Abstract
Ubiquitin (Ub) has a broad functional range that has been ascribed to the formation of an array of polymeric ubiquitin chains. Understanding the precise roles of ubiquitin chains, however, is difficult due to their complex chain topologies. Branched ubiquitin chains are particularly challenging, as multiple modifications on a single ubiquitin preclude the use of standard bottom-up proteomic approaches. Developing methods to overcome these challenges is crucial considering evidence suggesting branched chains regulate the stability of proteins. In this study, we employ Ubiquitin Chain Enrichment Middle-down Mass Spectrometry (UbiChEM-MS) to identify branched chains that cannot be detected using bottom-up proteomic methods. Specifically, we employ tandem ubiquitin binding entities (TUBEs) and the K29-selective Npl4 Zinc Finger 1 (NZF1) domain from the deubiquitinase TRABID to enrich for chains from human cells. Minimal trypsinolysis followed by high resolution mass spectrometric analysis reveals that Ub chain branching can indeed be detected using both Ub binding domains (UBDs) tested at endogenous levels. We find that ∼1% of chains isolated with TUBEs contain Ub branch points, with this value rising to ∼4% after proteasome inhibition. Electron-transfer dissociation (ETD) analysis indicates the presence of K48 in these branched chains. The use of the NZF1 domain reveals that ∼4% of the isolated chains contain branch points with no apparent dependence on proteasome inhibition. Our results demonstrate an effective strategy for detecting and characterizing the dynamics of branched conjugates under different cellular conditions.
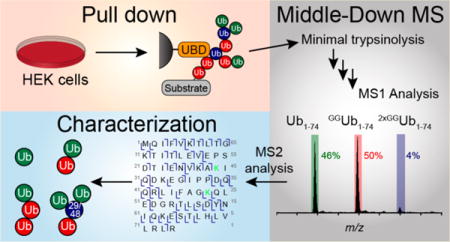
Graphical abstract
Over the last few decades, it has become widely appreciated that ubiquitin (Ub) plays a major role in regulating nearly every cellular pathway in eukaryotes. The cell cycle, for example, is driven by Ub-mediated degradation;1 the cellular response to DNA damage2 and our ability to ward off pathogens are also both intricately linked to Ub signaling.3 How Ub regulates the dynamics of such diverse biochemical pathways is thought to be a result of Ub chain formation.
Ub chains are generated after consecutive rounds of target protein modification by E1, E2, and E3 enzymes.4 After Ub is transferred to the ε-amino group of a target lysine, any of the eight amino groups of Ub (M1, K6, K11, K27, K29, K33, K48, and K63) can be attached to the C-terminus of another Ub leading to the formation of a Ub chain.5 During chain extension, there can be tremendous variability in linkage, length, and configuration (i.e., linear versus branched). This versatility is what likely gives rise to the multitude of cellular functions associated with Ub. Mounting evidence suggests that the type of ubiquitin chain determines the cellular fate of a modified protein. For instance, K11- and K48-linked chains signal for protein degradation through the proteasome, whereas M1- and K63-linked chains promote proteasome-independent activities.6–15
While there has been significant interest in understanding the functional impact of different chain linkages, much less attention has been paid to the importance of chain configuration. Initial reports on branched chains focused on bifurcations at consecutive lysines of Ub (e.g., K6/K11, K27/K29, or K29/K33) since these modifications can be detected using bottom-up proteomic approaches.16,17 On the basis of in vitro proteasome assays, bifurcated chains were found to be less effective at promoting protein degradation compared to chains composed of a single linkage.16,18 Whether these observations are solely due to chain branching, however, is unclear given the heterogeneity of the systems under investigations. More recently, there has been evidence suggesting K11/K48 branched chains produced by the anaphase-promoting complex/cyclosome (APC/C) are capable of triggering proteasomal degradation.19,20 In this case, K11/K48 branched chains were detected on APC/C substrates by expressing nonnative Ub variants in cells. Other branched chains unamenable to tryptic fragment mass spectrometric analyses have also been implicated in several fundamental cellular pathways. For example, M1/K63 chains have been proposed to regulate NF-κB signaling;21,22 K11/K63 conjugates are thought to play a role in receptor internalization,23,24 and K29/K48 branched chains have been linked to proteasomal degradation.25,26 Further defining the roles of branched chains in regulating protein stability and other cellular pathways requires new methods of detection and characterization.
In this study, we demonstrate the utility of Ubiquitin Chain Enrichment Middle-down Mass Spectrometry (UbiChEM-MS) in characterizing branched Ub chains present in human cell extracts. Previous reports from our lab and others have shown that, when integrated with MS analysis, minimal trypsinolysis and limited acid cleavage retain information on the connectivity of Ub chains, particularly branched chains.27–31 When Ub is subjected to trypsinolysis under nondenaturing conditions, a single cleavage occurs between Arg74 and Gly75 leaving behind an intact Ub1–74 fragment (calcd 8450.57 Da) as well as a Gly-Gly remnant.32 In the event of minimal trypsinolysis of Ub chains, this Gly-Gly remnant remains attached to the modified lysine resulting in a Ub1–74 fragment modified with a single Gly-Gly (GGUb1–74, calcd 8564.62 Da). If a branch point is present within the Ub chain, the Ub moiety bearing the branch point will be modified with two Gly-Gly modifications (2xGGUb1–74, calcd 8678.66 Da), thus enabling characterization of multiple modifications on a single Ub protein. Although useful for characterizing the products of E2–E3 enzymatic activity in vitro, these middle-down methods have not led to the identification of branched chains in complex mixtures such as cell extracts. We envisioned that, by lysing cells grown under different conditions and enriching particular Ub chains using linkage-selective Ub binding domains (UBDs),5,33 branched conjugates could be detected and characterized by middle-down MS. Here, we show this strategy is brought to fruition using both the nonselective tandem ubiquitin binding entities (TUBEs) and the K29 linkage-selective Npl4 Zinc Finger 1 (NZF1) domain from the deubiquitinase TRABID. Our results reveal that UbiChEM-MS can be used to not only detect previously undetectable branched chains but also monitor their dynamics. We surmise this same approach can readily be adapted for the detection and characterization of many other branched chains, provided a linkage specific binding domain exists.
EXPERIMENTAL SECTION
Preparation of Halo-NZF1 Resin
HaloLink resin (1.6 mL, Promega) was pelleted at 800g for 2 min, and the supernatant was removed. The resin was washed four times with binding buffer (2 mL; 50 mM Tris, 150 mM NaCl, 10% glycerol, 0.05% IGEPAL CA-630) and then finally resuspended in binding buffer to yield a 50% slurry. This slurry was then incubated with 800 μL of Halotag-NZF1 fusion protein (115 μM), and the resulting suspension was rocked overnight at 4 °C. The suspension was then pelleted at 800g for 2 min; the supernatant was discarded, and the resin was further washed four times with 5 mL of binding buffer.
Isolation of Ubiquitin Chains from HEK Cell Lysate
Resin bearing the appropriate ubiquitin binding domain (100 μL of TUBEs or 200 μL of NZF1) was incubated with 45 mg of cell lysate on a revolver overnight at 4 °C, after which the resin was pelleted at 800g for 2 min. The resin was then washed three times with 2 mL of binding buffer and twice with 2 mL of minimal buffer (50 mM Tris, 150 mM NaCl, pH 7.5). The resin was then resuspended in 100 μL of minimal buffer. 6× Laemmli loading buffer was added to aliquots of each step, separated on a 15% SDS-PAGE gel, and analyzed by Western blot with anti-Ub antibody (P4D1).
On-Resin Minimal Trypsinolysis of Isolated Ub Chains
Minimal trypsinolysis was performed on-resin at room temperature for 16 h using empirically determined ratios of lysate to trypsin (Table S1). These on-resin samples were then acidified to pH 2 with acetic acid to deactivate trypsin. These acidified solutions were incubated at 4 °C for 15–30 min and then centrifuged for 5 min at 13 000g, 4 °C. For samples analyzed by LC MS/MS, the supernatant was concentrated ∼5–7-fold using a speedvac. Samples were centrifuged for 30 min at 16 000g, 4 °C, prior to injection into the mass spectrometer. For samples analyzed by direct infusion into the mass spectrometer, the supernatant was pipetted into a 100 mg SeP-Pak C18 column (Waters) which was washed with 10 mL of equilibration solution (0.1% TFA in water). This column was then washed with 3 mL of equilibration solution, followed by collecting 1.5 mL fractions of 10%, 20%, 30%, 40%, 50%, 60%, 70%, and 100% acetonitrile in 0.1% TFA solutions. The 0–60% fractions were lyophilized. Lyophilized samples from the 30% and 40% fractions were dissolved in 200 μL of a water/acetonitrile/acetic acid (45:45:10) solution and analyzed by MALDI-TOF. Then, the samples containing Ub species were further analyzed by high resolution mass spectrometry.
Middle-Down Mass Spectrometry and Quantitative Analysis with Orbitrap Fusion
Samples were directly infused into an Orbitrap Fusion Tribrid mass spectrometer (Thermo Scientific Inc.), where the resolving power of the mass analyzer was set at 60 000. All spectra were processed with in-house software (MASH Suite34) using a signal-to-noise (S/N) threshold of 3 and a fit factor of 70% and then validated manually; all reported calculated and experimental values correspond to the most abundant molecular weights. Quantitative analysis of the relative abundance of the three Ub1–74 species was performed as described previously.35,36 The relative abundances of each Ub species in the 700–1300 m/z range were averaged to obtain the relative percentage. Percent distribution was calculated from three biological replicates, and data were represented as mean ± standard error of the mean (SEM).
Liquid Chromatography Middle-Down Mass Spectrometry and Quantitative Analysis with Bruker Maxis Impact LCMS System
Minimally digested products were separated using a nanoAcquity LC system (Waters) equipped with a replacement capillary custom packed trap (nanoLCMS Solutions LLC) followed by a home-packed PLRP-S column and a gradient from 5% B to 95% B over 43 min (solvent A, 0.1% formic acid in water; solvent B, 0.1% formic acid in 1:1 solution of acetonitrile and ethanol). The LC system was coupled online with a Bruker Impact II Q-TOF mass spectrometer, where the resolving power of the mass analyzer was set at 50 000. All spectra were processed with in-house software (MASH Suite34) using a signal-to-noise (S/N) threshold of 3 and a fit factor of 70% and then validated manually; all reported calculated and experimental values correspond to the most abundant molecular weights. Quantitative analysis of the relative abundance of the three Ub1–74 species was performed as described previously.35,36 The relative abundances of each Ub species in the 600–1250 m/z range were averaged to obtain the relative percentage. The percent distribution was calculated from three biological replicates, each with three technical replicates, and data were represented as the mean ± standard error of the mean (SEM).
Electron-Transfer Dissociation (ETD) Analysis of Ub Chain Linkages on Orbitrap Fusion
For tandem mass spectrometry (MS/MS) experiments, individual charge states of protein molecular ions were isolated and then dissociated by ETD using a 10 ms reaction time, with a 2.0 × 105 reagent ion target. ETD was also supplemented by collision induced decay (CID) using a collision energy of 10%. Spectra were recorded in the Orbitrap analyzer over an m/z range of 150–2000 with a resolution setting of 60 000 and AGC setting of 4 × 105 charges. All spectra were processed with the MASH suite software34 using a S/N threshold of 3 and a fit factor of 70% and then validated manually. The resulting mass lists were further assigned on the basis of the protein sequence of Ub with or without the diglycine (GG) modification at each lysine using a tolerance of 20 ppm for precursor and fragment ions. All reported calculated (calcd) and experimental (exptl) values correspond to the monoisotopic molecular weight.
ETD Analysis of Ub Chain Linkages on Bruker Maxis Impact LCMS System
Minimally digested products were separated using a nanoAcquity LC system (Waters) equipped with replacement capillary custom packed trap (nanoLCMS Solutions LLC) followed by a home-packed PLRP-S column and a gradient from 5% B to 95% B over 43 min (solvent A: 0.1% formic acid in water; solvent B: 0.1% formic acid in 1:1 solution of acetonitrile and ethanol). The LC system was coupled online with a Bruker maXis II ETD Q-TOF mass spectrometer. For tandem mass spectrometry (MS/MS) experiments, individual charge states of protein molecular ions were isolated; then, the ions were dissociated by ETD. The accumulation time, reagent time, and reaction time for ETD were determined on a case-by-case basis to achieve optimal fragmentation. The ranges for accumulation time, reagent time, and reaction time were 1000–4000, 4–20, and 1–10 ms, respectively. All spectra were processed with the MASH suite software34 using a S/N threshold of 3 and a fit factor of 70% and then validated manually. The resulting mass lists were further assigned on the basis of the sequence of Ub with or without the diglycine (GG) modification at each lysine using a tolerance of 20 ppm for precursor and fragment ions. All reported calculated (calcd) and experimental (exptl) values correspond to the monoisotopic molecular weight.
RESULTS AND DISCUSSION
A general characterization method is needed to understand the various roles that branched Ub chains play in cell signaling. To this end, we sought to develop an approach based on middle-down MS.27,28 We envisioned an experimental workflow commencing with the isolation of Ub chains from cells followed by minimal trypsinolysis and MS analysis (Figure 1). While there are several methods to enrich Ub chains using Ub binding domains (UBDs), we focused on two UBDs in particular: the nonselective tandem ubiquitin binding entity TUBE237 and the NZF1 domain from the K29/K33 selective deubiquitinase TRABID.38 We reasoned these two UBDs would allow us to monitor both global changes in Ub chain branching and changes in specific Ub chains bearing branched linkages.
Figure 1.
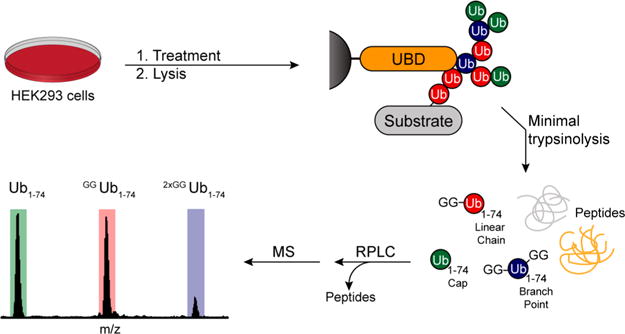
General workflow for isolation and detection of branched Ub chains by UbiChEM-MS. Cells are grown under different treatments and lysed under nondenaturing conditions. Clarified lysates are subsequently incubated with a resin bearing ubiquitin binding domains, and the isolated chains are then subjected to minimal trypsinolysis under nondenaturing conditions yielding Ub1–74 fragments containing Gly-Gly modifications. These products are enriched for using reverse-phase chromatography and characterized by high resolution mass spectrometry.
TUBE2- and NZF1-Based UbiChEM-MS
To gain an understanding of the global landscape of Ub chain architecture, we captured polyubiquitin chains using the commercially available agarose bound TUBE2. This tandem ubiquitin binding entity is based on the UBA1 domain of Rad23a and has been shown to enrich for Ub chains with little linkage selectivity, thus allowing us to monitor global changes in the Ub pool.37,39 Incubation of TUBEs with clarified lysate from HEK293 cells did indeed result in the isolation of high molecular weight chains (Figure S1). After washing, trypsin was added to achieve minimal digestion of the immobilized chains. The resulting digests were then subjected to reverse-phase column chromatography to remove salts and smaller peptides prior to high resolution MS analysis. With this strategy, we could detect all three Ub1–74 species in untreated HEK293 cell lysates (Figure 2A). Relative quantification reveals that 57% of Ub1–74 species exist as either caps or monoUb, 41% are GGUb1–74, and 2% are 2xGGUb1–74.
Figure 2.

Middle-down MS analysis of Ub chains isolated from HEK cells using agarose bound TUBE2 shows dynamics in Ub chain branching upon proteasome inhibition. (A) ESI TOF MS spectra showing the presence of all three minimally digested products: Ub1–74 (green box), GGUb1–74 (red box), and 2xGGUb1–74 (blue box). The spectra correspond to the Ub11+ charge state for chains isolated from either untreated or MG132 treated HEK lysate. (B) Quantification of Ub1–74 species at different MG132 time points. The percent distribution is calculated by averaging relative abundance of each Ub1–74 species to total abundance of all three species from three biological replicates for each treatment (Table S2). Error bars represent standard error of the mean (SEM) for each data set: *, p < 0.05; **, p < 0.01; ***, p < 0.001 (Student’s t test). Calcd, calculated most abundant weight; exptl, experimental most abundant molecular weight.
Having established a method that enables characterization of the Ub chain landscape, we wanted to explore whether UbiChEM-MS could be used to monitor chain dynamics. Branched Ub chains have been implicated as potent signals for proteasomal degradation, suggesting that proteasome inhibition would lead to an accumulation of these conjugates.20 To test this, cells were treated with MG132 for 2, 4, 8, and 16 h and chains were enriched using TUBE2 (Figures 2 and S3 and Table S2). The most significant changes occur after 16 h. Caps/monoUb decreases from 57% to 44%, while GGUb1–74 increases from 41% to 52% and 2xGGUb1–74 rises from 1% to 4% (Figure 2B). These results indicate that long chains harboring branch points accumulate after prolonged proteasome impairment, supporting the idea that branched chains are involved in proteasomal degradation.
We then shifted our attention to a UBD with linkage selectivity to capture a specific pool of Ub chains. The NZF1 domain from the deubiquitinase TRABID selectively binds K29-linked Ub chains and has been shown to enrich heterotypic Ub chains from cell lysates.38 On the basis of these results, we reasoned that a percentage of mixed linkage chains would contain branch points and the dynamics of a specifically linked chain could be monitored. To test this, the NZF1 domain was expressed with an N-terminal fusion to HaloTag and covalently tethered to HaloLink resin as previously reported.38,40 The immobilized NZF1 domain was then used to isolate Ub chains from HEK293. As with TUBE2 enrichment, Western blot analysis shows the NZF1 domain enables isolation of high molecular weight chains (Figures 3, S1, and S2). Moreover, all three Ub1–74 species are detected by MS. Intriguingly, the Ub chain profile differs from that of the TUBE2 pull-downs. In unstimulated cells, approximately 4% of the NZF1-enriched chains contain branch points (2xGGUb1–74) compared to only 1% in TUBE2 pull-downs (Figure 3B). These differences are indeed significant based on the consistency of results obtained using different mass spectrometers (see below) and the number of biological and technical replicates. Narrowing the scope of chains that can be pulled down thus enables detection of chain architectures that might normally be lost in the noise.
Figure 3.
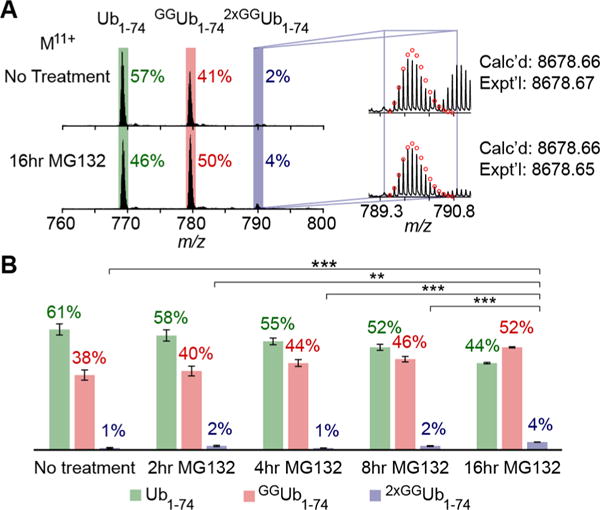
Middle-down MS analysis of Ub chains isolated from HEK cells using Halo-NZF1 resin. (A) ESI MS spectra showing the MS spectra of chains isolated using Halo-NZF1 resin. The spectra shows the presence of all three minimally digested products: Ub1–74 (green box), GGUb1–74 (red box), and 2xGGUb1–74 (blue box). The spectra correspond to the Ub11+ charge state for chains isolated from either untreated or MG132 treated HEK lysate. (B) Quantification of Ub1–74 species after different lengths of proteasome inhibition. The percent distribution is calculated by averaging relative abundance of each Ub1–74 species to total abundance of all three species from three biological replicates for each treatment (Table S3). Error bars represent the SEM for each data set. Calcd, calculated most abundant weight; exptl, experimental most abundant molecular weight.
Next, we sought to determine if NZF1-enriched chains are involved in proteasomal degradation. Again, HEK293 cells were treated with the proteasome inhibitor MG132 for 2, 4, 8, and 16 h (Figures 3 and S4 and Table S3).20 Interestingly, even after 16 h, no significant changes in any of the three Ub1–74 species are detected. These results suggest chains enriched by the NZF1 domain do not target substrates to the proteasome.
MS2 Analysis of Branched Chains
We sought to characterize the linkages in chains isolated from TUBE2 and NZF1 pull-downs. To this end, we employed electron-transfer dissociation (ETD) fragmentation to generate c- and z•-type ions.41,42 ETD analysis of the GGUb1–74 species points to chains primarily composed of K48 linkages (Figure S5). Analysis of the 2xGGUb1–74 species, however, proved much more difficult, precluding us from identifying specific lysines that comprise the branch points (Figure S6). This could be due to the nonselective nature of TUBE2, which is likely to enrich for chains composed of multiple linkages.
As the NZF1 domain of TRABID has been shown to selectively isolate K29-linked Ub chains, we hoped to confirm the presence of K29 and identify other linkages within the branch points. Using the sample from MG132 treated cells, we isolated the M11+ charge state for 2xGGUb1–74 and subjected it to ETD fragmentation (Figure 4A). Accounting for all possible linkage types, the fragmentation pattern best fits a K29/K48 branch point. With these linkages, we identified 31 c and 31 z• ions (Figures 4B,C, S7, and S8). The most important ions for characterization are c31 and z27• (Figure 4B). The presence of these two indicates that the Gly-Gly modifications are on K29 and K48. These results are consistent with the previously reported observation that NZF1 enriches heterotypic chains comprised mainly of K29 and K48 linkages.38
Figure 4.
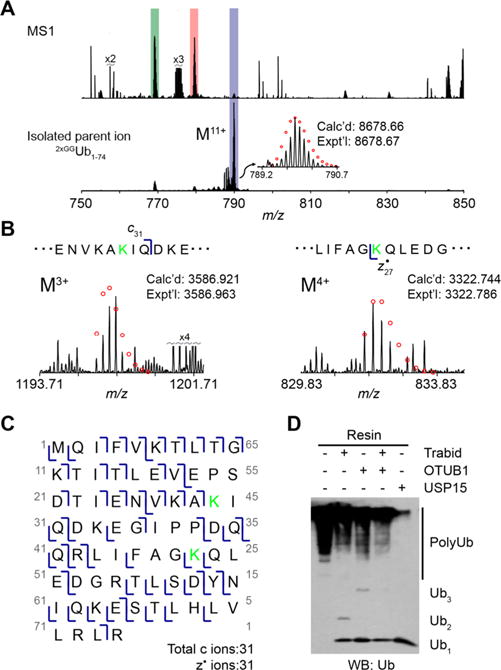
Characterization of Ub chains isolated with Halo-NZF1 by ETD and DUB restriction analysis uncovering K29/K48 branched chains. (A) ESI MS analysis of the minimally digested Ub chains isolated from HEK cells treated with 10 μM MG132 for 2 h. The spectra correspond to the Ub11+ charge state, where the top spectrum shows all ions present in the mass range and the bottom spectrum shows the isolated 2xGGUb1–74 M11+ parent ion. The red circles represent theoretical isotopic abundance distributions of the isotopomer peaks. calcd, calculated most abundant weight; exptl, experimental most abundant molecular weight. (B) Analysis of ETD fragments showing the presence of a di-Gly modification at K29 and K48. The green K shows the position of a modified lysine. (C) Observed ETD fragments (c and z• ions) mapped onto the sequence of Ub containing a di-Gly modification at K29 and K48. (D) DUB restriction analysis of Ub chains isolated from MG132 treated cells using Halo-NZF1 resin. Aliquots of this resin were incubated with the indicated DUBs overnight at 37 °C. The reaction mixtures were quenched with 6× Laemmli loading dye, separated on a 15% SDS-PAGE gel, and analyzed by Western blot using anti-Ub antibody (P4D1).
To provide additional support for the presence of Gly-Gly modifications on K29 and K48, we used Ub chain restriction analysis (UbiCrest).43,44 NZF1-enriched chains were digested with the K29/K33 linkage-selective DUB TRABID and the K48 specific DUB OTUB1.45,46 As a control, we also employed the nonselective DUB USP15.47 Western blot analysis shows chains are sensitive to TRABID and OTUB1 with the greatest amount of cleavage occurring when both DUBs are added (Figure 4D). These data confirm that NZF1-enriched chains contain K29 and K48 linkages. Not all high-molecular weight species, however, are removed by TRABID and OTUB1. This suggests that proteins enriched by NZF1 are modified with multiple mono-Ub molecules or chains with linkages other than K29 and K48.
We then performed ETD analysis on the M11+ charge state of GGUb1–74 to identify other possible linkages. From the fragmentation patterns, it is evident that K48 linkages predominate (Figure S9), suggesting many of the isolated ubiquitylated proteins are modified with multiple mono Ub that are poor substrates for the linkage selective DUBs employed in our chain restriction digests. On the basis of these results along with the MS data, we propose that most of the isolated proteins are carrying multimono-Ub modifications along with short chains linked through K29 and K48. Our middle-down MS approach therefore informs on the overall landscape of Ub modifications and chain configuration.
Ensuring Maximal Detection of the Ub Chain Population by UbiChEM-MS
Optimizing the conditions for restricted trypsinolysis is critical for detecting all three Ub1–74 species. For instance, the use of denaturants commonly used to elute Ub chains from UBD resins must be avoided to ensure minimal cleavage.48 In our case, this was achieved by performing trypsinolysis reactions on resin. Another important issue is the amount of lysate required to observe low abundant modifications such as branching. We empirically found a ratio of 3–4 μL of resin per mg of lysate consistently leads to the detection of branched chains. Overdigestion of resin-bound chains is another problem we encountered. To avoid this, we performed a time course analysis to find the optimal trypsin concentration. As evidenced by the formation of di-Ub and mono-Ub, we observe complete conversion of high molecular weight chains within 16 h using 1 μg of trypsin per 18 mg of lysate for untreated cells and 1 μg of trypsin per 6 mg of lysate for cells treated with MG132 (Figures S1 and S10 and Table S1).
Pull-down efficiency is also a concern. This was assessed through recapture experiments in which fractions from a flow through were further incubated with a fresh batch of resin. We performed these experiments with cell lysates from both the 0 and 16 h MG132 treatments. Western blot analysis shows that for the untreated cells (0 h) most of the available Ub chains are captured (Figure S11). However, with cell lysates from the 16 h MG132 treatment, an appreciable amount of Ub species escape the first pull-down and are recaptured by fresh resin (Figure S11). This is not surprising considering MG132 treatment leads to a dramatic boost in the global levels of Ub chains, which will affect the Ub to resin ratio.12 Despite the inability to capture all the Ub chains in the initial round of pull-downs, middle-down MS shows the chains isolated from the second round have the same architecture as those from the first. These results indicate that our pull-down conditions are sufficient to yield a complete picture of the Ub landscape.
UbiChEM-MS Is Not Limited by Instrumental Setup
For the UbiChEM-MS workflow, we used different separation and characterization methods. With the TUBEs, online RPLC separation was coupled to a TOF mass spectrometer. With the NZF1 domain, RPLC was performed offline and analytes were then directly introduced into an Orbitrap mass spectrometer. Despite these differences, similar Ub architectures are observed from experiment-to-experiment and instrument-to-instrument. These results highlight the reproducibility and accessibility of our method.
CONCLUSION
Although a number of studies have implicated heterotypic Ub chains in the regulation of fundamental cellular pathways, the precise configuration of these chains has largely remained unknown.1–3,5,13 In this study, we established Ubiquitin Chain Enrichment Middle-down Mass Spectrometry (UbiChEM-MS), as a method for detecting and quantifying the relative abundance of heterotypic branched chains in cell lysates. Using two different UBDs, we demonstrate the dynamics of Ub branched chains containing K29 linkages as well as the global Ub pool in response to MG132 treatment. From these experiments, we found that, while chain branching does indeed increase in response to proteasome inhibition, branched chains containing K29 linkages are likely to be a nondegradative signal. Our work thus potentially expands the known role of branched Ub chains beyond just proteasome targeting signals.
Our strategy of using UBDs coupled with middle-down MS to detect and quantify relative amounts of branched chains has several advantages. First, it enables unambiguous detection of branching using endogenous levels of Ub chains. This avoids the use of Ub mutants, which may provide misleading results. Second, it exploits the selectivity of certain UBDs to examine the architecture of chains harboring specific linkages. Third, it distinguishes between mono-Ub/chain end-caps, the linear portion of a chain, and branch points. This is powerful because the relative changes in each of these species can be monitored under different cellular conditions.
Supplementary Material
Acknowledgments
This work was supported by research grant RO1GM110543 from the National Institutes of Health (NIH) and the National Science Foundation (NSF) Graduate Research Fellowship Program (GRFP, award number: 1451512). The purchase of the Bruker ULTRAFLEX III in 2008 was partially funded by NIH NCRR 1S10RR024601-01 Award to the University of Wisconsin-Madison Department of Chemistry. The data described herein were acquired on an Orbitrap Fusion mass spectrometer funded by National Institutes of Health grant 1S10OD010645-01A1.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.analchem.6b03675.
Optimization of ubiquitin chain enrichments, additional MG132 treated experiments used to generate bar graphs in Figures 2 and 3, additional MS2 fragments supporting the assignment of K29 and K48 modifications, and supporting experimental methods (PDF)
ORCID
Ying Ge: 0000-0001-5211-6812
Eric R. Strieter: 0000-0003-3447-3669
Notes
The authors declare no competing financial interest.
References
- 1.Nakayama KI, Nakayama K. Nat Rev Cancer. 2006;6:369–381. doi: 10.1038/nrc1881. [DOI] [PubMed] [Google Scholar]
- 2.Jackson SP, Durocher D. Mol Cell. 2013;49:795–807. doi: 10.1016/j.molcel.2013.01.017. [DOI] [PubMed] [Google Scholar]
- 3.Liu YC. Annu Rev Immunol. 2004;22:81–127. doi: 10.1146/annurev.immunol.22.012703.104813. [DOI] [PubMed] [Google Scholar]
- 4.Hershko A, Ciechanover A. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 5.Komander D, Rape M. Annu Rev Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 6.Yau R, Rape M. Nat Cell Biol. 2016;18:579–586. doi: 10.1038/ncb3358. [DOI] [PubMed] [Google Scholar]
- 7.Swatek KN, Komander D. Cell Res. 2016;26:399–422. doi: 10.1038/cr.2016.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jin L, Williamson A, Banerjee S, Philipp I, Rape M. Cell. 2008;133:653–665. doi: 10.1016/j.cell.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chau V, Tobias J, Bachmair A, Marriott D, Ecker D, Gonda D, Varshavsky A. Science. 1989;243:1576–1583. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- 10.Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacobson AD, Zhang N-Y, Xu P, Han K-J, Noone S, Peng J, Liu C-W. J Biol Chem. 2009;284:35485–35494. doi: 10.1074/jbc.M109.052928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J, Rush J, Hochstrasser M, Finley D, Peng J. Cell. 2009;137:133–145. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haglund K, Dikic I. EMBO J. 2005;24:3353–3359. doi: 10.1038/sj.emboj.7600808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerlach B, Cordier SM, Schmukle AC, Emmerich CH, Rieser E, Haas TL, Webb AI, Rickard JA, Anderton H, Wong WWL, Nachbur U, Gangoda L, Warnken U, Purcell AW, Silke J, Walczak H. Nature. 2011;471:591–596. doi: 10.1038/nature09816. [DOI] [PubMed] [Google Scholar]
- 15.Ikeda F, Deribe YL, Skanland SS, Stieglitz B, Grabbe C, Franz-Wachtel M, van Wijk SJL, Goswami P, Nagy V, Terzic J, Tokunaga F, Androulidaki A, Nakagawa T, Pasparakis M, Iwai K, Sundberg JP, Schaefer L, Rittinger K, Macek B, Dikic I. Nature. 2011;471:637–641. doi: 10.1038/nature09814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim HT, Kim KP, Lledias F, Kisselev AF, Scaglione KM, Skowyra D, Gygi SP, Goldberg AL. J Biol Chem. 2007;282:17375–17386. doi: 10.1074/jbc.M609659200. [DOI] [PubMed] [Google Scholar]
- 17.Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. Nat Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- 18.Kim HT, Kim KP, Uchiki T, Gygi SP, Goldberg AL. EMBO J. 2009;28:1867–1877. doi: 10.1038/emboj.2009.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grice GL, Lobb IT, Weekes MP, Gygi SP, Antrobus R, Nathan JA. Cell Rep. 2015;12:545–553. doi: 10.1016/j.celrep.2015.06.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meyer HJ, Rape M. Cell. 2014;157:910–921. doi: 10.1016/j.cell.2014.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Emmerich CH, Ordureau A, Strickson S, Arthur JSC, Pedrioli PGA, Komander D, Cohen P. Proc Natl Acad Sci U S A. 2013;110:15247–15252. doi: 10.1073/pnas.1314715110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Emmerich CH, Bakshi S, Kelsall IR, Ortiz-Guerrero J, Shpiro N, Cohen P. Biochem Biophys Res Commun. 2016;474:452–461. doi: 10.1016/j.bbrc.2016.04.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boname JM, Thomas M, Stagg HR, Xu P, Peng J, Lehner PJ. Traffic. 2010;11:210–220. doi: 10.1111/j.1600-0854.2009.01011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goto E, Yamanaka Y, Ishikawa A, Aoki-Kawasumi M, Mito-Yoshida M, Ohmura-Hoshino M, Matsuki Y, Kajikawa M, Hirano H, Ishido S. J Biol Chem. 2010;285:35311–35319. doi: 10.1074/jbc.M110.112763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saeki Y, Tayama Y, Toh-e A, Yokosawa H. Biochem Biophys Res Commun. 2004;320:840–845. doi: 10.1016/j.bbrc.2004.05.216. [DOI] [PubMed] [Google Scholar]
- 26.Koegl M, Hoppe T, Schlenker S, Ulrich HD, Mayer TU, Jentsch S. Cell. 1999;96:635–644. doi: 10.1016/s0092-8674(00)80574-7. [DOI] [PubMed] [Google Scholar]
- 27.Xu P, Peng J. Anal Chem. 2008;80:3438–3444. doi: 10.1021/ac800016w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Valkevich EM, Sanchez NA, Ge Y, Strieter ER. Biochemistry. 2014;53:4979–4989. doi: 10.1021/bi5006305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee AE, Castañeda CA, Wang Y, Fushman D, Fenselau C. J Mass Spectrom. 2014;49:1272–1278. doi: 10.1002/jms.3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee AE, Geis-Asteggiante L, Dixon EK, Kim Y, Kashyap TR, Wang Y, Fushman D, Fenselau C. J Mass Spectrom. 2016;51:315–321. doi: 10.1002/jms.3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee AE, Geis-Asteggiante L, Dixon EK, Miller M, Wang Y, Fushman D, Fenselau C. J Mass Spectrom. 2016;51:629–637. doi: 10.1002/jms.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilkinson KD, Audhya TK. J Biol Chem. 1981;256:9235–9241. [PubMed] [Google Scholar]
- 33.Husnjak K, Dikic I. Annu Rev Biochem. 2012;81:291–322. doi: 10.1146/annurev-biochem-051810-094654. [DOI] [PubMed] [Google Scholar]
- 34.Guner H, Close PL, Cai W, Zhang H, Peng Y, Gregorich ZR, Ge Y. J Am Soc Mass Spectrom. 2014;25:464–470. doi: 10.1007/s13361-013-0789-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peng Y, Gregorich ZR, Valeja SG, Zhang H, Cai W, Chen Y-C, Guner H, Chen AJ, Schwahn DJ, Hacker TA, Liu X, Ge Y. Mol Cell Proteomics. 2014;13:2752–2764. doi: 10.1074/mcp.M114.040675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen YC, Ayaz-Guner S, Peng Y, Lane NM, Locher MR, Kohmoto T, Larsson L, Moss RL, Ge Y. Anal Chem. 2015;87:8399–8406. doi: 10.1021/acs.analchem.5b01745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hjerpe R, Aillet F, Lopitz-Otsoa F, Lang V, England P, Rodriguez MS. EMBO Rep. 2009;10:1250–1258. doi: 10.1038/embor.2009.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kristariyanto YA, Rehman SAA, Campbell DG, Morrice NA, Johnson C, Toth R, Kulathu Y. Mol Cell. 2015;58:83–94. doi: 10.1016/j.molcel.2015.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scott D, Oldham NJ, Strachan J, Searle MS, Layfield R. Proteomics. 2015;15:844–861. doi: 10.1002/pmic.201400341. [DOI] [PubMed] [Google Scholar]
- 40.Los GV, Encell LP, McDougall MG, Hartzell DD, Karassina N, Zimprich C, Wood MG, Learish R, Ohana RF, Urh M, Simpson D, Mendez J, Zimmerman K, Otto P, Vidugiris G, Zhu J, Darzins A, Klaubert DH, Bulleit RF, Wood KV. ACS Chem Biol. 2008;3:373–382. doi: 10.1021/cb800025k. [DOI] [PubMed] [Google Scholar]
- 41.Zubarev RA, Kelleher NL, McLafferty FW. J Am Chem Soc. 1998;120:3265–3266. [Google Scholar]
- 42.Zubarev RA, Horn DM, Fridriksson EK, Kelleher NL, Kruger NA, Lewis MA, Carpenter BK, McLafferty FW. Anal Chem. 2000;72:563–573. doi: 10.1021/ac990811p. [DOI] [PubMed] [Google Scholar]
- 43.Mevissen TET, Hospenthal MK, Geurink PP, Elliott PR, Akutsu M, Arnaudo N, Ekkebus R, Kulathu Y, Wauer T, El Oualid F, Freund SMV, Ovaa H, Komander D. Cell. 2013;154:169–184. doi: 10.1016/j.cell.2013.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hospenthal MK, Mevissen TET, Komander D. Nat Protoc. 2015;10:349–361. doi: 10.1038/nprot.2015.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Virdee S, Ye Y, Nguyen DP, Komander D, Chin JW. Nat Chem Biol. 2010;6:750–757. doi: 10.1038/nchembio.426. [DOI] [PubMed] [Google Scholar]
- 46.Wang T, Yin L, Cooper EM, Lai MY, Dickey S, Pickart CM, Fushman D, Wilkinson KD, Cohen RE, Wolberger C. J Mol Biol. 2009;386:1011–1023. doi: 10.1016/j.jmb.2008.12.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ritorto MS, Ewan R, Perez-Oliva AB, Knebel A, Buhrlage SJ, Wightman M, Kelly SM, Wood NT, Virdee S, Gray NS, Morrice NA, Alessi DR, Trost M. Nat Commun. 2014;5:4763. doi: 10.1038/ncomms5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Emmerich CH, Cohen P. Biochem Biophys Res Commun. 2015;466:1–14. doi: 10.1016/j.bbrc.2015.08.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


