Abstract
Hsp70 chaperones mediate folding of proteins and prevent their misfolding and aggregation. We report here on a new kind of Hsp70 interacting protein in mitochondria, Hep1. Hep1 is a highly conserved protein present in virtually all eukaryotes. Deletion of HEP1 results in a severe growth defect. Cells lacking Hep1 are deficient in processes that need the function of mitochondrial Hsp70s, such as preprotein import and biogenesis of proteins containing FeS clusters. In the mitochondria of these cells, Hsp70s, Ssc1 and Ssq1 accumulate as insoluble aggregates. We show that it is the nucleotide-free form of mtHsp70 that has a high tendency to self-aggregate. This process is efficiently counteracted by Hep1. We conclude that Hep1 acts as a chaperone that is necessary and sufficient to prevent self-aggregation and to thereby maintain the function of the mitochondrial Hsp70 chaperones.
Keywords: chaperone, Hsp70, mitochondria, protein aggregation
Introduction
The chaperones of the Hsp70 family have a number of essential functions. They assist in the de novo folding of proteins, in the refolding of unfolded proteins; they prevent aggregation of misfolded proteins and promote disaggregation of aggregated proteins (Morimoto et al, 1994; Polissi et al, 1995; Bukau and Horwich, 1998; Frydman, 2001; Hartl and Hayer-Hartl, 2002; Young et al, 2004). Hsp70s play a role in the degradation of proteins, in regulating enzyme activities, and are required for the translocation of proteins across intracellular membranes (Hohfeld et al, 2001; Voos and Rottgers, 2002).
These various functions of Hsp70s rely on their ability to bind to unfolded segments of proteins in an ATP-dependent, reversible manner. All Hsp70s in prokaryotic and eukaryotic cells consist of an ATP-binding domain and a peptide-binding domain (PBD) (Hartl and Hayer-Hartl, 2002). The two domains cooperate and in doing so Hsp70s undergo substantial conformational changes. It is generally agreed upon that the PBD of Hsp70 in its ATP form has an open binding pocket which recognizes unfolded segments of polypeptides (Bukau and Horwich, 1998; Mayer et al, 2001; Hartl and Hayer-Hartl, 2002). Upon hydrolysis of ATP by Hsp70, the peptide-binding pocket closes so that the ADP form holds tight the substrate. For their various functions Hsp70s require the cooperation with a set of cochaperones which help in binding the polypeptide substrates and which support the ATP/ADP cycle (Bukau and Horwich, 1998; Kelley, 1998; Fan et al, 2003).
Mitochondria of yeast contain three different Hsp70s (Craig and Marszalek, 2002; Voos and Rottgers, 2002). MtHsp70/Ssc1p is by far the most abundant form. It is involved in the functions of Hsp70s listed above. Furthermore, it is a constituent of the import motor of the mitochondria, which brings proteins from the cytosol into the matrix and the inner membrane. MtHsp70 is an essential protein (Craig et al, 1989). There are two other, minor, forms of Hsp70 in the mitochondria, which are not essential for the viability of yeast cells, Ssq1 and Ecm10 (Schilke et al, 1996; Baumann et al, 2000). Ssq1 has a role in the biogenesis of proteins that contain FeS clusters (Lill and Kispal, 2000; Craig and Marszalek, 2002). The function of Ecm10 is still not clear.
In this report, we describe a new type of Hsp70 interacting protein, Hep1. Homologs of Hep1 are present in the mitochondria of virtually all eukaryotic cells. The mitochondrial matrix protein Hep1 interacts in a specific manner with mtHsp70 and inhibits the self-aggregation of mtHsp70 and Ssq1 in mitochondria. We suggest that Hep1 is required for the maintenance of mtHsp70 function.
Results
Hep1 forms a specific complex with mtHsp70
In a systematic approach, we searched for the interaction partners of mtHsp70. Mitochondria from a yeast strain that contains mtHsp70 carrying a His-tag at the C terminus were lysed in Triton X-100. The lysate was passed over an NiNTA column; the proteins retained were eluted with SDS-containing buffer and subjected to SDS–PAGE (Figure 1A). Besides large amounts of mtHsp70, a few additional bands were detected. These were analysed by mass spectrometry. As expected, the known interaction partners Mge1 and Tim44 were found. One of the proteins with an apparent molecular mass of 17 kDa was further analysed. The gene for this protein was found in the yeast genome and identified as reading frame YNL310c. This reading frame was recently shown to encode a mitochondrial protein whose depletion led to a reduced rate in mitochondrial preprotein import (Burri et al, 2004). We termed the open reading frame HEP1, for mtHsp70 escort protein, as the present study shows this to be the function of the protein. To confirm the identity and specificity of the interaction with mtHsp70, mitochondria from wild-type (WT) cells and from cells harbouring His-tagged mtHsp70 were lysed and binding to NiNTA beads was allowed to occur. Supernatant (SN) and bound material were analysed by SDS–PAGE and immunoblotting (Figure 1B). Hep1 was recovered specifically, together with His-tagged mtHsp70 on the affinity beads, whereas the abundant mitochondrial protein AAC was not detected in the bound material.
Figure 1.
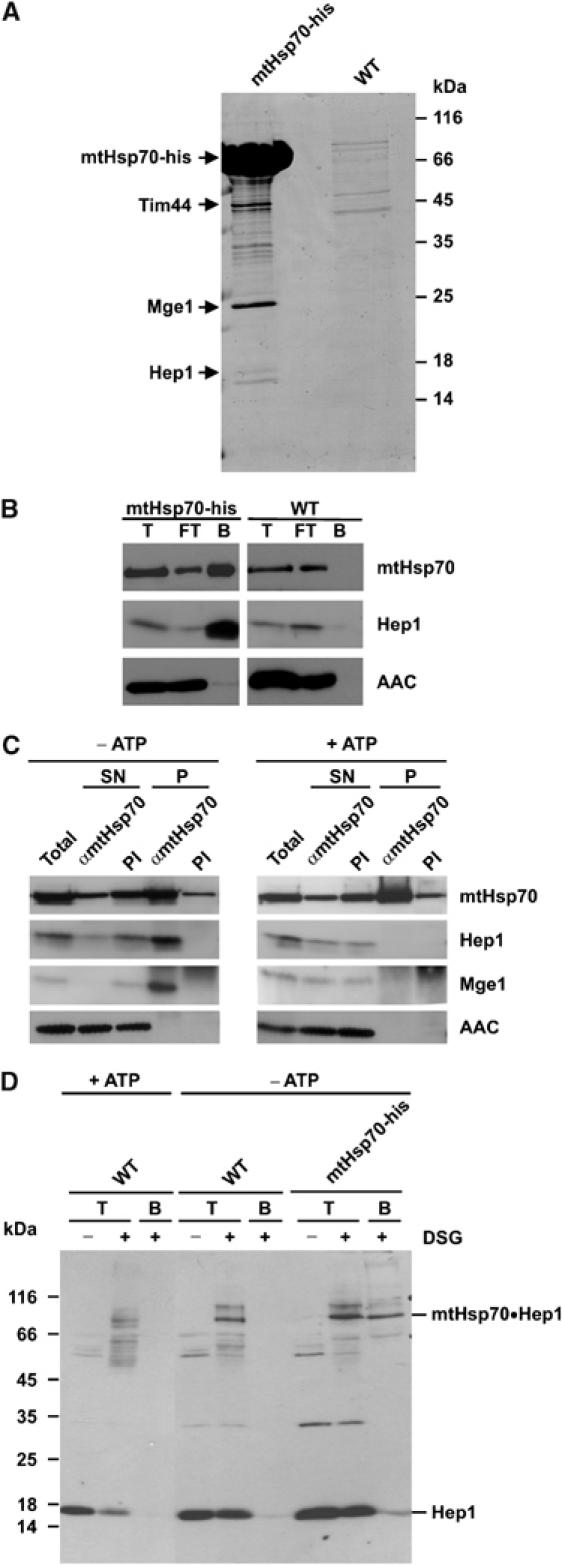
Hep1 interacts with mtHsp70. (A, B) Co-isolation of Hep1 with His-tagged mtHsp70. Isolated mitochondria from WT or from a yeast strain expressing Hsp70 with a hexahistidinyl tag were solubilized in Triton X-100. (A) Mitochondrial lysates were passed over a NiNTA column. Bound material was eluted and analysed by SDS–PAGE and Coomassie staining. Tim44, Mge1 and Hep1 were identified by mass spectrometry. (B) Mitochondrial lysates were incubated with NiNTA beads and the beads were isolated. Total proteins, T (10%), unbound material, FT (10%) and bound material, B (100%), were analysed by SDS–PAGE and immunodecoration for the indicated proteins. (C) Co-immunoprecipitation of Hep1 with mtHsp70. WT mitochondria were either depleted of ATP by incubation with apyrase and oligomycin (−ATP) or incubated with an ATP regenerating system (+ATP) for 10 min. Following lysis in digitonin, mitochondrial proteins were incubated with antibodies against mtHsp70 or with preimmune serum (PI). Immunoprecipitates were analysed by SDS–PAGE and immunodecoration with antibodies against the indicated proteins. The total and supernatant (SN) represent 20% of the material of the precipitate (P). (D) Hep1 can be crosslinked to mtHsp70. Mitochondria from WT cells and cells expressing His-tagged mtHsp70 were treated as in (C) and then subjected to crosslinking with DSG. Control samples without crosslinking reagent and aliquots after crosslinking were directly subjected to SDS–PAGE (T); the other aliquots were solubilized and incubated with NiNTA beads. Bound material was eluted and subjected to SDS–PAGE. Proteins were analysed by Western blotting using antibodies directed against Hep1.
Homologs of HEP1 were found in virtually all eukaryotes from yeast to human (Supplementary data, Figure S1A). Hep1 and its homologs contain two CXXC motifs which are conserved in all species analysed. They are part of the proposed zinc-finger (Zn-finger) domain zf-DNL (PFAM database accession PF05180), which is named after a short C-terminal motif of D(N/H)L (Bateman et al, 2004). In yeast and the other species, the protein sequence specifies a hydrophilic protein with a mitochondrial targeting signal (MTS) (Supplementary data, Figure S1A).
The protein starting with the first methionine of the putative reading frame was synthesized in a cell-free system and incubated with isolated mitochondria. The protein was imported and processed to a species of ca. 17 kDa (Supplementary data, Figure S1B). This suggests that Hep1 is indeed a mitochondrial protein with a cleavable MTS. To determine the N terminus of the mature protein after processing in mitochondria, the chromosomal copy of the HEP1 gene was replaced by a C-terminally His-tagged version using homologous recombination. This His-tagged Hep1 protein was functional. The protein was purified by NiNTA chromatography and analysed by Edman degradation. The N-terminal sequence obtained was NEVKK. Subcellular and submitochondrial fractionation of WT cells and immunoblotting with an antibody against Hep1 confirmed the location of endogenous Hep1 as a soluble protein in the mitochondrial matrix (Supplementary data, Figure S1C, D and E).
The 5′-untranslated region of the HEP1 gene contains a sequence, starting at nucleotide −140, resembling the heat shock element (HSE) and HSE-like sequence typical for yeast mitochondrial chaperones and cochaperones such as Hsp60, Hsp10 and Mdj1 (Supplementary data, Figure S1F). These elements were shown to be responsible for the heat-dependent upregulation of these proteins (Tachibana et al, 2002).
In order to study the interaction of Hep1 with mtHsp70, we performed co-immunoprecipitation and chemical crosslinking experiments in organello. To this end, mitochondria from WT cells were either incubated in buffer containing ATP and an ATP-regenerating system to keep intramitochondrial ATP high, or were treated with apyrase and oligomycin to deplete ATP in the matrix (Mokranjac et al, 2003b). Mitochondria were lysed with Triton X-100 and immunoprecipitation with antibodies against mtHsp70 was performed (Figure 1C). When ATP was depleted, the Hep1 protein was largely co-precipitated with mtHsp70. In contrast, when intramitochondrial ATP was high Hep1 was not found in the precipitate. The nucleotide exchange factor for mtHsp70, Mge1, was found in association with mtHsp70 when ATP was low, but not when the ATP level was high, as described previously (Horst et al, 1997). The abundant mitochondrial AAC was not detected in the precipitate, indicating that antibodies against mtHsp70 specifically co-precipitated Hep1.
In a crosslinking approach, mitochondria from WT cells were pretreated with ATP or with apyrase and then incubated with the chemical crosslinker disuccinimidyl glutarate (DSG). In addition, mitochondria from a strain harbouring a His-tagged variant of mtHsp70 were depleted of matrix ATP and treated with the crosslinking reagent. The mitochondria were subsequently analysed by SDS–PAGE and immunoblotting with antibodies against Hep1 (Figure 1D). When ATP was low, a band corresponding to an adduct of Hep1 and mtHsp70 was seen. In the presence of high ATP, this band was strongly reduced in its intensity. This adduct could be specifically adsorbed to NiNTA-agarose beads in case of the mtHsp70-his6 mitochondria, verifying that it is indeed a crosslinked adduct of Hep1 to mtHsp70. Thus, Hep1 interacts in a specific manner with mtHsp70 when ATP levels are low in the mitochondria.
We determined the relative concentration of Hep1 in the mitochondria in comparison to mtHsp70 by quantitative immunoblotting. In terms of protein mass, Hep1 was present at about 5% of the mass of Hsp70 (data not shown). Therefore, only a fraction of matrix mtHsp70 can be present in a complex with Hep1.
Does Hep1 stimulate the ATPase activity or the nucleotide exchange rate of mtHsp70? We addressed this question in an in vitro approach using purified proteins. A variant of Hep1 that contained a hexahistidinyl tag at the N terminus was expressed in Escherichia coli and purified. The protein was obtained as a stable soluble component (Figure 2A). The purified Hep1 did not affect the ATPase activity of mtHsp70 in the absence or the presence of the cochaperones Tim14 and Mge1 (Figure 2B). This argues against a function of Hep1 as a cochaperone which stimulates the ATPase activity of mtHsp70. To test for nucleotide exchange factor activity, a single turnover ATPase assay was used (Miao et al, 1997). MtHsp70 preloaded with radiolabelled ATP was incubated with excess unlabelled ATP in the presence of recombinant Hep1 or Mge1. The rate of hydrolysis of the prebound radiolabelled ATP was determined. If the nucleotide release is facilitated, the rate of hydrolysis of radiolabelled ATP will decrease, as it will be replaced by unlabelled ATP. In the presence of Mge1, a strong decrease of the rate of hydrolysis was observed. In contrast, the addition of Hep1 did not affect the rate of hydrolysis (Figure 2C). Thus, Hep1 does not have an activity as a nucleotide exchange factor.
Figure 2.
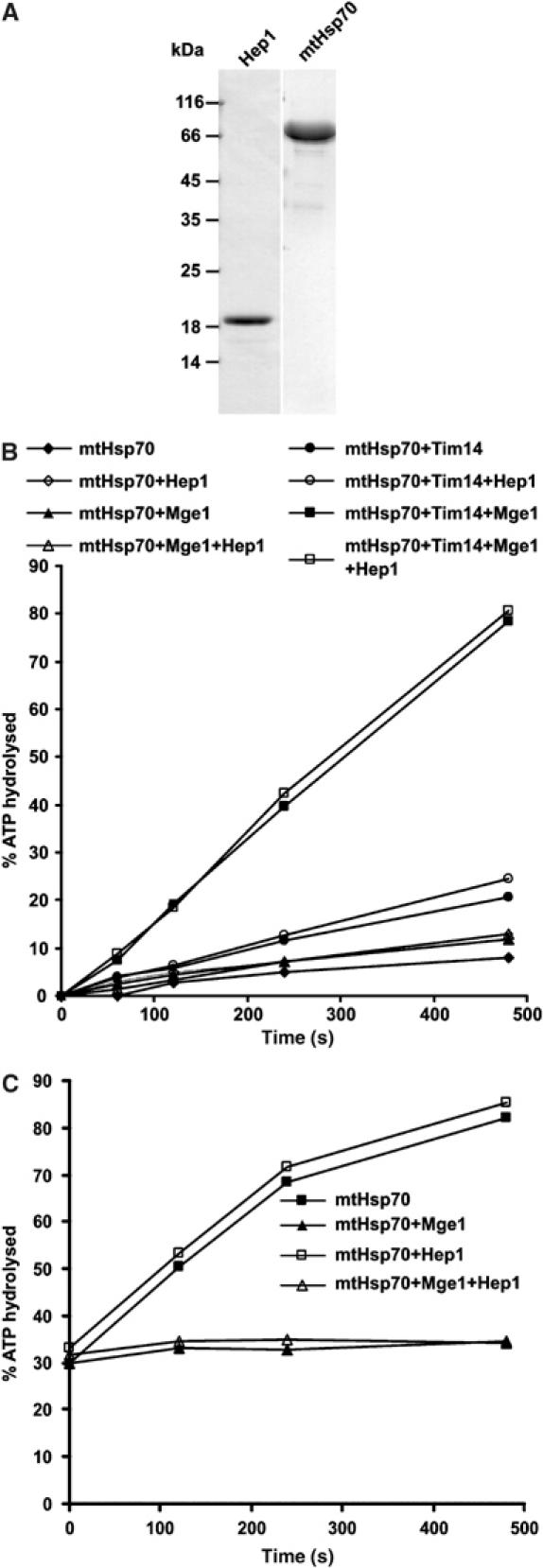
Hep1 does not increase the ATPase activity and the nucleotide exchange rate of mtHsp70. (A) Coomassie-stained gel of purified proteins Hep1-His6 and mtHsp70. (B) The ATPase activity of mtHsp70 was determined by the rate of conversion of 32P-ATP to ADP and 32P. Samples containing the indicated purified proteins were incubated for the indicated times, subjected to thin layer chromatography and the percentage of hydrolysed ATP was quantified by phosphoimaging. A fusion protein of MBP and Tim14 was used (Tim14). (C) Preformed complex of mtHsp70 with 32P-ATP was incubated at 30°C with excess of unlabelled ATP in the presence of Hep1, Mge1 or both Mge1 and Hep1 as indicated. The fraction of hydrolysed 32P-ATP was determined at the indicated time points as in (B).
MtHsp70s aggregate in cells lacking Hep1
In order to address the function of Hep1, we then analysed a yeast strain in which the gene for HEP1 was deleted. Deletion of HEP1 in yeast was reported to lead to death of cells (Giaever et al, 2002); yet a yeast strain, available from the Euroscarf strain collection, in which the HEP1 reading frame was disrupted, was able to grow on fermentable (YPD) medium. Growth, however, was slow at 24°C as compared to WT, and above 30°C no growth was observed (Figure 3A). On nonfermentable medium (YPG) the deletion strain did not grow.
Figure 3.
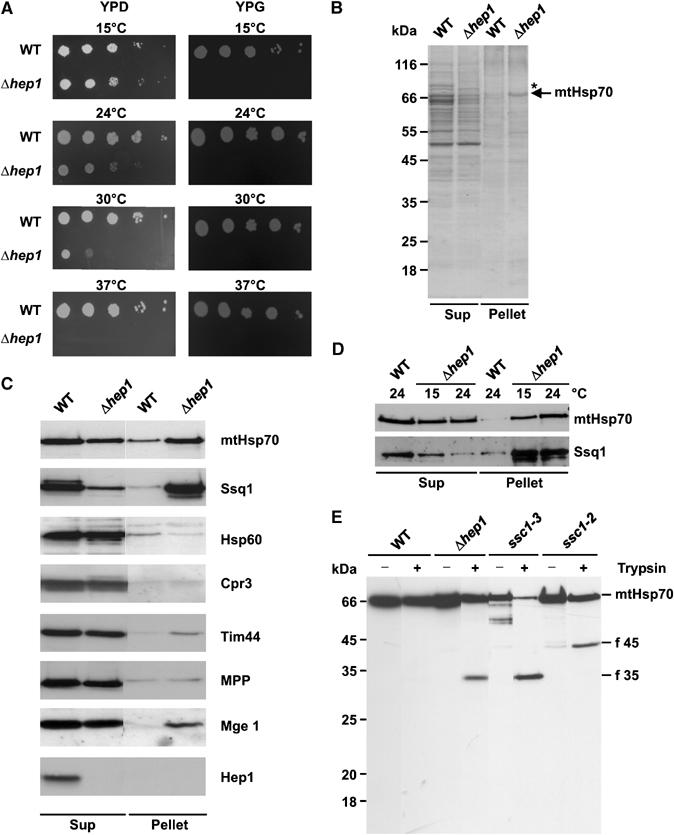
Deletion of HEP1 leads to aggregation of mitochondrial Hsp70 proteins. (A) Drop dilution test of WT and a HEP1 deletion strain (Δhep1). Dilutions, 10-fold, were plated on YPD and YPG plates and incubated at the indicated temperatures. (B, C) Aggregation of mtHsp70 in cells lacking Hep1. Mitochondria isolated from WT and Δhep1 cells were lysed with 1% digitonin and aggregated material was pelleted by centrifugation (Pellet). The SN fraction (Sup) was precipitated with TCA. Pellets and Sups were analysed by SDS–PAGE and Coomassie staining (B) or immunodecoration with antibodies against mtHsp70, Ssq1 and the other indicated proteins (C). (D) Aggregation of Hsp70 proteins depends on the growth temperature of cells. WT and Δhep1 cells were grown at 15°C and 24°C. Then, mitochondria were isolated and the aggregation state of mtHsp70 and Ssq1 was tested as above. Equal amounts of protein from the Pellet and Sup fraction were analysed. (E) Increased protease sensitivity of mtHsp70 in the absence of Hep1. Mitochondria were isolated from WT, from Δhep1 cells and from temperature-sensitive mutants, ssc1–3 and ssc1–2, grown at permissive temperature. Mitochondria were lysed in the presence of Triton X-100 and treated with trypsin for 5 min on ice. Samples were analysed by SDS–PAGE and immunodecoration with antibodies against full-length mtHsp70. The obtained fragments are indicated: f 45, N-terminal fragment; f 35, C-terminal fragment.
We analysed the aggregation of proteins in cells lacking Hep1. To this end, mitochondria from these cells grown at 24°C on YPD were solubilized in digitonin and insoluble material was separated from soluble material by centrifugation. For control the same was carried out with mitochondria from the corresponding WT. Both fractions were analysed by SDS–PAGE and staining with Coomassie blue (Figure 3B). A reproducible background of proteins was observed in WT cells, which represented either proteins insoluble in detergent or aggregates formed under the conditions applied. With the Δhep1 mitochondria on top of the background, a strong additional band was observed. Mass spectrometry analysis revealed that it represented mtHsp70 (data not shown). This result was also confirmed by immunoblotting with antibodies against mtHsp70 (Figure 3C). About 50% of mtHsp70 was found in the insoluble fraction.
We analysed the distribution between soluble and insoluble fractions of other mitochondrial proteins in Δhep1 mitochondria by immunodecoration (Figure 3C). No or very little aggregation was seen with these proteins, with the exception of Ssq1, another member of the mtHsp70 family. Ssq1 was largely recovered in the pellet fraction (Figure 3C).
Since the growth defect of cells lacking Hep1 was more pronounced at higher temperature, we asked whether the degree of aggregation of Hsp70s was related to the temperature during growth of the Δhep1 strain. The cells were grown at 15°C and at 24°C, mitochondria were prepared, and the aggregation of mtHsp70 and Ssq1 was analysed. The amounts of both mtHsp70 and Ssq1 were decreased in the supernatant fraction at 24°C compared to 15°C (Figure 3D). Thus, the aggregation of both proteins is enhanced at 24°C compared to 15°C. Interestingly, while the amount of mtHsp70 was increased in the pellet fraction, the amount of Ssq1 was decreased. This decrease in the pellet fraction may be due to rapid degradation as a consequence of misfolding. In a further experiment, isolated mitochondria were incubated at 37°C and then aggregation of mtHsp70 was determined. Mitochondria from WT and from a Δhep1 strain showed no substantial increase of aggregated mtHsp70 (data not shown).
In summary, we conclude that an important consequence of the deletion of HEP1 is the aggregation of mitochondrial Hsp70s. These aggregates accumulate in vivo during growth of the cells. They consist of mitochondrial Hsp70s, yet the presence of other components cannot be excluded.
The aggregation of mtHsp70 might be due to an altered conformation. Therefore, we compared the protease sensitivity of mtHsp70 in mitochondrial lysates of WT cells, cells lacking Hep1 and cells carrying mutant forms of mtHsp70, Ssc1-3 and Ssc1-2. At the chosen concentrations of trypsin, mtHsp70 in lysates of WT mitochondria was not cleaved. With mitochondria from Δhep1 cells, a fragment of ca. 35 kDa was formed (Figure 3E). The same fragment was observed after protease treatment of mitochondrial lysate from the ssc1-3 mutant, which has a mutation in the ATPase domain. The 35 kDa fragment could be decorated with an antibody specific for the C terminus of mtHsp70 (data not shown). Obviously, a stable PBD was present in mitochondria lacking Hep1, whereas the N-terminal part of mtHsp70 was sensitive to protease. In contrast, Ssc1-2, which carries a mutation in the PBD, was less prone to degradation and yielded a different, larger fragment of ca. 45 kDa (Figure 3E). This fragment could be decorated with an antibody against the N terminus of mtHsp70, and therefore represented the ATPase domain (data not shown) (Voisine et al, 1999).
In conclusion, Hep1 is required to keep mtHsp70 in a conformation that has a relative resistance to protease attack. These findings support the view that mtHsp70, by binding to Hep1, changes its conformation and thereby is prevented from self-aggregation.
Deletion of Hep1 results in impaired biogenesis of mitochondria
The aggregation of mtHsp70s may affect processes within the mitochondria which are dependent on the function of mtHsp70s. We analysed the TIM23 complex-dependent protein import and the biogenesis of FeS proteins, processes which both require the action of mitochondrial Hsp70 proteins. To assess the import of preproteins, mitochondria were isolated from Δhep1 and WT cells grown at 15°C and incubated with various radiolabelled preproteins (Figure 4A). In the absence of Hep1, the import rate of preproteins that use the TIM23 translocase was reduced to different degrees, but not the import of those which use the TIM22 translocase. In conclusion, mitochondria without any Hep1 are defective in preprotein import via the TIM23 complex. On the other hand, immunoprecipitation experiments revealed all known components of the TIM23 complex to be assembled in the complex in these mitochondria (Figure 4B). In addition, Hep1 itself was not found in the TIM23 complex in WT mitochondria (Figure 4B). This argues against a direct function of Hep1 in the TIM23 preprotein translocase.
Figure 4.
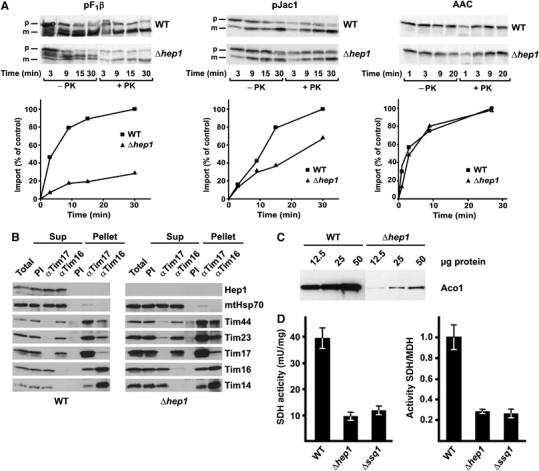
Cells lacking Hep1 are defective in import of mitochondrial preproteins and in the biogenesis of FeS proteins. (A) Mitochondria isolated from Δhep1 cells and from WT were incubated with radiolabelled preproteins. The substrates pF1β (subunit β of the F1Fo-ATPase) and pJac1 of the TIM23 complex and the substrate of the TIM22 complex, ATP/ADP carrier (AAC), were used as preproteins. Mitochondria were treated with proteinase K, reisolated and analysed by SDS–PAGE and autoradiography. The mature forms of the proteins were quantified. Import into WT mitochondria at the longest time point was set to 100%. p, precursor form; m, mature form. (B) Hep1 is not part of the TIM23 complex. Mitochondria were isolated from WT cells (left panel) and cells lacking Hep1 (right panel). After solubilization with digitonin, the lysate was subjected to immunoprecipitation with antibodies against Tim17, Tim16 and with preimmune serum (PI). Samples were analysed by SDS–PAGE and immunodecoration with antibodies against the indicated proteins of the TIM23 complex and Hep1. ‘Total' and ‘SNs' represent 20% of material present in ‘Pellets'. (C) Mitochondrial proteins were analysed by SDS–PAGE and immunodecoration with antibodies against aconitase. (D) The activities of SDH and malate dehydrogenase (MDH) were measured in mitochondria from WT, Δhep1 and Δssq1 cells. The SDH activities are given per mg of mitochondrial protein (left panel) and relative to the activity of malate dehydrogenase (right panel). The ratio obtained for WT mitochondria was set as 1. Error bars show the standard deviation of three independent experiments.
We noted that in the Coomassie-stained gel a band of about 85 kDa was almost missing in both soluble and aggregated fractions of cells lacking Hep1 (see asterisk, Figure 3B). Mass spectrometry analysis demonstrated that this band represented the quite abundant enzyme aconitase. Immunoblotting using antibodies against aconitase confirmed that its endogenous levels were strongly reduced in cells lacking Hep1 compared to those in WT cells (Figure 4C). Aconitase is an FeS cluster-containing protein. We therefore determined the activity of another FeS protein, succinate dehydrogenase (SDH). This was also strongly reduced in mitochondria lacking Hep1 (Figure 4D). Thus, like in the absence of Ssq1, the biogenesis of proteins containing FeS clusters is affected, when Hep1 is absent in mitochondria.
In conclusion, various processes requiring different mitochondrial Hsp70s were defective in mitochondria from cells lacking Hep1. Although specific functions of Hep1 in different mitochondrial processes cannot be excluded, the observed defects are consistent and can be explained with the observed compromised functional levels of mtHsp70 and Ssq1.
Hep1 prevents self-aggregation of mtHsp70 in vitro
We addressed the role of Hep1 in the self-aggregation of mtHsp70 using purified components (see Figure 2A). First, oligomerization of mtHsp70 was analysed by subjecting it to crosslinking with glutaraldehyde and subsequent analysis by SDS–PAGE and silver staining (Azem et al, 1997) (Figure 5A). In the absence of added nucleotides, massive formation of oligomers was observed. Beside dimers and trimers, substantial amounts of higher oligomers were seen in particular at the origin of the separation gel and in the stacking gel. Notably, the bands, in particular the monomer band, were diffuse. This suggests that the mtHsp70 was in a rather loose conformation that allowed access and extensive modification of side chains by the crosslinker, including formation of intramolecular links. In contrast, in the presence of ATP, virtually no oligomers were observed and the glutaraldehyde treatment altered the band of mtHsp70 only in a marginal manner. Minor bands were produced that apparently represent internal crosslinks. This suggests that addition of ATP to the purified mtHsp70 induces a tight folding and prevents self-aggregation. In the presence of ADP dimers and trimers of mtHsp70 were still formed, but higher oligomeric forms were not observed. As in the presence of ATP, the monomeric band of mtHsp70 was not substantially modified by glutaraldehyde treatment, indicating a tightly folded conformation of mtHsp70 in the ADP state.
Figure 5.
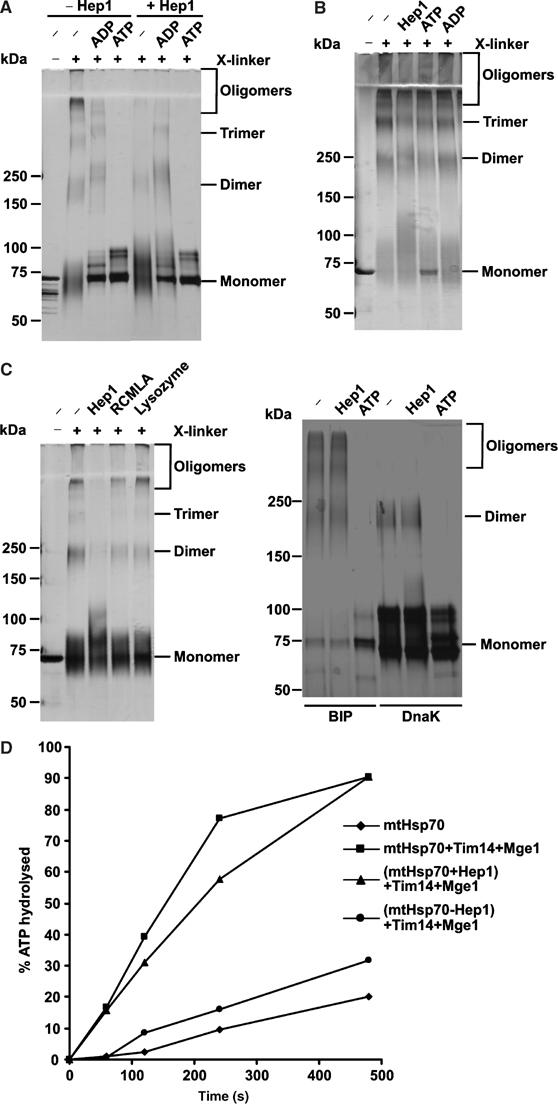
Hep1 prevents self-aggregation of mtHsp70. (A) Hep1 inhibits aggregation of mtHsp70. Purified mtHsp70 was incubated for 10 min at 30°C in the absence or presence of the indicated nucleotides and Hep1. The samples were subjected to crosslinking with glutaraldehyde and analysed by SDS–PAGE (6–14% gradient gel) and silver staining. (B) Self-aggregation of mtHsp70 is not reversible. MtHsp70 was first incubated for 10 min at 30°C to allow oligomerization and then kept for 10 min in the presence of nucleotides and Hep1 as indicated. Further treatment as in (A). (C) Prevention of self-aggregation is specific for Hep1 and mtHsp70. Left panel: mtHsp70 (left panel) in the presence of lysozyme or RCMLA and purified BiP and DnaK (right panel) were treated and analysed as in (A). (D) Hep1 keeps mtHsp70 functional. The ATPase activity of mtHsp70 in the presence of the indicated proteins was determined as in Figure 2B. Where indicated, mtHsp70 was first preincubated for 30 min at 30°C either in the absence (Hsp70−Hep1) or presence (Hsp70+Hep1) of equimolar amounts of Hep1.
Next, we asked whether Hep1 affects the oligomerization of mtHsp70. Purified Hep1 and mtHsp70 were mixed prior to incubation. This led to virtually complete inhibition of formation of higher oligomeric forms and to a decrease in the formation of dimers and trimers of mtHsp70 (Figure 5A). In the presence of ATP or ADP, when no higher oligomers of mtHsp70 were formed, Hep1 had no effect on mtHsp70. In the absence of nucleotides, the monomeric mtHsp70 still appeared as a rather diffuse band, now with a slightly higher molecular mass. As demonstrated by immunodecoration with antibodies against Hep1, it represents an adduct of mtHsp70 and Hep1 (data not shown). This indicates that Hep1, in contrast to ATP and ADP, did not confer a tightly folded conformation upon mtHsp70.
Does Hep1 cause dissociation of oligomers after they are formed? MtHsp70 was first allowed to form the oligomers and then Hep1 was added (Figure 5B). No dissociation of oligomers was observed. Interestingly, also addition of ATP did not lead to disaggregation. Even after incubation for 4 h ATP did not dissolve the oligomers (data not shown). Thus, both ATP and Hep1 can prevent, but cannot reverse, aggregation of Hsp70.
We performed a series of controls for the specificity of action of Hep1. The presence of other folded or unfolded proteins, such as lysozyme or reduced carboxymethylated lactalbumin (RCMLA), had no preventive effect on the oligomerization (Figure 5C). The same observation was made with the known mtHsp70-binding peptide P5 (Liu et al, 2001) which was used in a concentration range of 1–100-fold molar excess over mtHsp70 (data not shown). Other Hsp70 proteins, BiP from the ER compartment and DnaK from E. coli showed a similar oligomerization behaviour, though with less formation of higher oligomers; yet this was not affected by added Hep1 (Figure 5C).
In summary, we conclude that Hep1 is able to specifically prevent oligomerization of mtHsp70 that is in its nucleotide-free form in vitro.
Is Hep1 required to maintain the activity of mtHsp70? To address this, we measured the ATPase activity of mtHsp70. Preincubation of mtHsp70 at 30°C for 30 min resulted in a decrease in its ATPase activity, stimulated by Tim14 and Mge1, by roughly 80%. The presence of Hep1 during the preincubation completely prevented the loss of the ATPase activity (Figure 5D). These results confirm that the oligomeric form of mtHsp70 is not functional and that the role of Hep1 is to maintain mtHsp70 in an active state.
Since the experimental conditions in this reconstituted system differ from the in vivo conditions, we asked whether similar observations can be made in the context of the intact cell. To this end, E. coli cells were transformed with constructs that encoded either mtHsp70 or both mtHsp70 and His6-Hep1 on a single plasmid. The distribution of the proteins between soluble and insoluble fractions was determined in total cell lysates. When mtHsp70 was expressed alone, it was completely recovered in the insoluble fraction (Figure 6). In contrast, upon simultaneous expression of mtHsp70 and Hep1, both proteins were completely recovered in the soluble fraction. MtHsp70 purified from this fraction was active when tested for its ATPase activity (unpublished data). These data show that the presence of Hep1 in the environment of a bacterial cell can prevent mtHsp70 from aggregation.
Figure 6.
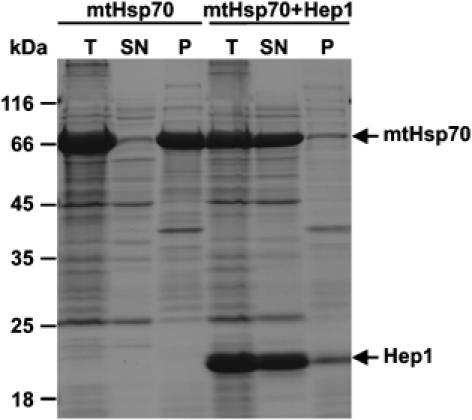
Hep1 prevents aggregation of mtHsp70 in E. coli cells. MtHsp70 was expressed in E. coli either alone (mtHsp70) or together with a His-tagged form of Hep1 (mtHsp70+Hep1). Both proteins lacked their matrix targeting sequences. Following induction of protein expression cells were lysed and one aliquot was removed (total, T). The other aliquot was separated by centrifugation into insoluble protein (P) and soluble protein fractions (SN). Samples were analysed by SDS–PAGE and Coomassie staining.
Discussion
We report here on the discovery that a chaperone requires another chaperone for maintenance of its structure and function. In the absence of this chaperone, Hep1, the mitochondrial Hsp70 chaperones, mtHsp70 and Ssq1, aggregate in the cell. They form self-aggregates, a process that leads to loss of their multiple essential functions such as in protein import and biogenesis of FeS proteins.
We identified this chaperone Hep1 by virtue of its interaction with mtHsp70. Complex formation of Hep1 with mtHsp70 was also observed in vitro, using the purified protein components. Hep1 binds to mtHsp70 and prevents its aggregation. It cannot, however, reverse aggregation once it has occurred. Furthermore, virtually complete self-aggregation of mtHsp70 takes place when it is expressed in E. coli, but co-expression of Hep1 leads to soluble mtHsp70. Thus, Hep1 is necessary to keep mtHsp70 in its native folding state. MtHsp70 aggregated in cells lacking Hep1 could not be dissociated by the addition of Hep1 to detergent lysed mitochondria from these cells (data not shown). The irreversibility of aggregation suggests that oligomeric mtHsp70 is not in a physiological state, for example, as an inactive storage form. Rather, it appears to be the product of an adverse process which is normally prevented by the action of Hep1. This is underlined by the essential nature of Hep1 for the life of cells growing under respiring conditions. Overexpression of mtHsp70 in cells lacking Hep1 did not suppress the observed growth defect of these cells (data not shown and Burri et al, 2004).
The details of the self-aggregation of mtHsp70 are presently not entirely clear. Higher oligomeric forms were not formed in the presence of ATP. Rather, the nucleotide-free mtHsp70 by itself displayed the tendency to form aggregates. A loose folding of mtHsp70 in the nucleotide-free state was suggested by a massive modification upon glutaraldehyde treatment. Hep1 might recognize a conformer of mtHsp70 that is constantly formed in the normal course of the reaction cycle in an off-pathway reaction. Other Hsp70-binding proteins, such as the nucleotide exchange protein Mge1, which also binds to the nucleotide-free form, could also have a stabilizing effect on Hsp70. Clearly, however, if such a role does exist, it would not be sufficient to prevent self-aggregation of mtHsp70s. Hep1 might thus be imagined to bind to Hsp70 cycle intermediates that are prone to aggregation. In this way, it would perform a chaperone function according to the classical definition of chaperones. Such a catalytic function is consistent with the observation that Hep1 is not present in stoichiometric amounts to the mitochondrial Hsp70s. Why just a chaperone, Hsp70, would be prone to formation of dangerous conformers in the normal reaction cycle under normal conditions is currently an open question. It is possible that similar Hsp70 folding intermediates are formed during de novo folding of mitochondrial Hsp70s. Hep1 might prevent aggregation of these intermediates and thereby mediates also de novo folding of mitochondrial Hsp70s.
A similar situation could possibly exist for other nucleotide-hydrolysing proteins such as members of the small G-protein family. The protein Dss4 was proposed to interact with and stabilize the transient nucleotide-free intermediate form of Rab proteins (Collins et al, 1997; Nuoffer et al, 1997; Esters et al, 2001).
The necessity for a chaperone to maintain the function of mtHsp70s has so far only been observed in yeast. However, homologs of Hep1 appear to be present in virtually all eukaryotic cells. Furthermore, the propensity for aggregation has been described for various nonmitochondrial Hsp70s. Cytosolic Hsc70 self-associates to dimers, trimers and likely to higher oligomers in vitro and in vivo (Schlossman et al, 1984; Benaroudj et al, 1996; Angelidis et al, 1999). Oligomeric forms have also been observed for BiP/Grp78 and the E. coli DnaK (Carlino et al, 1992; Freiden et al, 1992; Blond-Elguindi et al, 1993; Schonfeld et al, 1995). However, the mechanism of self-aggregation of these Hsp70s may differ from that of mtHsp70. It has been reported for some of them that self-aggregation can be reversed by unfolded proteins and ATP (Blond-Elguindi et al, 1993; Benaroudj et al, 1996). Thus, self-aggregation may mimic substrate binding of Hsp70 in these cases. It remains to be determined how general the requirement for a Hep1 type chaperone is. We have found in a database search only homologs of Hep1 which are predicted to be present in mitochondria. However, there might be proteins in other cellular compartments with similar or related function. A member of the Nm23/nucleoside diphosphate kinase family, the 16 kDa protein p16, has been reported which monomerizes Hsc70 (Leung and Hightower, 1997).
How does Hep1 interact with mtHsp70? Our data provide some hints as to which domains of mtHsp70 might be in contact with Hep1. The Ssc1–3 protein which carries a mutation in the ATPase domain (AD) showed reduced interaction with Hep1, however, not Ssc1-2 protein, with a mutation in the PBD (data not shown). MtHsp70 can be expressed as a soluble protein in E. coli, but only in the presence of Hep1. The PBD when expressed in E. coli without Hep1 folded to a soluble protein; the AD was neither soluble when expressed alone nor when co-expressed with Hep1 (unpublished data). These observations suggest that Hep1 cannot stabilize the AD alone, but in the context of the PBD. Thus, an interaction with the AD seems quite possible, but the PBD is apparently also necessary, in particular in the context of intact mtHsp70.
Hep1 contains two pairs of cysteine residues which are conserved among all Hep1 homologs found in databases. It has been suggested that they represent a Zn-finger motif and Hep1 might bind metal ions. However, this suggestion needs experimental support. The name Zim17 (for zinc-finger motif) has been proposed (Burri et al, 2004). We now determined the function of the protein in the cell. Based on these data, we termed it Hep1 for mtHsp70 escort protein expressing the function of the protein.
It was recently proposed that Zim17/Hep1 is a component of the mitochondrial import motor (Burri et al, 2004). When the endogenous level of Hep1 was downregulated, the import rates of substrates of the TIM23 complex were decreased. Our data using the deletion strain of HEP1 confirm these experimental findings, but also demonstrate that import occurs even in the complete absence of Hep1. Thus, our results do not exclude such an additional more specialized function of Hep1. However, the previous study and our experiments were unable to detect Hep1/Zim17 in the TIM23 complex. Furthermore, the following observations support the notion that the import of precursors is compromised in cells lacking Hep1 due to the massive aggregation of mtHsp70 and therefore strongly reduced levels of functional mtHsp70. First, Hep1 exerts its specific antiaggregation effect on mtHsp70 both in a reconstituted system comprised of the purified components and in the environment of the E. coli cytosol. Second, two structurally closely related but functionally quite distinct forms of mitochondrial Hsp70, mtHsp70 and Ssq1 are affected by depletion of Hep1. Thus, depletion of functional Hsp70s in cells lacking Hep1 is likely to lead to deficiencies in a number of processes which are dependent on the large variety of Hsp70 functions in mitochondria.
Materials and methods
Yeast strains and cell growth
The strain expressing mtHsp70 with a C-terminal hexahistidinyl tag under an endogenous promoter was described before (Bolliger et al, 1994). The haploid deletion strain of HEP1 (Δhep1) and the corresponding WT were obtained from Euroscarf (accession numbers 10179B and 10000R, respectively). The Hep1-7His strain was generated by inserting the nucleotides encoding the heptahistidinyl tag after the coding sequence of Hep1 on the chromosome in YPH499 by homologous recombination according to published methods (Knop et al, 1999). The Δhep1 strain and the control WT were grown in YPD medium at 24°C unless otherwise stated. Otherwise, cells were grown on lactate medium containing 0.1% glucose.
Purification of His-Hep1 and generation of antibodies
The nucleotide sequence encoding amino-acid residues 74–205 of Saccharomyces cerevisiae Hep1 was cloned into the pQE-30 vector (Qiagen), thus generating a protein with an N-terminal His6-tag. The protein was expressed in E. coli XL1-Blue cells and purified on a Ni-NTA-agarose column according to the manufacturer's instructions (Qiagen). For in vitro experiments, the buffer was changed to 20 mM HEPES, 100 mM KCl, 10 mM MgCl2 (pH 7.4) on a PD-10 column (Amersham). The purified protein was also injected into rabbits for generation of antibodies.
Purification of mtHsp70-His
Mitochondria from WT yeast or yeast cells containing C-terminally His-tagged mtHsp70 (24 mg protein) were incubated for 10 min at 25°C in the import buffer (0.05% bovine serum albumin (w/v), 500 mM sorbitol, 50 mM HEPES-KOH, 80 mM KCl, 10 mM MgAc2, 2.5 mM EDTA, 2 mM potassium phosphate and 1 mM MnCl2, pH 7.2) in the presence of apyrase (2 U/ml) and oligomycin (20 μM) to deplete ATP from mitochondria. Upon reisolation, mitochondria were solubilized at 1 mg/ml with 1% Triton X-100 (w/v) in buffer A (20 mM Tris, 80 mM KAc, 20 mM imidazole, 10% glycerol (v/v), 1 mM PMSF, pH 8.0) for 30 min at 4°C. After a clarifying spin, solubilized material was loaded on a 1 ml NiNTA-agarose column. The column was first washed with 10 ml buffer A containing 0.05% Triton X-100 and bound proteins were then eluted with buffer A containing 300 mM imidazole and 0.05% Triton X-100. Protein-containing fractions were pooled, precipitated with trichloroacetic acid (TCA) and analysed by SDS–PAGE and Coomassie brilliant blue staining.
Limited proteolysis of mtHsp70
Mitochondria (100 μg) were solubilized at 0.5 mg/ml with 1% (w/v) Triton X-100 in 20 mM HEPES, 100 mM KCl, 10 mM MgCl2, pH 7.4. After 15 min at 4°C, samples were split in two. Trypsin (0.5 μg) was added to one aliquot and the samples were incubated for additional 5 min on ice. Proteolysis was stopped by addition of TCA. Samples were analysed by SDS–PAGE and immunodecoration.
ATPase assay and nucleotide exchange factor assay
ATPase assay was modified after Horst et al (1997). The ATP-hydrolysing activity of mtHsp70 was measured at 25°C in 50 mM HEPES (pH 7.4), 100 mM KCl, 5 mM MgCl2, 66 μM ATP and 0.1 mM γ-32P-ATP. In all, 7.4 μg of mtHsp70 was used per reaction. Where indicated 2 μg Mge1 (Horst et al, 1997), 4.9 μg maltose-binding protein (MBP)-Tim14 and 2.5 μg Hep1 were added. Aliquots were removed at the indicated time points and the reactions were stopped by addition of 50 mM EDTA, 5 mM ATP and 5 mM ADP. Samples were spotted on PEI-cellulose FTLC plates and developed with 1 M formic acid, 0.5 M LiCl. The chromatogram was analysed by phosphoimaging. Activity was depicted as the % of total radiolabelled ATP which was hydrolysed at the indicated time point.
MtHsp70/ATP complex formation and subsequent single turnover experiments were essentially performed as described in Miao et al (1997). Briefly, 50 μg mtHsp70 was incubated with 10 μCi of α-32P-ATP in buffer C (50 mM Tris–HCl (pH 7.4), 100 mM KCl, 10 mM MgCl2, 2 mM DTT) containing 25 μM ATP at 4°C for 10 min. The reaction mixture was then loaded on an NICK column (Amersham Biosciences) pre-equilibrated with buffer C at 4°C. A volume of 10 μl of the elution fractions containing roughly 4 μg of the mtHsp70/ATP complex was used for single turnover experiments performed in the presence of 250 μM cold ATP. When indicated, 4 μM Mge1 and 4 μM Hep1 were added.
Glutaraldehyde crosslinking of mtHsp70
Crosslinking of purified mtHsp70 with glutaraldehyde was carried out essentially as described in Azem et al (1997). Briefly, mtHsp70 (2 μg) was incubated in 50 mM HEPES/KOH (pH 7.4), 100 mM KCl, 5 mM MgCl2, for 10 min at 30°C before glutaraldehyde was added to a final concentration of 1 mM. Crosslinking was performed for 30 min at 30°C and stopped by addition of 200 mM glycine in Laemmli buffer. Samples were analysed on 6–14% gradient SDS gels, followed by silver staining. Where indicated, 1 mM nucleotides and proteins in a molar ratio to Hsp70 of 1:1 were present in the reaction.
Co-expression of mtHsp70 and Hep1 in E. coli
The pETDuet-1 system (Novagen) was used for co-expression of mtHsp70 and Hep1 in E. coli. To express mtHsp70 alone, nucleotide sequence encoding the mature form of Ssc1 (amino acids 24–654) was cloned as an NdeI/XhoI fragment into the pETDuet-1 vector. For co-expression of mtHsp70 and Hep1, the nucleotide sequence coding for the mature form of Hep1 (amino acids 74–205) was cloned as a BamHI/HindIII fragment into the pETDuet-1 plasmid containing the SSC1 fragment. In this construct, Hep1 protein contains an N-terminal His6-tag. Plasmids were transformed into E. coli BL21(DE3) cells and protein expression was induced by addition of IPTG. Cells were lysed on ice with 1 mg/ml lysozyme in 20 mM Tris, 80 mM KCl, 10 mM imidazole, 1 mM PMSF, pH 7.4. Sonication was used to completely break the cells. The soluble fraction was obtained by centrifugation for 15 min at 15 000 r.p.m. in JA-20 rotor (Beckman).
Miscellaneous
Published procedures were used for subcellular and submitochondrial fractionations (Rowley et al, 1994), import of preproteins into isolated mitochondria (Mokranjac et al, 2003a), manipulation of ATP levels inside mitochondria (Mokranjac et al, 2003b), chemical crosslinking in intact mitochondria and affinity purification of crosslinking adducts by NiNTA-agarose beads (Mokranjac et al, 2003b). Aggregation assay was carried out essentially as described in Rowley et al (1994), with the modification that digitonin was used for solubilization of mitochondria. For coimmunoprecipitation experiments, mitochondria (1 mg/ml) were solubilized in 20 mM Tris (pH 7.5), 80 mM KAc, 10% glycerol, 1 mM PMSF with 1% digitonin for 30 min on ice, and further processed as described (Mokranjac et al, 2003b). Enzyme activities were measured as described in Lutz et al (2001). The mature form of Mge1 was expressed and purified as described in Horst et al (1997). Tim14 was expressed as a fusion protein of MBP and Tim14 and purified according to the manufacturer's instructions (NEB, Beverly).
Supplementary Material
Supplementary Data
Acknowledgments
We are very grateful to Ulrike Gärtner, Heiko Germeroth and Marica Malesic for excellent technical assistance. We thank Professor Ulrich Hartl and Professor Richard Zimmermann for providing purified DnaK and BiP, respectively. This work was supported by grants from the Deutsche Forschungsgemeinschaft, SFB 594 (B3, B13), the German–Israeli Foundation, the German–Israeli Project Cooperation DIP F.5.1 and the Fonds der Chemischen Industrie.
References
- Angelidis CE, Lazaridis I, Pagoulatos GN (1999) Aggregation of hsp70 and hsc70 in vivo is distinct and temperature-dependent and their chaperone function is directly related to non-aggregated forms. Eur J Biochem 259: 505–512 [DOI] [PubMed] [Google Scholar]
- Azem A, Oppliger W, Lustig A, Jeno P, Feifel B, Schatz G, Horst M (1997) The mitochondrial hsp70 chaperone system. Effect of adenine nucleotides, peptide substrate, and mGrpE on the oligomeric state of mhsp70. J Biol Chem 272: 20901–20906 [DOI] [PubMed] [Google Scholar]
- Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer EL, Studholme DJ, Yeats C, Eddy SR (2004) The Pfam protein families database. Nucleic Acids Res 32, (database issue): D138–D141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann F, Milisav I, Neupert W, Herrmann JM (2000) Ecm10, a novel hsp70 homolog in the mitochondrial matrix of the yeast Saccharomyces cerevisiae. FEBS Lett 487: 307–312 [DOI] [PubMed] [Google Scholar]
- Benaroudj N, Triniolles F, Ladjimi MM (1996) Effect of nucleotides, peptides, and unfolded proteins on the self-association of the molecular chaperone HSC70. J Biol Chem 271: 18471–18476 [DOI] [PubMed] [Google Scholar]
- Blond-Elguindi S, Fourie AM, Sambrook JF, Gething MJ (1993) Peptide-dependent stimulation of the ATPase activity of the molecular chaperone BiP is the result of conversion of oligomers to active monomers. J Biol Chem 268: 12730–12735 [PubMed] [Google Scholar]
- Bolliger L, Deloche O, Glick BS, Georgopoulos C, Jeno P, Kronidou N, Horst M, Morishima N, Schatz G (1994) A mitochondrial homolog of bacterial GrpE interacts with mitochondrial hsp70 and is essential for viability. EMBO J 13: 1998–2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukau B, Horwich AL (1998) The Hsp70 and Hsp60 chaperone machines. Cell 92: 351–366 [DOI] [PubMed] [Google Scholar]
- Burri L, Vascotto K, Fredersdorf S, Tiedt R, Hall MN, Lithgow T (2004) Zim17, a novel zinc-finger protein essential for protein import into mitochondria. J Biol Chem 279: 50243–50249 [DOI] [PubMed] [Google Scholar]
- Carlino A, Toledo H, Skaleris D, DeLisio R, Weissbach H, Brot N (1992) Interactions of liver Grp78 and Escherichia coli recombinant Grp78 with ATP: multiple species and disaggregation. Proc Natl Acad Sci USA 89: 2081–2085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins RN, Brennwald P, Garrett M, Lauring A, Novick P (1997) Interactions of nucleotide release factor Dss4p with Sec4p in the post-Golgi secretory pathway of yeast. J Biol Chem 272: 18281–18289 [DOI] [PubMed] [Google Scholar]
- Craig EA, Kramer J, Shilling J, Werner-Washburne M, Holmes S, Kosic-Smithers J, Nicolet CM (1989) SSC1, an essential member of the yeast HSP70 multigene family, encodes a mitochondrial protein. Mol Cell Biol 9: 3000–3008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig EA, Marszalek J (2002) A specialized mitochondrial molecular chaperone system: a role in formation of Fe/S centers. Cell Mol Life Sci 59: 1658–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esters H, Alexandrov K, Iakovenko A, Ivanova T, Thoma N, Rybin V, Zerial M, Scheidig AJ, Goody RS (2001) Vps9, Rabex-5 and DSS4: proteins with weak but distinct nucleotide-exchange activities for Rab proteins. J Mol Biol 310: 141–156 [DOI] [PubMed] [Google Scholar]
- Fan CY, Lee S, Cyr DM (2003) Mechanisms for regulation of Hsp70 function by Hsp40. Cell Stress Chaperones 8: 309–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freiden PJ, Gaut JR, Hendershot LM (1992) Interconversion of three differentially modified and assembled forms of BiP. EMBO J 11: 63–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frydman J (2001) Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu Rev Biochem 70: 603–647 [DOI] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, Arkin AP, Astromoff A, El-Bakkoury M, Bangham R, Benito R, Brachat S, Campanaro S, Curtiss M, Davis K, Deutschbauer A, Entian KD, Flaherty P, Foury F, Garfinkel DJ, Gerstein M, Gotte D, Guldener U, Hegemann JH, Hempel S, Herman Z, Jaramillo DF, Kelly DE, Kelly SL, Kotter P, LaBonte D, Lamb DC, Lan N, Liang H, Liao H, Liu L, Luo C, Lussier M, Mao R, Menard P, Ooi SL, Revuelta JL, Roberts CJ, Rose M, Ross-Macdonald P, Scherens B, Schimmack G, Shafer B, Shoemaker DD, Sookhai-Mahadeo S, Storms RK, Strathern JN, Valle G, Voet M, Volckaert G, Wang CY, Ward TR, Wilhelmy J, Winzeler EA, Yang Y, Yen G, Youngman E, Yu K, Bussey H, Boeke JD, Snyder M, Philippsen P, Davis RW, Johnston M (2002) Functional profiling of the Saccharomyces cerevisiae genome. Nature 418: 387–391 [DOI] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M (2002) Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 295: 1852–1858 [DOI] [PubMed] [Google Scholar]
- Hohfeld J, Cyr DM, Patterson C (2001) From the cradle to the grave: molecular chaperones that may choose between folding and degradation. EMBO Rep 2: 885–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horst M, Oppliger W, Rospert S, Schonfeld HJ, Schatz G, Azem A (1997) Sequential action of two hsp70 complexes during protein import into mitochondria. EMBO J 16: 1842–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley WL (1998) The J-domain family and the recruitment of chaperone power. Trends Biochem Sci 23: 222–227 [DOI] [PubMed] [Google Scholar]
- Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, Nasmyth K, Schiebel E (1999) Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15: 963–972 [DOI] [PubMed] [Google Scholar]
- Leung SM, Hightower LE (1997) A 16-kDa protein functions as a new regulatory protein for Hsc70 molecular chaperone and is identified as a member of the Nm23/nucleoside diphosphate kinase family. J Biol Chem 272: 2607–2614 [DOI] [PubMed] [Google Scholar]
- Lill R, Kispal G (2000) Maturation of cellular Fe–S proteins: an essential function of mitochondria. Trends Biochem Sci 25: 352–356 [DOI] [PubMed] [Google Scholar]
- Liu Q, Krzewska J, Liberek K, Craig EA (2001) Mitochondrial Hsp70 Ssc1: role in protein folding. J Biol Chem 276: 6112–6118 [DOI] [PubMed] [Google Scholar]
- Lutz T, Westermann B, Neupert W, Herrmann JM (2001) The mitochondrial proteins Ssq1 and Jac1 are required for the assembly of iron sulfur clusters in mitochondria. J Mol Biol 307: 815–825 [DOI] [PubMed] [Google Scholar]
- Mayer MP, Brehmer D, Gassler CS, Bukau B (2001) Hsp70 chaperone machines. Adv Protein Chem 59: 1–44 [DOI] [PubMed] [Google Scholar]
- Miao B, Davis JE, Craig EA (1997) Mge1 functions as a nucleotide release factor for Ssc1, a mitochondrial Hsp70 of Saccharomyces cerevisiae. J Mol Biol 265: 541–552 [DOI] [PubMed] [Google Scholar]
- Mokranjac D, Paschen SA, Kozany C, Prokisch H, Hoppins SC, Nargang FE, Neupert W, Hell K (2003a) Tim50, a novel component of the TIM23 preprotein translocase of mitochondria. EMBO J 22: 816–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokranjac D, Sichting M, Neupert W, Hell K (2003b) Tim14, a novel key component of the import motor of the TIM23 protein translocase of mitochondria. EMBO J 22: 4945–4956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto RI, Tissieres A, Georgopoulos C (1994) The Biology of Heat Shock Proteins and Molecular Chaperones. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press [Google Scholar]
- Nuoffer C, Wu SK, Dascher C, Balch WE (1997) Mss4 does not function as an exchange factor for Rab in endoplasmic reticulum to Golgi transport. Mol Biol Cell 8: 1305–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polissi A, Goffin L, Georgopoulos C (1995) The Escherichia coli heat shock response and bacteriophage lambda development. FEMS Microbiol Rev 17: 159–169 [DOI] [PubMed] [Google Scholar]
- Rowley N, Prip-Buus C, Westermann B, Brown C, Schwarz E, Barrell B, Neupert W (1994) Mdj1p, a novel chaperone of the DnaJ family, is involved in mitochondrial biogenesis and protein folding. Cell 77: 249–259 [DOI] [PubMed] [Google Scholar]
- Schilke B, Forster J, Davis J, James P, Walter W, Laloraya S, Johnson J, Miao B, Craig E (1996) The cold sensitivity of a mutant of Saccharomyces cerevisiae lacking a mitochondrial heat shock protein 70 is suppressed by loss of mitochondrial DNA. J Cell Biol 134: 603–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlossman DM, Schmid SL, Braell WA, Rothman JE (1984) An enzyme that removes clathrin coats: purification of an uncoating ATPase. J Cell Biol 99: 723–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonfeld HJ, Schmidt D, Schroder H, Bukau B (1995) The DnaK chaperone system of Escherichia coli: quaternary structures and interactions of the DnaK and GrpE components. J Biol Chem 270: 2183–2189 [DOI] [PubMed] [Google Scholar]
- Tachibana T, Astumi S, Shioda R, Ueno M, Uritani M, Ushimaru T (2002) A novel non-conventional heat shock element regulates expression of MDJ1 encoding a DnaJ homolog in Saccharomyces cerevisiae. J Biol Chem 277: 22140–22146 [DOI] [PubMed] [Google Scholar]
- Voisine C, Craig EA, Zufall N, von Ahsen O, Pfanner N, Voos W (1999) The protein import motor of mitochondria: unfolding and trapping of preproteins are distinct and separable functions of matrix Hsp70. Cell 97: 565–574 [DOI] [PubMed] [Google Scholar]
- Voos W, Rottgers K (2002) Molecular chaperones as essential mediators of mitochondrial biogenesis. Biochim Biophys Acta 1592: 51–62 [DOI] [PubMed] [Google Scholar]
- Young JC, Agashe VR, Siegers K, Hartl FU (2004) Pathways of chaperone-mediated protein folding in the cytosol. Nat Rev Mol Cell Biol 5: 781–791 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data


