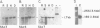Abstract
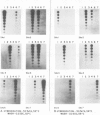
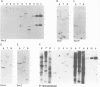
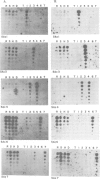
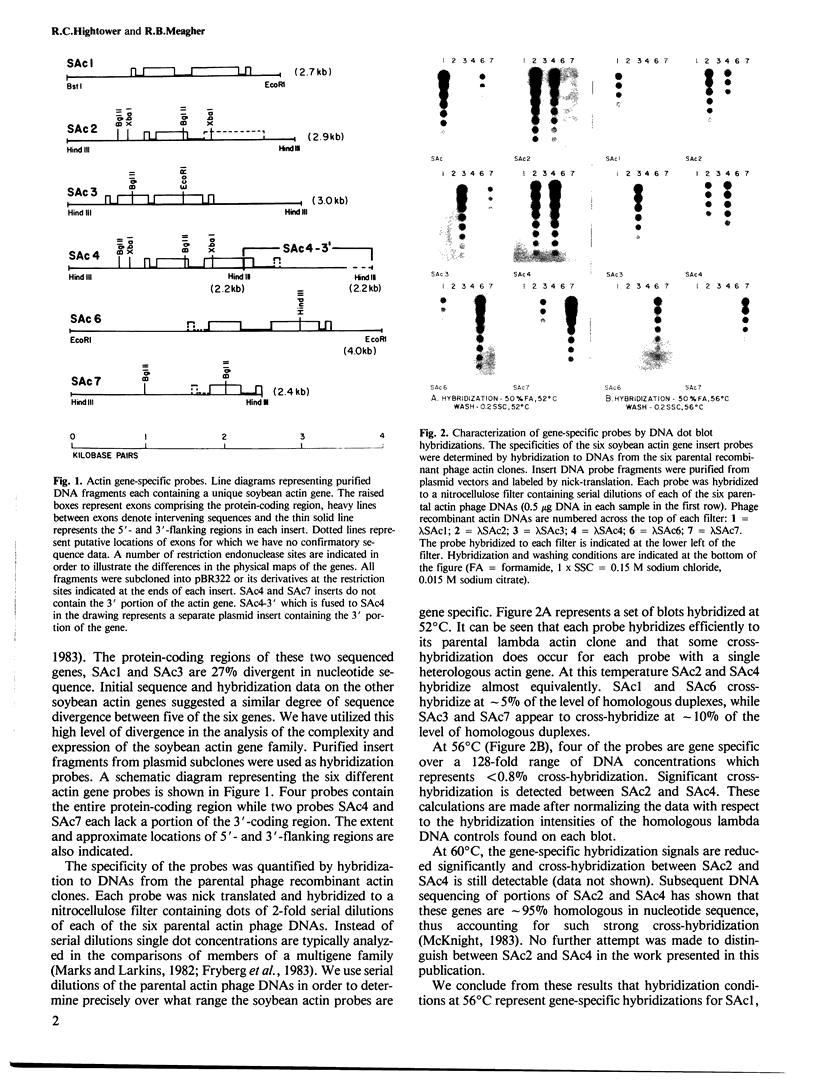
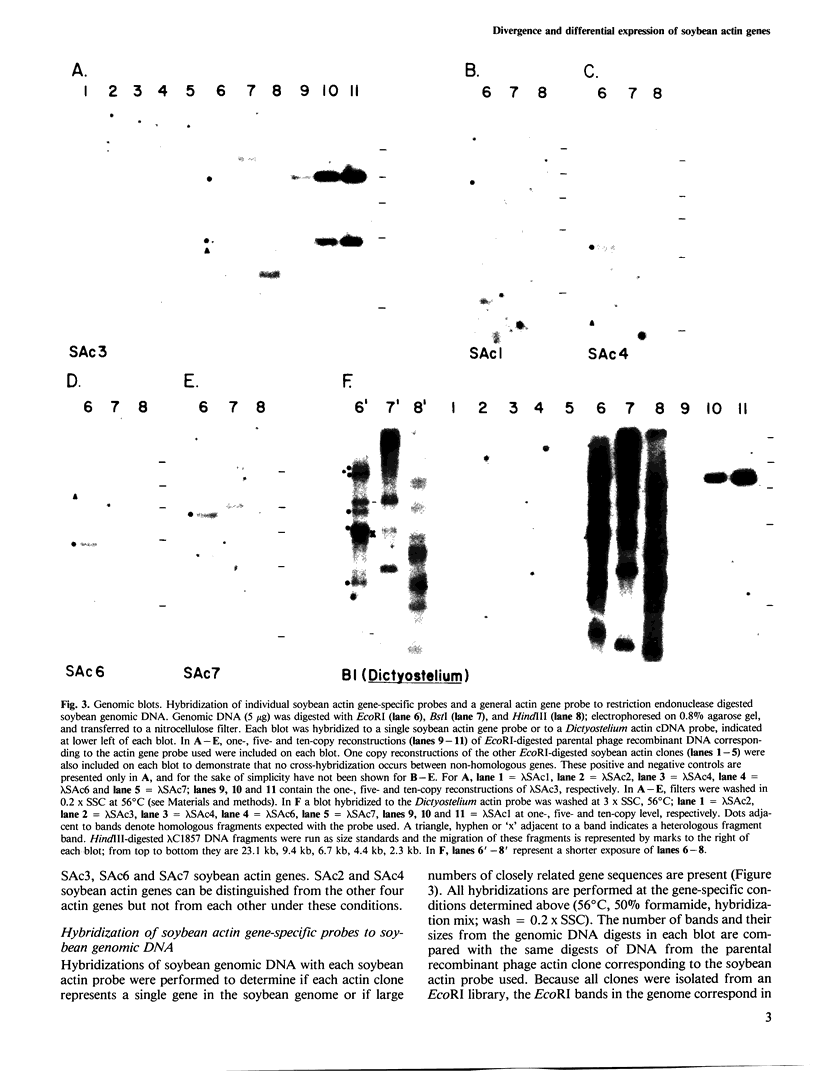
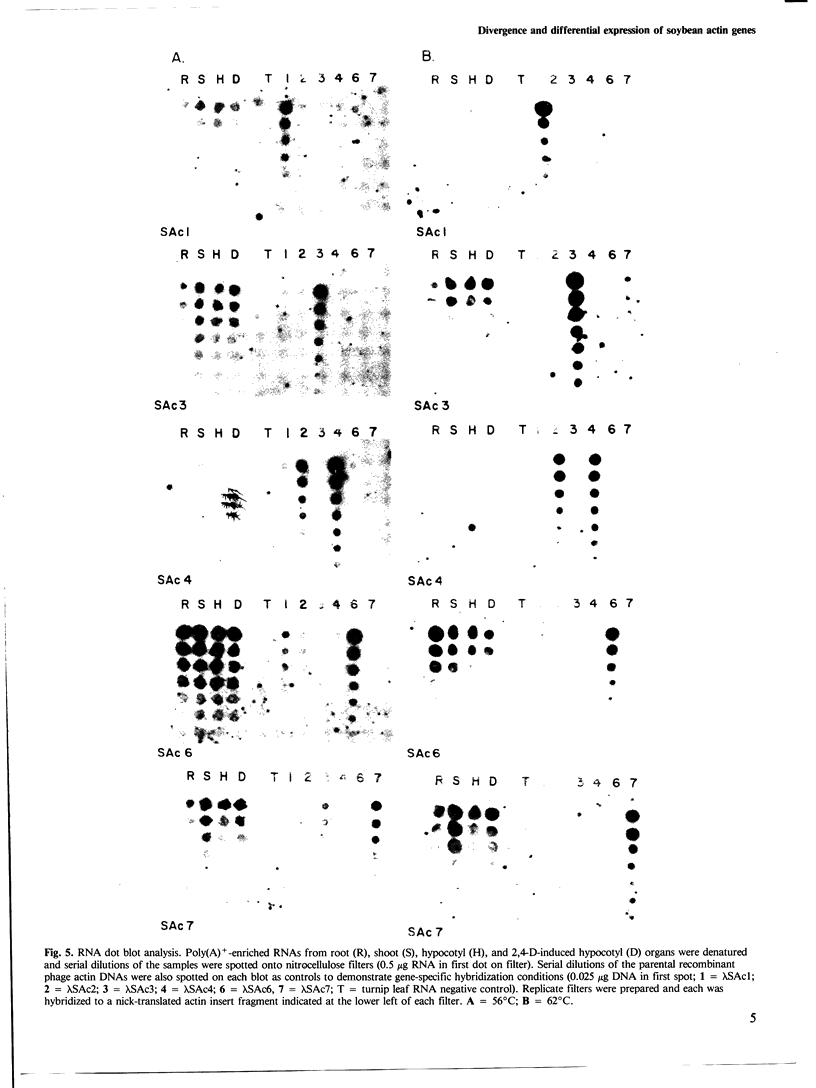
DNA sequence analysis as well as genomic blotting experiments using cloned soybean actin DNA sequences as probes show that large sequence heterogeneity exists among members of the soybean actin multigene family. This heterogeneity suggested that the members of this family might be diverged in function and/or regulation. Five of the six soybean actin gene family members examined are shown to be significantly more diverged from one another than members of other known actin gene families. This high level of divergence was utilized in the preparation of actin gene-specific probes in the analysis of the complexity and expression of these members of the soybean actin gene family. Hybridization studies indicate that the six soybean actin genes fall into three classes with a pair of genes in each class. These six genes account for all but two actin gene fragments detected in the soybean genome. We have compared the relative steady state mRNA levels of these classes of soybean actin genes in three organs of soybean. We find that actin genes SAc6 and SAc7 are most highly expressed accounting for 80% of all actin mRNA with respect to the six soybean actin genes examined. Actin genes SAc3 and SAc1 are expressed at intermediate and low levels respectively; and SAc2 and SAc4 are expressed at barely detectable levels. Four of the six soybean actin genes appear to be expressed at the same level in root, shoot and hypocotyl. SAc3 and SAc7 genes appear to be more highly expressed in shoot and 2,4-dichlorophenoxyacetic acid-induced hypocotyl than in root and hypocotyl.(ABSTRACT TRUNCATED AT 250 WORDS)
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beltz G. A., Jacobs K. A., Eickbush T. H., Cherbas P. T., Kafatos F. C. Isolation of multigene families and determination of homologies by filter hybridization methods. Methods Enzymol. 1983;100:266–285. doi: 10.1016/0076-6879(83)00061-0. [DOI] [PubMed] [Google Scholar]
- Carreira L. H., Carlton B. C., Bobbio S. M., Nagao R. T., Meagher R. B. Construction and application of a modified "gene machine": a circular concentrating preparative gel electrophoresis device employing discontinuous elution. Anal Biochem. 1980 Aug;106(2):455–468. doi: 10.1016/0003-2697(80)90548-5. [DOI] [PubMed] [Google Scholar]
- Casey J., Davidson N. Rates of formation and thermal stabilities of RNA:DNA and DNA:DNA duplexes at high concentrations of formamide. Nucleic Acids Res. 1977;4(5):1539–1552. doi: 10.1093/nar/4.5.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condit C., Hagen T. J., McKnight T. D., Meagher R. B. Characterization and preliminary mapping of cauliflower mosaic virus transcripts. Gene. 1983 Nov;25(1):101–108. doi: 10.1016/0378-1119(83)90172-5. [DOI] [PubMed] [Google Scholar]
- Files J. G., Carr S., Hirsh D. Actin gene family of Caenorhabditis elegans. J Mol Biol. 1983 Mar 5;164(3):355–375. doi: 10.1016/0022-2836(83)90056-6. [DOI] [PubMed] [Google Scholar]
- Fornwald J. A., Kuncio G., Peng I., Ordahl C. P. The complete nucleotide sequence of the chick a-actin gene and its evolutionary relationship to the actin gene family. Nucleic Acids Res. 1982 Jul 10;10(13):3861–3876. doi: 10.1093/nar/10.13.3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fyrberg E. A., Bond B. J., Hershey N. D., Mixter K. S., Davidson N. The actin genes of Drosophila: protein coding regions are highly conserved but intron positions are not. Cell. 1981 Apr;24(1):107–116. doi: 10.1016/0092-8674(81)90506-7. [DOI] [PubMed] [Google Scholar]
- Fyrberg E. A., Mahaffey J. W., Bond B. J., Davidson N. Transcripts of the six Drosophila actin genes accumulate in a stage- and tissue-specific manner. Cell. 1983 May;33(1):115–123. doi: 10.1016/0092-8674(83)90340-9. [DOI] [PubMed] [Google Scholar]
- Kafatos F. C., Jones C. W., Efstratiadis A. Determination of nucleic acid sequence homologies and relative concentrations by a dot hybridization procedure. Nucleic Acids Res. 1979 Nov 24;7(6):1541–1552. doi: 10.1093/nar/7.6.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks M. D., Larkins B. A. Analysis of sequence microheterogeneity among zein messenger RNAs. J Biol Chem. 1982 Sep 10;257(17):9976–9983. [PubMed] [Google Scholar]
- McKeown M., Firtel R. A. Evidence for sub-families of actin genes in Dictyostelium as determined by comparisons of 3' end sequences. J Mol Biol. 1981 Oct 5;151(4):593–606. doi: 10.1016/0022-2836(81)90425-3. [DOI] [PubMed] [Google Scholar]
- Meagher R. B., Shepherd R. J., Boyer H. W. The structure of cauliflower mosaic virus. I. A restriction endonuclease map of cauliflower mosaic virus DNA. Virology. 1977 Jul 15;80(2):362–375. doi: 10.1016/s0042-6822(77)80012-3. [DOI] [PubMed] [Google Scholar]
- Nagao R. T., Shah D. M., Eckenrode V. K., Meagher R. B. Multigene family of actin-related sequences isolated from a soybean genomic library. DNA. 1981;1(1):1–9. doi: 10.1089/dna.1.1981.1.1. [DOI] [PubMed] [Google Scholar]
- Nudel U., Katcoff D., Zakut R., Shani M., Carmon Y., Finer M., Czosnek H., Ginsburg I., Yaffe D. Isolation and characterization of rat skeletal muscle and cytoplasmic actin genes. Proc Natl Acad Sci U S A. 1982 May;79(9):2763–2767. doi: 10.1073/pnas.79.9.2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesacreta T. C., Carley W. W., Webb W. W., Parthasarathy M. V. F-actin in conifer roots. Proc Natl Acad Sci U S A. 1982 May;79(9):2898–2901. doi: 10.1073/pnas.79.9.2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez F., Tobin S. L., Rdest U., Zulauf E., McCarthy B. J. Two Drosophila actin genes in detail. Gene structure, protein structure and transcription during development. J Mol Biol. 1983 Feb 5;163(4):533–551. doi: 10.1016/0022-2836(83)90111-0. [DOI] [PubMed] [Google Scholar]
- Schliwa M. Proteins associated with cytoplasmic actin. Cell. 1981 Sep;25(3):587–590. doi: 10.1016/0092-8674(81)90166-5. [DOI] [PubMed] [Google Scholar]
- Shah D. M., Hightower R. C., Meagher R. B. Complete nucleotide sequence of a soybean actin gene. Proc Natl Acad Sci U S A. 1982 Feb;79(4):1022–1026. doi: 10.1073/pnas.79.4.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah D. M., Hightower R. C., Meagher R. B. Genes encoding actin in higher plants: intron positions are highly conserved but the coding sequences are not. J Mol Appl Genet. 1983;2(1):111–126. [PubMed] [Google Scholar]
- Shott R. J., Lee J. J., Britten R. J., Davidson E. H. Differential expression of the actin gene family of Strongylocentrotus purpuratus. Dev Biol. 1984 Feb;101(2):295–306. doi: 10.1016/0012-1606(84)90143-x. [DOI] [PubMed] [Google Scholar]
- Soriano P., Szabo P., Bernardi G. The scattered distribution of actin genes in the mouse and human genomes. EMBO J. 1982;1(5):579–583. doi: 10.1002/j.1460-2075.1982.tb01211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varmus H. E., Quintrell N., Ortiz S. Retroviruses as mutagens: insertion and excision of a nontransforming provirus alter expression of a resident transforming provirus. Cell. 1981 Jul;25(1):23–36. doi: 10.1016/0092-8674(81)90228-2. [DOI] [PubMed] [Google Scholar]
- Weeds A. Actin-binding proteins--regulators of cell architecture and motility. Nature. 1982 Apr 29;296(5860):811–816. doi: 10.1038/296811a0. [DOI] [PubMed] [Google Scholar]
- Yang R., Lis J., Wu R. Elution of DNA from agarose gels after electrophoresis. Methods Enzymol. 1979;68:176–182. doi: 10.1016/0076-6879(79)68012-6. [DOI] [PubMed] [Google Scholar]