Abstract
Carotenoid cleavage oxygenases (CCOs) are non-heme iron enzymes that catalyze scission of alkene groups in carotenoids and stilbenoids to form biologically important products. CCOs possess a rare four-His iron center whose resting-state structure and interaction with substrates are incompletely understood. Here, we address this knowledge gap through a comprehensive structural and spectroscopic study of three phyletically diverse CCOs. The crystal structure of a fungal stilbenoid-cleaving CCO, CAO1, reveals strong similarity between its iron center and those of carotenoid-cleaving CCOs, but with a markedly different substrate-binding cleft. These enzymes all possess a five-coordinate high-spin Fe(II) center with resting-state Fe-His bond lengths of ~2.15 Å. This ligand set generates an iron environment more electropositive than those of other non-heme iron dioxygenases as observed by Mössbauer isomer shifts. Dioxygen (O2) does not coordinate iron in the absence of substrate. Substrates bind away (~4.7 Å) from the iron and have little impact on its electronic structure, thus excluding coordination-triggered O2 binding. However, substrate binding does perturb the spectral properties of CCO Fe–NO derivatives, indicating proximate organic substrate and O2-binding sites, which might influence Fe–O2 interactions. Together, these data provide a robust description of the CCO iron center and its interactions with substrates and substrate mimetics that illuminates commonalities as well as subtle and profound structural differences within the CCO family.
Graphical abstract

CCOs constitute a family of non-heme iron enzymes, present in all life domains, that typically catalyze oxidative cleavage of carotenoid substrates (Figure 1A,B). This activity is critical for a number of biological processes, including vitamin metabolism, hormone production, and generation of the retinaldehyde chromophore for visual and non-visual light detection.1–6 The CCO family also encompasses enzymes expressed in bacteria and fungi that catalyze cleavage of stilbenoids such as resveratrol (Figure 1A,C).7–9 At present, the physiological functions of stilbenoid-cleaving CCOs are poorly understood. Retinal pigment epithelium 65 kDa protein (RPE65) is a vertebrate CCO that exhibits retinoid isomerase rather than alkene cleavage activity, which is necessary for visual cycle function.10
Figure 1.
Phylogeny and enzymatic activities of carotenoid and stilbenoid-cleaving CCOs. (A) Maximal likelihood phylogeny of biochemically characterized stilbenoid-cleaving CCOs (azure box) and two cyanobacterial apocarotenoid cleavage enzymes used as an outgroup to root the tree (gray box). Proteins colored red were examined in the study presented here. The scale bar represents the average number of amino acid substitutions over the indicated branch length. Numbers on the branches are bootstrap percentages from 2000 pseudoreplicates. (B) Carotenoid cleavage reaction catalyzed by ACO. (C) Stilbenoid cleavage reaction catalyzed by CAO1 and NOV2. The red wavy line indicates the position of the scissile bond in panels B and C.
The diversity of important biological processes in which alkene-cleaving CCOs are involved has prompted interest in characterizing their enzymatic properties, including substrate specificity and mode of catalysis. CCOs catalyzing cleavage of mono-, bi-, and acyclic carotenoids as well as various stilbenoids have been identified (e.g., refs 4, 5, 8, 9, 11, and 12). Despite this variability in substrate specificity, individual CCO enzymes are often highly stereo- and regioselective. Crystal structures of apocarotenoid oxygenase from Synechocystis (ACO),13 RPE65 from Bos taurus,14 and 9-cis-epoxy carotenoid dioxygenase from Zea mays, viviparous-14 (VP14),15 have revealed plasticity in the CCO substrate-binding cavity that has resulted from the need to accommodate the diverse substrates of these enzymes and control active site accessibility.
In contrast to this substrate and substrate-binding cleft diversity, all CCOs contain a conserved metal center consisting of an iron cofactor coordinated by four His residues.3,16 Two cis-localized sites in the iron coordination sphere remain unoccupied by protein ligands and are potentially available for ligand binding, although one is rendered inaccessible to many ligands by the presence of a nearby Val or Thr residue in most CCOs.13,17,18 Three conserved and functionally critical Glu residues serve as a second coordination sphere for the iron cofactor by forming hydrogen bonding interactions with the His residues. This mode of iron coordination is rare and differs from that of the well-characterized facial-triad non-heme iron dioxygenases wherein iron is bound by a 2-histidine, 1-carboxylate (2H-1C) motif with three sites open for ligand binding.19,20
Because CCOs require Fe(II) for their catalytic activity, it is assumed that the iron center mediates dioxygen (O2) activation as opposed to substrate activation.21 CCOs were previously thought to function as monooxygenases, but recent data obtained using improved assay systems suggest that dioxygenase activity is common to the entire family.17,22,23 The carotenoid and stilbenoid substrates of CCOs are electron-rich, redox-active molecules that provide all reducing equivalents needed for their oxidative cleavage. However, little is known about the specific mechanisms of O2 activation by CCOs. Crystal structures of VP14 and Novosphingobium aromaticivorans oxygenase (NOV) 1 have suggested that the iron center can form stable adducts with O2 in the absence of an organic substrate.15,24 This contrasts with the gating mechanism of O2 activation by oxygenases wherein organic substrate and O2 binding are tightly coupled to avoid generation of detrimental reactive oxygen species.25 In the 2H-1C dioxygenases, this synchronization is often accomplished by direct coordination of organic substrate to the iron center, which lowers the iron redox potential, allows electron transfer between the substrate and O2, and positions the scissile bond for cleavage.20,25,26 In the case of pterin-dependent dioxygenases, the iron–O2 interaction is facilitated by organic cofactor binding near but not in direct coordination with the Fe(II) ion.20,27 As typical CCO substrates lack functionalities for metal coordination (Figure 1B,C), it is clear that an alternative mechanism of O2 activation must be used by these enzymes.
Further progress in elucidating the CCO alkene cleavage mechanism has been slowed by an incomplete understanding of the CCO non-heme iron center and its interaction with substrates. Studies addressing this knowledge deficit were not previously feasible because of a lack of suitable high-yield expression systems for these enzymes. Recently, we developed methods for producing quantities of ACO, a prototypical carotenoid-cleaving CCO, sufficient for structural and spectro-scopic studies.28 These methods have also been applicable to a stilbenoid-cleaving CCO, which we anticipated would be more amenable to structural and spectroscopic characterization because of the greater aqueous solubility of its substrates relative to that of carotenoids.17 Carotenoids and stilbenoids are both redox-active polyenes whose CCO-catalyzed cleavage reactions are closely analogous, both occurring at nonaromatic alkene groups (Figure 1B,C). Therefore, information gleaned from stilbenoid-cleaving CCOs could be applicable to the broader group of alkene-cleaving CCOs.
Here, we characterize the four-His iron centers of ACO and two stilbenoid-cleaving CCOs, NOV2 and Neurospora crassa carotenoid oxygenase 1 (CAO1), using a combination of X-ray crystallography, X-ray absorption spectroscopy (XAS), and 57Fe Mössbauer and electron paramagnetic resonance (EPR) spectroscopies. The data allow detailed comparisons of these enzymes in terms of their substrate-binding clefts, iron center resting-state structures, and interactions with substrates and substrate analogues. Synthesis of these high-resolution data provides new insights into catalysis by CCOs as well as their biological functions.
MATERIALS AND METHODS
Reagents
Except as noted below, chemical reagents were purchased from Sigma-Aldrich (St. Louis, MO) in the highest-purity form available. LB medium was purchased from USB (Cleveland, OH). Iron-57 metal (95.93% isotope enrichment) was obtained from Cambridge Isotope Laboratories, Inc. (Andover, MA). Water from a Milli-Q purification system (resistivity of 18.2 mΩ cm) (ED Millipore, Billerica, MA) was used to prepare all reagents and buffer solutions.
Synthesis of β-Fluororesveratrol
β-Fluororesveratrol was prepared as described previously (Scheme 1).29,30 Further purity was achieved by prep-HPLC [Luna 15 µm PREP silica (2) 100 Å column, 75:25 n-hexanes/ethyl acetate, 20 mL/min, 280 nm; retention time of 46.0 min (Z-isomer)]: 1H NMR (500 MHz, acetone-d6) δ 8.67 (s, 1H), 8.20 (s, 2H), 7.55 (d, J = 7.8 Hz, 2H), 6.91 (d, J = 8.3 Hz, 2H), 6.68 (s, 2H), 6.28 (s, 1H), 6.24 (d, J = 41.5 Hz, 1H); 19F NMR (470 MHz, acetone-d6) δ – 114.24 (d, J = 41.3 Hz); 13C NMR (125 MHz, acetone-d6) δ 159.45 (s), 158.30 (d, 1JCF = 256.2 Hz), 136.79 (d, 3JCF = 2.6 Hz), 126.84 (d, 3JCF = 7.7 Hz), 125.20 (d, 2JCF = 28.4 Hz), 116.51 (d, 4JCF = 2.2 Hz), 108.35 (d, 4JCF = 8.1 Hz), 104.71 (d, 2JCF = 9.6 Hz), 102.69 (d, J = 1.9 Hz).31,32
Scheme 1.
Synthetic Route for Production of β-Fluororesveratrol
Protein Production
Cloning, Expression, and Purification of CAO1
The coding sequence of CAO1 (GI:759001907) from N. crassa OR74A was synthesized (GenScript, Piscataway, NJ) and cloned into the pET3a expression vector without fusion tags. The integrity of the expression plasmid was confirmed by sequencing.
The plasmid was transformed into the T7 express BL21 Escherichia coli strain (New England Biolabs, Ipswich, MA) for protein expression. One-liter cultures of LB medium containing 100 µg of ampicillin/mL were grown at 37 °C to an A600 of 0.5– 0.6, at which time protein expression was induced by the addition of isopropyl β-d-1-thiogalactopyranoside (IPTG) (Roche Applied Science, Indianapolis, IN) to a final concentration of 0.1 mM. The temperature was decreased to 28 °C, and the culture was supplemented with an additional 100 µg of ampicillin/mL. After overnight growth (12–14 h) at 230 rpm, cells were harvested by centrifugation, suspended in 20 mM Tris-HCl (pH 8.5), and either stored at −80 °C or used immediately.
All purification procedures were conducted at 4 °C. Harvested cells were lysed by two passes through a French press. The lysate was clarified by centrifugation at 186000g for 30 min. The supernatant was then loaded onto a HiTrap Q HP 5 mL anion-exchange column (GE Healthcare, Chicago, IL) equilibrated with buffer A consisting of 20 mM HEPES-NaOH (pH 7.0). The column was then washed with 60 mL of buffer A, followed by a linear increase in buffer B containing 20 mM HEPES-NaOH (pH 7) and 1 M NaCl. Fractions containing CAO1 were pooled, concentrated, and further purified by gel filtration chromatography. Specifically, the sample was injected into a 120 mL Superdex 200 gel filtration column (GE Healthcare) equilibrated with gel filtration buffer consisting of 20 mM HEPES-NaOH (pH 7.0) and 200 mM NaCl. Fractions containing purified CAO1 (≥90% pure as judged by sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis) were pooled, concentrated to 20 mg/mL, flash-frozen, and stored in liquid nitrogen or placed on ice for immediate use.
Production of Cobalt-Substituted CAO1 and Fe-CAO1 in Minimal Medium
Cobalt-substituted CAO1 (Co-CAO1) and Fe-CAO1 were produced by cultivating E. coli cultures in M9 minimal medium [1× M9 salts, 2 mM MgSO4, 0.4% (w/v) glucose, and 0.1 mM CaCl2] in the presence of cobalt or iron. E. coli colonies freshly transformed with the pET3a–CAO1 plasmid were grown on an LB agar plate containing 100 µg/mL ampicillin, inoculated into 2 mL of LB medium containing the same concentration of antibiotic, and grown for ~5 h at 37 °C. Cells were collected by centrifugation at 3220g for 15 min at 37 °C. Cell pellets were then suspended in 1 mL of M9 minimal medium and centrifuged again to remove residual LB medium. The cell pellet was then suspended in 1 mL of minimal medium and transferred into 1 L of M9 minimal medium containing 100 µg/mL ampicillin. The cell culture was grown at 37 °C with 230 rpm shaking to an A600 of 0.5–0.6, at which time IPTG was added to a final concentration of 0.1 mM to induce protein expression at 28 °C. Simultaneously, 15 or 30 mg of CoCl2·6H2O or (NH4)2Fe(SO4)2·6H2O was added to the culture, the former quantity for activity studies and the latter for crystallization studies. After overnight growth, cells were harvested, and protein purification was conducted as described for the native protein from LB culture. All glassware and protein purification instrumentation were washed with Milli-Q water containing 1 mM EDTA to reduce adventitious metal contamination. The purity of CAO1 expressed in minimal medium was similar to that of native protein expressed in LB culture.
Expression and Purification of ACO and NOV2
Synechocystis ACO and Novosphingobium NOV2 were expressed and purified as previously described.17,28
Expression and Purification of 57Fe-Labeled CAO1, NOV2, and ACO
E. coli cells (T7 express BL21 strain, New England Biolabs) harboring the pET3a–NOV217 or pET3a–ACO28 plasmids were grown for 3 h at 37 °C in LB medium containing 100 µg/mL ampicillin. Cells were pelleted by centrifugation at 37 °C and used to inoculate 2 L of M9 minimal medium containing 0.16% (v/v) glycerol, 100 µg/mL ampicillin, and 5 mg of 57Fe (delivered as a 25 mg/mL solution in 6 N HCl). The culture was grown at 37 °C for 9 h, at which time the culture optical density at 600 nm was ~0.6–0.8. Protein expression then was induced by the addition of IPTG to a final concentration of 100 µM, and the culture was supplemented with an additional 100 mg of ampicillin/mL and 5 mg of 57Fe. Protein expression was conducted overnight at 28 °C with 235 rpm shaking. Cells were pelleted by centrifugation the next morning and suspended in 50 mL of 20 mM Bis-Tris-HCl (pH 7) in the case of ACO and 20 mM Tris-HCl (pH 7.5) in the case of CAO1 and NOV2. Protein purification was conducted either as previously described17,28 or as described above for CAO1.
Protein Crystallization and Formation of Enzyme-Substrate Complexes
Crystallization of both Fe-CAO1 and Co-CAO1 was performed by the hanging drop vapor diffusion method by mixing a purified protein sample at 20 mg/mL with a crystallization cocktail containing 42–43% (w/v) sodium polyacrylate 2100 and 0.1 M HEPES-NaOH (pH 6.5) in a 1:1 ratio at room temperature and incubating the drops over 0.5 mL of the same crystallization solution at 8 °C. Rod-shaped crystals, hexagonal in cross section, appeared within 1–2 weeks and continued to grow to full size within an additional 1–2 weeks. Crystals of Fe-CAO1 in complex with β-fluororesveratrol were prepared by addition of a 200 mM dimethyl sulfoxide (DMSO) stock solution of the purified protein samples to a final concentration of 3 mM followed by an overnight incubation at 4 °C. Samples were centrifuged at 71680g for 15 min to remove any aggregates prior to initiation of crystallization trails. Crystals of Co-CAO1 in complex with resveratrol and piceatannol were obtained in a similar manner except that the crystals were soaked in synthetic mother liquor containing 10 mM target ligand overnight at 8 °C prior to being harvested to ensure full ligand occupancy. Mature crystals were harvested using Mitegen dual thickness loops (Mitegen, Ithaca, NY) and flash-cooled in liquid nitrogen before being exposed to X-rays.
Diffraction Data Collection, Phasing, and Structural Refinement
Data Collection and Processing
Diffraction data sets were collected at the NE-CAT 24-ID-C/E beamlines of the Advanced Photon Source and beamline 12–2 of the Stanford Synchrotron Radiation Lightsource. Data for cobalt-substituted CAO1 crystals were collected at X-ray energies above and below the cobalt K-edge based on an X-ray fluorescence scan to assess the active site cobalt occupancy. Data reduction was conducted with XDS33 (Table S1).
Phasing and Structural Refinement
The structure of CAO1 was determined by molecular replacement augmented by single-wavelength anomalous diffraction data (MR-SAD) and density modification. A 2.16 Å resolution data set from a ligand-free Fe-CAO1 crystal (Table S1) was submitted to the MrBUMP molecular replacement pipeline34 through the CCP4 Online web server. The top solution in space group P3221 with four monomers in the asymmetric unit was identified with a Sculptor-modified version of Synechocystis apocarotenoid oxygenase ACO (Protein Data Bank entry 4OU9) in PhaserMR.35 The solution featured a final translational Z score of 21.5, firmly indicating its correctness, but was not readily amenable to direct refinement efforts with an Rfree after REFMAC refinement of 48.7%.36 This MR solution was then applied as a partial model in the PhaserEP phasing pipeline by using a high-multiplicity Fe-CAO1 data set collected just above the iron K-edge.37 Four iron atoms and four sulfur atoms were located in the Phaser combined real– imaginary log likelihood gradient maps. The MR-SAD phases were then improved by solvent flipping in SOLOMON.38 The coordinates of the MR solution were then refined against the iron SAD data set in REFMAC using experimental phase restraints giving an Rfree value of 43.8%. The updated coordinates were then fed back into PhaserEP for a second round of anomalous scatterer identification and SAD phasing, followed by density modification in Parrot.39 The coordinates and updated phases were used for automated model building in Buccaneer40 with interspersed cycles of REFMAC refinement with Hendrickson– Lattman coefficients used as phase restraints, resulting in an Rfree of 34%. The updated model was then refined against the 2.16 Å resolution data set used originally for molecular replacement, resulting in an Rfree of 24.9%. ARP/wARP41 was then used to correct and extend several regions of the model, resulting in an Rfree of 21.8%. The model was further improved by multiple rounds of refinement in REFMAC using local NCS restraints together with manual adjustments performed in COOT42 based on difference map searches and MOLPROBITY43 geometry analyses. Metal ion temperature factors were refined anisotropi-cally in REFMAC. No distance restraints were applied to the metal–ligand bond lengths during refinement. Subsequent structures were determined either by direct refinement or by MR. Residues 1–26 or 1–30, depending on the subunit and specific structure, were not resolved in the electron density maps and hence were omitted. Identification of anomalous scatterers was accomplished through the use of imaginary LLG maps produced by Phaser.37 Residues Phe91 and Lys164 were harmonically restrained in the unliganded structures to prevent the side chains from being pulled into the unidentified active site electron density found in these structures. Atomic coordinates and geometric restraints for the ligands were generated using the GRADE server (Global Phasing Ltd.). Atomic models were validated using Molprobity and the wwPDB structure validation server.44 A summary of the X-ray data collection and refinement statistics is given in Table S1. All structure figures were prepared with PyMOL (Schrödinger).
CCO Multiple-Sequence Alignment
Protein sequences were retrieved from the NCBI protein database. A structure-based amino acid sequence alignment of CAO1 and ACO was generated with the DaliLite server using the pairwise option.45 Amino acid sequence alignment for the other resveratrol-cleaving CCOs was performed with MUSCLE.46 These two multiple-sequence alignments and the NsACO sequence were successively aligned in MUSCLE using the profile option.
Phylogenetic Analysis
Protein sequences for Synechocystis sp. PCC 6803 ACO (P74334.1), Nostoc sp. 7120 ACO (BAB75983.1), Ustilago maydis RCO1 (UM05084), Botryotinia fuckeliana RCO1 (XP_001548426), Chaetomium globosum RCO1 (XP_001219451), N. crassa CAO1 (NCU07008), Aspergillus f umigatus RCO1 (XP_746307), No. aromaticivorins NOV2 (YP_498079), Pseudomonas nitroreducens isoeugenol monooxygenase (IEM) (ACP17973.1), No. aromaticivorins NOV1 (YP_496081), Sphingomonas paucimobilis lignostilbene dioxygenase isoenzyme 1 (LSD1) (AAC60447), and S. paucimobilis lignostilbene dioxygenase isoenzyme 3 (LSD3) (AAB35856) were retrieved from the NCBI protein database and aligned with Clustal Omega.47 Phylogeny inference was performed with PhyML48 using the LG substitution matrix with empirical equilibrium frequencies along with the estimated proportion of invariant sites and gamma distribution parameter (six categories). The starting neighbor-joining tree topology was improved by surface pruning and regrafting. Branch lengths also were optimized. Tree robustness was assessed with 2000 bootstrap pseudoreplicates. The maximal likelihood tree had a log likelihood value of −12232 with good bootstrap support for most branches (Figure 1A). The tree was rooted with the Synechocystis and Nostoc ACO sequences used as outgroups. Visualization of the tree was performed with Interactive Tree of Life (iTOL) version 3.49
Enzyme Kinetics
Stilbenoid Cleavage Activity Assay
The enzymatic activity of CAO1 and NOV2 was assessed as previously described using either resveratrol or piceatannol as a substrate.17 For both substrates, the absorbance at 305 nm was monitored over time using a FlexStation 3 plate reader. Activity levels were determined using the initial linear regions of each reaction progress curve.
Determination of the Enzyme Transition Metal Content and Enzyme:Metal Stoichiometry
The transition metal content of the enzyme samples was measured by inductively coupled plasma optical emission spectroscopy (Soil Research Analytical Laboratory, University of Minnesota, St. Paul, MN). Samples for ICP-OES analysis were prepared by digesting each enzyme sample in 1% (v/v) HNO3. The proteins precipitated immediately upon addition of HNO3, and the samples were incubated for 3 h at room temperature while being gently rocked. Protein precipitates were then removed by filtration through a 0.22 µm filter (Millipore), and the aqueous solutions containing dissociated metals were analyzed by ICP-OES. The buffer solution was treated identically to determine the background metal content, which was below the limits of detection for all metals of interest. Amino acid analysis of the purified enzyme samples was conducted to allow accurate determination of the enzyme concentrations (Protein Chemistry Laboratory, Texas A&M University, College Station, TX). The molar absorptivities of ACO, NOV2, and CAO1 at 280 nm were 75248, 69786, and 64924 M−1 cm−1, respectively.
X-ray Absorption Spectroscopy
Sample Preparation
Samples of ACO at ~58 mg/mL (equal to ~1.2 mM) in 50 mM Tris-HCl (pH 8), 200 mM NaCl, 1 mM DTT, and 20% glycerol, NOV2 at ~97 mg/mL (1.7 mM) in 20 mM Tris-HCl (pH 8), 200 mM NaCl, and 20% (v/v) glycerol, and CAO1 at ~95 mg/ mL (1.2 mM) in 16 mM HEPES-NaOH (pH 7), 160 mM NaCl, and 20% (v/v) glycerol were used for XAS experiments. Samples were flash-cooled in liquid nitrogen and stored in dry ice before use. Samples were loaded into copper sample cells (NSLS) or Delrin cuvettes (SSRL) wrapped in Kapton tape and flash-frozen in liquid nitrogen immediately prior to XAS measurements.
Data Collection
XAS data were obtained at the National Synchrotron Light Source (NSLS) beamline X3B (ACO) and the Stanford Synchrotron Radiation Lightsource beamline 9–3 (NOV2 and CAO1). At NSLS, the storage ring operated at 2.8 GeV and 180–300 mA. A Si(111) double-crystal monochromator with sagittal focusing of the second crystal provided energy selection and horizontal focusing, with a downstream Ni-coated cylindrically bent mirror rejecting higher harmonics and vertically focusing the beam. At SSRL, the storage ring operated at 3.0 GeV and 500 mA in top-off mode. A Si(220) double-crystal monochromator provided energy selection, with an upstream Rh-coating collimating mirror rejecting higher harmonics prior to the monochromator and a downstream Rh-coated toroidal mirror focusing the monochromatic radiation at the sample. Temperature control was provided by a He Displex cryostat (NSLS, 15–20 K sample temperature) or Oxford liquid He cryostat (SSRL, 10 K sample temperature). XAS data were acquired in fluorescence mode using Canberra solid-state germanium detectors (NSLS, 31 element discrete; SSRL, 100 pixel monolithic) with XIA digital data acquisition electronics. Data were typically acquired in 10 eV steps in the pre-edge region (1s acquisition time), 0.3 eV steps along the edge (2 s acquisition time), and 0.05k steps in the EXAFS up to 14–15k (acquisition time increasing from 2 to 9 s in k2 -weighted fashion). Samples were monitored for evidence of radiation damage as indicated by red-shifts in either the pre-edge or edge energies, and new spots were exposed as needed. An iron metal foil placed between ion chambers downstream of the sample was used for energy calibration, with the first inflection point of the edge set to 7112.0 eV.
Data Analyses
XAS data were processed and averaged using Athena50 for NSLS X3B data, whereas EXAFSPAK51 was used for SSRL 9–3 data. XANES peak fitting was conducted using Fityk52 with the rising edge modeled using pseudo-Voigt functions and the pre-edge modeled with Gaussian functions. Fits were performed over several different energy ranges spanning 7104–7118 eV, and reported pre-edge peak fit parameters are averages of at least three fits. EXAFS analysis was performed with Artemis.50 Theoretical phase and amplitude parameters were calculated for simple models of the ACO active site using FEFF6L. From this, relevant paths were incorporated into the fitting model and evaluated for the significance of their contributions. For a given shell in all simulations, coordination number n was fixed while r and σ2 were allowed to float. Amplitude reduction factor S02 was fixed at 0.9, while edge shift parameter ΔE0 was allowed to float at a single common value for all shells. Histidine multiple scattering was evaluated using a method in which the imidazole moiety is represented by four sets of grouped paths representing the dominant scattering contributions.53 The fit was evaluated in k3-weighted R-space, and fit quality was judged by the reported R-factor and reduced χ2. Bond valence sums were used as an additional test to identify the best fit to the EXAFS data, using reported parameters for Fe(II).54
Sample Preparation for EPR and Mössbauer Spectroscopy
E and ES Sample Preparation
Samples of ACO [0.5 mM ACO in 20 mM HEPES-NaOH (pH 7) containing 20% glycerol and 0.02–0.05% (v/v) Triton X-100], CAO1 [1.2 mM CAO1 in 16 mM HEPES-NaOH (pH 7.5), 160 mM NaCl, and 20% (v/v) glycerol], and NOV2 [1.9 mM NOV2 in 20 mM Tris-HCl (pH 7.5 or 8.5), 200 mM NaCl, and 20% glycerol with or without 1 mM tris(carboxylethyl)phosphine] were made anaerobic by passing Ar(g) over the top of the sample as it was stirred at 5 °C for 30 min in a sealed vial. Following this, the protein vials were transferred into an anaerobic N2 gas-filled glovebox The vials were opened in the glovebox and stirred while open to the glovebox environment for 1 h. Resveratrol and piceatannol substrate stock solutions (200 mM) were prepared by preweighing an amount of solid substrate and transferring the powder into a sealed vial. The vial was then transferred into the glovebox to equilibrate with the glovebox environment overnight. Prior to its addition to enzyme samples, the substrate was dissolved in degassed DMSO that had also been stored in the glovebox overnight. Typically, 1–5 µL of substrate in DMSO was delivered to the protein solution, to a maximal concentration of 5% (v/v) DMSO, which did not affect protein activity. Stock solutions of 4 mM all-trans-8′-apocarotenol, generated by reduction of all-trans-8′-apocarotenal as previously described,28 were prepared by dissolving solid apocarotenoid in a solution of degassed ethanol. For O2 binding experiments, a 1 mM sample of 57Fe-enriched NOV2 in a Mössbauer cell at 4 °C was exposed to pure O2 gas at a pressure of 40 psi for 5 min. While the pressure was maintained at 40 psi and 4 °C, the sample was frozen in liquid nitrogen.
Preparation of Fe-Nitrosyl Complexes
Samples of ACO [0.2 mM protein in 20 mM HEPES-NaOH (pH 7), 20% glycerol, and 0.02–0.05% Triton X-100] and NOV2 [0.1 mM protein in 20 mM Tris-HCl (pH 7.5) and 20% glycerol] and substrates were prepared anaerobically as described above. A stock solution of diethylammonium salts (DEA NONOate) was prepared by dissolving a preweighed amount of NONOate powder in degassed 5 mM NaOH, to give a DEA NONOate concentration of 10–15 mM. The vial containing dissolved DEA NONOate was transferred to the glovebox and equilibrated in the glovebox overnight. This solution was stored in the glovebox for ≤24 h before use. Over this period, the DEA NONOate is stable in the NaOH stock solutions. An aliquot of substrate stock solution was added to the protein solutions in the glovebox (when applicable) and left to incubate for 2 min. An aliquot of the DEA NONOate solution then was added to each protein solution. The equivalents of NO delivered were determined on the basis of the calculated rate of decay of the DEA NONOate (half-life of 16 min at pH 7.4 and 22 °C) and were generally in excess (≥3 equiv) relative to the protein concentration. Following a 5 min incubation, the samples were transferred to either an EPR tube or a Mössbauer cup, and the vessels were sealed and quickly removed from the glovebox and the samples frozen in liquid nitrogen.
EPR and Mössbauer Spectroscopy
X-Band EPR spectra were recorded on a Bruker Elexsys spectrometer equipped with an Oxford ESR 910 cryostat and a Bruker bimodal cavity. The modulation amplitude and frequency were 1 mT and 100 kHz, respectively, for all spectra. The microwave frequency was measured with a frequency counter, and the magnetic field was calibrated with a NMR gaussmeter. The temperature was calibrated with a carbon glass resistor (Lakeshore CGR-1–1000). EPR signals were quantified relative to a 1 mM Cu(II)EDTA standard in 10% glycerol. The Cu(II) concentration was quantified by inductively coupled plasma mass spectrometry. Species concentrations were determined with the SpinCount software developed by M. P. Hendrich.55 The software diagonalizes the spin Hamiltonian H = βeB·ge·S + S·D·S, where all parameters have their usual definitions. The quantitative simulations are least-squares fits of the experimental spectra generated considering all intensity factors, which allows the computation of simulated spectra for a specified sample concentration.
57Fe Mössbauer spectra were recorded with two spectrometers using Janis Research dewars. Isomer shifts are reported relative to iron metal at 298 K. The simulations of Mössbauer spectra were calculated with least-squares fitting using the SpinCount program and the standard spin Hamiltonian:55
RESULTS
Crystal Structure of a Fungal Stilbenoid-Cleaving CCO
To provide a structural framework for our spectroscopic investigations, we aimed to elucidate the structure of a stilbenoid-cleaving CCO. We focused our search on those enzymes with defined substrate specificity. In our expression system, the originally characterized stilbenoid-cleaving CCO from Pseudomonas paucimobilis, lignostilbene-α,β-dioxygenase (LSD) 1,7 was found in inclusion bodies. NOV2, by contrast, was soluble and highly expressed and could be purified in an active form17 but failed to crystallize after extensive trials. A third stilbenoid-cleaving CCO from the fungus N. crassa, termed CAO1,12 was also active when purified (Figure S1) and readily formed rod-shaped trigonal crystals that diffracted X-rays to ~2Å resolution (Table S1). The crystal structure of this enzyme was determined in space group P3221 by molecular replacement in combination with anomalous scattering data.
CAO1, like other CCOs, adopts a seven-blade β-propeller covered on its top face by a set of loops and α-helices that form a dome structure housing the active site (Figure 2A). The iron cofactor resides on the top face of the propeller coordinated by His residues 197, 248, 313, and 510 (Figure 2B). Despite the low sequence identity between CAO1 and other CCOs of known structure, their overall three-dimensional similarity is relatively high with root-mean-square (RMS) differences between matched Cα atoms of 1.6, 2.1, 2.3, and 2.6 Å for NOV1, ACO, VP14, and RPE65, respectively.45 The asymmetric unit of the CAO1 crystals is composed of a nearly D2 symmetric dimer of dimers. The four monomers are practically identical in structure except for residues 265–285 and 334–355, which differ between subunits by a small rigid body displacement. One of the assemblies is a 2-fold symmetric dimer with a buried surface area of ~1415 Å2 per monomer (Figure S2A). The dimer interface is comprised of an antiparallel β-strand interaction formed by residues 51–54 of the outer β-strand of blade VII together with polar and nonpolar interactions involving residues 35–37 and 47–62 within the N-terminal segment of the protein along with residues 80, 81, 105–108, 126, and 500–504 (Figure S2B). Both the positive calculated ΔG of interface association56 and its modest shape complementary statistic of 0.61357 suggest the dimer may not be stable. However, CAO1 migrates faster on size exclusion medium than expected on the basis of its calculated molecular weight, which could be attributable to dimer formation in solution (Figure S1A,C). LSD isozymes also form homo- and heterodimeric assemblies in solution.58 Interestingly, a dimeric assembly like that of CAO1 is also present in the NOV1 crystal structure, suggesting a conserved interface.24
Figure 2.
Crystal structure of CAO1 and its comparison to ACO. (A) Cartoon representation of CAO1 (left) and ACO (right). The protein chain is colored blue to red from the N- to C-termini, respectively. The first- and second-sphere His and Glu iron ligands, respectively, are shown as sticks with the bound iron shown as a brown sphere. (B) Close-up of the CAO1 (left) and ACO (right) iron centers. (C) Comparison of the cavities leading to the iron center in CAO1 (left) and ACO (right). The prominent apolar tunnel (indicated by the arrow) present in ACO is absent in CAO1. The CAO1 active site instead can be accessed through a narrow tunnel that is found in roughly the same location as the hydrophilic tunnel found in ACO. The entrances to these tunnels are marked with orange asterisks. (D) CAO1 (left) and ACO (right) in surface representation as viewed down the axes of the arrows shown in panel C. The entrance to the hydrophobic tunnel in ACO (delineated by a dashed line) is surrounded by a cluster of hydrophobic residues (represented as sticks and ribbons) that allow the protein to interact with membranes where hydrophobic carotenoids are dissolved. The corresponding region of CAO1 is more polar because of multiple amino acid substitutions. Additionally, segments of CAO1 that are homologous to the hydrophobic patch residues adopt conformations that occlude the potential tunnel and thus prevent active site accessibility from this region of the protein.
The most notable differences between CAO1 and structurally characterized carotenoid/retinoid-metabolizing CCOs, for which ACO is considered the prototype, occur in the substrate-binding cavity and region corresponding to the membrane-interacting nonpolar patch (Figure 2C,D). These CCOs all possess a hydrophobic patch on their surfaces surrounding the opening to an apolar cavity that serves as both a passageway and a binding site for lipophilic carotenoid/retinoid substrates.3 The hydrophobic patch on the surface allows the enzymes to interact with membranes and extract their lipophilic substrates, which then traverse the apolar cavity to reach the catalytic center. In CAO1, this route is absent because of multiple structural alterations. The metal center is instead accessible only through a single narrow entry port whose opening is located in an alternative region of the protein surface (Figure 2C). This tunnel is topographically equivalent to the hydrophilic tunnel identified in ACO through which one of its two apocarotenoid products is thought to exit the active site.2 Similar structural features were observed in the bacterial stilbenoid-cleaving CCO, NOV1, although this enzyme has an entry port structure broader than that of CAO1 due in part to an unstructured loop in this region of the protein.
Many of the residues in CAO1 equivalent to those comprising the hydrophobic patch of ACO are either charged or polar or adopt non-solvent-exposed conformations. This together with the lack of a hydrophobic patch around the mouth of the substrate entry port demonstrates that membrane-integrating capability is absent in the stilbene-cleaving CCO lineage. The structure reveals a striking rearrangement of residues equivalent to those of the hydrophobic patch in ACO such that active site accessibility from this region of the enzyme is completely occluded (Figure 2D and Figure S3). The tunnel is further blocked by residues Arg166, Asp166, Glu213, and His277 whose charged side chains protrude into the potential cavity to form ionic or dipolar interactions.
Crystallographic Description of the CCO Iron Resting State
In contrast to these marked differences in the substrate-binding tunnel and active site accessibility, the CAO1 four-His iron center closely resembles that of other structurally characterized CCOs. His residues 197, 248, 313, and 510 directly coordinate the iron cofactor with bond lengths of 2.4, 2.2, 2.3, and 2.3 Å, respectively, averaged over the four monomers (Figure 2B). Notably, the iron B-factors in this structure were somewhat elevated as compared to those of the surrounding Nε atoms, suggesting submaximal iron occupancy. The His ligands are arranged with the Nε–Fe–Nε angles all near 90°. A solvent molecule is bound to the iron trans to His197 with a bond length of ~2.4 Å, which suggests it is a coordinated water, whereas the sixth site trans to His304 is unoccupied by ligand due to the nearby Thr151 methyl group. The iron is thus five-coordinate with a distorted square pyramidal geometry, similar to that of ACO (Figure 2B) but unlike those of VP1415 and NOV124 that contain O2 modeled in place of the solvent atom. Glu residues 165, 383, and 453 hydrogen bond with His 248, 313, and 510, respectively, forming an anionic second coordination sphere similar to that of ACO.
XAS Structures of the CCO Four-His Iron Center Resting State
Crystal structures of CCOs provide a general description of the four-His iron center, but they lack the ability to provide high-precision metal–ligand bond lengths, detect subtle differences in active site geometry, and may exhibit structural differences that arise from the conditions required for crystallization. To characterize the CCO four-His Fe(II) structure in greater detail and under a more uniform environment, we performed iron K-edge X-ray absorption spectroscopy studies on ACO, NOV2, and CAO1.
X-ray absorption near-edge spectroscopy (XANES) provides insight into the metal oxidation state and site symmetry. All three as-isolated enzymes exhibit similar edge shapes, with inflections observed at 7121.4–7121.8 and 7125.3–7126.1 eV, consistent with previous observations for Fe(II)-RPE6559 (Figure 3A and Table S2). There was minimal to no photoreduction in the X-ray beam upon extended exposure. Both observations are consistent with an Fe(II) center in all three as-isolated enzymes. The pre-edge peaks, associated with a parity-forbidden 1s–to-3d transition that gains intensity by mixing with 4p orbitals, provide additional understanding of metal site structure. Fe(II)-ACO exhibits a single, broad pre-edge peak centered at ~7113.0 eV with an area of 12.7(1) units (Table S2 and Figure S4). The other enzymes, measured with a higher-resolution Si(220) monochromator, show evidence of two pre-edge peaks with an approximate energy splitting of 1.9 eV. Specifically, Fe(II)-NOV2 has two pre-edge peaks at 7112.6 and 7114.5 eV with areas of 9.5(7) and 4.7(3) units, respectively, while the Fe(II)-CAO1 pre-edge peaks are found at 7112.8 eV [10.6(5) units] and 7114.6 eV [5.9(9) units] (Table S2 and Figure S4). The energies of these peaks are consistent with the expected Fe(II) state. The peak areas, on the high side for Fe(II), are most consistent with an Fe coordination number of four or five.60,61 We note that the Fe(II)-NOV2 pre-edge spectrum and intensity pattern are quite similar to those of previously reported five-coordinate, C3v Fe(II) sites in a model complex62 and the mineral grandidierite.63 We favor a five-coordinate assignment based on the EXAFS and Mössbauer analyses described below as well as the ACO13,28 and CAO1 crystal structures, which both feature five-coordinate iron.
Figure 3.
XAS analysis of as-isolated CCO iron centers. (A) Normalized XANES spectra of as-isolated Fe(II)-ACO (—), Fe(II)-NOV2 (– – –), and Fe(II)-CAO1 (⋯). The inset shows an expansion of the pre-edge region. The first-derivative XANES spectrum is shown in the bottom panel. (B) Representative best fits (bold entries in Tables S3-S5) to k3-weighted EXAFS data of Fe(II)-ACO (top), Fe(II)-NOV2 (middle), and Fe(II)-CAO1 (bottom). Experimental data are shown as a dotted line, whereas the best fit is shown as a solid line.
Extended X-ray absorption fine structure (EXAFS) analyses provided additional details about the iron coordination environment. All three Fe(II)-enzyme preparations can be fit with a single shell of five N/O scatterers at 2.12–2.13 Å (Figure 3B and Table S3–Table S5). For ACO, this shell can be further split into two subshells consisting of four N/O scatterers at 2.15 Å and one O/ N scatterer at 1.98–2.01 Å, the latter being attributable to an Fe(II)-hydroxo unit. These distances are in good agreement with the bond lengths determined by crystallography.28 CAO1 required a more elaborate scattering model consisting of four NHis scatterers at 2.15 Å, a shorter N/O scatterer at 1.99 Å, and two additional light atom (C/N/O) scatterers at 2.5 Å. In the CAO1 crystal structure, the short scatterer is not apparent, possibly because of the lower pH of the crystallization solution compared to that of the solution used for XAS studies, which could alter the bound solvent protonation state. The more distant scattering is at least partially attributed to the ~2.4 Å solvent–iron interaction observed crystallographically (Figure 2B). Notably, the relatively low Debye–Waller factor suggests a stable association of these longer-range scatterers with the iron center and is consistent with nearly complete occupancy of these sites by bound ligand. The requirement for 2.5 Å Fe–C/N/O units in the CAO1 scattering model bears resemblance to the scattering observed for RPE65.59 In the latter enzyme, the longer scatterer was attributed to the bridging carbon of a bound carboxylate. Our inability to resolve a short Fe–O moiety in Fe(II)-NOV2 would suggest that solvent water is not deprotonated in the as-isolated form of the enzyme. These five-coordinate solutions from EXAFS are consistent with the XANES analyses and are also supported by bond valence sum calculations.54 In addition, multiple scattering analyses provide evidence of three to four His-derived imidazole units bound to the iron center in each enzyme, consistent with the known four-His coordination motif of CCOs.
Characterization of the CCO Iron Center by 57Fe Mössbauer Spectroscopy
Mössbauer spectroscopy is sensitive to the primary coordination sphere of iron active sites in enzymes and provides information about the electronic structure of the metal center. As the natural abundance of 57Fe is low (2%), enrichment with 57Fe significantly improves the spectral content. We developed methods for high-yield expression of each CCO in iron-deficient M9 minimal medium in which 57Fe can be added to produce enriched enzyme samples. Of the three enzymes, NOV2 was expressed at the highest levels, which allowed the most detailed spectroscopic characterization for this protein. The Mössbauer spectrum of NOV2 recorded at 4.2 K (Figure 4A) showed a broad doublet with parameters listed in Table 1 that are indicative of non-heme S = 2 Fe(II). The width of the doublet (Γ = 0.5 mm/s) is compatible with the presence of two or more Fe(II) species. Approximately 20% of iron in the sample was adventitious Fe(III), indicated by the broad paramagnetic features extending to ±8 mm/s. This adventitious Fe(III) was unreactive to reductants or substrate turnover, and the amount was preparation-dependent. The Mössbauer spectrum of CAO1 recorded at 4.2 K in a small applied field (Figure S5C) showed a non-heme Fe(II) quadrupole doublet similar to that of NOV2 with parameters listed in Table 1. As with NOV2, the broad line width (Γ = 0.5 mm/s) indicates multiple conformations of the active site.
Figure 4.
Mössbauer spectra recorded at 4.2 K in an applied field of 45 mT: (A) NOV2, (B) NOV2 in the presence of 10 equiv of piceatannol, (C) NOV2 after turnover of 10 equiv of piceatannol, (D) NOV2 after turnover of 10 equiv of piceatannol and subsequent anaerobic addition of 10 equiv of piceatannol, and (E) NOV2 in the presence of 10 equiv of resveratrol. Protein (~2 mM) was prepared in 20 mM Tris-HCl (pH 7.5), 200 mM NaCl, and 20% (v/v) glycerol. The experimental and simulated spectra are shown as blue and black lines, respectively.
Table 1.
Mössbauer Parameters of Non-Heme Fe(II) Complexes
| sample | δ (mm/s) | ΔEq (mm/s) |
% | ref |
|---|---|---|---|---|
| ACO | 1.21 | 3.05 | 100 | twa |
| ACO and apocarotenal | 1.18 | 2.92 | 100 | twa |
| NOV2 | 1.09 | 3.47 | 100 | twa |
| NOV2 and resveratrol | 1.08 | 2.90/3.47 | 30/70 | twa |
| NOV2 and piceatannol | 1.08 | 2.27/3.42 | 30/70 | twa |
| CAO1 | 1.09 | 3.45 | >90b | twa |
| CAO1 and piceatannol | 1.09 | 3.45 | >90b | twa |
| 4,5-protocatechuate dioxygenase (PCD) | 1.28 | 2.23 | 66 | |
| 4,5-PCD and S | 1.27/1.22 | 2.33/2.8 | 60/40 | |
| 3,4-PCD | 1.21 | 3.13 | 67 | |
| taurine: α-ketoglutarate dioxygenase (TauD) | 1.27 | 3.06 | 68 | |
| TauD, α-ketoglutarate, and taurine | 1.16 | 2.76 | ||
| cysteine dioxygenase | 1.22 | 2.76 | 69 | |
| IPNS | 1.3 | 2.7 | 70 | |
| IPNS and ACV | 1.3/1.11 | 2.69/3.43 | 35/65 | |
| Rhodobacter sphaeroides R-26 | 1.16 | 2.16 | 71 | |
| FeEDTA | 1.28 | 3.45 | 72 |
This work.
A minor ferric impurity may also be present.
Notably, the isomer shifts of these two enzymes are comparable to each other, suggesting similar active site environments, but significantly lower than those of other non-heme iron enzymes (Table 1), including bacterial and plant reaction centers that possess four-His non-heme iron centers similar in structure to CCOs. The small isomer shifts for NOV2 and CAO1 indicate a comparatively more electropositive Fe(II) environment. The four-His, one-water/hydroxide iron coordination of CCOs contrasts with the more common two-His, one-carboxylate, and three-water coordination of 2H-1C dioxyge-nases. The difference in coordination of two additional His residues versus a carboxylate and two waters is expected to lower the level of ligand charge donation to the iron, thus causing the observed smaller isomer shift.
The Mössbauer spectrum of ACO recorded at 4.2 K in a small applied field (Figure S5A) showed a broad (Γ = 0.5 mm/s) quadrupole doublet, similar to that of NOV2 CAO1, with parameters listed in Table 1. However, the isomer shift of ACO is significantly larger than that of CAO1 and NOV2 and comparable to those of other non-heme iron proteins. This finding indicates a different primary coordination sphere for the ACO Fe(II) compared to those of NOV2 and CAO1. Considering the nearly uniform Fe–His bond lengths and coordination geometry observed for these three enzymes, the isomer shift difference may arise from differences in ligands occupying exchangeable sites, such as a change in the protonation state of water or an additional water coordinating to the iron.
In the absence of a substrate, the Fe(II) sites of the CCOs studied here were stable in air at room temperature. Previous crystallographic studies of VP14 and NOV1 reported a stable O2-iron species.15,64 In an attempt to generate such a species, a sample of uncomplexed NOV2 was exposed to pure O2 gas at the highest pressure that could be tolerated by the sample cell and preparatory equipment (40 psi). Coordination of O2 to Fe(II) centers results in significant and characteristic changes in the Mössbauer parameters. However, the Mössbauer spectrum before and after O2 exposure was unchanged. Assuming a minimal detection level of 10% of the total sample iron for an iron-oxy species gives a dissociation constant for O2 of >50 mM in the absence of a substrate. This result, in conjunction with the structural data presented above, demonstrates that the enzymes of this study do not have O2 bound to iron in the absence of an organic substrate.
Mössbauer Analysis of the Interaction of the Fe(II) Center with Substrates
With the resting state of the CCO Fe(II) center structurally and electronically defined, we next turned our attention to its interactions with substrates and substrate mimetics. We used previously established substrates for enzymes in these studies: resveratrol and piceatannol for NOV2 and CAO18,12 and all-trans-apo-8′-carotenol for ACO. The direct interaction of these organic substrates with the metal of each enzyme was first investigated by 57Fe Mössbauer spectroscopy, which probes ligand-induced perturbations in the iron electronic structure. For non-heme Fe(II) enzymes in which the substrate binds to the iron, the addition of a substrate causes a distinct change in the Mössbauer parameters (Table 1).
The anaerobic addition of 4 equiv of piceatannol to NOV2 produced an enzyme—substrate (ES) Mössbauer spectrum characterized by two doublets (Table 1 and Figure 4B). The doublet of the majority species (70%) was equal within experimental error to that of uncomplexed NOV2, suggesting partial occupation of the substrate in the active site. However, as discussed below, parallel EPR samples measuring substrate binding in the presence of NO indicated that nearly all active sites have piceatannol bound. Furthermore, addition of 10 equiv of piceatannol did not change the relative amounts of the two doublets, indicating that the new species is not under a titration equilibrium for substrate at ≥4 equiv. This speciation was the same at pH 8.5 and 7.5, suggesting that it does not arise from different protonation states of piceatannol, whose lowest calculated pKa value is ~8.5. The susceptibility of the new species to cleavage was investigated by incubating the ES sample at room temperature in air for 5 min with stirring to allow substrate turnover followed by refreezing. After turnover, the minority species (inner Mössbauer doublet of Figure 4B) nearly vanishes, consistent with the loss of the substrate-bound complex and return to the uncomplexed Fe(II) enzyme state for the minority species and presumably for the majority species (Figure 4C). As with the sample of uncomplexed NOV2 treated with O2, there was no change or increase in Fe(III) to indicate binding of O2 or oxidation of the active site upon O2 exposure. Ten equivalents of additional piceatannol was then added to the sample that had been turned over. The corresponding Mössbauer spectrum (Figure 4D) was unchanged relative to the spectrum of NOV2 prior to substrate addition; specifically, the inner doublet from the minority species did not reappear. From UV spectra, an assay of the enzyme from this Mössbauer sample showed that the enzyme was still active in converting piceatannol to product. These results suggest that turnover causes a structural rearrangement that reduces the heterogeneity of the species.
The results for the substrate resveratrol were similar to those with piceatannol. The Mössbauer spectrum of NOV2 in the presence of 4 equiv of resveratrol showed two sets of doublets in approximately the same ratio as in piceatannol (Table 1 and Figure 4E). The new doublet that appeared upon addition of resveratrol had parameters different from those of piceatannol. Like piceatannol, the majority species had parameters that were the same as those of uncomplexed NOV2. An amount of adventitious Fe(III) (~20%) was observed for this sample. Like the addition of piceatannol, the addition of 10 equiv of resveratrol showed the same Mössbauer spectrum as the 4 equiv sample, indicating that the new doublet is not under a titration equilibrium with the substrate for these amounts of substrate. These data indicate that a majority of the Fe(II) centers of these enzymes are unaffected by the addition of substrate, consistent with the substrates binding near but not in direct coordination with the iron. The same conclusion can also be drawn for the CAO1 and ACO Fe(II) centers whose Mössbauer parameters were only subtly affected by substrate binding (Table 1 and Figure S5B,D). In summary, indications of substrate binding based on Mössbauer spectroscopy were ambiguous, which necessitated alternative methods of probing active site substrate binding.
EPR and Mössbauer Characterization of CCO Fe-NO Adducts
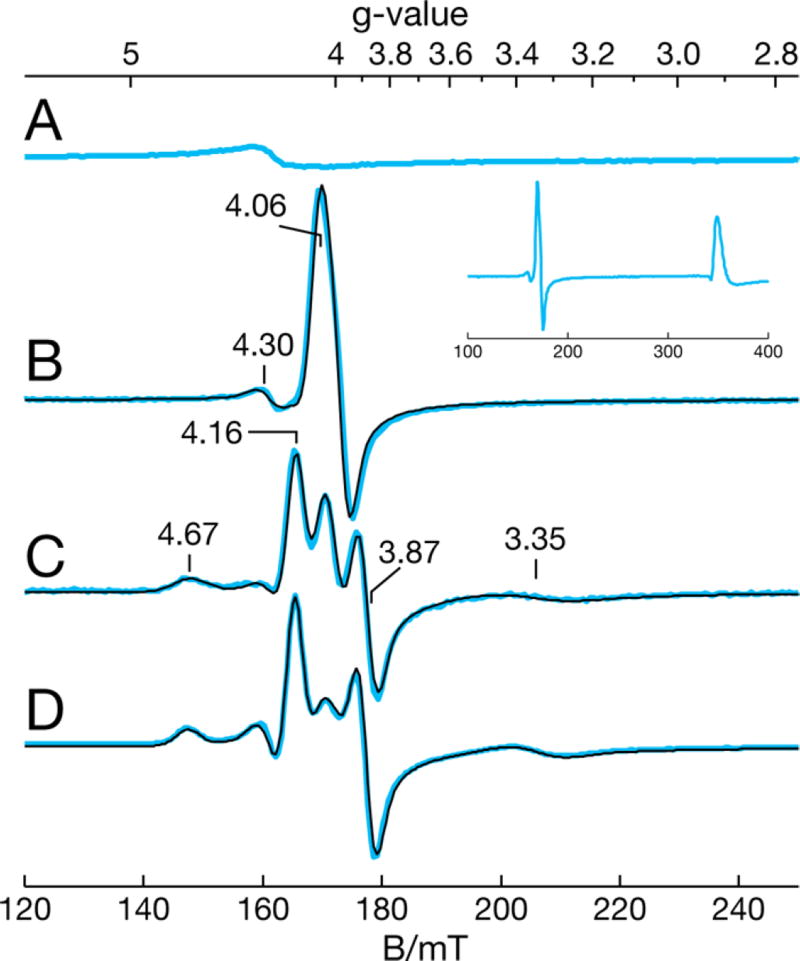
EPR spectroscopy is more sensitive to perturbations surrounding the active site, but high-spin non-heme Fe(II) sites do not necessarily show EPR signals. The coordination of NO to non-heme Fe(II) enzymes (E-NO) provides an additional electron to produce EPR-active complexes with NO acting as an O2 surrogate.73 EPR spectra of NO adducts of non-heme Fe(II) proteins show resonances near g = 4 and 2, indicative of {FeNO}7 metal centers (Enemark–Feltham notation).74 Perturbations to the active site, such as substrate binding, cause symmetry changes in the iron coordination as indicated by the rhombicity parameter E/D. A nonaxial symmetry (E/D > 0) splits the g = 4 signal.
EPR spectra of NOV2 and ACO samples in the presence of excess NO, plotted for equal protein concentrations, are shown in Figure 5 and Figure S6, respectively, for the g = 4 region. Quantitative simulations of these spectra gave the E/D values and relative concentrations listed in Table 2. The g = 4.3 signal is a preparation-dependent, adventitious Fe(III) species of ~5% of the protein concentration, corresponding to the minor paramagnetic Fe(III) species of the Mössbauer spectra, that was present in the samples prior to NO addition (Figure 5A and Figure S6A). The NOV2 E-NO adduct showed g = 4.0 and 2.0 signals (Figure 5B), indicative of a near-axial non-heme {FeNO}7 species with an E/D of 0.007, which was in quantitative agreement with the total amount of iron in the sample determined by inductively coupled plasma optical emission spectroscopy analysis. The spectrum of the ACO E-NO adduct (Figure S6B) showed an {FeNO}7 species with coordination geometry (E/D = 0.010) slightly more rhombic than that of NOV2. The spectrum was unperturbed by detergent addition, which is required for substrate delivery described below (Figure S6C). These spectra and E/D values are similar to those of E-NO species from other non-heme enzymes. All samples with NO addition show an axial S = ½ species in minority amounts (<5%) with g values of 2.22 and 2.03, and a signal from free NO near g = 1.97. The amounts of these species were preparation-dependent but not substrate-dependent. The g values of 2.22 and 2.03 indicate an unknown low-spin Fe(III) complex that does not fit within parameters for {Fe(NO)2}9 complexes.75
Figure 5.
X-Band EPR spectra of NOV2 alone and in the presence of excess NO from NONOate: (A) enzyme only as isolated, (B) enzyme with NO, (C) enzyme with 10 equiv of piceatannol and NO, and (D) enzyme with 4 equiv of resveratrol and NO. Spectra were recorded at 16 K. The experimental and simulated spectra are shown as blue and black lines, respectively. The inset depicts the full spectrum of panel B, showing excess NO as evidenced by the large characteristic signal near g = 1.94. Samples were prepared in 20 mM Tris-HCl (pH 8.5) in 200 mM NaCl and 20% (v/v) glycerol. The spectra have been scaled to represent an equal protein concentration of 0.15 mM NOV2.
Table 2.
E/D Values and Amounts of NO Adducts of Enzymes with Substrates
| E-NO
|
ES-NO (1)
|
ES-NO (2)
|
|||||
|---|---|---|---|---|---|---|---|
| E/D | % | E/D | % | E/D | % | ref | |
| NOV2 | 0.007 | 100 | twa | ||||
| NOV2 and piceatannol | 0.007 | 11 | 0.024 | 60 | 0.108 | 29 | twa |
| NOV2 and resveratrol | 0.007 | 7 | 0.024 | 66 | 0.112 | 27 | twa |
| ACO | 0.010 | 100 | twa | ||||
| ACO and apocarotenoid | 0.010 | 63 | 0.024 | 37 | twa | ||
| 4,5-PCD | 0.016 | 100 | 66 | ||||
| 4,5-PCD and protocatechuate | 0.031 | 70 | 0.065 | 30 | |||
| 2,3-homoprotocatechuate dioxygenase (2,3-HPCD) | 0.005 | 100 | 76 | ||||
| 2,3-HPCD and homoprotocatechuate | 0.005 | minor | 0.025 | major | |||
| IPNS | 0.015 | 100 | 70 | ||||
| IPNS and ACVb | 0.035 | 100 | |||||
This work.
(l-α-Amino-δ-adipoyl)-l-cysteinyl-d-valine.
The NOV2 E-NO complex was further characterized by Mössbauer spectroscopy in variable magnetic fields and temperatures (Figure S7). This sample had approximately 15% adventitious Fe(III). The simulations include this Fe(III) species, and the red lines are simulations for the two species that gave the sum displayed on the 45 mT spectrum. The electronic and nuclear parameters listed in Table S6 were determined from the displayed least-squares simulations. A comparison of corresponding parameters of characterized non-heme {FeNO}7 centers is shown in Table S6. The interaction with NO dominates the electronic properties of iron, and thus, the parameters are comparable to those from the literature. Given the limited set of characterized complexes available, the extent to which the four-His coordination affects these parameter values is unclear. Variable field Mössbauer spectra of the NOV2 ES-NO complex were not explored because of the multiple species in the sample. Additionally, acquisition of Mössbauer spectra for the NO adduct of ACO was not attempted because of difficulties in obtaining sufficient quantities of 57Fe-enriched ACO.
EPR Characterization of ES-NO Complexes of NOV2 and ACO
Next, we investigated the influence of substrate binding on the E-NO complex EPR spectra of NOV2 and ACO. For these studies, ES complexes were formed under anaerobic conditions followed by addition of NO in solution. The EPR spectrum of NOV2 reacted with 4 equiv of piceatannol followed by NO showed three species (Figure 5C) that are listed in Table 2. The substrate-free E-NO complex was from a minority of the iron present, indicating that nearly all active sites had substrate bound. Two new species formed with E/D values (0.024 and 0.10) different from that of E-NO (0.007), indicating different conformations or modes of binding of the substrate or NO to the active site. The relative concentrations of the two substrate-bound forms closely match the relative amounts of the two doublets observed in the Mössbauer spectrum of NOV2 with piceatannol. This suggests a common physical origin for the substrate species observed via Mössbauer spectroscopy with the ES-NO species of the EPR spectra. The additions of 4 and 10 equiv of substrate relative to the protein concentration did not change the ratio of these two species. Thus, full occupation with substrate is achieved at 4 equiv, and the minor amount of E-NO remaining presumably consists of inactive iron sites. The resveratrol ES+NO of NOV2 showed EPR spectra (Figure 5D) similar to those with piceatannol with approximately the same ratios of species. For a substoichiometric addition of 0.4 equiv of resveratrol, the levels of the two ES-NO species decrease by an amount (~30%) consistent with the amount of added resveratrol.
Addition of NO to the ACO-apocarotenoid complex resulted in the appearance of a more rhombic species (E/D = 0.024) as compared to the E-NO spectrum (E/D = 0.010). The amount of the new rhombic species increases with an increasing substrate concentration, and the new rhombic species was interpreted as a substrate-bound species that is similar to the majority ES-NO species of NOV2. The spectra indicated that for 1 (Figure S6D) and 3 (Figure S6E) equiv of substrate, 20 and 40%, respectively, of the active sites were bound with substrate. The titration could not be continued because 3 equiv of substrate was near the solubility limit of the compound. The second ES-NO species of NOV2 with an E/D of 0.1 was not observed for ACO.
The speciation observed by Mössbauer spectroscopy in the presence of substrates could possibly be interpreted as a low substrate binding constant or a low substrate solubility such that even 10 equiv of substrate produces only a minority occupation of the substrate-binding site. In this scenario, the addition of NO increases the binding constant for the substrate and all sites show bound substrate. Such a result has been reported for IPNS; however, the results presented here differ from those described for IPNS.70 For IPNS, the NO adduct without a substrate showed a single species that wholly changed to a new species upon substrate addition. In contrast, now focusing specifically on NOV2 for which the data are the clearest, we found the addition of ≥4 equiv of substrate did not change the ratio of species, and the concentration ratio of the two species was independent of NO.
Trapping CAO1-Substrate Complexes for Crystallo-graphic Studies
To clarify the atomic interactions underlying these spectroscopic findings, we sought to determine crystal structures of CCOs in complex with substrates. Previously, we showed that the apocarotenoid substrate modeled in ACO13 was mistaken for another component of the crystallization mixture.28 Our efforts to determine a genuine ACO-substrate complex were also unsuccessful likely because of the water-insoluble nature of its carotenoid substrates. We thus focused on trapping stilbenoids within the CAO1 active site, which have more favorable aqueous solubility properties. Two strategies were used to stably trap substrates in the CAO1 active site for structural characterization; one aimed to produce catalytically inert but structurally intact CAO1, and the other employed a modified substrate with slowed cleavage kinetics.
In the first case, we substituted Co(II) for the native Fe(II) cofactor. Because of its elevated standard reduction potential of 1.92 V compared to a potential of 0.77 V for iron, the +2 to +3 redox change associated with O2 binding and activation is disfavored, and cobalt-substituted non-heme iron oxygenases are often catalytically inert 77,78 with at least one exception.79 Cobalt substitution was accomplished by expressing CAO1 in minimal medium supplemented with Co(II). The protein was purified and crystallized in the same manner used for Fe-CAO1. Stilbenoid cleavage activity was observed when the cobalt-substituted protein was used at a higher than normal concentration in the assay mixture (Figure S8). However, this residual activity could be attributed to a small amount of Fe-CAO1 in the sample (Table S7). The presence of cobalt in the CAO1 metal-binding site was verified with Co–K edge anomalous scattering data (Figure S9A,B). Despite this difference in metal content, the crystal structure of Co-CAO1 was similar to that of Fe-CAO1 (Figure 6A) with a five-coordinate metal center containing a water molecule bound trans to His197 at a distance of ~2.4 Å (Figure 6A).
Figure 6.
Structures of CAO1 in complex with stilbenoids. (A) Active site structure of Co-substituted CAO1. The cobalt ion and coordinated solvent are shown as salmon and red spheres, respectively. (B and C) Structures of Co-CAO1 in complex with resveratrol (orange sticks) and piceatannol (yellow-orange sticks), respectively. (D) Structure of Fe-CAO1 in complex with β-fluororesveratrol (light purple sticks). The iron ion is shown as a brown sphere.
We also synthesized a resveratrol derivative, β-fluororesveratrol, in which the β-carbon hydrogen atom is replaced with fluorine. Substitution of an electronegative fluorine atom at the β-carbon position was expected to block the oxidative cleavage reaction by inductive withdrawal of electron density from the scissile alkene, thus weakening its ability to react with O2. It was shown previously that the β-fluoro derivative of 4-hydroxys-tilbene is a potent inhibitor of LSD80. We found that β-fluororesveratrol actually can be cleaved by CAO1, albeit slowly as compared to that with resveratrol, with cleavage only detectable at a high enzyme concentration with prolonged incubation (Figure S8).
Crystal Structures of CAO1 in Complex with Stilbenoid Substrates
Using these trapping methods, we generated crystals of CAO1 in complex with resveratrol, piceatannol, and β-fluororesveratrol and determined their structures to resolutions of ~1.9 Å. In all cases, the difference maps unambiguously demonstrated the presence of the target ligands in the active site with all their atoms resolved (Figure S9C–G).
Resveratrol and piceatannol are bound to Co-CAO1 with their β-carbon-associated rings located deepest within the substrate-binding pocket. The 4′-hydroxyl moiety forms hydrogen bonding interactions with Tyr133 and Lys164, which are conserved among functionally characterized stilbenoid-cleaving CCOs but not carotenoid-cleaving members (Figure 6B,C and Figure S3). Piceatannol forms an additional hydrogen bonding interaction with Thr151 through its 3’-hydroxyl group (Figure 6C). The 3,5-dihydroxy ring is near the pre-active site-binding region where it hydrogen bonds with Glu383. The other hydroxyl group of this ring is oriented toward the active site entrance. Substrate binding induces minimal active site conformational changes. The largest difference occurs at Phe384, which becomes more mobile in the presence of a substrate adopting at least two distinct conformations. There is a notable departure from the trans planar geometry expected for these ligands in solution with Cβ–Cα–C1–C2 and Cα–Cβ– C1′–C2′ dihedral angle deviations of ~17° and ~17° for resveratrol and ~10° and ~20° for piceatannol, respectively. The closest approach these substrates make to the metal is through their α-carbon atoms at a distance of ~4.7 Å. This lack of a direct interaction with iron is consistent with the minimal Mössbauer spectral changes observed upon CCO-substrate complexation. Although the substrates do not directly coordinate the metal cofactor, they do form interactions with the metal-binding ligands. In both cases, the substrate a-carbon atom makes an ~3 Å contact with the metal-coordinated solvent atom. This solvent ligand is located ~2.5 Å from the metal, suggesting it is bound water. The positioning of the metal-bound solvent is similar to that of the substrate-free Co-CAO1 structure, although it is more localized in the presence of the substrate. Notably, the metal coordination site trans to His313 remains unoccupied in the ligand-bound structures. Given the proximity of the solvent coordination site to the scissile double bond and the inaccessibility of the vacant sixth coordination position, O2 probably binds trans to His197, displacing the solvent molecule during catalysis. The strong perturbation of the CCO Fe-NO EPR spectra in the presence of the substrate suggests that NO also binds at this position, which would place the NO oxygen atom in van der Waals contact with the bound substrate, leading to a change in the symmetry of the Fe-NO adduct. The substrates also make ~3.3 Å contacts with His248Cε1 and His313Cε1 through their Cβ/C1′ and C2/C3 atoms, respectively. Both structures feature a second substrate molecule bound within the pre-active site-binding cleft. In both cases, the 4′-hydroxyl group formed a hydrogen bonding interaction with the 5-hydroxyl moiety of the active site-bound substrate molecule. The simultaneous binding of ligands in the active site cavity as well as the entry port is attributable to the high concentrations of substrate used to form the ES crystals.
During preparation of the manuscript, a structure of NOV1 in complex with resveratrol and O2 was reported.24 Despite the similar orientation and configuration of resveratrol in NOV1, this ligand has less clear electron density support than the CAO1 complex structures do (Figure S9H). The NOV1 structure was obtained with the active iron enzyme, making it likely that a mixture of substrate, product, and intermediates was present in the active site, resulting in the weak and broken density for the resveratrol ligand.
Binding of β-fluororesveratrol to Fe-CAO1 is similar to that of the natural ligands, but the orientation of the interphenyl alkene group is rotated by ~180° relative to that of resveratrol and piceatannol (Figure 6D). This arrangement places the Cβ-F moiety close to the iron center with the fluorine atom partially occupying the solvent-binding site of the iron. The fluorine atom resides ~3.9 Å from the iron, excluding a direct coordinate bonding interaction. However, the residual electron density observed after modeling the β-fluororesveratrol suggested the presence of a partially occupied solvent molecule between fluorine and iron with interaction distances of 2.2 and 2 Å, respectively. Organic fluorine typically does not engage in polar interactions,81 making the close F–O interaction unusual. It is possible that solvent is bound to iron only in the absence of β-fluororesveratrol and that β-fluororesveratrol only partially occupies the active site as suggested by the elevated B-factors of its atoms relative to those of the surrounding protein. Irrespective of these details, it is clear that the fluorine atom partially occludes the solvent-binding site and therefore may interfere with binding of O2 to iron, thus contributing to its slow rate of cleavage. The β-fluororesveratrol dihedral angles exhibit even greater deviation from planarity compared to the natural substrates.
DISCUSSION
Implications for CCO Biology and Substrate Specificity
Here we have shown that the four-His iron centers of alkene-cleaving CCOs are nearly indistinguishable with respect to their metal-protein interactions. This structural similarity extends to RPE65, a retinoid isomerase/ester-cleaving CCO, that we characterized previously.59 The preserved four-His Fe(II) structure across distantly related CCO enzymes suggests that their differences in activity are not due to an intrinsic difference in the reactivity of their iron centers toward O2 activation versus ester hydrolysis. Mössbauer spectroscopy did, however, reveal differences in apparent charge at the Fe(II) center between stilbenoid- and carotenoid-cleaving CCOs that could be relevant to their substrate specificity. The substrate-binding site is more polar in CAO1 than in ACO. Substrate recognition in ACO mainly involves nonpolar and π-stacking interactions, whereas hydrogen bonding plays a more prominent role in CAO1. Electrostatic differences in substrate-binding regions could influence the donation of charge to the iron and thereby affect solvent protonation or coordination and the metal redox potential. Stilbenoids such as resveratrol as antioxidants are stronger than carotenoids and may require a less powerful iron-oxidizing species for cleavage, consistent with the more positive iron environment in stilbenoid-cleaving CCOs.82
The crystal structures of fungal and bacterial stilbenoid-cleaving CCOs, CAO1 and NOV1,24 respectively, demonstrate that enzymes of this subfamily lack hydrophobic patches that are characteristic of carotenoid/retinoid-metabolizing CCOs. Re-sveratrol has an aqueous solubility greater than that of carotenoids but is still relatively hydrophobic and known to partition into membranes.83 The absence of a hydrophobic patch would appear to be maladaptive if resveratrol is in fact a biologically relevant substrate for these enzymes as has been proposed.12 This finding suggests the existence of substrates more biologically relevant than resveratrol or mechanisms of substrate access other than direct extraction from membranes. These structures also exemplify the malleability of the CCO substrate-binding site to allow the metabolism of diverse substrates. Such plasticity was first evident through comparison of the RPE6514 and ACO13 structures that showed loss of active site accessibility through a hydrophilic tunnel in RPE65. The stilbenoid-cleaving CCOs possess a contrasting structure with the active site entry port located within a distinct region of the protein and only vestiges of the hydrophobic tunnel present in carotenoid-cleaving CCOs and RPE65.
We also describe the active site-bound structure of a fluorinated substrate derivative that acts as a CCO inhibitor because of slow reaction kinetics. The mechanism of inhibition possibly involves both steric blockade of binding of O2 to the iron center as well as withdrawal of an electron from the scissile alkene, thus reducing its reactivity toward O2. Introduction of fluorine groups into the scissile double bonds of CCO substrates could be a generally effective strategy for producing inhibitors that are useful for studying the physiology of these enzymes.
Implications for the CCO Catalytic Mechanism
We have characterized the resting state of the CCO four-His iron center and its interactions with substrates in both carotenoid- and stilbenoid-cleaving members of the family. These enzymes are all Fe(II) proteins in their as-isolated states and are resistant to autoxidation during prolonged exposure to atmospheric oxygen, a behavior similar to that of some catechol dioxygenases.84 Crystallographic studies of VP14 and NOV1 have suggested that these enzymes stably bind O2 in the absence of substrate. Our crystallographic, XAS, and Mössbauer spectroscopy data demonstrate that this is not the case for the three enzymes examined in this study, which raises questions about the identity of the iron-bound ligands in the VP14 and NOV1 structures.
Nearly all characterized non-heme Fe(II) dioxygenases exhibit sequential binding of substrate and O2.20 O2 binding can be gated by substrate binding directly to the iron85 or by interactions of the substrate and/or cofactor with ligands bound to the metal,27,86 both of which alter the electronic properties of the iron. Alternatively, as in naphthalene 1,2-dioxygenase, the substrate has no interaction with the metal but its binding causes significant conformational changes that can gate O2 binding.26,87 For the CCOs studied here, these gating mechanisms do not seem applicable. Substrate binding did not induce a significant electronic change in the iron as observed by Mössbauer spectroscopy, nor did it induce significant protein structural changes. We observed an O2 dissociation constant for NOV2 of >50 mM, and it remains unclear how the substrate could influence this low affinity. The more electropositive iron environment presumably reduces the O2 affinity to lower than that of the 2H-1C dioxygenases. Whether such a low O2 affinity persists in the presence of substrate and its influence on enzymatic reactivity requires further kinetic measurements outside the scope of this study. However, it is possible that the substrate stabilizes binding of O2 to the metal by direct interaction of the substrate with O2 while bound to the metal. This could increase the binding affinity of O2 for iron, by disfavoring binding of the solvent molecule or from a weak aliphatic hydrogen bond between O2 and the substrate, although the structure indicates that the nearest substrate CH bond is pointed away from the O2 binding position. Consistent with this proposal, the EPR data presented here show a direct interaction of the substrate with the Fe-NO adduct, which may structurally mimic the iron-oxy complex.
Our EPR studies of the ES-NO complexes of NOV2 and ACO demonstrate that the CCO active site can bind NO in the presence and absence of the bound substrate. Similar findings were reported for NOV1, and the formation of an Fe-NO complex in the absence of substrate was taken as evidence of the similar reactivity with O2.24 However, such a conclusion is problematic because of the higher Fe(II) binding affinity of NO relative to O2. The literature provides several examples of non-heme iron oxygenases with known sequential binding of substrate and O2 that are nevertheless reactive toward NO without substrate.26,66,88
We speculate that upon formation of the CCO-O2-substrate complex, π electron density from the scissile double bond is redistributed to the iron-oxy complex to form an Fe(II)-peroxo-substrate cation intermediate, similar to that proposed for catechol dioxygenases.65,89 In the case of hydroxystilbenoids, the substrate cationic charge could be neutralized via deprotonation of the 4’-hydroxyl group to form a dienone intermediate, whereas other mechanisms of cation stabilization, including π-cation and quadrupole interactions, must be used by most carotenoids that lack stabilizing hydroxyl groups. These initial steps of the reaction are essentially consistent with mechanisms described for ACO based on density functional theory (DFT) calculations18. We also show that substitution of Co(II) for Fe(II) in CCOs does not afford catalytically active protein. This is in contrast to an extradiol dioxygenase in which Co(II) and Mn(II) support enzymatic activity.79,84,90 For this enzyme, the additional negative charge presented to metal upon substrate coordination probably overcomes the elevated redox barriers of Co and Mn to allow O2 binding and activation. Such substrate-triggered metal redox potential changes are apparently not possible in CCOs because of their lack of direct substrate–metal interactions.
DFT studies of the reaction mechanism of ACO have suggested that following O2 activation the cleavage reaction could proceed along two pathways. One involves dioxetane formation via substrate reactions with an Fe(III)-O2− species. In the second, the iron-superoxo adduct is converted to an Fe(IV)═O species with formation of an epoxide intermediate.18 Although the data presented here do not address these aspects of the catalytic mechanism, the methods we have developed along with the experimentally well-supported crystallographic and spectroscopic structures of the CCO active site resting state and its complex with substrates set the stage for future detailed mechanistic studies.
Supplementary Material
Acknowledgments
We thank Dr. Leslie T. Webster Jr. for helpful comments on the manuscript.
Funding
This research was supported in part by grants from the National Institutes of Health (NIH) (EY009339 to K.P. and P.D.K., EY020551 to J.v.L. and K.P., CA157735 to G.P.T., and GM77387 to M.P.H.), the Department of Veterans Affairs (IK2BX002683 to P.D.K.), and the Burroughs Wellcome Fund (1015187 to P.D.K.). M.P.H. recognizes National Science Foundation Grant CHE1126268 for funding of the EPR spectrometer. This work is based in part upon research conducted at the APS Northeastern Collaborative Access Team beamlines supported by Grants GM103403, RR029205, and DE-AC02-06CH11357. Use of NSLS beamline X3B was made possible by NIH Grant P30-EB-009998 (Case Western Reserve University Center for Synchrotron Biosciences) and the U.S. Department of Energy (DOE) Office of Science under Contract DE-AC02-98CH10886 to Brookhaven National Laboratory. Use of SSRL beamline 9–3 was made possible by the U.S. DOE Office of Science under Contract DE-AC02-76SF00515 to SLAC National Accelerator Laboratory and NIH Grant P41-GM-103393 (SSRL Structural Molecular Biology Program).
ABBREVIATIONS
- 2H-1C
2-histidine, 1-carboxylate
- ACO
Synechocystis sp. PCC6803 apocarotenoid oxygenase
- BCO1
β-carotene oxygen-ase 1
- CAO1
N. crassa carotenoid oxygenase 1
- CCO
carotenoid cleavage oxygenase
- DFT
density functional theory
- E-NO
enzyme-nitric oxide
- EPR
electron paramagnetic resonance
- ES
enzyme-substrate
- EXAFS
extended X-ray absorption fine structure
- IPNS
isopenicillin-N-synthase
- LSD
lignostilbene-α,β-dioxygenase
- NCED
9-cis-epoxycarotenoid dioxygenase
- NO
nitric oxide
- NOV
N. aromaticivorans oxygenase
- O2
dioxygen
- RMS
root-mean-square
- RPE65
retinal pigment epithelium 65 kDa protein
- VP14
viviparous-14
- XANES
X-ray absorption near-edge spectroscopy
- XAS
X-ray absorption spectroscopy
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.bio-chem.7b00251.
Accession Codes
Crystal structures reported here have been deposited in the Protein Data Bank as entries 5U8X (Fe-CAO1), 5U8Y (Co-CAO1), 5U8Z (Fe-CAO1 in complex with β-fluororesveratrol), 5U90 (Co-CAO1 in complex with resveratrol), and 5U97 (Co-CAO1 in complex with piceatannol).
Author Contributions
P.D.K., E.R.F., and M.P.H. designed research. X.S. performed protein expression, purification, and crystallization, collected X-ray diffraction data, and contributed to crystal structure refinements. ACW. performed EPR and Mössbauer spectros-copy sample preparation, data collection, and data analysis. E.R.F. performed XAS sample preparation, data collection, and data analysis. M.B. and G.P.T. synthesized β-fluororesveratrol. S.B. collected X-ray diffraction data and assisted with crystal structure refinements. K.P. and J.v.L. assisted with enzymology studies. M.P.H. oversaw the EPR and Mössbauer spectroscopy experiments and data analysis. P.D.K. contributed to and oversaw the biochemical experiments and crystal structure determinations. P.D.K., E.R.F., and M.P.H. wrote the paper with contributions from all authors. All authors have given approval to the final version of the manuscript. X.S., A.C.W., and E.R.F. contributed equally.
The authors declare no competing financial interest.
References
- 1.Auldridge ME, McCarty DR, Klee HJ. Plant carotenoid cleavage oxygenases and their apocarotenoid products. Curr. Opin. Plant Biol. 2006;9:315–321. doi: 10.1016/j.pbi.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 2.Kloer DP, Schulz GE. Structural and biological aspects of carotenoid cleavage. Cell. Mol. Life Sci. 2006;63:2291–2303. doi: 10.1007/s00018-006-6176-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sui X, Kiser PD, Lintig J, Palczewski K. Structural basis of carotenoid cleavage: from bacteria to mammals. Arch. Biochem. Biophys. 2013;539:203–213. doi: 10.1016/j.abb.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.von Lintig J, Vogt K. Filling the gap in vitamin A research. Molecular identification of an enzyme cleaving beta-carotene to retinal. J. Biol. Chem. 2000;275:11915–11920. doi: 10.1074/jbc.275.16.11915. [DOI] [PubMed] [Google Scholar]
- 5.Schwartz SH, Tan BC, Gage DA, Zeevaart JA, McCarty DR. Specific oxidative cleavage of carotenoids by VP14 of maize. Science. 1997;276:1872–1874. doi: 10.1126/science.276.5320.1872. [DOI] [PubMed] [Google Scholar]
- 6.Al-Babili S, Bouwmeester HJ. Strigolactones, a novel carotenoid-derived plant hormone. Annu. Rev. Plant Biol. 2015;66:161–186. doi: 10.1146/annurev-arplant-043014-114759. [DOI] [PubMed] [Google Scholar]
- 7.Kamoda S, Saburi Y. Cloning, expression, and sequence analysis of a lignostilbene-alpha,beta-dioxygenase gene from Pseudomonas paucimobilis TMY1009. Biosci., Biotechnol., Biochem. 1993;57:926–930. doi: 10.1271/bbb.57.926. [DOI] [PubMed] [Google Scholar]
- 8.Marasco EK, Schmidt-Dannert C. Identification of bacterial carotenoid cleavage dioxygenase homologues that cleave the interphenyl alpha,beta double bond of stilbene derivatives via a monooxygenase reaction. ChemBioChem. 2008;9:1450–1461. doi: 10.1002/cbic.200700724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brefort T, Scherzinger D, Limon MC, Estrada AF, Trautmann D, Mengel C, Avalos J, Al-Babili S. Cleavage of resveratrol in fungi: characterization of the enzyme Rco1 from Ustilago maydis. Fungal Genet. Biol. 2011;48:132–143. doi: 10.1016/j.fgb.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 10.Redmond TM, Yu S, Lee E, Bok D, Hamasaki D, Chen N, Goletz P, Ma JX, Crouch RK, Pfeifer K. Rpe65 is necessary for production of 11-cis-vitamin A in the retinal visual cycle. Nat. Genet. 1998;20:344–351. doi: 10.1038/3813. [DOI] [PubMed] [Google Scholar]
- 11.Ruch S, Beyer P, Ernst H, Al-Babili S. Retinal biosynthesis in Eubacteria: in vitro characterization of a novel carotenoid oxygenase from Synechocystis sp. PCC 6803. Mol. Microbiol. 2005;55:1015–1024. doi: 10.1111/j.1365-2958.2004.04460.x. [DOI] [PubMed] [Google Scholar]
- 12.Diaz-Sanchez V, Estrada AF, Limon MC, Al-Babili S, Avalos J. The oxygenase CAO-1 of Neurospora crassa is a resveratrol cleavage enzyme. Eukaryotic Cell. 2013;12:1305–1314. doi: 10.1128/EC.00084-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kloer DP, Ruch S, Al-Babili S, Beyer P, Schulz GE. The structure of a retinal-forming carotenoid oxygenase. Science. 2005;308:267–269. doi: 10.1126/science.1108965. [DOI] [PubMed] [Google Scholar]
- 14.Kiser PD, Golczak M, Lodowski DT, Chance MR, Palczewski K. Crystal structure of native RPE65, the retinoid isomerase of the visual cycle. Proc. Natl. Acad. Sci. U. S. A. 2009;106:17325–17330. doi: 10.1073/pnas.0906600106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Messing SA, Gabelli SB, Echeverria I, Vogel JT, Guan JC, Tan BC, Klee HJ, McCarty DR, Amzel LM. Structural insights into maize viviparous14, a key enzyme in the biosynthesis of the phytohormone abscisic acid. Plant Cell. 2010;22:2970–2980. doi: 10.1105/tpc.110.074815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tan BC, Schwartz SH, Zeevaart JA, McCarty DR. Genetic control of abscisic acid biosynthesis in maize. Proc. Natl. Acad. Sci. U. S. A. 1997;94:12235–12240. doi: 10.1073/pnas.94.22.12235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sui X, Golczak M, Zhang J, Kleinberg KA, von Lintig J, Palczewski K, Kiser PD. Utilization of Dioxygen by Carotenoid Cleavage Oxygenases. J. Biol. Chem. 2015;290:30212–30223. doi: 10.1074/jbc.M115.696799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Borowski T, Blomberg MR, Siegbahn PE. Reaction mechanism of apocarotenoid oxygenase (ACO): a DFT study. Chem. - Eur. J. 2008;14:2264–2276. doi: 10.1002/chem.200701344. [DOI] [PubMed] [Google Scholar]
- 19.Hegg EL, Que L., Jr The 2-His-1-carboxylate facial triad-an emerging structural motif in mononuclear non-heme iron(II) enzymes. Eur. J. Biochem. 1997;250:625–629. doi: 10.1111/j.1432-1033.1997.t01-1-00625.x. [DOI] [PubMed] [Google Scholar]
- 20.Kovaleva EG, Lipscomb JD. Versatility of biological non-heme Fe(II) centers in oxygen activation reactions. Nat. Chem. Biol. 2008;4:186–193. doi: 10.1038/nchembio.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abu-Omar MM, Loaiza A, Hontzeas N. Reaction mechanisms of mononuclear non-heme iron oxygenases. Chem. Rev. 2005;105:2227–2252. doi: 10.1021/cr040653o. [DOI] [PubMed] [Google Scholar]
- 22.dela Sena C, Riedl KM, Narayanasamy S, Curley RW, Jr, Schwartz SJ, Harrison EH. The human enzyme that converts dietary provitamin A carotenoids to vitamin A is a dioxygenase. J. Biol. Chem. 2014;289:13661–13666. doi: 10.1074/jbc.M114.557710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Babino D, Golczak M, Kiser PD, Wyss A, Palczewski K, von Lintig J. The Biochemical Basis of Vitamin A3 Production in Arthropod Vision. ACS Chem. Biol. 2016;11:1049–1057. doi: 10.1021/acschembio.5b00967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McAndrew RP, Sathitsuksanoh N, Mbughuni MM, Heins RA, Pereira JH, George A, Sale KL, Fox BG, Simmons BA, Adams PD. Structure and mechanism of NOV1, a resveratrol-cleaving dioxygenase. Proc. Natl. Acad. Sci. U. S. A. 2016;113:14324–14329. doi: 10.1073/pnas.1608917113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Costas M, Mehn MP, Jensen MP, Que L., Jr Dioxygen activation at mononuclear nonheme iron active sites: enzymes, models, and intermediates. Chem. Rev. 2004;104:939–986. doi: 10.1021/cr020628n. [DOI] [PubMed] [Google Scholar]
- 26.Wolfe MD, Parales JV, Gibson DT, Lipscomb JD. Single turnover chemistry and regulation of O2 activation by the oxygenase component of naphthalene 1,2-dioxygenase. J. Biol. Chem. 2001;276:1945–1953. doi: 10.1074/jbc.M007795200. [DOI] [PubMed] [Google Scholar]
- 27.Andersen OA, Stokka AJ, Flatmark T, Hough E. 2.0A resolution crystal structures of the ternary complexes of human phenylalanine hydroxylase catalytic domain with tetrahydrobiopterin and 3-(2-thienyl)-L-alanine or L-norleucine: substrate specificity and molecular motions related to substrate binding. J. Mol. Biol. 2003;333:747–757. doi: 10.1016/j.jmb.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 28.Sui X, Kiser PD, Che T, Carey PR, Golczak M, Shi W, von Lintig J, Palczewski K. Analysis of carotenoid isomerase activity in a prototypical carotenoid cleavage enzyme, apocarotenoid oxygenase (ACO) J. Biol. Chem. 2014;289:12286–12299. doi: 10.1074/jbc.M114.552836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eddarir S, Abdelhadi Z, Rolando C. Fluorinated resveratrol and pterostilbene. Tetrahedron Lett. 2001;42:9127–9130. [Google Scholar]
- 30.Park BH, Lee YR, Lyoo WS. Efficient and General Preparation of Pyranostilbenes: First Total Synthesis of Artocarbene and Pawhuskin B. Synthesis. 2009;2009:2146–2154. [Google Scholar]
- 31.Tsai HJ, Lin KW, Ting TH, Burton DJ. A general and efficient route for the preparation of phenyl-substituted vinyl fluorides. Helv. Chim. Acta. 1999;82:2231–2239. [Google Scholar]
- 32.Eddarir S, Francesch C, Mestdagh H, Rolando C. Regio- and stereoselective synthesis of fluorinated enynes and dienes via 1,1- or 1,2-halofluoroalkenes. Bull. Soc. Chim. Fr. 1997;134:741–755. [Google Scholar]
- 33.Kabsch W. Xds. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keegan RM, Winn MD. Automated search-model discovery and preparation for structure solution by molecular replacement. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2007;63:447–457. doi: 10.1107/S0907444907002661. [DOI] [PubMed] [Google Scholar]
- 35.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Read RJ, McCoy AJ. Using SAD data in Phaser. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2011;67:338–344. doi: 10.1107/S0907444910051371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abrahams JP, Leslie AG. Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1996;52:30–42. doi: 10.1107/S0907444995008754. [DOI] [PubMed] [Google Scholar]
- 39.Cowtan K. Recent developments in classical density modification. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010;66:470–478. doi: 10.1107/S090744490903947X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 41.Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 2008;3:1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Read RJ, Adams PD, Arendall WB, 3rd, Brunger AT, Emsley P, Joosten RP, Kleywegt GJ, Krissinel EB, Lutteke T, Otwinowski Z, Perrakis A, Richardson JS, Sheffler WH, Smith JL, Tickle IJ, Vriend G, Zwart PH. A new generation of crystallographic validation tools for the protein data bank. Structure. 2011;19:1395–1412. doi: 10.1016/j.str.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Holm L, Laakso LM. Dali server update. Nucleic Acids Res. 2016;44:W351–W355. doi: 10.1093/nar/gkw357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guindon S, Lethiec F, Duroux P, Gascuel O. PHYML Online-a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 2005;33:W557–W559. doi: 10.1093/nar/gki352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ravel B, Newville M. ATHENA, ARTEMIS, HEPHAESTUS: data analysis for X-ray absorption spectroscopy using IFEFFIT. J. Synchrotron Radiat. 2005;12:537–541. doi: 10.1107/S0909049505012719. [DOI] [PubMed] [Google Scholar]
- 51.George GN. EXAFSPAK, Stanford Synchrotron Radiation Lightsource. SLAC National Accelerator Laboratory; Stanford, CA: 2000. [Google Scholar]
- 52.Wojdyr M. Fityk: a general-purpose peak fitting program. J. Appl. Crystallogr. 2010;43:1126–1128. [Google Scholar]
- 53.Costello A, Periyannan G, Yang KW, Crowder MW, Tierney DL. Site-selective binding of Zn(II) to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia. JBIC, J. Biol. Inorg. Chem. 2006;11:351–358. doi: 10.1007/s00775-006-0083-z. [DOI] [PubMed] [Google Scholar]
- 54.Liu WT, Thorp HH. Bond Valence Sum Analysis of Metal-Ligand Bond Lengths in Metalloenzymes and Model Complexes 0.2. Refined Distances and Other Enzymes. Inorg. Chem. 1993;32:4102–4105. [Google Scholar]
- 55.Golombek AP, Hendrich MP. Quantitative analysis of dinuclear manganese(II) EPR spectra. J. Magn. Reson. 2003;165:33–48. doi: 10.1016/j.jmr.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 56.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 57.Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J. Mol. Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- 58.Kamoda S, Terada T, Saburi Y. Production of heterogeneous dimer lignostilbenedioxygenase II from lsdA and lsdB in Escherichia coli cells. Biosci., Biotechnol., Biochem. 2005;69:635–637. doi: 10.1271/bbb.69.635. [DOI] [PubMed] [Google Scholar]
- 59.Kiser PD, Farquhar ER, Shi W, Sui X, Chance MR, Palczewski K. Structure of RPE65 isomerase in a lipidic matrix reveals roles for phospholipids and iron in catalysis. Proc. Natl. Acad. Sci. U. S. A. 2012;109:E2747–E2756. doi: 10.1073/pnas.1212025109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Randall CR, Shu LJ, Chiou YM, Hagen KS, Ito M, Kitajima N, Lachicotte RJ, Zang Y, Que L. X-Ray-Absorption Pre-Edge Studies of High-Spin Iron(Ii) Complexes. Inorg. Chem. 1995;34:1036–1039. [Google Scholar]
- 61.Westre TE, Kennepohl P, DeWitt JG, Hedman B, Hodgson KO, Solomon EI. A multiplet analysis of Fe K-edge 1s->3d pre-edge features of iron complexes. J. Am. Chem. Soc. 1997;119:6297–6314. [Google Scholar]
- 62.England J, Farquhar ER, Guo YS, Cranswick MA, Ray K, Munck E, Que L. Characterization of a Tricationic Trigonal Bipyramidal Iron(IV) Cyanide Complex, with a Very High Reduction Potential, and Its Iron(II) and Iron(III) Congeners. Inorg. Chem. 2011;50:2885–2896. doi: 10.1021/ic102094d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wilke M, Farges F, Petit PE, Brown GE, Martin F. Oxidation state and coordination of Fe in minerals: An FeK-XANES spectroscopic study. Am. Mineral. 2001;86:714–730. [Google Scholar]
- 64.McAndrew RP, Sathitsuksanoh N, Mbughuni MM, Heins RA, Pereira JH, George A, Sale KL, Fox BG, Simmons BA, Adams PD. Structure and mechanism of NOV1, a resveratrol-cleaving dioxygenase. Proc. Natl. Acad. Sci. U. S. A. 2016;113:14324–14329. doi: 10.1073/pnas.1608917113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mbughuni MM, Chakrabarti M, Hayden JA, Bominaar EL, Hendrich MP, Munck E, Lipscomb JD. Trapping and spectroscopic characterization of an FeIII-superoxo intermediate from a nonheme mononuclear iron-containing enzyme. Proc. Natl. Acad. Sci. U. S. A. 2010;107:16788–16793. doi: 10.1073/pnas.1010015107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Arciero DM, Lipscomb JD, Huynh BH, Kent TA, Munck E. EPR and Mössbauer studies of protocatechuate 4,5-dioxygenase. Characterization of a new Fe2+ environment. J. Biol. Chem. 1983;258:14981–14991. [PubMed] [Google Scholar]
- 67.Zimmermann R, Huynh BH, Munck E, Lipscomb JD. High-Field Mössbauer Studies of Reduced Protocatechuate 3,4 -Dioxygenase. J. Chem. Phys. 1978;69:5463–5467. [Google Scholar]
- 68.Price JC, Barr EW, Tirupati B, Bollinger JM, Jr, Krebs C. The first direct characterization of a high-valent iron intermediate in the reaction of an alpha-ketoglutarate-dependent dioxygenase: a high-spin FeIV complex in taurine/alpha-ketoglutarate dioxygenase (TauD) from Escherichia coli. Biochemistry. 2003;42:7497–7508. doi: 10.1021/bi030011f. [DOI] [PubMed] [Google Scholar]
- 69.Tchesnokov EP, Wilbanks SM, Jameson GN. A strongly bound high-spin iron(II) coordinates cysteine and homo-cysteine in cysteine dioxygenase. Biochemistry. 2012;51:257–264. doi: 10.1021/bi201597w. [DOI] [PubMed] [Google Scholar]
- 70.Orville AM, Chen VJ, Kriauciunas A, Harpel MR, Fox BG, Munck E, Lipscomb JD. Thiolate ligation of the active site Fe2+ of isopenicillin N synthase derives from substrate rather than endogenous cysteine: spectroscopic studies of site-specific Cys— —Ser mutated enzymes. Biochemistry. 1992;31:4602–4612. doi: 10.1021/bi00134a010. [DOI] [PubMed] [Google Scholar]
- 71.Boso B, Debrunner P, Okamura MY, Feher G. Mössbauer-Spectroscopy Studies of Photosynthetic Reaction Centers from Rhodopseudomonas-Sphaeroides R-26. Biochim. Biophys. Acta, Bioenerg. 1981;638:173–177. [Google Scholar]
- 72.Hendrich MP, Debrunner PG. Integer-spin electron paramagnetic resonance of iron proteins. Biophys. J. 1989;56:489–506. doi: 10.1016/S0006-3495(89)82696-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang Y, Pavlosky MA, Brown CA, Westre TE, Hedman B, Hodgson KO, Solomon EI. Spectroscopic and Theoretical Description of the Electronic-Structure of the S = 3/2 Nitrosyl Complex of Nonheme Iron Enzymes. J. Am. Chem. Soc. 1992;114:9189–9191. [Google Scholar]
- 74.Enemark JH, Feltham RD. Principles of Structure, Bonding, and Reactivity for Metal Nitrosyl Complexes. Coord. Chem. Rev. 1974;13:339–406. [Google Scholar]
- 75.Butler AR, Megson IL. Non-heme iron nitrosyls in biology. Chem. Rev. 2002;102:1155–1166. doi: 10.1021/cr000076d. [DOI] [PubMed] [Google Scholar]
- 76.Miller MA, Lipscomb JD. Homoprotocatechuate 2,3-dioxygenase from Brevibacterium fuscum. A dioxygenase with catalase activity. J. Biol. Chem. 1996;271:5524–5535. doi: 10.1074/jbc.271.10.5524. [DOI] [PubMed] [Google Scholar]
- 77.D’Ordine RL, Rydel TJ, Storek MJ, Sturman EJ, Moshiri F, Bartlett RK, Brown GR, Eilers RJ, Dart C, Qi YL, Flasinski S, Franklin SJ. Dicamba Monooxygenase: Structural Insights into a Dynamic Rieske Oxygenase that Catalyzes an Exocyclic Monooxygenation. J. Mol. Biol. 2009;392:481–497. doi: 10.1016/j.jmb.2009.07.022. [DOI] [PubMed] [Google Scholar]
- 78.Kalliri E, Grzyska PK, Hausinger RP. Kinetic and spectroscopic investigation of Co-II, Ni-II, and N-oxalylglycine inhibition of the Fe-II/alpha-ketoglutarate dioxygenase, TauD. Biochem. Biophys. Res. Commun. 2005;338:191–197. doi: 10.1016/j.bbrc.2005.08.223. [DOI] [PubMed] [Google Scholar]
- 79.Fielding AJ, Kovaleva EG, Farquhar ER, Lipscomb JD, Que L. A hyperactive cobalt-substituted extradiol-cleaving catechol dioxygenase. JBIC, J. Biol. Inorg. Chem. 2011;16:341–355. doi: 10.1007/s00775-010-0732-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Han S, Inoue H, Terada T, Kamoda S, Saburi Y, Sekimata K, Saito T, Kobayashi M, Shinozaki K, Yoshida S, Asami T. Design and synthesis of lignostilbene-alpha,beta-dioxygenase inhibitors. Bioorg. Med. Chem. Lett. 2002;12:1139–1142. doi: 10.1016/s0960-894x(02)00126-9. [DOI] [PubMed] [Google Scholar]
- 81.Dunitz JD, Taylor R. Organic fluorine hardly ever accepts hydrogen bonds. Chem. - Eur. J. 1997;3:89–98. [Google Scholar]
- 82.Pulido R, Bravo L, Saura-Calixto F. Antioxidant activity of dietary polyphenols as determined by a modified ferric reducing/antioxidant power assay. J. Agric. Food Chem. 2000;48:3396–3402. doi: 10.1021/jf9913458. [DOI] [PubMed] [Google Scholar]
- 83.Neves AR, Nunes C, Reis S. New Insights on the Biophysical Interaction of Resveratrol with Biomembrane Models: Relevance for Its Biological Effects. J. Phys. Chem. B. 2015;119:11664–11672. doi: 10.1021/acs.jpcb.5b05419. [DOI] [PubMed] [Google Scholar]
- 84.Emerson JP, Kovaleva EG, Farquhar ER, Lipscomb JD, Que L., Jr Swapping metals in Fe- and Mn-dependent dioxygenases: evidence for oxygen activation without a change in metal redox state. Proc. Natl. Acad. Sci. U. S. A. 2008;105:7347–7352. doi: 10.1073/pnas.0711179105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kovaleva EG, Lipscomb JD. Crystal structures of Fe2+ dioxygenase superoxo, alkylperoxo, and bound product intermediates. Science. 2007;316:453–457. doi: 10.1126/science.1134697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Elkins JM, Ryle MJ, Clifton IJ, Dunning Hotopp JC, Lloyd JS, Burzlaff NI, Baldwin JE, Hausinger RP, Roach PL. X-ray crystal structure of Escherichia coli taurine/alpha-ketoglutarate dioxygenase complexed to ferrous iron and substrates. Biochemistry. 2002;41:5185–5192. doi: 10.1021/bi016014e. [DOI] [PubMed] [Google Scholar]
- 87.Ohta T, Chakrabarty S, Lipscomb JD, Solomon EI. Near-IR MCD of the nonheme ferrous active site in naphthalene 1,2-dioxygenase: correlation to crystallography and structural insight into the mechanism of Rieske dioxygenases. J. Am. Chem. Soc. 2008;130:1601–1610. doi: 10.1021/ja074769o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wolgel SA, Dege JE, Perkins-Olson PE, Jaurez-Garcia CH, Crawford RL, Munck E, Lipscomb JD. Purification and characterization of protocatechuate 2,3-dioxygenase from Bacillus macerans: a new extradiol catecholic dioxygenase. J. Bacteriol. 1993;175:4414–4426. doi: 10.1128/jb.175.14.4414-4426.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lipscomb JD. Mechanism of extradiol aromatic ring-cleaving dioxygenases. Curr. Opin. Struct. Biol. 2008;18:644–649. doi: 10.1016/j.sbi.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gunderson WA, Zatsman AI, Emerson JP, Farquhar ER, Que L, Jr, Lipscomb JD, Hendrich MP. Electron paramagnetic resonance detection of intermediates in the enzymatic cycle of an extradiol dioxygenase. J. Am. Chem. Soc. 2008;130:14465–14467. doi: 10.1021/ja8052255. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.