ABSTRACT
While copper is an essential trace element in biology, pollution of groundwater from copper has become a threat to all living organisms. Cellular mechanisms underlying copper toxicity, however, are still not fully understood. Previous studies have shown that iron-sulfur proteins are among the primary targets of copper toxicity in Escherichia coli under aerobic conditions. Here, we report that, under anaerobic conditions, iron-sulfur proteins in E. coli cells are even more susceptible to copper in medium. Whereas addition of 0.2 mM copper(II) chloride to LB (Luria-Bertani) medium has very little or no effect on iron-sulfur proteins in wild-type E. coli cells under aerobic conditions, the same copper treatment largely inactivates iron-sulfur proteins by blocking iron-sulfur cluster biogenesis in the cells under anaerobic conditions. Importantly, proteins that do not have iron-sulfur clusters (e.g., fumarase C and cysteine desulfurase) in E. coli cells are not significantly affected by copper treatment under aerobic or anaerobic conditions, indicating that copper may specifically target iron-sulfur proteins in cells. Additional studies revealed that E. coli cells accumulate more intracellular copper under anaerobic conditions than under aerobic conditions and that the elevated copper content binds to the iron-sulfur cluster assembly proteins IscU and IscA, which effectively inhibits iron-sulfur cluster biogenesis. The results suggest that the copper-mediated inhibition of iron-sulfur proteins does not require oxygen and that iron-sulfur cluster biogenesis is the primary target of anaerobic copper toxicity in cells.
IMPORTANCE Copper contamination in groundwater has become a threat to all living organisms. However, cellular mechanisms underlying copper toxicity have not been fully understood up to now. The work described here reveals that iron-sulfur proteins in Escherichia coli cells are much more susceptible to copper in medium under anaerobic conditions than they are under aerobic conditions. Under anaerobic conditions, E. coli cells accumulate excess intracellular copper, which specifically targets iron-sulfur proteins by blocking iron-sulfur cluster biogenesis. Since iron-sulfur proteins are involved in diverse and vital physiological processes, inhibition of iron-sulfur cluster biogenesis by copper disrupts multiple cellular functions and ultimately inhibits cell growth. The results from this study illustrate a new interplay between intracellular copper toxicity and iron-sulfur cluster biogenesis in bacterial cells under anaerobic conditions.
KEYWORDS: copper toxicity, iron-sulfur proteins, iron-sulfur cluster biogenesis, cell growth inhibition, anaerobic conditions
INTRODUCTION
Copper is an essential element required by at least 30 enzymes in living organisms (1). However, excess copper is highly toxic (2). In U.S. groundwater, copper concentrations range from undetectable to approximately 1 mM, with most samples in the nanomolar range (3). In industrial and mining waste streams, micromolar to millimolar concentrations of copper have also been found (4). Copper at high concentrations in water has become a threat to all living organisms (5). As a transition metal, copper was believed to produce reactive oxygen species via the Fenton reaction under aerobic conditions (6, 7). However, an elevated intracellular copper content did not damage DNA in Escherichia coli cells under aerobic conditions (8). In fact, copper is more toxic to E. coli cells under anaerobic conditions than under aerobic conditions (9, 10), indicating that other cellular mechanisms could be responsible for copper toxicity. Because copper needs only two thiol groups to form a linear biscysteinate coordination in proteins (11), excess copper in cells may directly compete with iron or iron-sulfur cluster binding sites in proteins to disrupt iron-sulfur clusters (12, 13) or block iron-sulfur cluster biogenesis (14–16). Since iron-sulfur proteins are involved in diverse and vital physiological processes ranging from energy metabolism to DNA replication and repair (17, 18), inactivation of iron-sulfur proteins by copper will have a broad impact on multiple physiological functions in cells.
Recent studies have also established that formation of iron-sulfur clusters in proteins is not a spontaneous process in cells. A group of dedicated proteins are responsible for iron-sulfur cluster biogenesis (19). In E. coli, there are two iron-sulfur cluster assembly systems, the housekeeping iscSUA-hscBA-fdx gene cluster (20), which is conserved from prokaryotes (bacteria) to eukaryotes (18, 21), and the inducible sufABCDSE gene cluster (22), which represents a redundant activity of iscSUA-hscBA-fdx and is found only in plastids in plants, archaea, and some bacteria (23). Among the proteins encoded by iscSUA-hscBA-fdx, IscS is a pyridoxal 5′ phosphate-dependent cysteine desulfurase that provides sulfide for iron-sulfur cluster assembly (24). IscU is a scaffold protein that hosts transient iron-sulfur clusters (25) and transfers the assembled clusters to target proteins (26, 27). IscA is characterized as an alternative scaffold (28, 29). However, unlike the scaffold IscU, IscA has strong iron binding activity (30, 31) and is able to transfer the bound iron for iron-sulfur cluster assembly in target proteins in vitro (31–33). Based on these and other studies, we have postulated that IscA is an iron chaperone that recruits intracellular iron and delivers it for iron-sulfur cluster biogenesis (32, 33) and that IscS, IscU, and IscA may constitute the core members of the iron-sulfur cluster assembly machinery in cells (32, 34).
In previous studies, we reported that excess intracellular copper competes with ferrous iron for binding in the iron-sulfur cluster assembly protein IscA and effectively blocks iron-sulfur cluster biogenesis in E. coli cells under aerobic conditions (15). Here, we find that under anaerobic conditions, iron-sulfur proteins in E. coli cells are even more susceptible to copper in growth medium. It appears that, under anaerobic conditions, as reported by Outten et al. (9), E. coli cells accumulate more intracellular copper, which effectively inhibits iron-sulfur cluster biogenesis by binding to the iron-sulfur cluster assembly proteins IscA and IscU and the IscA paralog SufA. The results from this study suggest that the copper-mediated inactivation of iron-sulfur proteins does not require oxygen and that iron-sulfur cluster biogenesis is the primary target of anaerobic copper toxicity in cells.
RESULTS
Iron-sulfur proteins in E. coli cells are highly susceptible to copper under anaerobic conditions.
Figure 1A shows that the addition of 1.0 mM CuCl2 to LB medium had very little or no effect on cell growth of wild-type E. coli under aerobic conditions. However, the same copper treatment completely inhibited cell growth under anaerobic conditions. The results are consistent with those in a previous report by Outten et al. (9) showing that E. coli cells are more sensitive to copper in medium under anaerobic conditions than they are under aerobic conditions. Nevertheless, to date, the cellular mechanism underlying anaerobic copper toxicity is not understood.
FIG 1.
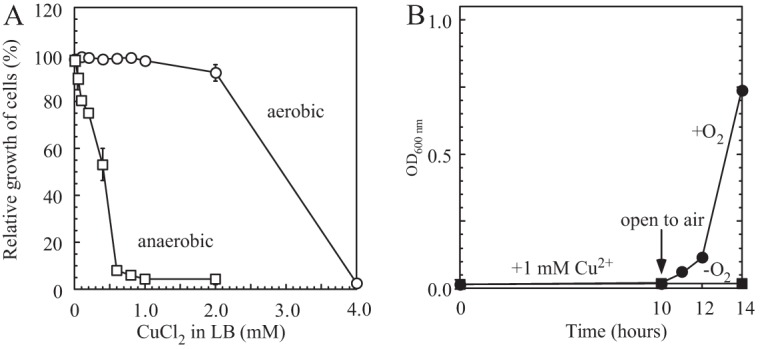
Cell growth inhibition of E. coli by copper under aerobic and anaerobic conditions. (A) Cell growth of E. coli in LB medium supplemented with the indicated concentrations of copper under aerobic and anaerobic conditions. Wild-type E. coli cells (MC4100) were grown in LB medium supplemented with the indicated concentrations of CuCl2 at 37°C for 4 h under aerobic (open circles) or anaerobic (open squares) conditions. Cell growth was determined by measuring turbidity at 600 nm. The data represent the averages ± standard deviations from three independent experiments. (B) Reversal of copper-mediated cell growth inhibition. E. coli cells grown in LB medium supplemented with 1.0 mM CuCl2 were incubated at 37°C for 10 h. Half of the cell culture (closed circles) was returned to aerobic conditions, while the other half (closed squares) remained under anaerobic conditions. Cell growth was monitored by OD determination at 600 nm.
When the E. coli cells grown in LB medium supplemented with 1.0 mM CuCl2 were shifted from anaerobic to aerobic conditions, the cell growth was immediately restored (Fig. 1B), indicating that the copper-mediated inhibition of cell growth under anaerobic conditions was reversible. Since iron-sulfur proteins are the primary targets of copper toxicity in E. coli cells under aerobic conditions (12–16) and iron-sulfur clusters can be reversibly assembled in proteins without de novo protein synthesis (35), we sought to explore the effect of copper on iron-sulfur proteins in E. coli cells under anaerobic conditions. In our experiments, recombinant iron-sulfur protein was expressed in wild-type E. coli cells grown in LB medium supplemented with 0.2 mM CuCl2 under aerobic and anaerobic conditions. The concentration of 0.2 mM CuCl2 was used because it significantly inhibited the cell growth under anaerobic conditions but not under aerobic conditions (Fig. 1A). We chose dihydroxyacid dehydratase (IlvD), a key enzyme in the branched-chain amino acid biosynthesis pathway (36), as an example of a recombinant iron-sulfur protein expressed in E. coli cells. IlvD contains a [4Fe-4S] cluster which is essential for its enzyme activity (36). We then isolated the recombinant iron-sulfur protein from the E. coli cells, purified it, and subjected it to the UV-visible absorption spectrum measurements and enzyme activity analyses.
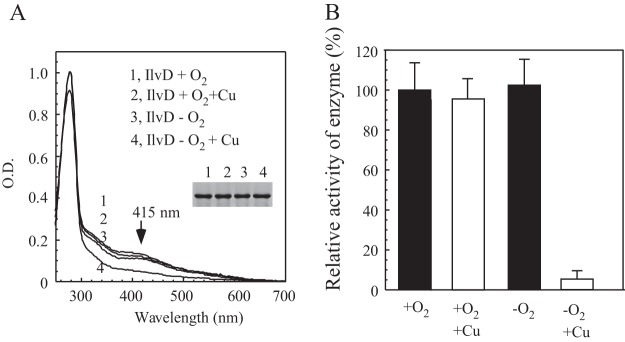
Figure 2A shows that addition of 0.2 mM CuCl2 to LB medium had very little or no effect on the UV-visible absorption spectrum of recombinant IlvD expressed in the E. coli cells grown under aerobic conditions. While the yield of recombinant IlvD expressed in E. coli cells under anaerobic conditions was about one-third of that under aerobic conditions, the 0.2 mM CuCl2 treatment did not significantly affect the protein expression in the cells under aerobic or anaerobic conditions. Nevertheless, the copper treatment significantly decreased the absorption peak at 415 nm of the [4Fe-4S] cluster (Fig. 2A) and the specific enzyme activity (Fig. 2B) of recombinant IlvD expressed in E. coli cells under anaerobic conditions. The iron and sulfide content analyses further showed that the anaerobic copper treatment had largely eliminated iron-sulfur clusters in IlvD in E. coli cells (data not shown). The enzyme activity of the purified IlvD was fully restored when iron-sulfur clusters were reassembled in vitro (15), further suggesting that the anaerobic copper treatment eliminated iron-sulfur clusters in IlvD and inactivated the enzyme activity of the protein in E. coli cells.
FIG 2.
Effect of copper on the iron-sulfur protein IlvD in E. coli cells under aerobic and anaerobic conditions. (A) UV-visible absorption spectra of recombinant IlvD purified from the E. coli cells grown in LB medium supplemented with or without 0.2 mM CuCl2 under aerobic or anaerobic conditions. Purified IlvD was dissolved in buffer containing 500 mM NaCl and 20 mM Tris (pH 8.0). The concentration of IlvD was 28 μM. (Inset) Photograph of SDS-PAGE gel of purified proteins. Samples: 1, IlvD under aerobic conditions; 2, IlvD under aerobic conditions with 0.2 mM CuCl2; 3, IlvD under anaerobic conditions; 4, IlvD under anaerobic conditions with 0.2 mM CuCl2. (B) Enzyme activity of IlvD. Recombinant IlvD was purified as described in panel A. For the enzyme activity assay, 1 μM IlvD was used. The highest enzyme activity (100%) for purified IlvD was 1.2 ± 0.1 mM/μM/min.
Similar results were also obtained when the recombinant iron-sulfur proteins aconitase B (37), endonuclease III (38), and phosphogluconate dehydratase (39) were expressed in E. coli cells grown in LB medium supplemented with 0.2 mM CuCl2 under aerobic and anaerobic conditions (data not shown). Thus, copper is able to inactivate multiple iron-sulfur proteins by eliminating iron-sulfur clusters in the proteins in E. coli cells under anaerobic conditions.
Copper inhibits the activities of iron-sulfur proteins in E. coli cells without affecting the proteins that do not have iron-sulfur clusters under anaerobic conditions.
The E. coli genome encodes three fumarases, fumarase A and fumarase B, each of which contains a [4Fe-4S] cluster (40), and fumarase C, which does not have iron-sulfur clusters (41). Each fumarase was expressed in the E. coli cells grown in LB medium supplemented with 0.2 mM CuCl2 under aerobic or anaerobic conditions. While addition of CuCl2 to LB medium had very little or no effect on iron-sulfur clusters in fumarase A and fumarase B expressed in E. coli cells under aerobic conditions (data not shown), the same copper treatment largely eliminated the iron-sulfur cluster content (Fig. 3A and B) and decreased the specific enzyme activity (Fig. 3D) of recombinant fumarase A and fumarase B expressed in the E. coli cells under anaerobic conditions. In contrast, the copper treatment did not affect the enzyme activity of recombinant fumarase C in E. coli cells under anaerobic conditions (Fig. 3D), suggesting that CuCl2 at a concentration of 0.2 mM may specifically target iron-sulfur proteins in cells under anaerobic conditions.
FIG 3.
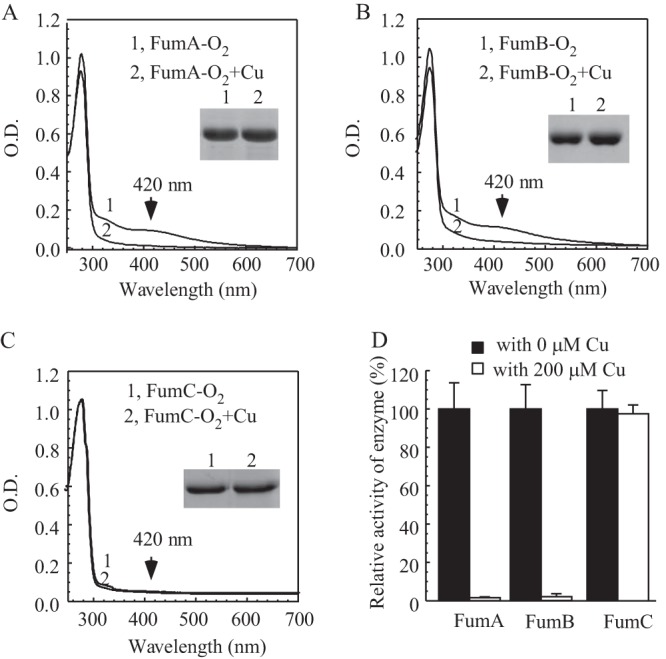
Effect of copper on fumarase A, B, and C in E. coli cells under anaerobic conditions. (A) UV-visible absorption spectra of recombinant fumarase A (20 μM) purified from E. coli cells grown in LB medium supplemented with 0 mM (spectrum 1) or 0.2 mM (spectrum 2) CuCl2 under anaerobic conditions. (B) UV-visible absorption spectra of recombinant fumarase B (20 μM) purified from E. coli cells grown in LB medium supplemented with 0 mM (spectrum 1) or 0.2 mM (spectrum 2) CuCl2 under anaerobic conditions. (C) UV-visible absorption spectra of recombinant fumarase C (30 μM) purified from E. coli cells grown in LB medium supplemented with 0 mM (spectrum 1) or 0.2 mM (spectrum 2) CuCl2 under anaerobic conditions. (Insets) Photograph of SDS-PAGE gels of purified proteins. (D) The enzyme activities of fumarases A, B, and C purified from E. coli cells grown in LB medium supplemented with 0 or 0.2 mM CuCl2 under anaerobic conditions. For the enzyme activity assay, 10 nM each fumarase was used. The highest enzyme activities (100%) for purified fumarase A, fumarase B, and fumarase C were 19.2 ± 0.3, 21.1 ± 0.1, and 16.6 ± 0.3 mM/μM/min, respectively. The results represent averages ± standard deviations from three independent experiments.
Copper inhibits iron-sulfur cluster biogenesis in E. coli cells under anaerobic conditions.
One possible explanation for the copper-mediated elimination of iron-sulfur clusters in proteins is that copper may directly disrupt iron-sulfur clusters in proteins in E. coli cells, as proposed by Macomber et al. (12). Alternatively, copper may block iron-sulfur cluster biogenesis (14–16), resulting in inactive apo-proteins in E. coli cells. To elucidate the possible effects, 0.2 mM CuCl2 was added to LB medium before and after recombinant iron-sulfur protein was expressed in the E. coli cells under anaerobic conditions.
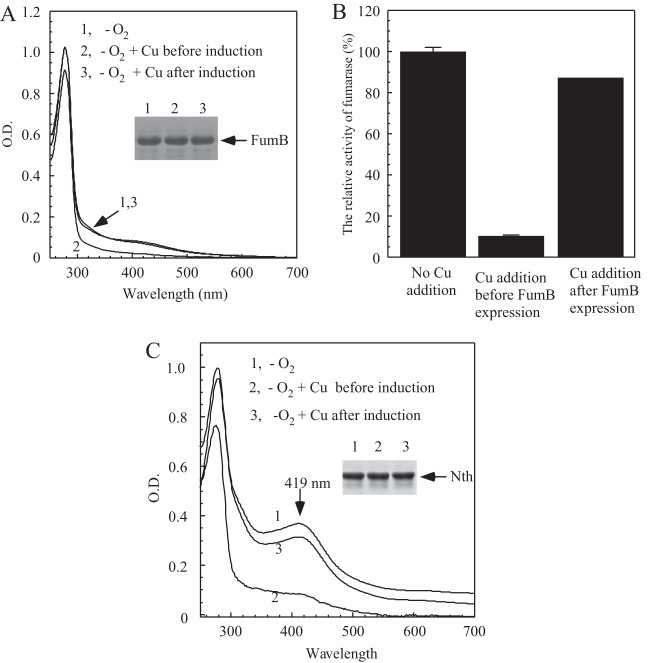
Figure 4A shows that, while addition of copper to LB medium before recombinant fumarase B was expressed largely eliminated iron-sulfur clusters in the protein, addition of copper to LB medium after fumarase B was expressed did not significantly affect iron-sulfur clusters in the protein under anaerobic conditions. The enzyme activity measurements further showed that copper blocked iron-sulfur cluster assembly without significantly affecting the assembled iron-sulfur clusters in fumarase B in the E. coli cells under anaerobic conditions (Fig. 4B).
FIG 4.
Copper inhibits iron-sulfur cluster assembly in E. coli cells under anaerobic conditions. (A) UV-visible absorption spectra of recombinant fumarase B purified from E. coli cells. Spectrum 1, fumarase B was expressed in the exponentially growing E. coli cells in LB medium without copper addition under anaerobic conditions for 2 h; spectrum 2, fumarase B was expressed in the exponentially growing E. coli cells in LB medium supplemented with 0.2 mM CuCl2 under anaerobic conditions for 2 h; spectrum 3, fumarase B was expressed in the exponentially growing E. coli cells in LB medium under anaerobic conditions for 2 h, followed by addition of 0.2 mM CuCl2 and an additional 2 h of growth under anaerobic conditions. Without addition of copper, fumarase B expressed in E. coli cells for 4 h had an absorption spectrum very similar to that of spectrum 3. The protein concentration of fumarase B was 20 μM. (Inset) Photograph of SDS-PAGE gel of purified proteins. (B) Relative activities of purified fumarase B from panel A. For enzyme activity assay, 10 nM fumarase B was used. The highest enzyme activity (100%) for fumarase B was 22.7 ± 0.8 μΜ/μM/min. (C) UV-visible absorption spectra of recombinant endonuclease III (Nth) purified from E. coli cells. Spectrum 1, endonuclease III was expressed in the exponentially growing E. coli cells in LB medium without copper addition under anaerobic conditions for 2 h; spectrum 2, endonuclease III was induced in the exponentially growing E. coli cells in LB medium supplemented with 0.2 mM CuCl2 under anaerobic conditions for 2 h; spectrum 3, endonuclease III was expressed in the exponentially growing E. coli cells in LB medium under anaerobic conditions for 2 h, followed by addition of 0.2 mM CuCl2 and an additional 2 h of growth under anaerobic conditions. Without addition of copper, endonuclease III expressed in E. coli cells for 4 h had an absorption spectrum very similar to that of spectrum 3. The protein concentration of endonuclease III was 40 μM. (Inset) Photograph of the SDS-PAGE gel of purified proteins.
The DNA repair enzyme endonuclease III hosts a stable [4Fe-4S] cluster (38). Unlike the solvent-exposed [4Fe-4S] clusters in IlvD, fumarase A, and fumarase B, the [4Fe-4S] cluster in endonuclease III is buried within the protein structure (38). Here, we used endonuclease III as another example of the effect of copper on iron-sulfur cluster biogenesis in E. coli cells under anaerobic conditions. Figure 4C shows that while addition of 0.2 mM CuCl2 to LB medium before endonuclease III was expressed eliminated the iron-sulfur cluster in the recombinant protein, addition of copper after endonuclease III was expressed did not significantly affect the assembled iron-sulfur clusters in the protein under anaerobic conditions. Altogether, these results show that copper appears to inhibit iron-sulfur cluster biogenesis without disrupting the assembled iron-sulfur clusters in proteins in E. coli cells under anaerobic conditions.
Copper binding in the iron-sulfur cluster assembly proteins IscA and IscU in E. coli cells under aerobic and anaerobic conditions.
In E. coli cells, the proteins encoded by iscSUA-hscBA-fdx are the major source of the activity responsible for iron-sulfur cluster biogenesis (20). If copper inhibits iron-sulfur cluster biogenesis, we would expect that copper may directly bind to the iron-sulfur cluster assembly proteins. To test this idea, we expressed the core iron-sulfur cluster assembly proteins, cysteine desulfurase IscS (24, 34), scaffold protein IscU (27), and iron chaperone IscA (15), in E. coli cells grown in LB medium supplemented with 0.2 mM CuCl2 under aerobic and anaerobic conditions.
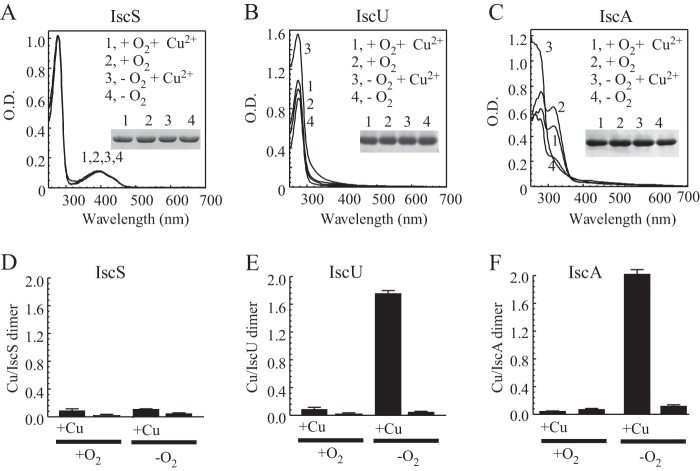
Figure 5A shows that addition of 0.2 mM CuCl2 to LB medium under aerobic or anaerobic conditions had very little or no effect on the UV-visible absorption spectrum of cysteine desulfurase IscS expressed in E. coli cells. The specific enzyme activity of purified IscS was also not affected by the copper treatment under aerobic or anaerobic conditions (data not shown). In contrast, when the E. coli cells were grown in LB medium supplemented with 0.2 mM CuCl2 under anaerobic conditions, purified IscU and IscA had an increased absorption peak of 260 nm (Fig. 5B and C), indicative of copper binding in the proteins (15). A copper content analysis further confirmed that IscU and IscA bind about 1.8 to 2.0 copper atoms per protein dimer (Fig. 5E and F).
FIG 5.
Copper binding in IscS, IscU, and IscA in E. coli cells under anaerobic conditions. Iron-sulfur cluster assembly proteins IscS, IscU, and IscA were expressed in E. coli cells grown in LB medium supplemented with 0 or 0.2 mM CuCl2 under aerobic or anaerobic conditions. UV-visible absorption spectra of purified IscS (25 μM) (A), IscU (100 μM) (B), and IscA (300 μM) (C) are shown. Spectrum 1, with 0.2 mM CuCl2 under aerobic conditions; spectrum 2, with no CuCl2 under aerobic conditions; spectrum 3, with 0.2 mM CuCl2 under anaerobic conditions; spectrum 4, with no CuCl2 under anaerobic conditions. (Insets) Photographs of SDS-PAGE gels of purified proteins. (D, E, and F) Copper contents of purified IscS, IscU, and IscA, respectively. The samples are the same as those in panels A through C. Copper contents in protein samples were determined as described in Materials and Methods and are presented as ratios of copper per protein dimer. The results are averages ± standard deviations from three independent experiments.
SufA is a member of the inducible sufABCDSE gene cluster for iron-sulfur cluster biogenesis (22) and a paralog of IscA (42). In this context, we also assessed the copper binding activity of SufA in E. coli cells grown in LB medium supplemented with 0.2 mM CuCl2 under aerobic or anaerobic conditions. As shown in Fig. S1 in the supplemental material, SufA, like IscA, was able to bind copper in E. coli cells under anaerobic conditions but not under aerobic conditions. Since IscA, IscU, and SufA, each with bound copper, fail to assemble iron-sulfur clusters in target proteins (15), the bound copper in IscA, IscU, and SufA effectively blocks iron-sulfur cluster biogenesis in E. coli cells under anaerobic conditions. Interestingly, under anaerobic conditions, the gene cluster iscSUA-hscBA-fdx is significantly repressed in E. coli cells (43), which makes the iron-sulfur cluster assembly proteins more sensitive to copper toxicity in the cells under anaerobic conditions.
Since the copper binding in IscA or IscU is not affected by oxygen in vitro (15), the observed copper binding in IscA and IscU in E. coli cells under anaerobic conditions (Fig. 5) could be due to the increased intracellular copper concentration, as reported by Outten et al. (9). Because different E. coli strains and growth media were used in the experiments by Outten et al. (9) and in the experiments described in this study, we reevaluated the intracellular copper content in E. coli cells grown in LB medium supplemented with 0.2 mM CuCl2 under aerobic and anaerobic conditions using whole-cell electron paramagnetic resonance (EPR) measurements (15). As copper in E. coli cells exists as Cu(I) (9), which does not yield an EPR signal, the cells were washed twice and then treated with 2.5% (vol/vol) nitric acid to oxidize Cu(I) to Cu(II) for the EPR measurements (44). Quantification of the Cu(II) by EPR determination (Fig. 6) showed that E. coli cells grown in LB medium supplemented with 0.2 mM CuCl2 contained about 10 to 15 times more intracellular copper content under anaerobic conditions than under aerobic conditions. We concluded that an elevated copper content in the E. coli cells (9) may effectively inhibit iron-sulfur cluster biogenesis by binding to the iron-sulfur cluster assembly proteins IscA, SufA, and IscU under anaerobic conditions.
FIG 6.
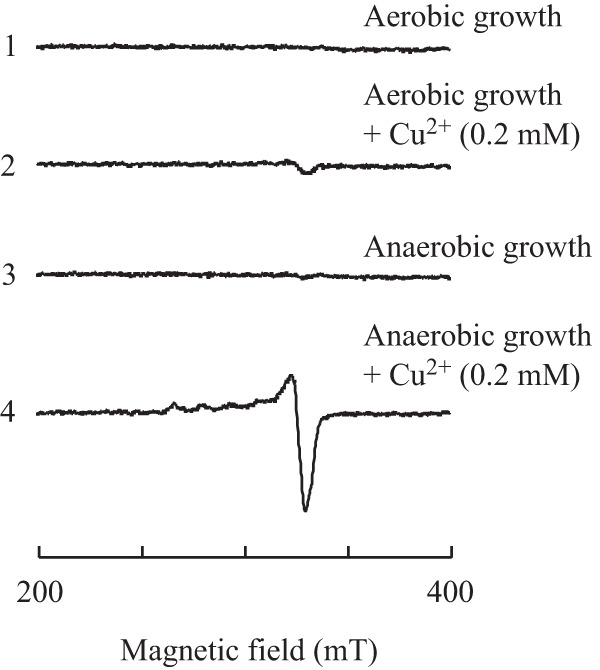
Accumulation of intracellular copper content in E. coli cells under anaerobic conditions. E. coli cells were grown in LB medium supplemented with or without 0.2 mM CuCl2 for 4 h under anaerobic or aerobic conditions. The E. coli cells were then washed with buffer containing 20 mM Tris (pH 8.0) and 500 mM NaCl twice and treated with 2.5% (vol/vol) nitric acid. The samples were subjected to EPR measurements. Spectrum 1, E. coli cells grown in LB medium under aerobic conditions; spectrum 2, E. coli cells grown in LB medium supplemented with 0.2 mM CuCl2 under aerobic conditions; spectrum 3, E. coli cells grown in LB medium under anaerobic conditions; spectrum 4, E. coli cells grown in LB medium supplemented with 0.2 mM CuCl2 under anaerobic conditions.
DISCUSSION
Previous studies have shown that iron-sulfur proteins are the primary targets of copper toxicity in E. coli (12, 14, 15), Bacillus subtilis (13), and mitochondria (16) under aerobic conditions. Here, we report that iron-sulfur proteins in E. coli cells are much more susceptible to copper in medium under anaerobic conditions. While addition of 0.2 mM CuCl2 to LB medium has very little or no effect on iron-sulfur proteins in E. coli cells under aerobic conditions, the same copper treatment largely inhibits the enzyme activity of iron-sulfur proteins by inhibiting iron-sulfur cluster biogenesis under anaerobic conditions. It appears that under anaerobic conditions, E. coli cells accumulate extra intracellular copper, which blocks iron-sulfur cluster biogenesis by binding to the iron-sulfur cluster assembly proteins IscA and IscU and the IscA analog SufA. We also find that while copper inhibits iron-sulfur proteins, it does not affect the proteins that do not have iron-sulfur clusters in E. coli cells under anaerobic conditions. Since iron-sulfur proteins are involved in diverse and vital physiological processes (17, 18), inactivation of iron-sulfur cluster biogenesis by copper would have a broad effect on cellular functions under anaerobic conditions.
Cu(I) requires only two cysteine residues to form a linear biscysteinate coordination geometry, as seen in the Parkinsonism-associated DJ-1 protein (11). The thiol ligands in the iron-sulfur cluster assembly proteins IscA (45), SufA (42), and IscU (27) may facilitate strong copper binding. Previously, we found that copper can compete with iron for binding in IscA in vitro and in E. coli cells (15). While we did not observe significant copper binding in IscU in E. coli cells under aerobic conditions (15), we found copper binding in both IscA and IscU in E. coli cells grown in LB medium supplemented with 0.2 mM CuCl2 under anaerobic conditions (Fig. 5). Perhaps the accumulation of extra copper in E. coli cells under anaerobic conditions results in copper binding in IscU and IscA and its paralog SufA. Since IscA, SufA, and IscU bearing bound copper fail to assemble iron-sulfur clusters in proteins (15), copper may effectively block iron-sulfur cluster biogenesis in cells under anaerobic conditions. It should be pointed out that the repressed expression of the gene cluster iscSUA-hscBA-fdx in E. coli cells under anaerobic conditions (43) may further contribute to the increased susceptibility of iron-sulfur proteins to intracellular copper content. In this context, we propose that iron-sulfur cluster biogenesis is the primary target of copper toxicity in E. coli cells under anaerobic conditions.
Anaerobic copper toxicity in E. coli cells was first reported in 1976 (10). Accumulation of intracellular copper is likely responsible for increased copper toxicity under anaerobic conditions (9). In E. coli, intracellular copper content is regulated by three major copper homeostasis systems, CopA, a P-type ATPase that pumps copper ion out of the cytoplasm (46), copper oxidase (CueO), a periplasmic copper oxidase that oxidizes Cu(I) to Cu(II) in the periplasm to prevent adventitious entry into cytoplasm (47), and a copper pump (CusCBA) that directly transports copper ions from the cytoplasm to the extracellular environment (48). Under anaerobic conditions, CueO would be inactive because the enzyme requires oxygen to oxidize Cu(I), thus partially contributing to accumulation of intracellular Cu(I) (9). Expression of the genes copA and cusCBA also has distinct responses to copper under aerobic and anaerobic conditions (9), which could further enhance copper accumulation in cells under anaerobic conditions. Nevertheless, the molecular mechanism underlying copper accumulation in E. coli cells under anaerobic conditions is not yet fully understood. Interestingly, Saccharomyces cerevisiae cells also accumulate intracellular copper under anaerobic conditions (49). Under anaerobic conditions, perhaps microorganisms have to promote the influx of copper or other metal ions for critical physiological functions, and accumulation of excess copper in cells will inadvertently inhibit iron-sulfur cluster biogenesis, as reported in this study. Nevertheless, the anaerobic copper toxicity appears to be reversible, as cell growth was almost immediately restored when the E. coli cells grown in LB medium supplemented with 1.0 mM CuCl2 were shifted from anaerobic to aerobic conditions (Fig. 1). Furthermore, when the E. coli cells grown in LB medium supplemented with 0.2 mM CuCl2 under anaerobic conditions were transferred to aerobic conditions, the iron-sulfur clusters and the enzyme activity of recombinant FumA expressed in E. coli cells were largely restored (see Fig. S2A and B in the supplemental material) with a concomitant decrease in the intracellular copper content (see Fig. S2C), suggesting a robust ability of bacteria to survive in response to copper toxicity.
While copper and iron are essential trace metals for living organisms, intracellular concentrations of both metals must be tightly controlled in order to balance their biological functions. In the presence of toxic concentrations of copper, bacteria may have to slow down or inactivate multiple physiological processes by blocking iron-sulfur cluster biogenesis in order to avoid potential damage to cells. On the other hand, excess iron may also interfere with components of the copper homeostasis and sensitize the bacteria to copper toxicity under anaerobic conditions (5). Since polluted water often contains different toxic metals (3, 4), the interplay between copper and iron/iron-sulfur clusters in microorganisms will require further investigation.
MATERIALS AND METHODS
Cell growth and protein purification.
The DNA fragments encoding fumarases A, B, and C were amplified from wild-type E. coli genomic DNA using PCR. The PCR products were ligated to an expression vector, pBAD (Novagen). The identity of the cloned DNA fragment was confirmed by direct sequencing. Each plasmid was then introduced into the wild-type E. coli strain (MC4100). For protein preparation, overnight cultures of E. coli containing expression plasmids were diluted (1:100) in freshly prepared LB medium. Cells were grown at 37°C with aeration to an optical density at 600 nm (OD600) of 0.6. The protein expression was induced by adding 0.2% (wt/vol) l-arabinose for 2 h. To express proteins under anaerobic conditions, the E. coli cells were purged with pure argon gas for 40 min before l-arabinose was added to the sealed flasks. The cells were grown for an additional 2 h before being harvested and washed twice with the protein purification buffer (20 mM Tris-HCl [pH 8.0] and 500 mM NaCl). Recombinant E. coli fumarase A, fumarase B, fumarase C, dihydroxyacid dehydratase (50), endonuclease III, IscS, IscU, and IscA were purified as described (15), and proteins were purified to a single band on SDS-PAGE gel. The UV-visible absorption spectra of purified proteins were recorded in a Beckman DU640 UV-visible absorption spectrometer equipped with a temperature controller. The extinction coefficients for purified E. coli IlvD, endonuclease III, fumarase A, fumarase B, fumarase C, IscS, IscU, and IscA are 35.2, 19.3, 51.4, 51.4, 34.3, 39.8, 11.2, and 1.6 mM−1cm−1, respectively.
Copper content analyses.
The copper contents of protein samples were determined by following the procedure described in reference 15, with slight modifications. Briefly, protein samples (160 μl) were incubated with 10 μl neocuproine (5 mM in H2O-ethanol [1:1]), 20 μl SDS (20%), and 10 μl sodium ascorbate (5 mM). Mixtures were incubated at room temperature for 20 min, followed by centrifugation. The amplitude of the absorption peak at 450 nm of the supernatant was used for quantification of copper content. Freshly prepared CuCl2 (10 μM) was used as a standard. The copper contents in protein samples were also analyzed by inductively coupled plasma mass spectrometry (ICP-MS) (Agilent 8800). The two metal content analyses produced similar results.
Enzyme activity measurements.
The enzyme activity of dihydroxyacid dehydratase was measured using the substrate dl-2,3-dihydroxy-isovalerate, as described previously (15). Briefly, the enzyme activity was measured using the substrate dl-2,3-dihydroxy-isovalerate. The reaction solution contained 1 μM dihydroxyacid dehydratase (IlvD), 50 mM Tris (pH 8.0), 10 mM MgCl2, and 10 mM dl-2,3-dihydroxy-isovalerate. The reaction product (keto acid) was monitored at 240 nm using an extinction coefficient of 0.19 mM−1 cm−1. Fumarase activity was measured using malic acid as the substrate (51). The reaction solution contained 10 nM fumarase, 50 mM sodium phosphate (pH 7.4), and 50 mM malic acid (pH 7.4). The reaction product (fumaric acid) was monitored at 250 nm using an extinction coefficient of 1.48 mM−1 cm−1. The cysteine desulfurase activity of IscS was measured as described previously (34). The reaction solution contained 1 μM IscS, 3 mM dithiothreitol, 20 mM Tris (pH 8.0), and 200 mM NaCl. The reaction solution was preincubated at 37°C for 5 min before 2 mM l-cysteine was added to initiate the reaction. The product sulfide in the reaction solution was determined by adding 20 mM N,N-dimethyl-p-phenylene-diamine sulfate (in 7.2 M HCl) and 30 mM FeCl3 (in 1.2 M HCl), as described previously (52). The sulfur production in the reaction solution was used to represent the relative enzyme activity of IscS.
EPR measurements.
For the EPR (electron paramagnetic resonance) measurements, the E. coli cells were harvested, washed twice with buffer containing 500 mM NaCl and 20 mM Tris (pH 8.0), and treated with 2.5% (vol/vol) nitric acid to oxidize Cu(I) in cells as described previously (15, 44). The EPR spectra were recorded at X-band on a Bruker ESP-300 spectrometer equipped with an Oxford Instruments 910 continuous flow cryostat. The routine EPR conditions were a microwave frequency of 9.45 GHz, microwave power of 10 mW, modulation frequency of 100 kHz, modulation amplitude of 2.0 mT, sample temperature of 30 K, and receive gain of 1.0 × 105. A solution containing Cu(II)-EDTA (10 μM) was used as a standard for quantification of Cu(II) in the protein.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the American Heart Association (grant 13GRNT16890014), the National Natural Science Foundation of China (grants 31629003, 81671124, and 81500440), and the National High Technology Research and Development Program of China (863 Program) (grant 2014AA06A514).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00867-17.
REFERENCES
- 1.Solomon EI, Heppner DE, Johnston EM, Ginsbach JW, Cirera J, Qayyum M, Kieber-Emmons MT, Kjaergaard CH, Hadt RG, Tian L. 2014. Copper active sites in biology. Chem Rev 114:3659–3853. doi: 10.1021/cr400327t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Besold AN, Culbertson EM, Culotta VC. 2016. The Yin and Yang of copper during infection. J Biol Inorg Chem 21:137–144. doi: 10.1007/s00775-016-1335-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Newcomb WD, Rimstidt JD. 2002. Trace element distribution in US groundwaters: a probabilistic assessment using public domain data. Appl Geochem 17:49–57. doi: 10.1016/S0883-2927(01)00089-0. [DOI] [Google Scholar]
- 4.Kabir E, Ray S, Kim KH, Yoon HO, Jeon EC, Kim YS, Cho YS, Yun ST, Brown RJ. 2012. Current status of trace metal pollution in soils affected by industrial activities. ScientificWorldJournal 2012:916705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bird LJ, Coleman ML, Newman DK. 2013. Iron and copper act synergistically to delay anaerobic growth of bacteria. Appl Environ Microbiol 79:3619–3627. doi: 10.1128/AEM.03944-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pham AN, Xing G, Miller CJ, Waite TD. 2013. Fenton-like copper redox chemistry revisited: hydrogen peroxide and superoxide mediation of copper-catalyzed oxidant production. J Catalysis 301:54–64. doi: 10.1016/j.jcat.2013.01.025. [DOI] [Google Scholar]
- 7.Gaetke LM, Chow CK. 2003. Copper toxicity, oxidative stress, and antioxidant nutrients. Toxicology 189:147–163. doi: 10.1016/S0300-483X(03)00159-8. [DOI] [PubMed] [Google Scholar]
- 8.Macomber L, Rensing C, Imlay JA. 2007. Intracellular copper does not catalyze the formation of oxidative DNA damage in Escherichia coli. J Bacteriol 189:1616–1626. doi: 10.1128/JB.01357-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Outten FW, Huffman DL, Hale JA, O'Halloran TV. 2001. The independent cue and cus systems confer copper tolerance during aerobic and anaerobic growth in Escherichia coli. J Biol Chem 276:30670–30677. doi: 10.1074/jbc.M104122200. [DOI] [PubMed] [Google Scholar]
- 10.Beswick PH, Hall GH, Hook AJ, Little K, McBrien DC, Lott KA. 1976. Copper toxicity: evidence for the conversion of cupric to cuprous copper in vivo under anaerobic conditions. Chem Biol Interact 14:347–356. doi: 10.1016/0009-2797(76)90113-7. [DOI] [PubMed] [Google Scholar]
- 11.Puno MR, Patel NA, Moller SG, Robinson CV, Moody PC, Odell M. 2013. Structure of Cu(I)-bound DJ-1 reveals a biscysteinate metal binding site at the homodimer interface: insights into mutational inactivation of DJ-1 in parkinsonism. J Am Chem Soc 135:15974–15977. doi: 10.1021/ja406010m. [DOI] [PubMed] [Google Scholar]
- 12.Macomber L, Imlay JA. 2009. The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc Natl Acad Sci U S A 106:8344–8349. doi: 10.1073/pnas.0812808106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chillappagari S, Seubert A, Trip H, Kuipers OP, Marahiel MA, Miethke M. 2010. Copper stress affects iron homeostasis by destabilizing iron-sulfur cluster formation in Bacillus subtilis. J Bacteriol 192:2512–2524. doi: 10.1128/JB.00058-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fung DK, Lau WY, Chan WT, Yan A. 2013. Copper efflux is induced during anaerobic amino acid limitation in Escherichia coli to protect iron-sulfur cluster enzymes and biogenesis. J Bacteriol 195:4556–4568. doi: 10.1128/JB.00543-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan G, Cheng Z, Pang Y, Landry AP, Li J, Lu J, Ding H. 2014. Copper binding in IscA inhibits iron-sulphur cluster assembly in Escherichia coli. Mol Microbiol 93:629–644. doi: 10.1111/mmi.12676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brancaccio D, Gallo A, Piccioli M, Novellino E, Ciofi-Baffoni S, Banci L. 2017. [4Fe-4S] Cluster assembly in mitochondria and its impairment by copper. J Am Chem Soc 139:719–730. doi: 10.1021/jacs.6b09567. [DOI] [PubMed] [Google Scholar]
- 17.Johnson DC, Dean DR, Smith AD, Johnson MK. 2005. Structure, function, and formation of biological iron-sulfur clusters. Annu Rev Biochem 74:247–281. doi: 10.1146/annurev.biochem.74.082803.133518. [DOI] [PubMed] [Google Scholar]
- 18.Paul VD, Lill R. 2015. Biogenesis of cytosolic and nuclear iron-sulfur proteins and their role in genome stability. Biochim Biophys Acta 1853:1528–1539. doi: 10.1016/j.bbamcr.2014.12.018. [DOI] [PubMed] [Google Scholar]
- 19.Roche B, Aussel L, Ezraty B, Mandin P, Py B, Barras F. 2013. Iron/sulfur proteins biogenesis in prokaryotes: formation, regulation and diversity. Biochim Biophys Acta 1827:455–469. doi: 10.1016/j.bbabio.2012.12.010. [DOI] [PubMed] [Google Scholar]
- 20.Zheng L, Cash VL, Flint DH, Dean DR. 1998. Assembly of iron-sulfur clusters. Identification of an iscSUA-hscBA-fdx gene cluster from Azotobacter vinelandii. J Biol Chem 273:13264–13272. [DOI] [PubMed] [Google Scholar]
- 21.Rouault TA. 2015. Mammalian iron-sulphur proteins: novel insights into biogenesis and function. Nat Rev Mol Cell Biol 16:45–55. [DOI] [PubMed] [Google Scholar]
- 22.Takahashi Y, Tokumoto U. 2002. A third bacterial system for the assembly of iron-sulfur clusters with homologs in archaea and plastids. J Biol Chem 277:28380–28383. doi: 10.1074/jbc.C200365200. [DOI] [PubMed] [Google Scholar]
- 23.Xu XM, Moller SG. 2008. Iron-sulfur cluster biogenesis systems and their crosstalk. Chembiochem 9:2355–2362. doi: 10.1002/cbic.200800384. [DOI] [PubMed] [Google Scholar]
- 24.Cupp-Vickery JR, Urbina H, Vickery LE. 2003. Crystal structure of IscS, a cysteine desulfurase from Escherichia coli. J Mol Biol 330:1049–1059. doi: 10.1016/S0022-2836(03)00690-9. [DOI] [PubMed] [Google Scholar]
- 25.Marinoni EN, de Oliveira JS, Nicolet Y, Raulfs EC, Amara P, Dean DR, Fontecilla-Camps JC. 2012. (IscS-IscU)2 complex structures provide insights into Fe2S2 biogenesis and transfer. Angew Chem Int Ed Engl 51:5439–5442. doi: 10.1002/anie.201201708. [DOI] [PubMed] [Google Scholar]
- 26.Unciuleac MC, Chandramouli K, Naik S, Mayer S, Huynh BH, Johnson MK, Dean DR. 2007. In vitro activation of apo-aconitase using a [4Fe-4S] cluster-loaded form of the IscU [Fe-S] cluster scaffolding protein. Biochemistry 46:6812–6821. doi: 10.1021/bi6026665. [DOI] [PubMed] [Google Scholar]
- 27.Yan R, Kelly G, Pastore A. 2014. The scaffold protein IscU retains a structured conformation in the Fe-S cluster assembly complex. Chembiochem 15:1682–1686. doi: 10.1002/cbic.201402211. [DOI] [PubMed] [Google Scholar]
- 28.Brancaccio D, Gallo A, Mikolajczyk M, Zovo K, Palumaa P, Novellino E, Piccioli M, Ciofi-Baffoni S, Banci L. 2014. Formation of [4Fe-4S] clusters in the mitochondrial iron-sulfur cluster assembly machinery. J Am Chem Soc 136:16240–16250. doi: 10.1021/ja507822j. [DOI] [PubMed] [Google Scholar]
- 29.Sheftel AD, Wilbrecht C, Stehling O, Niggemeyer B, Elsasser HP, Muhlenhoff U, Lill R. 2012. The human mitochondrial ISCA1, ISCA2, and IBA57 proteins are required for [4Fe-4S] protein maturation. Mol Biol Cell 23:1157–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ding H, Clark RJ. 2004. Characterization of iron binding in IscA, an ancient iron-sulphur cluster assembly protein. Biochem J 379:433–440. doi: 10.1042/bj20031702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mapolelo DT, Zhang B, Naik SG, Huynh BH, Johnson MK. 2012. Spectroscopic and functional characterization of iron-bound forms of Azotobacter vinelandii (Nif)IscA. Biochemistry 51:8056–8070. doi: 10.1021/bi300664j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang J, Bitoun JP, Ding H. 2006. Interplay of IscA and IscU in biogenesis of iron-sulfur clusters. J Biol Chem 281:27956–27963. doi: 10.1074/jbc.M601356200. [DOI] [PubMed] [Google Scholar]
- 33.Landry AP, Cheng Z, Ding H. 2013. Iron binding activity is essential for the function of IscA in iron-sulphur cluster biogenesis. Dalton Trans 42:3100–3106. doi: 10.1039/C2DT32000B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang J, Tan G, Zhang T, White RH, Lu J, Ding H. 2015. Deletion of the proposed iron chaperones IscA/SufA results in accumulation of a red intermediate cysteine desulfurase IscS in Escherichia coli. J Biol Chem 290:14226–14234. doi: 10.1074/jbc.M115.654269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bandyopadhyay S, Chandramouli K, Johnson MK. 2008. Iron-sulfur cluster biosynthesis. Biochem Soc Trans 36:1112–1119. doi: 10.1042/BST0361112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Flint DH, Emptage MH, Finnegan MG, Fu W, Johnson MK. 1993. The role and properties of the iron-sulfur cluster in Escherichia coli dihydroxy-acid dehydratase. J Biol Chem 268:14732–14742. [PubMed] [Google Scholar]
- 37.Varghese S, Tang Y, Imlay JA. 2003. Contrasting sensitivities of Escherichia coli aconitases A and B to oxidation and iron depletion. J Bacteriol 185:221–230. doi: 10.1128/JB.185.1.221-230.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cunningham RP, Ahern H, Xing D, Thayer MM, Tainer JA. 1994. Structure and function of Escherichia coli endonuclease III. Ann N Y Acad Sci 726:215–222. doi: 10.1111/j.1749-6632.1994.tb52818.x. [DOI] [PubMed] [Google Scholar]
- 39.Jang S, Imlay JA. 2007. Micromolar intracellular hydrogen peroxide disrupts metabolism by damaging iron-sulfur enzymes. J Biol Chem 282:929–937. doi: 10.1074/jbc.M607646200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Vugt-Lussenburg BM, van der Weel L, Hagen WR, Hagedoorn PL. 2013. Biochemical similarities and differences between the catalytic [4Fe-4S] cluster containing fumarases FumA and FumB from Escherichia coli. PLoS One 8:e55549. doi: 10.1371/journal.pone.0055549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weaver T. 2005. Structure of free fumarase C from Escherichia coli. Acta Crystallogr D Biol Crystallogr 61:1395–1401. doi: 10.1107/S0907444905024194. [DOI] [PubMed] [Google Scholar]
- 42.Lu J, Yang J, Tan G, Ding H. 2008. Complementary roles of SufA and IscA in the biogenesis of iron-sulfur clusters in Escherichia coli. Biochem J 409:535–543. doi: 10.1042/BJ20071166. [DOI] [PubMed] [Google Scholar]
- 43.Giel JL, Nesbit AD, Mettert EL, Fleischhacker AS, Wanta BT, Kiley PJ. 2013. Regulation of iron-sulphur cluster homeostasis through transcriptional control of the Isc pathway by [2Fe-2S]-IscR in Escherichia coli. Mol Microbiol 87:478–492. doi: 10.1111/mmi.12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ve T, Mathisen K, Helland R, Karlsen OA, Fjellbirkeland A, Røhr Å Andersson KK, Pedersen RB, Lillehaug JR, Jensen HB. 2012. The Methylococcus capsulatus (Bath) secreted protein, MopE*, binds both reduced and oxidized copper. PLoS One 7:e43146. doi: 10.1371/journal.pone.0043146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bilder PW, Ding H, Newcomer ME. 2004. Crystal structure of the ancient, Fe-S scaffold IscA reveals a novel protein fold. Biochemistry 43:133–139. doi: 10.1021/bi035440s. [DOI] [PubMed] [Google Scholar]
- 46.Fan B, Rosen BP. 2002. Biochemical characterization of CopA, the Escherichia coli Cu(I)-translocating P-type ATPase. J Biol Chem 277:46987–46992. doi: 10.1074/jbc.M208490200. [DOI] [PubMed] [Google Scholar]
- 47.Stoyanov JV, Hobman JL, Brown NL. 2001. CueR (YbbI) of Escherichia coli is a MerR family regulator controlling expression of the copper exporter CopA. Mol Microbiol 39:502–511. doi: 10.1046/j.1365-2958.2001.02264.x. [DOI] [PubMed] [Google Scholar]
- 48.Munson GP, Lam DL, Outten FW, O'Halloran TV. 2000. Identification of a copper-responsive two-component system on the chromosome of Escherichia coli K-12. J Bacteriol 182:5864–5871. doi: 10.1128/JB.182.20.5864-5871.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Strain J, Culotta VC. 1996. Copper ions and the regulation of Saccharomyces cerevisiae metallothionein genes under aerobic and anaerobic conditions. Mol Gen Genet 251:139–145. doi: 10.1007/BF02172911. [DOI] [PubMed] [Google Scholar]
- 50.Duan X, Yang J, Ren B, Tan G, Ding H. 2009. Reactivity of nitric oxide with the [4Fe-4S] cluster of dihydroxyacid dehydratase from Escherichia coli. Biochem J 417:783–789. doi: 10.1042/BJ20081423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Djaman O, Outten FW, Imlay JA. 2004. Repair of oxidized iron-sulfur clusters in Escherichia coli. J Biol Chem 279:44590–44599. doi: 10.1074/jbc.M406487200. [DOI] [PubMed] [Google Scholar]
- 52.Siegel LM. 1965. A direct microdetermination of sulfide. Anal Biochem 11:126–132. doi: 10.1016/0003-2697(65)90051-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.