Abstract
Colorectal cancer is the second-leading cause of cancer-related deaths in the United States and fourth-leading cause of cancer-related deaths worldwide. While cancer is largely considered to be a disease of genetic and environmental factors, increasing evidence has demonstrated a role for the microbiota (the microorganisms associated with the human body) in shaping inflammatory environments and promoting tumor growth and spread. Herein, we discuss both human data from meta’omics analyses and data from mechanistic studies in cell culture and animal models that support specific bacterial agents as potentiators of tumorigenesis—including Fusobacterium nucleatum, enterotoxigenic Bacteroides fragilis, and colibactin-producing Escherichia coli. Further, we consider how microbes can be used in diagnosing colorectal cancer and manipulating the tumor environment to encourage better patient outcomes in response to immunotherapy treatments.
Keywords: colorectal cancer, inflammation, colitis, mucosal immunology, microbiota/microbiome, Fusobacterium
INTRODUCTION
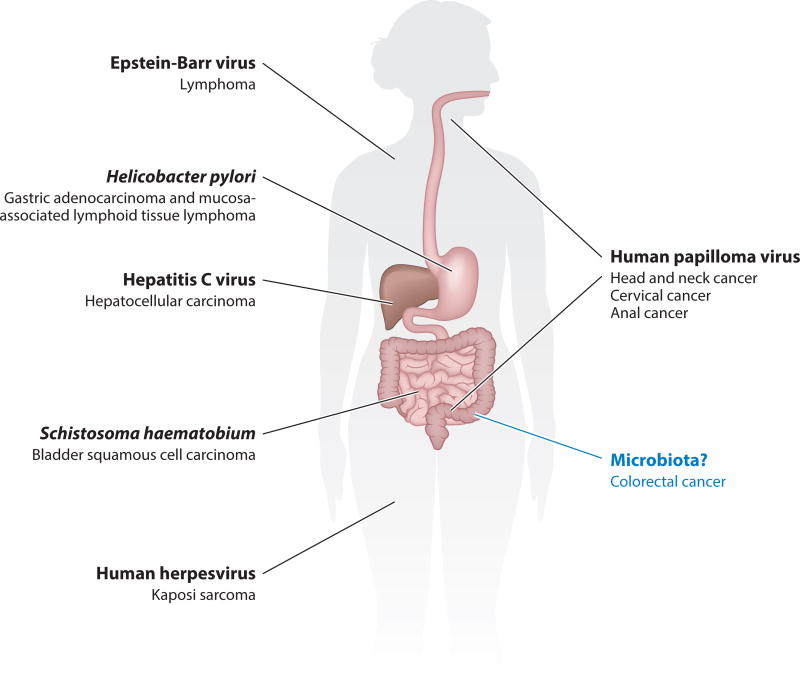
Cancer is a multifaceted disease influenced by both environmental and genetic factors. The microorganisms associated with the human body—collectively referred to as the microbiota—lie at the intersection of these factors. Aging and dietary patterns not only influence cancer susceptibility but also have profound effects on microbiota composition (17, 18, 21, 41, 47, 90). The microbiota plays a myriad of roles in human health and disease, from entraining immune system development and maintaining homeostasis to influencing autoimmune diseases and allergies, that cannot simply be parsed into strict pathogenesis and commensalism (9). How these organisms may influence a disease like cancer, which can develop over the course of decades, is similarly unclear. To survive in a human body tissue over the time frame in which solid tumors develop and influence cancer progression, a bacterium would need to identify metabolic substrates (carbon sources) to sustain growth, avoid immune-mediated destruction, and effectively compete with other microbes (if any) in that environment. These requirements necessitate features or effectors that shape a developing tumor microenvironment. Despite the seemingly insurmountable selective pressures exerted by evolving tumors, the concept of microbe-driven cancer is longstanding, given the well-described roles of Helicobacter pylori in gastric cancer (70) and human papillomavirus in cervical cancer (13), among others (Figure 1).
Figure 1.
Microbes with well-characterized roles in the development of specific cancers (34, 73, 74).
The gut microbiota is particularly well suited to influence cancer, as it has already evolved to survive and thrive in the intestinal environment. The gut microbiota, either as individual microbes (34) or as a microbial community exerting a collective effect, may potentiate or mitigate colorectal cancer (CRC) risk. The high bacterial density in the colon and the observation that bacteremias with certain microbes like Streptococcus gallolyticus can be clinical indicators of occult colonic adenomas (precancerous tumors) and CRC underscore the importance of studying the roles of gut microbes in CRC (11). The mechanisms by which microbes influence tumorigenesis in the intestine, a particularly microbially rich and immunologically complex environment in the human body, remain to be fully clarified.
DELINEATING HOW INFLAMMATION AND THE MICROBIOTA INFLUENCE CRC PROGRESSION
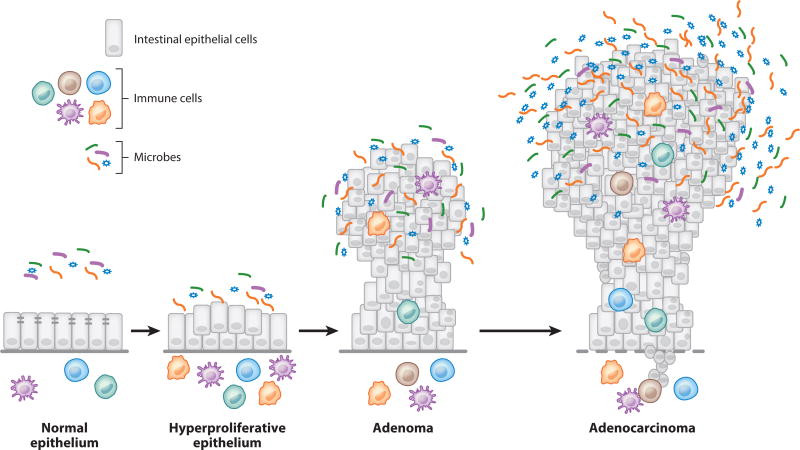
Tumor formation in the colon begins with the transition of a normal epithelium to a state of hyperplasia, in which cell proliferation is increased (Figure 2). As this occurs, epithelial architecture loses its characteristic shape and organization and becomes dysplastic. This dysplasia has the potential to develop into a nonmalignant adenoma, which usually is a polyp that grows from this region of hyperproliferative epithelium and protrudes into the colonic lumen. In response to other changes in the tumoral genetic and immunological microenvironment, adenomas can invade into the submucosa and become cancerous. With continued malignant growth, these tumors develop the potential to spread beyond the colon. For those interested in the microbial world, the microbial communities of the colonic lumen and of tumors offer rich opportunities for discovery efforts, ranging from whether microbes can block CRC development to how microbes can contribute to colorectal carcinogenesis.
Figure 2.
Progression of colorectal cancer development from normal epithelium to an invasive carcinoma.
The development of CRC from normal colonic epithelia requires a series of genetic and inflammatory-immunological factors to enable and shape a tumorigenic milieu. The initial formation of regions of hyperplasia and polyps can occur in response to the loss of tumor-suppressor genes like APC (adenomatous polyposis coli), a component of the Wnt/β-catenin pathway that is important for controlling cell proliferation. In addition, mutations in genes that encode the mahinery for DNA repair, such as hMSH2, can also contribute to colorectal tumorigenesis. These genetic alterations can be inherited, as in familial adenomatous polyposis or in Lynch syndrome, respectively. Such hereditary forms of CRC account for approximately 5–10% of all cases. Furthermore, the development of dysplasia and CRC is strongly influenced by the inflammatory state of the colon. In patients with inflammatory bowel disease (IBD), chronic, severe inflammation of the colon increases the likelihood of developing CRC (6). More subtle inflammation in otherwise healthy colonic tissues plays a major role in the conversion of a healthy colon to a dysplastic colon as well. As crypts become dysplastic, the barriers between the epithelium that aid in separating the microbiota from the immune cells in the lamina propria begin to break down. Barrier disruption facilitates bacterial translocation and, ultimately, exposure of immunogenic microbial compounds to both epithelial cells and antigen-presenting cells.
Activation of immune signaling pathways by bacterial stimuli results in a loss of homeostasis that drives a proneoplastic inflammatory environment. The role of bacterial products and their recognition by host cells in carcinogenesis has been thoroughly reviewed elsewhere (53). Below, we briefly revisit how this recognition results in inflammation and a protumorigenic milieu in CRC. Inflammatory signatures implicated in colorectal carcinogenesis studies include inflammasome activation (28) and activation of the NF-κB pathway (46), both of which can occur by changes in the mutational landscape or in response to either microbial stimuli or cytokines. NF-κB pathway activation mediates production of proinflammatory cytokines like IL-6, which has a pathogenic role in CRC by allowing survival and proliferation of intestinal epithelial cells, especially in colitis-associated cancer. The NF-κB pathway also serves as an important regulator of the genes encoding tumor necrosis factor (TNF) and cyclooxygenase 2 (COX-2), which are often highly overexpressed in inflammatory bowel disease as well as in colorectal adenomas and adenocarcinomas (7, 26, 54, 91). TNF is a cytokine that can drive activation of the NF-κB pathway, thereby driving a feed-forward loop that promotes cell proliferation and survival. COX-2 is an enzyme that produces prostaglandins, bioreactive lipids that influence both colonic inflammatory state and tumor progression through multiple mechanisms. Other key innate components of the inflammatory response that contribute to CRC progression include reactive nitrogen species (RNS) and reactive oxygen species (ROS), which serve as genotoxic compounds promoting the accumulation of mutations within proliferating epithelial cells. Beyond the innate immune system, both regulatory T cells and a subset of T helper cells known as Th17 cells modulate inflammation within the colon and contribute to inflammation-associated CRC. The development and function of these cells are influenced by microbes or microbial products, highlighting the dependence of inflammation and the microbiota in shaping the pretumor environment.
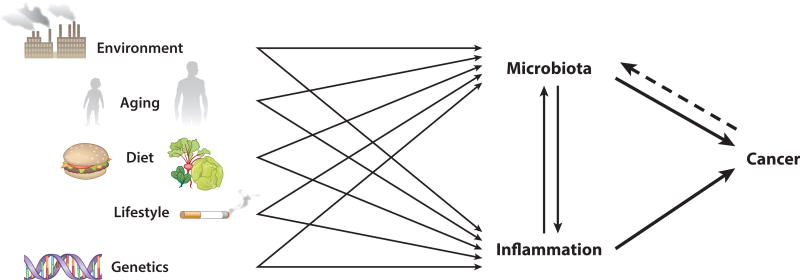
The microbiota and specific constituents and/or functions thereof are important drivers of the immune response. Supporting data come from clinical observations of patients with severe, protracted intestinal inflammation, as can occur in IBD, from molecular epidemiologic studies (64), and from rodent models. In the absence of microbes (i.e., germfree), mouse and rat models of intestinal tumorigenesis display reduced tumor loads as compared to those reared under conventional conditions (25, 52, 86). Such studies raise the question of whether specific microorganisms are necessary for the development of the proinflammatory tumor environment or the mere presence of bacterial products produced by any microorganisms is sufficient. Recent metagenomic analyses provided robust evidence not only that microbial communities in CRC tissues differ from the microbiota of healthy host tissues, but also that specific members of the microbiota may contribute to the development of a proinflammatory milieu and CRC. Inflammation, diet, and host genetics, among other considerations, further complicate this interpretation, as these factors can influence microbiota composition and function. Additionally, the tumor environment is populated by immune cells, which serve to provide both pro- and antitumor immunity and can be shaped by the resident microbiota even after progression to CRC. Rather than a straight-forward causal relationship, the interactions between the microbiota, immune system, and CRC are a multifactorial web that merits deep consideration, as their connectivities remain to be fully disentangled (Figure 3).
Figure 3.
Cross talk among environmental and genetic factors with the microbe-inflammation-cancer axis.
USING BIG DATA TO IDENTIFY POTENTIAL MICROBIAL DRIVERS OF CRC
Given the difficulties of parsing out the tripartite exchange between host genetics, environmental exposures, and the microbiota, it is important to identify the microbial members that may be a part of the conversation. As CRC develops over the course of 30 years or more, the microbial community that shaped the pretumor environment from a state of hyperplasia no longer exists and is rarely, if ever, obtainable from human sampling in sporadic colorectal cancer, given the need for longitudinal sampling over many decades. However, in order to have an effect on driving neoplasia, a microbe would need to localize to the region of interest (the evolving tumor) and remain for a long enough period of time for any procancerous functions to influence the colonic environment. This requirement for extended colonization enables the use of samples from later stages of CRC progression, for which human samples are more readily available from colonoscopic examinations and resections.
Following this logic, unbiased meta’omic analyses examined the microbial communities of human colonic adenocarcinoma samples and nearby normal colonic tissues. CRC tissues had decreased microbial diversity, including a reduction of certain bacterial genera like Clostridium and Bacteroides (49). This major shift in microbial community structure may be due to the inhospitable tumor environment, in which the rapidly growing tumor cells are competing for nutrients and the infiltrating immune cells are producing inflammatory compounds, like RNS and ROS, that can be toxic to microbes. In independent studies, CRC-involved tissues were specifically enriched in Fusobacterium spp., predominantly Fusobacterium nucleatum, relative to adjacent non-neoplastic tissue (15, 49, 55). F. nucleatum is a gram-negative bacterium and a normal constituent of the human oral cavity. As a resident member of the oral microbiota, F. nucleatum has been largely studied for its role in periodontal health (75) and its many adhesins that mediate binding to abiotic surfaces, host cells or other microorganisms (19, 40, 45). While Fusobacterium spp. are rarely detected in the gut microbiota of healthy individuals, they can be isolated from patients with IBD (80), further supporting a link between fusobacteria and an inflamed colonic environment.
Establishing direct relationships between the presence of F. nucleatum and increased CRC risk is a challenge in the absence of longitudinal data. However, identification of F. nucleatum enrichments during the premalignant stage of colorectal carcinogenesis begins to build a case for F. nucleatum as a biomarker for colonic pathology. Several studies now show F. nucleatum enrichments in colorectal tissues with high-grade dysplasia and adenomas (30, 55). Further, so-called big data studies are not limited to the microbial side of the equation, as cancer genomic and epigenetic research use similar techniques to define many molecular features of CRC. If F. nucleatum were mediating specific changes to the pretumor milieu, F. nucleatum enrichments might correlate with specific molecular phenotypes. Indeed, such correlations have been observed between fusobacterial enrichment and both genomic and epigenetic subsets of CRC, including microsatellite instability (MSI; a marker of mismatch repair)-high lesions and CpG island methylator phenotype (CIMP) high lesions (42, 59, 82). While several data sets support CRC enrichments of F. nucleatum, such data are not proof positive that F. nucleatum is a direct protagonist or lone actor in CRC. Deep sequencing efforts in human CRC samples have revealed that Fusobacterium species often co-occur with other gram-negative anaerobes, including Campylobacter species (94). Given F. nucleatum’s role as an important organizer of biofilms in the oral cavity, it may be a pioneer microbe that creates physical and metabolic scaffolds that support broader polymicrobial shifts in evolving tumors over time. Indeed, recent work examining microbial community changes across CRC progression has sought to identify strong microbial networks that might function together at the different stages of tumor formation (63).
Although meta’omic surveys have provided vast amounts of data that were unimaginable ten years ago, these approaches have important limitations that require an understanding of the methods used in these studies (see sidebar about Who’s Doing What in the Tumor Microenvironment?). Because of recent data supporting F. nucleatum as an important CRC-associated microbe in numerous independent studies, we have focused our attention on it here. However, other microbes that have been identified as potential drivers of CRC in both humans and mouse studies—enterotoxigenic Bacteroides fragilis (ETBF) (35, 38, 96) and colibactin-producing Escherichia coli (2)—have been highlighted in some (63, 89), but not all, of these studies. For colibactin-producing E. coli, species-level enrichment may not be necessary—the oncopathogenicity of this organism depends specifically on levels of the pks island that encodes the colibactin toxin rather than the total E. coli abundance as measured in the 16S rRNA surveys that underlie most metagenomic studies. Direct measurement of the levels of the pks island has demonstrated an enrichment of these bacteria in both IBD and CRC colonic mucosa samples (2). Similarly, for ETBF, the presence of the enterotoxin-encoding gene bft, rather than B. fragilis 16S rRNA levels, is the more relevant assessment; measurements of its prevalence have suggested enrichment of this microorganism in CRC tissues (10, 84, 89).
SHAPING THE TUMOR MICROENVIRONMENT: MECHANISMS USED BY MICROBES TO POTENTIATE CRC
Several models of microbe-mediated carcinogenesis have provided insight into how different bacteria may influence tumor formation (Table 1). While these models have limitations in their applicability to human CRC (e.g., using a microbe that, while able to induce tumor formation in a mouse model, is not prevalent in human colonic tumor tissues), they inform our understanding of the different mechanisms by which microbes influence the pretumor environment, including mediating DNA damage, inducing specific signaling pathways, promoting immune cell infiltration, and blocking antitumor immunity (recently reviewed in 34, 37, 53, 73, 74). Arguably the most important part of CRC development is the accumulation of multiple mutations within the epithelial cells, which results in uncontrolled proliferation. Some microbes, like Enterococcus faecalis, are able to indirectly influence DNA damage in the epithelium by eliciting high levels of ROS (92, 93), the same compounds produced by host cells during inflammation. Colibactin-producing E. coli attacks host DNA more directly, by introducing double-stranded DNA breaks that give rise to genomic instability and increased mutation frequency (20, 66). In the absence of the pks island, monoassociated Il10-deficient mice treated with the genotoxic agent azoxymethane develop comparable levels of inflammation, but fewer intestinal tumors than similarly treated mice monoassociated with pks+ E. coli (2). This observation highlights the importance of DNA damage by microbes in contributing directly to the proneoplastic environment, independent of inflammation. A less obvious example of the microbiota promoting CRC through DNA damage comes from work in mice with two genetic susceptibilities for intestinal tumorigenesis (ApcMin/+ and loss of Msh2; see sidebar about Addressing Causation: Animal Models of CRC). In this study, the authors found that in the absence of a functional mismatch repair system in the ApcMin/+ intestine, the fiber-derived microbial metabolite butyrate promoted cell hyperproliferation and resulted in increased tumor abundance (8). Antibiotic treatment or a low-carbohydrate dietary intervention was sufficient to disrupt this phenotype, highlighting that, even indirectly, the microbiota can have strong effects on the tumor environment.
Table 1.
Colorectal cancer–associated microbes
| Organism | Natural reservoir |
Evidence of association with CRCa | Mechanisms identified in modelsb |
Effectorsb | ||
|---|---|---|---|---|---|---|
| Epidemiology | Microbial enrichment |
Immune responses | ||||
| Streptococcus gallolyticus | GI tract | + | − | + | Unknown | Unknown |
| Enterococcus faecalis | GI tract | − | − | − | ROS-mediated DNA damage | Unknown |
| Colibactin-producing Escherichia coli | GI tract | + | + | − | Toxin-mediated DNA damage | Colibactin (Pks) |
| Enterotoxigenic Bacteroides fragilis | GI tract | + | + | − | Inflammation and immune-cell infiltration | Bft toxin |
| Fusobacterium nucleatum | Oral cavity | + | + | − | Inflammation and immune-cell infiltration; disruption of antitumor immunity | FadA, Fap2 |
Abbreviations: CRC, colorectal cancer; GI, gastrointestinal; ROS, reactive oxygen species.
Evidence of association with CRC is based on microbial enrichment as described in the main text and epidemiological and human immune responses as described and classified in Reference 74.
Beyond microbe-influenced DNA damage or microbial effects on cell proliferation in genetically susceptible hosts, intratumoral microbes can have effects on the tumor immune microenvironment that influence tumor growth and spread. In ApcMin/+ mice, exposure to F. nucleatum was sufficient to drive increased small intestinal and colonic adenoma formation and accelerate small intestinal adenocarcinoma development (48). Concurrent with an increase in adenoma formation, F. nucleatum treatment was associated with myeloid cell infiltration (predominantly dendritic cells, macrophages, and myeloid-derived suppressor cells) and an NF-κB proinflammatory transcriptional profile within mouse intestinal tumors, consistent with the progression of human CRC. Taken together, these data support that F. nucleatum serves to contribute to the development of the tumor environment itself. However, the molecular mechanisms of action underpinning F. nucleatum’s effects on intratumoral myeloid cell populations remain unclear. Other work using a xenograft model of tumorigenesis and in vitro carcinoma cell culture lines demonstrated that F. nucleatum can activate the Wnt/β-catenin pathway, which in turn led to NF-κB activation and stimulated tumor cell proliferation (72). This observation was dependent on the presence of FadA, an adhesin unique to fusobacteria, suggesting the importance of F. nucleatum adherence to and invasion of host cells in its promotion of CRC.
A microorganism may not only shape the tumor immune environment where it lives, but also teach us something about immune system function in cancer. The roles for many T cells in cancer are well characterized; for example, T regulatory cells function to suppress tumor immunity, while T helper type 1 cells promote antitumor immunity (33, 65). How T helper type 17 (Th17) cells, which function in inflammation and protection against extracellular microorganisms, influence tumorigenesis remained unclear until 2009. An important breakthrough came from work with enterotoxigenic B. fragilis, which demonstrated that ETBF activated the Stat3 transcription factor in the colon in ApcMin/+ mice (96). In a toxin-dependent manner, ETBF induced Th17 cell infiltration into the colon, and these Th17 cells mediated tumor formation. These data suggested that Th17 cells, at least in this model of CRC, were involved in the development of a protumorigenic microenvironment. More recent studies have further linked this important cell type to the promotion of CRC (22, 79), demonstrating the importance of microbiota research not just for elucidating how microbes contribute to carcinogenesis but also for defining cancer immunology principles.
By developing effective strategies to avoid immune-mediated destruction, microbes can enable tumor growth and spread by configuring an evolving tumor microenvironment into a milieu that is permissive to the survival of not only bacteria but also tumor cells. We term this the “live and let live” hypothesis. F. nucleatum manipulates the tumor microenvironment by using its Fap2 adhesin to engage the immune system and block natural killer (NK) cell–mediated killing (39). In doing so, F. nucleatum blocks a potent arm of antitumor immunity. F. nucleatum cells were shown to bind to tumor cells and inhibit killing of these cells by NK cells. Interaction of F. nucleatum with the NK cells occurs through interactions between Fap2 and human TIGIT [T cell immunoreceptor with Ig and ITIM (immunoreceptor tyrosine-based inhibitory motif) domains], an immune receptor found on NK and T cells and expressed by these immune cells in CRC. By engaging TIGIT, F. nucleatum avoids immune-mediated killing, as do the tumor cells to which they are bound. Thus, an interaction that likely evolved to protect F. nucleatum from the immune system is coopted by tumor cells to evade antitumor immunity.
EXPLOITING INTERACTIONS BETWEEN THE MICROBIOTA AND CRC FOR CANCER THERANOSTICS
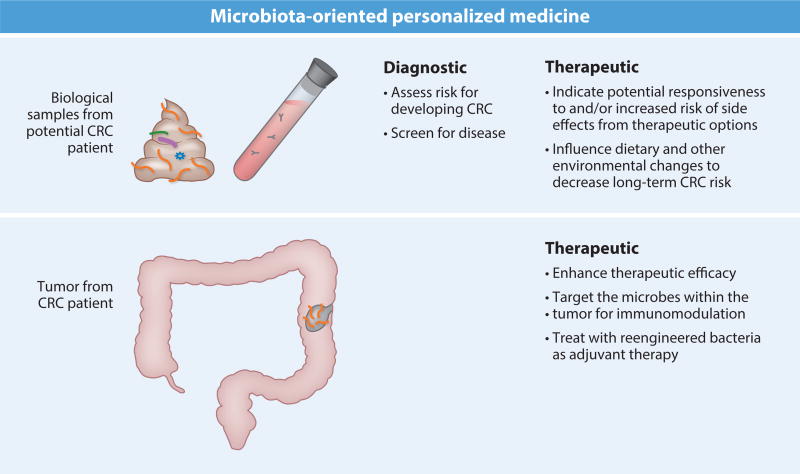
Much of the focus on the microbiota in CRC has been on microbes as pathogenic drivers of CRC, and hopefully these efforts can be leveraged for preventive, diagnostic, and therapeutic purposes (Figure 4); however, microbes may also reduce CRC risk. Several microbes, including some used widely in the food industry or as supplements, are able to reduce chronic inflammation under some conditions (5, 71, 87). Since inflammation drives CRC in a subset of cases, such beneficial microbes are one possibility for manipulating the intestinal environment that leads to CRC progression. Additionally, given the increasingly recognized role for members of the human body’s microbiota and their metabolites in shaping immune development and altering immune response (1, 3, 4, 43, 61, 62, 78), human gastrointestinal tracts may offer a pharmacopoeia of bacterial cancer immunotherapies ripe for development. Microbes are also being mined for their xenobiotic and vaccination potentials as they relate to cancer treatment. Furthermore, research like that in the Msh2-deficient mice mentioned above suggests that simple dietary interventions are sufficient to block the microbiota from shaping a proneoplastic environment (see sidebar about Feeding the Gut: The Effect of Diet on the Microbiota and CRC). Below, we use F. nucleatum as a model organism to discuss some potential avenues by which the microbiota can be used in cancer theranostics in more detail.
Figure 4.
The future of microbiota theranostics.
From a diagnostics perspective, the identification of novel biomarkers—that is, reliable noninvasive indicators of cancer within the body—is a challenging endeavor. Fecal samples, which can provide a periscopic view of the luminal and mucosal microbial environments (97) without the need for invasive procedures like colonoscopies, seem like an ideal approach (98). Studies have already shown increasing F. nucleatum levels in the fecal samples of patients with CRC (30, 32, 48, 98). However, in these studies fusobacterial levels are observed along a gradient, rather than being present at a standard level or else absent, thereby limiting efficacy of immediate usage of fecal fusobacterial load as a biomarker. Furthermore, the differences are far more subtle between adenoma patients and healthy subjects (no adenoma, CRC, or other pathology detected) than between CRC patients and healthy subjects. Earlier identification (i.e., at the adenoma stage) would be more valuable from a cancer prevention standpoint. Use of F. nucleatum as a prognostic marker in CRC may hold potential, given a recent study demonstrating a negative association between F. nucleatum levels and survival (58). However, these findings require further validation and a greater understanding of the mechanisms by which F. nucleatum shapes a tumorigenic milieu. Consideration of how F. nucleatum functions in its normal capacity (i.e., in the oral cavity) as compared to in a preneoplastic or neoplastic colon will provide insight into other biomarker targets, such as detection of tumor-specific gene products. Similarly, other screening approaches, such as detection of antibodies that may arise in response to F. nucleatum–specific antigens, may allow greater differentiation among F. nucleatum present in its distinct niches.
Another way to utilize an individual patient’s microbiota profiles—whether acquired broadly by 16S rRNA sequencing methods or directly by assessing the presence of a specific microbial marker with a directed assay like quantitative PCR—is in the treatment, rather than identification, of CRC. Many of the microbes associated with tumorigenesis in the colon do so by shaping the immune cell environment within the tumor. With F. nucleatum, one of these mechanisms is Fap2 engagement of TIGIT to protect against NK cell killing, thereby subverting antitumor immunity and allowing unrestricted tumor growth. Development of anti-Fap2 antibodies that could be used to treat an F. nucleatum–positive tumor may allow restoration of antitumoral immune detection and response. Alternatively, one could take a more general approach to correcting the intratumoral immune cell dysregulation in patients known to have F. nucleatum–positive tumors. As F. nucleatum–enriched tumors demonstrate increased myeloid cells, a treatment that would block myeloid cell migration and differentiation, such as an inhibitor of the chemokine CCL2 that can drive myeloid infiltration and intratumoral function (16), would be another approach. Personalized medicine honing in on immune-microbiota interactions could be used extensively once there are more data on the mechanisms by which microbes influence responsiveness to different cancer therapies.
Another therapeutic avenue to consider is using other, non-CRC-associated microbes to alter the tumor microenvironment response to immunotherapy treatments, which stimulate one’s own immune system to fight tumor cells but have had limited efficacy in CRC (see sidebar about Treating Colorectal Cancer). Recent advances in CRC treatment have focused on immune checkpoint blockade inhibitors that target the cytotoxic T lymphocyte–associated protein 4 (CTLA-4) and programmed death protein 1 (PD-1) pathways to enable T cell–mediated antitumor immunity. In two recent studies, particular microbes in the intestinal microbiota were shown to mediate efficacy of immunotherapies targeting these pathways in mouse models of cancer (77, 88). Using germfree mouse models, Vétizou and colleagues (88) demonstrated that the microbiota—specifically, Bacteroides spp.—was required for the therapeutic effects of the anti-CTLA-4 treatment to drive antitumor immunity in sarcomas. Similarly, Sivan et al. (77) identified Bifidobacterium as an important mediator of the effects of anti-PD-L1 therapy in melanoma. Such effects certainly have potential in CRC, where targeting microbes directly to the tumor could shape antitumor immunity and immunotherapy responses on a more local level. Much like the microbial drivers of CRC discussed earlier, such an organism would require a mechanism for localizing to a tumor and surviving in it long enough to affect the immunological milieu; then, ideally, it could be genetically or pharmacologically disarmed. An ideal microbe for such an approach would have much in common with a protumorigenic organism like F. nucleatum, further confounding the line between microbes as positive or negative actors in shaping immune response in the tumor microenvironment.
While such possibilities may influence translational approaches to CRC prevention and treatment, understanding the underlying biology of microbe-mediated CRC is essential before considering diagnostics or therapeutic strategies based on using the microbiota to manipulate the tumor microenvironment.
WHO’S DOING WHAT IN THE TUMOR MICROENVIRONMENT?
Understanding the limitations of methods used in microbiome research is important for interpreting such studies. The approaches used in Kostic et al. (49) and Castellarin et al. (15) demonstrate these differences. Bacterial 16S ribosomal amplicon DNAseq (14) and Pathseq (50) can be used to define the microbial DNA present but furnish no information regarding viability or function. RNAseq (60, 95) can be used to identify transcriptionally active microbial constituents, but it can be limited in providing functional information because of the preponderance of both bacterial and host ribosomal RNA sequences. Ideally, metagenomic approaches would be coupled to a metatranscriptomic analysis (31) of the microbes in colonic adenomas and adenocarcinomas to provide the greatest insight, but the methods to do so are currently limited by sequencing depth and the low microbial abundance relative to host cell number. To improve the bacterial transcriptional information provided by such studies, many technical development efforts focus on approaches to differentially isolate or selectively target host versus microbial nucleic acids (29, 36, 57). Additionally, tumor-associated microorganisms may modulate their functions based on their precise localization and structure within the colon. From a spatial perspective, the organization of the luminal microbiota in patients with CRC appears distinct from that of individuals without CRC (23), which may further affect how these microbes function and are able to influence the host epithelium and CRC development (44).
ADDRESSING CAUSATION: ANIMAL MODELS OF CRC.
To determine mechanisms underlying CRC, researchers use many different animal models of CRC that differ in their genetic basis and based on the investigators’ scientific objectives. The ApcMin/+ model, in which mice bear a point mutation in one copy of the Apc tumor suppressor gene, spontaneously forms adenomas along the intestinal tract, but most frequently in the small intestine (81). This model most resembles familial adenomatous polyposis. In comparison, Il10-deficient mice lack an important anti-inflammatory cytokine and develop spontaneous colitis; when these mice are treated with the carcinogen azoxymethane, they develop tumors that resemble the pathology seen in colitis-associated CRC (85). Finally, xenograft and allograft models of CRC (56) are generated when either primary or immortalized cancer cells from humans or mice are injected into recipient mice that are often immunocompromised to allow tumor growth. These injections can be either orthotopic, in this case into the distal colon or rectum, or subcutaneous. These models have utility in their ability to use primary human cancer cells and the relatively short time frame over which tumors develop.
FEEDING THE GUT: THE EFFECT OF DIET ON THE MICROBIOTA AND CRC.
The gut microbiota is greatly affected by dietary changes, in a matter of days (21, 90). Both the innate properties of the microbiota (i.e., its capacity to break down food into secondary metabolites with both pro- and anti-inflammatory properties) and the shifts that occur in these communities after dietary intervention can contribute to CRC (67–69). Dietary fiber provides perhaps the most interesting example of the relationship among diet, microbiota, immune system, and CRC. The microbiota can convert fiber to short-chain fatty acids, including acetic acid, propionic acid, and butyric acid, that then (a) shape immune system function and protect against colitis (1, 78), (b) drive anti-inflammatory responses and tumor-suppression (24, 27, 76), and (c) promote tumorigenesis in some models of CRC (8). Such seemingly contradictory effects underlie the need to better understand these multipartite associations.
TREATING COLORECTAL CANCER—WHAT’S THE RIGHT APPROACH?
CRC has proven to be a difficult disease to treat. Surgery remains the most common treatment, with radiotherapy and/or chemotherapy administered to patients with invasive tumors or metastatic disease. Recent advances in immunotherapies, such as immune checkpoint blockade inhibitors, have provided new hope to patients who are not adequately responding to chemotherapy (83). Despite the success of immunotherapies in treating other cancers, to date PD-1 blockade has had limited success in CRC treatment (12), mostly in a small trial of 41 patients that focused on one subset of CRC notable for a mismatch repair deficiency (51). Anti-CTLA-4 immunotherapy and checkpoint inhibitor combinations are in clinical trials for CRC. As such, the potential for personalized medicine to improve CRC outcomes depends on consideration of all relevant information at play, including cancer genomics and epigenetics as well as the microbial constituents that may be shaping the tumor immune environment.
SUMMARY POINTS.
Members of the intestinal microbiota are ideally suited to influence CRC, as the tools used by these microorganisms to survive, thrive, and avoid immune detection in the colonic mucosa are capable of becoming tumor-promoting weapons in a dysplastic precancerous environment.
CRC development is a sequential process with multiple stages at which both inflammation and the microbiota have important roles that are difficult to disentangle because of their intimate and intertwined relationship.
Meta’omics studies provide great insight into the microbes associated with CRC progression, but better tools are needed to parse the individual functions of these microbes in the tumor microenvironment.
Modulation of the tumor immune environment and promotion of DNA damage are common proneoplastic mechanisms by the best-studied CRC-associated microbes, including F. nucleatum, enterotoxigenic B. fragilis, and colibactin-producing E. coli.
Both the microbiota as a whole and specific CRC-associated microbes have enormous potential to enable much-needed new approaches to diagnose and treat CRC.
FUTURE ISSUES.
What about the pretumor environment (e.g., surface molecules/lectins, available metabolites, abiotic factors) leads to specific microbial targeting and/or enrichment?
What are the microbial molecules and/or pathways that influence the distinct stages of tumorigenesis?
How do the interactions between co-occurring microorganisms in colonic tumors shape the tumor microenvironment?
How can understanding the microbiota’s role in colorectal cancer direct personalized medicine?
Acknowledgments
W.S.G. and C.A.B thank S. Lavoie and C. Gallini for careful reading of this manuscript. Supported by NIH grant R01CA154426, a Burroughs Wellcome Career in Medical Sciences Award, and a research grant from Hoffman-LaRoche to W.S.G. C.A.B. is the Dennis and Marsha Dammerman fellow of the Damon Runyon Cancer Research Foundation (DRG-2205-14).
Glossary
- IBD
inflammatory bowel disease; includes ulcerative colitis and Crohn disease
- RNS/ROS
reactive nitrogen/oxygen species that cause nitrosative and oxidative stress, respectively, that can damage cells; they include such molecules as nitric oxide and superoxide
- Meta’omics
high-throughput, unbiased research approaches targeting DNA (metagenomics), RNA (metatranscriptomics) and metabolites (metabolomics), among others, from a microbial community
- Fusobacterium nucleatum
a CRC-associated microbe whose normal reservoir is the human oral cavity
- Immune checkpoint
important molecules modulating the level of immune system response; frequently uncontrolled in tumors
Footnotes
DISCLOSURE STATEMENT
W.S.G. is a member of the scientific advisory boards of Evelo Biosciences and Synlogic.
References
- 1.Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504(7480):451–55. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arthur JC, Perez-Chanona E, Mühlbauer M, Tomkovich S, Uronis JM, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338(6103):120–23. doi: 10.1126/science.1224820. Distinguished the roles of colibactin-producing E. coli in inflammation and tumorigenesis using animal models. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atarashi K, Tanoue T, Ando M, Kamada N, Nagano Y, et al. Th17 cell induction by adhesion of microbes to intestinal epithelial cells. Cell. 2015;163(2):367–80. doi: 10.1016/j.cell.2015.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331(6015):337–41. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ballal SA, Veiga P, Fenn K, Michaud M, Kim JH, et al. Host lysozyme-mediated lysis of Lactococcus lactis facilitates delivery of colitis-attenuating superoxide dismutase to inflamed colons. Proc. Natl. Acad. Sci. USA. 2015;112(25):7803–8. doi: 10.1073/pnas.1501897112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beaugerie L, Itzkowitz SH. Cancers complicating inflammatory bowel disease. N. Engl. J. Med. 2015;372(15):1441–52. doi: 10.1056/NEJMra1403718. [DOI] [PubMed] [Google Scholar]
- 7.Beissert S, Bergholz M, Waase I, Lepsien G, Schauer A, et al. Regulation of tumor necrosis factor gene expression in colorectal adenocarcinoma: in vivo analysis by in situ hybridization. Proc. Natl. Acad. Sci. USA. 1989;86(13):5064–68. doi: 10.1073/pnas.86.13.5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belcheva A, Irrazabal T, Robertson SJ, Streutker C, Maughan H, et al. Gut microbial metabolism drives transformation of MSH2-deficient colon epithelial cells. Cell. 2014;158(2):288–99. doi: 10.1016/j.cell.2014.04.051. [DOI] [PubMed] [Google Scholar]
- 9.Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157(1):121–41. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boleij A, Hechenbleikner EM, Goodwin AC, Badani R, Stein EM, et al. The Bacteroides fragilis toxin gene is prevalent in the colon mucosa of colorectal cancer patients. Clin. Infect. Dis. 2015;60(2):208–15. doi: 10.1093/cid/ciu787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boleij A, van Gelder MMHJ, Swinkels DW, Tjalsma H. Clinical importance of Streptococcus gallolyticus infection among colorectal cancer patients: systematic review and meta-analysis. Clin. Infect. Dis. 2011;53(9):870–78. doi: 10.1093/cid/cir609. [DOI] [PubMed] [Google Scholar]
- 12.Brahmer JR, Drake CG, Wollner I, Powderly JD, Picus J, et al. Phase I study of single-agent anti-programmed death-1 (MDX-1106) in refractory solid tumors: safety, clinical activity, pharmacodynamics, and immunologic correlates. J. Clin. Oncol. 2010;28(19):3167–75. doi: 10.1200/JCO.2009.26.7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burd EM. Human papillomavirus and cervical cancer. Clin. Microbiol. Rev. 2003;16(1):1–17. doi: 10.1128/CMR.16.1.1-17.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6(8):1621–24. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22(2):299–306. doi: 10.1101/gr.126516.111. With Kostic et al. (49), first to identify an association between F. nucleatum rRNA levels and human CRC tissues. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chun E, Lavoie S, Michaud M, Gallini CA, Kim J, et al. CCL2 promotes colorectal carcinogenesis by enhancing polymorphonuclear myeloid-derived suppressor cell population and function. Cell Rep. 2015;12(2):244–57. doi: 10.1016/j.celrep.2015.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Claesson MJ, Cusack S, O’Sullivan O, Greene-Diniz R, de Weerd H, et al. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc. Natl. Acad. Sci. USA. 2011;108(Suppl 1):4586–91. doi: 10.1073/pnas.1000097107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Claesson MJ, Jeffery IB, Conde S, Power SE, O’Connor EM, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488(7410):178–84. doi: 10.1038/nature11319. [DOI] [PubMed] [Google Scholar]
- 19.Coppenhagen-Glazer S, Sol A, Abed J, Naor R, Zhang X, et al. Fap2 of Fusobacterium nucleatum is a galactose-inhibitable adhesin involved in coaggregation, cell adhesion, and preterm birth. Infect. Immun. 2015;83(3):1104–13. doi: 10.1128/IAI.02838-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cuevas-Ramos G, Petit CR, Marcq I, Boury M, Oswald E, Nougayrède J-P. Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proc. Natl. Acad. Sci. USA. 2010;107(25):11537–42. doi: 10.1073/pnas.1001261107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505(7484):559–63. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Simone V, Pallone F, Monteleone G, Stolfi C. Role of TH17 cytokines in the control of colorectal cancer. Oncoimmunology. 2013;2(12):e26617. doi: 10.4161/onci.26617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dejea CM, Wick EC, Hechenbleikner EM, White JR, Mark Welch JL, et al. Microbiota organization is a distinct feature of proximal colorectal cancers. Proc. Natl. Acad. Sci. USA. 2014;111(51):18321–26. doi: 10.1073/pnas.1406199111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Donohoe DR, Holley D, Collins LB, Montgomery SA, Whitmore AC, et al. A gnotobiotic mouse model demonstrates that dietary fiber protects against colorectal tumorigenesis in a microbiota- and butyrate-dependent manner. Cancer Discov. 2014;4(12):1387–97. doi: 10.1158/2159-8290.CD-14-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dove WF, Clipson L, Gould KA, Luongo C, Marshall DJ, et al. Intestinal neoplasia in the ApcMin mouse: independence from the microbial and natural killer (beige locus) status. Cancer Res. 1997;57(5):812–14. [PubMed] [Google Scholar]
- 26.Eberhart CE, Coffey RJ, Radhika A, Giardiello FM, Ferrenbach S, Dubois RN. Up-regulation of cyclooxygenase 2 gene expression in human colorectal adenomas and adenocarcinomas. Gastroenterology. 1994;107(4):1183–88. doi: 10.1016/0016-5085(94)90246-1. [DOI] [PubMed] [Google Scholar]
- 27.Elangovan S, Pathania R, Ramachandran S, Ananth S, Padia RN, et al. The niacin/butyrate receptor GPR109A suppresses mammary tumorigenesis by inhibiting cell survival. Cancer Res. 2014;74(4):1166–78. doi: 10.1158/0008-5472.CAN-13-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elinav E, Nowarski R, Thaiss CA, Hu B, Jin C, Flavell RA. Inflammation-induced cancer: crosstalk between tumours, immune cells and microorganisms. Nat. Rev. Cancer. 2013;13(11):759–71. doi: 10.1038/nrc3611. [DOI] [PubMed] [Google Scholar]
- 29.Feehery GR, Yigit E, Oyola SO, Langhorst BW, Schmidt VT, et al. A method for selectively enriching microbial DNA from contaminating vertebrate host DNA. PLOS ONE. 2013;8(10):e76096. doi: 10.1371/journal.pone.0076096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flanagan L, Schmid J, Ebert M, Soucek P, Kunicka T, et al. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. Eur. J. Clin. Microbiol. Infect. Dis. 2014;33(8):1381–90. doi: 10.1007/s10096-014-2081-3. [DOI] [PubMed] [Google Scholar]
- 31.Franzosa EA, Morgan XC, Segata N, Waldron L, Reyes J, et al. Relating the metatranscriptome and metagenome of the human gut. Proc. Natl. Acad. Sci. USA. 2014;111(22):E2329–38. doi: 10.1073/pnas.1319284111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fukugaiti MH, Ignacio A, Fernandes MR, Ribeiro Júnior U, Nakano V, Avila-Campos MJ. High occurrence of Fusobacterium nucleatum and Clostridium difficile in the intestinal microbiota of colorectal carcinoma patients. Braz. J. Microbiol. 2015;46(4):1135–40. doi: 10.1590/S1517-838246420140665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313(5795):1960–64. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 34.Garrett WS. Cancer and the microbiota. Science. 2015;348(6230):80–86. doi: 10.1126/science.aaa4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Geis AL, Fan H, Wu X, Wu S, Huso DL, et al. Regulatory T-cell response to enterotoxigenic Bacteroides fragilis colonization triggers IL17-dependent colon carcinogenesis. Cancer Discov. 2015;5(10):1098–1109. doi: 10.1158/2159-8290.CD-15-0447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Giannoukos G, Ciulla DM, Huang K, Haas BJ, Izard J, et al. Efficient and robust RNA-seq process for cultured bacteria and complex community transcriptomes. Genome Biol. 2012;13(3):R23. doi: 10.1186/gb-2012-13-3-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goldszmid RS, Dzutsev A, Viaud S, Zitvogel L, Restifo NP, Trinchieri G. Microbiota modulation of myeloid cells in cancer therapy. Cancer Immunol Res. 2015;3(2):103–9. doi: 10.1158/2326-6066.CIR-14-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goodwin AC, Destefano Shields CE, Wu S, Huso DL, Wu X, et al. Polyamine catabolism contributes to enterotoxigenic Bacteroides fragilis-induced colon tumorigenesis. Proc. Natl. Acad. Sci. USA. 2011;108(37):15354–59. doi: 10.1073/pnas.1010203108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. 2015;42(2):344–55. doi: 10.1016/j.immuni.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han YW, Ikegami A, Rajanna C, Kawsar HI, Zhou Y, et al. Identification and characterization of a novel adhesin unique to oral fusobacteria. J. Bacteriol. 2005;187(15):5330–40. doi: 10.1128/JB.187.15.5330-5340.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoeijmakers JHJ. DNA damage, aging, and cancer. N. Engl. J. Med. 2009;361(15):1475–85. doi: 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- 42.Ito M, Kanno S, Nosho K, Sukawa Y, Mitsuhashi K, et al. Association of Fusobacterium nucleatum with clinical and molecular features in colorectal serrated pathway. Int. J. Cancer. 2015;137(6):1258–68. doi: 10.1002/ijc.29488. [DOI] [PubMed] [Google Scholar]
- 43.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139(3):485–98. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johnson CH, Dejea CM, Edler D, Hoang LT, Santidrian AF, et al. Metabolism links bacterial biofilms and colon carcinogenesis. Cell Metab. 2015;21(6):891–97. doi: 10.1016/j.cmet.2015.04.011. First metabolomics study of primary human CRC-associated tissues with microbes and tumorigenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaplan CW, Lux R, Haake SK, Shi W. The Fusobacterium nucleatum outer membrane protein RadD is an arginine-inhibitable adhesin required for inter-species adherence and the structured architecture of multispecies biofilm. Mol. Microbiol. 2009;71(1):35–47. doi: 10.1111/j.1365-2958.2008.06503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karin M, Greten FR. NF-kappaB: linking inflammation and immunity to cancer development and progression. Nat. Rev. Immunol. 2005;5(10):749–59. doi: 10.1038/nri1703. [DOI] [PubMed] [Google Scholar]
- 47.Key TJ, Schatzkin A, Willett WC, Allen NE, Spencer EA, Travis RC. Diet, nutrition and the prevention of cancer. Public Health Nutr. 2004;7(1A):187–200. doi: 10.1079/phn2003588. [DOI] [PubMed] [Google Scholar]
- 48.Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14(2):207–15. doi: 10.1016/j.chom.2013.07.007. Showed F. nucleatum was sufficient for tumor formation and myeloid cell infiltration without macroscopic inflammation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22(2):292–98. doi: 10.1101/gr.126573.111. With Castellarin et al. (15), first to identify an association between F. nucleatum rDNA levels and human CRC tissues. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kostic AD, Ojesina AI, Pedamallu CS, Jung J, Verhaak RGW, et al. PathSeq: software to identify or discover microbes by deep sequencing of human tissue. Nat. Biotechnol. 2011;29(5):393–96. doi: 10.1038/nbt.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N. Engl. J. Med. 2015;372(26):2509–20. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li Y, Kundu P, Seow SW, de Matos CT, Aronsson L, et al. Gut microbiota accelerate tumor growth via c-jun and STAT3 phosphorylation in APCMin/+ mice. Carcinogenesis. 2012;33(6):1231–38. doi: 10.1093/carcin/bgs137. [DOI] [PubMed] [Google Scholar]
- 53.Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat. Rev. Microbiol. 2014;12(10):661–72. doi: 10.1038/nrmicro3344. [DOI] [PubMed] [Google Scholar]
- 54.Masuda H, Iwai S, Tanaka T, Hayakawa S. Expression of IL-8, TNF-alpha and IFN-gamma m-RNA in ulcerative colitis, particularly in patients with inactive phase. J. Clin. Lab. Immunol. 1995;46(3):111–23. [PubMed] [Google Scholar]
- 55.McCoy AN, Araújo-Pérez F, Azcárate-Peril A, Yeh JJ, Sandler RS, Keku TO. Fusobacterium is associated with colorectal adenomas. PLoS ONE. 2013;8(1):e53653. doi: 10.1371/journal.pone.0053653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McIntyre RE, Buczacki SJA, Arends MJ, Adams DJ. Mouse models of colorectal cancer as preclinical models. Bioessays. 2015;37(8):909–20. doi: 10.1002/bies.201500032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Melnikov A, Galinsky K, Rogov P, Fennell T, Van Tyne D, et al. Hybrid selection for sequencing pathogen genomes from clinical samples. Genome Biol. 2011;12(8):R73. doi: 10.1186/gb-2011-12-8-r73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mima K, Nishihara R, Qian ZR, Cao Y, Sukawa Y, et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut. 2015 doi: 10.1136/gutjnl-2015-310101. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mima K, Sukawa Y, Nishihara R, Qian ZR, Yamauchi M, et al. Fusobacterium nucleatum and T cells in colorectal carcinoma. JAMA Oncol. 2015;1(5):653–61. doi: 10.1001/jamaoncol.2015.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moore RA, Warren RL, Freeman JD, Gustavsen JA, Chénard C, et al. The sensitivity of massively parallel sequencing for detecting candidate infectious agents associated with human tissue. PLoS ONE. 2011;6(5):e19838. doi: 10.1371/journal.pone.0019838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Naik S, Bouladoux N, Linehan JL, Han S-J, Harrison OJ, et al. Commensal-dendritic-cell interaction specifies a unique protective skin immune signature. Nature. 2015;520(7545):104–8. doi: 10.1038/nature14052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Naik S, Bouladoux N, Wilhelm C, Molloy MJ, Salcedo R, et al. Compartmentalized control of skin immunity by resident commensals. Science. 2012;337(6098):1115–19. doi: 10.1126/science.1225152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nakatsu G, Li X, Zhou H, Sheng J, Wong SH, et al. Gut mucosal microbiome across stages of colorectal carcinogenesis. Nat. Commun. 2015;6:8727. doi: 10.1038/ncomms9727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ng K, Meyerhardt JA, Chan AT, Sato K, Chan JA, et al. Aspirin and COX-2 inhibitor use in patients with stage III colon cancer. J. Natl. Cancer Inst. 2015;107(1):345–45. doi: 10.1093/jnci/dju345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nomura T, Sakaguchi S. Naturally arising CD25+CD4+ regulatory T cells in tumor immunity. Curr. Top. Microbiol. Immunol. 2005;293:287–302. doi: 10.1007/3-540-27702-1_13. [DOI] [PubMed] [Google Scholar]
- 66.Nougayrède J-P, Homburg S, Taieb F, Boury M, Brzuszkiewicz E, et al. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science. 2006;313(5788):848–51. doi: 10.1126/science.1127059. [DOI] [PubMed] [Google Scholar]
- 67.O’Keefe SJD, Li JV, Lahti L, Ou J, Carbonero F, et al. Fat, fibre and cancer risk in African Americans and rural Africans. Nat. Commun. 2015;6:6342. doi: 10.1038/ncomms7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.O’Keefe SJD, Ou J, Aufreiter S, O’Connor D, Sharma S, et al. Products of the colonic microbiota mediate the effects of diet on colon cancer risk. J. Nutr. 2009;139(11):2044–48. doi: 10.3945/jn.109.104380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ou J, Carbonero F, Zoetendal EG, DeLany JP, Wang M, et al. Diet, microbiota, and microbial metabolites in colon cancer risk in rural Africans and African Americans. Am. J. Clin. Nutr. 2013;98(1):111–20. doi: 10.3945/ajcn.112.056689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Polk DB, Peek RM. Helicobacter pylori: gastric cancer and beyond. Nat. Rev. Cancer. 2010;10(6):403–14. doi: 10.1038/nrc2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rooks MG, Veiga P, Wardwell-Scott LH, Tickle T, Segata N, et al. Gut microbiome composition and function in experimental colitis during active disease and treatment-induced remission. ISME J. 2014;8(7):1403–17. doi: 10.1038/ismej.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14(2):195–206. doi: 10.1016/j.chom.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schwabe RF, Jobin C. The microbiome and cancer. Nat. Rev. Cancer. 2013;13(11):800–812. doi: 10.1038/nrc3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sears CL, Garrett WS. Microbes, microbiota, and colon cancer. Cell Host Microbe. 2014;15(3):317–28. doi: 10.1016/j.chom.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Signat B, Roques C, Poulet P, Duffaut D. Fusobacterium nucleatum in periodontal health and disease. Curr. Issues Mol. Biol. 2011;13(2):25–36. [PubMed] [Google Scholar]
- 76.Singh N, Gurav A, Sivaprakasam S, Brady E, Padia R, et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity. 2014;40(1):128–39. doi: 10.1016/j.immuni.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350(6264):1084–89. doi: 10.1126/science.aac4255. With Vétizou et al. (88), implicated the microbiota in effective responses to (PD-1) immunotherapy against CRC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341(6145):569–73. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Song X, Gao H, Lin Y, Yao Y, Zhu S, et al. Alterations in the microbiota drive interleukin-17C production from intestinal epithelial cells to promote tumorigenesis. Immunity. 2014;40(1):140–52. doi: 10.1016/j.immuni.2013.11.018. [DOI] [PubMed] [Google Scholar]
- 80.Strauss J, Kaplan GG, Beck PL, Rioux K, Panaccione R, et al. Invasive potential of gut mucosa-derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm. Bowel Dis. 2011;17(9):1971–78. doi: 10.1002/ibd.21606. [DOI] [PubMed] [Google Scholar]
- 81.Su LK, Kinzler KW, Vogelstein B, Preisinger AC, Moser AR, et al. Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene. Science. 1992;256(5057):668–70. doi: 10.1126/science.1350108. [DOI] [PubMed] [Google Scholar]
- 82.Tahara T, Yamamoto E, Suzuki H, Maruyama R, Chung W, et al. Fusobacterium in colonic flora and molecular features of colorectal carcinoma. Cancer Res. 2014;74(5):1311–18. doi: 10.1158/0008-5472.CAN-13-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Topalian SL, Drake CG, Pardoll DM. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell. 2015;27(4):450–61. doi: 10.1016/j.ccell.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Toprak NU, Yagci A, Gulluoglu BM, Akin ML, Demirkalem P, et al. A possible role of Bacteroides fragilis enterotoxin in the aetiology of colorectal cancer. Clin. Microbiol. Infect. 2006;12(8):782–86. doi: 10.1111/j.1469-0691.2006.01494.x. [DOI] [PubMed] [Google Scholar]
- 85.Uronis JM, Mühlbauer M, Herfarth HH, Rubinas TC, Jones GS, Jobin C. Modulation of the intestinal microbiota alters colitis-associated colorectal cancer susceptibility. PLoS ONE. 2009;4(6):e6026. doi: 10.1371/journal.pone.0006026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vannucci L, Stepankova R, Kozakova H, Fiserova A, Rossmann P, Tlaskalova-Hogenova H. Colorectal carcinogenesis in germ-free and conventionally reared rats: different intestinal environments affect the systemic immunity. Int. J. Oncol. 2008;32(3):609–17. [PubMed] [Google Scholar]
- 87.Veiga P, Gallini CA, Beal C, Michaud M, Delaney ML, et al. Bifidobacterium animalis subsp. lactis fermented milk product reduces inflammation by altering a niche for colitogenic microbes. Proc. Natl. Acad. Sci. USA. 2010;107(42):18132–37. doi: 10.1073/pnas.1011737107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Vétizou M, Pitt JM, Daillère R, Lepage P, Waldschmitt N, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350(6264):1079–84. doi: 10.1126/science.aad1329. With Sivan et al. (77), implicated the microbiota in effective responses to (CTLA-4) immunotherapy against CRC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Viljoen KS, Dakshinamurthy A, Goldberg P, Blackburn JM. Quantitative profiling of colorectal cancer-associated bacteria reveals associations between Fusobacterium spp., enterotoxigenic Bacteroides fragilis (ETBF) and clinicopathological features of colorectal cancer. PLoS ONE. 2015;10(3):e0119462. doi: 10.1371/journal.pone.0119462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Walker AW, Ince J, Duncan SH, Webster LM, Holtrop G, et al. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J. 2011;5(2):220–30. doi: 10.1038/ismej.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang D, Dubois RN. The role of COX-2 in intestinal inflammation and colorectal cancer. Oncogene. 2010;29(6):781–88. doi: 10.1038/onc.2009.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang X, Allen TD, May RJ, Lightfoot S, Houchen CW, Huycke MM. Enterococcus faecalis induces aneuploidy and tetraploidy in colonic epithelial cells through a bystander effect. Cancer Res. 2008;68(23):9909–17. doi: 10.1158/0008-5472.CAN-08-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang X, Yang Y, Moore DR, Nimmo SL, Lightfoot SA, Huycke MM. 4-hydroxy-2-nonenal mediates genotoxicity and bystander effects caused by Enterococcus faecalis-infected macrophages. Gastroenterology. 2012;142(3):543–47. doi: 10.1053/j.gastro.2011.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Warren RL, Freeman DJ, Pleasance S, Watson P, Moore RA, et al. Co-occurrence of anaerobic bacteria in colorectal carcinomas. Microbiome. 2013;1(1):16. doi: 10.1186/2049-2618-1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Weber G, Shendure J, Tanenbaum DM, Church GM, Meyerson M. Identification of foreign gene sequences by transcript filtering against the human genome. Nat. Genet. 2002;30(2):141–42. doi: 10.1038/ng818. [DOI] [PubMed] [Google Scholar]
- 96.Wu S, Rhee K-J, Albesiano E, Rabizadeh S, Wu X, et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat. Med. 2009;15(9):1016–22. doi: 10.1038/nm.2015. Demonstrated ETBF as driver of Th17 responses in a Bft-dependent manner in colitis-associated tumorigenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yasuda K, Oh K, Ren B, Tickle TL, Franzosa EA, et al. Biogeography of the intestinal mucosal and lumenal microbiome in the rhesus macaque. Cell Host Microbe. 2015;17(3):385–91. doi: 10.1016/j.chom.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zackular JP, Rogers MA, Ruffin MT, 4th, Schloss PD. The human gut microbiome as a screening tool for colorectal caner. Cancer Prev. Res. 2014;7(11):1112–21. doi: 10.1158/1940-6207.CAPR-14-0129. [DOI] [PMC free article] [PubMed] [Google Scholar]