ABSTRACT
Streptococcus pneumoniae is a main cause of child mortality worldwide, but strains also asymptomatically colonize the upper airways of most children and form biofilms. Recent studies have demonstrated that ∼50% of colonized children carry at least two different serotypes (i.e., strains) in the nasopharynx; however, studies of how strains coexist are limited. In this work, we investigated the physiological, genetic, and ecological requirements for the relative distribution of densities, and spatial localization, of pneumococcal strains within biofilm consortia. Biofilm consortia were prepared with vaccine type strains (i.e., serotype 6B [S6B], S19F, or S23F) and strain TIGR4 (S4). Experiments first revealed that the relative densities of S6B and S23F were similar in biofilm consortia. The density of S19F strains, however, was reduced to ∼10% in biofilm consortia, including either S6B, S23F, or TIGR4, in comparison to S19F monostrain biofilms. Reduction of S19F density within biofilm consortia was also observed in a simulated nasopharyngeal environment. Reduction of relative density was not related to growth rates, since the Malthusian parameter demonstrated similar rates of change of density for most strains. To investigate whether quorum sensing (QS) regulates relative densities in biofilm consortia, two different mutants were prepared: a TIGR4ΔluxS mutant and a TIGR4ΔcomC mutant. The density of S19F strains, however, was similarly reduced when consortia included TIGR4, TIGR4ΔluxS, or TIGR4ΔcomC. Moreover, production of a different competence-stimulating peptide (CSP), CSP1 or CSP2, was not a factor that affected dominance. Finally, a mathematical model, confocal experiments, and experiments using Transwell devices demonstrated physical contact-mediated control of pneumococcal density within biofilm consortia.
IMPORTANCE Streptococcus pneumoniae kills nearly half a million children every year, but it also produces nasopharyngeal biofilm consortia in a proportion of asymptomatic children, and these biofilms often contain two strains (i.e., serotypes). In our study, we investigated how strains coexist within pneumococcal consortia produced by vaccine serotypes S4, S6B, S19F, and S23F. Whereas S6B and S23F shared the biofilm consortium, our studies demonstrated reduction of the relative density of S19F strains, to ∼10% of what it would otherwise be if alone, in consortial biofilms formed with S4, S6B, or S23F. This dominance was not related to increased fitness when competing for nutrients, nor was it regulated by quorum-sensing LuxS/AI-2 or Com systems. It was demonstrated, however, to be enhanced by physical contact rather than by a product(s) secreted into the supernatant, as would naturally occur in the semidry nasopharyngeal environment. Competitive interactions within pneumococcal biofilm consortia regulate nasopharyngeal density, a risk factor for pneumococcal disease.
KEYWORDS: Streptococcus pneumoniae, carriage, biofilm consortia, dominance, physical contact
INTRODUCTION
Streptococcus pneumoniae, commonly known as pneumococcus, is a Gram-positive opportunistic pathogen that is the leading cause of bacterial pneumonia and acute otitis media (1, 2). Despite its propensity for causing severe diseases, pneumococcus is a common commensal that quiescently colonizes the upper respiratory tract, forming biofilms that adhere to the epithelium of the nasopharynx, ear epithelium, and lungs, rather than as planktonic cells that are associated with septicemia and meningitis (3–7). In the nasopharyngeal environment, replication slows and bacteria show reduced expression of virulence factors, such as the polysaccharide capsule; instead, they form a biofilm structure made of extracellular DNA, proteins, lipids, and polysaccharides that facilitates asymptomatic carriage. This ability to colonize the upper respiratory tract and persist via biofilms, without causing disease, makes carriage of pneumococcal strains common (4, 6, 7).
To date, over 90 pneumococcal serotypes have been described, and while widespread immunization with conjugate pneumococcal vaccines (PCVs) targeting 7, 10, or 13 serotypes has been effective at reducing mortality, carriage of targeted serotypes still occurs (1, 8). With the widespread use of new methodologies for pneumococcal serotyping, carriage of multiple S. pneumoniae strains (i.e., serotypes) was demonstrated to be as common as carriage of a single strain (9–12). For example, a high prevalence of multiple pneumococcal strain carriage was reported by Turner et al., who used both a sweep-latex agglutination method and microarray studies to demonstrate that 43% or 48.8%, respectively, of nasopharyngeal (NP) swabs from Thai children carried more than one pneumococcal serotype (9). A similar prevalence of multiple serotype carriage (∼40%) was observed recently in Spain; serotypes were detected based on a combination of Quellung reactions, latex serotyping, and multiplex PCRs, and in another study serotypes carried by Peruvian children were identified by using serotype-specific quantitative PCRs (qPCRs) (10, 11). Therefore, simultaneous carriage of multiple serotypes is relatively common, meaning that these genetically distinct pneumococcal strains must compete not only with resident microflora and other opportunistic pathogens, such as Haemophilus influenzae and Staphylococcus aureus, but also with members of their own species (10–13).
Monostrain pneumococcal biofilms and biofilms made by pneumococcus and other species, such as H. influenzae or Moraxella catarrhalis, have been extensively studied during the last few years (4, 5, 13–16). During early stages of formation of monostrain biofilms (i.e., within 8 h), the pneumococcus utilizes the quorum-sensing (QS) LuxS/AI-2 and Com systems as a proxy of population density to begin forming the biofilm structure (6, 17–19). Supporting the role of QS in the control of monostrain pneumococcal biofilms, Carrolo et al. (17) demonstrated that strains expressing competence-stimulating peptide 1 (CSP1), a quorum-sensing pheromone, produced denser biofilms than strains producing CSP2. When pneumococcal monostrain biofilms are formed in an enclosed system (i.e., in a polystyrene plate), quorum-sensing-regulated fratricidal factors, including some bacteriocins, accumulate in the microenvironment and cause irreversible death of biofilm cells that begins after ∼12 h of incubation (18, 19). Biofilm lysis did not occur in a biofilm bioreactor with cultures of human pharyngeal cells (18) or in a plate biofilm model with immobilized pharyngeal cells where the culture medium was changed every 4 h (20, 21). With appropriate modifications, these biofilm models can be utilized to investigate chronic colonization or pneumococcal disease involving the upper organs (e.g., ear) or lower airways (e.g., lungs).
Despite compelling evidence of how monostrain pneumococcal biofilms are formed and how the biofilm structure is regulated, little is known about the coexistence and relative densities of pneumococcal strains within nasopharyngeal biofilm consortia. In this work, we investigated the physiological, genetic, and ecological requirements for the relative distribution densities and spatial localizations of different pneumococcal serotypes during the formation of biofilm consortia. Mixtures of invasive strains recently isolated from pneumococcal disease cases and belonging to vaccine serotypes 6B (S6B), S19F, and S23F were utilized to produce biofilm consortia. The relative densities of individual strains forming biofilm consortia were investigated by serotype-specific qPCR and CFU counts. These studies demonstrated higher relative densities (i.e., dominance) of strains S6B, S23F, and TIGR4 in forming biofilm consortia with S19F strains. The relative densities were similar (i.e., tolerant) when S6B and S23F formed pneumococcal biofilms. The spatial arrangement of pneumococcal strains within biofilm consortia was investigated by using confocal microscopy imaging. Our data revealed that the QS systems LuxS/AI-2 and Com play no role in controlling dominance within a biofilm consortium. Moreover, a secreted molecule appeared not to be involved in the dominance of the relative densities, while spatial physical contact of pneumococcal strains within biofilm consortia was required for dominance.
RESULTS
Population dynamics of pneumococcal biofilm consortia produced by vaccine strains.
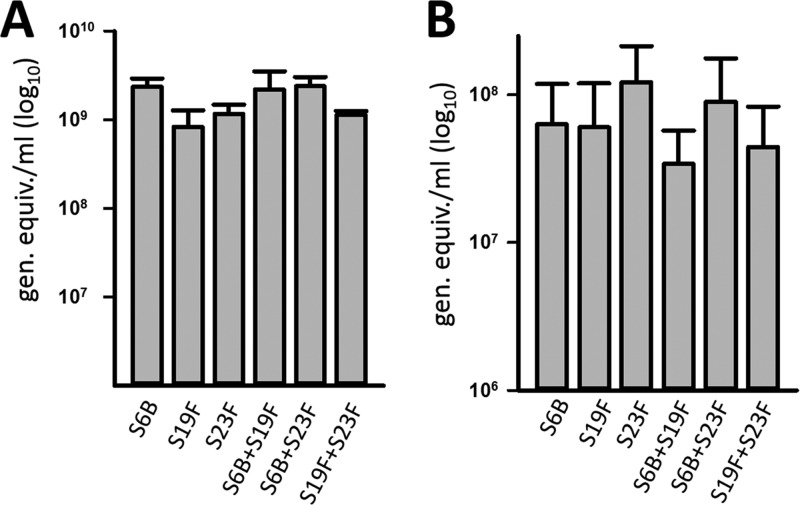
S. pneumoniae persists in the human nasopharynx in a biofilm state, and highly sensitive methods to investigate pneumococcal colonization include quantitative PCR assays (11, 22–24). Therefore, we first evaluated by qPCR the density of monostrain pneumococcal biofilms produced by vaccine strains and compared the densities to those produced by a mixture of two strains of different serotypes. Pneumococcal strains chosen for these studies belonged to vaccine types (e.g., 6B, 19F, and 23F) (Table 1) and were recently isolated from invasive pneumococcal disease (IPD) cases in the United States. They had not been genetically modified since their isolation from patients. Experiments showed that vaccine-type strains inoculated alone formed robust biofilms (>8 × 108 genome equivalents/ml) at 8 h postinoculation on both abiotic substrates (Fig. 1A) and human pharyngeal cells (Fig. 1B). These biofilms produced by S6B, S19F, and S23F were similar, i.e., not statistically significantly different, on either substrate (Fig. 1). Almost identical results were obtained when the densities of planktonic bacteria or of biofilms were obtained by culturing (see Fig. S1 in the supplemental material). When S6B was coinoculated in the same well with either S19F or S23F, the relative density was similar to that produced in wells inoculated only with S6B. Similarly, biofilms produced by a mixture of S19F and S23F were similar to those produced in wells inoculated with only S19F or S23F (Fig. 1). These results indicated that in vitro formation of pneumococcal biofilm consortia reaches a plateau, and they led us to hypothesize that strains may partition a biofilm consortium to coexist.
TABLE 1.
Streptococcus pneumoniae strains utilized in this study
| Streptococcus pneumoniae strain | Serotype | Descriptiona | Source or reference |
|---|---|---|---|
| D39 | 2 | Avery's virulent strain, isolated in 1916, CSP1 | 46 |
| TIGR4 | 4 | Invasive isolate from blood of 30-yr-old male in Norway, CSP2 | 25 |
| SPJV16 | 4 | TIGR4ΔluxS::erm(B), Eryr | This study |
| SPJV20 | 4 | TIGR4ΔcomC::erm(B), Eryr | This study |
| SPJV21 | 4 | SPJV16 transformed with Ami9 DNA, Eryr Strr | This study |
| ATCC 49619 | 19F | Reference strain recommended by CLSI for antimicrobial susceptibility testing | 42 |
| R6Ami9 | 2 | R6 derivative, Strr | D. Zanher |
| 6B | 6B | Invasive isolate, utilized as qPCR control | 11 |
| 8655 | 6B | Invasive isolate, serotype 6B, CSP2; Penr Eryr Clir Trmr Tetr | CDC |
| 3875 | 6B | Invasive isolate, serotype 6B, CSP1 | CDC |
| 3179 | 19F | Invasive isolate, utilized as qPCR control | 11 |
| 5131 | 19F | Invasive isolate, serotype 19F, CSP2; Penr Trmr | CDC |
| 4924 | 19F | Invasive isolate, serotype 19F, CSP1; Penr Eryr Clir Trmr Tetr | CDC |
| 8064 | 23F | Invasive isolate, CSP1; Trmr | CDC |
| 23F | 23F | Invasive isolate, utilized as qPCR control | 11 |
Abbreviations: Ery, erythromycin; Str, streptomycin; Pen, penicillin; Cli, clindamycin; Trm, trimethoprim; Tet, tetracycline.
FIG 1.
Formation of pneumococcal biofilm consortia by invasive strains. S. pneumoniae S6B (3875), S19F (5131), or S23F (8064) was inoculated alone or coinoculated with the indicated strain into 24-well plates (A) or immobilized human pharyngeal cells (B) and incubated for 8 h, after which biofilms (one strain) and biofilm consortia (mixtures) were harvested and DNA was extracted. DNA was used as the template in qPCRs targeting the pan-pneumococcus lytA gene, and the number of genome equivalents per milliliter was calculated. The error bars represent the standard errors of the means and were calculated using data from at least three independent experiments.
Relative densities of pneumococcal strains within biofilm consortia: dominance and tolerance.
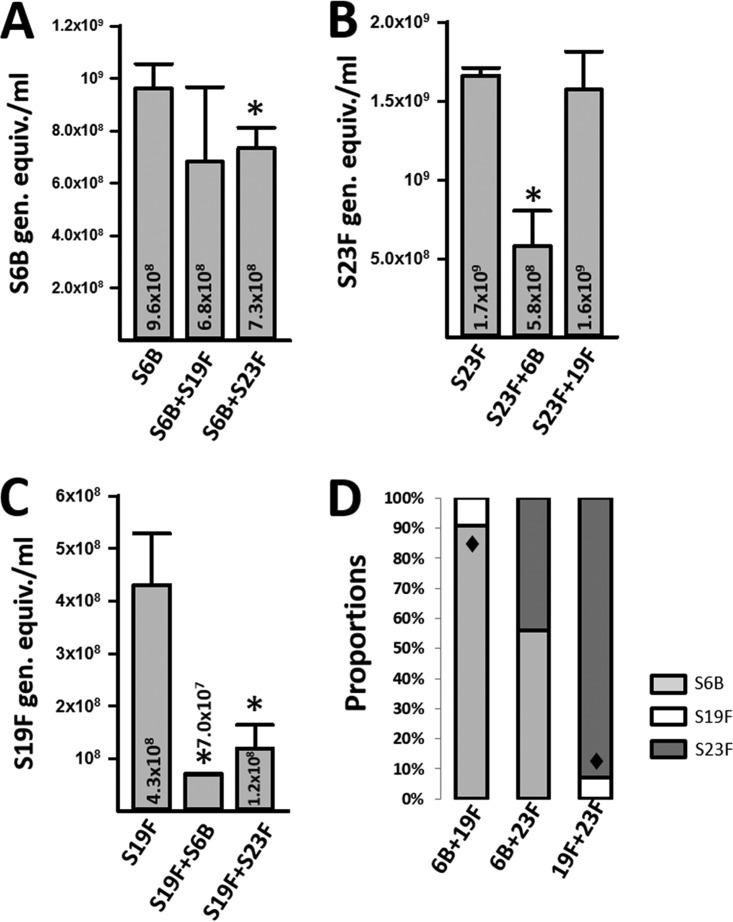
To compare the relative density of each strain forming early biofilm consortia, we utilized serotype-specific quantitative reactions (11, 24). The density of each strain within a consortium was compared with biofilms formed by individual strains at 8 h postinoculation. As shown in Fig. 2A, the relative density of S6B was similar (P = 0.57) whether incubated alone or coincubated with S19F, whereas it significantly decreased when forming biofilm consortia with 23F. Similarly, biofilms made by S23F were significantly reduced when coincubated with S6B, in comparison to biofilms obtained in wells inoculated with S23F alone, but S23F density did not change when coinoculated with S19F (Fig. 2B). In contrast, when inoculated alone, the density of S19F was significantly higher than that of S19F coincubated with S6B or S23F (P < 0.05 for both cases) (Fig. 2C).
FIG 2.
Dominance and tolerance within pneumococcal biofilm consortia. S. pneumoniae S6B (3875), S19F (5131), or S23F (8064) was inoculated alone or coinoculated with the indicated strain into 24-well plates and incubated for 8 h, after which biofilms were harvested and DNA was extracted. Serotype-specific qPCRs targeting serotype 6B (A), serotype 23F (B), or serotype 19F (C) were used to quantify the specific bacterial load of each strain. The error bars reflect the standard errors of the means and were calculated using data from three independent experiments. Numbers inside and above the bars are medians. *, statistically significant reduction (P < 0.05) in biomass in comparison to biomass of the strain inoculated alone. (D) The observed proportion was compared to the null proportion of 0.5, as the same amounts of each strain were coinoculated together. Shown are the average proportions obtained from three independent experiments. ◆, statistically different (P < 0.05) proportion of strains were incubated together.
The observed proportion of each strain within consortial biofilms was analyzed next. The observed proportion was compared to the null proportion of 0.5, as the same amounts of each strain were coinoculated together. When S6B and S19F were coinoculated, the proportion of biofilms made by S6B (0.91) was significantly higher than that observed for S19F (0.09) (Fig. 2D). As expected, a significantly different proportion was observed when S19F and S23F were coinoculated: 0.07 and 0.93, respectively. The proportions of biofilms made by S6B and S23F, however, were similar: 0.56 and 0.44, respectively. Overall similar proportions were obtained when early biofilm consortia were produced on human pharyngeal cells (data not shown). Together, these results demonstrated dominance of S6B, or of S23F, within pneumococcal biofilm consortia produced along with S19F. Data also indicated that relative densities of S6B and S23F within biofilm consortia were similar (i.e., strains shared the niche equally).
Rate of change of density of pneumococcal strains.
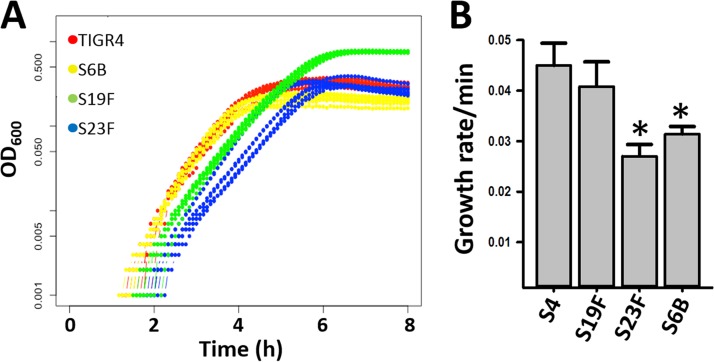
To investigate if the increased relative densities of S6B, or of S23F, in consortial biofilms with S19F were related to an accelerated consumption of resources, the Malthusian parameter, which evaluates the rate of change in density per minute, was obtained. Growth curves (based on the optical density at 600 nm [OD600]) first showed a delayed growth of S6B and S23F in comparison to S19F at 5 h postinoculation (Fig. 3A). Accordingly, the calculated Malthusian parameter (i.e., growth rate per minute) was significantly lower for S23F and S6B than for S19F (Fig. 3B), thus indicating that faster consumption of resources by dominant S6B and S23F strains did not account for the observed increased relative densities.
FIG 3.
Rate of change of density of pneumococcal strains. (A) S4 (TIGR4), S6B (3875), S19F (5131), and S23F (8064) strains were cultured in THY until they reached early log phase, and then bacteria were diluted to ∼2.5 × 105 CFU/ml. An aliquot (300 μl) from each strain was added to seven different wells of a BioScreen plate, and the plate was incubated at 37°C for 24 h. OD values were recorded by the plate reader every 5 min. (B) The Malthusian parameter (the growth rate per minute) was calculated as described in Materials and Methods. *, statistically significant difference (P < 0.001) in comparison to S19F.
Spatial localization of S19F strains within pneumococcal biofilm consortia.
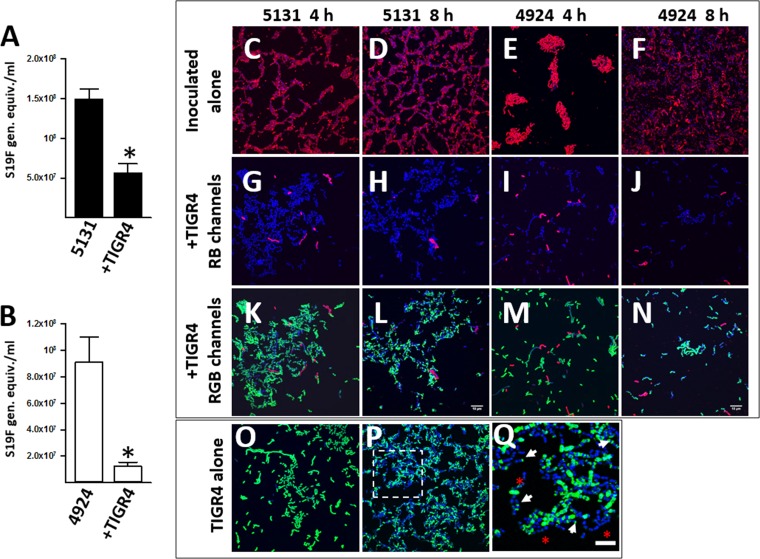
Strains utilized in the above-described experiments were recently isolated from patients with pneumococcal disease, and therefore genetic information was not available. To gain insights into the potential mechanism controlling relative densities of biofilm consortia, we incubated S19F along with a reference genome sequenced strain, TIGR4 serotype 4 (vaccine type) (25). These experiments showed that, in biofilm consortia including TIGR4 and S19F strain 5131, the relative density of S19F decreased to the same extent as that observed when S19F was coinoculated with S6B or S23F (Fig. 4A). The density of a different S19F strain, 4924, in consortial biofilms with TIGR4 was tested with similar results (Fig. 4B). This TIGR4-induced reduction of density of S19F strains was specific, since the relative density of reference strain D39 (serotype 2) was similar in both culture types when incubated alone or in biofilm consortia with TIGR4 (data not shown).
FIG 4.
Biofilm consortia formed by invasive strains TIGR4 and S19F. (A and B) S19F (strain 5131) (A) or S19F (strain 4924) (B) was inoculated alone or with TIGR4 and incubated for 8 h, after which biofilms were harvested and DNA was extracted. Serotype-specific qPCRs targeting serotype 19F were used to quantify the specific bacterial load of S19F strains. (C to Q). Strains were incubated (as described for panels A and B) for 4 or 8 h, after which pneumococcal strains were stained with an anti-S19 antibody conjugated to Alexa 555 (red channel) or an anti-S4 antibody conjugated to Alexa 488 (green channel). Pneumococcal DNA was stained with DAPI (blue channel). Shown are confocal micrographs of S19F stains inoculated alone (C to F) or with TIGR4 and only showing red and blue channels (RB) (G to J) or showing all channels (RGB) (K to N). Results for strain TIGR4 incubated alone for 4 h (O) or 8 h (P) are also shown. (Q) An enlargement of the indicated area in panel P. Arrows point to zones where capsule staining concentrated in poles, and asterisks show pneumococci that lost their capsule. Bars, 10 μm (shown in panels L and N and valid for all other panels except panel Q, in which the bar equals 5 μm).
To visualize the spatial localization of each strain within biofilm consortia, we stained their capsule by fluorescence, using serotype-specific anticapsule antibodies and confocal microscopy. Micrographs first revealed that attachment to the glass substratum varied between S19F strains. At 4 h postinoculation, strain 5131 attached and formed a net-like pattern (Fig. 4C), whereas 4924 formed biofilm aggregates (Fig. 4E). After 8 h of incubation, however, the aggregates formed by 4924 had apparently disaggregated, covering just 70% of the substrate (Fig. 4F). A similar biofilm phenotype was produced by strain 5131 8 h postincubation (Fig. 4B). As expected, as we and others have reported that decreased transcription of capsule genes when biofilms mature (26, 27), the pneumococcal capsule was clearly observed 4 h postinoculation but capsule staining decreased after 8 h of incubation. Selection of unencapsulated pneumococci binding to the substrate and then increasing in density after 8 h could also be a possibility. In the case of TIGR4 (Fig. 4O and P), the capsule aggregated in bacterial poles (Fig. 4Q, arrows) or was apparently lost (Fig. 4Q, asterisks). Due to this effect, to further confirm the presence of bacterial cells, the DNA was also stained. When TIGR4 was incubated with S19F strains, there was a dramatic reduction in S19F bacteria at both 4 h and 8 h postinoculation (Fig. 4, red bacteria [S19F] inoculated alone versus those inoculated with TIGR4). The density of TIGR4 bacteria in these consortia was not affected.
Density of S19F decreases in nasopharyngeal biofilm consortia with TIGR4.
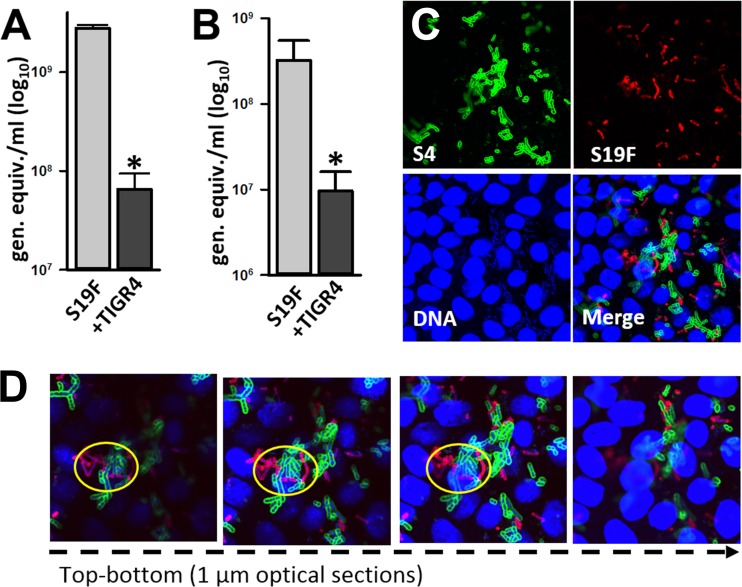
To investigate whether the relative density of S19F decreases in mature nasopharyngeal biofilm consortia along with TIGR4, we incubated both strains in a bioreactor containing living cultures of human pharyngeal cells for 24 h. Mature biofilms already form after 24 h of incubation in a biofilm bioreactor (18). The density of S19F biofilms and numbers of S19F planktonic cells coming off the bioreactor chamber were significantly reduced when S19F was coincubated with TIGR4, in comparison with bioreactor chambers incubated only with S19F (Fig. 5A and B). Confocal experiments additionally demonstrated that pneumococcal strains were in close proximity when incubated together (Fig. 5C); physical contact was confirmed through optical sections of confocal micrographs (Fig. 5D, yellow circles).
FIG 5.
TIGR4 reduces biofilms of S19F on consortia formed on human nasopharyngeal cells. (A and B) S19F (strain 4924) was inoculated with TIGR4 and incubated for 24 h, after which biofilm consortia (A) or planktonic bacteria (B) were harvested and DNA was extracted. Serotype 19-specific qPCRs were used to quantify the number of genome equivalents per milliliter. The error bars reflect the standard errors of the means and were calculated using data from three independent experiments. *, statistically significant difference (P < 0.05) in comparison to S19F incubated alone. (C and D) Confocal micrographs of biofilm consortia formed by TIGR4 and S19F on human nasopharyngeal cells. Panel C shows the projection and panel D shows optical sections. Bacteria were stained with an anti-S19F antibody conjugated to Alexa 555 (red channel) or an anti-S4 antibody conjugated to Alexa 488 (green channel). Cells and pneumococcal DNA were stained with TO-PRO-3 (blue channel).
The factor(s) allowing reduction of S19F relative density is not regulated by quorum-sensing systems LuxS/AI-2 and Com.
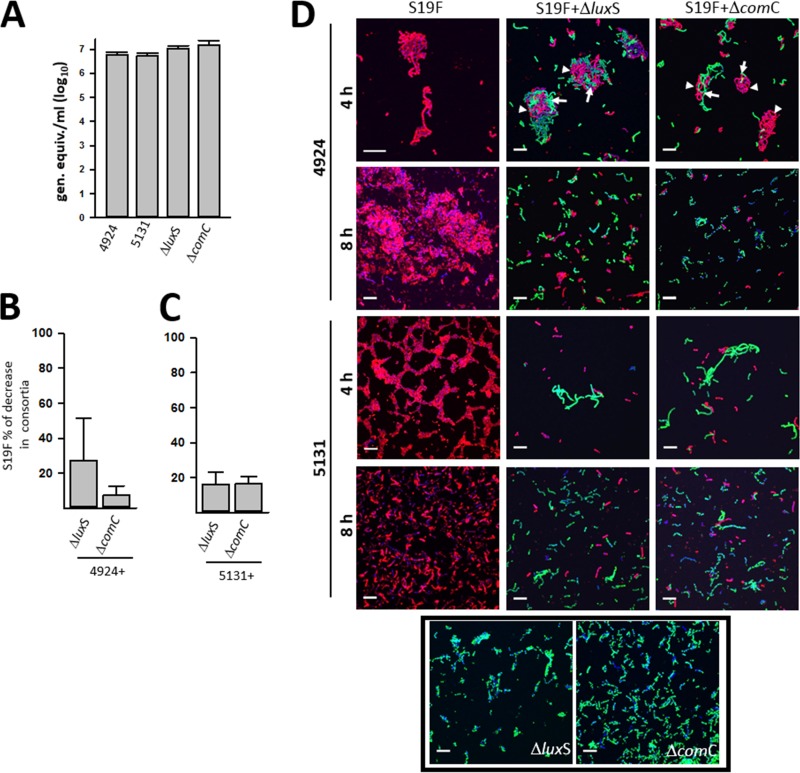
Given that the quorum-sensing systems LuxS/AI-2 and Com have been implicated in formation of monostrain pneumococcal biofilms with strain D39 and are key regulators of fratricide and competence (18, 26, 27), TIGR4ΔluxS and TIGR4ΔcomC mutant strains were prepared and evaluated for regulating the factor(s) that allows dominance of S19F biomass in biofilm consortia. Densities of monostrain biofilms were similar when 4924, 5131, mutant strain TIGR4ΔluxS, or mutant strain TIGR4ΔcomC were compared (P > 0.05 for all comparisons) (Fig. 6A).
FIG 6.
QS systems LuxS/AI-2 and Com do not regulate the dominant phenotype within biofilm consortia. (A to C) S19F strains (5131 and 4924) or mutant TIGR4ΔluxS or TIGR4ΔcomC strains were inoculated alone (A) or in mixtures containing strain 4924 and either TIGR4 derivative (B) or strain 5131 and either TIGR4 derivative (C). Biofilms were incubated for 8 h and then removed, diluted, and plated onto BAP containing ampicillin (for strain 5131), tetracycline (for strain 4929), erythromycin, or streptomycin (for the TIGR4ΔluxS and TIGR4ΔcomC mutant strains) when incubated with 5131 or 4929, respectively (i.e., 4929 is resistant to erythromycin). In panels B and C, the percent biomass decrease of S19F strains is presented for biofilm consortia, in comparison to strains inoculated alone. The error bars reflect the standard errors of the means and were calculated from three independent experiments. (D) S19F strains 5131 and 4924 were inoculated alone or with mutant strain TIGR4ΔluxS or TIGR4ΔcomC and incubated for 4 or 8 h, after which S19F strains and TIGR4 derivatives were stained with an anti-S19 antibody conjugated to Alexa 555 or an anti-S4 antibody conjugated to Alexa 488, respectively. Pneumococcal DNA was stained with DAPI. Arrows indicate where TIGR4 derivatives localized within the biofilm consortium with S19F strain 4924, for which localization is indicated with an arrowhead. Bars, 10 μm.
When biofilm consortia were produced with either the TIGR4ΔluxS or TIGR4ΔcomC mutant strains, the relative density of S19F strains 4924 or 5131 decreased ∼20% in comparison to the biomass produced by these strains when incubated alone (Fig. 6B and C). This reduction of S19F density was similar to that induced by the wild-type (wt) TIGR4 strain (data not shown). Therefore, the TIGR4 factor(s) that allows reduction of S19F density is not regulated by the QS systems LuxS/AI-2 and Com. Biofilms of the mutant strains TIGR4ΔluxS and TIGR4ΔcomC were not affected by the presence of S19F strains (P > 0.05 for all comparisons) (data not shown).
Confocal micrographs were also obtained. As expected, at 4 h postinoculation S19F 4924 formed aggregates (Fig. 6D). In wells incubated for 4 h with 4924 and mutant strain TIGR4ΔluxS or TIGR4ΔcomC, these aggregates were formed by a mixture of both strains (Fig. 6D, first row [red shows 4924 and green shows TIGR4 derivatives]). After 8 h of incubation, however, S19F aggregates had disappeared from wells incubated with either TIGR4 derivative QS mutant, reducing the population of 4924 bacteria. In contrast, S19F 4924 incubated alone formed robust biofilms. A similar reduction of 5131 biofilms attached to the substratum was observed when mutant strain TIGR4ΔluxS, or TIGR4ΔcomC was incubated with 5131, while the strain growing alone was able to form robust biofilms at 8 h postinoculation (Fig. 6D). The biomass of mutant strain TIGR4ΔluxS or TIGR4ΔcomC, whether incubated alone or with S19F, did not change.
Physical contact is required for dominance within pneumococcal biofilm consortia.
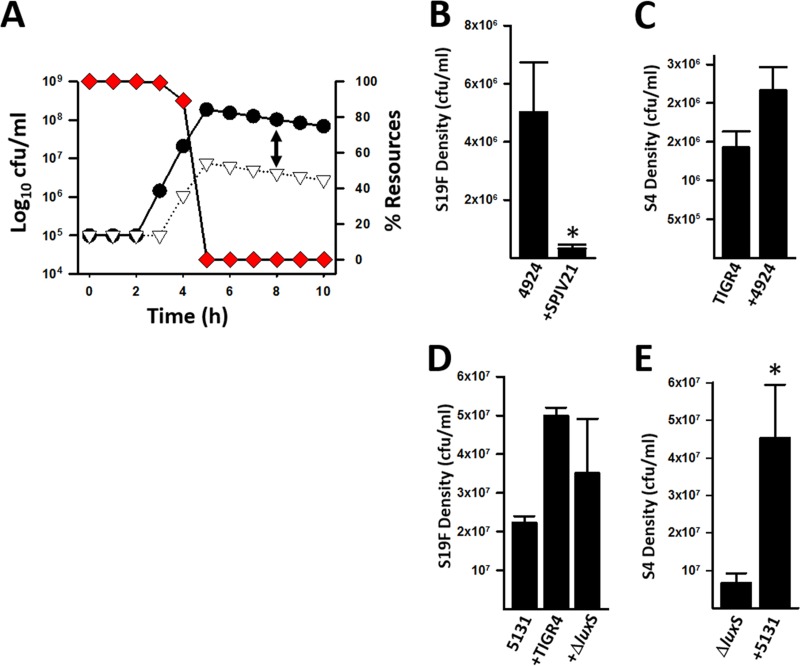
To investigate whether physical contact is required to dominate a biofilm consortium, we simulated physical interaction of an S4 strain and an S19F strain. (Parameters utilized to simulate the interactions are described in Materials and Methods.) The simulation showed that after 4 h of coincubation, the S4 strain limited the growth of the S19F strain by about 1 order of magnitude, 2.1 × 107 versus 1.1 × 106 log10 CFU/ml (Fig. 7A). Biofilm dominance was not related to an increased consumption of resources by the dominant S4 strain, since the hourly growth rates of individual strains were similar (Fig. 3). Moreover, at 4 h, when S19F inhibition was observed, we detected a ∼90% level of resources necessary to produce new cells (Fig. 7A). At 8 h postinoculation, the difference in densities of individual strains within a biofilm consortium made of S4 and S19F strains was about 2 orders of magnitude (Fig. 7A). Contact-dependent reduction of S19F density was verified by colony counts. To do this, we utilized S19F strain 4924, which is naturally resistant to erythromycin, and generated a TIGR4 strain encoding resistance to streptomycin, SPJV21. Figure 7B shows a statistically significant, ∼2-log reduction of the S19F strain when incubated with TIGR4, whereas the density of TIGR4 was not affected by coincubation with the S19F strain (Fig. 7C).
FIG 7.
Physical contact-mediated dominance of pneumococcal biofilm consortia. (A) Simulation of biofilm physical contact-required dominance during a 10-h incubation period. Circles, S4; triangles, S19F; diamonds, resources. Parameters and formulas utilized to construct the model and prepare the graphic are described in Materials and Methods. The arrow shows a 2-log difference in densities of S4 and S19F at 8 h. (B and C) S19F strain 4924 and a TIGR4 derivative streptomycin-resistant strain (SPJV21) were incubated alone or coincubated for 8 h, after which biofilms were harvested, serially diluted, and plated onto BAP with erythromycin to count S19F cells (B) or BAP with streptomycin to obtain counts of SPVJ21 cells (C). Error bars represent the standard errors of the means and were calculated using data from three independent experiments. *, statistically significant reduction (P < 0.05) of biomass in comparison to biomass of the strain inoculated alone. (D and E) TIGR4 wt or TIGR4ΔluxS was inoculated into the top chamber of a Transwell device, and S19F strain 5131 was inoculated into the bottom chamber. As a control, 5131 was incubated alone. Biofilms were incubated for 8 h and then harvested, and counts for S19F (D) and TIGR4ΔluxS (E) were obtained by dilution and plating. The error bars reflect the standard errors of the means and were calculated from three independent experiments. *, statistically significant increase in biomass (P < 0.05) in comparison to the strain inoculated alone.
To experimentally demonstrate whether reduction of the relative density of S19F required physical contact or whether it was mediated by a soluble factor(s), strain TIGR4 and S19F strain 4924, or S19F strain 5131, were inoculated into the same well but the strains were separated with a Transwell system device, i.e., TIGR4 was inoculated in the top chamber and the S19F strain (either 4924 or 5131) was inoculated in the bottom. Experiments with the TIGR4ΔluxS mutant were also included. The biomass of 5131 incubated with either TIGR4 strain (i.e., wt or the TIGR4ΔluxS mutant) was similar or actually increased after 8 h of incubation in comparison to wells where strain 5131 was inoculated alone (Fig. 7D). Similar results were obtained with S19F strain 4924. These data demonstrated that physical contact is required to reduce the density of S19F strains in consortia with TIGR4. A similar density of TIGR4 was observed in incubation wells that also contained 5131, in comparison with wells where only TIGR4 was inoculated (data not shown). As expected, given that the TIGR4ΔluxS mutant does not produce AI-2, in Transwell experiments where the TIGR4ΔluxS mutant and 5131 were inoculated together, the density of the TIGR4ΔluxS mutant increased, perhaps due to LuxS/AI-2 produced by 5131 (Fig. 7E).
DISCUSSION
In this work, we recreated the dynamics of competitive interactions within pneumococcal nasopharyngeal biofilm consortia and demonstrated a dominance of the relative density of S19F strains compared to all other strains tested, i.e., S6B, S23F, and TIGR4. Dominance and tolerance have been observed in recent epidemiological studies where the specific bacterial density of pneumococcal serotypes was obtained (explained in detail below) (11, 28). In this study, dominance against S19F strains within consortia required physical contact, since in experiments conducted in Transwell devices, the TIGR4 strain was not able to decrease the biomass of S19F strains. Dominance of pneumococcal cocolonization through a mechanism requiring physical contact recapitulates that it may naturally occur in the human nasopharynx, where pneumococcal strains cocolonize a nonaqueous microenvironment. Strain dominance was recently described in a mouse model of pneumococcal cocolonization (with use of serotype-specific qPCRs to evaluate serotype density) (29). Pneumococcal colonization has also been inhibited in vitro by incubating pneumococcal strains with probiotic bacteria (30, 31).
Vaccine serotypes 6B, 19F, and 23F evaluated in this study are the most common serotypes isolated in IPD in countries where the pneumococcal vaccine is not available (i.e., India and China); together, they cause ∼30% of IPD cases (32). Unlike strains belonging to serotype 1 or 5, which are highly prevalent in IPD cases but display low prevalence in carriage studies, serotype 6B, 19F, and 23F strains are also the most prevalent serotypes carried in the nasopharynx of children (6, 32, 33). While strains were able to form biofilms on both abiotic and biotic surfaces (i.e., human pharyngeal cells), only S6B and S23F dominated biofilm consortia when S19F colonized the same niche.
Recent studies have investigated nasopharyngeal colonization by multiple pneumococcal serotypes, showing that up to 50% of children are cocolonized by 2 or more strains (i.e., serotypes) (11, 34, 35). In Malawian children, for example, more than 75% of cocolonization events included a vaccine type (35), whereas our study using single-plex qPCR demonstrated a vaccine type in 80% of cocolonized, nonvaccinated Peruvian children (11). The study from our laboratory also quantified the specific bacterial load of pneumococcal serotypes in the cocolonized children and demonstrated dominance of a pneumococcal strain in ∼85% of cocolonization events (11). A semiquantitative microarray approach resulted in similar evidence (11, 34). We define strain dominance, in children cocolonized by two or more strains, as those cocolonization events where the bacterial load (i.e., biomass) of a specific strain accounts for at least 60% of the total pneumococcal load. Therefore, based on new recent epidemiological evidence and this study's findings, dominance appears to be the most common event during nasopharyngeal cocolonization by multiple pneumococcal strains.
Experiments comparing biofilm biomass formed by single strains versus the biomass of biofilm consortia suggest that dominance and tolerance within biofilm consortia are limited by the substrate. For example, whether S6B and S23F strains were inoculated separately or together, the total biomass was very similar (i.e., did not double in size). The in vivo situation in the human nasopharynx and oropharynx may be similar, although the host immune response should play an additional role in limiting the biomass of certain types. Experiments we have described here also demonstrated that when two strains cohabit a biofilm consortium produced on abiotic surfaces, the biomass of both strains will be proportionally similar (∼50%). S19F strains, however, were dominated by all strains tested, whether grown on abiotic substrates or on human pharyngeal cells. Dominance of S6B, and S23F, against 19F correlates with the findings of an epidemiological study with Spanish children that demonstrated that S6B strains were more likely to cocolonize children with strains from other serotypes (10), and recent unpublished studies from our lab have shown that serogroup 6 strains and 23F are the most prevalent strains in cocolonized, nonvaccinated Peruvian children.
The specific mechanism(s) for dominance, or tolerance (if any), that is common to all pneumococci is under investigation by our laboratory and others. A candidate for modulating densities of pneumococcal types had been the blp locus, which encodes a bacteriocin system that produces a potent bacteriocin, BlpC, but it has been recently demonstrated that BlpC plays a minor role in cocolonization (36, 37). A secreted factor might not be involved in dominance of S19F strains by the S4 strain TIGR4, as demonstrated in experiments using Transwell systems, although our experimental design did not allow us to test for a factor(s) that could be only released when in close proximity. The factor(s) appears not to be regulated by the quorum-sensing LuxS/AI-2 and Com systems, since individual mutants prepared in the TIGR4 background were still able to reduce the population of S19F strains in biofilm consortia with S4 mutants. The possibility exists that a minimum amount of a QS molecule produced by the coincubated wt strain may complement the mutant strain. In the case of the TIGR4ΔluxS mutant, the absence of LuxS/AI-2 in TIGR4 could be supplied by that produced by S19F strains. This may be the same case for the absence of production of CSP (i.e., encoded by the comC gene) in the TIGR4ΔcomC mutant when incubated along with S19F 5131, since both produced CSP2 but not in biofilm consortia with 4924, as this strain produced a different variant (i.e., CSP1).
It is likely that strains that dominate biofilm consortia induce cell lysis of the dominated strain, and therefore a source of DNA for recombination might be available. A study by Marks et al. (20) demonstrated higher frequencies of recombination in pneumococcal biofilms produced on human pharyngeal cells than on their planktonic counterparts. However, neither the recombination direction (i.e., when strains were incubated together) nor specific bacterial densities were obtained to allow us to determine whether, in mixtures of two different pneumococcal strains, the strain acquiring DNA from the other one dominates the biofilm consortium (20). We hypothesize that the strains which dominate biofilm consortia have an advantage, not just for colonization but also for acquiring DNA from other pneumococci. Studies in our laboratory are currently focused on investigating this hypothesis.
A very recent article from our laboratory demonstrated killing of Staphylococcus aureus biofilms and planktonic cells by TIGR4 and other pneumococci (38). The unknown factor required physical contact and completely eradicated preformed biofilms made by the S. aureus reference strain and methicillin-resistant S. aureus strain USA300-0114; whether the factor(s) that allows TIGR4 to limit the biomass of S19F strains is similar to that eradicating S. aureus strains needs to be investigated. The need for physical contact might correlate with the in vivo situation of the upper airways, where pneumococcal strains have a limited liquid environment in which to secrete products and must instead release and acquire these products when in close physical proximity to each other.
MATERIALS AND METHODS
Strains and bacterial culture media.
S. pneumoniae strains utilized in this study are listed in Table 1. Strains were routinely cultured on blood agar plates (BAP) or grown in Todd-Hewitt broth containing 0.5% (wt/vol) yeast extract (THY). When indicated, ampicillin (1 μg/ml), streptomycin (100 μg/ml), or erythromycin (0.5 μg/ml) was added to the culture medium. Antibiotics were purchased from Sigma.
Preparation of TIGR4 derivative strains.
SPJV16 and SPJV20, with a deletion within the luxS or comC gene, respectively, were prepared essentially as described for a previous study in our laboratory (18). Mutation was confirmed by PCR [i.e., different PCR product sizes compared to wt products, due to deletion within the target gene and insertion of the erm(B) gene] and by sequencing (data not shown). SPJV21 was prepared by transforming SPJV16 with DNA from R6Ami9, which confers resistance to streptomycin, and plated onto BAP with the antibiotic. Transformation was done by standard methods (26, 39) (Table 1).
Cell cultures.
Human pharyngeal Detroit 562 cells (ATCC CCL-138) were cultured in Eagle's minimum essential medium (EMEM; Lonza, Walkersville, MD) supplemented with non-heat-inactivated 10% fetal bovine serum (FBS; Atlanta Biologicals), 1% nonessential amino acids (Sigma), 1% glutamine (Sigma), penicillin (100 U/ml), and streptomycin (100 μg/ml), and the pH was buffered with HEPES (10 mM; Gibco). Cells were harvested with 0.25% trypsin (Gibco), resuspended in the cell culture medium, and incubated at 37°C in a 5% CO2 humidified atmosphere.
Preparation of inocula.
S. pneumoniae strains were streaked on BAP and incubated overnight at 37°C in a 5% CO2 atmosphere. Bacterial suspensions (OD600 of 0.05) were made in THY and further grown until they reached an OD600 of ∼0.2; a 10% (vol/vol) final solution of glycerol was added to this culture, which was then stored at −80°C until used. Some aliquots were removed from the freezer and then diluted and plated to obtain the CFU per milliliter data for inoculants.
Production of early biofilm consortia on abiotic surfaces.
Pneumococcal early biofilm consortia were generated by thawing inocula on ice, and then ∼7 × 105 CFU/ml of each strain was inoculated into a CellBIND surface 24-well polystyrene plate (Corning) containing THY. Biofilm consortia and biofilm controls were incubated for 8 h in 5% CO2, and after extensive phosphate-buffered saline (PBS) washes, biofilms were harvested by sonication for 15 s in a Bransonic ultrasonic water bath (Branson, Danbury, CT), followed by extensive pipetting to remove all attached bacteria. Biofilms were either counted by dilution and plating or frozen at −80°C for DNA extraction.
Production of early biofilm consortia on immobilized human pharyngeal cells.
The biofilm model using immobilized pharyngeal cells was developed by Marks et al. (21) and has been utilized in pneumococcal biofilm research by different laboratories (18, 21, 40). Detroit 562 cells were grown until confluent (∼5 days) on either 8-well glass slides (Lab-Tek), tissue culture treated 6-well polystyrene plates, or CellBIND surface 24-well polystyrene plates (Corning). Once confluent, cells were immobilized by fixation with 2% paraformaldehyde (PFA; Sigma) for 15 min at room temperature. After extensive washes with sterile PBS, immobilized pharyngeal cells were supplemented with cell culture medium without antibiotics and then inoculated with an aliquot containing ∼7 × 105 CFU/ml of each strain. After 8 h of incubation, biofilms were harvested and counted.
Transwell experiments to physically separate biofilms.
A Transwell support (Corning) was installed in each well of a 6-well plate, creating a bottom and a top compartment within the same well. The Transwell system has a permeable membrane (pore size, ∼0.4 μm) that allows the exchange of small molecules between compartments. The bottom compartment was inoculated with an S19F strain, whereas strain TIGR4 or the TIGR4ΔluxS mutant strain was inoculated directly into the Transwell device. The total volume was brought to 4 ml by addition of THY, and plates were incubated for 8 h in 5% CO2. Then, the Transwell support containing a TIGR4 strain (top compartment) was removed, and S19F biofilms formed on the bottom of the well were harvested and quantified by colony counts.
DNA extraction.
DNA was extracted from 200 μl of the harvested biofilm samples with the QIAamp DNA minikit according to the manufacturer's instructions. Final elution was done with 100 μl of elution buffer. DNA preps were quantified using a nanodrop spectrophotometer and stored at −80°C until used.
Sequencing of the comC gene from clinical pneumococcal isolates.
Downstream and upstream sequences spanning the comC gene were amplified by PCR using primers JVS71L and JVS72R, and the amplified PCR product (∼455 bp) was sequenced at Eurofins (Atlanta, GA). Sequences were analyzed with Lasergene software version 10.1.1 (DNASTAR) (3).
Calculation of the Malthusian parameter.
S4 (TIGR4), S6B (8655), S19F (5131), and S23F (8064) strains were cultured in THY at 37°C in a 5% CO2 atmosphere until they reached the early log phase, and then bacteria were diluted to ∼2.5 × 105 CFU/ml. An aliquot (300 μl) from each strain was added into seven different wells of a BioScreen C plate (Lab Systems, Helsinki, Finland), and the plate was incubated at 37°C in a BioScreen C reader. OD600 values were recorded by a BioScreen C plate reader every 5 min. A growth curve graphic was prepared using the R language and environment for statistical computing and graphics (http://www.gnu.org/). The Malthusian parameter, the rate of change in density, was finally calculated using the GrowthRates software (version 2.1) (41).
Antibiotic susceptibility testing.
Antibiotic resistance of invasive strains was investigated in order to count, using BAP with the appropriate antibiotic, individual strains when forming biofilm consortia. The Kirby-Bauer disc-diffusion method was used according to the Clinical and Laboratory Standards Institute (CLSI) guidelines (42). The following antibiotic discs (Becton-Dickinson, East Rutherford, NJ) were used: oxacillin (1 μg), erythromycin (15 μg), clindamycin (2 μg), chloramphenicol (30 μg), trimethoprim-sulfamethoxazole (1.25/23.75 μg), and tetracycline (30 μg). Isolates were regarded as susceptible, intermediate, or resistant by using the breakpoints set described by CLSI (42). Quality control was done with S. pneumoniae reference strain ATCC 49619.
Quantification of biofilm biomass by quantitative PCR.
Primers, probes, and the concentrations utilized are listed in Table 2. Total pneumococcal density was quantified using the pan-pneumococcus lytA assay (43), and densities of individual serotypes were quantified using primers and probes targeting serotype-specific sequences within the capsule (cps) locus (11, 43). Reactions were run along serially diluted DNA standards corresponding to 4.29 × 105, 4.29 × 104, 4.29 × 103, 4.29 × 102, 4.29 × 101, and 2.14 × 101 genome equivalents per reaction mixture (44). Reactions were carried out using a Bio-Rad CFX96 Touch real-time PCR detection system (Bio-Rad, Hercules, CA) and the following cycling parameters: 50°C for 2 min, 95°C for 2 min, and 40 cycles of 95°C for 15 s and 60°C for 1 min. The final number of genome equivalents per milliliter data were calculated using the CFX software (Bio-Rad, Hercules, CA).
TABLE 2.
qPCR primers and probes utilized in this study
| Target | Primer or probe sequence (5′–3′)a | Reference |
|---|---|---|
| Pan-pneumococcus lytA | F, ACGCAATCTAGCAGATGAAGCA | 43 |
| R, TCGTGCGTTTTAATTCCAGCT | ||
| FAM-GCCGAAAACGCTTGATACAGGGAG | ||
| Serotype 4 | F, TGGGATGACATTTCTACGCACTA | 47 |
| R, CCGTCGCTGATGCTTTATCA | ||
| FAM-TCCTATTGGATGGTTAGTTGGTGA | ||
| Serogroup 6 | F, AAGTTTGCACTAGAGTATGGGAAGGT | 47 |
| R, ACATTATGTCCRTGTCTTCGATACAAG | ||
| FAM-TGTTCTGCCCTGAGCAACTGG | ||
| Serotype 19F | F, GGTCATGCGAGATACGACAGAA | 47 |
| R, TCCTCATCAGTCCCAACCAATT | ||
| FAM-ACCTGAAGGAGTAGCTGCTGGAACGTTG | ||
| Serotype 23F | F, TGCTATTTGCGATCCTGTTCAT | 47 |
| R, AGAGCCTCCGTTGTTTCGTAAA | ||
| FAM-TTTCTCCGGCATCAAACGTTAAG | ||
| comC gene (JVS71L/JVS72R) | F, CCTTTACAAATAAAATGGTAACTGTG | This study |
| R, AATGCTCTATCCAGCTGAGCTAT |
qPCR was conducted using primer and probe concentrations of 100 nM each for lytA qPCR and 200 nM each for serotype-specific qPCR. Forward (F) and reverse (R) primer sequences are shown for each target, followed by the specific probe sequence. All probes were labeled at the 5′ end with 6-carboxyfluorescein (FAM) and also with black hold quencher 1 dye at the 3′ end.
Visualization of pneumococcal biofilms.
Biofilms inoculated into 8-well glass slides (Lab-Tek) were incubated for 8 h at 37°C in a 5% CO2 atmosphere. Biofilms were then washed twice with PBS and fixed with 2% PFA for 15 min at room temperature. Once PFA was removed, biofilms were washed with PBS and blocked with 2% bovine serum albumin (BSA) for 1 h at 37°C. These biofilms were then incubated with serotype-specific polyclonal antibodies (∼40 μg/ml; Statens Serum Institute, Denmark) for 1 h at room temperature. Antibodies had been previously labeled with Alexa 488 (anti-S4) or Alexa 555 (anti-S19) following the manufacturer's recommendations (Molecular Probes). Stained preparations were finally washed two times with PBS and mounted with ProLong Diamond antifade mountant with 4′,6-diamidino-2-phenylindole (DAPI; Molecular Probes). Biofilms produced on human pharyngeal cells were fixed with 2% PFA and washed three times with PBS. Nucleic acids were stained with TO-PRO-3 (1 μM), a carbocyanine monomer nucleic acid stain (Molecular Probes), for 15 min; S4 and S19F strains were stained as mentioned above. Confocal images were obtained using an Olympus FV1000 confocal microscope and analyzed with ImageJ version 1.49k (National Institutes of Health, USA) or the Imaris software (Bitplane, South Windsor, CT).
Production of biofilm consortia in a bioreactor with cultures of human pharyngeal cells.
S. pneumoniae strains were inoculated as indicated above into a biofilm bioreactor, which simulated human airways. The bioreactor includes a continuous flow of nutrients to feed both the cell cultures and pneumococci but also to wash off toxic products, thus avoiding pneumococcus-induced cell cytotoxicity (18). Briefly, human pharyngeal cells were grown on Snapwells (Corning) with a polyester membrane (0.4 μm) supported by a detachable ring. Once confluent (4 to 5 days), cells were inoculated with bacteria and immediately placed in sterile vertical diffusion chambers. The apical side (inner chamber) was perfused with sterile minimal essential medium with no antibiotics, using a Master Flex L/S precision pump system (Cole-Parmer, Vernon, IL). To avoid the accumulation of toxic substances but allow biofilm formation, a low flow rate (0.20 ml/min) was applied. Bioreactor chambers containing biofilm consortia on human pharyngeal cells were incubated for 24 h at 37°C under a sterile environment. At the end of the incubation period, chambers of the bioreactor were opened and biofilm bacteria were harvested as described above. Planktonic specimens were also collected from the outflow of the bioreactor. DNA from biofilms or planktonic cells was purified as described and was used as a template in qPCRs targeting serotype-specific sequences (22, 44). Final genome equivalents (CFU) per milliliter data were obtained as described above.
Model for contact-mediated killing.
To run the contact-mediated killing model, we considered two populations with densities of N1 and N2 cells per milliliter, respectively. These populations grew at a rate proportional to the concentration of a limiting resource, R (in micrograms per milliliter), and parameter k (also in micrograms per milliliter), the Monod constant, which is the concentration of the resource when the population is growing at half its maximum rate, v1 and v2 (per cell per hour) for populations 1 and 2, respectively. As described by Levin and Udekwu (45), resources are consumed at a rate proportional to the growth rate and a conversion efficiency parameter, e (in micrograms), which is the amount of resource necessary to produce a new cell. There are lags L1 and L2 (in hours) for populations 1 and 2, respectively. We assumed population 2 kills population 1 at a rate proportional to the product of their densities and kill rate constant. The killing commences when the time t exceeds that of the lag for population 2 and is proportional to the concentration of the limiting resource. When the concentration of the limiting resource reaches a lower threshold, RMIN (in micrograms per milliliter), autolysis sets in and populations 1 and 2 die off at rates of d1 and d2 (per cell per hour).
With these definitions and assumptions, the rates of change in the concentration of the limiting resource are given by the following equations.
where
To solve these equations numerically (to simulate the dynamics), we used the Berkeley Madonna program.
Statistical analysis.
The means of two pneumococcal densities were analyzed by a two-sample independent t test. For each t test, equality of variance was tested and, based on the result, equal or unequal variance was assumed for the corresponding t test. The means of more than two independent pneumococcal densities were analyzed by analysis of variance (ANOVA). Among samples that had two serotypes, proportions of a given serotype were compared using the Z test. The null hypothesis used was a proportion of 0.5, because equal amounts of each sample were added for the experiment. Two-tailed P values of <0.05 were considered statistically significant. Statistical analyses were performed using OpenEpi (Open Source Epidemiologic Statistics for Public Health [http://www.openepi.com/Menu/OE_Menu.htm]) and the software SigmaPlot version 12.0 (Systat Software, Inc.).
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by a grant from the National Institutes of Health (NIH; 5R21AI112768-02 to J.E.V.).
The content is solely the responsibility of the authors and does not necessarily represent the official view of the National Institutes of Health.
Confocal studies were supported in part by funds from the Integrated Cellular Imaging (ICI) pediatric core and the Emory + Children's Pediatric Research Center (J.E.V.).
We give special thanks to Lesley McGee and Bernard Beall from the Centers for Disease Control and Prevention (CDC) for providing pneumococcal invasive isolates. We appreciate the assistance of Neil Anthony, Emory University School of Medicine, with confocal microscopy and Veronique Parrot for her assistance on preparing some of the figures.
Footnotes
Present address: Preston Palm, College of Medicine, University of Tennessee Health Science Center, Memphis, Tennessee, USA.
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00953-17.
REFERENCES
- 1.Rodgers GL, Klugman KP. 2011. The future of pneumococcal disease prevention. Vaccine 29(Suppl 3):C43–C48. doi: 10.1016/j.vaccine.2011.07.047. [DOI] [PubMed] [Google Scholar]
- 2.Weiser JN. 2010. The pneumococcus: why a commensal misbehaves. J Mol Med (Berl) 88:97–102. doi: 10.1007/s00109-009-0557-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hall-Stoodley L, Nistico L, Sambanthamoorthy K, Dice B, Nguyen D, Mershon WJ, Johnson C, Hu FZ, Stoodley P, Ehrlich GD, Post JC. 2008. Characterization of biofilm matrix, degradation by DNase treatment and evidence of capsule downregulation in Streptococcus pneumoniae clinical isolates. BMC Microbiol 8:173. doi: 10.1186/1471-2180-8-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gilley RP, Orihuela CJ. 2014. Pneumococci in biofilms are non-invasive: implications on nasopharyngeal colonization. Front Cell Infect Microbiol 4:163. doi: 10.3389/fcimb.2014.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chao Y, Marks LR, Pettigrew MM, Hakansson AP. 2014. Streptococcus pneumoniae biofilm formation and dispersion during colonization and disease. Front Cell Infect Microbiol 4:194. doi: 10.3389/fcimb.2014.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shak JR, Vidal JE, Klugman KP. 2013. Influence of bacterial interactions on pneumococcal colonization of the nasopharynx. Trends Microbiol 21:129–135. doi: 10.1016/j.tim.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simell B, Auranen K, Kayhty H, Goldblatt D, Dagan R, O'Brien KL. 2012. The fundamental link between pneumococcal carriage and disease. Expert Rev Vaccines 11:841–855. doi: 10.1586/erv.12.53. [DOI] [PubMed] [Google Scholar]
- 8.Geno KA, Gilbert GL, Song JY, Skovsted IC, Klugman KP, Jones C, Konradsen HB, Nahm MH. 2015. Pneumococcal capsules and their types: past, present, and future. Clin Microbiol Rev 28:871–899. doi: 10.1128/CMR.00024-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turner P, Hinds J, Turner C, Jankhot A, Gould K, Bentley SD, Nosten F, Goldblatt D. 2011. Improved detection of nasopharyngeal cocolonization by multiple pneumococcal serotypes by use of latex agglutination or molecular serotyping by microarray. J Clin Microbiol 49:1784–1789. doi: 10.1128/JCM.00157-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ercibengoa M, Arostegi N, Marimon JM, Alonso M, Perez-Trallero E. 2012. Dynamics of pneumococcal nasopharyngeal carriage in healthy children attending a day care center in northern Spain. Influence of detection techniques on the results. BMC Infect Dis 12:69. doi: 10.1186/1471-2334-12-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sakai F, Chochua S, Satzke C, Dunne EM, Mulholland K, Klugman KP, Vidal JE. 2015. Single-plex quantitative assays for the detection and quantification of most pneumococcal serotypes. PLoS One 10:e0121064. doi: 10.1371/journal.pone.0121064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Satzke C, Dunne EM, Porter BD, Klugman KP, Mulholland EK, PneuCarriage Project Group. 2015. The PneuCarriage project: a multi-centre comparative study to identify the best serotyping methods for examining pneumococcal carriage in vaccine evaluation studies. PLoS Med 12:e1001903. doi: 10.1371/journal.pmed.1001903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Domenech M, Garcia E, Moscoso M. 2012. Biofilm formation in Streptococcus pneumoniae. Microb Biotechnol 5:455–465. doi: 10.1111/j.1751-7915.2011.00294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tikhomirova A, Kidd SP. 2013. Haemophilus influenzae and Streptococcus pneumoniae: living together in a biofilm. Pathog Dis 69:114–126. doi: 10.1111/2049-632X.12073. [DOI] [PubMed] [Google Scholar]
- 15.Perez AC, Pang B, King LB, Tan L, Murrah KA, Reimche JL, Wren JT, Richardson SH, Ghandi U, Swords WE. 2014. Residence of Streptococcus pneumoniae and Moraxella catarrhalis within polymicrobial biofilm promotes antibiotic resistance and bacterial persistence in vivo. Pathog Dis 70:280–288. doi: 10.1111/2049-632X.12129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weimer KE, Armbruster CE, Juneau RA, Hong W, Pang B, Swords WE. 2010. Coinfection with Haemophilus influenzae promotes pneumococcal biofilm formation during experimental otitis media and impedes the progression of pneumococcal disease. J Infect Dis 202:1068–1075. doi: 10.1086/656046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carrolo M, Pinto FR, Melo-Cristino J, Ramirez M. 2014. Pherotype influences biofilm growth and recombination in Streptococcus pneumoniae. PLoS One 9:e92138. doi: 10.1371/journal.pone.0092138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vidal JE, Howery KE, Ludewick HP, Nava P, Klugman KP. 2013. Quorum-sensing systems LuxS/autoinducer 2 and Com regulate Streptococcus pneumoniae biofilms in a bioreactor with living cultures of human respiratory cells. Infect Immun 81:1341–1353. doi: 10.1128/IAI.01096-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei H, Havarstein LS. 2012. Fratricide is essential for efficient gene transfer between pneumococci in biofilms. Appl Environ Microbiol 78:5897–5905. doi: 10.1128/AEM.01343-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marks LR, Reddinger RM, Hakansson AP. 2012. High levels of genetic recombination during nasopharyngeal carriage and biofilm formation in Streptococcus pneumoniae. mBio 3:e00200-12. doi: 10.1128/mBio.00200-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marks LR, Parameswaran GI, Hakansson AP. 2012. Pneumococcal interactions with epithelial cells are crucial for optimal biofilm formation and colonization in vitro and in vivo. Infect Immun 80:2744–2760. doi: 10.1128/IAI.00488-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanke CR, Grijalva CG, Chochua S, Pletz MW, Hornberg C, Edwards KM, Griffin MR, Verastegui H, Gil AI, Lanata CF, Klugman KP, Vidal JE. 2016. Bacterial density, serotype distribution and antibiotic resistance of pneumococcal strains from the nasopharynx of Peruvian children before and after pneumococcal conjugate vaccine 7. Pediatr Infect Dis J 35:432–439. doi: 10.1097/INF.0000000000001030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shak JR, Cremers AJ, Gritzfeld JF, de Jonge MI, Hermans PW, Vidal JE, Klugman KP, Gordon SB. 2014. Impact of experimental human pneumococcal carriage on nasopharyngeal bacterial densities in healthy adults. PLoS One 9:e98829. doi: 10.1371/journal.pone.0098829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pimenta FC, Roundtree A, Soysal A, Bakir M, du Plessis M, Wolter N, von Gottberg A, McGee L, Carvalho MDG, Beall B. 2013. Sequential triplex real-time PCR assay for detecting 21 pneumococcal capsular serotypes that account for a high global disease burden. J Clin Microbiol 51:647–652. doi: 10.1128/JCM.02927-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tettelin H, Nelson KE, Paulsen IT, Eisen JA, Read TD, Peterson S, Heidelberg J, DeBoy RT, Haft DH, Dodson RJ, Durkin AS, Gwinn M, Kolonay JF, Nelson WC, Peterson JD, Umayam LA, White O, Salzberg SL, Lewis MR, Radune D, Holtzapple E, Khouri H, Wolf AM, Utterback TR, Hansen CL, McDonald LA, Feldblyum TV, Angiuoli S, Dickinson T, Hickey EK, Holt IE, Loftus BJ, Yang F, Smith HO, Venter JC, Dougherty BA, Morrison DA, Hollingshead SK, Fraser CM. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498–506. doi: 10.1126/science.1061217. [DOI] [PubMed] [Google Scholar]
- 26.Vidal JE, Ludewick HP, Kunkel RM, Zahner D, Klugman KP. 2011. The LuxS-dependent quorum-sensing system regulates early biofilm formation by Streptococcus pneumoniae strain D39. Infect Immun 79:4050–4060. doi: 10.1128/IAI.05186-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trappetti C, Potter AJ, Paton AW, Oggioni MR, Paton JC. 2011. LuxS mediates iron-dependent biofilm formation, competence, and fratricide in Streptococcus pneumoniae. Infect Immun 79:4550–4558. doi: 10.1128/IAI.05644-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodrigues F, Danon L, Morales-Aza B, Sikora P, Thors V, Ferreira M, Gould K, Hinds J, Finn A. 2016. Pneumococcal serotypes colonise the nasopharynx in children at different densities. PLoS One 11:e0163435. doi: 10.1371/journal.pone.0163435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trzcinski K, Li Y, Weinberger DM, Thompson CM, Cordy D, Bessolo A, Malley R, Lipsitch M. 2015. Effect of serotype on pneumococcal competition in a mouse colonization model. mBio 6:e00902-15. doi: 10.1128/mBio.00902-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wong SS, Quan Toh Z, Dunne EM, Mulholland EK, Tang ML, Robins-Browne RM, Licciardi PV, Satzke C. 2013. Inhibition of Streptococcus pneumoniae adherence to human epithelial cells in vitro by the probiotic Lactobacillus rhamnosus GG. BMC Res Notes 6:135. doi: 10.1186/1756-0500-6-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manning J, Dunne EM, Wescombe PA, Hale JD, Mulholland EK, Tagg JR, Robins-Browne RM, Satzke C. 2016. Investigation of Streptococcus salivarius-mediated inhibition of pneumococcal adherence to pharyngeal epithelial cells. BMC Microbiol 16:225. doi: 10.1186/s12866-016-0843-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson HL, Deloria-Knoll M, Levine OS, Stoszek SK, Freimanis Hance L, Reithinger R, Muenz LR, O'Brien KL. 2010. Systematic evaluation of serotypes causing invasive pneumococcal disease among children under five: the Pneumococcal Global Serotype Project. PLoS Med 7:e1000348. doi: 10.1371/journal.pmed.1000348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adegbola RA, DeAntonio R, Hill PC, Roca A, Usuf E, Hoet B, Greenwood BM. 2014. Carriage of Streptococcus pneumoniae and other respiratory bacterial pathogens in low and lower-middle income countries: a systematic review and meta-analysis. PLoS One 9:e103293. doi: 10.1371/journal.pone.0103293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Valente C, Hinds J, Gould KA, Pinto FR, de Lencastre H, Sa-Leao R. 2016. Impact of the 13-valent pneumococcal conjugate vaccine on Streptococcus pneumoniae multiple serotype carriage. Vaccine 34:4072–4078. doi: 10.1016/j.vaccine.2016.06.017. [DOI] [PubMed] [Google Scholar]
- 35.Kamng'ona AW, Hinds J, Bar-Zeev N, Gould KA, Chaguza C, Msefula C, Cornick JE, Kulohoma BW, Gray K, Bentley SD, French N, Heyderman RS, Everett DB. 2015. High multiple carriage and emergence of Streptococcus pneumoniae vaccine serotype variants in Malawian children. BMC Infect Dis 15:234. doi: 10.1186/s12879-015-0980-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Valente C, Dawid S, Pinto FR, Hinds J, Simoes AS, Gould KA, Mendes LA, de Lencastre H, Sa-Leao R. 2016. The blp locus of Streptococcus pneumoniae plays a limited role in the selection of strains that can cocolonize the human nasopharynx. Appl Environ Microbiol 82:5206–5215. doi: 10.1128/AEM.01048-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dawid S, Roche AM, Weiser JN. 2007. The blp bacteriocins of Streptococcus pneumoniae mediate intraspecies competition both in vitro and in vivo. Infect Immun 75:443–451. doi: 10.1128/IAI.01775-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khan F, Wu X, Matzkin GL, Khan MA, Sakai F, Vidal JE. 2016. Streptococcus pneumoniae eradicates preformed Staphylococcus aureus biofilms through a mechanism requiring physical contact. Front Cell Infect Microbiol 6:104. doi: 10.3389/fcimb.2016.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shak JR, Ludewick HP, Howery KE, Sakai F, Yi H, Harvey RM, Paton JC, Klugman KP, Vidal JE. 2013. Novel role for the Streptococcus pneumoniae toxin pneumolysin in the assembly of biofilms. mBio 4:e00655-13. doi: 10.1128/mBio.00655-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wholey WY, Kochan TJ, Storck DN, Dawid S. 2016. Coordinated bacteriocin expression and competence in Streptococcus pneumoniae contributes to genetic adaptation through neighbor predation. PLoS Pathog 12:e1005413. doi: 10.1371/journal.ppat.1005413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hall BG, Acar H, Nandipati A, Barlow M. 2014. Growth rates made easy. Mol Biol Evol 31:232–238. doi: 10.1093/molbev/mst187. [DOI] [PubMed] [Google Scholar]
- 42.Clinical and Laboratory Standards Institute. 2013. Performance standards for antimicrobial susceptibility testing; twenty-third informational supplement. CLSI document M100-S23. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 43.Carvalho MDG, Tondella ML, McCaustland K, Weidlich L, McGee L, Mayer LW, Steigerwalt A, Whaley M, Facklam RR, Fields B, Carlone G, Ades EW, Dagan R, Sampson JS. 2007. Evaluation and improvement of real-time PCR assays targeting lytA, ply, and psaA genes for detection of pneumococcal DNA. J Clin Microbiol 45:2460–2466. doi: 10.1128/JCM.02498-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sakai F, Talekar SJ, Klugman KP, Vidal JE, Respira Peru Group. 2013. Expression of virulence-related genes in the nasopharynx of healthy children. PLoS One 8:e67147. doi: 10.1371/journal.pone.0067147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Levin BR, Udekwu KI. 2010. Population dynamics of antibiotic treatment: a mathematical model and hypotheses for time-kill and continuous-culture experiments. Antimicrob Agents Chemother 54:3414–3426. doi: 10.1128/AAC.00381-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lanie JA, Ng WL, Kazmierczak KM, Andrzejewski TM, Davidsen TM, Wayne KJ, Tettelin H, Glass JI, Winkler ME. 2007. Genome sequence of Avery's virulent serotype 2 strain D39 of Streptococcus pneumoniae and comparison with that of unencapsulated laboratory strain R6. J Bacteriol 189:38–51. doi: 10.1128/JB.01148-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Azzari C, Moriondo M, Indolfi G, Cortimiglia M, Canessa C, Becciolini L, Lippi F, de Martino M, Resti M. 2010. Real-time PCR is more sensitive than multiplex PCR for diagnosis and serotyping in children with culture negative pneumococcal invasive disease. PLoS One 5:e9282. doi: 10.1371/journal.pone.0009282. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.