Abstract
Differential expression of long non-coding RNAs (lncRNAs) during differentiation and their misregulation in cancer highlight their potential as cell fate regulators. While some example lncRNAs have been characterized in great detail, the functional in vivo relevance of others has been called into question. Finding functional lncRNAs will most probably require a combination of complementary approaches that will greatly vary depending on their mode of action. In this review, we discuss the different tools available to dissect genetically lncRNA requirements and how each is best suited to studies in particular contexts. Moreover, we review different strategies used to select candidate lncRNAs and give an overview of lncRNAs described to regulate development and cancer through different mechanisms.
Keywords: long non-coding RNAs, development, cancer
1. Introduction
Regulated gene expression is the basis for the extensive variety of cell types our bodies generate from the same set of DNA instructions. Specific gene programmes are transcribed in particular cells providing them with their molecular identity and the protein products that underlie their functions. Together with coding genes, thousands of long non-coding RNAs (lncRNAs) are also expressed in a cell-type-specific manner during differentiation and in certain cancers. This has been extensively reported in many organisms and cell types [1–4], yet demonstrating that these molecules play functional roles has not been easy.
The now ever-expanding catalogue of lncRNAs first became apparent from efforts to annotate the functional features of the human genome, which showed that the vast majority of the genome was transcribed [5]. Currently, lncRNAs are defined as capped transcripts longer than 200 nucleotides, which coincides with the cut-off for many RNA extraction protocols [6]. They can be spliced and, in most of the published studies, are also polyadenylated. LncRNAs have little or no coding potential, although some do bind to ribosomes [7–9]. They were originally described to have equivalent chromatin features to protein-coding genes [10]. However, more recent work has highlighted differences in the abundance of particular histone marks [11,12] and splicing efficiency [12,13] between lncRNAs and coding genes, as well as subsets of lncRNAs that differ in their chromatin signatures [4,14].
Hundreds of thousands of lncRNAs have been annotated in different species and tissues [15]. Of those, only a handful have been shown to be critical for organism development [16–18] or cancer progression [19], and the mechanisms by which they act have been established in just a few cases. For some, biochemical partners have been carefully identified, yet in vivo evidence for their function is missing or questions have been raised regarding the relevance of the previously reported mechanisms of action [20,21]. Bridging this gap is essential for building a solid body of knowledge of how lncRNAs function in cell fate choices and the mechanisms by which they act. In this review, we will focus on the different strategies to select and identify functional lncRNAs and some mechanistic examples of lncRNA acting in differentiation and cancer.
2. Cell-type-specific expression of long non-coding RNAs: cause or consequence?
The cell-type-specific expression observed for lncRNAs has provoked much excitement, as it implies that they might function during cell fate decisions. In accord with this notion, deregulation of lncRNAs has also been widely observed across human cancers [22–25]. Such disease-associated expression changes suggested a potential role for these lncRNAs in driving cancer or at least contributing to maintaining an aberrant transcriptional landscape.
2.1. Long non-coding RNA expression during development
Differential expression of lncRNAs has been reported between regions of the mammalian brain [26,27], and lncRNA dynamics have been analysed in more detail during corticogenesis [28]. Several studies have shown differential expression of lncRNAs in in vitro differentiation models of haematopoiesis [29,30] and in freshly isolated cell populations [31,32], as well as during mammalian adipogenesis [33,34]. This tissue and cell-type-specific regulation is observed across species, including during development of zebrafish [2], Caenorhabditis elegans [3], and even during the life cell cycle of our close unicellular relative, Capsaspora owczarzaki [4].
If lncRNAs are to regulate key developmental genes, a very appealing possibility is that they do so in cis. Correlated expression of lncRNAs and their neighbouring genes has been reported in embryonic stem (ES) cell differentiation to endoderm [35] and to embryoid bodies [36], as well as in human B and T cell lineages [37]. However, neighbour correlation is not a special property of lncRNA–gene pairs, as the expression levels of neighbouring genes are often correlated. This is thought to be due to shared regulatory elements affecting each neighbour [38] and to the general neighbourhood or chromosomal domain around them [39,40].
This leaves us with several possibilities to consider, and a myriad of experimental challenges to distinguish between them. The genomic location of lncRNAs and their neighbouring regulatory elements could determine their cell-type-specific expression, with the RNA being a mere by-product of the regulatory mechanisms already in place [41]. On the other hand, lncRNAs could be critical for expression of those developmental genes, orchestrating chromatin changes by specific RNA–protein interactions, or by increasing the local concentration of transcriptional machinery regardless of the actual RNA sequence transcribed. A general mechanism by which lncRNA transcription in specific cell types could reorganize nuclear architecture, and thus contribute to the new transcription landscape, has even been proposed [15].
2.2. Long non-coding RNA misregulation in cancer
Along the same lines, cancer-specific lncRNA expression could simply be a by-product of aberrant gene expression in cancer. However, genetic mutations can directly affect lncRNA expression, with the lncRNAs themselves playing a causal role in specific scenarios. LncRNA CCAT2, for example, encompasses a cancer-associated SNP. The risk allele correlates with a higher expression of the lncRNA, which in turn promotes proliferation in colorectal cancer [42]. This lncRNA is part of the 8q24.21 region, where many cancer-associated mutations and amplifications have been reported. Several disease-associated SNPs and translocations including the lncRNA PVT1 have drawn researchers' attention to this very complex locus [43]. One of these amplifications is that of PVT1 and its neighbouring gene c-MYC. In breast cancer models PVT1 RNA levels correlate with MYC protein, yet PVT1 and c-MYC are not always co-amplified. This suggests that amplification of this lncRNA alone (even without MYC) can promote tumorigenesis in breast cancer by increasing MYC expression [44], which has been proposed to occur by protein stabilization. However, it is unlikely that this is the only mechanism of action for this lncRNA as this multi-exonic transcript encoding over 20 different isoforms is itself under the control of c-MYC and harbours multiple microRNAs within its locus [45].
While these are a few well-characterized examples, a much greater number of functional studies will be required to tease apart collaterally expressed lncRNAs from those with important roles both in development and in cancer.
3. Finding functional long non-coding RNAs
Many experimental strategies have been used to dissect genetically lncRNA requirements in differentiation and cancer. Powerful in their own ways, each of these techniques has its own drawbacks. Therefore, a combination of complementary approaches will probably be required to reveal the biological impact of lncRNAs. The choice of approach also strongly depends on the biological question, whether it is the identification of lncRNAs important for a differentiation or disease process, the study of specific types of regulatory mechanisms—cis versus trans, or an in-depth analysis of a particular lncRNA.
3.1. Different tools for different questions
The main consideration is that, in a particular lncRNA locus, the act of transcription itself could be key to establishing or maintaining the chromatin state of the surrounding area, while, in this scenario, the actual sequence of the RNA would be irrelevant. Or the RNA itself could be the functional unit, having some sequence-dependent interactions with proteins, RNAs or DNA elements. It could even be that both these mechanisms apply for the same locus. Therefore, it is very important to understand each experimental set-up and what it tells us about each particular lncRNA.
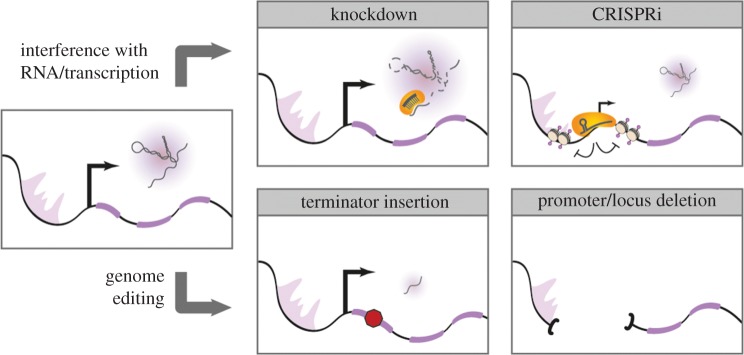
Several studies have taken advantage of RNA interference (RNAi) approaches, either transduced shRNAs or transfected siRNAs [46,47]. This strategy has been coupled with a phenotypic readout, such as viability or differentiation, to identify lncRNAs where the RNA molecule itself is important (figure 1). However, many worry about potential off-target effects (though this is no different from shRNA studies with protein-coding genes). There are additional concerns regarding the difficulty of knocking down lncRNAs that are chromatin-associated versus cytoplasmic, given that small RNA loading into the RISC complex takes places in the cytoplasm. While there is some evidence for differences in knockdown efficiency depending on subcellular location [48], this concern would apply only to lncRNAs that are never exported to the cytoplasm. LncRNAs that function in the nucleus but in trans could very well be exported just like other RNAs and then re-imported. Undoubtedly, the main advantage of knockdown is that it allows for high-throughput screens that could yield a list, though potentially incomplete, of lncRNAs with functions in the phenotypic assay of our choice.
Figure 1.
Different approaches for disrupting lncRNAs. Methods such as knockdown and CRISPRi affect the RNA itself or reduce the transcription of the lncRNA. Knockdown can be achieved in a variety of ways (siRNA, shRNA, LNA, ASO). CRISPRi is most efficient if Cas9 is fused to repressor domains (e.g. KRAB). These methods can also be transient. Insertion of an early terminator sequence or complete deletion of the locus or promoter are achieved via genome engineering and are non-reversible.
To circumvent possible subcellular localization biases, other researchers have taken advantage of alternative knockdown techniques, independent of the RNAi machinery, that do not require processing in any specific cell compartment. Morpholinos targeting splice junctions or conserved regions have been used in zebrafish for identification of functional lncRNAs in vivo [16]. Locked nucleic acids (LNAs) have also been used in mammalian cells [49,50]. Both of these approaches rely on annealing a synthetic nucleic acid to the lncRNA and blocking its function or its splicing. Antisense oligos (ASOs) are another alternative that takes advantage of RNase-H activity. ASOs have been used in a variety of systems, both delivered to cultured cells and administered in vivo in mice [19,51]. Several ASOs have now been approved for clinical use and, although their targets so far have been coding genes, this opens up the path towards therapeutic targeting of lncRNAs. These approaches, although incredibly useful because they exclusively target the RNA, can only be deployed where these molecules can be injected or otherwise delivered. This would not allow for pooled high-throughput screening and misses out on the advantages of genetically encoding knockdown, which can be conditionally induced for in vivo studies.
The ultimate proof of functionality is a genetic knockout. They allow for the study of in vivo function and reduce the possibility of off-target effects. Discrepancies between the hypothesized mechanisms for some lncRNAs based on the in vitro data and the absence of or very mild phenotypes observed in knockout animals [20,52–54] have resulted in scepticism regarding broad regulatory roles for lncRNAs [55].
Two recent examples of lncRNA knockouts emphasize how, in some cases, phenotypes might be more context-specific than anticipated. Malat1 is a very abundant lncRNA that localizes to nuclear speckles. Although it was hypothesized that this RNA was required for speckle or paraspeckle formation and maintenance and regulated alternative splicing through interaction with SR proteins [56], three independent mouse knockout models showed that Malat1 was dispensable for viability [52–54]. Furthermore, Malat1 was shown not to be required for nuclear speckle formation and its deletion did not affect SR protein phosphorylation [54]. However, when crossed with the MMTV-PyMT mouse model of human breast cancer, Malat1 deletion impaired tumour progression as evidenced by a severe reduction in metastatic burden [19].
LincRNA-EPS was shown to have an anti-apoptotic role and be required for red blood cell development in tissue culture models of erythroid development [57]. The knockout mouse model for this lncRNA showed no defects in blood development. However, LincRNA-EPS controls expression of immune response genes in macrophages and proved essential for the animals to respond to endotoxin challenge [21].
Although their molecular mechanisms still remain to be elucidated, these RNAs are representative examples of how, just as for coding genes, some lncRNAs could play roles under particular stress or disease conditions. This could potentially be the case for other lncRNAs, whose proposed roles have met with controversy, such as HOTAIR, where knockout animals seem to be viable and healthy [20,58].
When removing a DNA locus to generate a knockout—especially when dealing with large deletion—any phenotype observed could be due either to loss of an encoded RNA or to deletion of DNA sequences that might include regulatory elements. For this reason, full transcript knockouts can be combined with complementary strategies to dissect roles of RNA from those of DNA elements. The possibilities of genome engineering are not limited to full locus knockout, but instead allow more subtle modifications, such as polyadenylation (poly-A) signal insertions for premature termination (figure 1). Taking advantage of CRISPR/Cas9, a recent study looked at whether a set of 12 lncRNAs regulated the expression of their neighbouring genes in cis. Although expression defects were observed upon promoter knockout for five lncRNAs, only one had the same effect when a poly-A signal was inserted, suggesting that only the regulatory elements in the DNA surrounding the promoter and not the RNAs were required for these cis effects [59]. An equivalent mechanism has been proposed to explain the differences between knockout and poly-A insertions for the lncRNA Lockd and its neighbour gene Cdkn1b [60].
Two independent models for Fendrr knockout showed that it was required for mouse development [17,18]. Interestingly, the phenotypes differed, one being embryonic lethal with a presumed requirement for lateral plate mesoderm [17], while the other was perinatal lethal [18]. These differences could be the consequence of the distinct genetic strategies, one being a triple polyadenylation insertion and the other one a whole gene replacement, emphasizing the need for complementary approaches that distinguish between DNA and RNA elements.
The scenario is substantially more complicated when the lncRNA and its target gene have overlapping transcripts. Airn overlaps in antisense with the imprinted gene Igfr2. Through a series of polyadenylation cassette insertions, it was shown that transcriptional termination of Airn only leads to Igfr2 de-repression when the non-coding transcript no longer overlapped the Igfr2 promoter. This work concluded that transcription of Airn, rather than the final transcript, is responsible for promoter silencing [61]. Combinations of promoter, exon knockouts and termination signals created using CRISPR/Cas9 have helped dissect the relationship between Haunt (also known as linc1547 or linc-Hoxa5) and the HoxA locus. Knockdown, termination or deletions of the first exons lead to increased expression of HoxA genes during retinoid acid-induced differentiation of ES cells, supporting a repressive role for Haunt at the HoxA genes. However, deletion of the whole Haunt locus prevents expression of HoxA, presumably due to deletion of some regulatory DNA elements required for HoxA induction [62].
While more accessible owing to the advancements in genome engineering, mouse model generation is still not amenable to high-throughput studies, and therefore requires careful selection of lncRNA candidates. Genome-scale strategies for lncRNA CRISPR/Cas9 deletion are being developed [63], and variations in the ever-expanding CRISPR/Cas9 toolkit could help identify functional lncRNAs. Cas9 fused to repressors [64] or activators [65] now allows for the manipulation of expression levels at the loci themselves (figure 1). Using the former approach, researchers have identified human lncRNAs essential for cell growth in a diverse set of cell lines [64]. This technology is scalable and could help identify lncRNAs required in a variety of contexts by performing loss-of-function studies. The main limitation is that altering the chromatin state of the lncRNA promoter could directly affect nearby genes, complicating the interpretation of the phenotype.
3.2. Which long non-coding RNAs to study
The primary approach will depend on the biological question being asked. Some studies directly focus on a particular lncRNA of interest, while others aim at the unbiased identification of lncRNAs important for a process. Different strategies have been employed to select subsets of lncRNAs for study based on their expression level, dynamic regulation, tissue expression and even conservation.
Historically, highly abundant lncRNAs were chosen as representatives of this RNA class, with the hope of identifying possible mechanisms by which lncRNAs might act generally. This was, in part, due to experimental convenience and technical limitations, and this class includes the best characterized lncRNA to date, Xist, as well as Malat1, and Neat1. Xist orchestrates X chromosome inactivation. Expressed from the silenced allele, this lncRNA acts in cis to inactivate the expression of that X chromosome copy [66]. Focusing on a particular lncRNA has allowed researchers to channel all their efforts towards a mechanistic understanding of its mode of action, while also developing in vivo tools for its study. While in vivo functional validation is everyone's dream, placing all eggs in one basket is always risky and can lead to disappointing outcomes [20,52,53].
The opposite approach to studying a single lncRNA is genome-wide unbiased screening of lncRNAs. This can be an excellent filter to identify potential functional lncRNAs, building a resource for further mechanistic studies, although this approach is best suited to easily measured phenotypes, such as proliferation. Genome-wide screens have been used to identify lncRNAs essential for human cancer cell growth or survival [64], or those required in mouse ES cell self-renewal [47]. In ES cells, this approach identified TUNA, a lncRNA required for neural specification from ES cells that had already been described to have neural phenotype in zebrafish (named Megamind) [16], validating this strategy.
LncRNA annotation can be intersected with expression levels if one wishes to reduce the number of targeted lncRNAs. This can be necessary for more elaborate phenotypic assays that are not as scalable. A straightforward approach is to assess only the lncRNAs expressed in the cell type or tissue by setting some minimal expression cut-off [46]. However, for some strategies further reduction of lncRNA candidates is necessary. By analysing expression in different related tissues, especially in developmental systems, several groups have focused only on differentially regulated lncRNAs. The rationale behind this is that a transcript dynamically induced or silenced during a cell fate transition, for example, is more likely to be important for that process. Differential expression helped in the selection of candidates in epidermal differentiation [67,68], cardiac differentiation [69] and haematopoiesis [32].
Another extremely useful layer of filtering is evolutionary conservation. Although their sequence is not broadly conserved, lncRNAs can often be found in syntenic positions in different species. These ‘syntelogs’ can even share some small conserved domains [70]. This approach has been used to identify several conserved lncRNAs that act during zebrafish development [16] and drew researchers’ attention to NORAD, a conserved lncRNA in mammals that modulates Pumilio proteins [71,72]. Combined with differential expression, conservation can help focus the candidate list on lncRNAs with key functions in developmental or disease processes. As ‘syntenic conservation’ is a rather loose criterion and the presence of a lncRNA does not necessarily indicate that it will have the same function in a different organism, complementary strategies will be very helpful in identifying orthologous lncRNAs. If they are functionally conserved, lncRNA ‘syntelogs’ might share structure similarities, even if they do not share much sequence identity. Some studies have approached this by looking at predicted secondary RNA structure [73]. Although RNA structure predictions for long RNAs might not be particularly useful, new experimental approaches to globally identify structure features in lncRNAs could aid in this task [74].
When dealing with lncRNA annotation, it is important to be aware of the limitations. Although most assemblies set up stringent coding potential cut-offs, lncRNAs often contain very short open reading frames (ORFs). The functionality of these micropeptides is hard to assess unless one addresses it experimentally. Three different short proteins have been found to play a role in muscle function or regeneration [75–77], which emphasizes the importance of testing for RNA-mediated rather than protein-mediated effects.
Overall, the ability to modulate the expression of lncRNAs or disrupt it altogether now allows an assessment of lncRNA requirements in many developmental and cancer contexts. This, combined with some clever candidate selection strategies, has identified a number of lncRNAs important in these processes. The level of current mechanistic understanding for each of these lncRNAs is variable, yet the techniques available and being developed hint at a promising future.
4. Long non-coding RNAs shape development and cancer
Even for well-studied lncRNAs, our mechanistic understanding has deepened only in the last few years. The poster child for lncRNA researchers, Xist, orchestrates X chromosome inactivation. The functional properties of Xist and the order of events it directs have been known for decades (reviewed in [66]). However, it has taken until very recently to better understand the X inactivation at the molecular level. We now know the protein partners Xist requires for X chromosome silencing [78,79], how Xist spreads [80] and takes advantage of the chromosome's three-dimensional structure to initiate silencing [81], and how that chromosomal conformation changes during transcriptional silencing [82,83]. Additionally, this RNA is modified with N6-methyladenosines, which contribute to its transcriptional repressive activity [84]. Xist illustrates not only the detailed mechanistic understanding to which we can aspire for other lncRNAs of interest but also the tremendous amount of effort required to understand even a single lncRNA. Of note, Xist is also highly abundant when it is expressed, and that induction takes place in a cell type we can culture in large amounts (ES cells). Greater challenges can be expected with lncRNAs expressed to lower levels in very specific cell types.
Some hints at how other lncRNAs exert their functions in development and cancer have been reported. Although the field is still maturing, lncRNAs have been described to play a myriad of roles, from regulating gene expression to regulating mRNA processing or affecting protein stability (figure 2). There are also several examples of lncRNA loci where the act of transcription but not the RNA itself seems to be of functional relevance [59,85] or where transcription is even dispensable [59]. Here, we focus on the RNAs themselves as the functional units.
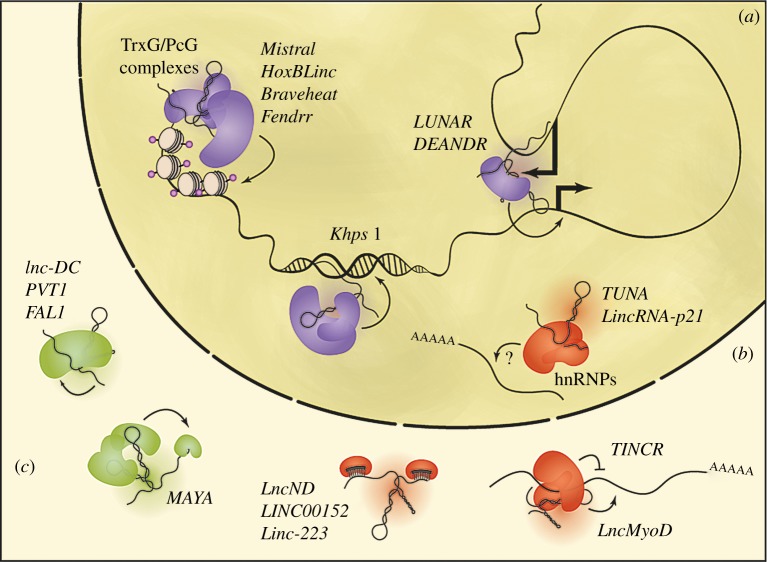
Figure 2.
Themes in lncRNA functions. LncRNAs have been described to play multiple roles affecting gene expression at the transcriptional level via interactions with chromatin remodelling complexes, direct binding to the DNA as an RNA–DNA triplex, or facilitating chromatin looping (a), interacting with RNA processing machinery or affecting mRNA stability (b) or directly regulating protein function (c).
4.1. Effects on chromatin and DNA interactions
Following Xist's example, many researchers have focused on potential chromatin regulatory roles of lncRNAs. Mistral (Mira) is a lncRNA expressed in ES cells that is reported to interact with MLL1 to recruit this protein to Hoxa6 and Hoxa7, leading to their activation. Consequently, siRNA-mediated knockdown of Mistral leads to reduced transcription of these genes and negation of the overall germ-cell specification programme [86]. Also expressed in ES cells, HoxBlinc binds to the same complex to promote HoxB transcription and mesoderm specification [87].
Two lncRNAs have been shown to be required for heart development. Braveheart, required for the production of contracting embryoid bodies from ES cells, interacts with the Polycomb factor SUZ12 [69], while Fendrr, a lncRNA essential for mouse development [17,18], binds to SUZ12 as well as EZH2 and WDR5 [17]. These interactions with members of the TrxG/MLL and Polycomb complexes place these lncRNAs in a position to direct chromatin modifications to particular DNA loci in a sequence-specific manner (figure 2a).
Following this hypothesized mechanism, it was shown that Fendrr interacts, at least in vitro, with the promoter regions of Pitx2 and Foxf1, both of which are expressed during heart development [17]. In HeLa cells, Khps1 regulates the promoter of its antisense gene, the proto-oncogene SPHK1, by forming a DNA–RNA triplex at its promoter while recruiting p300/CBP [88] (figure 2a). TARID also regulates its antisense gene, TFC21, by guiding GADD45A to the locus and promoting demethylation, which leads to gene activation. TARID and TCF21 are silent and heavily covered by DNA methylation in non-small cell lung cancer (NSCLC), head and neck squamous cell carcinomas (HNSCC) and ovarian cancers (OVC) [49]. And although direct RNA–DNA binding has not been shown, lncRNA SLNCR1 is required for bringing androgen receptor to the MMP9 promoter, which increases MMP9 expression and leads to melanoma invasion [89].
The ultraconserved lncRNA Megamind/TUNA showed brain development phenotypes upon knockdown in zebrafish [16] and is required for ES cell pluripotency and neuronal differentiation from mouse ES cells [47]. In the mouse, this lncRNA binds three RNA-binding proteins and interacts with the Sox2 promoter [47]. Sox2 is a key transcription factor in neuronal differentiation, so its regulation could explain the resulting phenotype. And while the interaction seems to be indirect, Dali has been shown to localize globally to active promoters in the N2A neuronal differentiation model [90], as shown by CHART-seq [91].
Interaction with transcription factors themselves is another plausible mechanism to promote expression of specific gene programmes. RMST, for example, is a lncRNA up-regulated during neurogenesis that interacts with Sox2. Knockdown of RMST leads to a reduction in Sox2 ChIP-seq peaks in this model, which suggests that this lncRNA is somehow facilitating the binding of this transcription factor [92].
Additionally, LUNAR and DEANR1 both seem to function by facilitating DNA looping between the lncRNA locus and their target gene to promote activation. LUNAR is a Notch-regulated lncRNA that activates the IGFR1 gene in T-cell acute lymphoblastic leukaemia [51], while DEANR1 functions via a similar mechanism in endoderm development, activating FOXA2 expression through the recruitment of SMAD2/3 [93] (figure 2a).
4.2. Effects on mRNA stability and processing
Conceptually, the next level of regulation would be for lncRNAs to negatively or positively affect the stability or processing of coding mRNAs. By having cell-type or tumour-specific expression, lncRNAs would effectively control the output levels for these genes. In accord with this model, a few exemplar lncRNAs have been shown to interact with heterogeneous nuclear ribonucleoproteins (hnRNPs) (figure 2b). LincRNA-p21, induced in response to DNA damage, interacts with hnRNP-K, and it is regulated by p53. Knockdown of this lncRNA leads to up-regulation of genes normally repressed by p53 and also reduces apoptosis similarly to p53 knockdown [94]. This suggests a model whereby this lncRNA acts as a repressor of p53-dependent genes.
In neural differentiation, Pnky knockdown leads to progenitor expansion, and mass spectroscopy of lncRNA-interacting proteins followed by immunoblotting validation revealed PTBP1 as one of its interaction partners. Knockdown of this lncRNA leads to misexpression and altered splicing of many key genes [95]. Both PTPB1 and hnRNP-K also bind TUNA during in vitro neuronal differentiation [47]. Being highly expressed and broadly acting proteins, it is only reasonable to wonder whether these functions are truly specific. Only more detailed biochemical studies will be able to clarify this.
In a complementary approach to mass spectroscopy, protein microarrays identified STAU1 as the interacting partner of lncRNA TINCR [68]. Combined knockdown of TINCR and STAU1 seems to affect the stability of important epidermal differentiation genes such as Krt80 [68], which would explain its essential role in skin differentiation. During muscle differentiation, LncMyoD binds IGF2 mRNA-binding protein 2 (IMP2), which leads to enhanced translation of mRNAs involved in proliferation. Interestingly, this is a conserved lncRNA and the mouse and human sequence can rescue each other's knockdown [96] (figure 2b).
Rather than binding elements of the RNA processing machinery as a way to regulate the fate of coding mRNAs, other lncRNAs have been shown to act as endogenous competitors for microRNAs, thus dampening the silencing of microRNA targets. LncND, for example, is a primate-conserved lncRNA expressed in neural progenitors and down-regulated in neurons. This lncRNA competes for miR-143-3p, which would normally target Notch. Relieving Notch silencing promotes neuronal differentiation [97].
In the cancer context, LINC00152 acts as an oncogenic lncRNA, competing with HIF1-α for miR-138. Expression of this lncRNA promotes invasion in gall bladder cancer [98]. As an additional example, linc-223 would usually bind to miR-125-5p but it is down-regulated in acute myeloid leukaemia, leading to increased repression of IRF4, a target of miR-125-5p [99] (figure 2b).
The range of action of lncARSR extends even further because, apart from competing with mir-34 and miR-449 thus promoting stability of AXL and c-MET, this lncRNA can be packaged into exosomes to be secreted. Down-regulation of microRNA target genes renders renal cancer cells resistant to sunitinib, and secretion of the lncRNA can disseminate this property to neighbouring cells [50].
4.3. Effects on protein stability and function
Rather than affecting the mRNAs of genes important for differentiation or malignant proliferation, lncRNAs can also directly bind proteins essential for a signalling pathway and modulate their function. Lnc-DC, for example, is induced during dendritic differentiation from human monocytes. This lncRNA interacts with STAT3 and, when knocked down, leads to a reduction in Y705 phosphorylation of STAT3, decreasing its nuclear translocation. For that reason, expression of lnc-DC is required for differentiation of dendritic cells [100] (figure 2c).
Similar protein–lncRNA relationships have been observed in different cancer models. FAL1 and PVT1 are amplified in ovarian and breast cancer, respectively. FAL1 associates with Bmi1, and FAL1 knockdown leads to a reduction in Bmi1 levels and misregulation of large numbers of genes involved in cell cycle progression [101]. PVT1 has a similar relationship with the oncogene C-MYC, promoting the stability of this protein [44] (figure 2c).
lncRNA-LET and LINK-A have opposing effects on HIF1-α in hepatocellular carcinoma. Enforced expression of lncRNA-LET leads to reduced HIF1-α and results in lower metastatic potential [102], while LINK-A interacts with tyrosine protein kinase 6 to promote stabilization of HIF1-α in triple negative breast cancer [103].
Other lncRNAs have specific roles in particular pathways, such as MAYA, a lncRNA recruited by HER3-ROR1 that then binds directly to NSUN6, preventing methylation of MST1. In breast cancer, MST1 is inactivated by this methylation resulting in YAP signalling target activation and increased bone metastasis [104] (figure 2c). SAMMSON is involved in regulating mitochondrial integrity by associating with p32. Knockdown of this lncRNA results in aberrant mitochondrial structures in melanoma, a cancer where SAMMSON is amplified [105].
By playing roles in essential cell functions, other lncRNAs also affect cancer progression. NORAD is a conserved lncRNA with repetitive regions that binds to PUMILIO proteins [71]. When this lncRNA is absent, PUMILIO proteins carry out their roles as negative regulators of mRNA stability and translation, and this results in aneuploidy [72]. LINP1 interacts with Ku80 and DNA-PKcs, coordinating the non-homologous end-joining (NHEJ) pathway. Apart from providing essential functions for any cell, this pathway is particularly required in triple negative breast cancer [106].
5. Concluding remarks
lncRNAs are being heavily studied in the context of development and cancer, as their unique properties could allow them to interact with multiple proteins via three-dimensional structures and also recognize other nucleic acids by base pairing. Their specific expression during differentiation and disease places them in an ideal position to play key regulatory roles.
Because of the added complexity in studying lncRNA loci, a combination of genetic approaches is often required to distinguish between the function of the RNA molecule and the regulatory activity from a DNA element in that locus. Many examples have been described for lncRNAs interacting with chromatin, regulating genes at the RNA or protein level, or interfering globally with splicing. These functions are diverse and expand the original hypothesis of a nuclear-specific function for most lncRNAs. It was also proposed that lncRNAs would mostly act in cis, as their expression mirrored that of their neighbour genes [36,37,107,108]. Although that is the case for some examples, it does not seem to be a general rule [59].
LncRNAs are diverse molecules that are not likely to fit in one functional class. Consequently, we should start thinking of them more like proteins, some functioning in the nucleus [109], others acting in the cytoplasm and others supporting the structure of cells [110,111]. What is becoming clearer with the development of in vivo models and our expanding mechanistic understanding is that there are lncRNAs with essential functions in development and others required for cancer progression, taking this class of RNAs out of the ‘junk DNA’ category once and for all. Not every annotated lncRNA will have an RNA-mediated function—or a function at all—but identifying the biologically relevant ones and understanding their mechanisms will certainly be a hotbed for future study.
Acknowledgements
We thank the members of the Hannon laboratory for helpful discussion and their critical reading of the manuscript.
Data accessibility
This article has no additional data.
Competing interests
The authors have no relevant competing interests.
Funding
M.J.D. was funded by a PhD fellowship from Boehringer Ingelheim Fonds, a graduate studies fellowship from ‘la Caixa’ Foundation and by the Watson School of Biological Sciences. G.J.H. is a Wellcome Trust Investigator, was an investigator of the Howard Hughes Medical Institute, and is supported by Cancer Research UK, The Royal Society (Wolfson Professorship) and a generous gift from Kathryn W. Davis.
References
- 1.Ponting CP, Oliver PL, Reik W. 2009. Evolution and functions of long noncoding RNAs. Cell 136, 629–641. (doi:10.1016/j.cell.2009.02.006) [DOI] [PubMed] [Google Scholar]
- 2.Pauli A, et al. 2012. Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res. 22, 577–591. (doi:10.1101/gr.133009.111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nam J-W, Bartel DP. 2012. Long noncoding RNAs in C. elegans. Genome Res. 22, 2529–2540. (doi:10.1101/gr.140475.112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sebé-Pedrós A, Ballaré C, Parra-Acero H, Chiva C, Tena JJ, Sabidó E, Gómez-Skarmeta JL, Di Croce L, Ruiz-Trillo I. 2016. The dynamic regulatory genome of Capsaspora and the origin of animal multicellularity. Cell 165, 1224–1237. (doi:10.1016/j.cell.2016.03.034) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.ENCODE Project Consortium et al. 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447, 799–816. (doi:10.1038/nature05874) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rinn JL, Chang HY. 2012. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 81, 145–166. (doi:10.1146/annurev-biochem-051410-092902) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ingolia NT, Lareau LF, Weissman JS. 2011. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147, 789–802. (doi:10.1016/j.cell.2011.10.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guttman M, Russell P, Ingolia NT, Weissman JS, Lander ES. 2013. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell 154, 240–251. (doi:10.1016/j.cell.2013.06.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruiz-Orera J, Messeguer X, Subirana JA, Alba MM, Tautz D. 2014. Long non-coding RNAs as a source of new peptides. Elife 3, e03523 (doi:10.7554/eLife.03523) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guttman M, et al. 2009. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458, 223–227. (doi:10.1038/nature07672) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alam T, Medvedeva YA, Jia H, Brown JB, Lipovich L, Bajic VB. 2014. Promoter analysis reveals globally differential regulation of human long non-coding RNA and protein-coding genes. PLoS ONE 9, e109443 (doi:10.1371/journal.pone.0109443) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Melé M., Mattioli K, Mallard W, Shechner DM, Gerhardinger C, Rinn JL. 2017. Chromatin environment, transcriptional regulation, and splicing distinguish lincRNAs and mRNAs. Genome Res. 27, 27–37. (doi:10.1101/gr.214205.116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schlackow M, Nojima T, Gomes T, Dhir A, Carmo-Fonseca M, Proudfoot NJ. 2017. Distinctive patterns of transcription and RNA processing for human lincRNAs. Mol. Cell 65, 25–38. (doi:10.1016/j.molcel.2016.11.029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marques AC, Hughes J, Graham B, Kowalczyk MS, Higgs DR, Ponting CP. 2013. Chromatin signatures at transcriptional start sites separate two equally populated yet distinct classes of intergenic long noncoding RNAs. Genome Biol. 14, R131 (doi:10.1186/gb-2013-14-11-r131) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Melé M, Rinn JL. 2016. ‘Cat's cradling’ the 3D genome by the act of lncRNA transcription. Mol. Cell 62, 657–664. (doi:10.1016/j.molcel.2016.05.011) [DOI] [PubMed] [Google Scholar]
- 16.Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. 2011. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell 147, 1537–1550. (doi:10.1016/j.cell.2011.11.055) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grote P, et al. 2013. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Cell 24, 206–214. (doi:10.1016/j.devcel.2012.12.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sauvageau M, et al. 2013. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. Elife 2, e01749 (doi:10.7554/eLife.01749) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arun G, et al. 2016. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. 30, 34–51. (doi:10.1101/gad.270959.115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amândio AR, Necsulea A, Joye E, Mascrez B, Duboule D. 2016. Hotair is dispensible for mouse development. PLoS Genet. 12, e1006232 (doi:10.1371/journal.pgen.1006232) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Atianand MK, et al. 2016. A long noncoding RNA lincRNA-EPS acts as a transcriptional brake to restrain inflammation. Cell 165, 1672–1685. (doi:10.1016/j.cell.2016.05.075) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan X, et al. 2015. Comprehensive genomic characterization of long non-coding RNAs across human cancers. Cancer Cell 28, 529–540. (doi:10.1016/j.ccell.2015.09.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iyer MK, et al. 2015. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet 47, 199–208. (doi:10.1038/ng.3192) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Du Z, Fei T, Verhaak RG. W., Su Z, Zhang Y, Brown M, Chen Y, Liu XS. 2013. Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nat. Struct. Mol. Biol. 20, 908–913. (doi:10.1038/nsmb.2591) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brunner AL, Beck AH, Edris B. 2012. Transcriptional profiling of long non-coding RNAs and novel transcribed regions across a diverse panel of archived human cancers. Genome Biol. 13, R75 (doi:10.1186/gb-2012-13-8-r75) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. 2008. Specific expression of long noncoding RNAs in the mouse brain. Proc. Natl Acad. Sci. USA 105, 716–721. (doi:10.1073/pnas.0706729105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goff LA, et al. 2015. Spatiotemporal expression and transcriptional perturbations by long noncoding RNAs in the mouse brain. Proc. Natl Acad. Sci. USA 112, 6855–6862. (doi:10.1073/pnas.1411263112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Molyneaux BJ, Goff LA, Brettler AC, Chen H.-H., Brown JR, Hrvatin S, Rinn JL, Arlotta P. 2015. DeCoN: genome-wide analysis of in vivo transcriptional dynamics during pyramidal neuron fate selection in neocortex. Neuron 85, 275–288. (doi:10.1016/j.neuron.2014.12.024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alvarez-Dominguez JR, Hu W, Yuan B, Shi J, Park SS, Gromatzky AA, van Oudenaarden A, Lodish HF. 2014. Global discovery of erythroid long noncoding RNAs reveals novel regulators of red cell maturation. Blood 123, 570–581. (doi:10.1182/blood-2013-10-530683) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paralkar VR, et al. 2014. Lineage and species-specific long noncoding RNAs during erythro-megakaryocytic development. Blood 123, 1927–1937. (doi:10.1182/blood-2013-12-544494) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cabezas-Wallscheid N, et al. 2014. Identification of regulatory networks in HSCs and their immediate progeny via integrated proteome, transcriptome, and DNA methylome analysis. Cell Stem Cell 15, 507–522. (doi:10.1016/j.stem.2014.07.005) [DOI] [PubMed] [Google Scholar]
- 32.Luo M, et al. 2015. Long non-coding RNAs control hematopoietic stem cell function. Cell Stem Cell 16, 426–438. (doi:10.1016/j.stem.2015.02.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alvarez-Dominguez JR, et al. 2015. De novo reconstruction of adipose tissue transcriptomes reveals long non-coding RNA regulators of brown adipocyte development. Cell Metab. 21, 764–776. (doi:10.1016/j.cmet.2015.04.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun L, et al. 2013. Long noncoding RNAs regulate adipogenesis. Proc. Natl Acad. Sci. USA 110, 3387–3392. (doi:10.1073/pnas.1222643110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sigova AA, et al. 2013. Divergent transcription of long noncoding RNA/mRNA gene pairs in embryonic stem cells. Proc. Natl Acad. Sci. USA 110, 2876–2881. (doi:10.1073/pnas.1221904110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dinger ME, et al. 2008. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 18, 1433–1445. (doi:10.1101/gr.078378.108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Casero D, Sandoval S, Seet CS, Scholes J, Zhu Y, Ha VL, Luong A, Parekh C, Crooks GM. 2015. Long non-coding RNA profiling of human lymphoid progenitor cells reveals transcriptional divergence of B cell and T cell lineages. Nat. Immunol. 16, 1282–1291. (doi:10.1038/ni.3299) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cohen BA, Mitra RD, Hughes JD, Church GM. 2000. A computational analysis of whole-genome expression data reveals chromosomal domains of gene expression. Nat. Genet 26, 183–186. (doi:10.1038/79896) [DOI] [PubMed] [Google Scholar]
- 39.Dowen JM, et al. 2014. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell 159, 374–387. (doi:10.1016/j.cell.2014.09.030) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gonzalez-Sandoval A, Gasser SM. 2016. On TADs and LADs: spatial control over gene expression. Trends Genet. 32, 485–495. (doi:10.1016/j.tig.2016.05.004) [DOI] [PubMed] [Google Scholar]
- 41.van Bakel H, Nislow C, Blencowe BJ, Hughes TR. 2010. Most ‘dark matter’ transcripts are associated with known genes. PLoS Biol. 8, e1000371 (doi:10.1371/journal.pbio.1000371) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ling H, et al. 2013. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res. 23, 1446–1461. (doi:10.1101/gr.152942.112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huppi K, Pitt JJ, Wahlberg BM, Caplen NJ. 2012. The 8q24 gene desert: an oasis of non-coding transcriptional activity. Front Genet. 3, 69 (doi:10.3389/fgene.2012.00069) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tseng Y-Y, et al. 2014. PVT1 dependence in cancer with MYC copy-number increase. Nature 512, 82–86. (doi:10.1038/nature13311) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carramusa L, Contino F, Ferro A, Minafra L, Perconti G, Giallongo A, Feo S. 2007. The PVT-1 oncogene is a Myc protein target that is overexpressed in transformed cells. J. Cell. Physiol. 213, 511–518. (doi:10.1002/jcp.21133) [DOI] [PubMed] [Google Scholar]
- 46.Guttman M, et al. 2011. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477, 295–300. (doi:10.1038/nature10398) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin N, et al. 2014. An evolutionarily conserved long noncoding RNA TUNA controls pluripotency and neural lineage commitment. Mol. Cell 53, 1005–1019. (doi:10.1016/j.molcel.2014.01.021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lennox KA, Behlke MA. 2016. Cellular localization of long non-coding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic Acids Res. 44, 863–877. (doi:10.1093/nar/gkv1206) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arab K, et al. 2014. Long noncoding RNA TARID directs demethylation and activation of the tumor suppressor TCF21 via GADD45A. Mol. Cell 55, 604–614. (doi:10.1016/j.molcel.2014.06.031) [DOI] [PubMed] [Google Scholar]
- 50.Qu L, et al. 2016. Exosome-transmitted lncARSR promotes sunitinib resistance in renal cancer by acting as a competing endogenous RNA. Cancer Cell 29, 653–668. (doi:10.1016/j.ccell.2016.03.004) [DOI] [PubMed] [Google Scholar]
- 51.Trimarchi T, Bilal E, Ntziachristos P, Fabbri G, Dalla-Favera R, Tsirigos A, Aifantis I. 2014. Genome-wide mapping and characterization of Notch-regulated long noncoding RNAs in acute leukemia. Cell 158, 593–606. (doi:10.1016/j.cell.2014.05.049) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nakagawa S, Ip JY, Shioi G, Tripathi V, Zong X, Hirose T, Prasanth KV. 2012. Malat1 is not an essential component of nuclear speckles in mice. RNA 18, 1487–1499. (doi:10.1261/rna.033217.112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eißmann M, et al. 2012. Loss of the abundant nuclear non-coding RNAMALAT1 is compatible with life and development. RNA Biol. 9, 1076–1087. (doi:10.4161/rna.21089) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang B, et al. 2012. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. 2, 111–123. (doi:10.1016/j.celrep.2012.06.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Selleri L, Bartolomei MS, Bickmore WA, He L, Stubbs L, Reik W, Barsh GS. 2016. A hox-embedded long noncoding RNA: is it all hot air? PLoS Genet. 12, e1006485 (doi:10.1371/journal.pgen.1006485) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tripathi V, et al. 2010. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 39, 925–938. (doi:10.1016/j.molcel.2010.08.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hu W, Yuan B, Flygare J, Lodish HF. 2011. Long noncoding RNA-mediated anti-apoptotic activity in murine erythroid terminal differentiation. Genes Dev. 25, 2573–2578. (doi:10.1101/gad.178780.111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li L, et al. 2013. Targeted disruption of hotair leads to homeotic transformation and gene derepression. Cell Rep. 5, 3–12. (doi:10.1016/j.celrep.2013.09.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Engreitz JM, Haines JE, Perez EM, Munson G, Chen J, Kane M, McDonel PE, Guttman M, Lander ES. 2016. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 539, 452–455. (doi:10.1038/nature20149) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Paralkar VR, et al. 2016. Unlinking an lncRNA from its associated cis element. Mol. Cell 62, 104–110. (doi:10.1016/j.molcel.2016.02.029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Latos PA, et al. 2012. Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science 338, 1469–1472. (doi:10.1126/science.1228110) [DOI] [PubMed] [Google Scholar]
- 62.Yin Y, et al. 2015. Opposing roles for the lncRNA haunt and its genomic locus in regulating HOXA gene activation during embryonic stem cell differentiation. Cell Stem Cell 16, 504–516. (doi:10.1016/j.stem.2015.03.007) [DOI] [PubMed] [Google Scholar]
- 63.Zhu S, et al. 2016. Genome-scale deletion screening of human long non-coding RNAs using a paired-guide RNA CRISPR-Cas9 library. Nat. Biotechnol. 34, 1279–1286. (doi:10.1038/nbt.3715) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu SJ, et al. 2017. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science 355, pii. aah7111 (doi:10.1126/science.aah7111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Konermann S, et al. 2014. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517, 583–588. (doi:10.1038/nature14136) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Augui S, Nora EP, Heard E. 2011. Regulation of X-chromosome inactivation by the X-inactivation centre. Nat. Rev. Genet. 12, 429–442. (doi:10.1038/nrg2987) [DOI] [PubMed] [Google Scholar]
- 67.Kretz M, et al. 2012. Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes Dev. 26, 338–343. (doi:10.1101/gad.182121.111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kretz M, et al. 2013. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature 493, 231–235. (doi:10.1038/nature11661) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Klattenhoff CA, et al. 2013. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell 152, 570–583. (doi:10.1016/j.cell.2013.01.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ulitsky I. 2016. Evolution to the rescue: using comparative genomics to understand long non-coding RNAs. Nat. Rev. Genet. 17, 601–614. (doi:10.1038/nrg.2016.85) [DOI] [PubMed] [Google Scholar]
- 71.Tichon A, Gil N, Lubelsky Y, Solomon TH, Lemze D, Itzkovitz S, Stern-Ginossar N, Ulitsky I. 2016. A conserved abundant cytoplasmic long noncoding RNA modulates repression by Pumilio proteins in human cells. Nat. Commun. 7, 1–10. (doi:10.1038/ncomms12209) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee S, Kopp F, Chang T-C, Sataluri A, Chen B, Sivakumar S, Yu H, Xie Y, Mendell JT. 2016. Noncoding RNA NORAD regulates genomic stability by sequestering PUMILIO proteins. Cell 164, 69–80. (doi:10.1016/j.cell.2015.12.017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Quinn JJ, Zhang QC, Georgiev P, Ilik IA, Akhtar A, Chang HY. 2016. Rapid evolutionary turnover underlies conserved lncRNA–genome interactions. Genes Dev. 30, 191–207. (doi:10.1101/gad.272187.115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lu Z, et al. 2016. RNA duplex map in living cells reveals higher-order transcriptome structure. Cell 165, 1267–1279. (doi:10.1016/j.cell.2016.04.028) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Matsumoto A, et al. 2016. mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide. Nature 541, 228–232. (doi:10.1038/nature21034) [DOI] [PubMed] [Google Scholar]
- 76.Anderson DM, et al. 2015. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell 160, 595–606. (doi:10.1016/j.cell.2015.01.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nelson BR, et al. 2016. A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science 351, 271–275. (doi:10.1126/science.aad4076) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chu C, Zhang QC, da Rocha ST, Flynn RA, Bharadwaj M, Calabrese JM, Magnuson T, Heard E, Chang HY. 2015. Systematic discovery of Xist RNA binding proteins. Cell 161, 404–416. (doi:10.1016/j.cell.2015.03.025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.McHugh CA, et al. 2015. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 521, 232–236. (doi:10.1038/nature14443) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Simon MD, et al. 2013. High-resolution Xist binding maps reveal two-step spreading during X-chromosome inactivation. Nature 504, 465–469. (doi:10.1038/nature12719) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Engreitz JM, et al. 2013. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science 341, 1237973 (doi:10.1126/science.1237973) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Giorgetti L, et al. 2016. Structural organization of the inactive X chromosome in the mouse. Nature 535, 575–579. (doi:10.1038/nature18589) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Darrow EM, et al. 2016. Deletion of DXZ4 on the human inactive X chromosome alters higher-order genome architecture. Proc. Natl Acad. Sci. USA 113, E4504–E4512. (doi:10.1073/pnas.1609643113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Patil DP, Chen C-K, Pickering BF, Chow A, Jackson C, Guttman M, Jaffrey SR. 2016. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537, 369–373. (doi:10.1038/nature19342) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Anderson KM, Anderson DM, McAnally JR, Shelton JM, Bassel-Duby R, Olson EN. 2016. Transcription of the non-coding RNA upperhand controls Hand2 expression and heart development. Nature 539, 433–436. (doi:10.1038/nature20128) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bertani S, Sauer S, Bolotin E, Sauer F. 2011. The noncoding RNA mistral activates Hoxa6 and Hoxa7 expression and stem cell differentiation by recruiting MLL1 to chromatin. Mol. Cell 43, 1040–1046. (doi:10.1016/j.molcel.2011.08.019) [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 87.Deng C, et al. 2016. HoxBlinc RNA recruits Set1/MLL complexes to activate Hox gene expression patterns and mesoderm lineage development. Cell Rep. 14, 103–114. (doi:10.1016/j.celrep.2015.12.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Postepska-Igielska A, Giwojna A, Gasri-Plotnitsky L, Schmitt N, Dold A, Ginsberg D, Grummt I. 2015. LncRNA Khps1 regulates expression of the proto-oncogene SPHK1 via triplex-mediated changes in chromatin structure. Mol. Cell 60, 626–636. (doi:10.1016/j.molcel.2015.10.001) [DOI] [PubMed] [Google Scholar]
- 89.Schmidt K, Joyce CE, Buquicchio F, Brown A, Ritz J, Distel RJ, Yoon CH, Novina CD. 2016. The lncRNA SLNCR1 mediates melanoma invasion through a conserved SRA1-like region. Cell Rep. 15, 2025–2037. (doi:10.1016/j.celrep.2016.04.018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chalei V, Sansom SN, Kong L, Lee S, Montiel JF, Vance KW, Ponting CP. 2014. The long non-coding RNA Dali is an epigenetic regulator of neural differentiation. Elife 3, e04530 (doi:10.7554/eLife.04530) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Simon MD, Wang CI, Kharchenko PV, West JA, Chapman BA, Alekseyenko AA, Borowsky ML, Kuroda MI, Kingston RE. 2011. The genomic binding sites of a noncoding RNA. Proc. Natl Acad. Sci. USA 108, 20 497–20 502. (doi:10.1073/pnas.1113536108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ng S-Y, Bogu GK, Soh B.-S, Stanton LW. 2013. The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol. Cell 51, 349–359. (doi:10.1016/j.molcel.2013.07.017) [DOI] [PubMed] [Google Scholar]
- 93.Jiang W, Liu Y, Liu R, Zhang K, Zhang Y. 2015. The lncRNA DEANR1 facilitates human endoderm differentiation by activating FOXA2 expression. Cell Rep. 11, 137–148. (doi:10.1016/j.celrep.2015.03.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Huarte M, et al. 2010. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 142, 409–419. (doi:10.1016/j.cell.2010.06.040) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ramos AD, et al. 2015. The long noncoding RNA Pnky regulates neuronal differentiation of embryonic and postnatal neural stem cells. Cell Stem Cell 16, 439–447. (doi:10.1016/j.stem.2015.02.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gong C, Li Z, Ramanujan K, Clay I, Zhang Y, Lemire-Brachat S, Glass DJ. 2015. A long non-coding RNA, LncMyoD, regulates skeletal muscle differentiation by blocking IMP2-mediated mRNA translation. Dev. Cell 34, 181–191. (doi:10.1016/j.devcel.2015.05.009) [DOI] [PubMed] [Google Scholar]
- 97.Rani N, Nowakowski TJ, Zhou H, Godshalk SE, Lisi V, Kriegstein AR, Kosik KS. 2016. A primate lncRNA mediates Notch signaling during neuronal development by sequestering miRNA. Neuron 90, 1174–1188. (doi:10.1016/j.neuron.2016.05.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cai Q, Wang Z, Wang S, Weng M, Zhou D, Li C, Wang J, Chen E, Quan Z. 2017. Long non-coding RNA LINC00152 promotes gallbladder cancer metastasis and epithelial–mesenchymal transition by regulating HIF-1α via miR-138. Open Biol. 7, 160247 (doi:10.1002/hep.24563) [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 99.Mangiavacchi A, Sorci M, Masciarelli S, Larivera S, Legnini I, Iosue I, Bozzoni I, Fazi F, Fatica A. 2016. The miR-223 host non-coding transcript linc-223 induces IRF4 expression in acute myeloid leukemia by acting as a competing endogenous RNA. Oncotarget 7, 60 155–60 168. (doi:10.18632/oncotarget.11165) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang P, et al. 2014. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science 344, 310–313. (doi:10.1126/science.1251456) [DOI] [PubMed] [Google Scholar]
- 101.Hu X, et al. 2014. A functional genomic approach identifies FAL1 as an oncogenic long noncoding RNA that associates with BMI1 and represses p21 expression in cancer. Cancer Cell 26, 344–357. (doi:10.1016/j.ccr.2014.07.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yang F, Huo X-S, Yuan S-X, Zhang L, Zhou W-P, Wang F, Sun S-H. 2013. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Mol. Cell 49, 1083–1096. (doi:10.1016/j.molcel.2013.01.010) [DOI] [PubMed] [Google Scholar]
- 103.Lin A, et al. 2016. The LINK-A lncRNA activates normoxic HIF1α signalling in triple-negative breast cancer. Nat. Cell Biol. 18, 213–224. (doi:10.1038/ncb3295) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Li C, et al. 2017. A ROR1–HER3–lncRNA signalling axis modulates the Hippo–YAP pathway to regulate bone metastasis. Nat. Cell Biol. 19, 106–119. (doi:10.1038/ncb3464) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Leucci E, et al. 2016. Melanoma addiction to the long non-coding RNA SAMMSON. Nature 531, 518–522. (doi:10.1038/nature17161) [DOI] [PubMed] [Google Scholar]
- 106.Zhang Y, et al. 2016. Long noncoding RNA LINP1 regulates repair of DNA double-strand breaks in triple-negative breast cancer. Nat. Struct. Mol. Biol. 23, 522–530. (doi:10.1038/nsmb.3211) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang KC, et al. 2011. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472, 120–124. (doi:10.1038/nature09819) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Amaral PP, et al. 2016. Genomic positional conservation identifies topological anchor point (tap)RNAs linked to developmental loci. Pre-print at bioRxiv. (doi:10.1101/051052) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Engreitz JM, Ollikainen N, Guttman M. 2016. Long non-coding RNAs: spatial amplifiers that control nuclear structure and gene expression. Nat. Rev. Mol. Cell Biol. 17, 756–770. (doi:10.1038/nrm.2016.126) [DOI] [PubMed] [Google Scholar]
- 110.Hacisuleyman E, et al. 2014. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat. Struct. Mol. Biol. 21, 198–206. (doi:10.1038/nsmb.2764) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. 2009. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell 33, 717–726. (doi:10.1016/j.molcel.2009.01.026) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.