Abstract
The microbiota — the collection of microorganisms that live within and on all mammals — provides crucial signals for the development and function of the immune system. Increased availability of technologies that profile microbial communities is facilitating the entry of many immunologists into the evolving field of host-microbiota studies. The microbial communities, their metabolites and components are not only necessary for immune homeostasis, they also influence the susceptibility of the host to many immune-mediated diseases and disorders. In this Review, we discuss technological and computational approaches for investigating the microbiome, as well as recent advances in our understanding of host immunity and microbial mutualism with a focus on specific microbial metabolites, bacterial components and the immune system.
Mammals harbour widely diverse and immensely active microbial communities. Through an enduring mutualistic partnership, the host has evolved to coordinate and integrate conserved metabolic signalling, microbial sensing and immune response pathways to ensure its survival in a microbially dominated world. This dynamic crosstalk between the host and its microbiota is important for achieving and maintaining homeostasis. If, or when, this dialogue goes awry — owing to perturbations to the microbiota or the host — dysbiosis can ensue. Studies continue to establish the role of an altered microbiome in human disorders and diseases, as well as in preclinical models, both within and outside the gastrointestinal tract (TABLE 1).
Table 1.
Microbial metabolites or components that are implicated in disease
| Human disease and preclinical models |
Microbial metabolites or components |
Refs |
|---|---|---|
| Allergic and immune disorders | ||
| Asthma | SCFAs | 40,45 |
| Inflammatory bowel disease | SCFAs | 42–44,53 |
| B vitamins | 53 | |
| Cancer | ||
| Colorectal cancer | SCFAs | 53,118,119 |
| B vitamins | 53 | |
| N1,N12-diacetylspermine | 80 | |
| Gynaecological and reproductive disorders | ||
| Bacterial vaginosis and other sexually transmitted infections | Polyamines | 120 |
| HBP | 100 | |
| Preterm labour | SCFAs | 50 |
| Metabolic disorders | ||
| Cardiovascular disease | TMAO | 121 |
| Kidney disease | SCFAs | 122 |
| p-Cresol | 123 | |
| Obesity and metabolic syndrome | TMAO | 121 |
| Type 2 diabetes | TMAO | 121 |
| Neurological disorders | ||
| Autism spectrum disorder | 4-EPS | 124 |
| Central nervous system dysfunction | SCFAs | 52,125 |
| Other gastrointestinal disorders | ||
| Infectious colitis (Clostridium difficile) | Bile acids | 126 |
4-EPS, 4-ethyl phenol sulfate; HBP, D-glycero-β-D-manno-heptose-1,7-biphosphate; SCFAs, short-chain fatty acids; TMAO, trimethylamine N-oxide.
Recent advances in characterizing the composition and function of individual microbial species and complex microbial communities are revealing the importance of microbial metabolism for the host immune system. A ‘healthy’ microbiome provides crucial molecular cues — through microbial surface antigens and metabolites — that are essential for the maturation of immune tissues and for the fine tuning of immune responses. Here, we review the technologies that are available for identifying symbiotic microbial strains, microbial-derived components and metabolic products, and discuss their role in shaping the host immune system.
Studying the microbiome
Characterizing microbiome composition
The advent of high-throughput DNA sequencing technology — initially by clustering reads based on bacterial and archaeal 16S ribosomal RNA amplicon sequences, and now by aligning whole genomes to all domains of life — allows the direct classification of samples without the need for culturing. These technological advances provide a robust method for profiling complex microbial communities from diverse environments and for analysing changes in community structure over time. Although microbiome composition differs between individuals and at times fluctuates markedly within individuals, core features exist in the microbial communities that colonize the human body1. Each body habitat is spatially distinct and differentially dominated by specific phyla, with niche-specific microbiomes present in varying abundance and distribution across anatomical sites2. In the gut, the quantity and diversity of microbial species increase longitudinally from the stomach to the colon3,4, with the colon being home to the most dense and metabolically active community (comprising more than 1013 individual microbial cells)5. An appreciation of the magnitude of microbial diversity within and across individuals is affecting how the microbiome is studied, changing from descriptive studies of members of the microbial communities to more mechanistic studies of the functional contribution of the microbiota to health and disease.
Investigating microbiome function
Whole metagenomic and metatranscriptomic sequencing endeavours (from cDNA libraries) are defining the functional potential and real-time activity of microbiomes, and revealing interactions between microbial metabolism and host development. The ability to profile the regulation and dynamics of the microbiome and the expression patterns of host genes is revealing how the functions of the microbial community can influence the host6 and, conversely, how host genetics shape the composition and function of the microbiome7. Moreover, simultaneous sequencing of the metagenomes and metatranscriptomes of the host and microbiome is providing insight into the mechanisms of host–microbiota mutualism and the differences between healthy and diseased individuals. In addition, the development of tools to identify and reconstruct genes into broader biological pathways has allowed functional features of the microbiome to be divided into distinct but related categories that are essential to the health of the host8 (BOX 1).
Box 1 | Technology and tools for studying host-microbiome interactions.
In-depth investigation of the microbiome has been made possible by advances in high-throughput sequencing together with the development of computational tools for the analysis of high-complexity data. Microbial DNA or RNA can be processed into next-generation sequencing (NGS) libraries for high-throughput sequencing, and subsequent analysis of microbial communities can be achieved using computational tools, such as quantitative insights into microbial ecology (QIIME; from the Knight laboratory), mothur (Schloss laboratory), the bioBakery of analysis tools (Huttenhower laboratory) and other software packages for analysing microbial ecology (Alm Lab) (see the figure).
To identify individual bacterial species, hybridization methods that selectively enrich for genomic targets of interest have been applied to diverse studies and have been used successfully to capture DNAs and cDNAs that are rare, ancient and difficult to obtain101–104. This technology involves array-based or solution-based capture of target homologous DNA fragments from an NGS library using synthesized oligonucleotide probes or PCR products with DNA sequences of interest as bait. Library fragments with homology to the bait sequences are selected and can then be sequenced and analysed. Although this technique improves the recovery of whole genomes and transcriptomes from rare species, it is still limited by the need for a priori knowledge of the target sequences105.
Characterization and quantification of individual microbial cells can be achieved using flow cytometry with tandem mass spectrometry to measure metabolic activity106. This single-cell strategy enables the analysis of cell-to-cell functional variation and the discovery of novel regulatory mechanisms. However, it remains difficult to sort individual cells from a heterogeneous community and sample at enough depth to retrieve biologically meaningful data. Despite these limitations, these sequencing strategies can minimize host sequence contamination, reduce the difficulty of assembling genes and individual genomes, and decrease amplification bias107, which often hinders the analysis of more diverse microbial communities.
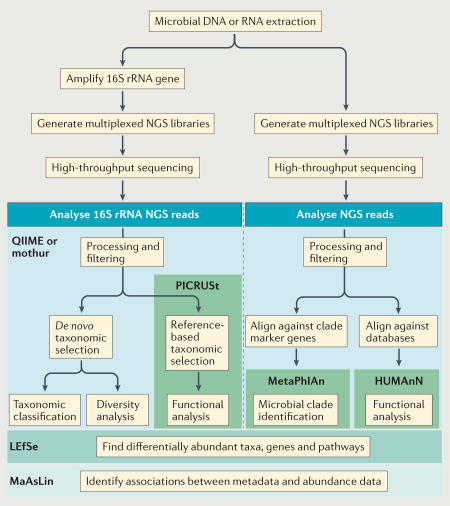
HUMAnN, The Human Microbiome Project Unified Metabolic Analysis Network; LEfSe, Linear Discriminate Analysis with Effect Size; MaAsLin, Multivariate Association with Linear Models; MetaPhlAn, Metagenomic Phylogenetic Analysis; PICRUSt, Phylogen e tic Investigation of Communities by Reconstruction of Unobserved States; rRNA, ribosomal RNA.
Attributing causality and directionality
Gnotobiotic mouse models — including germ-free mice and mice with defined microbial communities — are highly valuable systems for elucidating microbiome function in health and disease (BOX 2). Indeed, the use of gnotobiotic animals has led to key insights into the role of the gut microbiome in intra- and extra-intestinal immune-mediated diseases9–18. Furthermore, it is now possible to reconstitute a functional human immune system in these models by transplanting human liver and thymus tissue and by injecting human haematopoietic stem cells into immunodeficient mouse hosts19. Bone marrow–liver–thymus humanized mice (BLT humanized mice) develop gut-associated lymphoid tissue (GALT)-like structures that contain all the human haematopoietic lineages and thus provide an in vivo system to more accurately investigate human immune responses at the mucosal interface, as well as the development of inflammatory and autoimmune diseases20. As with all animal model systems, there are limitations to how well gnotobiotic and humanized animals recapitulate the developmental complexity of human disease, especially when considering the heterogeneity of the gut microbiome21. However, both gnotobiotic and humanized mouse models will continue to be important systems for understanding host–microbiome communication in the context of disease initiation and progression, and for evaluating therapeutic strategies that manipulate the microbiome to prevent and treat disease.
Box 2 | Gnotobiotics.
Germ-free animals have been used extensively to study the contribution of microorganisms — both individual species or defined communities — to host development and disease pathogenesis108. Gnotobiotic animals are generated by colonizing germ-free animals with a defined microbial community or consortia of bacteria from a mammalian host.
Germ-free animals have a distinct metabolic phenotype, including the tendency to consume more calories, excrete more lipids and weigh less than their conventional counterparts109. Microbial colonization of germ-free mice corrects defects in metabolic function and leads to more efficient energy extraction from the diet110.
Germ-free animals also have severe immune defects and a higher susceptibility to infection. These defects are most pronounced in the gut, where there is underdevelopment of gut-associated lymphoid tissue (GALT), including smaller and fewer Peyer’s patches and mesenteric lymph nodes, altered crypt morphology and reduced mucus thickness111,112. At a cellular level, B cells in germ-free mice produce decreased amounts of secretory IgA, which is an essential antibody at mucosal sites that maintains barrier integrity113. In addition, the balance between splenic CD4+ T helper cell subsets is skewed towards a T helper 2 (TH2) cell phenotype in germ-free mice, with fewer TH1 cells and a heightened allergic response83. Germ-free mice also have decreased numbers of peripheral CD4+ T cells — including TH17 cells114 and regulatory T cells115 — which are potent mediators of mucosal immunity owing to the production of pro-inflammatory interleukin-17 (IL-17) and anti-inflammatory IL-10, respectively. Colonization of germ-free mice rescues the TH1-TH2 cell imbalance and results in rapid increases in peripheral CD4+ T cell numbers to levels seen in conventional mice116, demonstrating that the microbiota is necessary for the maintenance of systemic and mucosal T cell populations.
Germ-free animals also show alterations in their behaviour and stress responses. Emerging evidence attributes these differences to reduced levels of molecules such as dopamine, noradrenaline and serotonin, which are important for the development of the enteric nervous system and for signalling by the hypothalamic-pituitary-adrenal (HPA) axis117. Through the use of gnotobiotic mice, HPA function is now considered to be dependent on microbiome composition and colonization at key periods in development.
Profiling biochemical activity and structure
Mass spectrometry-based and chromatography-based techniques have been available for over a century; however, they have only recently been applied to host– microbiome studies22–24. Both targeted and untargeted metabolomic and metaproteomic strategies hold promise for uncovering the chemical diversity and full biochemical capacity of synthetic, engineered and natural microbial communities. However, these techniques raise many experimental challenges that are related to the extraction and processing of samples, especially faecal material25,26. As technical hurdles are overcome, data analyses will continue to expand our knowledge of the widespread influences of the microbiome on host physiology, providing opportunities for developing and testing diagnostics and therapeutic approaches.
Targeting the microbiome
Given the abundance and diversity of microbial communities, it is important to analyse the individual species and strains within these communities and their associated functions, especially for uncultured or low-abundance microorganisms27. This can be achieved using tools that assign strain specificity and assemble individual genomes from whole metagenomic sequencing data28,29 or using hybrid capture and single-cell approaches that involve the isolation and sequencing of rare species or single microbial cells (BOX 1). The promise of these approaches has led to a push for improved microfluidics platforms and software applications that can more accurately capture and analyse microbiome diversity and provide an enhanced understanding of the genetic variation and functional contribution of individual members30.
Surveillance of host–microbiota interactions
New tools and technological advances have tremendously advanced our understanding of complex microbial communities and their interactions with a host over time, in relation to diet and lifestyle changes, and in health and disease. However, there is limited knowledge of how microorganisms interact with each other or with host cells in their preferred environmental niche in situ rather than in culture. Two recent studies have tackled this fundamental challenge. In the first study31, fluorescence in situ hybridization (FISH) was combined with single-cell imaging and quantitative analysis software to measure the spatial organization of the gut microbiota. Examination of the effects of dietary perturbations on the gut microbiota and host revealed that a fibre- deficient diet results in a reduced inner intestinal mucus layer in mice. Loss of this protective layer allowed bacteria to gain closer proximity to the epithelium and, in turn, triggered host production of the antimicrobial peptide (AMP) regenerating islet-derived protein 3β (REG3β). The absence of microbiota-accessible carbohydrates dramatically shifted the clustering pattern of distinct groups of bacteria. Although much remains to be understood about microbial spatial distribution and the factors influencing microbial community organization, this imaging and analysis approach provides a new way to investigate host–microbiota interactions. The second study32 used metabolic oligosaccharide engineering (MOE) and bio-orthogonal click chemistry (BCC) in combination with whole-body imaging to label and track bacteria in vivo. Combining these techniques enabled researchers to track the distribution of a symbiotic bacterium along the intestinal tract, its ability to compete with other species and its interaction with host cells. This approach holds promise for studying host–microbiota interactions in real time and acquiring ‘visual proof ‘ of microbial niche specificity, as well as how microbe-derived products modulate the host immune system.
Immune modulation by metabolites
Although host and microbial metabolism can occur in tandem, the host depends on its microbiome for an expanded collection of digestive and metabolic enzymes33. The gut microbiota produces an extremely diverse metabolite repertoire from the anaerobic fermentation of exogenous undigested dietary components that reach the colon, as well as endogenous compounds that are generated by microorganisms and the host. The single layer of epithelial cells that makes up the mucosal interface between the host and microorganisms allows microbial metabolic products to gain access to and interact with host cells, and thus influence immune responses and disease risk.
Short-chain fatty acids
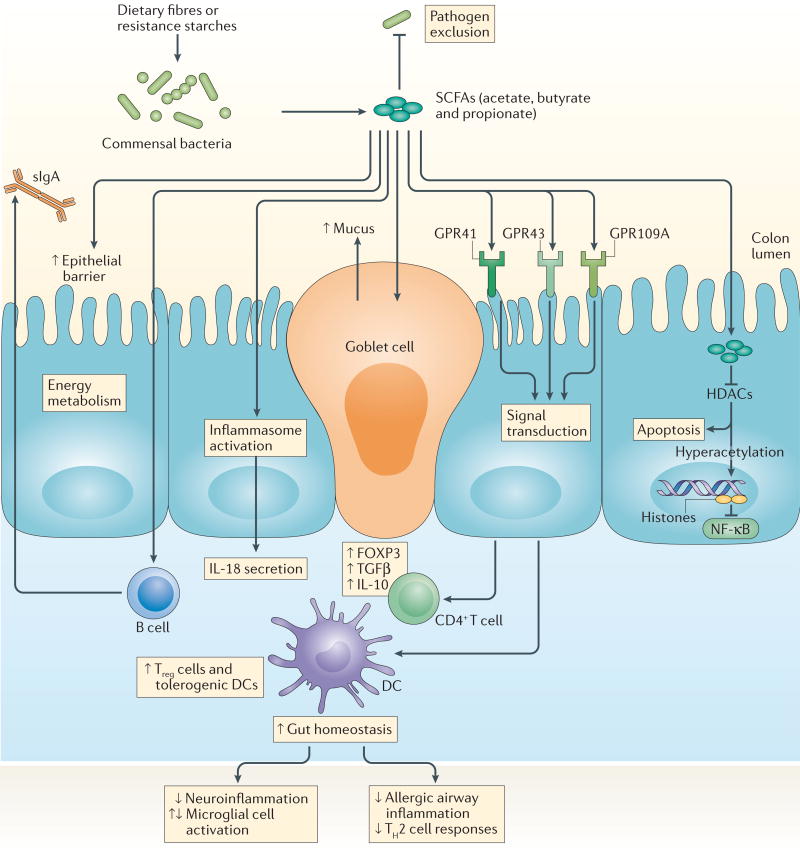
Undigested complex carbohydrates are abundant substrates for bacterial fermentation in the colon, and their main metabolic end products are short-chain fatty acids (SCFAs), including acetic acid, butyric acid and propionic acid. SCFA concentrations in the gut (which can be 20–140 mM)34 depend on microbiota composition, intestinal transit time, host– microbiota metabolic flux of SCFAs and the fibre content of the host diet. These microbiota-generated metabolites are important energy sources not only for the gut microbiota itself but also for intestinal epithelial cells (IECs). In addition to acting as local substrates for energy production, SCFAs have diverse regulatory functions, and their effects on host physiology and immunity continue to be revealed (FIG. 1).
Figure 1. SCFAs, GPCRs, host physiology and immunity.
Short-chain fatty acids (SCFAs)—such as butyric acid, propionic acid and acetic acid — are produced by colonic microbial fermentation of undigested or partially digested dietary fibres and have broad effects on host immune system development and function. SCFAs bind G protein-coupled receptors (GPCRs), such as GPR41, GPR43 and GPR109A, on the surface of epithelial cells and immune cells (not shown). Transport or diffusion of SCFAs into host cells results in their metabolism and/or inhibition of histone deacetylase (HDAC) activity. The effects of SCFAs are manifold and include enhanced epithelial barrier function and immune tolerance, which promote gut homeostasis through specific mechanisms: enhanced production of mucus by intestinal goblet cells; inhibition of nuclear factor-κB (NF-κB); activation of inflammasomes and subsequent production of interleukin-18 (IL-18); increased secretion of secretory IgA (sIgA) by B cells; reduced expression of T cell-activating molecules on antigen-presenting cells, such as dendritic cells (DCs); and increased number and function of colonic regulatory T (Treg) cells, including their expression of forkhead box P3 (FOXP3) and their production of anti-inflammatory cytokines (transforming growth factor-β (TGFβ) and IL-10). SCFAs also reach other organs, such as the brain and lungs, in which they directly or indirectly act on local or resident antigen-presenting cells to decrease inflammatory responses that are associated with neuroinflammation and allergic airway disease, respectively.
SCFAs are inhibitors of histone deacetylases (HDACs) and ligands for G protein-coupled receptors (GPCRs), and thereby act as signalling molecules that influence the expansion and function of haematopoietic and non-haematopoietic cell lineages. SCFA-driven inhibition of HDACs tends to promote a tolerogenic, anti- inflammatory cell phenotype that is crucial for maintaining immune homeostasis, and this activity supports the concept that the microbiota can function as an epigenetic regulator of host physiology. Exposure of peripheral blood mononuclear cells and neutrophils to SCFAs, similarly to their exposure to global HDAC inhibitors, inactivated nuclear factor-κB (NF-κB) and downregulated production of the pro-inflammatory cytokine tumour necrosis factor (TNF)35,36. Additional studies extended the anti-inflammatory effects of HDAC inhibition by SCFAs to macrophages37,38 and dendritic cells (DCs)39,40. Collectively, these results identify SCFA-induced HDAC inhibition as a crucial regulator of NF-κB activity and pro-inflammatory innate immune responses.
SCFAs also influence peripheral T cells, particularly regulatory T (Treg) cells, through HDAC inhibition. HDAC inhibitors can modify Treg cell frequency and function in vivo. Indeed, inhibition of HDAC9 increased forkhead box P3 (FOXP3) expression and Treg cell numbers, enhanced the suppressive function of FOXP3+ Treg cells under homeostatic conditions and amplified Treg cell-mediated attenuation of colitis in mice41. Studies have characterized the ability of specific SCFAs to regulate the size and function of the colonic FOXP3+ Treg cell pool and have shown that SCFAs induce FOXP3 expression in an HDAC-dependent manner to promote colonic homeostasis42–44. Putting mice on high-fibre or SCFA-supplemented diets not only suppressed colonic inflammation but also dampened allergic airway disease through increased suppressive activity of FOXP3+ Treg cells45. Maternal intake of SCFA-enriched diets allowed the suppressive effects to be passed on to offspring45, indicating the epigenetic potential of SCFAs in the development of the immune system and in protection from disease. The immunomodulatory effects and therapeutic benefits of HDAC inhibitors, in part through enhanced regulatory function of FOXP3+ Treg cells, have been described in several animal models of inflammatory disease46–48. These studies have also identified other mechanisms that mediate the effects of SCFAs on host immunity, including engagement of GPCRs.
Many regulatory properties of SCFAs require signalling through GPCRs, including GPR43 (also known as FFAR2), GPR41 (also known as FFAR3) and GPR109A (also known as HCAR2), which are expressed by numerous cell types, including immune cells and IECs. GPR43 expression is necessary for SCFA-induced neutrophil chemotaxis36 and for the expansion and suppressive function of Treg cells44. In the dextran sodium sulfate-induced injury model (DSS-induced injury model) and the T cell transfer colitis model, supplementing drinking water with SCFAs attenuated disease in wild-type mice but not in Gpr43−/− mice, through enhanced frequency and function of inducible FOXP3+ Treg cells44,49. Outside the gut, SCFA–GPR43 interactions reduced the risk of bacteria- induced preterm labour by decreasing chemotaxis and inflammatory gene expression in neutrophils50, and downregulated gout-associated inflammation by mediating inflamma some assembly and immune cell clearance of monosodium urate crystals51.
The GPR43-dependent effects of SCFAs on host physiology also extend to the central nervous system (CNS). The maturation and function of microglia, which are the resident macrophages of the CNS, were dependent on the gut microbiota, and the maintenance of microglia homeostasis required SCFAs and GPR43 (REF. 52). Other SCFA-sensing GPCRs are also crucial for host immune function. In wild-type mice but not in Gpr41−/− mice, SCFAs blocked DC maturation and ameliorated allergic airway inflammation40. SCFA-mediated activation of GPR109A, a receptor that responds to both niacin and butyric acid, prevented colitis and colon carcinogenesis through increased expression of anti-inflammatory effector molecules by monocytes and induced differentiation of Treg cells and interleukin-10 (IL-10)-producing T cells53. However, SCFAs can exacerbate disease. A study measuring SCFA concentrations in sputum from patients with cystic fibrosis found that SCFA-mediated recruitment and persistence of neutrophils aggravated inflammatory responses and promoted the outgrowth of Pseudomonas aeruginosa54. Thus, the immunomodulatory effects of SCFAs depend on the context and cell type under investigation. The presence of cell-specific and tissue-specific GPCRs and their varied metabolite- sensing capabilities allows the host to regulate inflammation to control infection or injury and maintain homeostasis.
SCFAs are also essential for the maintenance of mucosal immunity by fortifying IEC barrier function. In response to SCFAs, intestinal epithelial goblet cells increased their transcription of mucin genes55,56, and inoculation of germ-free mice with SCFA-producing Bacteroides thetaiotaomicron or Faecalibacterium praus-nitzii induced goblet cell differentiation and mucus production57. SCFAs also alter tight junction permeability of IECs. Colonization with a strain of Bifidobacterium longum that produces high levels of acetate conferred protection against enteropathogenic Escherichia coli O157:H7 infection, indicating that SCFAs can enhance IEC integrity and inhibit the translocation of lethal toxins from the gut lumen into the systemic circulation58. Binding of SCFAs to GPR43 and GPR109A on IECs also activated inflammasome assembly and increased production of the downstream inflammatory cytokine IL-18 (REF. 59), which prevented a colitogenic phenotype60. Collectively, these observations highlight the role of microbial-derived SCFAs in modulating local and systemic immune responses and in maintaining mucosal homeostasis.
As SCFAs are volatile compounds with short half-lives and rapid metabolism, their inhibition of HDACs is concentration dependent. Only high millimolar SCFA concentrations are sufficient to perturb HDAC function61, and their effects may require specific transporters39. However, SCFAs can also indirectly suppress HDACs through GPCR-dependent mechanisms62, with GPCRs differing in their SCFA specificity and SCFAs in turn having varying potencies. Thus, whether SCFAs directly or indirectly block HDAC activity is greatly influenced by several factors, including the concentration, transporters, receptors, and the cell and/or tissue type involved. Thus, more studies are needed to investigate the immunomodulatory functions and therapeutic potential of SCFAs in health and disease.
AHR ligands
Members of the gut microbiota and their metabolism of specific dietary components generate metabolites that can bind the aryl hydrocarbon receptor (AHR) on host cells. AHR is a ligand-inducible transcription factor that is expressed by immune cells, epithelial cells and some tumour cells. AHR was initially recognized for its role in the metabolism of xenobiotics, but evidence has also emerged for its role in regulating mucosal immune responses. Deficiency of AHR or lack of AHR ligands in mice resulted in perturbations to gut micro-biota composition (that is, increased bacterial lumen loads enriched with Bacteroides spp.) and reductions in AMP production, numbers of intestinal intraepithelial lymphocytes (IELs) and turnover of IECs63. Transfer of wild-type IELs to Ahr−/− mice restored IEC barrier function and normalized bacterial load63. In wild-type mice, an absence of AHR ligands increased the severity of DSS-induced colonic inflammation, which was attenuated when mice were administered diets supplemented with synthetic AHR ligands63. AHR activation is also necessary for postnatal expansion of intestinal lymphoid follicles and specific innate lymphoid cell (ILC) populations, particularly IL-22-producing retinoic acid receptor- related orphan receptor-γt (RORγt)+ group 3 ILCs (ILC3s), which were required for the clearance of Citrobacter rodentium infection64. Together, these studies demonstrate that intestinal immune cell subsets have an intrinsic requirement for AHR, as absence of AHR activity can leave a host susceptible to enhanced immune activation and immunopathology, and that microbial metabolism of specific dietary components is crucial for appropriate AHR signalling and host–microbial mutualism.
AHR activation is influenced both by diet and by gut microbiota composition. Only specific subsets of bacteria, particularly Lactobacilli spp., can metabolize dietary tryptophan and generate AHR ligands that can stimulate ILC3s65. ILC3-induced IL-22 production drives the expression of AMPs, which inhibit pathogen fitness by sequestering metal ions and thus reducing their availability for pathogens such as the opportunistic fungus Candida albicans65. Thus, endogenous microbe-derived tryptophan metabolites may provide cues to the host that are crucial for resistance to colonization and for protection from mucosal inflammation. Recent evidence indicates that AHR has species-dependent preferences for ligand binding66, suggesting a co-evolution between ligands that are common to the host and its microbiota. As ligands for AHRs, microbe-generated metabolites are crucial to host immunity, particularly for conferring protection from inflammation at mucosal interfaces, and warrant further research to realize their potential as treatments for infectious and inflammatory disorders.
Polyamines
Polyamines, such as putrescine, spermidine and spermine, are polycationic molecules that are found in almost all living cells and are integral to a wide range of biological functions, including gene transcription and translation, and cell growth and death. Mammalian poly-amine synthesis involves the enzyme arginase 1 (which converts arginine to ornithine), the rate-limiting enzyme ornithine decarboxylase (which synthesizes putrescine from ornithine) and the sequential enzymes that interconvert putrescine, spermidine and spermine. Given their reactive nature, intracellular polyamine concentrations are tightly regulated by the host through biosynthesis, catabo-lism, uptake and efflux mechanisms, as well as by the transcription, translation and degradation of biosynthetic enzymes. In contrast to host polyamine metabolism, bacteria use constitutive or inducible forms of amino acid decarboxylase enzymes to produce polyamines. Several bacterial pathogens depend on polyamines for virulence and survival within a host, including Helicobacter pylori, Salmonella enterica subsp. enterica serovar Typhimurium, Shigella spp., Staphylococcus aureus, Streptococcus pneumonia and Vibrio cholera67. Polyamines are also essential to host cell function, but dysregulated levels of polyamines and ornithine decarboxylase are linked to cell growth defects at low concentrations and to toxic effects and carcinogenesis at high concentrations.
The intestinal tract contains high levels of polyamines, which are derived from the diet and from de novo production by host and microbial cells. IECs depend on this reservoir to maintain their rapid turnover and high proliferation rates. Polyamines are also responsible for enhancing the integrity of the IEC barrier. In vitro studies have demonstrated that polyamines can stimulate the production of intercellular junction proteins, including occludin, zonula occludens 1 (ZO1; also known as TJP1) and E-cadherin (also known as cadherin 1)68,69, which are crucial for regulating paracellular permeability and reinforcing epithelial barrier function. Furthermore, administration of polyamines to rat pups induced the production of mucus and secretory IgA in the small intestine70,71, whereas feeding rats a polyamine-deficient diet led to intestinal mucosal hypoplasia72. These observations suggest that host–microbial polyamine synthesis is an important function of the gut microbiome that is acquired early in life and is necessary for postnatal development of the gastrointestinal tract.
Polyamine metabolism has a central role in regulating immunity and must be tightly controlled by the host. Arginase 1 and nitric oxide synthase (NOS) compete for arginine to produce either polyamines or nitric oxide, respectively, and are important enzymes that balance effector immune responses. Polarization of macrophages towards a classic (M1) pro- inflammatory phenotype leads to the activation of the inducible iso-form of NOS, the production of pro-inflammatory cytokines and increased cytotoxic activity. Spermine can inhibit M1 macrophage activation by suppressing the expression of ornithine decarboxylase and the synthesis of pro-inflammatory cytokines without altering the synthesis of anti- inflammatory transforming growth factor-β (TGFβ) and IL-10 (REF. 73). Administration of arginine in combination with Bifidobacterium animalis subsp. lactis LKM512 resulted in increased levels of circulating and colonicpoly amines, which correlated with decreased levels of colonic TNF and IL-6 (REF. 74). These findings are correlative and raise the hypothesis that manipulating the diet and providing beneficial bacteria may favourably alter colonic polyamine metabolism to benefit host health.
Polyamines also modulate systemic and mucosal adaptive immunity. Pups receiving polyamine-enriched breast milk showed an accelerated maturation of intra-epithelial CD8+ T cells and lamina propria CD4+ T cells, as well as an enhanced early appearance of B cells in the spleen75. Ageing and age-related shifts in cellular processes and impaired immune function are thought to be driven by and result from the accumulation of chronic low-grade inflammation. Studies looking at alterations in metabolism with age have found that polyamine levels tend to decrease and are altered in many neurodegener-ative diseases76. Remarkably, in healthy mice, B. lactis LKM512 induced resistance to oxidative stress and promoted longevity, which was dependent on enhanced microbial polyamine synthesis77. Taken together, shifts in host and microbial polyamine metabolism may alter the cytokine milieu and induce cellular processes in both acute and chronic inflammatory settings.
There is increasing evidence to support the role of aberrant polyamine biosynthesis in carcinogenesis and tumour immunity. Cancer cells, similarly to all highly proliferative cells, require polyamines to meet the demands for sustained rapid growth. Many patients with cancer have increased levels of polyamines in their urine and blood compared with healthy individuals78, and dysregulated poly amine metabolism by the host and gut microbiota may contribute to colorectal cancer79. A metabo lomics screen comparing paired colon cancer and normal tissue samples from patients with colo-rectal cancer revealed that host-generated polyamines in cancer cells could promote the growth of bacterial biofilms and, in turn, bacteria-generated polyamines in biofilms could promote and potentiate cancer development80. Following treatment with antibiotics, resected colo rectal cancer tissues harboured no biofilms or culturable bacteria and had decreased levels of a specific poly-amine metabolite, N1,N12-diacetylspermine, compared with biofilm- positive tissues80. Thus, host-derived and bacteria- derived polyamines may act synergistically to promote colorectal cancer and N1,N12-diacetylspermine may be a potential biomarker of biofilm-associated tumours. Polyamines are also implicated in skin cancer and hormone-related cancers, including breast and prostate cancers78. Preclinical tumour models indicate that polyamines suppress antitumour immune responses. Polyamine depletion through inhibition of ornithine decarboxylase activity could abrogate tumour growth in a T cell-dependent manner, which provides evidence that reducing intratumoural availability of polyamines could reverse immunosuppression in the tumour microenvironment81. Thus, the importance of polyamine metabolism in carcinogenesis makes the polyamine pathway a promising target for anticancer therapy and chemoprevention. The complex regulation of polyamine levels is essential to host and microbial cell function, and merits further investigation to understand how perturbations to host and microbial polyamine metabolism influence host health and disease.
Immune modulation by microbial components
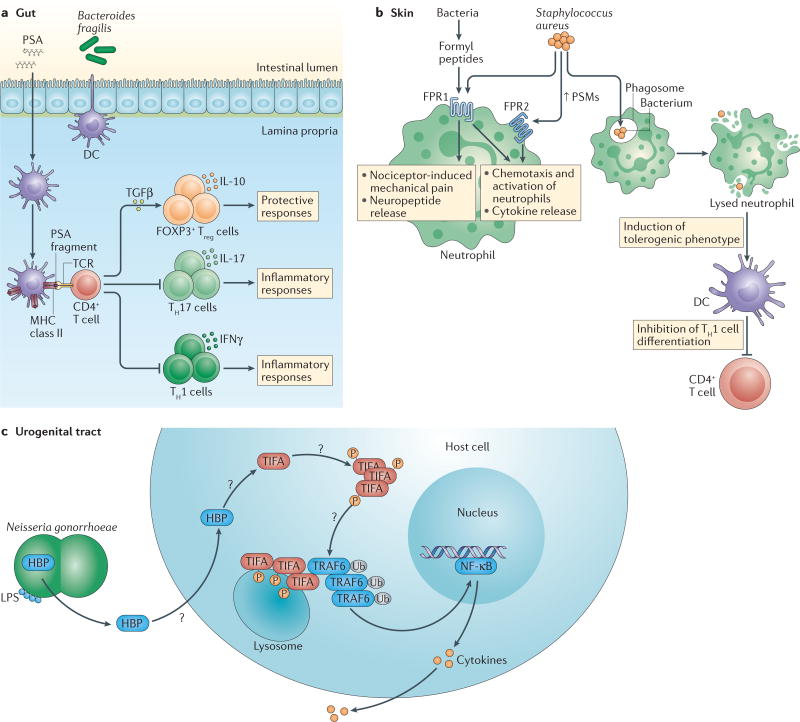
The innate immune system encounters abundant and diverse self and non-self antigens and is equipped with germline-encoded pattern recognition receptors (PRRs) to monitor, coordinate and respond to changes in the microbial landscape. PRRs detect microorganism-associated molecular patterns (MAMPs) of bacterial, fungal and viral origin, including lipopolysaccharide, flagellin, peptidoglycan, formyl peptides and unique nucleic acid structures. Transmembrane and cytoplasmic PRRs initiate conserved signalling cascades that drive stimulatory or regulatory effector responses that are crucial for host defence. The activation of PRR signalling pathways leads to the production of AMPs, cytokines, chemokines and apoptotic factors, and disruptions or alterations in signalling can contribute to disease pathogenesis. Elucidating how the numerous microbial products influence PRR-mediated responses is instrumental for understanding the development and maintenance of host–microbe homeostasis (FIG. 2). Here, we focus on three specific MAMPs — polysaccharide A (PSA), formyl peptides and D-glycero-β-D-manno-heptose-1,7-bisphosphate (HBP) — which are instructive examples for understanding host–microbe mutualism and for realizing new therapeutic opportunities from the study of host–microbiota interactions.
Figure 2. Immune modulation by the microbial components PSA, formyl peptides and HBP.
So far, several bacterial factors that engage non-canonical pattern recognition receptors (PRRs) have been identified, including polysaccharide A (PSA), formyl peptides and D-glycero-β-D-manno-heptose-1,7-biphosphate (HBP). a | PSA from Bacteroides fragilis can alter the CD4+ T helper 1 (TH1)-TH2 cell balance in the spleen (not shown) and shift the balance of effector T cell subsets in the periphery. In the gut, PSA is taken up by lamina propria dendritic cells (DCs), and processed and presented to naive CD4+ T cells. Activated transforming growth factor-β(TGFβ) induces the expansion of anti-inflammatory forkhead box P3 (FOXP3)+ regulatory T (Treg) cells and the production of interleukin-10 (IL-10), which suppress the activity of inflammatory TH1 and TH17 cells. b | Formyl peptides released by all bacteria bind formyl peptide receptors (FPRs), which are G protein-coupled receptors that are found on neutrophils and other immune cells. Formyl peptides from Staphylococcus aureus can signal through FPR1 and contribute to the activation of nociceptor-driven mechanical pain and the release of immunosuppressive neuropeptides. At high nanomolar concentrations, S. aureus-derived formyl peptides that are known as phenol-soluble modulins (PSMs) stimulate massive neutrophil influx to infection sites by binding to FPR2. Induced neutrophil activation leads to an oxidative burst. PSMs affect the adaptive immune system by inducing a tolerogenic phenotype in DCs and inhibiting the differentiation of TH1 cells. S. aureus can also use PSMs to escape from phagolysosomes, lyse host cells and disperse biofilms, and can also kill competing bacteria (not shown). c | HBP is a monosaccharide that is generated during lipopolysaccharide (LPS) biosynthesis by Gram-negative bacteria and detected in the cytosol of host cells. HBP that is secreted by the extracellular pathogen Neisseria gonorrhoeae induces innate and adaptive immune responses by phosphorylation-dependent oligomerization of TIFA (TRAF-interacting protein with FHA domain-containing protein A). Activation of TIFA and its subsequent interaction with TNF receptor-associated factor 6 (TRAF6) leads to TRAF6 ubiquitin (Ub)-dependent activation of nuclear factor-κB (NF-κB), which induces the expression of pro-inflammatory immune genes. IFNγ, interferon-γ; P, phosphate; TCR, T cell receptor.
PSA
PSA is one of eight structurally distinct capsular polysaccharides that are produced and exported by Bacteroides fragilis, a Gram-negative symbiont that is predominantly found in the outer mucus layer of the colon. This zwitterionic structure is essential for the growth and efficient colonization of B. fragilis and mediates its interactions with other microbiota members and the host. PSA has pleiotropic modulatory effects on innate and adaptive immune cells. PSA interacts with Toll-like receptor 2 (TLR2) on DCs82, and it is also sampled, processed and presented to T cells by CD11c+ DCs83,84; hence, PSA administration can correct the imbalance between T helper 1 (TH1) cells and TH2 cells that is observed in germ-free mice83. In preclinical models of abscess formation and colitis, PSA can suppress inflammation by driving IL-10 production by activated CD4+ T cells85 and by enhancing the population frequency and function of IL-10-producing CD25+FOXP3+ Treg cells86. Although PSA has been extensively studied in the spleen and gastrointestinal tract, its anti-inflammatory activities extend beyond these compartments. In neuroinflammation, PSA-driven effects on Treg cells require the induction of CD39 (also known as NTPDase 1) expression for Treg cell migration to the CNS87. CD39 is an important regulatory enzyme that limits inflammation by converting pro- inflammatory extracellular ATP into less-inflammatory ADP. CD39 surface expression is a marker that distinguishes human FOXP3+ Treg cells from naive T cells or other effector T cell populations, and upregulation of CD39 expression by human FOXP3+ Treg cells is necessary for their suppressive activity88. Recent in vitro studies with human peripheral blood mononuclear cells suggested that PSA can enhance the expansion and suppressive function of IL-10-producing CD4+CD39+FOXP3+ Treg cells89. Deficiency of CD39 in Treg cells was linked to an inability to suppress experimental colitis, and increased CD39 expression in patients with inflammatory bowel disease was associated with disease remission90. Collectively, mechanistic studies in preclinical models and in vitro experiments with human cells indicate that PSA may be a useful immunomodulatory MAMP for the treatment of human autoimmune diseases.
Formyl peptides
Conserved N-formyl peptide motifs that are recognized by formyl peptide receptors (FPRs) are found in bacteria, and their closely related motifs are found in mitochondria. Other non-formylated endogenous ligands are also sensed by FPRs, including serum amyloid A, the cathelicidin antimicrobial peptide LL-37 and the protein annexin A1. Stimulation of FPRs leads to the recruitment of leukocytes and the production of pro-inflammatory cytokines, enzymes and superoxides to combat infection. FPRs are expressed by innate immune cells, epithelial cells, endothelial cells, muscle cells and neural cells, and recent studies suggest that stimulation of FPRs on non-phagocytic cells is essential for achieving tissue homeostasis after infection or injury91. Given the diverse effects and expression profiles of FPRs, roles for aberrant FPR activation have been described in inflammation, autoimmune diseases, neurodegenerative disorders and cancer.
Pathogenic S. aureus produces formyl peptides that are known as phenol-soluble modulins (PSMs). Which FPRs are activated by PSMs depends on the length and secondary structure of PSMs, and the strength of FPR activation depends on PSM concentration92. At low levels, PSMs weakly signal through FPR1, but at high levels PSMs are potent activators of FPR2, inducing marked influx of neutrophils to infection sites and causing cytotoxic damage to host cells and competing microbial cells93. FPRs can also act with nociceptors to mediate S. aureus-induced inflammatory pain. S. aureus-derived formyl peptides signal through FPR1 to contribute to the activation of nociceptor-driven mechanical pain and the release of immunosuppressive neuropeptides94. S. aureus is also capable of secreting proteins that can inhibit FPRs and block leukocyte migration95,96. Together, these studies suggest that S. aureus suppresses the host immune system indirectly by stimulating nociceptors to release immunosuppressive neuropeptides, and directly by inhibiting or weakly signalling through FPRs, thereby allowing bacterial propagation in infected tissues. In later phases of infection, as PSMs accumulate, S. aureus can enhance the cytotoxic activity of neutrophils, causing further damage to host cells and tissues. Studies with pathogenic S. aureus highlight the importance of MAMP recognition by the host and the need for novel strategies that activate innate immune responses during early stages of disease.
Given the ability of formyl peptides to potently activate the innate immune system, peptide deformylase inhibitors such as actinonin are promising therapies for drug-resistant bacteria such as S. aureus97. Many bacteria encode peptide deformylases, and regulation of these enzymes is a mechanism by which bacteria can inactivate formyl peptides and block leukocyte chemotaxis during infection. Genetic inactivation or chemical inhibition of methionyl-tRNA formyltransferase — an enzyme involved in formylating the initiator methionyl-tRNA for protein synthesis — in S. aureus reduced its ability to mount a robust infection in vivo98. Although actinonin can drive loss-of-function mutations in methionyl-tRNA formyltransferase in bacterial pathogens and confer resistance to peptide deformylase inhibitors, these mutations resulted in substantial reductions in fitness in vitro and in vivo99, and thus offer a useful strategy for managing S. aureus infections.
HBP
Certain pathogens seem to evade detection by known PRRs, which raises the question of whether these pathogens are detected through other means that are yet to be determined. A recent study demonstrated that a bacteria-derived metabolite, HBP, can drive innate immune responses through a previously unidentified signalling axis. Using a combination of genetic and biochemical approaches, researchers showed that Neisseria gonorrhoeae, a Gram-negative bacterium that invades the urogenital tract, released the pro-inflammatory metabolite HBP into its extracellular environment100. Although HBP is an intermediate in the lipopolysaccharide biosynthesis pathway for other Gram-negative bacteria, the ability to release HBP without first requiring bacterial lysis is a phenomenon unique to N. gonorrhoeae. Thus, as an extracellular pathogen, N. gonorrhoeae can engage the innate immune system without first being engulfed and lysed. N. gonorrhoeae-derived HBP can enter a host cell by endocytosis and initiate conserved signalling cascades that converge to induce pro- inflammatory responses. Specifically, HBP activated the signalling molecule TIFA (TRAF-interacting protein with FHA domain- containing protein A), which stimulates its phosphorylation, oligomerization and relocation to lyso-somes, where it can interact with the adaptor molecule TRAF6 (TNF receptor-associated factor 6). Interaction of TIFA with TRAF6 triggered the canonical NF-κB pathway to induce the expression of genes involved in the innate immune response. These effects were observed with N. gonorrhoeae-derived HBP in the context of infection and with HBP injection alone. The mechanisms that regulate bacterial HBP production, as well as host transport and intracellular recognition of HBP, require further investigation. However, these findings establish that microbial metabolites can function as MAMPs that drive the expression of proinflammatory genes and may prime adaptive immune responses using a novel PRR pathway. Elucidation of the full signalling pathway of N. gonorrhoeae-derived HBP will not only provide important insights for controlling infection but will also serve as a model for identifying how microbial products signal to the host and influence host immunity.
Conclusions
The microbiota and the immune system are involved in a complex crosstalk that is influenced by innumerable environmental cues, and they interact both locally and across great distances within the body. Microbiota-generated metabolites and their cellular and molecular components are increasingly being recognized as an essential part of human physiology, with profound effects on immune function and dysfunction. Microbial metabolites are generated through microorganism– microorganism and host–microorganism interactions, and there is a growing appreciation of a role for this co-metabolism in human health and disease. These observations support the concept that mammals are holobionts that are dependent on both host and microbial genomes (that is, the hologenome) for optimal functioning. Meta-omics and evolving computational frameworks will hopefully lead to the systematic prediction and discovery of more microbial metabolites and components that are relevant to immune system function. However, it is also important to further probe how well-known microbial metabolites (such as SCFAs) and co-metabolites (such as polyamines and AHR ligands) influence immune cell subsets and their functions.
Acknowledgments
The authors thank the members of the Garrett laboratory for their helpful discussions. The work relevant to this Review is supported by the grants R01 CA154426 and R01 GM099531, a Burroughs Wellcome Career in Medical Sciences Award and a Searle Scholars Award to W.S.G.
Glossary
- Dysbiosis
An imbalance in the composition or function of the microbial species that are normally found in mammalian hosts. It is associated with alterations in immune function and susceptibility to inflammatory diseases, allergies and metabolic conditions
- Gnotobiotic mouse models
Experimental models in which germ-free mice are selectively colonized by defined microorganisms and kept in isolators to control their microbial colonization status
- Bone marrow–liver–thymus humanized mice
(BLT humanized mice). Immunodeficient mice that are engrafted with human fetal liver and thymus under the renal capsule. Three weeks later, mice are irradiated and then injected with a suspension of CD34+ cells from the same human fetal liver sample. These fetal liver cells seed to the mouse bone marrow
- Humanized mouse models
Experimental models in which mice carry functioning human genes, cells, tissues (including faecal material) or organs that are introduced by transgenesis, injection or transplantation. For example, an immunodeficient mouse transgenically expressing susceptibility genes for type 1 diabetes and reconstituted with T cells from a patient with type 1 diabetes and human islets of Langerhans can be used to study relevant autoimmune processes
- Fluorescence in situ hybridization
(FISH). A technique in which fluorescent probes are used to visually label specific DNA sequences in the nuclei of cells
- Metabolic oligosaccharide engineering
(MOE). A technology in which synthetic sugar analogues are exogenously supplied to living cells and biosynthetically incorporated into cell surface polysaccharides. An advantage of this technology is that it can be used for prokaryotic and eukaryotic cells that are grown under aerobic or anaerobic conditions
- Bio-orthogonal click chemistry
(BCC). A chemical reaction in a living cell that allows a labelled synthetic probe to be covalently linked to targeted cellular substrates without disrupting any native functions of the cell. This method can be used to tag and visualize biomolecules within cells of interest
- Anaerobic fermentation
The process of extracting energy from carbohydrates. Some bacteria are facultative anaerobes, meaning they can switch between aerobic respiration and anaerobic pathways, depending on the availability of oxygen or other electron acceptors. Other bacteria are obligate anaerobes, meaning they completely rely on anaerobic fermentation and can only survive in the absence of oxygen
- Histone deacetylases
(HDACs). Enzymes that remove the acetyl groups from lysine residues that are located at the amino termini of histones. In general, decreased levels of histone acetylation are associated with the repression of gene expression. The balance of histone acetylation is maintained by the interplay between HDACs and histone acetyltransferases
- Dextran sodium sulfate-induced injury model
(DSS-induced injury model). A commonly used experimental model of colonic injury and mucosal inflammation induced in mice by ingestion of the sulfated polysaccharide DSS. This model causes acute colonic epithelial damage and inflammation
- T cell transfer colitis model
A well-characterized model of chronic colitis that is induced by the transfer of CD4+CD45RBhi (naive) T cells from healthy wild-type mice into immunodeficient syngeneic recipients
- Xenobiotics
Compounds inclusive of drugs, food components and pollutants
- Innate lymphoid cell
(ILC). A lymphoid cell that is derived from the common lymphoid progenitor and does not express a recombined antigen receptor. ILCs have important roles in innate immune responses to infectious microorganisms, in epithelial homeostasis and in lymphoid tissue formation
Footnotes
Competing interests statement
The authors declare no competing interests.
FURTHER INFORMATION
Alm Lab: http://almlab.mit.edu/software.html
bioBakery: https://bitbucket.org/biobakery/biobakery/wiki/Home
HUMAnN: http://huttenhower.sph.harvard.edu/humann
LEfSe: http://huttenhower.sph.harvard.edu/galaxy/
MaAsLin: http://huttenhower.sph.harvard.edu/maaslin
MetaPhlAn: http://huttenhower.sph.harvard.edu/metaphlan
Mothur: http://www.mothur.org
PICRUSt: http://picrust.github.io/picrust/
QIIME: http://qiime.org/
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
References
- 1.Ding T, Schloss PD. Dynamics and associations of microbial community types across the human body. Nature. 2014;509:357–360. doi: 10.1038/nature13178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eckburg PB, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donaldson GP, Lee SM, Mazmanian SK. Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 2015;14:20–32. doi: 10.1038/nrmicro3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sender R, Fuchs S, Milo R. Are we really vastly outnumbered? Revisiting the ratio of bacterial to host cells in humans. Cell. 2016;164:337–340. doi: 10.1016/j.cell.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 6.Franzosa EA, et al. Sequencing and beyond: integrating molecular ‘omics’ for microbial community profiling. Nat. Rev. Microbiol. 2015;13:360–372. doi: 10.1038/nrmicro3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blekhman R, et al. Host genetic variation impacts microbiome composition across human body sites. Genome Biol. 2015;16:191. doi: 10.1186/s13059-015-0759-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morgan XC, Huttenhower C. Meta’omic analytic techniques for studying the intestinal microbiome. Gastroenterology. 2014;146:1437–1448. doi: 10.1053/j.gastro.2014.01.049. [DOI] [PubMed] [Google Scholar]
- 9.Dianda L, et al. T cell receptor-αβ-deficient mice fail to develop colitis in the absence of a microbial environment. Am. J. Pathol. 1997;150:91–97. [PMC free article] [PubMed] [Google Scholar]
- 10.Sellon RK, et al. Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infect. Immun. 1998;66:5224–5231. doi: 10.1128/iai.66.11.5224-5231.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reháková Z, et al. Germ-free mice do not develop ankylosing enthesopathy, a spontaneous joint disease. Hum. Immunol. 2000;61:555–558. doi: 10.1016/s0198-8859(00)00122-1. [DOI] [PubMed] [Google Scholar]
- 12.Sudo N, et al. Postnatal microbial colonization programs the hypothalamic-pituitary-adrenal system for stress response in mice. J. Physiol. 2004;558:263–275. doi: 10.1113/jphysiol.2004.063388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bohn E, et al. Host gene expression in the colon of gnotobiotic interleukin-2-deficient mice colonized with commensal colitogenic or noncolitogenic bacterial strains: common patterns and bacteria strain specific signatures. Inflamm. Bowel Dis. 2006;12:853–862. doi: 10.1097/01.mib.0000231574.73559.75. [DOI] [PubMed] [Google Scholar]
- 14.Wen L, et al. Innate immunity and intestinal microbiota in the development of type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu H-J, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garrett WS, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berer K, et al. Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature. 2011;479:538–541. doi: 10.1038/nature10554. [DOI] [PubMed] [Google Scholar]
- 18.Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc. Natl Acad. Sci. USA. 2011;108(Suppl. 1):4615–4622. doi: 10.1073/pnas.1000082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Melkus MW, et al. Humanized mice mount specific adaptive and innate immune responses to EBV and TSST-1. Nat. Med. 2006;12:1316–1322. doi: 10.1038/nm1431. [DOI] [PubMed] [Google Scholar]
- 20.Nochi T, Denton PW, Wahl A, Garcia JV. Cryptopatches are essential for the development of human GALT. Cell Rep. 2013;3:1874–1884. doi: 10.1016/j.celrep.2013.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laukens D, Brinkman BM, Raes J, De Vos M, Vandenabeele P. Heterogeneity of the gut microbiome in mice: guidelines for optimizing experimental design. FEMS Microbiol. Rev. 2016;40:117–132. doi: 10.1093/femsre/fuv036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lichtman JS, Marcobal A, Sonnenburg JL, Elias JE. Host-centric proteomics of stool: a novel strategy focused on intestinal responses to the gut microbiota. Mol. Cell Proteom. 2013;12:3310–3318. doi: 10.1074/mcp.M113.029967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marcobal A, et al. A metabolomic view of how the human gut microbiota impacts the host metabolome using humanized and gnotobiotic mice. ISME J. 2013;7:1933–1943. doi: 10.1038/ismej.2013.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sridharan GV, et al. Prediction and quantification of bioactive microbiota metabolites in the mouse gut. Nat. Commun. 2014;5:5492. doi: 10.1038/ncomms6492. [DOI] [PubMed] [Google Scholar]
- 25.Kolmeder CA, de Vos WM. Metaproteomics of our microbiome — developing insight in function and activity in man and model systems. J. Proteomics. 2014;97:3–16. doi: 10.1016/j.jprot.2013.05.018. [DOI] [PubMed] [Google Scholar]
- 26.Xiong W, Abraham PE, Li Z, Pan C, Hettich RL. Microbial metaproteomics for characterizing the range of metabolic functions and activities of human gut microbiota. Proteomics. 2015;15:3424–3438. doi: 10.1002/pmic.201400571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lasken RS, McLean JS. Recent advances in genomic DNA sequencing of microbial species from single cells. Nat. Rev. Genet. 2014;15:577–584. doi: 10.1038/nrg3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cleary B, et al. Detection of low-abundance bacterial strains in metagenomic datasets by eigengenome partitioning. Nat. Biotechnol. 2015;33:1053–1060. doi: 10.1038/nbt.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo C, et al. ConStrains identifies microbial strains in metagenomic datasets. Nat. Biotechnol. 2015;33:1045–1052. doi: 10.1038/nbt.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ufarté L, Potocki-Veronese G, Laville É. Discovery of new protein families and functions: new challenges in functional metagenomics for biotechnologies and microbial ecology. Front. Microbiol. 2015;6:563. doi: 10.3389/fmicb.2015.00563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Earle KA, et al. Quantitative imaging of gut microbiota spatial organization. Cell Host Microbe. 2015;18:478–488. doi: 10.1016/j.chom.2015.09.002. This work showcases a robust pipeline for measuring the localization and organization of specific microorganisms and defined microbial communities within the gut. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geva-Zatorsky N, et al. In vivo imaging and tracking of host-microbiota interactions via metabolic labeling of gut anaerobic bacteria. Nat. Med. 2015;21:1091–1100. doi: 10.1038/nm.3929. This paper describes an innovative strategy for labelling anaerobic microorganisms in vitro and tracking their location and function in vivo. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hooper LV, Midtvedt T, Gordon JI. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu. Rev. Nutr. 2002;22:283–307. doi: 10.1146/annurev.nutr.22.011602.092259. [DOI] [PubMed] [Google Scholar]
- 34.Cummings JH, Pomare EW, Branch WJ, Naylor CP, Macfarlane GT. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut. 1987;28:1221–1227. doi: 10.1136/gut.28.10.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Usami M, et al. Butyrate and trichostatin A attenuate nuclear factor κB activation and tumor necrosis factor α secretion and increase prostaglandin E2 secretion in human peripheral blood mononuclear cells. Nutr. Res. 2008;28:321–328. doi: 10.1016/j.nutres.2008.02.012. [DOI] [PubMed] [Google Scholar]
- 36.Vinolo MAR, et al. Suppressive effect of short-chain fatty acids on production of proinflammatory mediators by neutrophils. J. Nutr. Biochem. 2011;22:849–855. doi: 10.1016/j.jnutbio.2010.07.009. [DOI] [PubMed] [Google Scholar]
- 37.Kendrick SFW, et al. Acetate, the key modulator of inflammatory responses in acute alcoholic hepatitis. Hepatology. 2010;51:1988–1997. doi: 10.1002/hep.23572. [DOI] [PubMed] [Google Scholar]
- 38.Chang PV, Hao L, Offermanns S, Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc. Natl Acad. Sci. USA. 2014;111:2247–2252. doi: 10.1073/pnas.1322269111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singh N, et al. Blockade of dendritic cell development by bacterial fermentation products butyrate and propionate through a transporter (Slc5a8)-dependent inhibition of histone deacetylases. J. Biol. Chem. 2010;285:27601–27608. doi: 10.1074/jbc.M110.102947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trompette A, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med. 2014;20:159–166. doi: 10.1038/nm.3444. This study shows that gut microbiota-derived SCFAs from dietary fibre-enriched diets can dampen the severity of allergic inflammation in the lungs by signalling through GPR41 and generating tolerogenic DCs. [DOI] [PubMed] [Google Scholar]
- 41.Tao R, et al. Deacetylase inhibition promotes the generation and function of regulatory T cells. Nat. Med. 2007;13:1299–1307. doi: 10.1038/nm1652. [DOI] [PubMed] [Google Scholar]
- 42.Furusawa Y, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. This paper establishes that enhanced HDAC activity at the Foxp3 locus is one mechanism for how microbial-derived butyrate induces the differentiation and suppressive function of peripheral Treg cells and how colonic Treg cells can protect against the development of colitis. [DOI] [PubMed] [Google Scholar]
- 43.Arpaia N, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. This work demonstrates that butyrate specifically promotes the differentiation of peripheral, not thymus-derived, Treg cells and that the mode of delivery, local versus systemic administration, influences SCFA-mediated effects on colonic Treg cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith PM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. This paper illustrates that microbiota-derived SCFAs regulate Treg cell homeostasis in the colon and that SCFA-mediated effects on Treg cells are mediated in part by GPR43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thorburn AN, et al. Evidence that asthma is a developmental origin disease influenced by maternal diet and bacterial metabolites. Nat. Commun. 2015;6:7320. doi: 10.1038/ncomms8320. [DOI] [PubMed] [Google Scholar]
- 46.Maslowski KM, Mackay CR. Diet, gut microbiota and immune responses. Nat. Immunol. 2011;12:5–9. doi: 10.1038/ni0111-5. [DOI] [PubMed] [Google Scholar]
- 47.Wang L, de Zoeten EF, Greene MI, Hancock WW. Immunomodulatory effects of deacetylase inhibitors: therapeutic targeting of FOXP3+ regulatory T cells. Nat. Rev. Drug Discov. 2009;8:969–981. doi: 10.1038/nrd3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thorburn AN, Macia L, Mackay CR. Diet, metabolites, and ‘western-lifestyle’ inflammatory diseases. Immunity. 2014;40:833–842. doi: 10.1016/j.immuni.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 49.Maslowski KM, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–1286. doi: 10.1038/nature08530. This landmark paper provides a molecular link between diet, the gut microbiota and host immune function by establishing that GPR43 expression on immune cells is crucial for imparting the regulatory properties of SCFAs and for resolving inflammation in experimental models of colitis, arthritis and asthma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Voltolini C, et al. A novel antiinflammatory role for the short-chain fatty acids in human labor. Endocrinology. 2012;153:395–403. doi: 10.1210/en.2011-1457. [DOI] [PubMed] [Google Scholar]
- 51.Vieira AT, et al. A role for gut microbiota and the metabolite-sensing receptor GPR43 in a murine model of gout. Arthritis Rheumatol. 2015;67:1646–1656. doi: 10.1002/art.39107. [DOI] [PubMed] [Google Scholar]
- 52.Erny D, et al. Host microbiota constantly control maturation and function of microglia in the CNS. Nat. Neurosci. 2015;18:965–977. doi: 10.1038/nn.4030. This paper demonstrates that microbiota-generated SCFAs from the gut regulate microglia homeostasis, which has important implications for local and systemic consequences of microbial dysbiosis on CNS function. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Singh N, et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity. 2014;40:128–139. doi: 10.1016/j.immuni.2013.12.007. This study illustrates that the SCFA receptor GPR109A on immune and epithelial cells triggers the production of protective cytokines and promotes anti-inflammatory immune responses, which are crucial for preventing colitis and colitis-associated colorectal cancer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ghorbani P, et al. Short-chain fatty acids affect cystic fibrosis airway inflammation and bacterial growth. Eur. Respir. J. 2015;46:1033–1045. doi: 10.1183/09031936.00143614. [DOI] [PubMed] [Google Scholar]
- 55.Willemsen LEM, Koetsier MA, van Deventer SJH, van Tol EAF. Short chain fatty acids stimulate epithelial mucin 2 expression through differential effects on prostaglandin E1 and E2 production by intestinal myofibroblasts. Gut. 2003;52:1442–1447. doi: 10.1136/gut.52.10.1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gaudier E, et al. Butyrate specifically modulates MUC gene expression in intestinal epithelial goblet cells deprived of glucose. Am. J. Physiol. Gastrointest. Liver Physiol. 2004;287:G1168–G1174. doi: 10.1152/ajpgi.00219.2004. [DOI] [PubMed] [Google Scholar]
- 57.Wrzosek L, et al. Bacteroides thetaiotaomicron and Faecalibacterium prausnitzii influence the production of mucus glycans and the development of goblet cells in the colonic epithelium of a gnotobiotic model rodent. BMC Biol. 2013;11:61. doi: 10.1186/1741-7007-11-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fukuda S, et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543–547. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- 59.Macia L, et al. Metabolite-sensing receptors GPR43 and GPR109A facilitate dietary fibre-induced gut homeostasis through regulation of the inflammasome. Nat. Commun. 2015;6:6734. doi: 10.1038/ncomms7734. [DOI] [PubMed] [Google Scholar]
- 60.Elinav E, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schilderink R, Verseijden C, de Jonge WJ. Dietary inhibitors of histone deacetylases in intestinal immunity and homeostasis. Front. Immunol. 2013;4:226. doi: 10.3389/fimmu.2013.00226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu J, Zhou Z, Hu Y, Dong S. Butyrate-induced GPR41 activation inhibits histone acetylation and cell growth. J. Genet. Genom. 2012;39:375–384. doi: 10.1016/j.jgg.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 63.Li Y, et al. Exogenous stimuli maintain intraepithelial lymphocytes via aryl hydrocarbon receptor activation. Cell. 2011;147:629–640. doi: 10.1016/j.cell.2011.09.025. [DOI] [PubMed] [Google Scholar]
- 64.Kiss EA, et al. Natural aryl hydrocarbon receptor ligands control organogenesis of intestinal lymphoid follicles. Science. 2011;334:1561–1565. doi: 10.1126/science.1214914. [DOI] [PubMed] [Google Scholar]
- 65.Zelante T, et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39:372–385. doi: 10.1016/j.immuni.2013.08.003. This paper offers a mechanistic insight into the functional role of microbial-derived tryptophan metabolites in gut immune homeostasis through activation of the AHR in ILC3s, which provides colonization resistance to a fungal pathogen and protection from mucosal inflammation. [DOI] [PubMed] [Google Scholar]
- 66.Hubbard TD, et al. Adaptation of the human aryl hydrocarbon receptor to sense microbiota-derived indoles. Sci. Rep. 2015;5:12689. doi: 10.1038/srep12689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Di Martino ML, et al. Polyamines: emerging players in bacteria-host interactions. Int. J. Med. Microbiol. 2013;303:484–491. doi: 10.1016/j.ijmm.2013.06.008. [DOI] [PubMed] [Google Scholar]
- 68.Chen J, et al. Polyamines are required for expression of Toll-like receptor 2 modulating intestinal epithelial barrier integrity. Am. J. Physiol. Gastrointest. Liver Physiol. 2007;293:G568–G576. doi: 10.1152/ajpgi.00201.2007. [DOI] [PubMed] [Google Scholar]
- 69.Liu L, et al. Polyamines regulate E-cadherin transcription through c-Myc modulating intestinal epithelial barrier function. Am. J. Physiol. Cell Physiol. 2009;296:C801–C810. doi: 10.1152/ajpcell.00620.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dufour C, et al. Spermine and spermidine induce intestinal maturation in the rat. Gastroenterology. 1988;95:112–116. doi: 10.1016/0016-5085(88)90298-3. [DOI] [PubMed] [Google Scholar]
- 71.Buts JP, De Keyser N, Kolanowski J, Sokal E, Van Hoof F. Maturation of villus crypt cell functions in rat small intestine. Role of dietary polyamines. Dig. Dis. Sci. 1993;38:1091–1098. doi: 10.1007/BF01295726. [DOI] [PubMed] [Google Scholar]
- 72.Löser C, Eisel A, Harms D, Fölsch UR. Dietary polyamines are essential luminal growth factors for small intestinal and colonic mucosal growth and development. Gut. 1999;44:12–16. doi: 10.1136/gut.44.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang M, Wang H, Tracey KJ. Regulation of macrophage activation and inflammation by spermine: a new chapter in an old story. Crit. Care Med. 2000;28:N60–N66. doi: 10.1097/00003246-200004001-00007. [DOI] [PubMed] [Google Scholar]
- 74.Kibe R, et al. Upregulation of colonic luminal polyamines produced by intestinal microbiota delays senescence in mice. Sci. Rep. 2014;4:4548. doi: 10.1038/srep04548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Perez-Cano FJ, González-Castro A, Castellote C, Franch A, Castell M. Influence of breast milk polyamines on suckling rat immune system maturation. Dev. Comp. Immunol. 2010;34:210–218. doi: 10.1016/j.dci.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 76.Minois N, Carmona-Gutierrez D, Madeo F. Polyamines in aging and disease. Aging (Albany NY) 2011;3:716–732. doi: 10.18632/aging.100361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Matsumoto M, Kurihara S, Kibe R, Ashida H, Benno Y. Longevity in mice is promoted by probiotic-induced suppression of colonic senescence dependent on upregulation of gut bacterial polyamine production. PLoS ONE. 2011;6:e23652. doi: 10.1371/journal.pone.0023652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Miller-Fleming L, Olin-Sandoval V, Campbell K, Ralser M. Remaining mysteries of molecular biology: the role of polyamines in the cell. J. Mol. Biol. 2015;427:3389–3406. doi: 10.1016/j.jmb.2015.06.020. [DOI] [PubMed] [Google Scholar]
- 79.Gerner EW, Meyskens FL. Polyamines and cancer: old molecules, new understanding. Nat. Rev. Cancer. 2004;4:781–792. doi: 10.1038/nrc1454. [DOI] [PubMed] [Google Scholar]
- 80.Johnson CH, et al. Metabolism links bacterial biofilms and colon carcinogenesis. Cell. Metab. 2015;21:891–897. doi: 10.1016/j.cmet.2015.04.011. This work demonstrates a role for colonic mucosal biofilms in potentiating colonic carcinogenesis through the generation of polyamine metabolites that can enhance cell proliferation and cancer cell growth. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hayes CS, et al. Polyamine-blocking therapy reverses immunosuppression in the tumor microenvironment. Cancer Immunol. Res. 2014;2:274–285. doi: 10.1158/2326-6066.CIR-13-0120-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang Q, et al. A bacterial carbohydrate links innate and adaptive responses through Toll-like receptor 2. J. Exp. Med. 2006;203:2853–2863. doi: 10.1084/jem.20062008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 84.Dasgupta S, Erturk-Hasdemir D, Ochoa-Repáraz J, Reinecker H-C, Kasper DL. Plasmacytoid dendritic cells mediate anti-inflammatory responses to a gut commensal molecule via both innate and adaptive mechanisms. Cell Host Microbe. 2014;15:413–423. doi: 10.1016/j.chom.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 86.Round JL, et al. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332:974–977. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang Y, et al. A commensal bacterial product elicits and modulates migratory capacity of CD39+ CD4 T regulatory subsets in the suppression of neuroinflammation. Gut Microbes. 2014;5:552–561. doi: 10.4161/gmic.29797. [DOI] [PubMed] [Google Scholar]
- 88.Dwyer KM, et al. CD39 and control of cellular immune responses. Purinerg. Signal. 2007;3:171–180. doi: 10.1007/s11302-006-9050-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Telesford KM, et al. A commensal symbiotic factor derived from Bacteroides fragilis promotes human CD39+Foxp3+ T cells and Treg function. Gut Microbes. 2015;6:234–242. doi: 10.1080/19490976.2015.1056973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gibson DJ, et al. Heightened expression of CD39 by regulatory T lymphocytes is associated with therapeutic remission in inflammatory bowel disease. Inflamm. Bowel Dis. 2015;21:2806–2814. doi: 10.1097/MIB.0000000000000566. [DOI] [PubMed] [Google Scholar]
- 91.Liu M, et al. Formylpeptide receptors mediate rapid neutrophil mobilization to accelerate wound healing. PLoS ONE. 2014;9:e90613. doi: 10.1371/journal.pone.0090613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kretschmer D, Rautenberg M, Linke D, Peschel A. Peptide length and folding state govern the capacity of staphylococcal β-type phenol-soluble modulins to activate human formyl-peptide receptors 1 or 2. J. Leukoc. Biol. 2015;97:689–697. doi: 10.1189/jlb.2A0514-275R. [DOI] [PubMed] [Google Scholar]
- 93.Bloes DA, Kretschmer D, Peschel A. Enemy attraction: bacterial agonists for leukocyte chemotaxis receptors. Nat. Rev. Microbiol. 2015;13:95–104. doi: 10.1038/nrmicro3390. [DOI] [PubMed] [Google Scholar]
- 94.Chiu IM, et al. Bacteria activate sensory neurons that modulate pain and inflammation. Nature. 2013;501:52–57. doi: 10.1038/nature12479. This paper illuminates an unanticipated role for the CNS in mediating host-pathogen interactions, by showing that a bacterial pathogen can stimulate pain and influence inflammatory immune responses by directly activating peripheral sensory neurons. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Haas P-J, et al. N-terminal residues of the chemotaxis inhibitory protein of Staphylococcus aureus are essential for blocking formylated peptide receptor but not C5a receptor. J. Immunol. 2004;173:5704–5711. doi: 10.4049/jimmunol.173.9.5704. [DOI] [PubMed] [Google Scholar]
- 96.Prat C, et al. A homolog of formyl peptide receptorlike 1 (FPRL1) inhibitor from Staphylococcus aureus (FPRL1 inhibitory protein) that inhibits FPRL1 and FPR. J. Immunol. 2009;183:6569–6578. doi: 10.4049/jimmunol.0801523. [DOI] [PubMed] [Google Scholar]
- 97.Yang S, et al. Synthesis, antibacterial activity, and biological evaluation of formyl hydroxyamino derivatives as novel potent peptide deformylase inhibitors against drug-resistant bacteria. Eur. J. Med. Chem. 2014;86:133–152. doi: 10.1016/j.ejmech.2014.07.106. [DOI] [PubMed] [Google Scholar]
- 98.Lewandowski T, et al. Staphylococcus aureus formyl-methionyl transferase mutants demonstrate reduced virulence factor production and pathogenicity. Antimicrob. Agents Chemother. 2013;57:2929–2936. doi: 10.1128/AAC.00162-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Min S, et al. Frequency of spontaneous resistance to peptide deformylase inhibitor GSK1322322 in Haemophilus influenzae, Staphylococcus aureus, Streptococcus pyogenes, and Streptococcus pneumoniae. Antimicrob. Agents Chemother. 2015;59:4644–4652. doi: 10.1128/AAC.00484-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gaudet RG, et al. Cytosolic detection of the bacterial metabolite HBP activates TIFA-dependent innate immunity. Science. 2015;348:1251–1255. doi: 10.1126/science.aaa4921. This work identifies a novel innate immune signalling axis that is initiated by a pro-inflammatory bacterial-derived metabolite, an intermediate in lipopolysaccharide biosynthesis of Gram-negative bacteria. [DOI] [PubMed] [Google Scholar]
- 101.Maricic T, Whitten M, Pääbo S. Multiplexed DNA sequence capture of mitochondrial genomes using PCR products. PLoS ONE. 2010;5:e14004. doi: 10.1371/journal.pone.0014004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Burbano HA, et al. Targeted investigation of the Neandertal genome by array-based sequence capture. Science. 2010;328:723–725. doi: 10.1126/science.1188046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Duncavage EJ, et al. Hybrid capture and next-generation sequencing identify viral integration sites from formalin-fixed, paraffin-embedded tissue. J. Mol. Diagn. 2011;13:325–333. doi: 10.1016/j.jmoldx.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cabanski CR, et al. cDNA hybrid capture improves transcriptome analysis on low-input and archived samples. J. Mol. Diagn. 2014;16:440–451. doi: 10.1016/j.jmoldx.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lim SW, Tran TM, Abate AR. PCR-activated cell sorting for cultivation-free enrichment and sequencing of rare microbes. PLoS ONE. 2015;10:e0113549. doi: 10.1371/journal.pone.0113549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Maurice CF, Turnbaugh PJ. Quantifying the metabolic activities of human-associated microbial communities across multiple ecological scales. FEMS Microbiol. Rev. 2013;37:830–848. doi: 10.1111/1574-6976.12022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Segata N, et al. Computational meta’omics for microbial community studies. Mol. Syst. Biol. 2013;9:666. doi: 10.1038/msb.2013.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Faith JJ, et al. Creating and characterizing communities of human gut microbes in gnotobiotic mice. ISME J. 2010;4:1094–1098. doi: 10.1038/ismej.2010.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wostmann BS, Larkin C, Moriarty A, Bruckner-Kardoss E. Dietary intake, energy metabolism, and excretory losses of adult male germfree Wistar rats. Lab. Anim. Sci. 1983;33:46–50. [PubMed] [Google Scholar]
- 110.Rosenbaum M, Knight R, Leibel RL. The gut microbiota in human energy homeostasis and obesity. Trends Endocrinol. Metab. 2015;26:493–501. doi: 10.1016/j.tem.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Deplancke B, Gaskins HR. Microbial modulation of innate defense: goblet cells and the intestinal mucus layer. Am. J. Clin. Nutr. 2001;73:1131S–1141S. doi: 10.1093/ajcn/73.6.1131S. [DOI] [PubMed] [Google Scholar]
- 112.Smith K, McCoy KD, Macpherson AJ. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin. Immunol. 2007;19:59–69. doi: 10.1016/j.smim.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 113.Pabst O. New concepts in the generation and functions of IgA. Nat. Rev. Immunol. 2012;12:821–832. doi: 10.1038/nri3322. [DOI] [PubMed] [Google Scholar]
- 114.Ivanov II, et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4:337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Strauch UG, et al. Influence of intestinal bacteria on induction of regulatory T cells: lessons from a transfer model of colitis. Gut. 2005;54:1546–1552. doi: 10.1136/gut.2004.059451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Belkaid Y, Naik S. Compartmentalized and systemic control of tissue immunity by commensals. Nat. Immunol. 2013;14:646–653. doi: 10.1038/ni.2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Borre YE, et al. Microbiota and neurodevelopmental windows: implications for brain disorders. Trends Mol. Med. 2014;20:509–518. doi: 10.1016/j.molmed.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 118.Belcheva A, et al. Gut microbial metabolism drives transformation of MSH2-deficient colon epithelial cells. Cell. 2014;158:288–299. doi: 10.1016/j.cell.2014.04.051. [DOI] [PubMed] [Google Scholar]
- 119.Donohoe DR, et al. A gnotobiotic mouse model demonstrates that dietary fiber protects against colorectal tumorigenesis in a microbiota- and butyrate-dependent manner. Cancer Discov. 2014;4:1387–1397. doi: 10.1158/2159-8290.CD-14-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Srinivasan S, et al. Metabolic signatures of bacterial vaginosis. MBio. 2015;6:e00204–00215. doi: 10.1128/mBio.00204-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Koeth RA, et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 2013;19:576–585. doi: 10.1038/nm.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Pluznick JL, et al. Olfactory receptor responding to gut microbiota-derived signals plays a role in renin secretion and blood pressure regulation. Proc. Natl Acad. Sci. USA. 2013;110:4410–4415. doi: 10.1073/pnas.1215927110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Meijers BKI, et al. p-Cresol and cardiovascular risk in mild-to-moderate kidney disease. Clin. J. Am. Soc. Nephrol. 2010;5:1182–1189. doi: 10.2215/CJN.07971109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hsiao EY, et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013;155:1451–1463. doi: 10.1016/j.cell.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Braniste V, et al. The gut microbiota influences blood-brain barrier permeability in mice. Sci. Transl Med. 2014;6:263ra158. doi: 10.1126/scitranslmed.3009759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Buffie CG, et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 2015;517:205–208. doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]