Abstract
Pseudomonas aeruginosa is a pathogenic micro-organism responsible for many hospital-acquired infections. It is able to adhere to solid surfaces and develop an immobilized community or so-called biofilm. Many studies have been focusing on the use of specific materials to prevent the formation of these biofilms, but the reactivity of the bacteria in contact to surfaces remains unknown. The aim of this study was to evaluate the impact of the abiotic surface on the physiology of adherent bacteria. Three different materials, stainless steel (SS), glass (G), and polystyrene (PS) that were relevant to industrial or medical environments were characterized at the physicochemical level in terms of their hydrophobicity and roughness. We showed that SS was moderately hydrophilic and rough, potentially containing crevices, G was hydrophilic and smooth while PS was hydrophobic and smooth. We further showed that P. aeruginosa cells were more likely able to adhere to SS and G rather than PS surfaces under our experimental conditions. The physiological response of P. aeruginosa when adhering to each of these materials was then evaluated by global proteomic analysis. The abundance of 70 proteins was shown to differ between the materials suggesting that their abundance was modified as a function of the material to which bacteria adhered. Our data lead to enabling the identification of abundance patterns that appeared to be specific to a given surface. Taken together, our data showed that P. aeruginosa is capable of sensing and responding to a surface probably via specific programmes to adapt its physiological response accordingly.
Keywords: Pseudomonas aeruginosa, adhesion, abiotic material, physicochemical properties, hydrophobicity, roughness, proteome modification, porins
Introduction
Pseudomonas aeruginosa is an opportunistic pathogen responsible for a variety of human infections such as pneumonia, aggravated cystic fibrosis, septic shock, urinary tract infection, skin, and soft tissue infections. It is also involved in nosocomial infections due to the development of resistance mechanisms to disinfecting procedures, notably in healthcare environments. Various surfaces can be contaminated by this pathogen and may become new starting points for its dissemination (Dunne, 2002; Leibovitz et al., 2003; Zegans et al., 2005). The exceptional survivability properties of P. aeruginosa have been related to this strain's ability to colonize solid substrates and form three-dimensional (3D) structured aggregates of bacterial cells called biofilms (Costerton et al., 1995). In this state, the bacteria become immobilized and embedded into an exopolymer-containing matrix, leading to their increased resistance to antimicrobials, either via the chemical and mechanical protections provided by the matrix itself, or by the considerable physiological modifications that are encountered by the bacteria in these conditions compared to motile planktonic cells (Stewart and Costerton, 2001; Whiteley et al., 2001; Drenkard and Ausubel, 2002). Many studies have been performed on sessile cells at the proteomic level (Sauer et al., 2002) and showed that the bacterial protein content was markedly modified, depending on various parameters such as nutritional cues (carbon and nitrogen sources, calcium, or ferrous ion concentrations), temperature, oxygen availability, or even biofilm maturity (Whiteley et al., 2001; Sauer et al., 2002; Yoon et al., 2002; Arevalo-Ferro et al., 2003; Vilain et al., 2004; Southey-Pillig et al., 2005; Waite et al., 2005; Patrauchan et al., 2007). While many studies have focused on the physiology of the cells within biofilms, few have examined their behavior during the first stage of biofilm formation, i.e., adhesion. To date, the impact of the material on bacterial physiology during the adhesion stage is far from being known. Bacterial adhesion to inert surfaces has been shown to impact the viability of cells (Terada et al., 2006; Nejadnik et al., 2008), their resistance to antibiotics (Aaron et al., 2002) and their virulence (Siryaporn et al., 2014). Most studies of bacterial adhesion have hitherto only focused on the amount of adherent cells found on various abiotic surfaces (Yuan et al., 2011; Gadenne et al., 2013; Bedel et al., 2015) in relation to the surface energy, charge, topography, and stiffness of the materials employed (Song et al., 2015). Surface stiffness was recently shown to impact the proteome of a non-pathogenic strain of Pseudoalteromonas sp. when adhering to agar (Guégan et al., 2013), suggesting that bacteria are able to sense different surfaces (Busscher and van der Mei, 2012; Song et al., 2015). Remarkably, in P. aeruginosa the two SadC (surface attachment defective) and Wsp (wrinkly spreader phenotype) regulated circuits have been shown to be involved in surface sensing, leading to higher production of the second messenger bis-(3′–5′)-cyclic guanosine monophosphate (c-di-GMP) resulting in biofilm formation (Belas, 2014). However, the degree to which the physiology of adhering bacteria is affected by the physicochemical properties of the material involved still needs to be elucidated. This is a key question, particularly with respect to safety issues, because the answers would enable strategies to optimize the control of surface bio-contamination (choice of material, modification of the solid surface, etc.) and consequently to reduce associated risks. The aim of this study was therefore to evaluate the impact of the abiotic surface on the physiology of adherent bacteria. Three different materials (stainless steel, glass, and polystyrene) that were relevant to industrial or medical environments were first of all characterized at the physicochemical level in terms of their hydrophobicity and roughness. The physiological response of P. aeruginosa when adhering to each of these materials was then evaluated by means of global proteomic analysis relative to the physicochemical properties of these materials.
Materials and methods
Bacterial strain and medium
P. aeruginosa PAO1 strain was used during this study. Frozen bacterial stocks of the strain were sub-cultured three times in Luria-Bertani broth (LB) and then adjusted to pH 7.4 at 37°C with vigorous orbital shaking (180 rpm).
Materials
AISI 316 stainless steel (SS) (Goodfellow, Huntingdon, United Kingdom), borosilicate glass Petri dishes (G) (Schott, Mainz, Germany), and polystyrene Petri dishes (PS) (Gosselin, Borre, France) were used as the model surfaces. Before each experiment, these surfaces were cleaned as previously described (Kamgang et al., 2007). Briefly, all surfaces were soaked for 15 min in a 2% (v/v) solution of commercial RBS 35 detergent (Société des Traitements Chimiques de Surface, Lambersart, France), then rinsed five times for 5 min in sterile deionized water at 50°C and then a further five times for 5 min with sterile deionized water at room temperature (RT). The surfaces were stored for 24 h at RT in sterile deionized water to prevent any atmospheric carbon pollution before the experiment.
Physicochemical characterization of the materials
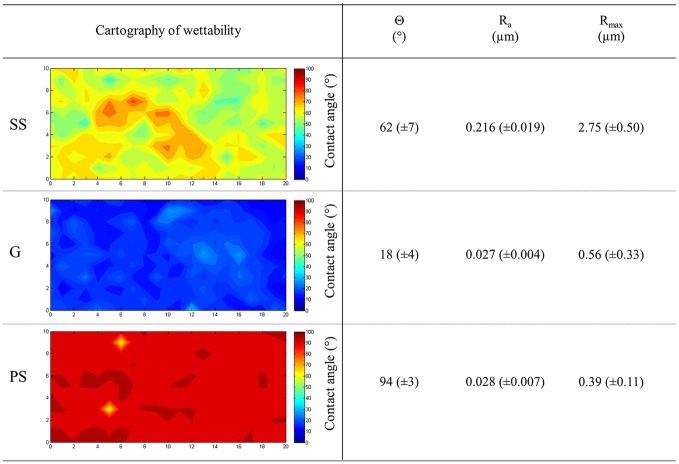
The homogeneity of surface hydrophobicity was investigated by water contact angle measurements using the sessile drop technique (DSA100M goniometer, Krüss, Palaiseau, France). This apparatus deposited a sterile deionized water droplet (300 pL) with a piezo dosing unit every 1 mm on an area of 10 × 20 mm2. The droplets were monitored for 2 s using a fast CCD (Charge Coupled Device) camera with 4x zoom and a 20x microscope objective. An automatic procedure was used for drop generation, image acquisition, contact angle determination, and the mapping of sample hydrophobicity. For each material, a cartography of wettability was constructed using Matlab 2011b software (Mathworks, Natick, USA) with a color map of visible colors from blue to red representing the contact angle to water (θ) from 0 to 100°. The average contact angle was determined from the 231 drops. All measurements were performed in triplicate.
Surface roughness was investigated as previously described (Poncin-Epaillard et al., 2013) using a surface profilometer (Perthometer M2, Mahr, Igny, France). The Ra (arithmetic average roughness) and Rmax (maximum peak to valley height) parameters were calculated from the altitude of a stylus on seven consecutive sections of 0.8 mm. The experiments were conducted six times on each sample.
Adhesion assays
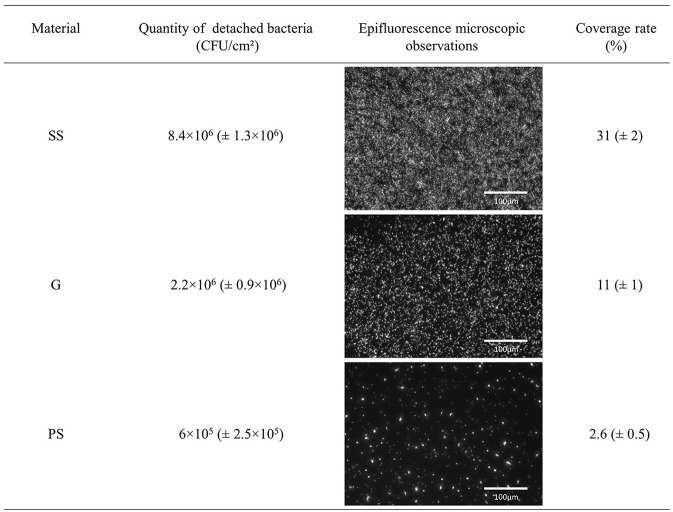
Adhesion assays were performed in 150 mM NaCl to prevent bacterial growth and a shift to a biofilm phenotype. The viability of P. aeruginosa in this medium was tested and confirmed for an extended time of 72 h. P. aeruginosa PAO1 cultures grown overnight were washed three times in NaCl 150 mM and their concentrations were adjusted to 108 CFU/mL (Colony Forming Unit per mL). The three model surfaces were covered with this bacterial suspension (to a depth of 0.5 cm) for 3 h at 37°C. Each material was then rinsed six times with 150 mM NaCl to remove any non- or weakly-attached bacteria. For direct observations, the fluorescent labeling of bacteria was achieved by immersing each contaminated material in distilled water containing SYTO9 1 μM (Molecular Probes, Life Technologies, Saint Aubin, France). Epifluorescence microscopic observations of the materials (Leica Microsystems, Nanterre, France) were performed using a 20X objective. The coverage rate of the bacteria adhering to the materials was determined using ImageJ (Rasband, 1997; Schneider et al., 2012). To enumerate the adherent cells, harvesting from the surfaces was performed with a cell scraper (Sarstedt, Newton, USA) in 150 mM NaCl. The cells were then centrifuged (10,000 g, 30 min, 4°C) and the pellets were stored at –80°C. Enumeration on LB agar plates enabled a determination of the number of bacteria that had thus been detached from each material.
Total protein extraction
Bacterial pellets (109 CFU) were lysed with 20 μL of 10% sodium dodecyl sulfate (SDS, Sigma-Aldrich, Lyon, France), sonicated three times for 5 min at 4°C and then centrifuged (10,000 g, 10 min, 4°C). Supernatants containing soluble proteins were recovered and mixed with an equivalent volume of Laemmli 2X running buffer (Biorad, Marnes-la-Coquette, France).
Protein digestion
The proteins thus extracted were reduced with 5 mM dithiothreitol (Sigma-Aldrich), and alkylated with 25 mM iodoacetamide for 45 min in the dark. The protein sample was mixed with SDS loading buffer (63 mM Tris–HCl, pH 6.8, 10 mM DTT, 2% SDS, 0.02% bromophenol blue, 10% glycerol), then loaded onto a SDS-PAGE stacking gel (7%) and subjected to a short period of electrophoresis (10 mA, 50 V, 5 min). After migration, the gels were stained with Coomassie blue and de-stained with a solution containing 50% ethanol, 10% acetic acid, and 40% deionized water. The protein band revealed was then excised, washed with water, and subjected to protein digestion with 300 ng trypsin (Promega France, Charbonnières, France) in 25 mM ammonium bicarbonate buffer at 37°C. After overnight hydrolysis, tryptic peptides were recovered by extraction (Shevchenko et al., 2006), dried in under a speed vacuum and finally suspended in 25 μL nano-LC loading buffer (0.08% trifluoroacetic acid, 2% acetonitrile; Sigma-Aldrich). The peptides were then dried and stored at −20°C.
Tandem mass spectrometry
Peptide analyses were performed on an Ultimate 3000 RSLC Nanosystem (Dionex, Voisins le Bretonneux, France) coupled to an LTQ-Orbitrap Discovery mass spectrometer (Thermo Fisher, Waltham, USA; PAPPSO proteomic platform, INRA, Jouy-en-Josas) with a nanoelectrospray interface. Briefly, 4 μL of tryptic digest were loaded onto a Pepmap100C18 pre-column (0.3 mm ID x 5 mm, 5 μm, Dionex) at a flow rate of 20 μL/min for 4 min. All peptides were separated onto a Pepmap100C18 column (0.075 mm ID × 500 mm, 2 μm, Dionex) with a gradient from 2 to 35% for 68 min at 300 nL/min and 40°C, for a total running time of 85 min which included the regeneration and equilibration steps of the column. Ionization was performed in positive mode (1.4 kV ionization potential) with a liquid junction and capillary probe (PicoTip Emiter, 10 μm ID; New Objective, Woburn, MA, USA). Data were acquired using Xcalibur (v2.07, Thermo Fisher Scientific) using a data-dependent method comprising two steps: first, a full scan MS in an Orbitrap analyser (from 300 to 1,400 in profile mode with a resolution of 15,000 at m/z 400) and second, fragmentation and the detection of daughter ions in the linear trap (qz = 0.22, activation time 50 ms and collision energy fixed at 35%) in centroid mode. The dynamic exclusion time was set at 45 s. To enhance mass accuracy, the lock mass option was activated on dimethylcyclosiloxan (m/z 445.120029).
Protein quantification
The raw files produced under Xcalibur were first converted to mzXML files with MSconvert (http://sashimi.sourceforge.net, version 3.0.3768) and in a second step, proteins were identified using X!tandem software (X!tandem Piledriver 2015.04.01.1; http://www.thegpm.org) (Craig and Beavis, 2004) against a P. aeruginosa PAO1 protein database (NCBInr, NC_002516.2; 5566 sequences) associated with a standard proteomic contaminant database. The X!Tandem search parameters were (i) trypsin specificity with three missed cleavages and (ii) variable oxidation states of methionine. Semi-tryptic peptide detection was included by activating the option in the data refine mode. The mass tolerance was fixed at 10 ppm for precursor ions and 0.5 Da for fragment ions. The final search results were filtered using a multiple threshold filter applied at the protein level and consisting of a Log10 protein E-value lower than −2.6 identified with a minimum of two different peptide sequences, detected in at least one analysis as having a peptide E-value lower than 0.05. These criteria led to false discovery rates estimated using the decoy database of 0.07 and 0.10% for peptide and protein identification, respectively.
X!TandemPipeline 3.3.5 analysis (Langella et al., 2017) (http://pappso.inra.fr/bioinfo/xtandempipeline/) was used to filter and group the peptide/protein identifications from MS/MS mass spectra.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Vizcaíno et al., 2016) partner repository with the dataset identifier PXD006881 and 10.6019/PXD006881. PRIDE Converter 2 was used to convert our data in PRIDE-xml format (Côté et al., 2012). The submission was achieved using ProteomeXchange (Vizcaíno et al., 2014).
The mass spectrometry proteomics data have been also deposited on the PROTICdb platform (http://moulon.inra.fr/protic/p_aeruginosa_adhesion).
Statistical analyses
Physicochemical results (contact angles and roughness) were considered to be significantly variable using ANOVA when the p-value was <0.05.
A spectral counting approach was used to identify proteins detected in statistically different quantities between the conditions. Data from the mass spectrometry protein quantification of five biological replicates were compared. First, all conditions were compared (70 proteins significantly different) and second, the conditions were compared between each other and the results were merged.
Filtering and statistical analysis were performed using MassChroqR, an R package developed by PAPPSO (http://pappso.inra.fr/). A small difference in the number of spectra may be noise-induced, so proteins displaying a minor variation between the conditions (whose mean difference was less than three spectra) were discarded from the analysis. A generalized linear mixed model (GLM) with a Poisson distribution was applied. This model is appropriate in the case of a counting such as spectral counting. The significance of protein abundance changes was determined by analysis of variance ANOVA using a Chi-square test. A protein was considered to be significantly variable when the p-value was <0.01. The p-values were adjusted for multiple comparisons using Benjamini and Hochberg's method (Benjamini and Hochberg, 1995).
Heat map representation
Heat maps were generated under MassChroqR using the heatplot function from the made4 package (http://www.bioconductor.org). The heat map revealed a gradient of colors assigned to the different numbers of spectra (centered and reduced) and the dendrogram provided a graphical representation of a hierarchical cluster with distances calculated using the Pearson correlation coefficient (default distfun argument of the heatplot function of the “made4” of the R package); the aggregation criterion was the average-linkage or UPGMA (Unweighted Pair Group Method with Arithmetic mean).
PseudoCAP functional classification and GO (gene ontology) term annotations
Functional classifications were performed using the facilitating community-based PseudoCAP (Pseudomonas Community Project) functional classes (http://www.pseudomonas.com) (Winsor et al., 2016). GO term annotations based on mappings to InterPro functional domain predictions provide additional information.
Results
Physicochemical properties of the surfaces of different materials
Surface hydrophobicity or hydrophilicity can be characterized by water surface wettability, measured using contact angles (θ) (Boulangé-Petermann et al., 1997). A cartography of wettability was constructed by measuring 231 contact angles per material. As shown in Figure 1, stainless steel (SS), glass (G), and polystyrene (PS) displayed homogeneous wettability which ensured the homogeneous adhesion of bacteria. Under our experimental conditions, SS was considered to be moderately hydrophobic (62° ± 7°), G as hydrophilic (18° ± 4°), and PS as hydrophobic (94° ± 3°) (Figure 1), providing an hydrophobic range for adhesion studies with materials exhibiting statistically different contact angles (p < 0.05). Concerning the topography of these surfaces, SS was shown to be rough, with an Ra of 0.216 μm, while G and PS were considered as smoother surfaces with Ra = 0.027 and 0.028 μm, respectively (Figure 1). The Rmax parameter (maximum peak to valley height) was clearly higher for SS than for G and PS, suggesting the presence of crevices on the SS surface (Boulangé-Petermann et al., 1997). SS roughness (Ra and Rmax) was shown to differ statistically from that of G and PS, while the roughness of G and PS was statistically similar (p < 0.05). Taken together, our data led us to conclude that the three studied surfaces presented different physicochemical properties, i.e., SS was moderately hydrophilic and rough, potentially containing crevices, G was hydrophilic and smooth while PS was hydrophobic and smooth. As the number of adherent bacteria could be related to the physicochemical properties of materials (Bellon-Fontaine et al., 1990), the adhesion of P. aeruginosa was investigated on these different surfaces.
Figure 1.
Physicochemical properties of the materials. Stainless steel (SS), glass (G), and polystyrene (PS) were assayed for their wettability. The average contact angles (Θ) and the roughness parameters Ra and Rmax were also reported. Standard deviations are given into brackets.
Materials impact the adhesion capability of P. aeruginosa
Bacteria were allowed to adhere to these three surfaces for 3 h before being recovered and observed or counted. Figure 2 shows that 8.4 × 106 (± 1.3 × 106) CFU/cm2, 2.2 × 106 (± 0.9 × 106) CFU/cm2, and 6 × 105 (± 2.5 × 105) CFU/cm2 (P < 0.05) were detached from SS, G, and PS, respectively, indicating that the material had an impact on the number of adherent cells. Direct epifluorescence microscopic observations of the contaminated materials (Figure 2) were correlated to the enumeration results by showing a higher coverage rate of bacteria on SS (31%), an intermediate rate on G (11%), and a lower rate on PS (2.6%). Taken together, these data suggest that P. aeruginosa cells were more likely able to adhere to SS and G rather than PS surfaces under our experimental conditions.
Figure 2.
Adhesion of P. aeruginosa PAO1 to stainless steel (SS), glass (G), and polystyrene (PS), 3 h at 37°C. Quantity of adherent P. aeruginosa PAO1 determined by enumeration and epifluorescence microscopic observations with a ×20 objective after SYTO9 staining.
P. aeruginosa displays varied global proteome patterns in response to adhesion on different materials
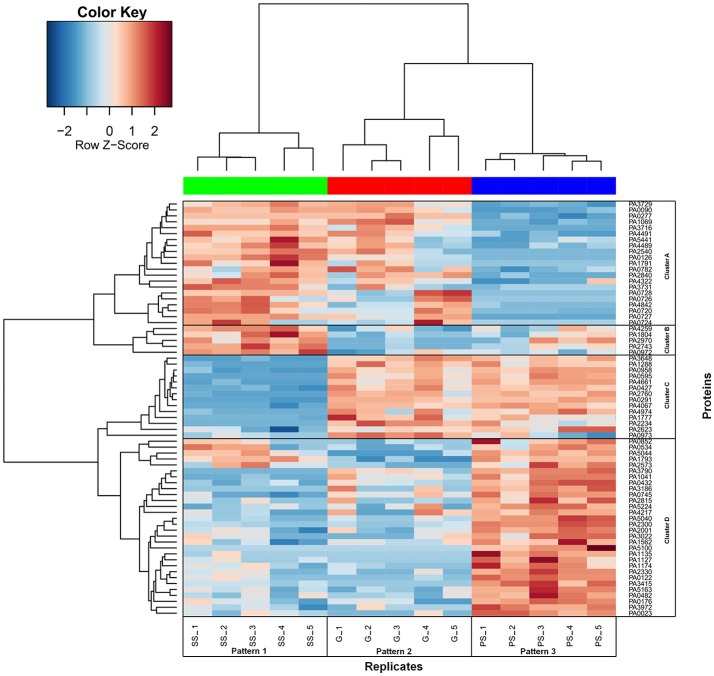
The total proteome of P. aeruginosa PAO1 adhering to SS, G and PS was analyzed using nano-LC-MS/MS and X!TandemPipeline. The mass spectra thus generated (Table S1) enabled the identification of 930 proteins (Table S2) out of the 5,570 predicted ORFs that are encoded by the P. aeruginosa PAO1 genome. Of these, 785 were identified on the three materials, 107 on two of them, and 38 were detected specifically on only one surface (Figure 3, Tables S2, S3). Using strong statistical validation assays, the abundance of 70 out of these 930 proteins was shown to differ between the materials compared by pairs (Table S4). P. aeruginosa displayed differential abundance of the greatest number of proteins (or a differential abundance of peptide ions) when adhesion was compared between SS and PS (57 proteins). Among the 834 proteins identified on these two materials (Figure 3), 57 proteins showed significant differences in their abundances: four were specifically present in this pair comparison, 8 in SS vs. G and 7 in PS vs. G. By contrast, comparing SS vs. G and PS vs. G led to 21 and 39 differentially abundant proteins (Table S4). Taken together, these data showed a strong bacterial response that was specific to the material used for adhesion. The data were then subjected to hierarchical clustering which focused on the 70 proteins that displayed differential content following adhesion on at least two materials. The abundance of these 70 proteins was compared using the normalized quantities of spectral counting from five biological replicates (Figure 4, Table S4). Protein levels were represented as a color map ranging from red to blue, indicating proteins produced at lower and higher levels, respectively. Clustering of the five replicates highlighted a strong coherence for each material in terms of protein abundance, which demonstrated the reproducibility of our assays (Figure 4, upper dendrogram). Clustering of the 70 proteins in terms of their abundance on a given material, followed by a comparison of these results on the three materials, enabled the identification of a further four clusters (Figure 4, left dendrogram, clusters A, B, C, and D). Under these conditions, each cluster enabled the grouping of proteins with a relatively similar level on a particular material compared to the two others. SS-adherent bacteria presented more proteins belonging to the A- and B- clusters, while G-adherent bacteria displayed more proteins from the A- and C- clusters. PS-adherent bacteria were characterized by an abundance of proteins belonging to the C- and D-clusters. Taken together, this clustering led to the definition of patterns on each material which reflected protein abundance profiles considering A to D clusters on a given material, vs. the other two (Figure 4). Remarkably, this global approach permitted the identification of three protein abundance patterns that were each relevant to a specific material.
Figure 3.
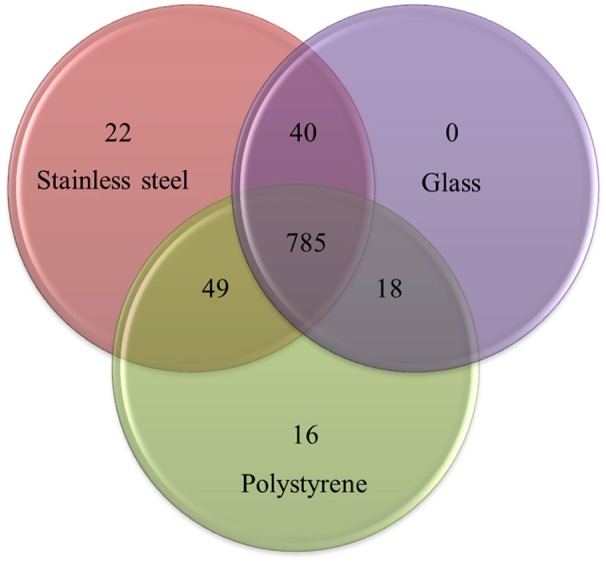
Venn diagram based comparisons indicating the number of total detected proteins (930) from P. aeruginosa adhering to stainless steel, glass, and polystyrene. The complete list of these proteins is available in Table S1.
Figure 4.
Normalized number of spectra of the 70 proteins with significant differences of quantities after adhesion of P. aeruginosa cells to stainless steel (SS), glass (G), and polystyrene (PS). Quantity of each protein is normalized from the number of spectra counted by mass spectrometry. The 70 proteins identified as significantly altered in abundance between SS, G, and PS were sorted by heatplot function. Replicates are indicated by numbers 1 to 5 and are referred to patterns 1 to 3 (1 corresponds to SS, 2 to G, and 3 to PS). Proteins are designated by their locus tag referenced in Table S4. Four clusters of proteins (A, B, C, and D) following the same abundance pattern are represented by black boxes.
Physiological response to the materials of adherent P. aeruginosa
To obtain further insights into the physiological response of P. aeruginosa to adhesion on each of the three materials, the functional classes of the 70 proteins whose abundance differed significantly between at least two materials were assigned according to the PseudoCAP classification (Winsor et al., 2016). The functional classes relative to all differentially abundant proteins are available in Table 1 and Table S5.
Table 1.
PseudoCAP-based functional classes of the significantly differentially abundant proteins between the materials
| Averageb | p adjc | Clusterd | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PseudoCAP-based functional classesa | Protein name | Locus tag | Gene name | SS | G | PS | SS/G | SS/PS | PS/G | A | B | C | D |
| Adaptation, Protection | RahU | PA0122 | rahU | 1.4 | 0.2 | 5.4 | 8.9E-04 | 3.1E-07 | |||||
| LPS-assembly protein LptD | PA0595 | lptD | 0.0 | 11.0 | 13.0 | 1.5E-16 | 8.1E-20 | ||||||
| MagD | PA4489 | magD | 11.6 | 7.8 | 2.6 | 1.3E-07 | 6.0E-04 | ||||||
| probable oxidoreductase | PA1127 | 0.8 | 0.0 | 4.2 | 8.5E-04 | 4.5E-07 | |||||||
| Chemotaxis | probable chemotaxis transducer | PA2573 | 2.8 | 0.4 | 4.0 | 1.1E-04 | |||||||
| aerotaxis transducer Aer2 | PA0176 | aer2 | 6.6 | 5.8 | 15.0 | 1.2E-04 | 1.7E-05 | ||||||
| Amino acid biosynthesis and metabolism | S-adenosyl-L-homocysteine hydrolase | PA0432 | sahH | 9.6 | 12.0 | 18.0 | 7.4E-04 | ||||||
| proline dehydrogenase PutA | PA0782 | putA | 17.0 | 18.6 | 10.4 | 8.6E-03 | 1.6E-03 | ||||||
| Central intermediary metabolism | malate synthase G | PA0482 | glcB | 13.0 | 12.4 | 20.4 | 7.7E-03 | 3.5E-03 | |||||
| Carbon compound catabolism | chitinase | PA2300 | chiC | 1.0 | 1.4 | 11.2 | 1.7E-11 | 6.5E-10 | |||||
| Energy metabolism | periplasmic nitrate reductase protein NapA | PA1174 | napA | 2.8 | 1.6 | 8.0 | 7.3E-04 | 7.7E-06 | |||||
| aconitate hydratase 1 | PA1562 | acnA | 12.6 | 15.6 | 21.2 | 1.9E-03 | |||||||
| urocanate hydratase | PA5100 | hutU | 0.0 | 0.0 | 3.2 | 8.6E-06 | 1.1E-05 | ||||||
| probable dihydrolipoamide acetyltransferase | PA3415 | 0.8 | 0.0 | 4.6 | 3.0E-04 | 1.5E-07 | |||||||
| quinone oxidoreductase | PA0023 | qor | 1.6 | 0.6 | 5.2 | 3.2E-03 | 1.7E-05 | ||||||
| isocitrate dehydrogenase | PA2623 | icd | 22.2 | 28.8 | 32.0 | 5.6E-03 | |||||||
| Fatty acid and phospholipid metabolism | acetyl-CoA acetyltransferase | PA2001 | atoB | 4.0 | 5.0 | 10.6 | 2.2E-04 | 3.0E-03 | |||||
| DNA replication, recombination, modification and repair | DNA-binding protein HU | PA1804 | hupB | 12.4 | 5.4 | 7.6 | 5.3E-04 | ||||||
| Transcription, RNA processing and degradation | probable ATP-dependent RNA helicase | PA2840 | 4.0 | 3.8 | 0.2 | 1.4E-05 | 2.8E-05 | ||||||
| Translation, post-translational modification, degradation | aminopeptidase P | PA5224 | pepP | 1.4 | 2.4 | 6.0 | 2.2E-04 | 8.6E-03 | |||||
| 50S ribosomal protein L32 | PA2970 | rpmF | 3.4 | 0.2 | 2.4 | 1.2E-04 | |||||||
| 30S ribosomal protein S19 | PA4259 | rpsS | 8.6 | 3.8 | 5.4 | 5.7E-03 | |||||||
| translation initiation factor IF-3 | PA2743 | infC | 5.6 | 1.2 | 3.0 | 2.8E-04 | |||||||
| peptidyl-prolyl cis-trans isomerase B | PA1793 | ppiB | 4.8 | 0.8 | 5.0 | 2.4E-04 | 1.1E-04 | ||||||
| Transport of small molecules | OprQ | PA2760 | oprQ | 0.2 | 10.6 | 11.2 | 1.7E-14 | 1.6E-15 | |||||
| glucose/carbohydrate outer membrane porin OprB precursor | PA3186 | oprB | 0.0 | 2.0 | 5.4 | 5.4E-09 | 8.1E-03 | ||||||
| basic amino acid, basic peptide and imipenem outer membrane porin OprD precursor | PA0958 | oprD | 1.0 | 13.6 | 16.0 | 1.7E-14 | 1.1E-17 | ||||||
| putative copper transport outer membrane porin OprC precursor | PA3790 | oprC | 0.0 | 7.4 | 14.4 | 7.8E-12 | 1.8E-21 | 1.7E-03 | |||||
| TolB protein | PA0972 | tolB | 15.4 | 6.0 | 6.6 | 1.7E-05 | 6.3E-05 | ||||||
| anaerobically-induced outer membrane porin OprE precursor | PA0291 | oprE | 0.6 | 11.2 | 11.6 | 3.7E-13 | 8.2E-14 | ||||||
| probable outer membrane protein precursor | PA1288 | 0.2 | 8.2 | 7.4 | 2.4E-11 | 3.6E-10 | |||||||
| outer membrane protein Opr86 | PA3648 | opr86 | 0.2 | 7.2 | 6.6 | 5.7E-10 | 4.2E-09 | ||||||
| probable outer membrane protein precursor | PA1041 | 0.2 | 3.6 | 6.4 | 6.7E-05 | 6.8E-09 | |||||||
| peptidoglycan associated lipoprotein OprL precursor | PA0973 | oprL | 2.6 | 7.2 | 2.8 | 2.4E-03 | 3.3E-03 | ||||||
| Membrane proteins | lipid A 3-O-deacylase | PA4661 | pagL | 1.2 | 6.4 | 7.6 | 4.2E-05 | 1.4E-06 | |||||
| outer membrane protein OprG precursor | PA4067 | oprG | 1.6 | 10.6 | 11.4 | 6.4E-09 | 8.0E-10 | ||||||
| major intrinsic multiple antibiotic resistance efflux outer membrane protein OprM precursor | PA0427 | oprM | 1.6 | 12.2 | 11.8 | 8.1E-11 | 3.2E-10 | ||||||
| major porin and structural outer membrane porin OprF precursor | PA1777 | oprF | 7.8 | 26.6 | 20.6 | 2.0E-12 | 2.0E-07 | ||||||
| Cell wall/LPS/capsule | PslD | PA2234 | pslD | 0.0 | 3.8 | 2.2 | 1.4E-06 | ||||||
| glucose-1-phosphate thymidylyltransferase | PA5163 | rmlA | 5.0 | 4.2 | 9.0 | 5.6E-03 | |||||||
| Motility and attachment | type 4 fimbrial biogenesis outer membrane protein PilQ precursor | PA5040 | pilQ | 1.2 | 2.2 | 8.8 | 5.5E-08 | 1.7E-05 | |||||
| type 4 fimbrial biogenesis protein PilM | PA5044 | pilM | 5.6 | 2.4 | 6.6 | 3.1E-03 | |||||||
| Protein secretion/export apparatus | probable outer membrane protein precursor | PA4974 | 0.2 | 6.0 | 7.2 | 2.8E-08 | 6.7E-10 | ||||||
| ClpV1 | PA0090 | clpV1 | 13.4 | 12.0 | 0.8 | 2.6E-15 | 5.6E-13 | ||||||
| Related to phages, transposon or plasmid | probable coat protein A of bacteriophage Pf1 | PA0724 | coaA | 3.6 | 3.0 | 0.0 | 2.2E-06 | ||||||
| hypothetical protein of bacteriophage Pf1 | PA0726 | 4.2 | 3.2 | 0.0 | 3.0E-07 | 1.1E-05 | |||||||
| hypothetical protein from bacteriophage Pf1 | PA0727 | 10.8 | 8.4 | 0.0 | 1.1E-16 | 5.6E-13 | |||||||
| helix destabilizing protein of bacteriophage Pf1 | PA0720 | 16.8 | 11.8 | 0.6 | 1.3E-20 | 2.6E-13 | |||||||
| probable bacteriophage integrase | PA0728 | 5.4 | 4.6 | 0.0 | 5.4E-09 | ||||||||
| Secreted factors (toxins, enzymes, alginate) | chitin-binding protein CbpD precursor | PA0852 | cbpD | 2.6 | 1.0 | 5.0 | 3.8E-04 | ||||||
| Putative enzymes | FAD-dependent oxidoreductase | PA0534 | pauB1 | 3.4 | 1.0 | 4.4 | 1.7E-03 | ||||||
| probable acyl-CoA dehydrogenase | PA2815 | 3.0 | 3.2 | 7.4 | 3.9E-03 | 6.7E-03 | |||||||
| probable acyl-CoA dehydrogenase | PA3972 | 2.0 | 2.2 | 5.8 | 3.8E-03 | 7.1E-03 | |||||||
| probable enoyl-CoA hydratase/isomerase | PA0745 | 2.4 | 5.6 | 8.0 | 1.8E-04 | ||||||||
| flavin-containing monooxygenase | PA4217 | phzS | 1.2 | 3.0 | 5.0 | 8.9E-04 | |||||||
| Hypothetical proteins | hypothetical protein | PA2330 | 1.6 | 1.0 | 6.2 | 3.5E-04 | 1.7E-05 | ||||||
| conserved hypothetical protein | PA1135 | 0.4 | 0.0 | 3.8 | 1.8E-04 | 1.7E-06 | |||||||
| hypothetical protein | PA3022 | 1.4 | 1.0 | 4.2 | 2.6E-03 | ||||||||
| hypothetical protein | PA0126 | 3.8 | 1.6 | 0.0 | 1.2E-06 | ||||||||
| conserved hypothetical protein | PA2540 | 6.4 | 4.2 | 0.0 | 2.7E-10 | 4.5E-07 | |||||||
| hypothetical protein | PA1791 | 5.8 | 2.6 | 0.0 | 1.5E-09 | ||||||||
| conserved hypothetical protein | PA3731 | 7.6 | 4.0 | 2.8 | 1.5E-03 | ||||||||
| hypothetical protein | PA5441 | 5.8 | 3.4 | 1.2 | 1.4E-04 | ||||||||
| hypothetical protein | PA3716 | 10.0 | 7.0 | 0.0 | 1.5E-15 | 5.9E-11 | |||||||
| conserved hypothetical protein | PA3729 | 22.4 | 21.4 | 9.6 | 1.2E-06 | 8.2E-06 | |||||||
| hypothetical protein | PA1069 | 8.6 | 10.8 | 2.0 | 8.6E-06 | 9.7E-08 | |||||||
| conserved hypothetical protein | PA0277 | 6.0 | 6.4 | 1.0 | 2.7E-05 | 1.2E-05 | |||||||
| conserved hypothetical protein | PA4322 | 7.2 | 4.6 | 1.6 | 3.5E-05 | ||||||||
| hypothetical protein | PA4842 | 6.4 | 3.0 | 0.2 | 6.8E-09 | ||||||||
| MagB | PA4491 | magB | 5.8 | 5.0 | 0.6 | 2.7E-06 | 2.8E-05 | ||||||
PseudoCAP-based functional classes of the proteins (p-adjust < 0.01) significantly differentially abundant showing a maximal variation of three spectra between the replicates of a same material.
average of peptide spectra number of the five replicates for SS, G and PS.
p adj values from ANOVA analyses were reported only for significant results (p < 0.01). SS was compared to G (SS/G) and PS (SS/PS). PS was compared to G (PS/G). Shaded blue p adj values corresponded to under-production of the protein on SS compared to PS or G and PS compared to G. Conversely, shaded red p adj values corresponded to over-production of SS compared to PS or G and PS compared to G.
Clusters A, B, C, and D determined from heatplot function (Figure 4). Each cluster allows the grouping of proteins with a relative similar abundance tendency on a given material, compared to the two other materials.
Proteomic response of P. aeruginosa adhering to SS compared to G (SS/G)
Based on PseudoCAP analysis, most proteins belonged to three functional classes (Table 1). These included the “Membrane proteins” and “Transport of small molecules” classes, in which the great majority of the affected proteins were less abundant when P. aeruginosa was allowed to adhere to SS/G. Conversely, more abundant proteins were mostly found in the “Translation, post-translational modification, degradation” class. Numerous outer membrane proteins (OMPs) were produced at markedly lower levels when the bacteria were allowed to adhere to SS/G (Table 1). This was notably the case for many major porins, including the non-specific major porin OprF, the specific channels OprD, OprE, OprQ, OprC, OprG, and the OprM OM component of the MexAB/OprM efflux pump. It was also the case for Opr86, which is involved in porin folding into the OM (Tashiro et al., 2008), and for the peptidoglycan associated lipoprotein (Pal) OprL, a member of the Tol-Pal complex that is important to maintaining OM integrity in Gram negative bacteria (Lazzaroni et al., 1999). In addition, lipid A deacylase (PagL), which is involved in lipopolysaccharide (LPS) modification, was also less abundant when P. aeruginosa was allowed to adhere to SS/G. Conversely, four proteins belonging to the “Translation” PseudoCAP functional class, such as the ribosomal proteins RpmF, RpsS, the translation initiation factor InfC, the peptidyl-prolyl cis-trans isomerase PpiB and the TolB protein belonging to the “Transport of small molecules” class, were found to be produced at higher levels when bacteria were allowed to adhere to SS/G.
Proteomic response of P. aeruginosa adhering to SS compared to PS (SS/PS)
Based on PseudoCAP analysis, most of the dysregulated proteins belonged to eight functional classes (Table 1). These included the “Transport of small molecules,” “Membrane proteins,” “Putative enzymes,” “Energy metabolism,” and “Adaptation, protection” classes, in which the great majority of affected proteins were less abundant when P. aeruginosa was allowed to adhere to SS/PS. By contrast, proteins included in the “Hypothetical proteins” and “Related to phages, transposon or plasmid” functional classes were found at higher abundance after the adhesion of P. aeruginosa to SS/PS (Table 1, Table S5). Numerous OMPs were less abundant when P. aeruginosa was allowed to adhere to SS/PS. Interestingly, many of them, including the previously described porins, the porin folding chaperone Opr86 and the LPS lipase PagL, were also found at lower levels when SS and G were compared. Remarkably, and as indicated elsewhere regarding the abundance of the 70 proteins that were differentially produced between the three materials, this led to grouping all of these OMPs in a single cluster (Figure 4, cluster C). In addition, the glucose-specific porin OprB, the OMP precursors PA1041, PA1288, and PA4974, the LPS-assembly protein LptD and the type 4 fimbrial biogenesis OMP PilQ (which is involved in the production of adhesive organelles type IV pili) were also produced less when bacteria were allowed to adhere to SS/PS. In addition, 10 proteins in the energy, amino acid, carbon compound, central, or fatty acid metabolism pathways were down-regulated when P. aeruginosa was allowed to adhere to SS/PS. For example, this was the case for the periplasmic nitrate reductase protein NapA, urocanate hydratase, aconitate hydratase 1, probable dihydrolipoamide acetyltransferase, quinone oxidoreductase and isocitrate dehydrogenase which are involved in energy metabolism. In addition, 3 proteins belonging to the “putative enzymes” class, PA2815, PA3972, and PA0745, were less abundant in this condition and might be involved in lipid homeostasis (Table S5). Conversely, several proteins were more abundant in the context of SS/PS. This was the case for PA0720, PA0724, PA0726, PA0727, and PA0728, which are components of the Pf1 bacteriophage. PA0724, PA0726, PA0727, and PA0728 were not detected on PS and PA0720 was about 30 times more abundant when the bacteria were allowed to adhere to SS/PS. Similarly, the type 6 secretion system ATPase ClpV1, and the Tol-Pal periplasmic TolB component that is involved in cell wall integrity maintenance, were found to be more abundant when the bacteria adhered to SS/PS. Finally, the ATP-dependent RNA helicase that is involved in transcription was more abundant on SS/PS. Interestingly, most of the hypothetical proteins were more abundant in this condition and for several their predicted activity is related to the envelope metabolism, i.e., PA2540 is a putative phospholipase, and PA1791, PA4842, and PA4491 are putative membrane proteins.
Proteome response of P. aeruginosa adhering to PS compared to G (PS/G)
Based on PseudoCAP analysis, most of the dysregulated proteins belonged to six functional classes (Table 1). These included the “Energy metabolism,” “Adaptation, protection,” “Putative enzymes,” and “Motility and attachment” classes, where the great majority of affected proteins were up-regulated when P. aeruginosa was allowed to adhere to PS/G. By contrast, down-regulated proteins were mostly found in the “Hypothetical proteins” and “Related to phages” classes. Several metabolic proteins were over-produced on PS/G, namely periplasmic nitrate reductase NapA, urocanate hydratase, dihydrolipoamide acetyltransferase, and quinone oxidoreductase, all involved in energy metabolism and malate synthase G related to central intermediary metabolism. Chitinase that is involved in carbon catabolism is regulated by quorum sensing (QS) in P. aeruginosa. The over-production of ChiC in response to adhesion on PS might thus reflect some activity of QS. Similarly, two proteins involved in chemotaxis (PA2573 and Aer2), and two proteins related to pili biogenesis (PilQ and PilM) were over-produced on PS/G. Other proteins were found in smaller quantities on PS/G, including MagB which is potentially involved in host-pathogen interactions (Robert-Genthon et al., 2013), and the three phage-related proteins PA0720, PA0726, and PA0727.
Taken together, these data showed that the abundance of numerous proteins was modified as a function of the material to which bacteria adhered, enabling the identification of abundance patterns that appeared to be specific to a given surface.
Discussion
This study was designed to use a proteomic approach to highlight the physiological responses of P. aeruginosa regarding its adhesion to three different surfaces that are commonly found in many environments, including hospitals and industry. It had previously been shown that bacterial adhesion to solid surfaces is dependent on a number of factors, e.g., physicochemical and mechanical interactions (Bos et al., 1999). Therefore, under our experimental conditions (static adhesion, high ionic strength), the number of adherent bacteria would mainly depend on the Lifshitz-van der Waals, Lewis acid-base and roughness characteristics of the materials, or in another words their hydrophobic/hydrophilic properties (Bruzaud et al., 2015). First of all, the three studied surfaces were shown to present different physicochemical properties: SS was moderately hydrophilic and rough, containing possible crevices, G was hydrophilic and smooth and PS was hydrophobic and smooth. Enumerations combined with microscopic assays revealed that P. aeruginosa PAO1, previously characterized in terms of its hydrophilic and electron-donor properties (Bruzaud et al., 2015), was able to adhere about 14- and 4-fold more to SS and G than to PS. This phenotype could at least partly explain the weaker interactions between P. aeruginosa and the hydrophobic material. Although SS was less hydrophilic than G, maximum adhesion was observed using SS. These observations may have resulted from the rougher surface of SS, potentially trapping the cells (Faille et al., 2000).
Given these results, which indicate different adhesion phenotypes as a function of the material involved, proteomic studies were performed to determine bacterial physiology in response to adhesion to these three materials. One technical problem was to recover sufficient adhered bacteria to be able to perform a proteomic study (at least 109 CFU). Indeed, whereas the concentration of cells recovered from biofilms is clearly sufficient (Walker et al., 2005), under our conditions the concentration of recovered adherent bacteria was 8.4 × 106 CFU/cm2 with SS, 2.2 × 106 CFU/cm2 with G, and 6.0 × 105 CFU/cm2 with PS. Large surface areas of the materials (120 cm2 for SS, 500 cm2 for G, and 1,700 cm2 for PS per replicate) were therefore used for the adhesion assays in order to recover 109 CFU of bacteria. Furthermore, because this study was trying to determine the effects of the abiotic material on bacterial physiology, proper contact between the cells and surfaces was necessary to ensure that the majority of harvested cells had been affected. The adherent cells were observed using epifluorescence microscopy, which confirmed that most of the adherent cells were indeed in contact with the surface.
Total proteins were then extracted from P. aeruginosa that had been allowed to adhere to the three materials. Nano-LC MS/MS analyses enabled the identification of 930 proteins, representing about 16% of the open reading frames encoded by the P. aeruginosa genome. Among these, 70 proteins were identified as displaying a different abundance between at least two materials, representing about 7% of the total identified proteome. These observations were consistent with previous studies that had compared planktonic and biofilm cells and revealed similar quantities of dysregulated proteins (Vilain et al., 2004; Southey-Pillig et al., 2005). Remarkably, clustering of these proteins enabled the identification of three protein abundance patterns, each relevant to a specific material, suggestive of complex molecular mechanisms as a function of the relationships between cells and the surfaces of materials, as had previously been observed for bacteria adhering to agar surfaces of differing stiffness (Belas, 2014). The abundance of these 70 P. aeruginosa proteins varied according to the substrate, indicating that adhesion was probably complex and involved more than one protein (as shown by the three abundance patterns). A broad diversity of cellular functions was affected differently by the type of material, including mainly the “Membrane proteins,” “Transport of small molecules,” and “Metabolism” PseudoCAP functional classes. Taken together, these data clearly suggest that P. aeruginosa may develop a specific physiological state in response to such abiotic surfaces. To our knowledge, this is the first proteomic study to have shown such an impact of a substrate on the bacterial response, suggesting that P. aeruginosa is capable of the differential sensing of surfaces and modulates its physiology accordingly. This is the case for the sensing system responsible for type IV pili biogenesis (Whitchurch et al., 2004) that may be activated when the bacteria are grown on PS rather than other surfaces as shown with the increase on PS of two proteins belonging to type IV pili biogenesis pathway. This sensing system is involved in the surface-associated behaviors of P. aeruginosa as previously shown (Luo et al., 2015) but this role might be modulated by the nature of the surfaces.
We detected only one bacteriophage Pf1 protein with a very low level on PS when compared to SS and G. Although these proteins play a key role in host interactions and are largely involved in the formation of P. aeruginosa biofilms (Webb et al., 2004; Secor et al., 2015), their production seems not systematically enhanced when bacteria adhere to inert materials.
One of the most striking findings of this study was the variations in abundance of major porins and of porin folding chaperone Opr86, a member of the Omp85 family that is involved in OM biogenesis (Tashiro et al., 2008). Porins are proteins which form water-filled channels across the OM of Gram-negative bacteria and mediate the uptake or efflux of a number of compounds such as ions, small nutrient molecules, antibiotics, and large iron-siderophore complexes (Tamber et al., 2006). Porins are thus involved in controlling OM permeability and selectivity (Hancock and Farmer, 1993), and are also important actors in maintaining cell wall structure and homeostasis (Tashiro et al., 2008). OprF belongs to the OmpA family, which is a group of genetically related, surface-exposed porin proteins that are present in high-copy numbers in the OM of Gram-negative bacteria. OprF is a general porin that is non-covalently linked to the peptidoglycan layer, contributing thus to maintenance of the cell wall structure (Hancock and Carey, 1979). It is involved in adhesion, biofilm formation and virulence (Azghani et al., 2002; Sriramulu et al., 2005; Fito-Boncompte et al., 2011; Bouffartigues et al., 2015). OprD, OprG, and OprQ are involved in the uptake of small peptides, amino acids, or carboxylic acids (Hancock and Brinkman, 2002; Kucharska et al., 2015). OprE and OprC are induced in response to anaerobiosis and copper, respectively (Yoneyama and Nakae, 1996; Jaouen et al., 2006), and OprM is the OM component of the general efflux pump MexAB/OprM that is involved in general cell detoxification (Hancock and Brinkman, 2002). It should be noted that the abundance of these porins was found to be lower in bacteria that adhered to SS comparatively to PS and G, suggesting a reduction in OM permeability. A similar reduction in porin content has already been described in the OM of P. aeruginosa in response to a copper-induced stress (Teitzel et al., 2006), potentially suggesting a specific response to the metals composing the SS (iron, chromium, molybdenum, and nickel).
In addition to porins, other proteins associated with cell wall homeostasis were identified being produced at lower levels on SS than on the other two materials. This was the case for OprL, LptD, and PagL. OprL is homologous to the Escherichia coli Pal major OM lipoprotein that is involved in anchoring the peptidoglycan to the OM (Lloubès et al., 2001; Lim et al., 2012). LptD is an OM protein that is involved in the assembly of LPS in the OM outer leaflet (Srinivas et al., 2010). Lipid A 3O-deacylase PagL removes the 3-OH C10 or 3-OH C14 β-hydroxy fatty acid from lipid A, thus modifying the lipopolysaccharide structure (Ernst et al., 2006). Because lipid A is the biologically active component of LPS, its remodeling is known to alter the integrity of the bacterium's OM (King et al., 2009; Shah et al., 2013).
We therefore found on the one hand that numerous OM proteins were found at a lower abundance after adhesion to SS than to the other two surfaces. On the other hand, we were able to show that P. aeruginosa was able to adhere about 3- or 10-fold more to SS than to G or PS, respectively. This was quite surprising since it has been shown that some porins are involved in adhesion. Indeed, because they are exposed to the cell surface, some porins have been found to be involved in adhesion to biotic and/or abiotic surfaces. For example, this is the case for OprF (Azghani et al., 2002; Rebiere-Huet et al., 2002; Bodilis et al., 2004; Hemery et al., 2007; Fito-Boncompte et al., 2011), OprQ (Arhin and Boucher, 2010), and OprD (Paulsson et al., 2015). Taken together, these data enabled to highlight the importance of physicochemical properties to adhesion to solid surfaces. In E. coli, the low abundance of OM proteins including porins and the major lipoprotein Lpp, is correlated to a complex pathway called the general envelop stress response (ESR), which is under control of the master sigma alternative factor RpoE (Ades, 2004; Guo et al., 2014). RpoE maintains OM homeostasis by inducing synthesis of proteins involved in membrane repair and small regulatory RNAs (sRNAs) that down-regulate synthesis of abundant membrane porins and Lpp (Guo et al., 2014). Noticeably, it has been recently suggested that stress pathways, among which ESR, can play a role in surface sensing, leading to adhesion and biofilm formation (O'Toole and Wong, 2016). The periplasmic stress pathways Rcs (Regulator of capsule synthesis) and Cpx, which are members of the global ESR in E. coli, may indeed act as surface sensors with perturbation of the membrane(s), cell wall, and/or periplasmic space probably triggering these canonical stress systems, thereby perhaps alerting the microbe to cell-to-substratum contact (Otto and Silhavy, 2002; Morgenstein and Rather, 2012). In Enterobacteriaceae, specific Cpx pathway may prevent damages to the cell envelope caused by interactions of the cell with a surface (Otto and Silhavy, 2002). In P. aeruginosa, ESR is far less studied than in E. coli, and to date, only the two ECF sigma factors AlgU and SigX have been shown to be involved in maintaining the cell wall integrity (Wood and Ohman, 2009; Duchesne et al., 2013). Remarkably, the OM proteome of a SigX deficient P. aeruginosa strain was shown to present a much lower abundance in OM porins (Duchesne et al., 2013). Moreover, the conditions leading to cell wall stress among which membrane perturbations, were shown to increase SigX activity resulting in enhancing adhesion and biofilm formation (Bouffartigues et al., 2014, 2015). It may be conceivable that perception of an SS surface may occur via membrane or cell wall perturbations, leading to activation of the cell wall stress response of P. aeruginosa.
Conclusion
This study showed that under our experimental conditions, the abiotic surface impacted the protein production of adherent cells of P. aeruginosa PAO1. About 7% of the proteome thus detected underwent quantitative changes that involved all cell compartments and particularly outer membrane proteins. To our knowledge, no such global proteomic study has previously been performed at such an early stage in the surface biocontamination process. This study showed that biological and physiological functions can be modified by the surface involved, suggesting that P. aeruginosa was able to sense the material to which it was adhering. However, the molecular mechanisms that lead to this surface sensing are probably numerous and complex, as highlighted by the various physiological responses of P. aeruginosa to adhesion on SS, G, or PS surfaces, and confirming the hypothesis that P. aeruginosa is capable of sensing and responding to a surface via specific programmes. These different proteomes found in P. aeruginosa adhering to surfaces with various physicochemical properties may also result in part from the different adhesion forces that exist between the surfaces of bacteria and materials. This physiological reactivity of bacteria to materials needs to be taken into account in sensitive environments where pathogenic strains may present a health threat, such as in hospitals and in the food industry. Further studies now need to investigate whether contact with the material can induce unwanted phenotypes for example cross-resistance to antibacterial products.
Author contributions
Performed adhesion assays and protein extraction: MG, JB, and EB. Performed physicochemical characterizations: J-MH. Performed tandem mass spectrometry: AG. Performed protein quantification, heatmap representation, and statistical analyses: AG, AA and VM. Wrote the manuscript: all authors. Coordinated the study: M-NB-F.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We wish to thank Mikkel Klausen (University of Copenhagen) for the gift of the PAO1 strain and Victoria Hawken for English revision of the manuscript.
Glossary
Abbreviations
- SS
stainless steel
- G
glass
- PS
polystyrene
- SS/G
SS compared to G
- SS/PS
SS compared to PS
- PS/G
PS compared to G
- RT
room temperature
- Ra
arithmetic average roughness
- Rmax
maximum peak to valley height
- SDS
sodium dodecyl sulfate
- PseudoCAP
Pseudomonas Community Annotation Project
- OM
outer membrane
- OMP
outer membrane proteins
- PagL
lipid A deacylase
- QS
quorum sensing
- SadC
surface attachment defective
- Wsp
wrinkly spreader phenotype
- LPS
lipopolysaccharide.
Footnotes
Funding. Agence Nationale de la Recherche (ANR) funded this work (ANR-2010-CD2I-002, SANBACT project) and the grant for JB.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01465/full#supplementary-material
References
- Aaron S. D., Ferris W., Ramotar K., Vandemheen K., Chan F., Saginur R. (2002). Single and combination antibiotic susceptibilities of planktonic, adherent, and biofilm-grown Pseudomonas aeruginosa isolates cultured from sputa of adults with cystic fibrosis. J. Clin. Microbiol. 40, 4172–4179. 10.1128/JCM.40.11.4172-4179.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ades S. E. (2004). Control of the alternative sigma factor σE in Escherichia coli. Curr. Opin. Microbiol. 7, 157–162. 10.1016/j.mib.2004.02.010 [DOI] [PubMed] [Google Scholar]
- Arevalo-Ferro C., Hentzer M., Reil G., Görg A., Kjelleberg S., Givskov M., et al. (2003). Identification of quorum-sensing regulated proteins in the opportunistic pathogen Pseudomonas aeruginosa by proteomics. Environ. Microbiol. 5, 1350–1369. 10.1046/j.1462-2920.2003.00532.x [DOI] [PubMed] [Google Scholar]
- Arhin A., Boucher C. (2010). The outer membrane protein OprQ and adherence of Pseudomonas aeruginosa to human fibronectin. Microbiology 156, 1415–1423. 10.1099/mic.0.033472-0 [DOI] [PubMed] [Google Scholar]
- Azghani A. O., Idell S., Bains M., Hancock R. E. (2002). Pseudomonas aeruginosa outer membrane protein F is an adhesin in bacterial binding to lung epithelial cells in culture. Microb. Pathog. 33, 109–114. 10.1006/mpat.2002.0514 [DOI] [PubMed] [Google Scholar]
- Bedel S., Lepoittevin B., Costa L., Leroy O., Dragoe D., Bruzaud J., et al. (2015). Antibacterial poly(ethylene terephthalate) surfaces obtained from thymyl methacrylate polymerization. J. Polym. Sci. Part A Polym. Chem. 53, 1975–1985. 10.1002/pola.27647 [DOI] [Google Scholar]
- Belas R. (2014). Biofilms, flagella, and mechanosensing of surfaces by bacteria. Trends Microbiol. 22, 517–527. 10.1016/j.tim.2014.05.002 [DOI] [PubMed] [Google Scholar]
- Bellon-Fontaine M. N., Mozes N., Van der Mei H. C., Sjollema J., Cerf O., Rouxhet P. G., et al. (1990). A comparison of thermodynamic approaches to predict the adhesion of dairy microorganisms to solid substrata. Cell Biophys. 17, 93–106. 10.1007/BF02989805 [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300. [Google Scholar]
- Bodilis J., Calbrix R., Guérillon J., Mérieau A. (2004). Phylogenetic relationships between environmental and clinical isolates of Pseudomonas fluorescens and related species deduced from 16S rRNA gene and OprF protein sequences. Syst. Appl. Microbiol. 27, 93–108. 10.1078/0723-2020-00253 [DOI] [PubMed] [Google Scholar]
- Bos R., Van der Mei H. C., Busscher H. J. (1999). Physico-chemistry of initial microbial adhesive interactions–its mechanisms and methods for study. FEMS Microbiol. Rev. 23, 179–230. 10.1111/j.1574-6976.1999.tb00396.x [DOI] [PubMed] [Google Scholar]
- Bouffartigues E., Duchesne R., Bazire A., Simon M., Maillot O., Dufour A., et al. (2014). Sucrose favors Pseudomonas aeruginosa pellicle production through the extracytoplasmic function sigma factor SigX. FEMS Microbiol. Lett. 356, 193–200. 10.1111/1574-6968.12482 [DOI] [PubMed] [Google Scholar]
- Bouffartigues E., Moscoso J. A., Duchesne R., Rosay T., Fito-Boncompte L., Gicquel G., et al. (2015). The absence of the Pseudomonas aeruginosa OprF protein leads to increased biofilm formation through variation in c-di-GMP level. Front. Microbiol. 6:630. 10.3389/fmicb.2015.00630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulangé-Petermann L., Rault J., Bellon-Fontaine M.-N. (1997). Adhesion of Streptococcus thermophilus to stainless steel with different surface topography and roughness. Biofouling 11, 201–216. 10.1080/08927019709378331 [DOI] [Google Scholar]
- Bruzaud J., Tarrade J., Coudreuse A., Canette A., Herry J.-M., Taffin de Givenchy E., et al. (2015). Flagella but not type IV pili are involved in the initial adhesion of Pseudomonas aeruginosa PAO1 to hydrophobic or superhydrophobic surfaces. Colloids Surf. B 131, 59–66. 10.1016/j.colsurfb.2015.04.036 [DOI] [PubMed] [Google Scholar]
- Busscher H. J., van der Mei H. C. (2012). How do bacteria know they are on a surface and regulate their response to an adhering state? PLoS Pathog. 8:e1002440. 10.1371/journal.ppat.1002440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton J. W., Lewandowski Z., Caldwell D. E., Korber D. R., Lappin-Scott H. M. (1995). Microbial biofilms. Annu. Rev. Microbiol. 49, 711–745. 10.1146/annurev.mi.49.100195.003431 [DOI] [PubMed] [Google Scholar]
- Côté R. G., Griss J., Dianes J. A., Wang R., Wright J. C., van den Toorn H. W. P., et al. (2012). The PRoteomics IDEntifications (PRIDE) Converter 2 framework: an improved suite of tools to facilitate data submission to the PRIDE database and the ProteomeXchange consortium. Mol. Cell. Proteomics 11, 1682–1689. 10.1074/mcp.O112.021543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig R., Beavis R. C. (2004). Tandem: matching proteins with tandem mass spectra. Bioinformatics 20, 1466–1467. 10.1093/bioinformatics/bth092 [DOI] [PubMed] [Google Scholar]
- Drenkard E., Ausubel F. M. (2002). Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature 416, 740–743. 10.1038/416740a [DOI] [PubMed] [Google Scholar]
- Duchesne R., Bouffartigues E., Oxaran V., Maillot O., Bénard M., Feuilloley M. G., et al. (2013). Proteomic approach of SigX function in Pseudomonas aeruginosa outer membrane composition. J. Proteomics 94, 451–459. 10.1016/j.jprot.2013.10.022 [DOI] [PubMed] [Google Scholar]
- Dunne W. M. (2002). Bacterial adhesion: seen any good biofilms lately? Clin. Microbiol. Rev. 15, 155–166. 10.1128/CMR.15.2.155-166.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst R. K., Adams K. N., Moskowitz S. M., Kraig G. M., Kawasaki K., Stead C. M., et al. (2006). The Pseudomonas aeruginosa lipid A deacylase: selection for expression and loss within the cystic fibrosis airway. J. Bacteriol. 188, 191–201. 10.1128/JB.188.1.191-201.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faille C., Membre J. M., Ussier J. P., Bellon-Fontaine M.-N., Carpentier B., Laroche M., et al. (2000). Influence of physicochemical properties on the hygienic status of stainless steel with various finishes. Biofouling 15, 261–274. 10.1080/08927010009386317 [DOI] [Google Scholar]
- Fito-Boncompte L., Chapalain A., Bouffartigues E., Chaker H., Lesouhaitier O., Gicquel G., et al. (2011). Full virulence of Pseudomonas aeruginosa requires OprF. Infect. Immun. 79, 1176–1186. 10.1128/IAI.00850-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadenne V., Lebrun L., Jouenne T., Thebault P. (2013). Antiadhesive activity of ulvan polysaccharides covalently immobilized onto titanium surface. Colloids Surf. B 112, 229–236. 10.1016/j.colsurfb.2013.07.061 [DOI] [PubMed] [Google Scholar]
- Guégan C., Garderes J., Le Pennec G., Gaillard F., Fay F., Linossier I., et al. (2013). Alteration of bacterial adhesion induced by the substrate stiffness. Colloids Surf. B 114, 193–200. 10.1016/j.colsurfb.2013.10.010 [DOI] [PubMed] [Google Scholar]
- Guo M. S., Updegrove T. B., Gogol E. B., Shabalina S. A., Gross C. A., Storz G. (2014). MicL, a new σE-dependent sRNA, combats envelope stress by repressing synthesis of Lpp, the major outer membrane lipoprotein. Genes Dev. 28, 1620–1634. 10.1101/gad.243485.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock R. E., Brinkman F. S. (2002). Function of Pseudomonas porins in uptake and efflux. Annu. Rev. Microbiol. 56, 17–38. 10.1146/annurev.micro.56.012302.160310 [DOI] [PubMed] [Google Scholar]
- Hancock R. E., Carey W. A. M. (1979). Outer membrane of Pseudomonas aeruginosa: heat- 2-mercaptoethanol-modifiable proteins. J. Bacteriol. 140, 902–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock R. E., Farmer S. W. (1993). Mechanism of uptake of deglucoteicoplanin amide derivatives across outer membranes of Escherichia coli and Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 37, 453–456. 10.1128/AAC.37.3.453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemery G., Chevalier S., Bellon-Fontaine M.-N., Haras D., Orange N. (2007). Growth temperature and OprF porin affect cell surface physicochemical properties and adhesive capacities of Pseudomonas fluorescens MF37. J. Ind. Microbiol. Biotechnol. 34, 49–54. 10.1007/s10295-006-0160-x [DOI] [PubMed] [Google Scholar]
- Jaouen T., Coquet L., Marvin-Guy L., Orange N., Chevalier S., Dé E. (2006). Functional characterization of Pseudomonas fluorescens OprE and OprQ membrane proteins. Biochem. Biophys. Res. Commun. 346, 1048–1052. 10.1016/j.bbrc.2006.06.013 [DOI] [PubMed] [Google Scholar]
- Kamgang J. O., Briandet R., Herry J.-M., Brisset J.-L., Naïtali M. (2007). Destruction of planktonic, adherent and biofilm cells of Staphylococcus epidermidis using a gliding discharge in humid air. J. Appl. Microbiol. 103, 621–628. 10.1111/j.1365-2672.2007.03286.x [DOI] [PubMed] [Google Scholar]
- King J. D., Kocíncová D., Westman E. L., Lam J. S. (2009). Review: lipopolysaccharide biosynthesis in Pseudomonas aeruginosa. Innate Immun. 15, 261–312. 10.1177/1753425909106436 [DOI] [PubMed] [Google Scholar]
- Kucharska I., Seelheim P., Edrington T., Liang B., Tamm L. K. (2015). OprG harnesses the dynamics of its extracellular loops to transport small amino acids across the outer membrane of Pseudomonas aeruginosa. Structure 23, 2234–2245. 10.1016/j.str.2015.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langella O., Valot B., Balliau T., Blein-Nicolas M., Bonhomme L., Zivy M. (2017). X!TandemPipeline: a tool to manage sequence redundancy for protein inference and phosphosite identification. J. Proteome Res. 16, 494–503. 10.1021/acs.jproteome.6b00632 [DOI] [PubMed] [Google Scholar]
- Lazzaroni J. C., Germon P., Ray M. C., Vianney A. (1999). The Tol proteins of Escherichia coli and their involvement in the uptake of biomolecules and outer membrane stability. FEMS Microbiol. Lett. 177, 191–197. 10.1111/j.1574-6968.1999.tb13731.x [DOI] [PubMed] [Google Scholar]
- Leibovitz A., Dan M., Zinger J., Carmeli Y., Habot B., Segal R. (2003). Pseudomonas aeruginosa and the oropharyngeal ecosystem of tube-fed patients. Emerg. Infect. Dis. 9, 956–959. 10.3201/eid0908.030054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C. K., Hassan K. A., Tetu S. G., Loper J. E., Paulsen T. (2012). The effect of iron limitation on the transcriptome and proteome of Pseudomonas aeruginosa Pf-5. PLoS ONE 7:e39139 10.1371/journal.pone.0039139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloubès R., Cascales E., Walburger A., Bouveret E., Lazdunski C., Bernadac A., et al. (2001). The Tol-Pal proteins of the Escherichia coli cell envelope: an energized system required for outer membrane integrity? Res. Microbiol. 152, 523–529. 10.1016/S0923-2508(01)01226-8 [DOI] [PubMed] [Google Scholar]
- Luo Y., Zhao K., Baker A. E., Kuchma S. L., Coggan K. A., Wolfgang M. C., et al. (2015). A hierarchical cascade of second messengers regulates Pseudomonas aeruginosa surface behaviors. MBio 6, e02456–e02414. 10.1128/mBio.02456-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenstein R. M., Rather P. N. (2012). Role of the Umo proteins and the Rcs phosphorelay in the swarming motility of the wild type and an O-antigen (waaL) mutant of Proteus mirabilis. J. Bacteriol. 194, 669–676. 10.1128/JB.06047-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nejadnik M. R., van der Mei H. C., Norde W., Busscher H. J. (2008). Bacterial adhesion and growth on a polymer brush-coating. Biomaterials 29, 4117–4121. 10.1016/j.biomaterials.2008.07.014 [DOI] [PubMed] [Google Scholar]
- O'Toole G. A., Wong G. C. (2016). Sensational biofilms: surface sensing in bacteria. Curr. Opin. Microbiol. 30, 139–146. 10.1016/j.mib.2016.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto K., Silhavy T. J. (2002). Surface sensing and adhesion of Escherichia coli controlled by the Cpx-signaling pathway. Proc. Natl. Acad. Sci. U.S.A. 99, 2287–2292. 10.1073/pnas.042521699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrauchan M. A., Sarkisova S. A., Franklin M. J. (2007). Strain-specific proteome responses of Pseudomonas aeruginosa to biofilm-associated growth and to calcium. Microbiology 153, 3838–3851. 10.1099/mic.0.2007/010371-0 [DOI] [PubMed] [Google Scholar]
- Paulsson M., Singh B., Al-Jubair T., Su Y. C., Høiby N., Riesbeck K. (2015). Identification of outer membrane porin D as a vitronectin-binding factor in cystic fibrosis clinical isolates of Pseudomonas aeruginosa. J. Cyst. Fibros. 14, 600–607. 10.1016/j.jcf.2015.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poncin-Epaillard F., Herry J.-M., Marmey P., Legeay G., Debarnot D., Bellon-Fontaine M.-N. (2013). Elaboration of highly hydrophobic polymeric surface—a potential strategy to reduce the adhesion of pathogenic bacteria? Mater. Sci. Eng. C Mater. Biol. Appl. 33, 1152–1161. 10.1016/j.msec.2012.12.020 [DOI] [PubMed] [Google Scholar]
- Rasband W. S. (1997). ImageJ. Bethesda, MD: US National Institutes of Health. [Google Scholar]
- Rebiere-Huet J., Guérillon J., Pimenta A. L., Di Martino P., Orange N., Hulen C. (2002). Porins of Pseudomonas fluorescens MFO as fibronectin-binding proteins. FEMS Microbiol. Lett. 215, 121–126. 10.1111/j.1574-6968.2002.tb11380.x [DOI] [PubMed] [Google Scholar]
- Robert-Genthon M., Casabona M. G., Neves D., Couté Y., Cicéron F., Elsen S., et al. (2013). Unique features of a Pseudomonas aeruginosa α2-macroglobulin homolog. MBio 4, e00309–e00313. 10.1128/mBio.00309-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer K., Camper A. K., Ehrlich G. D., Costerton J. W., Davies D. G. (2002). Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 184, 1140–1154. 10.1128/jb.184.4.1140-1154.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C. A., Rasband W. S., Eliceiri K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secor P. R., Sweere J. M., Michaels L. A., Malkovskiy A. V., Lazzareschi D., Katznelson E., et al. (2015). Filamentous bacteriophage promote biofilm assembly and function. Cell Host and Microbe 18, 549–559. 10.1016/j.chom.2015.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah N. R., Albitar-Nehme S., Kim E., Marr N., Novikov A., Caroff M., et al. (2013). Minor modifications to the phosphate groups and the C3′ acyl chain length of lipid A in two Bordetella pertussis strains, BP338 and 18-323, independently affect Toll-like receptor 4 protein activation. J. Biol. Chem. 288, 11751–11760. 10.1074/jbc.M112.434365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A., Tomas H., Havli J., Olsen J. V., Mann M. (2006). In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1, 2856–2860. 10.1038/nprot.2006.468 [DOI] [PubMed] [Google Scholar]
- Siryaporn A., Kuchma S. L., O'Toole G. A., Gitai Z. (2014). Surface attachment induces Pseudomonas aeruginosa virulence. Proc. Natl. Acad. Sci. U.S.A. 111, 16860–16865. 10.1073/pnas.1415712111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song F., Koo H., Ren D. (2015). Effects of material properties on bacterial adhesion and biofilm formation. J. Dent. Res. 94, 1027–1034. 10.1177/0022034515587690 [DOI] [PubMed] [Google Scholar]
- Southey-Pillig C. J., Davies D. G., Sauer K. (2005). Characterization of temporal protein production in Pseudomonas aeruginosa biofilms. J. Bacteriol. 187, 8114–8126. 10.1128/JB.187.23.8114-8126.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas N., Jetter P., Ueberbacher B. J., Werneburg M., Zerbe K., Steinmann J., et al. (2010). Peptidomimetic antibiotics targe outer-membrane biogenesis in Pseudomonas aeruginosa. Science 327, 1010–1013. 10.1126/science.1182749 [DOI] [PubMed] [Google Scholar]
- Sriramulu D. D., Lünsdorf H., Lam J. S., Römling U. (2005). Microcolony formation: a novel biofilm model of Pseudomonas aeruginosa for the cystic fibrosis lung. J. Med. Microbiol. 54, 667–676. 10.1099/jmm.0.45969-0 [DOI] [PubMed] [Google Scholar]
- Stewart P. S., Costerton J. W. (2001). Antibiotic resistance of bacteria in biofilms. Lancet 358, 135–138. 10.1016/S0140-6736(01)05321-1 [DOI] [PubMed] [Google Scholar]
- Tamber S., Ochs M. M., Hancock R. E. W., Robert E. W. (2006). Role of the novel OprD family of porins in nutrient uptake in Pseudomonas aeruginosa. J. Bacteriol. 188:45–54. 10.1128/JB.188.1.45-54.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro Y., Nomura N., Nakao R., Senpuku H., Kariyama R., Kumon H., et al. (2008). Opr86 is essential for viability and is a potential candidate for a protective antigen against biofilm formation by Pseudomonas aeruginosa. J. Bacteriol. 190, 3969–3978. 10.1128/JB.02004-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teitzel G. M., Geddie A., De Long S. K., Kirisits M. J., Whiteley M., Parsek M. R. (2006). Survival and growth in the presence of elevated copper: transcriptional profiling of copper-stressed Pseudomonas aeruginosa. J. Bacteriol. 188, 7242–7256. 10.1128/JB.00837-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada A., Yuasa A., Kushimoto T., Tsuneda S., Katakai A., Tamada M. (2006). Bacterial adhesion to and viability on positively charged polymer surfaces. Microbiology 152, 3575–3583. 10.1099/mic.0.28881-0 [DOI] [PubMed] [Google Scholar]
- Vilain S., Cosette P., Hubert M., Lange C., Junter G.-A., Jouenne T. (2004). Comparative proteomic analysis of planktonic and immobilized Pseudomonas aeruginosa cells: a multivariate statistical approach. Anal. Biochem. 329, 120–130. 10.1016/j.ab.2004.02.014 [DOI] [PubMed] [Google Scholar]
- Vizcaíno J. A., Csordas A., del-Toro N., Dianes J. A., Griss J., Lavidas I., et al. (2016). 2016 update of the PRIDE database and related tools. Nucleic Acids Res. 44, 447–456. 10.1093/nar/gkv1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaíno J. A., Deutsch E. W., Wang R., Csordas A., Reisinger F., Ríos D., et al. (2014). ProteomeXchange provides globally co-ordinated proteomics data submission and dissemination. Nat. Biotechnol. 30, 223–226. 10.1038/nbt.2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waite R. D., Papakonstantinopoulou A., Littler E., Curtis M. A. (2005). Transcriptome analysis of Pseudomonas aeruginosa growth: comparison of gene expression in planktonic cultures and developing and mature biofilms. J. Bacteriol. 187, 6571–6576. 10.1128/JB.187.18.6571-6576.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker T. S., Tomlin K. L., Worthen G. S., Poch K. R., Lieber J. G., Saavedra M. T., et al. (2005). Enhanced Pseudomonas aeruginosa biofilm development mediated by human neutrophils. Infect. Immun. 73, 3693–3701. 10.1128/IAI.73.6.3693-3701.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb J. S., Lau M., Kjelleberg S. (2004). Bacteriophage and phenotypic variation in Pseudomonas aeruginosa biofilm development. J. Bacteriol. 186, 8066–8073. 10.1128/JB.186.23.8066-8073.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitchurch C. B., Leech A. J., Young M. D., Kennedy D., Sargent J. L., Bertrand J. J., et al. (2004). Characterization of a complex chemosensory signal transduction system which controls twitching motility in Pseudomonas aeruginosa. Mol. Microbiol. 52, 873–893. 10.1111/j.1365-2958.2004.04026.x [DOI] [PubMed] [Google Scholar]
- Whiteley M., Bangera M. G., Bumgarner R. E., Parsek M. R., Teitzel G. M., Lory S., et al. (2001). Gene expression in Pseudomonas aeruginosa biofilms. Nature 413, 860–864. 10.1038/35101627 [DOI] [PubMed] [Google Scholar]
- Winsor G. L., Griffiths E. J., Lo R., Dhillon B. K., Shay J. A., Brinkman F. S. (2016). Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res. 44, D646–D653. 10.1093/nar/gkv1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood L. F., Ohman D. E. (2009). Use of cell wall stress to characterize σ22 (AlgT/U) activation by regulated proteolysis and its regulon in Pseudomonas aeruginosa. Mol. Microbiol. 72, 183–201. 10.1111/j.1365-2958.2009.06635.x [DOI] [PubMed] [Google Scholar]
- Yoneyama H., Nakae T. (1996). Protein C (OprC) of the outer membrane of Pseudomonas aeruginosa is a copper-regulated channel protein. Microbiology 142, 2137–2144. 10.1099/13500872-142-8-2137 [DOI] [PubMed] [Google Scholar]
- Yoon S. S., Hennigan R. F., Hilliard G. M., Ochsner U. A., Parvatiyar K., Kamani M. C., et al. (2002). Pseudomonas aeruginosa anaerobic respiration in biofilms: relationships to cystic fibrosis pathogenesis. Dev. Cell. 3, 593–603. 10.1016/S1534-5807(02)00295-2 [DOI] [PubMed] [Google Scholar]
- Yuan S., Wan D., Liang B., Pehkonen S. O., Ting Y. P., Neoh K. G., et al. (2011). Lysozyme-coupled poly(poly(ethylene glycol) methacrylate)-stainless steel hybrids and their antifouling and antibacterial surfaces. Langmuir 27, 2761–2774. 10.1021/la104442f [DOI] [PubMed] [Google Scholar]
- Zegans M. E., Shanks R. M. Q., O'Toole G. A. (2005). Bacterial biofilms and ocular infections. Ocul. Surf. 3, 73–80. 10.1016/S1542-0124(12)70155-6 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.