Abstract
Transcription and processing of mRNA precursors are coordinated events that require numerous complex interactions to ensure that they are successfully executed. We described previously an unexpected association between a transcription factor, PC4 (or Sub1 in yeast), and an mRNA polyadenylation factor, CstF-64 (Rna15 in yeast), and provided evidence that this was important for efficient transcription elongation. Here we provide insight into the mechanism by which this occurs. We show that Sub1 and Rna15 are recruited to promoters and present along the length of several yeast genes. Allele-specific genetic interactions between SUB1 and genes encoding an RNA polymerase II (RNAP II)-specific kinase (KIN28) and phosphatase (FCP1) suggest that Sub1 influences and/or is sensitive to the phosphorylation status of elongating RNAP II. Remarkably, we find that cells lacking Sub1 display decreased accumulation of Fcp1, altered RNAP II phosphorylation and decreased crosslinking of RNAP II to transcribed genes. Our data provide evidence that Rna15 and Sub1 are present along the length of several genes and that Sub1 facilitates elongation by influencing enzymes that modify RNAP II.
Keywords: CstF64/Rna15, Fcp1, PC4/Sub1, RNA polymerase II
Introduction
RNA polymerase II (RNAP II) primary transcripts undergo specific and extensive processing, often tightly coordinated with transcription, and which in many cases occurs as transcription proceeds. Additionally, the three major mRNA processing reactions, capping, splicing and polyadenylation, are all linked and can influence each other. Many studies support the idea that RNAP II coordinates these processing steps, and specifically that the carboxy-terminal domain of the RNAP II largest subunit (CTD) is important for targeting of capping enzymes, for enhancing an early step in spliceosome assembly and for activating the cleavage/polyadenylation complex at the 3′ processing site (reviewed by Hirose and Manley, 2000; Maniatis and Reed, 2002; Proudfoot et al, 2002; Zorio and Bentley, 2004). The CTD thus plays a critical, integrating role in essentially all steps of mRNA synthesis, and in keeping with this is subject to dynamic regulation during the transcription cycle.
The CTD consists of multiple repeats of a seven amino-acid consensus motif, YSPTSPS (Corden et al, 1985; Corden, 1990). This consensus is highly conserved in eukaryotes, although the number of repeats varies among species, from 26 in yeast to 52 in mammals. Five out of these seven residues can be phosphorylated, but serines (ser) 2 and 5 appear to be the major sites of phosphorylation (West and Corden, 1995; Bensaude et al, 1999). RNAP II is hypophosphorylated when recruited to a promoter (designated RNAP IIA), becoming hyperphosphorylated during transcription elongation (RNAP IIO). Experiments involving chromatin immunoprecipitation (ChIP) have provided evidence that differential phosphorylation of ser 2 and 5 coincides with recruitment of factors important for pre-mRNA processing and transcription elongation, and that the pattern of phosphorylation can change, from ser 5 to ser 2, during the course of transcription (Komarnitsky et al, 2000; Schroeder et al, 2000; Cho et al, 2001). Reversible phosphorylation of the CTD thus plays a significant role in modulating the transcription cycle and coordinating the steps of gene expression.
A number of potential kinases and phosphatases have been described that likely target the CTD (reviewed by Kobor and Greenblatt, 2002; Prelich, 2002). Among the kinases, CDK7, CDK8 and CDK9 are well characterized cyclin-dependent kinases that target ser 2 and/or ser 5 residues in the CTD. For example, CDK7 along with cyclin H (Kin28/Ccl1 in yeast) is part of the general transcription factor (GTF) TFIIH. Based initially on studies in yeast, CDK7/Kin28 was shown to phosphorylate the CTD following preinitiation complex formation, thereby positively regulating early steps in transcription (Feaver et al, 1994; Hengartner et al, 1998). Furthermore, both genetic and ChIP experiments in yeast have provided evidence that Kin28 targets principally ser 5 at the 5′ end of genes, facilitating recruitment of capping enzymes and perhaps functioning in coupling transcription and polyadenylation (Komarnitsky et al, 2000; Rodriguez et al, 2000; Schroeder et al, 2000). Several CTD phosphatases have also been described. The best studied of these is Fcp1, which is required for transcription and plays a role in recycling RNAP II for new rounds of transcription (Cho et al, 1999; Kobor et al, 1999). Fcp1 in yeast is chromatin associated both at the promoter and the coding region and has been suggested to function in ser 2 dephosphorylation (Cho et al, 2001). Fcp1 thus appears to play an important role during transcription elongation. More recently, another potential yeast CTD phosphatase, Ssu72, has been identified (Krishnamurthy et al, 2004). Ssu72 is an intriguing protein first discovered genetically via interactions with the gene encoding the GTF TFIIB (SUA7; Sun and Hampsey, 1996) and subsequently as a component of the mRNA 3′ end processing machinery (Dichtl et al, 2002; He et al, 2003; Nedea et al, 2003). Ssu72 may thus play a role in linking transcription with pre-mRNA 3′ processing.
Considerable data now exist that transcription by RNAP II is linked to the process of cleavage/polyadenylation, and furthermore that this can involve interactions that initiate at the promoter (reviewed by Calvo and Manley, 2003; Proudfoot, 2004). One protein that has the potential to play a significant role in this process is PC4 (or Sub1 in yeast). PC4/Sub1 (which we refer to here as Sub1) was initially discovered as a transcription factor, biochemically as a coactivator required for activated transcription in vitro (Henry et al, 1996) and genetically, analogous to Ssu72, as a suppressor of certain TFIIB mutations (Knaus et al, 1996). Indeed, in light of the fact that both these proteins now seem to be associated with the 3′ end machinery, it is noteworthy that the SUA7 allele specificity of the SSU72 and SUB1 interactions is nearly identical (Wu et al, 1999). The suggestion that Sub1 might function in 3′ end formation came from the discovery of a physical interaction, conserved from yeast to human, with the polyadenylation factor Rna15 (CstF-64 in humans). The physiological significance of this association was supported by the demonstration of genetic interactions between the two genes showing that deletion or overexpression of SUB1 suppresses or enhances, respectively, both growth and a transcription termination defect associated with a specific RNA15 mutant (Calvo and Manley, 2001). These results suggested a model by which Sub1, in association with Rna15, functions as an antitermination factor during transcription. But direct support for this model has been lacking. For example, it remains unclear whether Sub1 and/or Rna15 associate with the elongation complex throughout transcription, nor is it apparent how Sub1 could influence transcription elongation.
We describe here experiments aimed at elucidating more about how Sub1, together with Rna15, might function in transcription elongation. We first provide evidence that both Sub1 and Rna15 are present at the promoter and along the length of several genes. Although not essential, Sub1 enhances, directly or indirectly, recruitment of Rna15 to promoters. Analysis of genetic interactions involving SUB1 revealed synthetic phenotypes with genes encoding a CTD kinase (KIN28) and phosphatase (FCP1). Remarkably, this is reflected in a significant decrease in accumulation of Fcp1 in cells lacking Sub1, as well as changes in the phosphorylation status and accumulation of RNAP II. ChIP assays indicate a reduction in both Fcp1 and RNAP II along transcribed genes. These data provide new insights into how Sub1 can function to enhance transcription elongation.
Results
Sub1 and Rna15 are recruited to promoters and remain chromatin associated during transcription elongation
We have proposed a model in which the 3′ end processing factor CstF-64/Rna15 interacts with the transcriptional coactivator PC4/Sub1 at promoter regions, and the two then travel together with RNAP II during transcription elongation until the 3′ end processing signals. An important aspect of this model is that Rna15 and Sub1 be localized at active promoters and along the length of transcribed genes. This question can be addressed by ChIP assays, and indeed related experiments have already been performed (Komarnitsky et al, 2000; Licatalosi et al, 2002; Nedea et al, 2003; Kim et al, 2004). However, these studies were not aimed at the specific question we are concerned with, the interactions of Rna15 and Sub1, and in addition reached somewhat contradictory conclusions (see Discussion). For ChIP experiments, yeast strains bearing HA epitope-tagged Rpb3 (an RNAP II subunit), Kin28, Rna15 or Sub1 were utilized. HA-Rna15 was first introduced into rna15-1 (Bloch et al, 1978; Minvielle-Sebastia et al, 1991) mutant cells, which were grown at 37°C. Full complementation of the slow growth (25°C) and temperature-sensitive (37°C) phenotypes by HA-RNA15 was observed (data not shown). For others, the appropriate endogenous gene was disrupted by either LEU or URA and substituted with the HA-tagged version (see Materials and methods; Komarnitsky et al, 2000).
Crosslinked chromatin was isolated, processed, immunoprecipitated and analyzed by PCR. Figure 1A indicates the regions amplified initially from the ACT1 and ADH1 genes. HA-Rpb3 and Kin28-HA, which have been analyzed previously (Komarnitsky et al, 2000; Licatalosi et al, 2002), served as controls for the specificity of our assays. As expected, HA-Rpb3 was localized to both the 5′ and 3′ regions of ACT1, while Kin28-HA was predominantly localized to the 5′ (promoter) region of the gene (Figure 1B). The inactive silent mating type locus (HMR1) or an intergenic region on chromosome V (V) served as a negative control, and little or no crosslinking was detected. The pattern of HA-Rna15 and HA-Sub1 crosslinking to ACT1 was similar to that observed with HA-Rpb3, and significant crosslinking to three regions of ADH1, including the promoter, was also detected (Figure 1C, left and center panels, and results not shown). Essentially identical results were obtained when HA-Rna15 was expressed in a wild-type background (Figure 1C, right panel, and results not shown), ruling out the possibility that the mutant Rna15-1 protein may have influenced our results. Given the uncertainty as to whether polyadenylation factors, including Rna15, can be efficiently crosslinked to promoters, we repeated this experiment with primers spanning two additional promoters, from the PYK1 and RPS5 genes, and obtained nearly identical results (Figure 1C, right panel). These results indicate that both HA-Sub1 and HA-Rna15 can associate with active genes, both at the promoters and at sites within the transcribed regions.
Figure 1.
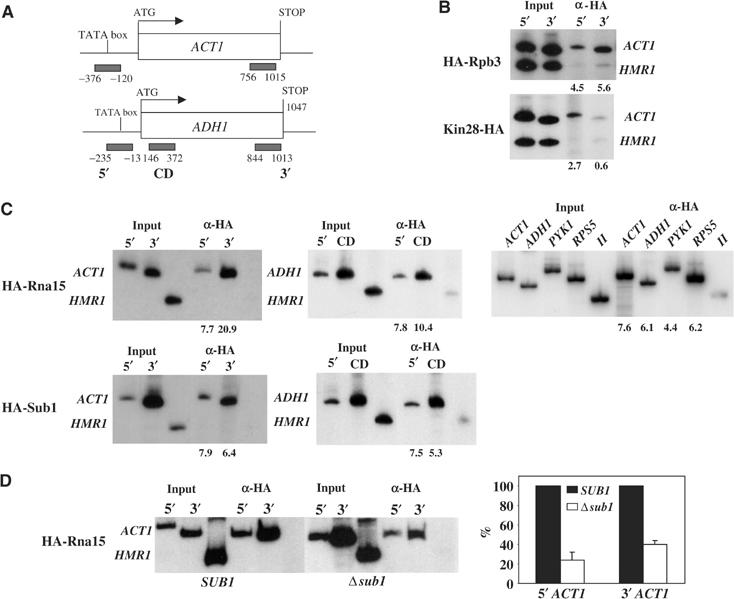
Sub1 and Rna15 are recruited to promoters and remain chromatin associated during transcription elongation. ChIP analysis of HA-tagged Rpb3, Kin28, Rna15 and Sub1 strains to test the presence of these proteins in promoter (5′), coding (CD) and 3′ regions of the ACT1 and ADH1 genes. (A) Schematic diagram of the ACT1 and ADH1 genes: open boxes represent open reading frames and black bars PCR products. (B) ChIP analyses of HA-Rpb3 and HA-Kin28, which serve as positive and negative controls for proteins associated with both 5′ and 3′ or only with 5′ regions, respectively. Primers that amplified nontranscribed regions (HMR and V) were used as background controls. (C) ChIP analysis of HA-Rna15 and HA-Sub1: the left panel shows Rna15 and Sub1 at 5′ and 3′ ends of ACT1, the middle panel at 5′ and coding region of ADH1 and the right panel 5′ regions of ACT1, ADH1, PYK1 and RPS5, all four in RNA15 background. Numbers below gels represent adjusted fold over background (see Materials and methods). (D) ChIP analysis of HA-Rna15 from wild-type (SUB1) and mutant (Δsub1) cells. Numbers on the y-axis of the graph represent the percentage of Rna15 localized to 5′ and 3′ regions of ACT1 in the Δsub1 mutant relative to wild-type cells, where Rna15 localization in wild type is considered as 100%.
Given the interactions we have described between Rna15 and Sub1 (Calvo and Manley, 2001), we next investigated whether Sub1 can function in recruitment of Rna15 to active genes. To this end, we introduced a SUB1 deletion into the HA-Rna15 expressing strain analyzed above, and compared crosslinking of HA-Rna15 to the ACT1 and ADH1 genes in the isogenic SUB1 and Δsub1 strains (results with ADH1 not shown). Significant crosslinking was detected at the 5′ and 3′ ends of both genes in each of the strains (Figure 1D, left), indicating that Sub1 is not essential for HA-Rna15 recruitment. However, quantitation of results from multiple experiments revealed a considerable decrease in crosslinking in the Δsub1 strain, by approximately 75% at the promoter and 60% in the 3′ region (Figure 1D, right). Although RNAP II loading at the promoter was also reduced by SUB1 deletion (see below), the reduction was relatively modest (∼40%) and not sufficient to explain fully the more significant effect on Rna15 recruitment.
The data described above suggest that Sub1 can associate with promoter and transcribed regions of two genes. The promoter association was not unexpected in light of the multiple functions suggested for Sub1/PC4 during transcription initiation, and the association with transcribed regions is consistent with our proposal that Sub1 functions to prevent premature transcription termination (see Introduction). Our results are also in general agreement with ChIP assays performed by Nedea et al (2003), who observed Sub1 association at the promoter and 3′ end regions of the PYK1 gene. However, these authors also observed that Sub1 was present at a reduced level at an internal site in this gene, and therefore suggested that Sub1 might have distinct functions at the 5′ and 3′ ends of the gene. In light of this, we measured HA-Sub1 distribution by ChIP over the length of a third gene, PMA1. We utilized six primer pairs, extending from the promoter to just downstream of the poly(A) signal, and the results are shown in Figure 2. Consistent with the results in Figure 1, significant and essentially equivalent Sub1-HA crosslinking was detected across the entire PMA1 gene. Together, our findings support the notion that Sub1 is associated with the transcription complex throughout elongation.
Figure 2.
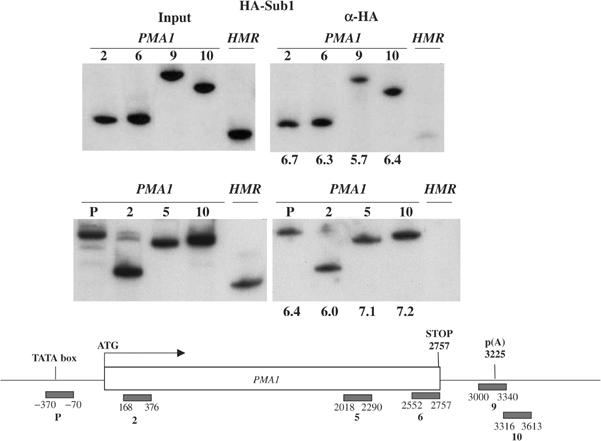
Sub1 is associated with the transcription complex throughout elongation. ChIP analysis of 5′, coding and 3′ sequences of the PMA1 gene using an HA-Sub1 expressing strain is shown. Primers that amplified HMR were used as background control. The bottom panel shows a schematic diagram of PCR primers used, and the black bars represent PCR products. Numbers below the gels were calculated as described in Figure 1 and Materials and methods.
Sub1 function is connected with CTD phosphorylation
The results we have described here and previously are consistent with the idea that Sub1 associates with the transcription complex and facilitates elongation by RNAP II. While this involves interaction with Rna15, precisely how Sub1 affects elongation is unclear. One possibility is that Sub1 interacts with factors that bind to or modify the CTD. To investigate this idea, we next performed genetic crosses involving SUB1 and several genes encoding CTD interacting factors.
Kin28 is an essential CTD kinase that functions early in the transcription cycle (e.g., Valay et al, 1995; Komarnitsky et al, 2000; Schroeder et al, 2000) and facilitates recruitment of RNA capping and perhaps polyadenylation factors to the transcription complex (Rodriguez et al, 2000). In light of these properties, we performed a series of genetic crosses with three different kin28 alleles (T17D, K36A and T162A) and a SUB1 deletion (Δsub1). The kin28-T17D and kin28-K36A mutants display slow growth at 25 and 37°C, and a decrease in kinase activity affecting CTD phosphorylation and capping enzyme recruitment, with kin28-T17D giving rise to the strongest phenotypes. The kin28-T162A allele, which does not display a slow growth phenotype, decreases kinase activity in vitro, but does not detectably affect CTD phosphorylation and capping enzymes recruitment (Espinoza et al, 1998; Kimmelman et al, 1999; Rodriguez et al, 2000). The three double mutants and corresponding parental strains (kin28 and Δsub1) were streaked on rich medium plates. The combination of kin28 mutants with Δsub1 resulted in either synthetic lethality at 25°C (T17D; Figure 3A), reduced growth at 25°C and lethality at 37°C (K36A; Figure 3B) or only slightly reduced growth at 37°C (T162A; Figure 3C). The synthetic phenotypes of these double mutants thus exactly parallel the decrease in CTD phosphorylation of the single kin28 mutants. The results establish an allele-specific genetic interaction between KIN28 and SUB1, and importantly show that Sub1 is essential under conditions of decreased CTD phosphorylation.
Figure 3.
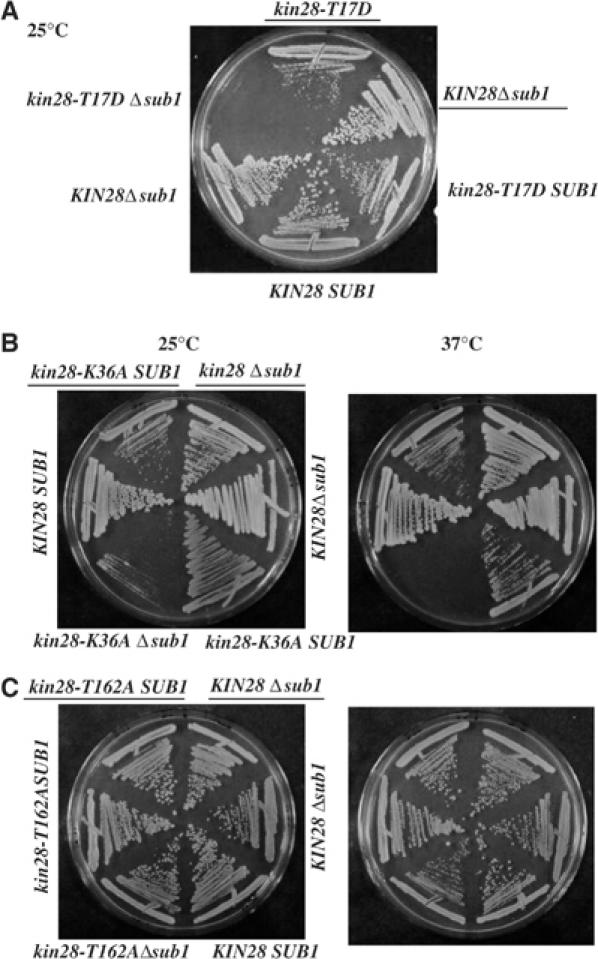
Sub1 function is connected with CTD phosphorylation. We generated kin28 Δsub1 double mutant strains by crossing a Δsub1 strain with different kin28 alleles: (A) Δsub1 × kin28-T17D (YSB592) cross; (B) Δsub1 × kin28-K36A (YSB609) cross; (C) Δsub1 × kin28-T162A (YSB595) cross. Parental strains (Δsub1, kin28-T17D and kin28-T162) and double mutants were streaked on rich medium plates at 25°C (A) or at 25°C (B) and 37°C (C).
The above results provide evidence for an interaction between KIN28 and SUB1. Given that a significant aspect of Kin28 activity is to facilitate recruitment and activation of the capping apparatus via the CTD, and that Sub1 interacts with and facilitates recruitment of another RNA processing factor, Rna15, to the transcription complex, we next tested whether SUB1 also interacts genetically with a component of the capping apparatus. To this end, we crossed the Δsub1 strain with a strain bearing a mutation in one of the subunits of the capping complex, the guanylyltransferase, Ceg1. The ceg1-250 mutation (Fresco and Buratowski, 1996) causes lethality at 37°C, and is localized to the C-terminal domain of Ceg1, which is involved in the interaction with the triphosphatase subunit of the capping enzyme, and with the RNAP II CTD (Cho et al, 1998). Additionally, ceg1-250 causes lethality when combined with certain kin28 mutants, including kin28-T17D (Rodriguez et al, 2000). The double mutant (ceg1-250 Δsub1) and parental strains (ceg1-250 and Δsub1) were streaked on rich medium plates. Double mutant cells grew indistinguishably from either single mutant alone (Figure 4). This negative result suggests that the SUB1–KIN28 interaction is not related to the kinase requirement for capping enzyme recruitment.
Figure 4.
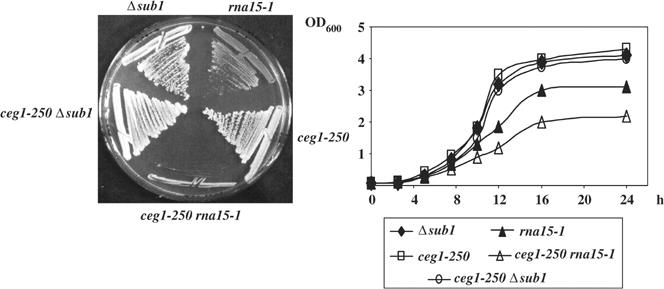
Rna15 and Ceg1 interact genetically. We crossed ceg1-250 strains (YSB625 or YSB491) with Δsub1 and rna15-1 strains. Single and double mutants from both crosses were streaked on rich medium plates at 25°C (left panel) and grown in liquid medium to perform the growth curve (right panel).
The partially overlapping but distinct interactions involving KIN28, CEG1, SUB1 and RNA15 suggested the possibility of an interaction between the genes encoding the capping enzyme, CEG1, and the polyadenylation factor, RNA15. Such an interaction would provide an intriguing genetic link between the processes of capping and 3′ end formation, as well as provide further support for Rna15 function at the promoter region, especially given that Ceg1 is specifically localized at the 5′ ends of genes and is released soon after transcription initiates (Schroeder et al, 2000). To investigate this, we crossed the rna15-1 mutant strain with the ceg1-250 mutant, and single and double mutants were grown on solid and liquid media at a permissive temperature of 25°C (Figure 4). Interestingly, ceg1-250, rna15-1 cells grew significantly slower than either single mutant, establishing a genetic interaction between capping and polyadenylation. This result is consistent with our ChIP data showing that Rna15 is present at the 5′ regions of transcribed genes.
The above data documenting allele-specific interactions between KIN28 and SUB1 provide evidence that Sub1 function is related to phosphorylation of the CTD by Kin28. If this is in fact the case, then SUB1 might show genetic interactions with genes encoding other factors that affect CTD phosphorylation. One such factor is the CTD phosphatase Fcp1. As we have shown here for Sub1, Fcp1 appears to remain chromatin associated along the length of transcribed genes (Cho et al, 2001), and to play a critical role in catalyzing the dynamic changes in CTD phosphorylation that occur during transcription elongation (Cho et al, 1999; Kobor et al, 1999; Lehman and Dahmus, 2000; Marshall and Dahmus, 2000).
In light of the above, we set out to determine whether we could detect a genetic interaction between SUB1 and FCP1. We generated two fcp1 Δsub1 double mutant strains by crossing the Δsub1 strain with two different fcp1 mutants, fcp1-1, with a mutation (R250A, P251A) in a region required for phosphatase activity, and fcp1-3, which encodes a protein lacking residues 2–134, which are not required for phosphatase activity (Kobor et al, 1999). Figure 5A shows a schematic of Fcp1 (top panel) and the growth phenotypes of the fcp1 mutants analyzed (bottom panel). The fcp1-1 mutation caused slow growth at 25°C and lethality at 37°C, while fcp1-3 mutant cells grew at both temperatures (Figure 5A). We tested the growth phenotypes of the double mutants and corresponding parental strains (fcp1-1 or fcp1-3 and Δsub1) on rich medium at 25 and/or 37°C (Figure 5B and C, respectively). We also performed growth curves on rich medium for FCP1, fcp1-1 and fcp1-1Δsub1 strains at 25°C (Figure 5B) and FCP1, fcp1-3 and fcp1-3 Δsub1 strains at 37°C (data not shown). Deletion of SUB1 in an fcp1-1 mutant significantly enhanced the slow growth phenotype at 25°C, but had no effect on the fcp1-3 mutant at any temperature. These results indicate an allele-specific interaction between FCP1 and SUB1 that, as with the kin28 mutants, correlates with effects on CTD phosphorylation. Together, our findings raise the possibility that Sub1 functions to facilitate RNAP II elongation by influencing CTD kinase and/or phosphatase activity.
Figure 5.
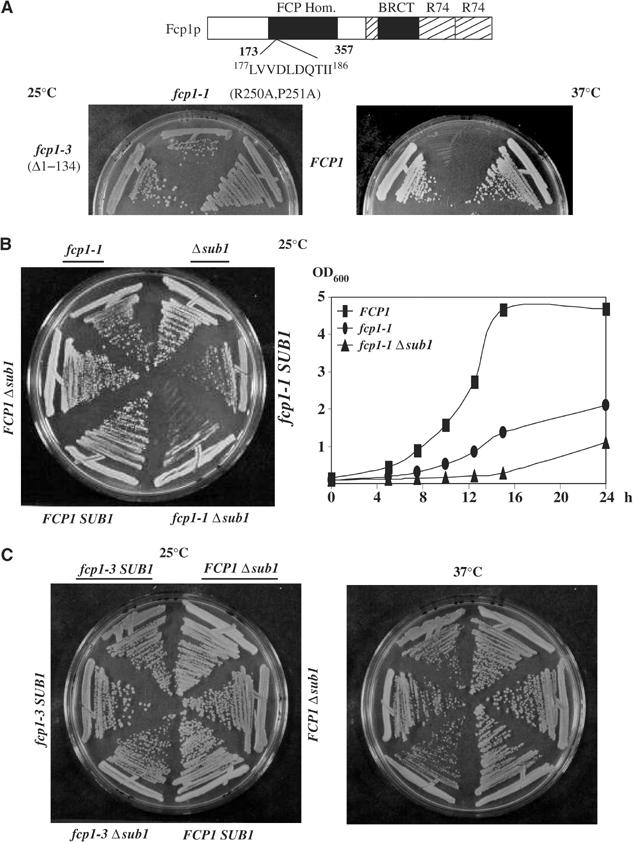
SUB1 deletion increases the slow growth defect of fcp1-1 mutant cells. (A) The upper panel is a schematic representation of Fcp1, showing the phosphatase motif (177 LVVDLDQTII 186) inside the FCP homology region (FCP Hom), the carboxy-terminal domains involved in the interaction with other components of the transcriptional machinery (TFIIB, TFIIF) (represented as R74) and the BRCT domain (Kobor et al, 2000). The lower panel shows the growth defects on rich medium for the fcp1 mutants (fcp1-1 and fcp1-3) at 25 and 37°C; Fcp1 mutated residues are indicated. (B, C) Two fcp1Δsub1 double mutant strains were generated by crossing a Δsub1 strain with either fcp1-1 strain (YMK20) or fcp1-3 (YMK28). Growth of double mutants and the parental strains (fcp1-1 or fcp1-3 and Δsub1) was tested by streaking them on rich medium at 25°C (B) or at 25 and 37°C (C).
Sub1 is a regulator of Fcp1 and influences RNAP II accumulation
The above genetic interactions involving SUB1 and genes encoding Kin28 and Fcp1 suggest that a function of Sub1 may involve modulating CTD phosphorylation. One possibility is that Sub1 helps maintain appropriate levels of one or more of these proteins. To test this, we prepared lysates from SUB1 and Δsub1 cells and performed Western blots with anti-RNAP II large subunit (LS), -Fcp1 and -HA (to detect Kin28-HA present in this strain) antibodies, as well as control antibodies (Figure 6A). No effect on Kin28-HA levels was detected. RNAP II LS levels were reduced very slightly, but more strikingly, Fcp1 accumulation decreased significantly. This was not due to an effect of SUB1 deletion on FCP1 transcription, as Northern blotting revealed equivalent levels of FCP1 mRNA in the presence and absence of Sub1 (Figure 6B).
Figure 6.
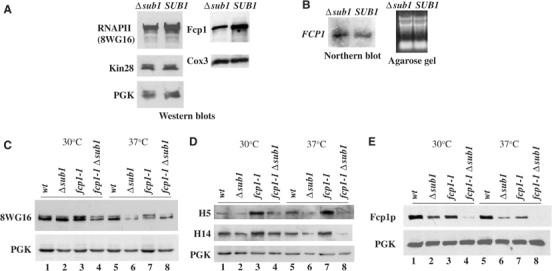
Sub1 is a regulator of Fcp1 and influences RNAP II accumulation. (A) Extracts prepared from isogenic Δsub1 and wild-type (SUB1) cells, YOC07 and YOC06 respectively, were subjected to Western blot and probed with anti-RNAP II, anti-Fcp1 and anti-HA (to detect Kin28p) antibodies. PGK and Cox3 proteins were used as loading controls. (B) Northern blot analysis of total RNAs prepared from Δsub1 and SUB1 cells (left panel), and the corresponding agarose gel showing equal amounts of RNA loaded (right panel). (C, D) Western blots of samples from four spores (same as those used in Figure 5B), grown at 30°C and then for 1 h at 37°C, were probed with three different anti-RNAPII CTD antibodies, 8WG16, H14 and H5 (as indicated). (E) Same samples as in panels C and D, but probed with anti-Fcp1 and anti-PGK (control) antibodies.
To investigate further the effects of Sub1 on RNAP II and Fcp1, we examined in more detail the status of RNAP II, and phosphorylation of the CTD, in wild-type cells and in Δsub1, fcp1-1 and Δsub1 fcp1-1 mutant cells. We utilized for Western blots three antibodies that recognize distinct epitopes in the CTD: 8WG16 recognizes an unphosphorylated epitope and thus detects the IIa isoform, but also recognizes IIo (Thompson et al, 1990; Patturajan et al, 1998), while the monoclonals H5 and H4 recognize preferentially phosphorylated ser 2- and ser 5-containing epitopes, respectively (Bregman et al, 1995; Patturajan et al, 1998; but see Jones et al, 2004). At 30°C, SUB1 deletion alone had little effect on total RNAP II levels or phosphorylation (Figure 6C and D, compare lanes 1 and 2). The fcp1-1 mutation also had no detectable effect on total RNAP II levels, but resulted in significant increases in H5 and H14 reactivity, suggesting enhanced phosphorylation of both ser 2 and ser 5 residues (Figure 6C and D, compare lanes 1 and 3). Although H5 reactivity (i.e., ser 2 phosphorylation) showed the greatest increase, these results suggest that Fcp1 can dephosphorylate both ser 2 and ser 5 (see Cho et al, 2001). The Δsub1 fcp1-1 double mutant (lane 4) had a striking effect: the amount of the IIa subunit was significantly reduced compared to the wild-type strain as well as both single mutants, while the levels of hyperphosphorylation due to the fcp1-1 mutation were reduced to wild-type levels. The results at 37°C (Figure 6C and D) were similar, although here Δsub1 alone revealed a small effect on both IIo and IIa accumulation (lane 6). SUB1 deletion entirely suppressed the hyperphosphorylation induced by the fcp1-1 mutation (lanes 7 and 8). The results indicate that Sub1 can influence accumulation and phosphorylation of RNAP II.
We also examined levels of Fcp1 in these strains (Figure 6E). SUB1 deletion reduced accumulation at both 30 and 37°C, somewhat more so than did the fcp1-1 mutation, previously shown to reduce Fcp1 accumulation by Kobor et al (1999) (lanes 1–3 and 5–7). Strikingly, Fcp1 levels were dramatically reduced in the double mutant cells (lanes 4 and 8), becoming undetectable at 37°C. These results indicate that a function of Sub1 is to facilitate accumulation of Fcp1, likely by directly or indirectly increasing its stability. It is noteworthy however that this is not sufficient to explain the effects of SUB1 deletion on RNAP II phosphorylation (see Discussion).
SUB1 deletion reduces RNAP II and Fcp1 crosslinking to promoter and coding regions
The above data provide evidence that Sub1 is required for maintaining optimal levels of Fcp1 and to a lesser extent RNAP II. To determine how this might reflect the presence of these factors on transcribed genes, we carried out ChIP experiments with antibodies against Fcp1 or RNAP II utilizing wild-type and Δsub1 isogenic strains. Fcp1 crosslinking was significantly reduced in the Δsub1 strain, as shown here on the ACT1 and ADH1 genes, with comparable reductions at both 5′ and 3′ ends of the gene (Figure 7A). When total RNAP II levels were measured (with 8WG16), crosslinking was also reduced, on both ACT1 and ADH1 (Figure 7B). Importantly, reductions were more severe, by roughly threefold, toward the 3′ end of each gene, consistent with impaired transcription elongation. However, when phosphorylated RNAP II was measured (with H5), an increase (∼2.5-fold) was detected on both genes (Figure 7C), indicating that elongating RNAP II is hyperphosphorylated in the absence of Sub1. Together, these results provide evidence that Sub1 is required for optimal transcription elongation, and that this is due, at least in part, to stabilization of the CTD phosphatase Fcp1.
Figure 7.
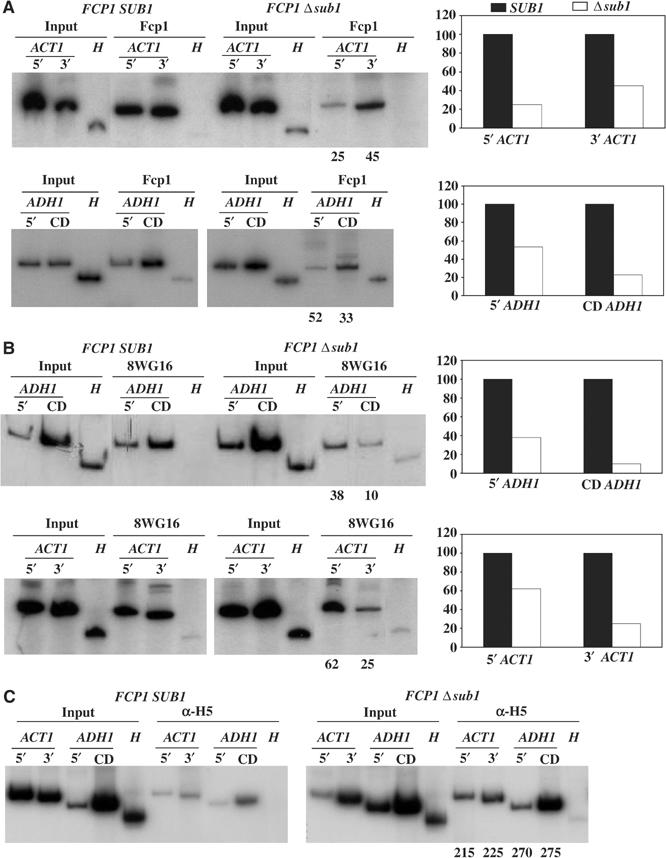
SUB1 deletion affects RNAP II and Fcp1 crosslinking to promoter and coding regions. ChIP analysis of Fcp1 (A) and RNAP II (B, C) in SUB1 and Δsub1 isogenic cells, using anti-Fcp1 (A), 8WG16 (B) or H5 (C) antibodies. PCR primers for ACT1 and ADH1 were the same as those used in Figure 1. Numbers under gels and/or graphs compare relative crosslinking of RNAP II or Fcp1 to the 5′ and 3′ of the ACT1 gene and 5′ and coding region (CD) of the ADH1 gene in Δsub1 cells relative to wild-type (SUB1) cells, taken as 100.
Discussion
The data we have presented here provide new insights into how factors initially described as participating in disparate aspects of gene expression can function together in unexpected ways. Specifically, we have provided considerable support for the idea that polyadenylation factors, such as Rna15, can stably associate with chromatin at or near gene promoters and remain associated along the length of transcribed genes. Our data strengthen the case that this is an important, evolutionarily conserved aspect of gene expression. Our findings that the transcriptional coactivator Sub1 is important for this to occur optimally strengthen the significance of the Rna15/Sub1 interaction we described previously. Our data also provide considerable support for our hypothesis that Sub1 functions to prevent premature transcription termination. The presence of Sub1 along the length of transcribed genes indicates a direct interaction between Sub1 and the elongating RNAP II complex, and the ability of Sub1 to interact with both a CTD kinase and phosphatase suggests a novel function: to regulate CTD modifiers during transcription.
Considerable data have accumulated over the last few years addressing the question of whether polyadenylation factors associate with transcription factors at promoters, and/or with elongating RNAP II. Perhaps the earliest evidence supporting this came from experiments utilizing human cell extracts, which showed that the polyadenylation factor CPSF could be immunopurified in a relatively stable complex with the GTF TFIID, and then in reconstituted transcription reactions was transferred to elongating RNAP II (Dantonel et al, 1997). This was consistent with other experiments indicating a direct role for the CTD in 3′ end formation (McCracken et al, 1997; Hirose and Manley, 1998; Additionally, immunostaining experiments examining lampbrush chromosomes in frog oocytes (Gall et al, 1999) and polytene chromosomes in flies (Osheim et al, 2002) revealed the presence of polyadenylation factors along the entire length of transcribed genes, coincident with RNAP II. To our knowledge, ChIP experiments to localize 3′ end factors in metazoan organisms have not been reported.
Experiments in yeast have also provided support for the presence of polyadenylation factors at the 5′ ends of genes, although the evidence has not been conclusive. Intriguingly, immunopurification of yeast TFIID also found CPSF polypeptides associated with it (Sanders et al, 2002). However, in these experiments, antibodies directed against multiple TFIID subunits were utilized, and the CPSF subunits were detected with only one. ChIP experiments have produced somewhat conflicting results. Initial experiments using antibodies directed against several different polyadenylation factors failed to detect chromatin crosslinking (Komarnitsky et al, 2000). However, Licatalosi et al (2002), also using antibodies directed against specific polyadenylation factors (Rna15 was not tested), detected crosslinking at both the 5′ and 3′ ends of genes, with somewhat stronger signals toward the 3′ ends. The authors suggested from this data that polyadenylation factors begin to be recruited at or near promoters and then throughout the length of the transcribed gene. More recently, Kim et al (2004) performed a thorough analysis of the association of a number of TAP-tagged polyadenylation factors (including Rna15) along the length of several genes. They observed low to modest levels of crosslinking at promoters and along the lengths of genes until the 3′ processing signal was reached, at which point dramatic increases in crosslinking were detected. The authors suggested that while their findings did not exclude recruitment of polyadenylation factors to initiating and/or elongating RNAP II complexes, a simpler explanation was that such factors were recruited only at the 3′ ends of nascent transcripts.
Our results provide strong support for a model in which certain polyadenylation factors, for example, Rna15, are in fact recruited to promoters and remain chromatin associated along the length of at least a subset of transcribed genes. By using HA-tagged proteins, we provided evidence not only that Rna15 is present at and downstream of promoters, but also, as judged by crosslinking efficiency, at levels at least comparable to RNAP II (i.e., Rpb3) itself. In each case, levels significantly above background crosslinking were detected. Notably, these values were only two- to three-fold greater than those observed by Kim et al (2004) and differences could be attributed to any number of variations in experimental conditions, for example, use of TAP versus HA epitope tags. What was most striking however was that these values appear small relative to the signals detected at and just downstream of 3′ end sites, which were as high as 60- to 70-fold above background (Kim et al, 2004; note that our experiments were not designed to detect interactions at the polyadenylation signals). These values are unusually high for ChIP assays and, rather than suggesting that the more typical upstream crosslinking may not be indicative of significant interactions, point to a major change in the polyadenylation complex that allows especially efficient detection by ChIP assays. This could include recruitment of additional factors necessary for 3′ processing that stabilize the complex, and/or dissociation of other factors present during elongation that cause the RNA processing factors to become more accessible. These changes likely involve the switch from ser 5 to ser 2 CTD phosphorylation that occurs during elongation. Ser 2 phosphorylation is important for one polyadenylation factor, Pcf11, to bind the CTD (Licatalosi et al, 2002; Meinhart and Cramer, 2004), and reductions in ser 2 phosphorylation both decrease 3′ end formation in yeast and flies (Ahn et al, 2004; Ni et al, 2004) and prevent the high-efficiency crosslinking at the 3′ processing site (Ahn et al, 2004).
Our data have provided evidence that Sub1 is also present across the length of several transcribed genes. Although consistent with our proposal that Sub1 can function as an antitermination factor, they differ from a recent suggestion that Sub1 may be present only at the 5′ and 3′ ends of genes (Nedea et al, 2003). This conclusion was based on an observed decreased ChIP signal at a single site within the PYK1 gene, and led to the suggestion that Sub1 has distinct functions at the 5′ and 3′ ends of genes (Nedea et al, 2003; Kim et al, 2004). This drop off in crosslinking could reflect a specific property of the PYK1 gene or the primers utilized in PCR. Nonetheless, it is in fact likely that Sub1/PC4 has multiple functions in gene expression (and perhaps other nuclear processes; see Wang et al, 2004), including its well-studied transcriptional coactivator function. Whether or not Sub1 has a distinct function at the 3′ ends of genes remains to be determined.
How does Sub1 function to enhance transcription elongation? The genetic and biochemical data presented here provide evidence that this involves interaction with CTD modifiers such as Kin28 and Fcp1. Most striking was the significant decrease in Fcp1 accumulation brought about by SUB1 deletion. This resulted in enhanced phosphorylation of RNAP II on two genes, which likely contributed to the observed elongation defects. But the fact that total RNAP IIO levels were not significantly enhanced suggests a complex role for Sub1 in influencing CTD modification, and we propose a model whereby Sub1 influences both CTD kinase and phosphatase activity. Our experiments showed that the fcp1-1 mutation enhanced CTD phosphorylation, a not unexpected result based on previous studies with related Fcp1-deficient strains (Kobor et al, 1999). In the simplest scenario, the further loss of Fcp1 activity brought about SUB1 deletion would have been expected to further enhance CTD phosphorylation. But since the opposite was observed, that is, CTD phosphorylation actually decreased, we propose that Sub1 affects CTD phosphorylation in multiple ways. An intriguing possibility would be that Sub1 might influence Ssu72 phosphatase activity. Although Sub1 and Ssu72 are related in several ways, there is no evidence of an interaction, genetic or biochemical, between them (e.g., Wu et al, 1999). But it is noteworthy that these two proteins, initially discovered by their similar interactions with TFIIB, have now each been shown to have activities related to both mRNA 3′ end formation and CTD phosphorylation. Another possibility is that Sub1 can block CTD kinase activity, and biochemical data in mammals in fact support this possibility: PC4 has been shown to be a substrate-specific inhibitor of CTD phosphorylation by CDKs, including CDK7/Kin28 (Schang et al, 2000). While additional studies are necessary to elucidate the exact mechanism(s), our work has provided strong evidence that Sub1 function involves control of CTD modifiers.
The ability of Sub1 to function as a transcription antiterminator likely involves more than its ability to influence CTD phosphorylation. We showed previously that Sub1 interacts with a region of Rna15 that appears to constitute a termination domain (Aranda and Proudfoot, 2001), and presented evidence that Sub1 downregulates this activity (Calvo and Manley, 2001). Although the underlying mechanism remains to be determined, this likely constitutes an important aspect of Sub1 antitermination activity. However, it is likely that Sub1 enhances elongation by another mechanism independent of CTD phosphorylation, which nonetheless involves Fcp1. Specifically, previous studies by Reinberg and colleagues, involving both biochemical experiments with human transcription factors and genetic experiments in yeast, indicate that Fcp1 possesses a positive elongation function that is independent of its phosphatase activity (Cho et al, 1999; Mandal et al, 2002). The reduced levels of Fcp1 detected in the absence of Sub1 provide at least a partial explanation for the observed defect in transcription elongation.
Materials and methods
Yeast strains
Genotypes of yeast strains used are as follows:
Δsub1 MATa or α ura3 trp1 ade2 leu2 his3 Δsub1∷URA3 (Calvo and Manley, 2001)
rna15-1 MATa ura3 trp1 ade2 leu2 his3 rna15-1 (Calvo and Manley, 2001)
YOC03 MATα sub1Δ∷URA3 [HASUB1 TRP CEN] (this study)
YOC04a MATa ura3 trp1 ade2 leu2 his3 [HARNA15 LEU2 CEN] (this study)
YOC04b MATa rna15-1 [HARNA15 LEU2 CEN] (this study)
YOC05 MATa rna15-1 sub1Δ∷URA3 [HARNA15 LEU2 CEN] (this study)
YOC06 and YOC07 strains are SUB1KIN28HA and Δsub1KIN28HA spores, respectively, constructed by crossing YSB756 (Komarnitsky et al, 2000) with Δsub1 (Calvo and Manley, 2001), and dissecting tetrads
YSB625 MATα ura3 leu2 trp1 his3 lys2 can1 ceg1-250 (Rodriguez et al, 2000)
YSB491 MATa ura3 leu2 trp1 his3 ade2 ade3 can1 ceg1-250 (Cho et al, 1998)
YF131 MATa, ura3 leu2 his3Δ200 rpb3-(HA)3∷LEU2 (Komarnitsky et al, 2000)
YSB756 MATα ura3 leu2 trp1 his3 ade2 ade3 can1 kin28Δ∷LEU2 [HAKIN28 TRP CEN] (Komarnitsky et al, 2000)
YSB592 MATα ura3 leu2 trp1 his3 ade2 ade3 can1 kin28Δ∷LEU2 [HAkin28-T17D TRP CEN] (Komarnitsky et al, 2000)
YSB595 MATα ura3 leu2 trp1 his3 ade2 ade3 can1 kin28Δ∷LEU2 [HAkin28-T162V TRP CEN] (Rodriguez et al, 2000)
YSB609 MATα ura3 leu2 trp1 his3 ade2 ade3 can1 kin28Δ∷LEU2 [HAkin28-K36A TRP CEN] (Rodriguez et al, 2000)
YMK18 MATα ura3 leu2 trp1 his3 ade2 can1 fcp1Δ∷LEU2 [FCP1 URA CEN] (Kobor et al, 1999)
YMK20 MATα ura3 leu2 trp1 his3 ade2 can1 fcp1Δ∷LEU2 [fcp1-1 (R250A, P251A) TRP CEN] (Kobor et al, 1999)
YMK28 MATα ura3 leu2 trp1 his3 ade2 can1 fcp1Δ∷LEU2 [fcp1-3 (Δ2-134) TRP CEN] (Kobor et al, 1999).
We also generated new yeast strains by crossing rna15-1 and/or Δsub1 mutants with ceg1-250, fcp1-1,3 or kin28-T17D, kin28-K36A and kin28-T162A mutants (see Results for details).
Yeast strain constructions and other genetic manipulations were performed by standard procedures (Guthrie and Fink, 1991). To obtain double mutants, the sub1 strain was crossed with the appropriate mutant and the corresponding zygotes were sporulated to obtain tetrads. Spores were dissected in selective medium to differentiate wild type, single and double mutants. Strains were grown at 25 and/or 37°C in either YPD (rich medium 1% yeast extract, 2% bactotryptone, 2% glucose) or YNB (minimal medium 0.67% yeast nitrogen base without amino acids, 2% glucose or 2% galactose/1% raffinose) supplemented with appropriate amino acids when necessary. Strains were transformed by the modified lithium acetate technique (Gietz et al, 1992) and plasmids were maintained by growth in selective medium.
Western blot analyses
Whole-cell extracts were prepared for Western blot as described previously (Patturajan et al, 1998). For experiments requiring heat shock, cells were grown at 30°C until OD600 reached 0.8. The culture was split in two: half was diluted 1:1 with fresh prewarmed medium and then incubated at 37°C for 60 min, and the other half was diluted 1:1 using fresh medium at room temperature and then incubated at 30°C for 60 min. Blots were probed with different antibodies for detection of phosphorylated and nonphosphorylated RNAP IIo and a: the monoclonal antibody 8WG16 that recognizes an unphosphorylated CTD epitope, and the monoclonals H14 and H5 directed against phosphorylated CTD Ser5 and Ser2 epitopes, respectively (BabCO Covance). Monoclonal anti-HA (12CA5) was from Roche), monoclonal anti-Pgk was from Molecular Probes, and monoclonal anti-Cox3 and polyclonal anti-Fcp1 were gifts from Dr T Barrientos and J Greenblatt, respectively.
Northern blot analyses
Total RNA was prepared as described (Schmitt et al, 1990); 10 μg was separated on a 1.2% agarose gel and then transferred to a nylon membrane. A 1.3 kb FCP1 fragment amplified by PCR from pMK86 plasmid bearing FCP1 wt gene (Kobor et al, 1999) was used as probe.
Chromatin immunoprecipitations
We performed ChIPs as previously described (Komarnitsky et al, 2000; Schroeder et al, 2000). All yeast strains were grown in minimal medium at 30°C, with the exception of rna15-1 HA-RNA15, which was grown at 37°C, to an OD600 of 0.5–1.0. We used anti-HA antibody to immunoprecipitate HA-tagged Rpb3, Kin28, Rna15 and Sub1; 8WG16 or H5 to immunoprecipitate RNAP II; and anti-Fcp1 to immunoprecipitate Fcp1. Radiolabeled PCR products were analyzed on agarose gels (routinely 25% of input and 100% of immunoprecipitate) and quantified by PhosphorImager. Numbers below gels were calculated essentially as described previously (Kim et al, 2004): signal intensities of immunoprecipitated PCR products were compared to those of the total input, and the ratio of values from each PCR product of transcribed genes to that of the nontranscribed HMR gene or an intergenic region on chromosome V was calculated. Therefore, numbers obtained are x-fold over HMR or V (background signal). Numbers on the y-axis of graphs are detailed in the corresponding figure legend. All ChIP assays were repeated 3–8 times, and in each case typical results are shown.
Acknowledgments
We thank S Buratowski, J Greenblatt, T Barrientos and A Tzagoloff for yeast strains, plasmids and antibodies; T Barrientos and A Tzagoloff for advice in yeast genetics; and J Viardell and A Pandiella for support and advice. This work was supported by grants from the NIH to JLM. OC was supported in part by a Ramon y Cajal grant from the Spanish Ministerio de Ciencia y Tecnologia.
References
- Ahn SH, Kim M, Buratowski S (2004) Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol Cell 13: 67–76 [DOI] [PubMed] [Google Scholar]
- Aranda A, Proudfoot NJ (2001) Transcriptional termination factors for RNA polymerase II in yeast. Mol Cell 7: 1003–1011 [DOI] [PubMed] [Google Scholar]
- Bensaude O, Bonnet F, Casse C, Dubois MF, Nguyen VT, Palancade B (1999) Regulated phosphorylation of the RNA polymerase II C-terminal domain (CTD). Biochem Cell Biol 77: 249–255 [PubMed] [Google Scholar]
- Bloch JC, Perrin F, Lacroute F (1978) Yeast temperature-sensitive mutants specifically impaired in processing of poly(A)-containing RNAs. Mol Gen Genet 165: 123–127 [DOI] [PubMed] [Google Scholar]
- Bregman DB, Du L, van der Zee S, Warren SL (1995) Transcription-dependent redistribution of the large subunit of RNA polymerase II to discrete nuclear domains. J Cell Biol 129: 287–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo O, Manley JL (2001) Evolutionarily conserved interaction between CstF-64 and PC4 links transcription, polyadenylation, and termination. Mol Cell 7: 1013–1023 [DOI] [PubMed] [Google Scholar]
- Calvo O, Manley JL (2003) Strange bedfellows: polyadenylation factors at the promoter. Genes Dev 17: 1321–1327 [DOI] [PubMed] [Google Scholar]
- Cho EJ, Kobor MS, Kim M, Greenblatt J, Buratowski S (2001) Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes Dev 15: 3319–3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho EJ, Rodriguez CR, Takagi T, Buratowski S (1998) Allosteric interactions between capping enzyme subunits and the RNA polymerase II carboxy-terminal domain. Genes Dev 12: 3482–3487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Kim TK, Mancebo H, Lane WS, Flores O, Reinberg D (1999) A protein phosphatase functions to recycle RNA polymerase II. Genes Dev 13: 1540–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corden JL (1990) Tails of RNA polymerase II. Trends Biochem Sci 15: 383–387 [DOI] [PubMed] [Google Scholar]
- Corden JL, Cadena DL, Ahearn JM Jr, Dahmus ME (1985) A unique structure at the carboxyl terminus of the largest subunit of eukaryotic RNA polymerase II. Proc Natl Acad Sci USA 82: 7934–7938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantonel JC, Murthy KG, Manley JL, Tora L (1997) Transcription factor TFIID recruits factor CPSF for formation of 3′ end of mRNA. Nature 389: 399–402 [DOI] [PubMed] [Google Scholar]
- Dichtl B, Blank D, Ohnacker M, Friedlein A, Roeder D, Langen H, Keller W (2002) A role for SSU72 in balancing RNA polymerase II transcription elongation and termination. Mol Cell 10: 1139–1150 [DOI] [PubMed] [Google Scholar]
- Espinoza FH, Farrell A, Nourse JL, Chamberlin HM, Gilead O, Morgan DO (1998) Cak1 is required for Kin28 phosphorylation and activation in vivo. Mol Cell Biol 18: 6365–6373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feaver WJ, Svejstrup JQ, Henry NL, Kornberg RD (1994) Relationship of CDK-activating kinase and RNA polymerase II CTD kinase TFIIH/TFIIK. Cell 79: 1103–1109 [DOI] [PubMed] [Google Scholar]
- Fresco LD, Buratowski S (1996) Conditional mutants of the yeast mRNA capping enzyme show that the cap enhances, but is not required for, mRNA splicing. RNA 2: 584–596 [PMC free article] [PubMed] [Google Scholar]
- Gall JG, Bellini M, Wu Z, Murphy C (1999) Assembly of the nuclear transcription and processing machinery: Cajal bodies (coiled bodies) and transcriptosomes. Mol Biol Cell 10: 4385–4402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz D, St. Jean A, Woods RA, Schiestl RH (1992) Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res 20: 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C, Fink GR (1991) Guide to yeast genetics and molecular biology. Methods Enzymol 194: 1–495 [PubMed] [Google Scholar]
- Hengartner CJ, Myer VE, Liao SM, Wilson CJ, Koh SS, Young RA (1998) Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Mol Cell 2: 43–53 [DOI] [PubMed] [Google Scholar]
- Henry NL, Bushnell DA, Kornberg RD (1996) A yeast transcriptional stimulatory protein similar to human PC4. J Biol Chem 271: 21842–21847 [DOI] [PubMed] [Google Scholar]
- He X, Khan AU, Cheng H, Pappas DL Jr, Hampsey M, Moore CL (2003) Functional interactions between the transcription and mRNA 3′ end processing machineries mediated by Ssu72 and Sub1. Genes Dev 17: 1030–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose Y, Manley JL (1998) RNA polymerase II is an essential mRNA polyadenylation factor. Nature 395: 93–96 [DOI] [PubMed] [Google Scholar]
- Hirose Y, Manley JL (2000) RNA polymerase II and the integration of nuclear events. Genes Dev 14: 1415–1429 [PubMed] [Google Scholar]
- Jones JC, Phatnani HP, Haystead TA, MacDonald JA, Alam SM, Greenleaf AL (2004) C-terminal repeat domain kinase I phosphorylates Ser2 and Ser5 of RNA polymerase II C-terminal domain repeats. J Biol Chem 279: 24957–24964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Ahn SH, Krogan NJ, Greenblatt JF, Buratowski S (2004) Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J 23: 354–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmelman J, Kaldis P, Hengartner CJ, Laff GM, Koh SS, Young RA, Solomon MJ (1999) Activating phosphorylation of the Kin28p subunit of yeast TFIIH by Cak1p. Mol Cell Biol 19: 4774–4787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knaus R, Pollock R, Guarente L (1996) Yeast SUB1 is a suppressor of TFIIB mutations and has homology to the human co-activator PC4. EMBO J 15: 1933–1940 [PMC free article] [PubMed] [Google Scholar]
- Kobor MS, Archambault J, Lester W, Holstege FC, Gileadi O, Jansma DB, Jennings EG, Kouyoumdjian F, Davidson AR, Young RA, Greenblatt J (1999) An unusual eukaryotic protein phosphatase required for transcription by RNA polymerase II and CTD dephosphorylation in S. cerevisiae. Mol Cell 4: 55–62 [DOI] [PubMed] [Google Scholar]
- Kobor MS, Greenblatt J (2002) Regulation of transcription elongation by phosphorylation. Biochim Biophys Acta 1577: 261–275 [DOI] [PubMed] [Google Scholar]
- Kobor MS, Simon LD, Omichinski J, Zhong G, Archambault J, Greenblatt J (2000) A motif shared by TFIIF and TFIIB mediates their interaction with the RNA polymerase II carboxy-terminal domain phosphatase Fcp1p in Saccharomyces cerevisiae. Mol Cell Biol 20: 7438–7449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarnitsky P, Cho EJ, Buratowski S (2000) Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev 14: 2452–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy S, He X, Reyes-Reyes M, Moore C, Hampsey M (2004) Ssu72 is an RNA polymerase II CTD phosphatase. Mol Cell 14: 387–394 [DOI] [PubMed] [Google Scholar]
- Lehman AL, Dahmus ME (2000) The sensitivity of RNA polymerase II in elongation complexes to C-terminal domain phosphatase: a protein phosphatase functions to recycle RNA polymerase II. J Biol Chem 275: 14923–14932 [DOI] [PubMed] [Google Scholar]
- Licatalosi DD, Geiger G, Minet M, Schroeder S, Cilli K, McNeil JB, Bentley DL (2002) Functional interaction of yeast pre-mRNA 3′ end processing factors with RNA polymerase II. Mol Cell 9: 1101–1111 [DOI] [PubMed] [Google Scholar]
- Mandal S, Cho C, Kim S, Cabane K, Reinberg D (2002) FCP1, a phosphatase specific for the heptapeptide repeat of the largest subunit of RNA polymerase II, stimulates transcription elongation. Mol Cell Biol 22: 7543–7552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T, Reed R (2002) An extensive network of coupling among gene expression machines. Nature 4: 499–506 [DOI] [PubMed] [Google Scholar]
- Marshall NF, Dahmus ME (2000) C-terminal domain phosphatase sensitivity of RNA polymerase II in early elongation complexes on the HIV-1 and adenovirus 2 major late templates. J Biol Chem 275: 32430–32437 [DOI] [PubMed] [Google Scholar]
- McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson SD, Wickens M, Bentley DL (1997) The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature 385: 357–361 [DOI] [PubMed] [Google Scholar]
- Meinhart A, Cramer P (2004) Recognition of RNA polymerase II carboxy-terminal domain by 3″-RNA-processing factors. Nature 430: 223–226 [DOI] [PubMed] [Google Scholar]
- Minvielle-Sebastia L, Winsor B, Bonneaud N, Lacroute F (1991) Mutations in the yeast RNA14 and RNA15 genes result in an abnormal mRNA decay rate; sequence analysis reveals an RNA-binding domain in the RNA15 protein. Mol Cell Biol 11: 3075–3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedea E, He X, Kim M, Pootoolal J, Zhong G, Canadien V, Hughes T, Buratowski S, Moore CL, Greenblatt J (2003) Organization and function of APT, a subcomplex of the yeast cleavage and polyadenylation factor involved in the formation of mRNA and small nucleolar RNA 3′-ends. J Biol Chem 278: 33000–33010 [DOI] [PubMed] [Google Scholar]
- Ni Z, Schwartz BE, Werner J, Suarez JR, Lis JT (2004) Coordination of transcription, RNA processing, and surveillance by P-TEFb kinase on heat shock genes. Mol Cell 13: 55–65 [DOI] [PubMed] [Google Scholar]
- Osheim YN, Sikes ML, Beyer AL (2002) EM visualization of Pol II genes in Drosophila: most genes terminate without prior 3′ end cleavage of nascent transcripts. Chromosoma 111: 1–12 [DOI] [PubMed] [Google Scholar]
- Patturajan M, Schulte RJ, Sefton BM, Berezney R, Vincen M, Bensaude O, Warren SL, Corden JL (1998) Growth-related changes in phosphorylation of yeast RNA polymerase II. J Biol Chem 273: 4689–4694 [DOI] [PubMed] [Google Scholar]
- Prelich G (2002) RNA polymerase II carboxy-terminal domain kinases: emerging clues to their function. Eukaryot Cell 1: 153–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot NJ (2004) New perspectives on connecting messenger RNA 3′ end formation to transcription. Curr Opin Cell Biol 16: 272–278 [DOI] [PubMed] [Google Scholar]
- Proudfoot NJ, Furger A, Dye MJ (2002) Integrating mRNA processing with transcription. Cell 108: 501–512 [DOI] [PubMed] [Google Scholar]
- Rodriguez CR, Cho EJ, Keogh MC, Moore CL, Greenleaf AL, Buratowski S (2000) Kin28, the TFIIH-associated carboxy-terminal domain kinase, facilitates the recruitment of mRNA processing machinery to RNA polymerase II. Mol Cell Biol 20: 104–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SL, Garbett KA, Weil PA (2002) Molecular characterization of Saccharomyces cerevisiae TFIID. Mol Cell Biol 22: 6000–6013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schang LM, Hwang GJ, Dynlacht BD, Speicher DW, Bantly A, Schaffer PA, Shilatifard A, Ge H, Shiekhattar R (2000) Human PC4 is a substrate-specific inhibitor of RNA polymerase II phosphorylation. J Biol Chem 275: 6071–6074 [DOI] [PubMed] [Google Scholar]
- Schmitt ME, Brown TA, Trumpower BL (1990) A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res 18: 3091–3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder SC, Schwer B, Shuman S, Bentley D (2000) Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev 14: 2435–2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun ZW, Hampsey M (1996) Synthetic enhancement of a TFIIB defect by a mutation in SSU72, an essential yeast gene encoding a novel protein that affects transcription start site selection in vivo. Mol Cell Biol 16: 1557–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson NE, Aronson DB, Burgess RR (1990) Purification of eukaryotic RNA polymerase II by immunoaffinity chromatography. Elution of active enzyme with protein stabilizing agents from a polyol-responsive monoclonal antibody. J Biol Chem 265: 7069–7077 [PubMed] [Google Scholar]
- Valay JG, Simon M, Dubois MF, Bensaude O, Facca C, Faye G (1995) The KIN28 gene is required both for RNA polymerase II mediated transcription and phosphorylation of the Rpb1p CTD. J Mol Biol 249: 535–544 [DOI] [PubMed] [Google Scholar]
- Wang JY, Sarker AH, Cooper PK, Volkert MR (2004) The single-strand DNA binding activity of human PC4 prevents mutagenesis and killing by oxidative DNA damage. Mol Cell Biol 24: 6084–6093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West ML, Corden JL (1995) Construction and analysis of yeast RNA polymerase II CTD deletion and substitution mutations. Genetics 140: 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu WH, Pinto I, Chen BS, Hampsey M (1999) Mutational analysis of yeast TFIIB. A functional relationship between Ssu72 and Sub1/Tsp1 defined by allele-specific interactions with TFIIB. Genetics 153: 643–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorio DA, Bentley DL (2004) The link between mRNA processing and transcription: communication works both ways. Exp Cell Res 296: 91–97 [DOI] [PubMed] [Google Scholar]


