Abstract
Background
Heroin’s analgesic, euphoric and dependence-producing effects are primarily mediated by the mu opioid receptor (MOR). A single gene, OPRM1, encodes the MOR. The functional polymorphism A118G, located in exon 1 of the OPRM1 gene, results in anatomically-specific reductions in MOR expression, which may alter an individual’s response to heroin. In prior studies 118G (rare allele) carriers demonstrated significantly greater opioid tolerance, overdose vulnerability, and pain sensitivity than 118AA homozygotes. Those findings suggest OPRM1 genotype may impact characteristics of heroin use.
Methods
The present pilot study characterized the impact of OPRM1 genotype (rs1799971, 118G allele carriers vs. 118AA homozygotes) on heroin-use phenotypes associated with heroin dependence severity in a sample of male, Caucasian chronic heroin users (n = 86).
Results
Results indicate that 118G allele carriers reported significantly more heroin use-related consequences and heroin-quit attempts, and were more likely to have sought treatment for their heroin use than 118AA homozygotes.
Conclusions
These preliminary findings, consistent with extant data, illustrate a role for OPRM1 allelic variation on heroin use characteristics, and provide support for considering genotype in heroin treatment and relapse prevention.
INTRODUCTION
Mu opioid receptors (MORs) mediate most of the clinically salient effects of heroin (MOR agonist), including analgesia, euphoria, and depressed respiration.1–3 MOR anatomical location and post-synaptic connectivity dictate the downstream effects of a bound agonist. A single gene, OPRM1, encodes the MOR.4 Variants in the OPRM1 gene can alter opioidergic function and may therefore impact an individual’s response to opioid administration. A widely-studied OPRM1 variant is a single nucleotide polymorphism (SNP) A118G, rs1799971, resulting in an Asn40Asp amino acid change.4 While the specific neurobiological effects of this SNP are not fully understood, data from rodent and human studies suggest the minor Asp allele is less physiologically functional, though findings to the contrary5 exist. The 118G variant is associated with diminished mRNA and protein levels in some brain regions.6,7 Using whole brain quantitative autoradiography, Wang et al.8 found diminished MOR expression in the ‘pain matrix’ (cingulate and insular cortices, amygdala and periaqueductal gray;9–11) and ‘reward centers’ (nucleus accumbens and ventral tegmental area) of the 112GG homozygote mouse brain (mouse equivalent of human 118GG homozygotes) compared to 112AA homozygotes. Mague et al.12 found less morphine-induced antinociception in 112GG homozygous mice than 112AA homozygotes. Further, human 118G carriers report less morphine-induced analgesia13–15 and hypersensitivity to nociceptive stimuli compared to 118AA homozygotes.15,16 Consistent with the above findings, Huang et al.17 demonstrated diminished MOR protein expression in a rodent model of the 118GG genotype, and Beyer et al.18 and Kroslak et al.19 each found human 118G-variant cell lines demonstrated lower cell-surface MOR agonist binding capacity (lower Bmax). Further, clinical studies using [11C] carfentanil PET imaging found the 118G* genotype was associated with diminished MOR receptor availability.20,21
Genetic association studies frequently attempt to link polymorphisms with initial vulnerability to drug dependence. Coller et al.22 conducted a meta-analysis of 16 studies examining opioid dependent and control subjects (> 5000 subjects) and found no link between OPRM1 rs1799971 genotype and opioid dependence. Arias et al.23 conducted a meta-analysis of 22 published studies investigating risk of substance dependence by OPRM1 rs1799971 genotype across ethnicities (>8000 subjects) and drugs of abuse (alcohol, heroin, opioids, and cocaine); they found no link between genotype and substance dependence, nor did they find any significant relationships between ethnicity, genotype and dependence. Haerian and Haerian24 conducted a meta-analysis of 18 studies (>8000 subjects) and found no association between OPRM1 rs1799971 genotype and opioid/cocaine or heroin dependence in African-American or Caucasian subjects, but opioid-dependent Asian subjects (among whom the 118G variant is more prevalent: 40–50% vs. 15–30% in Caucasians and 1–3% in African-Americans;25) were more likely to be 118G carriers than 118AA homozygotes.
However, genetic factors likely influence all phases of drug dependence, not solely vulnerability to dependence.26 Nikolov et al.27 hypothesized that the 118G variant may not impact risk for but, rather, severity of heroin dependence. In support of this hypothesis, heroin-dependent 118G carriers demonstrated enhanced heroin tolerance (in haplotype with IVS2 +31 G/A variant;28) and 5.3 × greater risk of cardiac or respiratory arrest29 than 118AA homozygotes. Diminished MOR expression in analgesia- and euphoria-mediating brain regions8,30 may explain 118G carriers’ enhanced opioid tolerance13–15,28 and pain sensitivity.15,16 Exaggerated pain sensitivity and enhanced opioid tolerance may lead users to self-administer heroin via a more pharmacokinetically-efficient route (e.g., injection vs. insufflation), use more frequently, or consume larger doses to achieve the desired effects, thereby increasing risk of overdose and respiratory/cardiac arrest.29
In the present study, we examined heroin use phenotypes associated with heroin dependence severity by A118G genotype in chronic, regular Caucasian male heroin users not currently seeking treatment. Consistent with Nikolov et al.27, we hypothesized 118G carriers would report heroin use characteristics indicative of greater severity of heroin dependence including: more lifetime heroin use consequences (specifically overdose), lifetime heroin quit attempts, and frequent current heroin use, and greater likelihood of having sought treatment for their heroin use and self-administering heroin via injection than 118AA homozygotes.
MATERIALS AND METHODS
Participants
This retrospective investigation utilized screening data obtained from four source studies approved by Institutional Review Boards at Wayne State University and the University of Michigan (for methodological details see: 31–34), and was conducted in accordance with the Declaration of Helsinki (1964).35 The National Institute on Drug Abuse issued certificates of confidentiality for these studies. Participants were recruited from the Detroit, Michigan metropolitan area using print media advertisements and word-of-mouth referral.
Individuals identifying themselves as regular heroin users and not seeking treatment completed a brief phone screen. Eligible individuals were scheduled for an in-person screening interview. Volunteers provided informed consent, demographic data, and comprehensive substance use and medical histories. Current heroin use was confirmed by opioid-positive urine sample (>300 ng/ml). Participants were sober (<.002% expired blood alcohol content) during the intake interview. Completion of the screening visit earned each participant a $30 check.
Genotyping
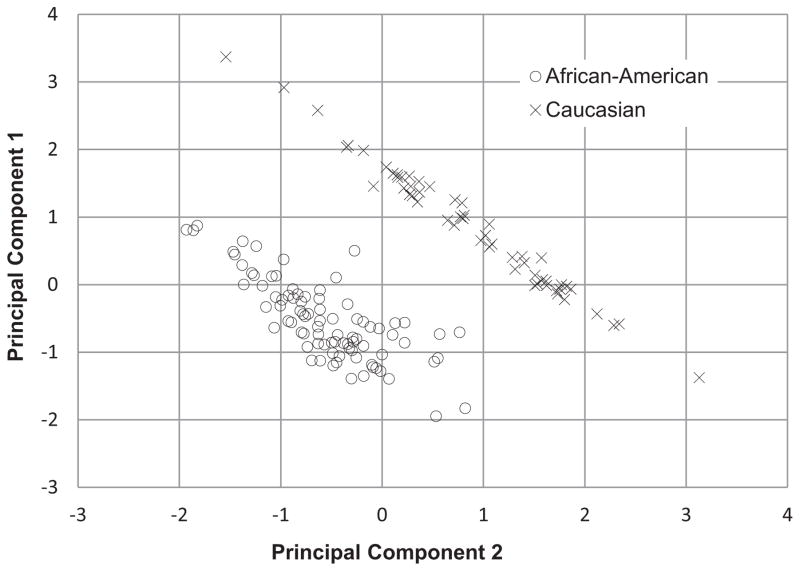
Plasma samples were collected (6 ml per participant) in EDTA tubes. Participant DNA was extracted using the Qiagen kit (formerly Gentra Puregene) and genotyped using the Taqman assay according to the manufacturer’s conditions and solutions (Applied Biosystems ABI, Foster City, CA). Additionally, a subset of participant DNA samples were genotyped using Illumina Golden Gate addiction panel.35 The two assays were in complete agreement for OPRM1 rs1799971 genotype for this sample. The Golden Gate panel has the advantage of including 186 ancestry informative markers (AIMs) that were used to evaluate racial stratification and confirm self-reported racial identity from the full sample of source study participants, which included African- Americans and Caucasians only (n =186). Principal component analyses were conducted (forcing a two-factor solution with a Varimax rotation) with the 150 AIMs containing at least 90% genotype data and a factor loading coefficient ≥.30 which revealed complete separation between races and no overlapping cases (Fig. 1). These analyses supported the self-reported racial identities and indicated the presence of genetically-distinct groups.
FIGURE 1.
Self-reported racial identification (African-American vs. Caucasian) stratified by principal component factor loading. Principal component analysis of the 150 ancestry informative markers (AIMs) with >90% genotype data for this sample and a factor loading coefficient ≥.30 revealed complete separation between races. These analyses indicated that self-reported racial identity agrees with AIMs and argued against racial stratification being a primary study confound.
A common OPRM1 SNP occurs at position 118 (rs1799971) in exon 1 (adenine-to-guanine substitution: A118G) encoding an asparagine-to-aspartic acid amino acid exchange (N40D) in the N-terminal domain.4 Due to scarcity of the OPRM1 118GG genotype and scarcity of the G allele in African-Americans, analyses presented herein contrasted Caucasian 118G carriers (118GG and 118AG) with 118AA homozygotes. Indeed, of the 131 African-American individuals available for analysis, only five were 118G carriers: insufficient for outcome analyses.
Phenotyping
Heroin-use phenotypes were derived from a comprehensive, standardized self-report substance use history questionnaire routinely used in our laboratory. Analyses focused on five dependent variables: current heroin administration route (injection vs. non-injection), past-month heroin use frequency, lifetime number of heroin use-related consequences, lifetime number of attempts to quit using heroin, and having ever sought treatment for heroin use. Current route of self-administration, lifetime treatment for heroin use, and lifetime number of attempts to quit (with and without treatment) using heroin were assessed via single-item face-valid self-report variables. Past-month heroin use frequency was calculated as a product score of past-week mean daily use multiplied by number of days using heroin in the past month. Heroin use consequences were assessed via a 21-item questionnaire (see Table 2). Items parallel SCID symptom checklists and encompass potential heroin use-related consequences. Participants were asked to indicate (present/absent) each consequence they experienced as a direct result of their heroin use. Endorsed consequences were summed for analysis of total consequences (outcome measure); however, individual item endorsement was also explored to better understand the effects of A118G genotype on heroin use consequences.
TABLE 2.
Heroin use consequences item endorsement
| Item (% Endorsed) | Overall (n =86) | 118AA (n =68) | 118G* (n =18) | χ2 |
|---|---|---|---|---|
| Instructions: In your opinion, have you had any of these problems because you took heroin? If you have, please mark each below: | ||||
| Unexpected reaction | 35.3% | 32.8% | 44.4% | 0.84 |
| Overdose | 40.7% | 33.8% | 66.7% | 6.36* |
| Memory lapse | 31.4% | 32.4% | 27.8% | 0.14 |
| Seizure/fits | 5.8% | 2.9% | 16.7% | 4.90* |
| Shakes/tremors | 31.8% | 32.4% | 29.4% | 0.05 |
| Couldn’t stop using | 88.4% | 85.3% | 100.0% | 3.00 |
| Arrested/legal problems | 45.3% | 45.6% | 44.4% | 0.01 |
| Accident/injury | 17.4% | 19.1% | 11.1% | 0.63 |
| Health problem | 30.2% | 30.9% | 27.8% | 0.07 |
| High at work | 81.2% | 76.1% | 100.0% | 5.30* |
| Lost job | 57.6% | 55.2% | 66.7% | 0.76 |
| Missed work | 67.4% | 64.7% | 77.8% | 1.11 |
| Got warning or disciplined at work | 43.0% | 41.2% | 50.0% | 0.45 |
| High at school | 25.6% | 22.1% | 38.9% | 2.12 |
| Missed school | 20.9% | 17.6% | 33.3% | 2.12 |
| Suspended or expelled from school | 10.5% | 10.3% | 11.1% | 0.01 |
| Fight or quarrel | 33.7% | 29.4% | 50.0% | 2.70 |
| Drove under influence | 87.2% | 85.3% | 94.4% | 1.07 |
| Family problems | 87.2% | 85.3% | 94.4% | 1.07 |
| Financial problems | 95.3% | 95.6% | 94.4% | 0.04 |
| Visited Emergency Room | 34.3% | 32.7% | 40.0% | 0.28 |
Indicates significant difference between OPRM1 118AA and 118G carrier genotypes (p <.05).
Data Analyses
Distributions of continuous variables were evaluated for skewness and kurtosis36 to assess normality prior to outcome analyses. The only non-normal distribution was lifetime heroin quit attempts (positively skewed), which was normalized using a square root transformation. Unequal sample sizes across genotype increased our risk of Type I error. To protect against Type I error inflation, Levene’s Test of Equality of Error Variances was used to confirm (all p values >.50) homogeneity of variance for each outcome variable in our analyses.
All analyses were conducted using SPSS version 22 with the criterion to reject the null hypothesis set at p ≤.05. Fisher’s Exact and Chi Square tests were performed to assess the distribution of genotype by route of heroin administration (injection vs. non-injection), seeking treatment for heroin use, and individual items in the heroin use consequences survey. Continuous variables (e.g., past-month heroin use frequency) were evaluated using one-way Analyses of Variance (ANOVAs) to compare genotype effects. Number of lifetime heroin use consequences and quit attempts are likely to accumulate with longer duration of heroin use, which could bias interpretation of results. If these variables were positively correlated, analysis of covariance (ANCOVA), with years of heroin use entered as a covariate, was used in analyses. Descriptive statistics are presented as mean (M) ± one standard deviation (SD).
RESULTS
Participant Characteristics
All participants (n =86 Caucasian males) were current (opioid positive urine sample; >300 ng/ml) and chronic [M±1 SD=14.2±10.2 years] heroin users who were not currently seeking treatment. The average participant was 37.7±10.0 years old with 12.3±1.5 years of formal education. All participants reported regular heroin use, defined as >3 uses during the past week. 118G carriers were significantly younger than 118AA homozygotes, F(1, 85)=6.28, p <.05 [32.6 ± 9.7 vs. 39.1 ± 9.8 years old], but did not differ (p >.25) by years of education [12.7 ± 1.6 vs. 12.2 ± 1.5 years old].
Genotype Associated With Heroin Use Characteristics
Heroin use data are presented in Table 1. Fisher’s Exact Test (FET) revealed current route of heroin administration (injection vs. non-injection) was not significantly associated with genotype [100% of 118G carriers vs. 86.8% of 118AA homozygotes injected heroin, p=.08; FET one-tailed; Cramer’s V =.19 (small-to-moderate effect size)].
TABLE 1.
Heroin use characteristics
| 118AA (n =68) M (± 1 SD) | 118G* (n =18) M (± 1 SD) | F or χ2 | Cohen’s d | |
|---|---|---|---|---|
| Age at First Use | 24.4 (7.2) | 20.9 (4.4) | 3.78 | 0.53 |
| Years of Use | 14.8 (10.4) | 11.7 (9.2) | 1.32 | 0.31 |
| Use Frequency | 101.6 (68.5) | 126.8 (66.1) | 1.95 | 0.37 |
| Use Consequences# | 9.2 (4.7) | 11.1 (4.2) | 4.47* | 0.42 |
| Quit Attempts# | 11.0 (20.9) | 19.1 (30.4) | 3.92* | 0.35 |
| Route (% inject) | 85.3% | 100.0% | 2.66 | – |
| Sought Treatment (%) | 61.8% | 88.9% | 4.77* | – |
Indicates ANCOVA was performed with duration of heroin use entered as covariate.
Significant differences indicated: *p≤.05.
Chi-square analyses indicated 118G carriers were significantly more likely (88.9% vs. 61.8%) to report having sought treatment for their heroin use at some point during their lifetime than 118AA homozygotes [χ2(1)=4.77, p <.05]. Two thirds of participants (67.4%) reported seeking treatment for their heroin use, which was unrelated to duration of their heroin use, F(1, 85)=0.93, p=.34.
Past-month heroin-use frequency was not correlated with duration of heroin use (r =−.15; p =.16) and thus, ANOVA was used to evaluate the effect of genotype on use frequency. 118G carriers did not report significantly different past-month heroin use than 118AA homozygotes, F(1, 84)=1.95, p =.17 [126.8 ± 66.1 vs. 101.6 ± 68.5].
As expected, number of heroin quit attempts and years using heroin (duration) were positively correlated (r=.35, p <.001); thus, number of years using heroin was entered as a covariate. ANCOVA indicated genotype was significantly associated with heroin quit attempts, F(1, 85) =3.92, p =.05; partial eta-squared [η2]=.045. 118G carriers reported nearly twice as many heroin quit attempts as AA homozygotes [19.1 ± 30.4 vs. 11.0 ± 20.9].
Lifetime heroin use consequences were positively correlated with number of years using heroin (r=.39, p<.01); again, as anticipated, the latter variable was entered as a covariate in analyses. ANCOVA revealed genotype was significantly associated with heroin-use consequences, F(1, 85)=4.47, p<.05; η2=.051. 118G carriers reported more lifetime heroin use consequences than AA homozygotes [11.1 ± 4.2 vs. 9.2 ± 4.7].
Item-level endorsements of lifetime heroin use consequences are presented in Table 2. Chi-square analyses revealed item-level differences between genotypes: 118G carriers endorsed ‘overdose’ (66.7% vs. 33.8%), ‘seizure/fits’ (16.7% vs. 2.9%), and ‘high at work’ (100.0% vs. 76.1%) significantly more frequently than 118AA homozygotes [χ2(1) =6.36, p<.05, χ2(1)=4.90, p<.05, and χ2(1)=5.30, p<.05; respectively]. Although every 118G carrier in this sample endorsed they ‘couldn’t stop using’ heroin, this item did not significantly differ by genotype [100.0% vs. 85.3%; χ2(1)=3.00, p=.08]. No other items approached significance (p values >.10).
Behavioral Associations Unrelated to Genotype
Lifetime quit attempts and heroin use consequences were positively correlated (r =.31, p <.01). Number of heroin use consequences (but not quit attempts or use frequency, ps >.10) differed by current heroin administration route, F(1, 85)=11.67, p <.001, while controlling for duration of heroin use. Those who currently injected heroin endorsed significantly more lifetime heroin use consequences than those who did not inject heroin [10.1 ± 4.5 vs. 5.7 ± 3.7].
DISCUSSION
The current study contrasted clinically-relevant heroin use characteristics by OPRM1 rs1799971 genotype (118G carriers vs. 118AA homozygotes) in a non-treatment-seeking sample of chronic, regular Caucasian male heroin users. We hypothesized 118G carriers would report heroin use characteristics in support of Nikolov and colleagues27 hypothesis that the 118G allele is associated with more ‘severe’ opioid dependence. 118G carriers reported more lifetime heroin-use consequences, quit attempts and were more likely to have sought treatment for their heroin use, relative to 118AA homozygotes. However, there were no genotype differences in reported past-month heroin use frequency or current route of self-administration. These preliminary findings provide initial evidence that the OPRM1 118G variant may be associated with more burdensome outcomes from chronic heroin use.
118G carriers reported more lifetime heroin-use consequences than 118AA homozygotes (endorsing ~11 vs. ~9 of the 21 possible consequences). Rather than being a simple count of potentially redundant episodes of dysfunctional behavior and adverse events, these data indicate 118G carriers experienced a more expansive range of heroin use-related consequences. Clinical and preclinical studies6–8,17,20,21,30 indicate the 118G allele may be less physiologically functional, resulting in anatomically-specific diminished MOR availability associated with increased pain sensitivity and opioid tolerance.13–16,28 Considering these findings, we hypothesized that heroin may be less potent or have lower intrinsic efficacy in 118G carriers, who may therefore self-administer larger doses to achieve desired effects, thereby enhancing risk of overdose. We could not collect accurate data on heroin dosage so we were unable to test this hypothesis directly. However, item-level analyses of heroin use consequences confirm that 118G carriers were more likely to report ‘overdose’ and ‘seizures/fits’ from heroin use than 118AA homozygotes, providing indirect support for this hypothesis. These data are consistent with Manini et al.29 indicating the 118G variant conveyed a 5.3-fold greater risk of cardiac or respiratory arrest from heroin overdose. 118G carriers reported having been ‘high at work’ more frequently, which was unexpected, but consistent with the working hypothesis. Finally, 118G carriers were not statistically (p =.08) more likely to report they ‘couldn’t stop using’ than 118AA homozygotes, though a ceiling effect is possible (100% of 118G carriers endorsed they ‘couldn’t stop using’ heroin).
118G carriers reported significantly more lifetime attempts to quit using heroin than 118AA homozygotes, controlling for years of heroin use. Given that study participants were current heroin users, these prior quit attempts were by definition unsuccessful. While these data cannot address the temporal (or causal) relationship between heroin-quit attempts and use consequences, these variables are significantly positively correlated. Moreover, 118G carriers were significantly more likely to have sought past treatment for their heroin use than 118AA homozygotes.
All 118G carriers (100%) reported current self-administration of heroin via injection (vs. non-injection), but not statistically more than 118AA homozygotes (86.8%; possible ceiling effect). Injection is a more efficient and dangerous drug delivery route compared to snorting or smoking, and is associated with more rapid progression to dependence,37 more severe heroin dependence, quicker relapse, and worse opioid withdrawal symptoms.38–40
Some limitations of the present study are important to note. First, this pilot investigation used a small convenience sample. Thus, the present results should be interpreted with caution and would benefit from replication in a larger sample. However, the specific a priori selection of the A118G polymorphism for analysis, our hypothesis-driven analytic approach, demographic homogeneity of the sample, and moderate effect sizes compensated for the limited statistical power and revealed several significant differences by genotype. Second, while some of the present findings support Nikolov and colleagues27 working hypothesis, these findings are not conclusive. Indeed, several of our hypotheses were not supported by the data: 118G carriers did not report more frequent heroin use or higher rates of injection (possibly due to a ceiling effect). Third, reliance on self-report data is a limitation in the current study. Fourth, while some participants did report poly-substance use, and previous findings indicate the A118G polymorphism is related to alcohol use phenotypes, this sample is composed of primary heroin users and thus analyses focused solely on heroin use-related phenotypes. Finally, due to scarcity of the 118G allele, we focused on a demographically-homogenous sample (Caucasian males) for three reasons. First, the A118G polymorphism is infrequent among African-Americans (only five 118G carriers in our sample; 3.8%), which is consistent with the literature (1–3%;25). Second, meta-analyses indicate ethnicity may be related to dependence.24 Third, extant research suggests there may be gender-specific effects of the A118G genotype on substance use characteristics,41–43 pain threshold,44 and opioid response.12 Despite our interest in examining possible gender effects in heroin use characteristics, there were too few female 118G carriers (n =7) available to address this issue. Finally, some findings presented here would not survive multiple comparison correction for the variants tested, and hence should be considered hypothesis-generating.
With these caveats in mind, the present findings offer initial support that A118G genotype may influence heroin use characteristics relevant to treatment efficacy in opioid dependent individuals. 118G carriers reported experiencing more heroin use consequences indicative of greater psychosocial dysfunction and possible health problems (e.g., ‘seizure/fits’, ‘high at work’, ‘overdose’), which may complicate and prolong treatment (i.e., necessitating a higher level of care). All 118G carriers reported currently injecting heroin, thereby increasing risk of contracting blood borne viruses or developing infections from shared and/or unclean syringes. Finally, 118G carriers were more likely to have sought treatment in the past and reported more lifetime attempts (i.e., prior failures) to abstain from heroin use, which may reflect greater susceptibility to relapse. Taken together, these findings highlight the importance of, and potential role for, considering OPRM1 genotype in opioid dependence treatment.
Acknowledgments
NIH grant R01 DA015462 from the National Institute on Drug Abuse (to MKG), a research grant (Joe Young, Sr./Helene Lycaki Funds) from the State of Michigan, and the Detroit Wayne Mental Health Authority supported this research. Data for this study were obtained under registered NIH clinical trials NCT00218309, NCT00218361, NCT00608504, and NCT00684840.
The authors thank Ken Bates for recruiting participants; and Debra Kish, Joi Moore and Lisa Sulkowski for data collection and management.
Footnotes
Author Contributions: E.W. was responsible for conceptualizing and performing this data analysis, and drafting the manuscript. L.H.L. contributed to psychiatric screening and edited the manuscript. M.B. conducted genotyping analysis and edited the manuscript. M.K.G. contributed to study design, data coordination, and edited the manuscript. All authors have reviewed content and approved the final version for publication.
Declaration of Interest
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this paper.
References
- 1.Raynor K, Kong H, Chen Y, et al. Pharmacological characterization of the cloned kappa-, delta-, and mu-opioid receptors. Mol Pharmacol. 1994;45:330–334. [PubMed] [Google Scholar]
- 2.Lalley PM. Opioidergic and dopaminergic modulation of respiration. Respir Physiol Neurobiol. 2008;164:160–167. doi: 10.1016/j.resp.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Z, Zhuang J, Zhang C, et al. Activation of opioid μ-receptors in the commissural subdivision of the nucleus tractus solitarius abolishes the ventilatory response to hypoxia in anesthetized rats. Anesthesiology. 2011;115:353–363. doi: 10.1097/ALN.0b013e318224cc1f. [DOI] [PubMed] [Google Scholar]
- 4.Bergen AW, Kokoszka J, Peterson R, et al. Mu opioid receptor gene variants: Lack of association with alcohol dependence. Mol Psychiatry. 1997;2:490–494. doi: 10.1038/sj.mp.4000331. [DOI] [PubMed] [Google Scholar]
- 5.Bond C, LaForge KS, Tian M, et al. Single-nucleotide polymorphism in the human mu opioid receptor gene alters β-endorphin binding and activity: Possible implications for opiate addiction. Proc Natl Acad Sci USA. 1998;95:9608–9613. doi: 10.1073/pnas.95.16.9608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Y, Wang D, Johnson AD, et al. Allelic expression imbalance of human mu opioid receptor (OPRM1) caused by variant A118G. J Biol Chem. 2005;280:32618–32624. doi: 10.1074/jbc.M504942200. [DOI] [PubMed] [Google Scholar]
- 7.Oertel BG, Doehring A, Roskam B, et al. Genetic-epigenetic interaction modulates μ-opioid receptor regulation. Hum Mol Genet. 2012;21:4751–4760. doi: 10.1093/hmg/dds314. [DOI] [PubMed] [Google Scholar]
- 8.Wang YJ, Huang P, Blendy JA, et al. Brain region- and sex-specific alterations in DAMGO-stimulated [(35) S]GTPgammaS binding in mice with OPRM1 A112G. Addict Biol. 2012a;19:354–361. doi: 10.1111/j.1369-1600.2012.00484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yaksh TL, Yeung JC, Rudy TA. Systematic examination in the rat of brain sites sensitive to the direct application of morphine: Observation of differential effects within the periaqueductal gray. Brain Res. 1976;114:83–103. doi: 10.1016/0006-8993(76)91009-x. [DOI] [PubMed] [Google Scholar]
- 10.Ingvar M. Pain and functional imaging. Philos Trans R Soc Lond B Biol Sci. 1999;354:1347–1358. doi: 10.1098/rstb.1999.0483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peyron R, Laurent B, Garcia-Larrea L. Functional imaging of brain responses to pain. A review and meta-analysis. Clin Neurophysiol. 2000;30:263–288. doi: 10.1016/S0987-7053(00)00227-6. [DOI] [PubMed] [Google Scholar]
- 12.Mague SD, Isiegas C, Huang P, et al. Mouse model of OPRM1 (A118G) polymorphism has sex-specific effects on drug-mediated behavior. Proc Natl Acad Sci USA. 2009;106:10847–10852. doi: 10.1073/pnas.0901800106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chou WY, Wang CH, Liu PH. Human opioid receptor A118G polymorphism affects intravenous patient-controlled analgesia morphine consumption after total abdominal hysterectomy. Anesthesiology. 2006a;105:334–337. doi: 10.1097/00000542-200608000-00016. [DOI] [PubMed] [Google Scholar]
- 14.Chou WY, Yang LC, Lu HF, et al. Association of mu-opioid receptor gene polymorphism (A118G) with variations in morphine consumption for analgesia after total knee arthroplasty. Acta Anaesthesiol Scand. 2006b;50:787–792. doi: 10.1111/j.1399-6576.2006.01058.x. [DOI] [PubMed] [Google Scholar]
- 15.Sia AT, Lim Y, Lim EC, et al. A118G single nucleotide polymorphism of human mu-opioid receptor gene infiuences pain perception and patient-controlled intravenous morphine consumption after intrathecal morphine for postcesarean analgesia. Anesthesiology. 2008;109:520–526. doi: 10.1097/ALN.0b013e318182af21. [DOI] [PubMed] [Google Scholar]
- 16.Tan EC, Lim EC, Teo YY, et al. Ethnicity and OPRM1 variant independently predict pain perception and patient-controlled analgesia usage for post-operative pain. Mol Pain. 2009;5:1–8. doi: 10.1186/1744-8069-5-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang P, Chen C, Mague SD, et al. A common single nucleotide polymorphism A118G of the mu opioid receptor alters its N-glycosylation and protein stability. Biochem J. 2012;441:379–386. doi: 10.1042/BJ20111050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beyer A, Koch T, Schroder H, et al. Effect of the A118G polymorphism on binding affinity, potency, and agonist-mediated endocytosis, desensitization, and resensitization of the human mu-opioid receptor. J Neurochem. 2004;89:553–560. doi: 10.1111/j.1471-4159.2004.02340.x. [DOI] [PubMed] [Google Scholar]
- 19.Kroslak T, LaForge KS, Gianotti RJ, et al. The single nucleotide polymorphism A118G alters functional properties of the human mu opioid receptor. J Neurochem. 2007;103:77–87. doi: 10.1111/j.1471-4159.2007.04738.x. [DOI] [PubMed] [Google Scholar]
- 20.Ray R, Ruparel K, Newberg A, et al. Human mu opioid receptor (OPRM1 A118G) polymorphism is associated with brain mu-opioid receptor binding potential in smokers. Proc Natl Acad Sci USA. 2011;108:9268–9273. doi: 10.1073/pnas.1018699108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weerts EM, McCaul ME, Kuwabara H, et al. Infiuence of OPRM1 Asn40Asp variant (A118G) on [11C]carfentanil binding potential: Preliminary findings in human subjects. Int J Neuropsychopharmacol. 2013;16:47–53. doi: 10.1017/S146114571200017X. [DOI] [PubMed] [Google Scholar]
- 22.Coller JK, Beardsley J, Bignold J, et al. Lack of association between the A118G polymorphism of the mu opioid receptor gene (OPRM1) and opioid dependence: A meta-analysis. Pharmacogenomics Pers Med. 2009;2:9–19. [PMC free article] [PubMed] [Google Scholar]
- 23.Arias A, Feinn R, Kranzler HR. Association of an Asn40Asp (A118G) polymorphism in the μ-opioid receptor gene with substance dependence: A meta analysis. Drug Alcohol Depend. 2006;83:262–268. doi: 10.1016/j.drugalcdep.2005.11.024. [DOI] [PubMed] [Google Scholar]
- 24.Haerian BS, Haerian MS. OPRM1 rs1799971 polymorphism and opioid dependence: Evidence from a meta-analysis. Pharmacogenomics. 2013;7:813–824. doi: 10.2217/pgs.13.57. [DOI] [PubMed] [Google Scholar]
- 25.Kreek MJ, Nielsen DA, Butelman ER, et al. Genetic infiuences on impulsivity, risk taking, stress responsivity and vulnerability to drug abuse and addiction. Nature Neuroscience. 2005;8:1450–1457. doi: 10.1038/nn1583. [DOI] [PubMed] [Google Scholar]
- 26.Khokhar JY, Ferguson CS, Zhu AZ, et al. Pharmacogenetics of drug dependence: role of gene variations in susceptibility and treatment. Annu Rev Pharmacol Toxicol. 2010;50:39–61. doi: 10.1146/annurev.pharmtox.010909.105826. [DOI] [PubMed] [Google Scholar]
- 27.Nikolov MA, Beltcheva O, Galabova A, et al. No evidence of association between 118A>G OPRM1 polymorphism and heroin dependence in a large Bulgarian case-control sample. Drug Alcohol Depend. 2011;117:62–65. doi: 10.1016/j.drugalcdep.2010.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi J, Hui L, Xu Y, et al. Sequence variations in the mu-opioid receptor gene (OPRM1) associated with human addiction to heroin. Human Mutat. 2002;19:459–460. doi: 10.1002/humu.9026. [DOI] [PubMed] [Google Scholar]
- 29.Manini AF, Jacobs MM, Vlahov D, et al. Opioid receptor polymorphism A118G associated with clinical severity in a drug overdose population. J Med Toxicol. 2013;2:148–154. doi: 10.1007/s13181-012-0286-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang YJ, Huang P, Ung A, et al. Reduced expression of the mu opioid receptor in some, but not all, brain regions in mice with OPRM1 A112G. Neuroscience. 2012b;205:178–184. doi: 10.1016/j.neuroscience.2011.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Greenwald MK, Hursh SR. Behavioral economic analysis of opioid consumption in heroin-dependent individuals: Effects of unit price and pre-session drug supply. Drug Alcohol Depend. 2006;85:35–48. doi: 10.1016/j.drugalcdep.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 32.Greenwald MK, Steinmiller CL. Behavioral economic analysis of opioid consumption in heroin-dependent individuals: Effects of alternative reinforcer magnitude and post-session drug supply. Drug Alcohol Depend. 2009;104:84–93. doi: 10.1016/j.drugalcdep.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Greenwald MK. Effects of experimental unemployment, employment and punishment analogs on opioid seeking and consumption in heroin-dependent volunteers. Drug Alcohol Depend. 2010;111:64–73. doi: 10.1016/j.drugalcdep.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Greenwald MK, Lundahl LH, Steinmiller CL. Yohimbine increases opioid-seeking behavior in heroin-dependent, buprenorphine-maintained individuals. Psychopharmacology. 2013;225:811–824. doi: 10.1007/s00213-012-2868-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rickham PP. Human experimentation. Code of ethics of the World Medical Association Declaration of Helsinki. Br Med J. 1964;2:177. doi: 10.1136/bmj.2.5402.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hodgkinson CA, Yuan Q, Xu K, et al. Addictions biology: Haplotype-based analysis for 130 candidate genes on a single array. Alcohol Alcohol. 2008;43:505–515. doi: 10.1093/alcalc/agn032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.West SG, Finch JF, Curran PJ. Structural equation modeling: Concepts, issues, and applications. Newberry Park, CA: Sage Publications; 1995. Structural equation models with nonnormal variables: Problems and remedies R.H. Hoyle; pp. 56–75. [Google Scholar]
- 38.Barrio G, De La Fuente L, Lew C, et al. Differences in severity of heroin dependence by route of administration: The importance of length of heroin use. Drug Alcohol Depend. 2001;63:169–177. doi: 10.1016/S0376-8716(00)00204-0. [DOI] [PubMed] [Google Scholar]
- 39.Gossop M, Griffiths P, Powis B, et al. Severity of dependence and route of administration of heroin, cocaine, and amphetamines. Br J Addict. 1992;87:1527–1536. doi: 10.1111/j.1360-0443.1992.tb02660.x. [DOI] [PubMed] [Google Scholar]
- 40.Smolka M, Schmidt LG. The influence of heroin dose and route of administration on the severity of the opiate withdrawal syndrome. Addiction. 1999;94:1191–1198. doi: 10.1046/j.1360-0443. [DOI] [PubMed] [Google Scholar]
- 41.Smyth BP, Barry J, Keenan E, et al. Lapse and relapse following treatment of opiate dependence. Ir Med J. 2010;103:176–179. [PubMed] [Google Scholar]
- 42.Ray R, Jepson C, Patterson F, et al. Association of OPRM1 A118G variant with the relative reinforcing value of nicotine. Psychopharmacology (Berl) 2006;188:355–363. doi: 10.1007/s00213-006-0504-2. [DOI] [PubMed] [Google Scholar]
- 43.Munafò MR, Elliot KM, Murphy MF, et al. Association of the mu-opioid receptor gene with smoking cessation. Pharmacogenomics J. 2007;7:353–361. doi: 10.1038/sj.tpj.6500432. [DOI] [PubMed] [Google Scholar]
- 44.Kim SG. Gender differences in the genetic risk for alcohol dependence -The results of a pharmacogenetic study in Korean alcoholics. Japanese Journal of Alcohol Studies & Drug Dependence. 2009;44:680–685. [PubMed] [Google Scholar]
- 45.Fillingim RB, Kaplan L, Staud R, et al. The A118G single nucleotide polymorphism of the μ-opioid receptor gene (OPRM1) is associated with pressure pain sensitivity in humans. J Pain. 2005;6:159–167. doi: 10.1016/j.jpain.2004.11.008. [DOI] [PubMed] [Google Scholar]