each other.
Figure 4.

Interaction of endogenous FHL2 and FOXO1 in the nucleus of PTEN-intact prostate cancer cells in the presence or absence of LPA. (A) DU145 cells were starved in serum-free medium for 36 h and treated with or without 20 μM LPA for 10 h. Nuclear and cytosolic extracts were immunoblotted with anti-FHL2, anti-FOXO1, anti-Hsp60, and anti-PARP antibodies as indicated. Hsp60 and PARP are cytosolic and nuclear proteins, respectively, and are used as controls for subfractionation. (B–D) DU145 (B), JCA1 (C), and 267B1 (D) cells were starved and treated with 20 mM LPA as in (A). Nuclear extracts were immunoprecipitated with rabbit anti-FOXO1 or rabbit anti-Flag (as a control) antibody. The immunoprecipitates were immunoblotted with anti-FHL2 or goat anti-FOXO1 antibody as indicated. Immunoblotting with anti-PARP antibody without prior immunoprecipitation was included as a control.
Co-precipitation experiments were also performed on control and LPA-treated JCA1 cells (Figure 4C) and 267B1 cells (Figure 4D), and results similar to those in Figure 4B were obtained. JCA1 cells are prostate cancer cells with an intact PTEN, as are DU145 cells, and 267B cells are immortalized but nontumorigenic prostate epithelial cells. These results show that enhancement of FOXO1–FHL2 interaction by LPA is not limited to DU145 cells.
To assess the effects of the LPA-induced nuclear localization of FHL2 on the transcriptional activity of FOXO1, we monitored FOXO1 activity in DU145 cells expressing 3 × IRSLuc and either vector alone or vector encoding siRNA to FHL2 or to GFP (negative control). As shown in Figure 5A, expression of FHL2 siRNA substantially reduced the abundance of endogenous FHL2, whereas expression of GFP siRNA had little effect. LPA reduced FOXO activity in cells expressing empty vector or GFP siRNA but did not reduce FOXO activity in cells expressing FHL2 siRNA (Figure 5B). Thus, depletion of FHL2 attenuates the inhibitory effects of LPA on FOXO1 activity. These experiments were repeated in JCA1 cells (Figure 5C) and 267B cells (Figure 5D), and similar results were obtained.
Figure 5.
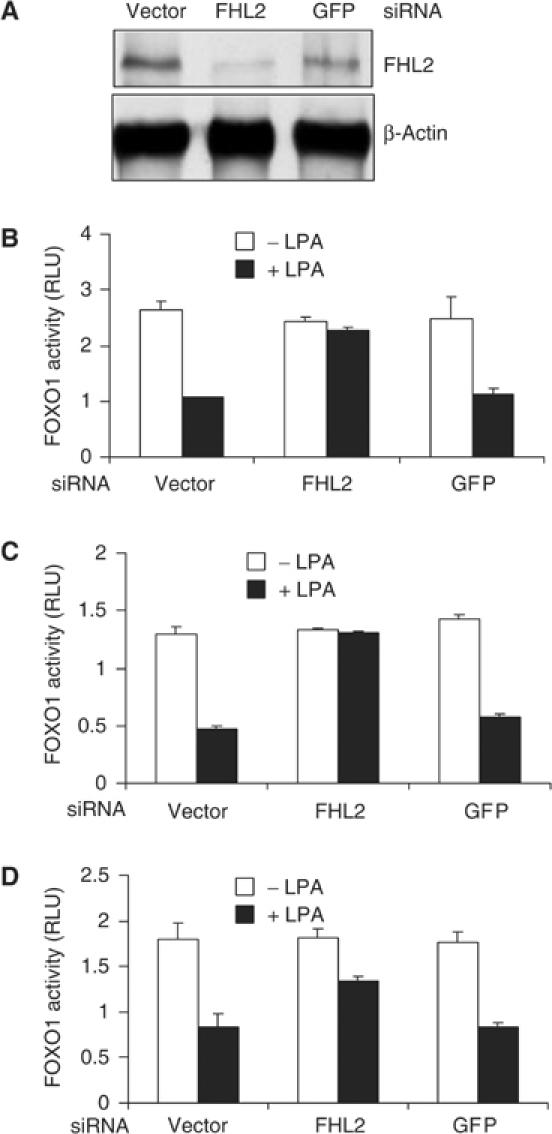
Inhibition of endogenous FOXO1 activity by LPA requires FHL2. (A) DU145 cells were transfected with 4 μg of pcDNA3 (vector) or the indicated siRNA vectors. Cell extracts were immunoblotted with anti-FHL2 or anti-β-actin antibody. (B–D) DU145 (B), JCA1 (C), and 267B1 (D) cells were transfected with 3 × IRSLuc (0.3 μg), pRL-null (0.01 μg), and 1 μg of the indicated siRNA vectors. Luciferase activity was assayed and normalized as in Figure 1D.
Overall, the results of the studies shown in Figures 4 and 5 suggest that LPA may promote prostate tumorigenesis by sending FHL2 to the nucleus where it interacts with and inhibits the antitumor activity of FOXO1.
Relief of FHL2-mediated suppression of FOXO1 activity by the inhibition or depletion of SIRT1
The inhibitory affects of FHL2 on FOXO1 activity suggest that FHL2 functions as a corepressor of FOXO1 activity. Because corepressors often recruit histone deacetylases (HDACs) to transcriptional complexes, we asked whether inhibition of FOXO1 activity by FHL2 requires HDAC activity. In these experiments, DU145 cells were transfected with 3 × IRSLuc and with plasmids encoding FHL2, FOXO1, or both; cells were subsequently treated with and without nicotinamide, an inhibitor of the SIRT family of HDACs. At 5 mM, nicotinamide partially blocked the inhibition of FOXO1 activity by FHL2; at 20 mM, nicotinamide completely blocked this process (Figure 6A). Nicotinamide also increased FOXO1 activity in cells ectopically expressing FOXO1 but not FHL2. These findings show that FHL2 (both exogenous and endogenous) inhibits FOXO1 activity in a manner dependent on SIRT activity.
Figure 6.
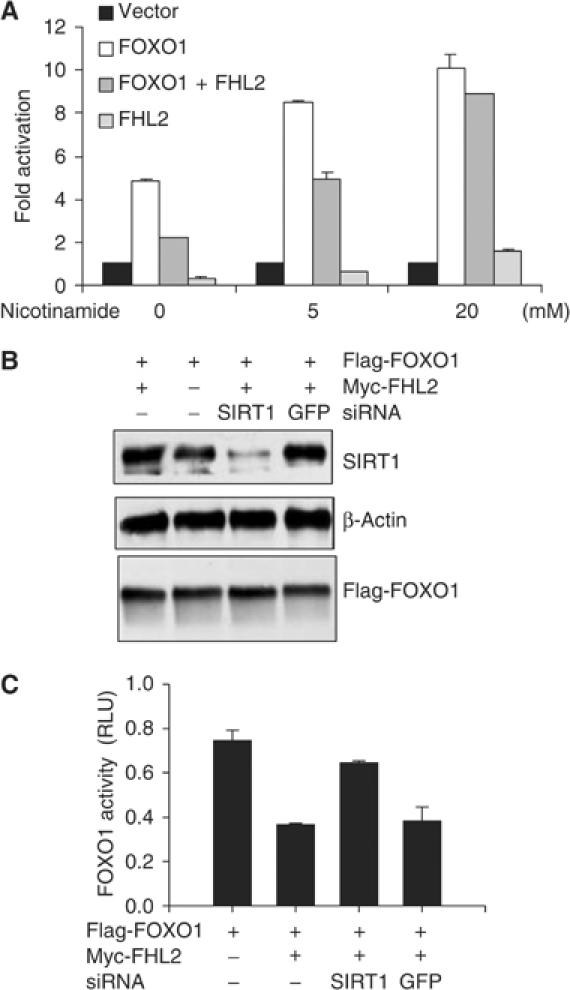
Inhibition of FOXO1 activity by FHL2 requires SIRT1 activity. (A) DU145 cells were transfected with 3 × IRSLuc (0.3 μg) and pCMVβGal (0.3 μg). As indicated, cells were cotransfected with Flag-FOXO1 (0.3 μg) and Myc-FHL2 (0.3 μg). Transfected cells received 5 or 20 mM nicotinamide for 24 h. Luciferase activity was determined and normalized to β-gal activity. Fold activation was calculated by dividing the normalized activity of cells expressing FOXO1 and FHL2 by that of the corresponding vector controls. (B) DU145 cells were transfected with Flag-FOXO1 (3 μg), Myc-FHL2 (3 μg), and 6 μg of SIRT1 or GFP siRNA. Cell extracts were immunoblotted with anti-SIRT1, anti-β-actin, or M2 antibody. (C) DU145 cells were transfected with 3 × IRSLuc (0.3 μg), pRL-null (0.01 μg), Flag-FOXO1 (0.3 μg), and 0.6 μg of the indicated siRNA vectors. Luciferase activity was assayed and normalized as in Figure 1D.
Among the members of the SIRT family, SIRT1 is the closest mammalian homolog of yeast Sir2. To assess the need for SIRT1 in the FHL2 inhibition of FOXO1 activity, we depleted DU145 cells of SIRT1 by RNA interference. Western blots showed substantial reductions in SIRT1 abundance in cells expressing SIRT1 siRNA but not in cells expressing GFP siRNA (Figure 6B). Depletion of SIRT1 had no effect on FOXO1 abundance. As monitored by reporter assays with 3 × IRSLuc, FHL2 inhibited FOXO1 activity in cells expressing GFP siRNA, but did not inhibit FOXO1 activity in cells expressing SIRT1 siRNA (Figure 6C). Thus, depletion of SIRT1 abrogates the inhibitory effects of FHL2 on FOXO1 activity.
SIRT1 interacts with and deacetylates FOXO1 and inhibits FOXO1 activity
To determine the mechanism by which SIRT1 represses FOXO1 activity, we first asked whether SIRT1 and FOXO1 interact. DU145 cells were transfected with plasmids encoding Flag-SIRT1, HA-FOXO1, or both, and co-precipitation experiments were performed. Antibody to anti-Flag co-precipitated HA-FOXO1 and antibody to HA co-precipitated Flag-SIRT1 from cells expressing both proteins but not either protein alone (Figure 7A). Thus, SIRT1 associates with FOXO1 in vivo.
Figure 7.
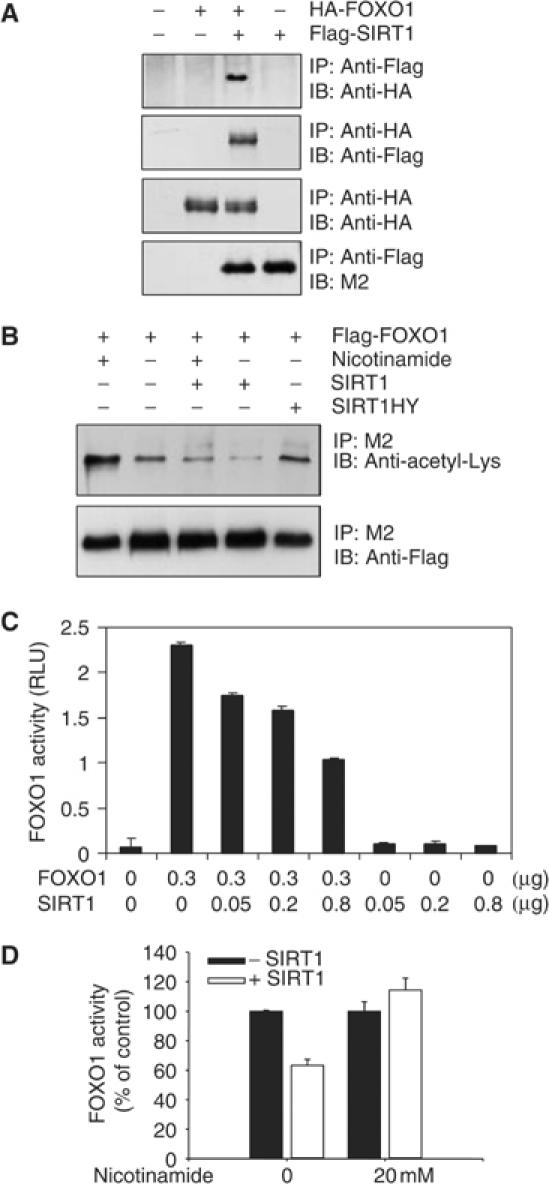
SIRT1 binds to FOXO1 and inhibits FOXO1 by deacetylating FOXO1. (A) DU145 cells were transfected with HA-FOXO1 (1 μg) and Flag-SIRT1 (1 μg). Cell extracts were immunoprecipitated with rabbit anti-Flag or mouse anti-HA antibody, and immunoprecipitates were immunoblotted with rabbit anti-HA or M2 anti-Flag antibody as indicated. (B) DU145 cells were transfected with 4 μg of either Flag-FOXO1, pcDNA-SIRT1, pcDNA-SIRT1HY, or pcDNA3 and treated with or without 5 mM nicotinamide as indicated. Cell extracts were immunoprecipitated with M2 anti-Flag antibody. The immunoprecipitates were immunoblotted with an anti-acetyl lysine antibody (upper panel) or rabbit anti-Flag antibody (lower panel). (C) DU145 cells were transfected with 3 × IRSLuc (0.3 μg), pRL-null (0.01 μg), and the indicated amounts of Flag-FOXO1 and pcDNA3 SIRT1. Luciferase activity was assayed and normalized as in Figure 1D. (D) DU145 cells were transfected with 3 × IRSLuc (0.3 μg), pRL-null (0.01 μg), Flag-FOXO1 (0.3 μg), and 0.3 μg of either pcDNA3 or pcDNA-SIRT1. Transfected cells were treated with or without nicotinamide for 24 h as indicated. Luciferase activity was assayed and normalized as in Figure 1D. FOXO1 activity is expressed as % of control and was calculated by normalizing the activity in cells transfected with SIRT1 to that of the vector controls (as 100%).
In addition to histones, SIRT1 also deacetylates non-histone proteins such as p53 (Luo et al, 2001; Vaziri et al, 2001). Therefore, in our next set of experiments, we asked whether SIRT1 deacetylates FOXO1. Flag-FOXO1 was expressed in DU145 cells with either empty vector or vector encoding SIRT1 or a SIRT1 mutant that lacks deacetylase activity (SIRT1HY) (Vaziri et al, 2001). Cell extracts were immunoprecipitated with antibody to Flag and immunoblotted with antibody to acetyl-lysine. Cells expressing SIRT1 contained less acetylated Flag-FOXO1 than did cells expressing empty vector or SIRTHY (Figure 7B). SIRT1 did not appreciably reduce amounts of acetylated Flag-FOXO1 in cells receiving 5 mM nicotinamide. These data show that SIRT1 reduces the abundance of acetylated FOXO1 in a manner dependent on its deacetylase activity. Consistent with its capacity to increase FOXO1 activity (see Figure 6A), nicotinamide also increased the abundance of acetylated FOXO1 in cells expressing FOXO1 in the absence of ectopic SIRT1.
A third set of experiments examined the effects of SIRT1 on the transcriptional activity of FOXO1 using the 3 × IRSLuc reporter construct. As shown in Figure 7C and D, ectopic expression of SIRT1 reduced FOXO activity in the absence but not in the presence of nicotinamide. These data are in agreement with recent reports by Motta et al (2004) and Brunet et al (2004), which show that SIRT1 deacetylates and inhibits the activity of FOXO factors.
Enhancement of the interaction of FOXO1 and SIRT1 by FHL2
Our data show that FHL2 does not inhibit FOXO1 activity in the absence of SIRT1 activity (Figure 6). Thus, FHL2 may recruit SIRT1 to FOXO1, thus allowing SIRT1 to interact with and deacetylate FOXO1 and inhibit FOXO1 activity (Figure 7). As demonstrated by co-precipitation assays of ectopically expressed proteins, Myc-FHL2 interacts with Flag-SIRT1 (Figure 8A). Co-precipitation was observed in cells expressing both tagged proteins but not either protein alone.
To determine whether FHL2 modulates the interaction of FOXO1 with SIRT1, we cotransfected DU145 cells with plasmids encoding Flag-FOXO1 and increasing amounts of Myc-FHL2, and co-precipitation assays were performed. Expression of Myc-FHL2 substantially increased the abundance of FOXO1-associated SIRT1 without affecting the total abundance of Flag-FOXO1 or endogenous SIRT1. Increased binding of SIRT1 to FOXO1 was paralleled by reductions in amounts of acetylated Flag-FOXO1 (Figure 8B).
In other experiments, Flag-FOXO1, Myc-FHL2, and SIRT1 were expressed in DU145 cells. Myc-FHL2 substantially reduced the abundance of acetylated Flag-FOXO1 in the absence but not in presence of SIRT1 siRNA (Figure 8C). More importantly, depletion of SIRT1 ablated the capacity of FHL2 to inhibit endogenous gene expression by FOXO1. As shown in Figure 8D, expression of FOXO1 increased the abundance of Fas ligand and Bim. Coexpression of FHL2 reduced the extent of increase, whereas coexpression of FHL2 and SIRT1 siRNA did not. These studies show that FHL2 suppresses FOXO1 activity by SIRT1-mediated deacetylation of FOXO1.
FHL2 inhibits gene expression regulated by the ‘tethering' of FOXO1 to other transcription factors
As mentioned in Introduction, the FOXO factors repress the transcription of the cyclin D1 gene by a mechanism that does not require the direct interaction of FOXO1 with the IRS (Ramaswamy et al, 2002; Schmidt et al, 2002). This finding suggests that FOXO1 represses cyclin D1 gene transcription by ‘tethering' to other sequence-specific transcription factors (Figure 9A). To determine whether FHL2 affects the capacity of FOXO1 to modulate gene expression by ‘tethering', we transfected LNCaP cells with a luciferase reporter driven by the cyclin D1 promoter and with plasmids encoding FHL2 and FOXO1. As shown in Figure 9B, expression of FOXO1 decreased the activity of the cyclin D1 promoter, and coexpression of FHL2 ablated its capacity to do so.
Figure 9.
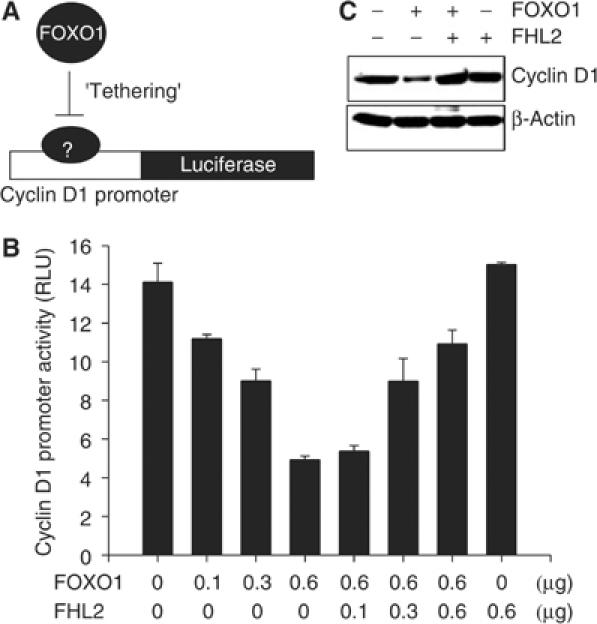
FHL2 suppresses the ‘tethering' mode of FOXO1 action. (A) Diagram depicting FOXO1 action through ‘tethering'. (B) LNCaP cells were transfected with the pGL2-CD1 reporter plasmid (0.3 μg), pRL-null (0.01 μg), and the indicated amounts of Flag-FOXO1 and Myc-FHL2. Luciferase activity was assayed and normalized as in Figure 1D. (C) DU145 cells were transfected with Flag-FOXO1 (4 μg) and 4 μg of either Myc-FHL2 or pcDNA3. Amounts of endogenous cyclin D1 protein were determined by immunoblotting with anti-cyclin D1 antibody. A β-actin immunoblot is included as a loading control.
We also monitored amounts of cyclin D1 protein in cells expressing Flag-FOXO1 and Myc-FHL2. Consistent with the results of the reporter assay, FOXO1 reduced the abundance of cyclin D1 in the absence but not in the presence of ectopic FHL2. Together, the data in Figures 2 and 9 show that FHL2 inhibits the transcriptional activity of FOXO1 regardless of whether FOXO1 directly binds DNA or associates with other transcription factors.
Discussion
Despite the need for FOXO1 for multiple biological processes, little is known about the cofactors that potentially regulate its transcriptional activity. Our study shows that FHL2 interacts with the amino terminus of FOXO1, a putative transactivation region (Puigserver et al, 2003). Several findings indicate that FHL2 negates the transcriptional activity of FOXO1 by promoting its deacetylation by SIRT1 (Figure 10). First, FHL2 colocalizes and interacts with FOXO1 in prostate cancer cells. The interaction of endogenous FHL2 and FOXO1 occurs in the nucleus and is enhanced by the serum growth factor LPA. Second, FHL2 inhibits the activation of an IRS-driven reporter gene by FOXO1 in the presence but not in the absence of SIRT1 activity. This was demonstrated by use of SIRT1 siRNA and the SIRT inhibitor nicotinamide. FHL2 also attenuates the expression of endogenous FOXO1-targeted genes by FOXO1 and inhibits FOXO1-induced apoptosis. Third, SIRT1 binds FOXO1, decreases its acetylation, and inhibits its transcriptional activity. Fourth, FHL2 interacts with SIRT1 and promotes its interaction with FOXO1. Finally, FHL2 decreases FOXO1 acetylation in a manner dependent on the deacetylase activity of SIRT1.
Figure 10.
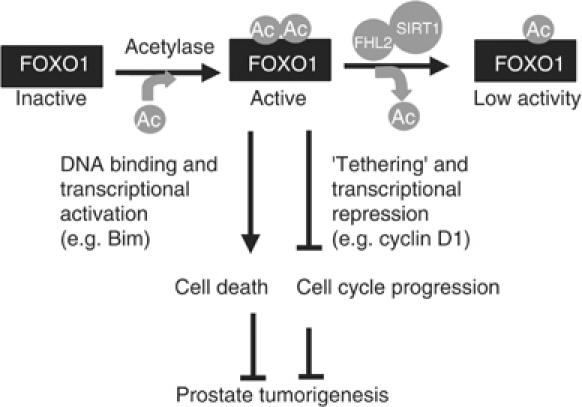
A model of how FHL2 regulates FOXO1 activity by SIRT1-mediated deacetylation. Active FOXO1 suppresses prostate tumorigenesis by inhibiting cell cycle progression and by inducing apoptosis. FOXO1 activation in prostate cancer cells requires acetylation at yet to be defined sites. These sites are essential for its modulation of gene expression by both direct DNA binding and ‘tethering'. FHL2 promotes prostate tumorigenesis by serving as an adaptor that couples SIRT1 and FOXO1 and thus inhibits FOXO1 action in prostate cells.
Our data in Figure 1D showed that the amino terminus of FOXO1 contains an intrinsic activation function that is suppressed by FHL2 binding. However, FHL2 also suppressed cyclin D1 repression by FOXO1. It is thus likely that the amino terminus is a region essential for both transcriptional activation and repression. Because FHL2 antagonizes both the activation and the repression functions of FOXO1, it differs from classical transcriptional corepressors. It functions more like an adaptor that couples SIRT1 to FOXO1 and thus allows SIRT1 to deacetylate FOXO1. In addition to SIRT1 recruitment, the decreases in the acetylation and activity of FOXO1 by FHL2 may also involve interference with the interaction between FOXO1 and histone acetyl transferases associated with transcriptional coactivators such as PGC-1, which interacts with the amino terminus of FOXO1 (Puigserver et al, 2003).
It has been well established that acetylation of histones generates novel adhesive surfaces required for the recruitment of transcription complexes (Agalioti et al, 2002). Acetylation is also known to regulate the DNA binding activity of transcription factors such as p53 (Gu and Roeder, 1997) and E2F1 (Martinez-Balbas et al, 2000). FOXO1 contains multiple lysine residues, the acetylation of which might regulate its direct transcriptional activity and ‘tethering' (Figure 10). FHL2–SIRT1 complex may deacetylate FOXO1 at two separate lysine residues, one of which is critical for DNA binding whereas the other for ‘tethering'. Alternatively, the complex may deacetylate FOXO1 at a single lysine residue that, at acetylated status, provides a ‘docking' site for interaction with a ‘ying-yang' transcriptional cofactor that is involved in FOXO1 action through either direct transcriptional activation or ‘tethering', depending on the promoter context. While we have presented multiple evidences for the involvement of SIRT1-mediated deacetylation in the suppression of direct transcriptional activity of FOXO1 by FHL2, it remains to be determined whether deacetylation is involved in the relief of FOXO1-mediated suppression of cyclin D1 expression by FHL2. For example, FHL2 has been shown to be a coactivator for β-catenin (Wei et al, 2003) that upregulates the expression of cyclin D1 (Tetsu and McCormick, 1999). It is possible that the effect of FHL2 on cyclin D1 involves its interaction with β-catenin. Although the data in Figure 9 show that the effect of FHL2 on cyclin D1 depends on FOXO1, it does not exclude the possibility that FOXO1 is ‘tethered' to β-catenin on cyclin D1 promoter and that FHL2 regulates the ‘tethering' through its ability to interact with both FOXO1 and β-catenin.
Although the current study focuses on FOXO1, it is possible that FHL2 also modulates the activities of other FOXO factors. The FOXO factors share a similar domain structure and mode of activation/inactivation by phosphorylation. Indeed, recent studies by Motta et al (2004) and Brunet et al (2004) show that SIRT1 deacetylates and inhibits the activity of FOXO3a, FOXO4, and FOXO1. On the other hand, in the studies by Fukuoka et al (2003) and Van der Horst et al (2004), acetylation of FOXO4 inhibited its activity. Thus, whether our findings on FOXO1 extend to other FOXO factors requires further study.
Previous studies (Nakamura et al, 2000; Li et al, 2001, 2003) have shown that FOXO1 acts downstream of PTEN to induce the apoptosis of prostate cancer cells. Our current work suggests that FHL2 may negate the antitumor activity of PTEN by inactivating FOXO1 in these cells. The enhanced interaction of FHL2 with FOXO1 in LPA-treated cells supports the existence of an additional pathway in prostate cancer cells that inhibits FOXO activity independently of PKB. Androgens promote prostate tumorigenesis, and FHL2 may enhance their tumor-promoting activity in two ways. First, FHL2 is a coactivator for the androgen receptor and colocalizes with the receptor in the nucleus of prostate epithelial cells (Muller et al, 2000). Second, FHL2 may cooperate with androgens to inhibit FOXO1 activity. We (Li et al, 2003) have shown that androgens inhibit FOXO1 activity by promoting the interaction of FOXO1 with the androgen receptor. Consistent with the idea that FHL2 promotes tumorigenesis, others have shown that (1) stimulation of the Rho signaling pathway induces the translocation of FHL2 to the nucleus and the subsequent activation of androgen receptor-dependent genes (Muller et al, 2002), (2) FHL2 is a serum-inducible transcriptional coactivator of AP-1 (Morlon and Sassone-Corsi, 2003); and (3) FHL2 is a coactivator of β-catenin (Wei et al, 2003). These proteins have been implicated in prostate tumorigenesis.
In yeast, the abundance of Sir2 increases in response to caloric restriction and sublethal levels of stress. Because extension of the lifespan of C. elegans by Sir2 requires the FOXO-related protein Daf-16 (Tissenbaum and Guarente, 2001), it may appear paradoxical that SIRT1 inhibited FOXO1 activity. Recently, SIRT1 was shown to suppress the induction of proapoptotic genes by FOXO3a but to enhance the induction of genes involved in growth inhibition (e.g. p27Kip1) and stress responses (e.g. GADD45) (Brunet et al, 2004). Moreover, deacetylation of FOXO1 by SIRT1 potentiates the FOXO1-dependent expression of p27Kip1 and manganese superoxide dismutase (Daitoku et al, 2004). Similarly, deacetylation of FOXO4 by SIRT1 relieves the inhibition of FOXO4 activity by CREB-mediated acetylation and enhances the expression of p27Kip1 (Van der Horst et al, 2004). These studies suggest that Sir2 and its mammalian homologs may increase organismal longevity by shifting the FOXO-dependent response away from apoptosis and toward stress resistance and growth inhibition, which would explain the need for Daf-16 in the increase of longevity by members of Sir2 family. However, SIRT1 has recently been shown to augment TNFα-induced apoptosis by inhibiting the transcriptional activity of NF-κB (Yeung et al, 2004), suggesting a positive role of SIRT1 in the apoptosis of mammalian cells. It remains to be determined whether the effect of FHL2 on the activity of FOXO1 is sensitive to stress conditions or depends on cell type, FOXO subtype, or the promoter context of specific FOXO target genes.
Because the members of Sir2 family and the FOXO factors increase longevity, our finding that FHL2 inhibits FOXO1 through SIRT1 reveals a potential novel function for this LIM protein in the control of longevity. Cancer is considered an aging disease. This is particularly true for prostate cancers, whose incidence increases with age more rapidly than does that of other types of cancer. The inhibition of apoptosis by interaction of FHL2, FOXO1, and SIRT1 in prostate cancer cells may represent a novel molecular pathway the interruption of which could lead to the development of strategies that prolong lifespan without increasing the risk of prostate cancer.
Although the current data were generated in prostate cancer cells, the functional interaction between FHL2, SIRT1, and FOXO1 may also occur in other human cancers, even though the nature of the interaction may vary. In this regard, it is interesting that FHL2 was initially identified as a protein that is downregulated in rhabdomyosarcoma (Genini et al, 1997), whereas FOXO1 was identified as a gene involved in chromosomal translocation in the same disease (Galili et al, 1993).
Materials and methods
Constructs
Flag-FKHR (Tang et al, 1999), 3 × IRSLuc (Guo et al, 1999; Tang et al, 1999), and pGL2-CD1 (Tetsu and McCormick 1999) have been described in previous studies. To generate the mammalian and bacterial expression vectors for FHL2, full-length FHL2 cDNA was amplified by polymerase chain reaction (PCR) using the positive clone identified from yeast two-hybrid screening as a template and was inserted into the BamH1 and XhoI sites of pCMV-tag-3B (Stratagene) and pGEX-4T-1 (Pharmacia), respectively. A mammalian vector expressing the amino terminus of FOXO1 (1–150) was constructed by inserting the BamH1/PstI fragment of pFA-CMV-FKHR (Li et al, 2003) into pcDNA4-V5-His(A) (Invitrogen). The HA-FOXO1 vector was constructed by inserting the BamH1/XbaI fragment from Flag-FKHR into pcDNA3-HA (Invitrogen). To generate SIRT1 expression vectors, wild-type SIRT1 and mutant SIRT1 were excised from retroviral vectors (Vaziri et al, 2001) and inserted into the BamH1 and BstX1 sites of pcDNA3.1 or pcDNA3-Flag. SIRT1 and FHL2 siRNAs were subcloned into pSilencer-Neo (Ambion). The corresponding oligonucleotides for generating the SIRT1 siRNA are 5′-GAT CCG CTT CAG TGG CTG GAA CAG TTT CAA GAG AAC TGT TCC AGC CAC TGA AGT TTT TTG GAA A-3′ (forward) and 5′-AGC TTT TCC AAA AAA CTT CAG TGG CTG GAA CAG TTC TCT TGA AAC TGT TCC AGC CAC TGA AGC G-3′ (reverse). The corresponding oligonucleotides for generating the FHL2 siRNA are 5′-GAT CCG TGA CTG AGC GCT TTG ACT GTT CAA GAG ACA GTC AAA GCG CTC AGT CAT TTT TTG GAA A-3′ (forward) and 5′-AGC TTT TCC AAA AAA TGA CTG AGC GCT TTG ACT GTC TCT TGA ACA GTC AAA GCG CTC AGT CAC G-3′ (reverse). The oligonucleotides for GFP siRNA were provided by Ambion.
Yeast two-hybrid screening
The Matchmaker Two-Hybrid System 3 (Clontech) was used to screen the human prostate MATCHMAKER cDNA library (Clontech). The bait plasmid pGBKT-FOXO1 (1–150) was constructed by inserting the BamH1–PstI fragment of pFA-CMV-FKHR (Li et al, 2003) into the NdeI–PstI site of GBKT7. BamH1 and NdeI were blunt ended with Klenow I before ligation. The screening was performed in AH109 yeast cells according to the protocol provided by the Clontech Matchmaker Two-Hybrid System.
Cell culture, transfections, and reporter assays. For reporter analysis, DU145 cells were plated in 12-well plates in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum at 1.5 × 105 cells/well. LNCaP, JCA1, and 267B1 cells were plated in RPMI 1640 at 2 × 105 cells/well. At 1 day after plating, cells were transfected by Lipofectamine Plus or Lipofectamine 2000 following the protocol provided by Gibco/BRL. Transfected cells were cultured in medium containing 5% charcoal-stripped FBS (sFBS) for 24 h. Then, cell lysates were prepared and luciferase activity was determined using the luciferase assay systems or the dual luciferase assay system from Promega Corporation (Madison, WI). β-Gal activity was determined as previously described (Lee et al 2000; Lee and Bai, 2002). Luciferase activity was normalized to Renilla luciferase activity or to β-gal activity.
GST pull-down assays. Plasmids expressing GST fused to full-length or truncated FHL2 were transformed into the BL21 (DE3) strain. Transformed bacteria were cultured at 37°C until OD600 had reached 0.8. IPTG (0.5 mM) was added and the culturing continued overnight at 30°C. Bacterial lysates containing GST fusion proteins were prepared by sonication in buffer containing 50 mM Tris, 10 mM NaCl, 1mM EDTA, 6 mM MgCl2, 1 mM DTT, and 1 mM PMSF and were incubated with extracts of DU145 cells transfected with His-FOXO1(1–150). FOXO–FHL2 complexes were precipitated with Glutathione Sepharose 4B Beads (Amersham Pharmacia, Piscataway, NJ). The beads were then washed with the binding buffer containing 0.5% NP-40 and heated for 5 min in SDS–PAGE sample buffer. After brief centrifugation, the supernatant was resolved on an 8% SDS gel. The presence of FOXO1(1–150) in the precipitates was detected by immunoblotting with goat anti-FOXO1 antibody (Santa Cruz, N18).
Immunological assays. For immunoprecipitations, cells in 100- or 60-mm dishes were transfected as described for reporter assays and were treated as indicated in the figures. Whole-cell extracts or nuclear extracts were incubated with 4 μg of antibody for 4 h at 4°C and subsequently with protein G-agarose beads for an additional 4 h. Beads were washed four times, and the immunoprecipitates were heated in SDS–PAGE sample buffer. After brief centrifugation, the supernatants were resolved on an 8% SDS gel for subsequent immunoblotting analysis. Commercially available antibodies including M2 anti-Flag (Sigma), rabbit anti-Flag (Sigma), anti-Myc (Santa Cruz, 9E10), rabbit anti-FOXO1 (Santa Cruz, H-128), and mouse anti-HA (Berkeley Antibody Laboratories, HA11) antibodies were used for immunoprecipitations.
For immunoblotting, cell extracts, nuclear extracts, or immunoprecipitates were separated on SDS–polyacrylamide gels and transferred to nitrocellulose membranes. Membranes were incubated with antibody, and proteins detected by the antibody were visualized with ECL. Besides the antibodies described above for immunoprecipitations, additional antibodies used for immunoblotting analyses include goat anti-FHL2 (Santa Cruz, C16), anti-Fas ligand (Santa Cruz, N-20), anti-β-actin (Sigma), anti-Hsp60 (Santa Cruz, H-1), anti-PARP (Santa Cruz, H-250), goat anti-FOXO1, anti-SIRT1 (Upstate Biotechnologies), rabbit anti-HA (Cell Signaling), anti-acetyl lysine (Upstate Biotechnologies), anti-Bim (BD Pharmingen), and anti-cyclin D1 (Santa Cruz, HD11) antibodies. Nuclear and cytosolic fractions were prepared according to a standard protocol (Dignam et al, 1983).
For immunofluorescence and deconvolution imaging, DU145 cells transfected with Myc-FHL2 and Flag-FOXO1 (wild type) were cultured in 5% FBS and stained with DAPI and goat anti-FHL2 or with M2 anti-Flag antibodies. FHL2 and FOXO1 were detected by chicken anti-goat IgG conjugated with Alexa Fluor 594 (red)- and fluorescein isothiocyanate (FITC; green)-conjugated anti-mouse IgG (Molecular Probes), respectively. Fluorescent microscopic images were obtained with a Nikon Diaphot microscope using a Photometrix PXL cooled CCD camera. The microscope was equipped with the appropriate filters for three-color imaging of cells and with a motorized stage for obtaining z-series images. Digital image files were processed and deconvolved using Oncor Image software (Oncor Inc.). High-resolution images of the deconvolved and 3-d reconstructed image z-series stacks were processed for presentation with Adobe Photoshop.
Caspase-3 assays. Caspase-3 activity of cell extracts normalized for protein abundance was determined by the Caspase-3/CPP32 Colorimetric Assay Kit (BD Biosciences, Pharmingen) following a protocol from the vendor. Absorption at 405 nm was measured in an MRX microplate reader (DYNEX Technologies, Chantilly, VA). Caspase-3 activity is expressed as OD405 per μg protein.
Cell sorting and apoptotic assays. The apoptotic index of transfected cells was determined with the Annexin V PE Apoptosis Detection Kit (BD Pharmingen). Cells grown in 100 mm dishes were cotransfected with CD20 plasmid that expresses a cell surface antigen. At 16 h post-transfection, the cells were collected in PBS containing 2.5 mM EDTA, washed twice with PBS, and incubated at 4°C for 60 min with FITC-conjugated mouse anti-CD20 antibody (Becton-Dickinson, 20 μl/106 cells). After staining with anti-CD20 antibody and washing, the cells were incubated with Annexin V-PE and 7-amino-actinomycin D (7-AAD) for 30 min at 25°C. CD20-positive cells were separated from the CD20-negative cells by FACS-based cell sorting. The apoptotic index of transfected cells was determined by flow cytometry. Cell sorting and flow cytometry analysis were performed on a Becton-Dickinson FACScan (Mountain View, CA).
Acknowledgments
We thank Dr FG Barr for the permission to use FKHR in the studies, Drs KL Guan and Terry G Unterman for FKHR plasmids, Dr Robert Weinberg for the SIRT1 retroviral plasmids, and Drs Nancy Olashaw and Paul Byvoet for reading the manuscript. FACS analyses were performed in the Flow Cytometry core facility at H Lee Moffitt Cancer Center and Research Institute. This work was supported by the Public Health Service grant CA79530 (WB) and a DOD Prostate Cancer grant DAMD17-02-1-0140 (WB).
References
- Agalioti T, Chen G, Thanos D (2002) Deciphering the transcriptional histone acetylation code for a human gene. Cell 111: 381–392 [DOI] [PubMed] [Google Scholar]
- Anderson MJ, Viars CS, Czekay S, Cavenee WK, Arden KC (1998) Cloning and characterization of three human forkhead genes that comprise an FKHR-like gene subfamily. Genomics 47: 187–199 [DOI] [PubMed] [Google Scholar]
- Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, Anderson MJ, Arden KC, Blenis J, Greenberg ME (1999) Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 96: 857–868 [DOI] [PubMed] [Google Scholar]
- Brunet A, Park J, Tran H, Hu LS, Hemmings BA, Greenberg ME (2001) Protein kinase SGK mediates survival signals by phosphorylating the forkhead transcription factor FKHRL1 (FOXO3a). Mol Cell Biol 21: 952–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, Hu LS, Cheng HL, Jedrychowski MP, Gygi SP, Sinclair DA, Alt FW, Greenberg ME (2004) Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science 303: 2011–2015 [DOI] [PubMed] [Google Scholar]
- Chu PH, Bardwell WM, Gu Y, Ross J Jr, Chen J (2000) FHL2 (SLIM3) is not essential for cardiac development and function. Mol Cell Biol 20: 7460–7462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daitoku H, Hatta M, Matsuzaki H, Aratani S, Ohshima T, Miyagishi M, Nakajima T, Fukamizu A (2004) Silent information regulator 2 potentiates Foxo1-mediated transcription through its deacetylase activity. Proc Natl Acad Sci USA 101: 10042–10047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res 11: 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowell P, Otto TC, Adi S, Lane MD (2003) Convergence of peroxisome proliferator-activated receptor gamma and Foxo1 signaling pathways. J Biol Chem 278: 45485–45491 [DOI] [PubMed] [Google Scholar]
- Fukuoka M, Daitoku H, Hatta M, Matsuzaki H, Umemura S, Fukamizu A (2003) Negative regulation of forkhead transcription factor AFX (Foxo4) by CBP-induced acetylation. Int J Mol Med 12: 503–508 [PubMed] [Google Scholar]
- Galili N, Davis RJ, Fredericks WJ, Mukhopadhyay S, Rauscher FJ III, Emanuel BS, Rovera G, Barr FG (1993) Fusion of a fork head domain gene to PAX3 in the solid tumour alveolar rhabdomyosarcoma. Nat Genet 5: 230–235 [DOI] [PubMed] [Google Scholar]
- Genini M, Schwalbe P, Scholl FA, Remppis A, Mattei MG, Schafer BW (1997) Subtractive cloning and characterization of DRAL, a novel LIM-domain protein down-regulated in rhabdomyosarcoma. DNA Cell Biol 16: 433–442 [DOI] [PubMed] [Google Scholar]
- Gu W, Roeder RG (1997) Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90: 595–606 [DOI] [PubMed] [Google Scholar]
- Guo S, Rena G, Cichy S, He X, Cohen P, Unterman T (1999) Phosphorylation of serine 256 by protein kinase B disrupts transactivation by FKHR and mediates effects of insulin on insulin-like growth factor-binding protein-1 promoter activity through a conserved insulin response sequence. J Biol Chem 274: 17184–17192 [DOI] [PubMed] [Google Scholar]
- Hu MC, Lee DF, Xia W, Golfman LS, Ou-Yang F, Yang JY, Zou Y, Bao S, Hanada N, Saso H, Kobayashi R, Hung MC (2004) IkappaB kinase promotes tumorigenesis through inhibition of forkhead FOXO3a. Cell 117: 225–237 [DOI] [PubMed] [Google Scholar]
- Jacobs FM, van der Heide LP, Wijchers PJ, Burbach JP, Hoekman MF, Smidt MP (2003) FoxO6, a novel member of the FoxO class of transcription factors with distinct shuttling dynamics. J Biol Chem 278: 35959–35967 [DOI] [PubMed] [Google Scholar]
- Kaestner KH, Knochel W, Martinez DE (2000) Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev 14: 142–146 [PubMed] [Google Scholar]
- Kaufmann E, Knöchel W (1996) Five years on the wings of fork head. Mech Dev 57: 3–20 [DOI] [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R (1993) A C. elegans mutant that lives twice as long as wild type. Nature 366: 461–464 [DOI] [PubMed] [Google Scholar]
- Kong Y, Shelton JM, Rothermel B, Li X, Richardson JA, Bassel-Duby R, Williams RS (2001) Cardiac-specific LIM protein FHL2 modifies the hypertrophic response to beta-adrenergic stimulation. Circulation 103: 2731–2738 [DOI] [PubMed] [Google Scholar]
- Lee H, Bai W (2002) Inhibition of estrogen receptor nuclear export by ligand-induced and p38-mediated receptor phosphorylation. Mol Cell Biol 22: 5835–5845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Jiang F, Wang Q, Nicosia SV, Yang J, Su B, Bai W (2000) MEKK1 activation of human estrogen receptor α and stimulation of the agonistic activity of 4-hydroxytamoxifen in endometrial and ovarian cancer cells. Mol Endocrinol 14: 1882–1896 [DOI] [PubMed] [Google Scholar]
- Li P, Lee H, Guo S, Unterman TG, Jenster G, Bai W (2003) AKT-independent protection of prostate cancer cells from apoptosis mediated through complex formation between the androgen receptor and FKHR. Mol Cell Biol 23: 104–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Nicosia SV, Bai W (2001) Antagonism between PTEN/Mmac1/Tep-1 and androgen receptor in growth and apoptosis of prostate cancer cells. J Biol Chem 276: 20444–20450 [DOI] [PubMed] [Google Scholar]
- Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W (2001) Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell 107: 137–148 [DOI] [PubMed] [Google Scholar]
- Martinez-Balbas MA, Bauer UM, Nielsen SJ, Brehm A, Kouzarides T (2000) Regulation of E2F1 activity by acetylation. EMBO J 19: 662–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morlon A, Sassone-Corsi P (2003) The LIM-only protein FHL2 is a serum-inducible transcriptional coactivator of AP-1. Proc Natl Acad Sci USA 100: 3977–3982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motta MC, Divecha N, Lemieux M, Kamel C, Chen D, Gu W, Bultsma Y, McBurney M, Guarente L (2004) Mammalian SIRT1 represses forkhead transcription factors. Cell 116: 551–563 [DOI] [PubMed] [Google Scholar]
- Muller JM, Isele U, Metzger E, Rempel A, Moser M, Pscherer A, Breyer T, Holubarsch C, Buettner R, Schule R (2000) FHL2, a novel tissue-specific coactivator of the androgen receptor. EMBO J 19: 359–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller JM, Metzger E, Greschik H, Bosserhoff AK, Mercep L, Buettner R, Schule R (2002) The transcriptional coactivator FHL2 transmits Rho signals from the cell membrane into the nucleus. EMBO J 21: 736–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N, Ramaswamy S, Vazquez F, Signoretti S, Loda M, Sellers WR (2000) Forkhead transcription factors are critical effectors of cell death and cell cycle arrest downstream of PTEN. Mol Cell Biol 20: 8969–8982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry P, Wei Y, Evans G (1994) Cloning and characterization of the t(X;11) breakpoint from a leukemic cell line identify a new member of the forkhead gene family. Genes Chromosomes Cancer 11: 79–84 [DOI] [PubMed] [Google Scholar]
- Puigserver P, Rhee J, Donovan J, Walkey CJ, Yoon JC, Oriente F, Kitamura Y, Altomonte J, Dong H, Accili D, Spiegelman BM (2003) Insulin-regulated hepatic gluconeogenesis through FOXO1–PGC-1alpha interaction. Nature 423: 550–555 [DOI] [PubMed] [Google Scholar]
- Purcell NH, Darwis D, Bueno OF, Muller JM, Schule R, Molkentin JD (2004) Extracellular signal-regulated kinase 2 interacts with and is negatively regulated by the LIM-only protein FHL2 in cardiomyocytes. Mol Cell Biol 24: 1081–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswamy S, Nakamura N, Sansal I, Bergeron L, Sellers WR (2002) A novel mechanism of gene regulation and tumor suppression by the transcription factor FKHR. Cancer Cell 2: 81–91 [DOI] [PubMed] [Google Scholar]
- Ramaswamy S, Nakamura N, Vazquez F, Batt DB, Perera S, Roberts TM, Sellers WR (1999) Regulation of G1 progression by the PTEN tumor suppressor protein is linked to inhibition of the phosphatidylinositol 3-kinase/Akt pathway. Proc Natl Acad Sci USA 96: 2110–2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M, Fernandez de Mattos S, van der Horst A, Klompmaker R, Kops GJ, Lam EW, Burgering BM, Medema RH (2002) Cell cycle inhibition by FoxO forkhead transcription factors involves downregulation of cyclin D. Mol Cell Biol 22: 7842–7852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuur ER, Loktev AV, Sharma M, Sun Z, Roth RA, Weigel RJ (2001) Ligand-dependent interaction of estrogen receptor-alpha with members of the forkhead transcription factor family. J Biol Chem 276: 33554–33560 [DOI] [PubMed] [Google Scholar]
- Seoane J, Le HV, Shen L, Anderson SA, Massague J (2004) Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell 117: 211–223 [DOI] [PubMed] [Google Scholar]
- Tang ED, Nunez G, Barr FG, Guan KL (1999) Negative regulation of the forkhead transcription factor FKHR by Akt. J Biol Chem 274: 16741–16746 [DOI] [PubMed] [Google Scholar]
- Tetsu O, McCormick F (1999) Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature 398: 422–426 [DOI] [PubMed] [Google Scholar]
- Tissenbaum HA, Guarente L (2001) Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature 410: 227–230 [DOI] [PubMed] [Google Scholar]
- van der Horst A, Tertoolen LG, de Vries-Smits LM, Frye RA, Medema RH, Burgering BM (2004) FOXO4 is acetylated upon peroxide stress and deacetylated by the longevity protein hSir2(SIRT1). J Biol Chem 279: 28873–28879 [DOI] [PubMed] [Google Scholar]
- Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA (2001) hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell 107: 149–159 [DOI] [PubMed] [Google Scholar]
- Wei Y, Renard CA, Labalette C, Wu Y, Levy L, Neuveut C, Prieur X, Flajolet M, Prigent S, Buendia MA (2003) Identification of the LIM protein FHL2 as a coactivator of beta-catenin. J Biol Chem 278: 5188–5194 [DOI] [PubMed] [Google Scholar]
- Woods YL, Rena G, Morrice N, Barthel A, Becker W, Guo S, Unterman TG, Cohen P (2001) The kinase DYRK1A phosphorylates the transcription factor FKHR at Ser329 in vitro, a novel in vivo phosphorylation site. Biochem J 355: 597–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung F, Hoberg JE, Ramsey CS, Keller MD, Jones DR, Frye RA, Mayo MW (2004) Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J 23: 2369–2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao HH, Herrera RE, Coronado-Heinsohn E, Yang MC, Ludes-Meyers JH, Seybold-Tilson KJ, Nawaz Z, Yee D, Barr FG, Diab SG, Brown PH, Fuqua SA, Osborne CK (2001) Forkhead homologue in rhabdomyosarcoma functions as a bifunctional nuclear receptor-interacting protein with both coactivator and corepressor functions. J Biol Chem 276: 27907–27912 [DOI] [PubMed] [Google Scholar]


