Abstract
Hypoxia-inducible factor 1 (HIF-1) activates the transcription of genes encoding proteins that enable cells to adapt to reduced O2 availability. Proteins encoded by HIF-1 target genes play a central role in mediating physiological processes that are dysregulated in cancer and heart disease. These diseases are also characterized by increased production of cyclic adenosine monophosphate (cAMP), the allosteric activator of cAMP-dependent protein kinase A (PKA). Using GST-pulldown, coimmunoprecipitation and mass spectrometry analyses, we demonstrated that PKA interacts with HIF-1α in HeLa cervical carcinoma cells and rat cardiomyocytes. PKA phosphorylated Thr63 and Ser692 on HIF-1α in vitro and enhanced HIF transcriptional activity and target gene expression in HeLa cells and rat cardiomyocytes. PKA inhibited the proteasomal degradation of HIF-1α in an O2-independent manner that required the phosphorylation of Thr63 and Ser692 and was not affected by prolyl hydroxylation. PKA also stimulated the binding of the coactivator p300 to HIF-1α to enhance its transcriptional activity and counteracted the inhibitory effect of asparaginyl hydroxylation on the association of p300 with HIF-1α. Furthermore, increased cAMP concentrations enhanced the expression of HIF target genes encoding CD39 and CD73, which are enzymes that convert extracellular ATP to adenosine, a molecule that enhances tumor immunosuppression and reduces heart rate and contractility. These data link stimuli that promote cAMP signaling, HIF-1α-dependent changes in gene expression, and increased adenosine, all of which contribute to the pathophysiology of cancer and heart disease.
INTRODUCTION
Hypoxia-inducible factor 1 (HIF-1) functions as a master regulator of cellular and systemic O2 homeostasis in all metazoan species by activating the transcription of target genes that are critical for adaptive responses to hypoxia. In mammals, proteins encoded by HIF-1 target genes stimulate vascular remodeling to increase blood flow in ischemic tissue and mediate the beneficial effects of ischemic preconditioning (1). HIF-1 also functions as a critical cardioprotective component of adaptive responses to left ventricular pressure overload. Cancer cells in hypoxic tumor microenvironments exploit adaptive responses that are mediated by HIF-1 to promote vascularization, metabolic reprogramming, immune evasion, cancer stem cell maintenance, invasion, and metastasis (1).
Protein kinase A (PKA) is a primary effector of cAMP-responsive cellular processes. Numerous intercellular messengers, such as catecholamines and adenosine, bind to G-protein coupled receptors (GPCRs), such as β1- and β2-adrenergic receptors or A2A and A2B adenosine receptors, respectively. These receptors are coupled to G-proteins that promote conversion of ATP into cAMP by activating transmembrane adenylyl cyclases (2). Increased intracellular cAMP concentrations activate PKA. The adenosine and β-adrenergic signaling pathways are dysregulated in cancer and heart disease, contributing to tumor progression and cardiac hypertrophy (2, 3, 4). Thus, there is considerable overlap in the physiologic and pathologic processes that are mediated by PKA and HIF-1.
O2-regulated HIF-1α and constitutively-expressed HIF-1β subunits comprise the functional HIF-1 heterodimer (5). Under normoxic conditions, prolyl-4-hydroxylase domain proteins (PHDs) hydroxylate Pro402 and Pro564, which is required for binding of the von Hippel-Lindau protein (VHL) to HIF-1α (1). VHL recruits an E3-ligase complex that ubiquitinates HIF-1α and targets it for proteasomal degradation. Hydroxylation of Asn803 by factor inhibiting HIF-1 (FIH-1) blocks binding of the co-activator p300 and transactivation by HIF-1α. Because PHDs and FIH-1 utilize O2 as a catalytic substrate, hypoxia inhibits hydroxylation, leading to stabilization of HIF-1α protein, dimerization with HIF-1β, and target gene transactivation (1).
PKA is a ubiquitously distributed Ser- and Thr-kinase that is present as an inactive heterotetramer composed of two regulatory and two catalytic subunits. cAMP binding to the regulatory subunits induces conformational changes that release active catalytic subunit monomers (6). There are four regulatory (R1a, R1b, R2a, and R2b) and catalytic (Ca, Cb, Cg and Prkx) subunits (7). Heterotetramers containing two different regulatory subunits are rare and the predominant isoforms are designated type 1 or 2, based on the presence of two R1 or R2 subunits, respectively. Type 1 isoforms are found in the cytosol, whereas type 2 isoforms are generally localized to organelles, such as the mitochondria and sarcoplasmic reticulum of muscle cells (8). The catalytic subunit monomers phosphorylate a multitude of targets in the cytosol, at subcellular locations, and in the nucleus, including the transcription factor cAMP-response element (CRE) binding protein (CREB) (6). Given the critical involvement of both PKA and HIF-1 in the pathogenesis of cancer and heart disease, we investigated whether PKA could directly interact with and regulate HIF-1 function in human HeLa cervical carcinoma cells and neonatal rat cardiomyocytes (NRCMs).
RESULTS
PKA is a HIF-1α-interacting protein
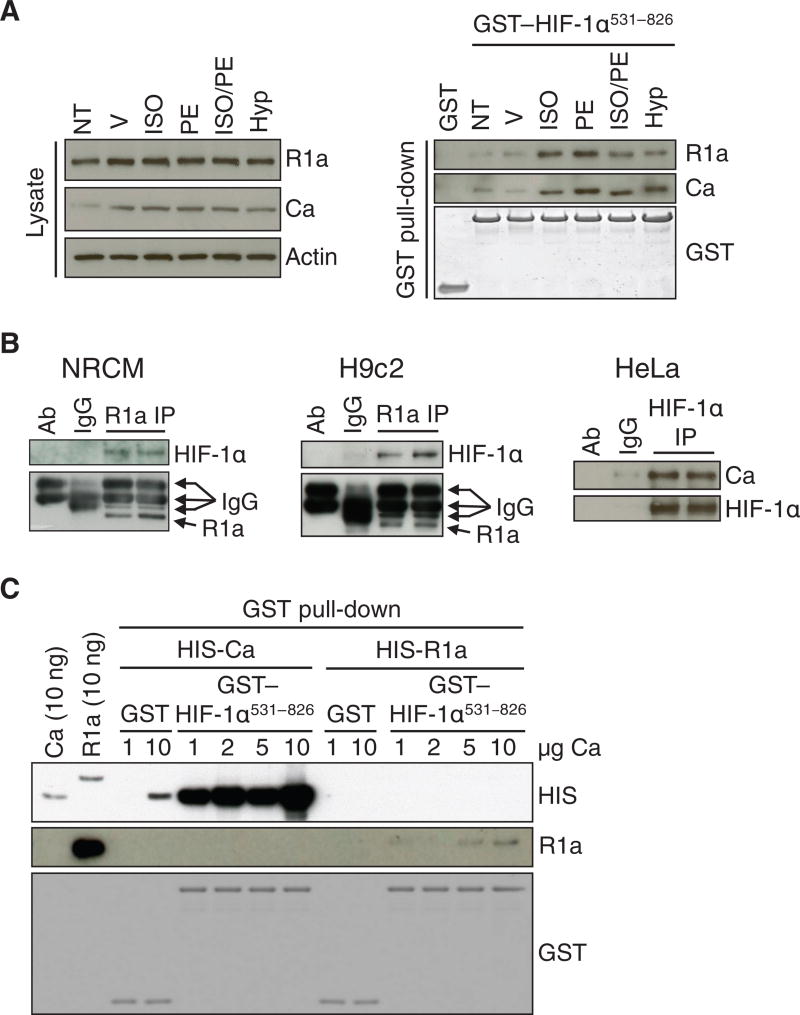
To investigate if PKA directly interacted with HIF-1α, we performed pull-down, immunoprecipitation, and in vitro binding assays. Both the R1a and Ca subunits of PKA were present in lysates of H9c2 rat cardiomyoblast cells and bound to GST-HIF-1α531-826, which is a fusion protein consisting of GST and amino acid residues 531-826 of HIF-1α, but not to GST alone (Fig. 1A). Binding of the R1a and Ca subunits of PKA to GST-HIF-1α531-826 was increased in lysates of H9c2 rat cardiomyoblast cells exposed to the β-adrenergic agonists isoproterenol, phenylephrine, or both, or to hypoxia (1% O2), compared to lysates isolated from untreated or vehicle-treated cells (Fig. 1A). An unbiased approach using label-free quantitative proteomic analyses of NRCM protein lysates purified over GST or GST-HIF-1α531-826 further demonstrated a higher abundance of the R1a subunit of PKA bound to GST-HIF-1α531-826 compared to GST alone (Data File 1). Coimmunoprecipitation experiments confirmed that endogenous HIF-1α interacted with R1a or Ca in NRCMs, H9c2 cells and HeLa cervical carcinoma cells (Fig. 1B). Recombinant protein binding assays demonstrated that purified recombinant Ca had a higher affinity for HIF-1α531-826 than purified R1a (Fig. 1C), suggesting that the interaction between HIF-1α and R1a observed in GST-pulldown and coimmunoprecipitation assays is due to PKA holoenzyme forming a complex with HIF-1α through direct binding of the Ca subunit to HIF-1α.
Fig. 1. PKA interacts with HIF-1α.
(A) Immunoblot assays of lysates (left) and proteins pulled down by GST or GST-HIF-1α531-826 from the same lysates (right), which were prepared from H9c2 cells subjected to no treatment (NT); treated with vehicle (V), ISO, PE, or ISO/PE; or exposed to 1% O2 (Hyp) (n ≥ 3 independent experiments). (B) Immunoprecipitation (IP) of R1a from NRCMs (left panel; n = 2 biological replicates, as shown) or H9c2 cells (middle panel; n = 2 biological replicates, as shown) and immunoprecipitation of HIF-1α from HeLa cells (right panel; n = 2 biological replicates, as shown) were performed, followed by immunoblot assays using antibodies against HIF-1α, R1a, or Ca. Primary antibody alone (ab) and lysate incubated with non-specific IgG (IgG) were included as negative controls. (C) Pull-down assays were performed using GST or GST-HIF-1α531-826 and recombinant His-tagged Ca or R1a, followed by immunoblot assays using the indicated antibodies (n = 2 independent experiments).
PKA stimulates HIF-1 transcriptional activity and inhibits HIF-1α degradation
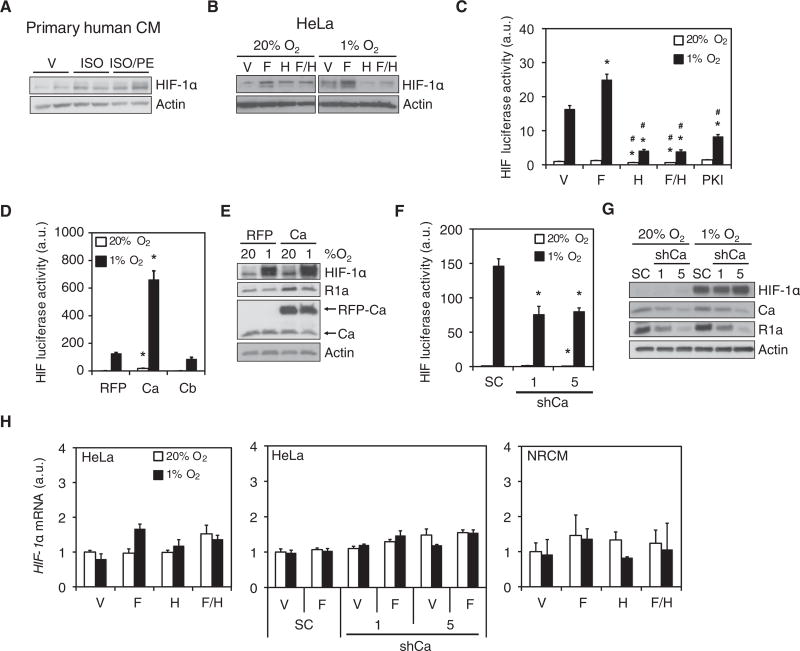
To determine if PKA regulates HIF-1, we assessed the effect of pharmacological activators and inhibitors of PKA on HIF-1α abundance and HIF-1-dependent transcription. Treatment of primary human cardiomyocytes under normoxic conditions (20% O2) with isoproterenol, or isoproterenol and phenylephrine, increased HIF-1α protein abundance (Fig. 2A). Similar effects were observed in HeLa cells treated with the adenylyl cyclase activator forskolin under normoxic or hypoxic conditions (Fig. 2B). Treatment of HeLa cells with the PKA inhibitor H89 blocked both forskolin- and hypoxia-induced increases in HIF-1α abundance (Fig. 2B). To test whether increased HIF-1α abundance led to increased HIF-1 transcriptional activity, we cotransfected HeLa cells with the HIF-1-dependent reporter plasmid p2.1, which contains a hypoxia response element upstream of SV40 promoter and firefly luciferase coding sequences (9), and pSV-Renilla, a control reporter containing the SV40 promoter upstream of Renilla luciferase coding sequences. The ratio of firefly:Renilla luciferase activity is a measure of HIF-1 transcriptional activity. Forskolin treatment increased HIF-1 transcriptional activity under hypoxic conditions, whereas HIF-1 activity was suppressed by H89 or myristoylated 14-22 amide (PKI), a cell-permeable R1a peptide-based PKA inhibitor (Fig. 2C). Thus, cAMP stimulates HIF-1 transcriptional activity and increases HIF-1α protein abundance through activation of PKA.
Fig. 2. PKA increases HIF-1α protein abundance and transcriptional activity.
(A and B) Immunoblot assays were performed using lysates from: primary human left-ventricle cardiomyocytes (CMs) exposed to vehicle (V), isoproterenol (ISO), or isoproterenol + phenylephrine (ISO/PE) (A; n = 2 biological replicates, as shown); or HeLa cells exposed to V, forskolin (F), H89 (H), or forskolin + H89 (F/H) at 20% or 1% O2 (B; representative of n = 3 independent experiments). (C) HeLa cells were co-transfected with p2.1 and pSV-Renilla, exposed to V, F, H, F/H, or PKI at 20% or 1% O2 and the ratio of firefly:Renilla luciferase was determined (HIF luciferase activity; mean ± SD, n = 9 biological replicates from 3 independent experiments). *p < 0.05 compared to V at same % O2; #p < 0.05 compared to forskolin at same % O2. (D) HeLa cells were co-transfected with p2.1, pSV-Renilla and vector encoding red fluorescent protein (RFP), RFP-Ca fusion protein, or HA-tagged Cb, exposed to 20% or 1% O2, and HIF luciferase activity was determined (mean ± SD, n = 9 biological replicates from 3 independent experiments). *p < 0.05 compared to RFP at the same % O2. (E) Immunoblot assays were performed using lysates of HeLa cells expressing RFP or RFP-Ca and exposed to 20% or 1% O2 (n = 3 independent experiments). (F and G) HIF-dependent luciferase reporter assays (F; mean ± SD, n = 9 biological replicates from 3 independent experiments; *p < 0.05 compared to SC at same % O2) and immunoblot assays (G; n = 3 independent experiments) were performed using lysates of HeLa cells expressing a scrambled control shRNA (SC) or either of two shRNAs targeting Ca (shCa-1 or shCa-5) and were exposed to 20% or 1% O2. (H) HIF-1α mRNA abundance was determined by RT-qPCR in parental HeLa cells (left panel), SC, shCa-1 and shCa-5 HeLa subclones (middle panel) or NRCMs (right panel), which were exposed to 20% or 1% O2 (mean ± SD, n = 6 biological replicates from 3 independent experiments; none of the differences were statistically significant). Data in C, D, F and H are graphed as fold change from respective controls at 20% O2. The Mann-Whitney U test was used to assess statistical significance for all data in this figure.
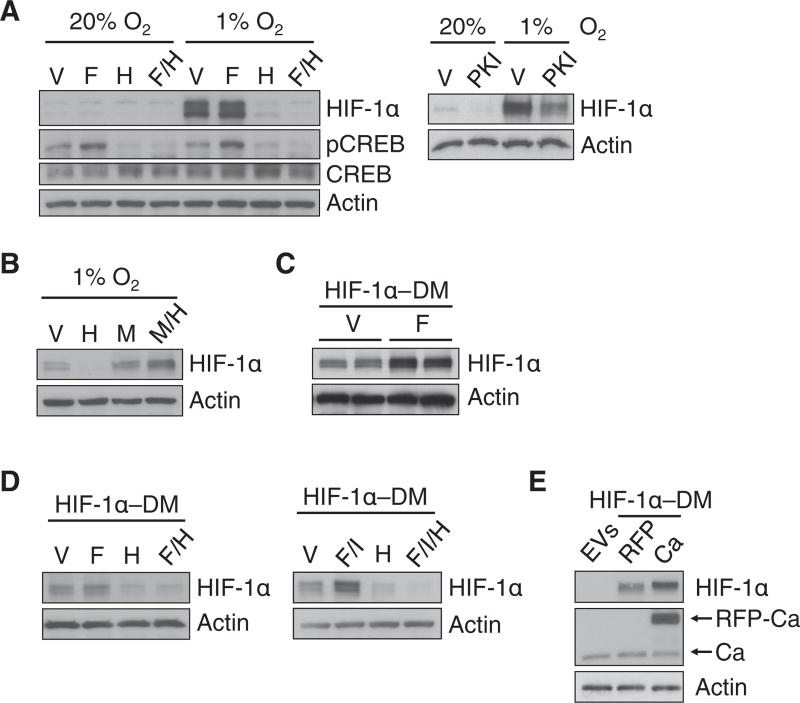
To confirm the pharmacological data, we analyzed the effect of transient overexpression or stable knockdown of PKA catalytic subunits on HIF-1 transcriptional activity and HIF-1α protein abundance. In HeLa cells under normoxic or hypoxic conditions, overexpression of the Ca subunit, but not Cb, increased HIF-1 transcriptional activity (Fig. 2D) and Ca overexpression enhanced HIF-1α protein abundance (Fig. 2E). Stable knockdown of Ca in HeLa cells decreased HIF-1 transcriptional activity (Fig. 2F), but to a lesser extent than H89 or PKI administration. Ca knockdown did not appear to alter the hypoxic induction of HIF-1α protein abundance (Fig. 2G). Ca knockdown induced a decrease in R1a, which together with incomplete knockdown of Ca (Fig. 2G), may explain the differences between pharmacologic and genetic PKA loss-of-function. HIF-1α mRNA abundance in HeLa cells or NRCMs was not affected by pharmacologic or genetic manipulation of PKA (Fig. 2H), suggesting that PKA may enhance HIF-1α protein stability. Indeed, H89 and PKI inhibited the hypoxia-induced increase in HIF-1α protein abundance (Fig. 3A), and the effect of H89 was reversed by the proteasome inhibitor MG132 (Fig. 3B). PKA mediates phosphorylation of CREB at Ser133, which was stimulated by forskolin and inhibited by H89, confirming the efficacy of these agents (Fig. 3A). Thus, PKA promotes HIF-1 transcriptional activity in part by inhibiting the proteasomal degradation of HIF-1α.
Fig. 3. PKA inhibits the proteasomal degradation of HIF-1α independently of prolyl hydroxylation.
(A and B) HeLa cells were exposed to: vehicle (V), forskolin (F), H89 (H), forskolin + H89 (F/H), or PKI at 20% or 1% O2 (A); or V, H, MG132 (M) or MG132 + H89 (M/H) at 1% O2 (B). (C) HEK293T cells expressing FLAG-tagged P402A/P564A double-mutant HIF-1α (HIF-1α-DM) were exposed to V or F. For each condition, two biological replicates are shown. (D) HeLa cells expressing FLAG-tagged HIF-1α-DM were exposed to V, F, H, F/H, forskolin + IBMX (F/I); or forskolin + IBMX + H89 (F/I/H). (E) HeLa cells transfected with empty vectors (EVs), or FLAG-tagged HIF-1α-DM and RFP or RFP-Ca. Data in all panels are representative of at least 3 independent experiments unless otherwise indicated.
To determine if PKA-induced HIF-1α stabilization involves prolyl hydroxylation, we analyzed the effect of forskolin, H89, 3-isobutyl-1-methylxanthine (IBMX, an inhibitor of phosphodiesterases that hydrolyze cAMP to AMP), or Ca overexpression on the abundance of P402A/P564A double-mutant HIF-1α (HIF-1α-DM), which cannot be prolyl hydroxylated and is resistant to oxygen-dependent proteasomal degradation (1). Forskolin increased HIF-1α-DM abundance in HEK293T cells (Fig. 3C) and HeLa cells (either alone or with IBMX), an effect that was negated by H89 (Fig. 3D). In addition, Ca overexpression increased HIF-1α-DM protein abundance in HeLa cells (Fig. 3E). Thus, PKA inhibits the proteasomal degradation of HIF-1α independently of prolyl hydroxylation.
PKA phosphorylates Thr63 and Ser692 to stabilize HIF-1α
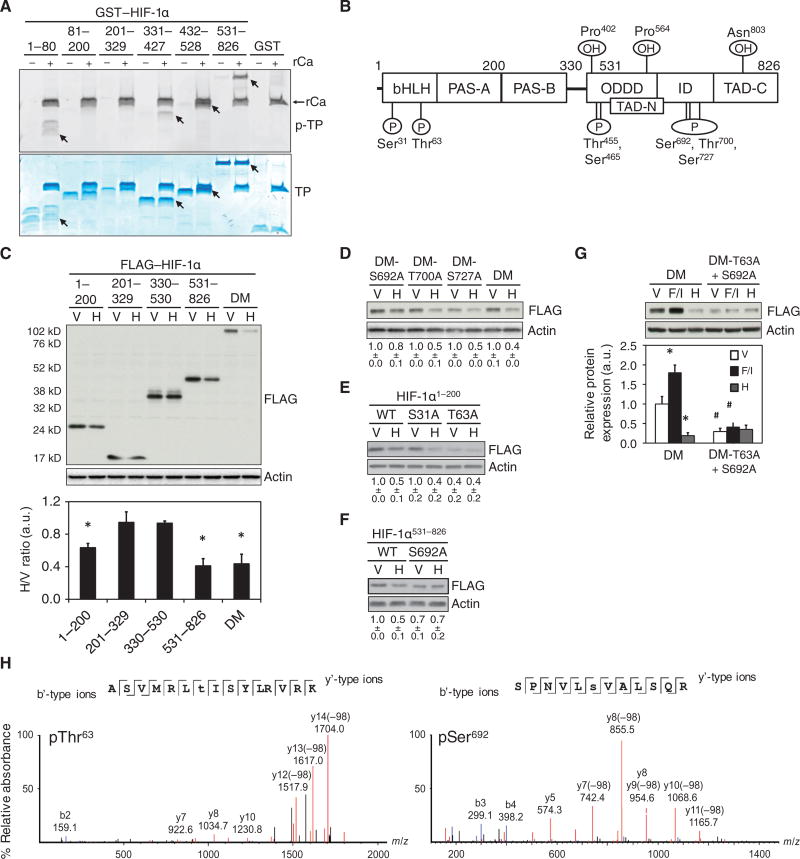
To determine if HIF-1α was a direct substrate of PKA, we performed in vitro kinase assays using GST-fusion proteins encompassing residues 1-80, 331-427, 432-528 or 531-826 of HIF-1α and the recombinant Ca subunit of PKA (rCa) (Fig. 4A). LC-MS/MS analyses of GST-HIF-1α fusion proteins indicated that Ser31, Thr63, Thr455, Ser465, Thr700, and Ser727 were phosphorylated by rCa in vitro (Fig. 4B). In HeLa cells expressing the HIF-1α deletion mutants, H89 decreased the abundance of the constructs encompassing HIF-1α residues 1-200 and 531-826, but not those encompassing 201-329 or 330-530 (Fig. 4C), suggesting that PKA may increase HIF-1α abundance by phosphorylating residues in the amino-terminal (residues 1-200) and carboxy-terminal (residues 531-826) regions of HIF-1α.
Fig. 4. PKA phosphorylates Thr63 and Ser692 to increase HIF-1α stability.
(A) GST fusion proteins encompassing the indicated HIF-1α residues were incubated with the recombinant Ca subunit of PKA (rCa) and analyzed for phospho-total protein (p-TP) by ProQ Diamond staining (top) or total protein (TP) by Coomassie Blue staining (bottom). Phosphorylated proteins are indicated (arrows) (representative of n = 2 independent experiments). (B) The schematic shows HIF-1α residues that were phosphorylated (P) by rCa in vitro as determined by LC-MS/MS analysis of LysC- or trypsin-digested peptides (n = 1 experiment per digestion). The location of the basic helix-loop-helix (bHLH), Per-Arnt-Sim homology (PAS-A and PAS-B), O2-dependent degradation (ODDD), inhibitory (ID), and amino- and carboxy-terminal transactivation (TAD-N and TAD-C) domains and sites of hydroxylation (OH) are shown. (C) Immunoblot assays were performed using lysates of HeLa cells expressing FLAG-tagged HIF-1α-DM or a deletion mutant containing the indicated HIF-1α residues and exposed to vehicle (V) or H89 (H). The bar graph shows the H/V ratio of FLAG:actin densitometry ratios for each fusion protein (mean ± SD, n = 3 independent experiments). *p < 0.05 compared to vehicle. (D-F) HeLa cells expressing FLAG-tagged HIF-1α-DM (DM) or S692A-, T700A- or S727A-mutant DM (D); FLAG-tagged HIF-1α1-200, which was either wild-type (WT) or contained an S31A or T63A mutation (E); or FLAG-tagged HIF-1α531-826, which was WT or contained an S692A mutation (F) were treated with V or H and subjected to immunoblot assays. (G) HeLa cells expressing DM or T63A+S692A-mutant DM, and exposed to V, forskolin + IBMX (F/I) or H were subjected to immunoblot assays. *p < 0.05 compared to V, #p < 0.05 compared to DM under same condition. FLAG:actin ratios displayed as a number below each blot (D-F) or as a bar graph (G) are the mean ± SD from 3 independent experiments normalized to V (D) or respective WT (E and F) or DM (G) vehicle treated controls. (H) MS/MS spectra of the PKA-phosphorylated HIF-1α tryptic peptides ASVMRLpTISYLRVRK (left) and SPNVLpSVALSQR (right) demonstrating phospho-Thr63 (pThr63) and phospho-Ser692 (pSer692), respectively. Monoisotopic b’- and y’-type fragment ion masses are displayed above the respective annotated ion peaks (n = 1 experiment per digestion). The Mann-Whitney U test was used to analyze statistical significance for all data in this figure.
To determine whether residues phosphorylated by rCa in vitro were biologically relevant, we tested if mutation of Ser692, Thr700 or Ser727 made HIF-1α-DM resistant to H89 treatment. H89 decreased the abundance of HIF-1α-DM and T700A-and S727A-mutant HIF-1α-DM by ~50–60%, but decreased that of the S692A-mutant HIF-1α-DM by only ~20% (Fig. 4D). H89 also decreased the abundance of S31A-mutant HIF-1α1-200 by 50-60%, but T63A-mutant HIF-1α1-200 was constitutively unstable regardless of whether H89 was added (Fig. 4E). Consistent with these findings, the abundance of S692A-mutant HIF-1α531-826 was decreased in vehicle-treated cells and resistant to further degradation induced by H89 (Fig. 4F). Lastly, the abundance of T63A/S692A-mutant HIF-1α-DM was markedly decreased compared to wild-type HIF-1α and was not affected by treatment with forskolin and IBMX or H89 (Fig. 4G). Thus, residues Thr63 and Ser692 of HIF-1α are phosphorylated by PKA in vitro, as demonstrated by mass spectrometry (Fig. 4H), and are required for PKA to increase HIF-1α abundance in HeLa cells.
PKA stimulates the binding of p300 to the carboxy-terminal transactivation domain of HIF-1α
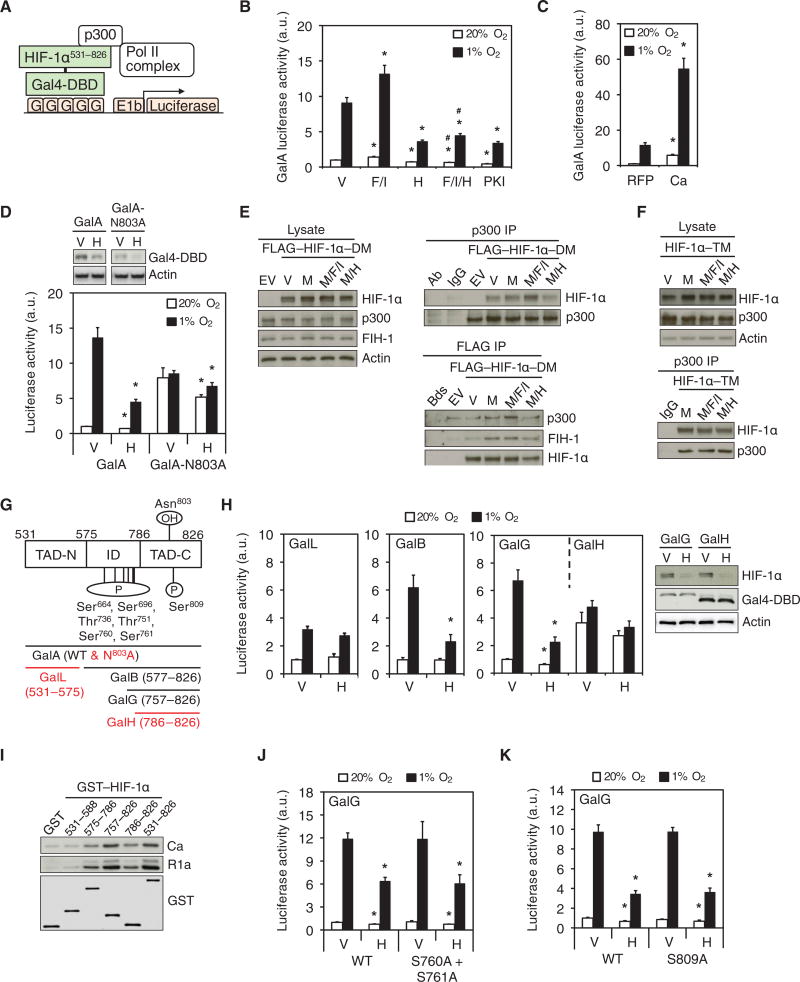
In addition to protein stability, the transcriptional activity of HIF-1α is increased by recruitment of coactivators, particularly p300, to the transactivation domains (10, 11). To measure HIF-1α transactivation domain function, we analyzed the ratio of firefly:Renilla luciferase activity in HeLa cells, which were cotransfected with: pGalA, a vector encoding a fusion protein (designated GalA) containing the Gal4 DNA-binding domain (DBD) and residues encompassing both HIF-1α transactivation domains [HIF-1α531-826] (10) (Fig. 5A); pG5-E1b-Luc, a reporter containing five Gal4 DNA binding sites upstream of the E1b promoter and firefly luciferase coding sequences; and pSV-Renilla (11). Treatment with H89 or PKI decreased GalA-dependent reporter activity. Forskolin and IBMX treatment increased GalA activity, an effect that was abolished by H89 (Fig. 5B). Coexpression of the Ca subunit of PKA markedly increased GalA activity (Fig. 5C). H89 had a reduced effect on transactivation mediated by N803A-mutant GalA, which encodes a protein that is constitutively active due to the loss of inhibitory Asn803 hydroxylation, compared to the GalA fusion protein containing the unmutated HIF-1α transactivation domain (Fig. 5D). Consistent with its effect on HIF-1α531-826 (Fig. 4C), H89 decreased the abundance of both forms of the GalA fusion protein (Fig. 5D), which explains why the N803A-mutant GalA was not completely resistant to the inhibitory effects of H89 on reporter activity.
Fig. 5. PKA stimulates the TAD function of HIF-1α.
(A) The schematic illustrates the HIF-1α transactivation domain function reporter assay (G, Gal4 binding sites). (B and C) Luciferase assays (mean ± SD, n = 9 biological replicates from 3 independent experiments) were performed using lysates from: HeLa cells treated as indicated (B); or HeLa cells expressing RFP or RFP-Ca and exposed to 20% or 1% O2 (C). *p < 0.05 compared to vehicle (V) or RFP at the same % O2; #p < 0.05 compared to F/I at the same % O2. (D) HeLa cells expressing GalA or GalA with an N803A mutation were exposed to 20% or 1% O2 and subjected to immunoblot (top) or luciferase (bottom) assays (mean ± SD, n = 9 biological replicates from 3 independent experiments). *p < 0.05 compared to V at the same % O2. (E) HeLa cells expressing empty vector (EV) or FLAG-tagged HIF-1α-DM were exposed to V, MG132 (M), MG132+forskolin+IBMX (M/F/I), or MG132+H89 (M/H) and subjected to immunoblot assays (left) or p300 (top right) or FLAG (bottom right) immunoprecipitation (IP) were performed (n = 3 independent experiments). (F) Immunoblot assays were performed using lysates and p300 immunoprecipitated from HeLa cells expressing FLAG-tagged P402A/P564A/N803A triple-mutant HIF-1α (HIF-1α-TM) and treated as in E (n = 3 independent experiments). (G) The schematic shows Gal4-HIF-1α fusion proteins and HIF-1α residues identified as phosphorylated (P) by the recombinant Ca subunit of PKA in vitro using LC-MS/MS on GluC-digested peptides (n = 1 experiment). Location of inhibitory domain (ID), amino- and carboxy-terminal transactivation domains (TAD-N and TAD-C), and the Asn hydroxylation (OH) site are shown. Fusion proteins with TAD function that was not inhibited by H89 are highlighted in red. (H) Luciferase assays were performed using lysates from HeLa cells, which were transfected with Gal4-DBD fused to HIF-1α residues 531-575 (GalL), 577-826 (GalB), 757-826 (GalG) or 786-826 (GalH), and incubated at 20% or 1% O2 (left) (mean ± SD, n = 6 biological replicates from 2 independent experiments). *p < 0.05 compared to V at the same % O2. Endogenous HIF-1α (right, top panel), GalG and GalH (right, middle panel), and Actin (right, bottom panel) were detected by immunoblot assays. (I) GST-HIF-1α fusion proteins were tested for their ability to capture the Ca or R1a subunits of PKA from HeLa cell lysates (representative of n = 3 independent experiments). (J and K) Luciferase assays were performed using lysates from HeLa cells expressing unmutated wild-type (WT) GalG, or GalG with S760A+S761A, or S809A mutations and exposed to V or H at 20% or 1% O2 (mean ± SD, n=6 biological replicates from 2 independent experiments); *p < 0.05 compared to V at same % O2. Data in B, C, D, G, J and K are graphed as fold change from respective controls at 20% O2. The Mann-Whitney U test was used to analyze statistical significance for all data in this figure.
FIH-1 is constitutively bound to HIF-1α, and FIH-1-dependent hydroxylation of Asn803 inhibits the binding of p300 to HIF-1α. We sought to determine whether PKA stimulated the recruitment of p300 to full-length HIF-1α under conditions in which HIF-1α abundance does not change. Treatment of HeLa cells with forskolin and IBMX or H89 did not affect FLAG-HIF-1α-DM, p300 or FIH-1 abundance in the presence of MG132 (Fig. 5E, left). Binding of p300 (but not that of FIH-1) to HIF-1α-DM was increased by forskolin and IBMX and decreased by H89 (Fig. 5E, right). Consistent with the GalA assays, forskolin and IBMX or H89 did not affect the binding of p300 to the triple-mutant (P402A/P564A/N803A) HIF-1α (HIF-1α-TM) (Fig. 5F). Thus, PKA stimulates p300 binding to HIF-1α by counteracting the inhibitory effect of Asn803 hydroxylation.
We have previously demonstrated (10) that residues 531-575 constitute the amino-terminal transactivation domain (TAD-N) and residues 786-826 constitute the carboxy-terminal transactivation domain (TAD-C) of HIF-1α. The activity of both TADs is attenuated by the presence of the inhibitory domain (ID), which encompasses residues 576-786 (Fig. 5G). Furthermore, residues 757-786 are required for FIH-1 binding and inhibition of TAD function (11). We analyzed HIF-1α transactivation function in HeLa cells expressing the Gal4-DBD fused to residues 531-575 (GalL; TAD-N), 577-826 (GalB; ID and TAD-C), 757-826 (GalG; TAD-C and FIH-1 binding site) or 786-826 (GalH; TAD-C) (Fig. 5G). H89 treatment inhibited transactivation mediated by GalB or GalG, but not by GalL or GalH, without affecting the protein abundance of GalG or GalH, which were detected using an antibody against the Gal4-DBD (Fig. 5H). Thus, PKA did not affect TAD-N activity and, similar to FIH-1 binding, residues 757-786 were required for the stimulatory effect of PKA on TAD-C. These results were consistent with the effects of PKA on p300 recruitment and indicate that PKA stimulated the transactivation domain function of HIF-1α by counteracting the inhibitory effect of Asn803 hydroxylation.
Pull-down assays using HeLa cell lysate and GST-HIF-1α proteins revealed strongest binding of both the Ca and R1a subunits of PKA to residues 757-826 and 531-826 (Fig. 5I). We next investigated whether phosphorylation of HIF-1α by PKA played a direct role in regulating TAD-C function. None of the residues in GST-HIF-1α531-826 that were phosphorylated in vitro (Fig. 4B) were located in the 757-826 region. However, tryptic digestion of residues 531-826 did not result in complete coverage of all Ser and Thr residues. We therefore performed LC-MS/MS analyses of peptides from PKA-phosphorylated GST-HIF-1α531-826 digested with GluC. An additional seven residues were identified as phosphorylated by PKA in vitro, three of which (Ser760, Ser761, and Ser809) were located in the 757-826 region (Fig. 5G). However, both S760A/S761A- and S809A-mutant GalG exhibited no differences in reporter transactivation compared to unmutated GalG (Fig. 5, J and K). Thus, PKA counteracts the inhibitory effect of Asn803 hydroxylation by stimulating the interaction of p300 with TAD-C through a mechanism that does not appear to involve phosphorylation of HIF-1α.
PKA promotes HIF-1 target gene expression
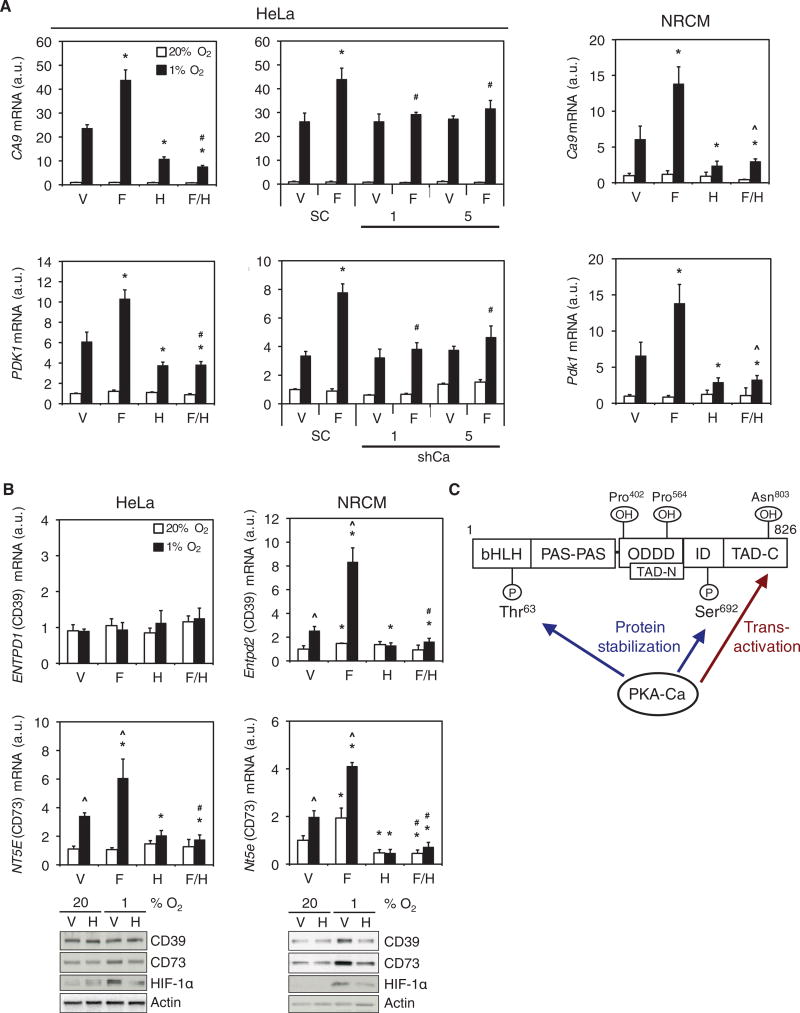
We next analyzed the effects of pharmacologic and genetic manipulation of PKA activity on the expression of HIF-1 target genes encoding carbonic anhydrase 9 (CA9) and pyruvate dehydrogenase kinase 1 (PDK1). H89 treatment of HeLa cells (Fig. 6A, left) or NRCMs (Fig. 6A, right) decreased CA9 and PDK1 mRNA abundance under hypoxic conditions as compared to vehicle-treated controls. In contrast, forskolin treatment increased CA9 and PDK1 mRNA abundance under hypoxic conditions, an effect that was inhibited by H89. Stable knockdown of the Ca subunit of PKA in HeLa cells inhibited the increase in CA9 and PDK1 mRNA abundance by forskolin (Fig. 6A, center). HIF-1 mediates the expression of ENTPD1, which encodes ectonucleoside triphosphate diphosphohydrolase 1, also known as CD39, and NT5E, which encodes ecto-5’-nucleotidase, also known as CD73 (12). CD39 converts extracellular ATP to AMP, which is then dephosphorylated by CD73 to produce adenosine (12). Hypoxia increased CD73 mRNA and protein abundance in HeLa cells and NRCMs, whereas CD39 mRNA and protein abundance increased only in NRCMs (Fig. 6B), and these effects of hypoxia were inhibited by H89 (Fig. 6B). Forskolin increased CD39 and CD73 mRNA abundance, and this induction was also blocked by H89. Thus, consistent with effects on HIF-dependent reporter activity, HIF-1α protein abundance, and HIF-1α transactivation domain function, PKA promotes endogenous HIF-1 target gene expression in HeLa cells and NRCMs.
Fig. 6. PKA stimulates HIF-1 target gene expression.
(A) CA9 and PDK1 mRNA expression was determined by RT-qPCR in: parental HeLa cells; scrambled shRNA control (SC), shCa-1 or shCa-5 HeLa subclones; and NRCMs exposed to vehicle (V), forskolin (F), H89 (H), or forskolin+H89 (F/H) at 20% or 1% O2 (mean ± SD, n = 6 biological replicates from 2 independent experiments). *p < 0.05 compared to V; #p < 0.05 compared to SC; ^ p < 0.05 compared to H. (B) CD39 and CD73 mRNA expression and the abundance of the corresponding proteins in HeLa cells or NRCMs exposed to V, F, H, or F/H at 20% or 1% O2 were determined by RT-qPCR (mean ± SD, n = 6 biological replicates from 2 independent experiments) and immunoblot assays (n = 3 independent experiments). ^p < 0.05 compared to 20% O2; *p < 0.05 compared to V; #p < 0.05 compared to H. Data in A and B are graphed as fold change from respective controls at 20% O2. The Mann-Whitney U test was used to assess statistical significance for all data in A and B. (C) PKA regulates HIF-1α protein stability and carboxy-terminal transactivation domain (TAD-C) function. Phosphorylation of Thr63 and Ser692 by PKA inhibits proteasomal degradation of HIF-1α. PKA also stimulates p300 binding to overcome the inhibitory effect of FIH-1-mediated Asn803 hydroxylation, thereby increasing transactivation by HIF-1α.
DISCUSSION
We have identified and characterized PKA as a protein that physically and functionally interacts with HIF-1α. PKA inhibited the proteasomal degradation and stimulated the transactivation domain function of HIF-1α, which together increased HIF-1 target gene expression (Fig. 6C). Thr63 and Ser692 were phosphorylated by the Ca catalytic subunit of PKA, and mutation of these residues blocked PKA-induced HIF-1α protein stabilization. PKA-induced p300 recruitment and transactivation did not appear to require phosphorylation of HIF-1α. Not only Ca but also the R1a regulatory subunit of PKA associated with HIF-1α, indicating that the PKA holoenzyme may be bound to HIF-1α before activation and is thus poised to respond to increased cAMP concentrations by stimulating HIF-1 activity. Surprisingly, PKA activators increased the association of both PKA subunits with HIF-1α, suggesting that phosphorylation may further increase the binding affinity of HIF-1α for PKA. A previous study suggested that HIF-1α might be stimulated by PKA in EAhy296 endothelial cells subjected to cyclic hypoxia-reoxygenation but there was no evidence that PKA phosphorylated HIF-1α directly and no effect of PKA on HIF-1α protein abundance was reported in that study (13). We demonstrated that PKA increased HIF-1 activity by increasing HIF-1α stability and transactivation domain function. However, the effects of modulating PKA activity in non-hypoxic cells were modest and variable in different assays, and may reflect differences in the rate of HIF-1α synthesis.
Several other interacting proteins inhibit the proteasomal degradation of HIF-1α independently of prolyl hydroxylation, including BCL2, HSP90, and septin 9 variant 1 (14–16). Phosphorylation of HIF-1α by ATM at Ser696 or by cyclin-dependent kinase 1 at Ser668 increases HIF-1α stability (17, 18), whereas phosphorylation by GSK-3β at Ser551, Thr555 and Ser589, or by polo-like kinase 3 at Ser576 and Ser657, increases HIF-1α degradation (19, 20). Thus, the sites in HIF-1α that are phosphorylated by PKA (Thr63 and Ser692) are different from those targeted by other kinases. Thr63 is located in the amino-terminal bHLH domain (Fig. 6C), suggesting that phosphorylation at this site stabilizes HIF-1α by a distinct molecular mechanism. PKA promoted the stability of both HIF-1α1-200 and HIF-1α531-826, indicating that the stabilizing effects of phosphorylation at Thr63 and Ser692 were independent of each other. In addition, PKA-dependent stabilization of HIF-1α protein was observed even when Pro402 and Pro564 were mutated, indicating that the mechanism of stabilization is independent of prolyl hydroxylation and VHL binding.
In addition to effects on HIF-1α protein stability, PKA stimulates p300 binding to the C-terminal transactivation domain of HIF-1α. The effect of PKA on HIF-1α-mediated transactivation required Asn803 and residues 757-786, which are critical for FIH-1 binding, but PKA did not affect the interaction between HIF-1α and FIH-1. These data suggest that PKA may inhibit the ability of FIH-1 to hydroxylate HIF-1α or may promote interaction of p300 with HIF-1α even when Asn803 is hydroxylated. Because PKA-mediated phosphorylation of Ser133 in CREB induces the binding of the coactivator CBP and transactivation through binding of CREB-CBP complexes to CREs in target genes (6), we analyzed whether phosphorylation of HIF-1α by PKA stimulates the p300-HIF-1α interaction. However, none of the sites that were phosphorylated by PKA in vitro were required for stimulation of HIF-1α transactivation domain function by PKA. Further study is required to determine whether PKA-catalyzed phosphorylation of FIH-1, p300, another coactivator, or an unidentified HIF-1α residue is responsible for the PKA-dependent increase in HIF-1α transactivation domain function. In this regard, it is worth noting that in PC12 pheochromocytoma cells, intermittent hypoxia stimulates HIF-1α transactivation domain function as a result of p300 phosphorylation by Ca2+/calmodulin-dependent kinase II (22).
Studies are also warranted to determine whether PKA interacts with, and phosphorylates, HIF-2α. Despite 87% sequence identity between HIF-1α and HIF-2α over residues 14-68, Thr63 is not conserved in HIF-2α. With respect to Ser692, HIF-1α residues 600-699 share only 20% sequence identity with the corresponding region of HIF-2α. It also remains to be determined whether PKA interacts primarily with the isolated HIF-1α subunit or with the HIF-1 heterodimer in vivo, in which case it is formally possible that PKA might also modify HIF-1β.
We focused on the interaction of HIF-1α with PKA because hypoxic, ischemic, and inflammatory states are associated with increased HIF-1 activity and increased production of extracellular adenosine and catecholamines, which signal through PKA (1, 4, 21). Cancer cells exploit adaptive responses mediated by HIF-1 in hypoxic tumor microenvironments to drive angiogenesis, metabolic reprogramming, cancer stem cell maintenance, immune avoidance, invasion and metastasis (1). Many cancer cells also bear receptors for neurotransmitters and can synthesize neurotransmitters (including catecholamines) that act in an autocrine or paracrine manner through PKA to promote proliferation, survival, migration, invasion and metastasis (4). HIF-1-dependent expression of the genes encoding the extracellular adenosine-generating enzymes CD39 and CD73, and subsequent binding of adenosine to A2A or A2B receptors increases intracellular cAMP synthesis and stimulates PKA signaling, which promotes immunosuppression in the context of ischemia, inflammation, and cancer (3, 4, 12). We demonstrated PKA-dependent regulation of HIF-1 target genes involved in metabolism (CA9 and PDK1) and adenosine production (ENTPD1 and NT5E, which encode CD39 and CD73, respectively). HIF-1-dependent adenosine production should increase PKA activity, resulting in a potential feed-forward effect in the tumor microenvironment, in which hypoxia induces PKA activity and PKA induces HIF-1. Exposure of A549 lung carcinoma cells to hypoxia induces the expression of PRKACA (which encodes the Ca subunit of PKA) in a HIF-dependent manner (23), providing another potential feed-forward mechanism.
In heart failure, β-adrenergic-mediated activation of PKA and subsequent phosphorylation of voltage-gated Ca2+ and K+ channels, the SERCA inhibitory protein phospholamban, ryanodine receptor, troponin I, myosin binding protein C, and other proteins is a major mechanism leading to altered left ventricular structure and function (2, 8). Decreased β1-adrenergic receptor density and increased β-adrenergic receptor uncoupling from GPCR-mediated signaling pathways, which lead to decreased PKA activity, are correlated with disease progression and increased risk of decompensated heart failure (2, 8). Left ventricular pressure overload induced by transaortic constriction in mice initially results in increased HIF-1α abundance in the heart, which is associated with increased expression of the HIF-1α target gene VEGF (which encodes vascular endothelial growth factor) and with enhanced vascularization, maintenance of tissue perfusion, and compensated cardiac hypertrophy. However, HIF-1α abundance eventually decreases in these mice and the reduced vascular density results in impaired left ventricular function. In addition, cardiomyocyte-specific HIF-1α knockout leads to left ventricular dysfunction after transaortic constriction (24). Future studies are required to determine whether loss of PKA-dependent regulation of HIF-1 may contribute to the progression of heart failure. Finally, HIF-1 also mediates protective responses in the context of acute lung injury, antimicrobial immunity, inflammatory bowel disease, lung transplantation, myocardial ischemia, peripheral arterial disease, and wound healing (25, 26). Prolyl hydroxylase inhibitors induce HIF-1 activity and provide therapeutic benefits in animal models of these conditions. Our results suggest that studies are warranted to determine whether drugs that induce cAMP synthesis or inhibit its degradation may also be useful for this purpose.
MATERIALS AND METHODS
Cell culture
NRCMs were isolated as described (27). Human cardiomyocytes were purchased from PromoCell. NRCMs, H9c2, HeLa and HEK293T cells were cultured in DMEM with 10% FBS, and penicillin-streptomycin at 37°C in a 5% CO2/95% air (20% O2) incubator. The β-adrenergic agonists isoproterenol and phenylephrine were purchased from Sigma-Aldrich. Forskolin, IBMX, H89 and MG132 were from Selleckchem. Myr-PKI was from R&D Systems. For pharmacological studies, cells were pre-incubated in serum-free media (SFM) for 30 min. For studies using H89 or MG132, cells were pre-incubated in SFM or SFM supplemented with H89 or MG132 for an additional 30 min or 2 h prior to subsequent treatments, respectively. Treatments were performed in SFM (supplemented with 0.1% FBS for studies longer than 20 h). To induce hypoxia, cells were placed in a modular incubator chamber (Billups-Rothenberg), which was flushed with a 1% O2/5% CO2/94% N2 gas mixture, and incubated at 37°C. Cells were transfected using PolyJet (SignaGen) and lysed or treated 24 h post-transfection. For the GST-HIF-1α pulldown assay (Fig. 1A), NRCMs were treated for 72 h. For Fig. 1B, H9c2 cells were treated for 72 h, or exposed to 1% O2 for 48 h. For Fig. 2A, primary human cardiomyocytes were treated for 48 h. For Fig. 2B, 2E, 2G, 5D and 5G, cells were treated in parallel under the same conditions as described for luciferase assays. For Fig. 3, 4 5E, and 5F, all treatments were for 4 h at 20% O2 (or 1% O2 as indicated). With the exception of protein analyses in Fig. 6B, the following concentrations were used for all treatments: isoproterenol (10 µM), phenylephrine (20 µM), forskolin (50 µM), H89 (10 µM), PKI (20 µM), IBMX (20 µM), MG132 (10 µM). For protein analyses in Fig. 6B, cells were treated with 3 µM H89 for 24 h (compared to 10 µM for 4 or 16 h) to compensate for increased exposure time.
Plasmid constructs and recombinant protein expression
rsTagRFP and rsTagRFP-Ca (in pCDNA3.1) were gifts from Jin Zhang (Johns Hopkins University). Human Cb (in pPM-C-HA) was purchased from Applied Biological Materials. Five shRNA vectors targeting Ca (MISSION shRNA, Sigma-Aldrich) were screened for knockdown efficiency and vectors encoding shCa-1 (TRCN0000233525; Table S1) and shCa-5 (TRCN0000356094; Table S1) were used to generate stable puromycin-resistant HeLa subclones. Missense mutants of HIF-1α were generated by PCR using the Q5 Site-Directed Mutagenesis Kit and primers (Table S1) designed by NEBaseChanger software (New England Biolabs). His6-R1a vector was generated by isolation of bovine R1a cDNA sequences from pRSETB (Addgene, plasmid 14922) as a HindIII/NdeI fragment and ligation to pET15b (Rapid DNA Ligation Kit, Thermo Scientific). Other plasmids have been previously described (28). GST, GST-HIF-1α, His-Ca and His-R1a recombinant proteins were expressed in Escherichia coli and purified using glutathione or Ni-NTA beads (28, 29).
Virus production and subclone generation
Lentivirus transduction was performed as previously described (28, 30). Briefly, lentivirus particles were generated by transfecting HEK293T cells with an shRNA transducing vector and the packaging vectors pCMVR8.91 and pVSVg. The supernatant containing virus particles was harvested after 48 h, centrifuged at 250 × G for 10 min, and passed through a 0.45-µm filter. HeLa cells were incubated in growth media supplemented with 10% lentiviral supernatant and 8 µg/ml polybrene (Sigma-Aldrich) for 24 h, rinsed, cultured for an additional 48 h in growth media, followed by selection and maintenance of resistant clones in growth media containing 1 µg/ml puromycin (Sigma-Aldrich).
GST-HIF-1α pulldown assay
Capture of HIF-1α interacting proteins from cell lysates was performed as described (28, 29), with minor modifications. Briefly, GST or GST-HIF-1α531-826 was covalently coupled to CNBr-activated Sepharose 4 Fast Flow beads (GE Healthcare) according to the manufacturer’s instructions. For each column, 2 mg of GST or GST-HIF-1α531-826 was coupled to CNBr beads and incubated at 4°C overnight with 15 mg of protein lysate from NRCMs. Columns were subsequently washed and protein was eluted with 8M urea, dialyzed, and concentrated to > 3 mg/mL using a 10-kDa-exclusion Vivaspin column (Sartorius). The concentrated protein samples were fractionated by SDS-PAGE, stained for total protein using G250 Coomassie Brilliant Blue (BioRad) and gel bands were excised for analysis by MS.
MS analysis of GST-HIF-1α-interacting proteins
Digestion conditions were described previously (29). Briefly, excised bands were pH equilibrated in 25 mM NH4HCO3; reduced and alkylated in 10 mM DTT, 55 mM iodoacetamide, and 25 mM NH4HCO3; and digested in 10 ng/µL trypsin (Promega) at 37°C for 24 h. Tryptic peptides were recovered by first incubating gel slices in 80% acetonitrile/0.1% trifluoroacetic acid for 30 min and then in acetonitrile/0.1% trifluoroacetic acid for 30 min on a shaker. Extracted peptides were desalted using Oasis HLB 1-cc Extraction Cartridges (Waters), dried to completion and stored at −80 °C until ready for MS. If necessary, dried peptides were resuspended in 0.1% trifluoroacetic acid and subjected to repetitive desalting using 0.6 µl C18 ZipTips (Millipore) until dried peptides were visibly clear.
Liquid chromatography (LC) and tandem MS (MS/MS) were performed in the Johns Hopkins Mass Spectrometry and Proteomics Core using an EASY-nLC 1000 (mobile phase A was 0.1% fluoroacetic acid in water and mobile phase B was 0.1% fluoroacetic acid in acetonitrile) connected to an Orbitrap Elite (Thermo Scientific) equipped with a nano-electrospray source (27). A portion of the peptide digest was pressure-loaded onto a Thermo Acclaim PepMap 100 trap precolumn (75 µm × 2 cm) packed with C18 reverse-phase resin (3 µm, 100 Å; Thermo Scientific) and separated on a Thermo Acclaim PepMap RSLC analytical column (50 µm × 15 cm) packed with C18 reverse-phase resin (2 µm, 100 Å; Thermo Scientific) at a flow rate of 300 nl/min using a linear gradient of 2–40% buffer B for 30 min, 40–98% buffer B for 3 min, then holding at 98% buffer B for 7 min. The nano-source capillary temperature was set to 275 °C with a spray voltage of 2 kV.
MS1 scans were acquired in the Orbitrap Elite at a resolution of 60,000 FWHM (400–1600 m/z) with an AGC target of 1×106 ions over a maximum of 500 ms. The 10 most intense peaks from MS1 scans, with signals exceeding 1000 counts, were set to trigger MS2 scans in the linear ion trap. The following MS/MS settings were used: AGC target = 5e4; MS/MS = 1 µscan, 100 ms max ion accumulation time, collision energy = 35% and rapid resolution scan rate. Monoisotopic precursor selection was enabled with +1 and unassigned charge states not selected for MS2 analyses. Dynamic exclusion was used with a repeat count of 2, repeat duration of 30 s and exclusion duration of 90 s.
Raw MS/MS data were converted to mzXML and mgf format using Msconvert version 3.0.3858 from ProteoWizard (31) for peaklist generation. Data were searched using the X! Tandem algorithm (32), version 2009.10.01.1, and OMSSA algorithm (33), version 2.1.9. The dataset was searched against the concatenated target/decoy Rat Uniprot (34) database as of October 9, 2013. The search parameters were as follows: fixed modification of carbamidomethyl (C) and variable modifications of carbamylation (K), oxidation (M), phosphorylation (STY); enzyme: trypsin with 2 maximum missed cleavages, parent tolerance: 0.08 Da; fragment tolerance: 1.00 Da. Post-search analysis was performed using Trans Proteomic Pipeline (35), version v4.6 rev 1, with protein group and peptide probability thresholds set to 95% and 99% respectively, with one or more peptides required for identification. PeptideProphet (36) was used for peptide validation and iProphet (37) was used to further refine identification probabilities. Lastly, ProteinProphet (38) was used to infer protein identifications from the resulting combined peptide list and to group ambiguous hits. Protein Group and Peptide False Discovery Rates were calculated automatically using a target-decoy method for the above probability thresholds. Protein isoforms were only reported if a peptide comprising an amino acid sequence that was unique to the isoform was identified.
In vitro binding and phosphorylation
For binding assays, 10 µg of GST or GST-HIF-1α531-826 bound to glutathione beads was incubated with His-Ca or His-R1a in 100 µl of binding buffer (20 mM Na-HEPES [pH 7.5], 50 mM NaF, 5 mM Na4P2O7, 1 mM EDTA, 1 mM EGTA, 0.1% (v/v) Triton X-100, 1 mM dithiothreitol, 1 mM phenylmethane sulfonyl fluoride (PMSF), 64 µM benzamidine, 2 µM leupeptin, 3 µM antipain, and 10 U/mL aprotinin) for 2 h at 30°C on an orbital shaker (300 rpm). Samples were washed, eluted, fractionated by SDS-PAGE, and immunoblot assays were performed. For phosphorylation assays, 2 µg of GST or GST-HIF-1α were incubated with or without 1.5 µg of recombinant human Ca (Active Motif) in 50 µl of reaction buffer (20 mM Tris-HCl [pH 7.5], 5 mM EGTA, 25 mM β-glycerol phosphate, 1 mM Na3VO4, 1 mM DTT, 200 µM ATP, and 10 mM MgCl2) for 3 h at 30°C on an orbital shaker (300 rpm). Reactions were stopped by addition of 4x Laemmli loading buffer and proteins were fractionated by SDS-PAGE. Phospho-proteins were stained using Pro-Q Diamond Phosphoprotein Gel Stain (Thermo-Fisher) and imaged on a Typhoon 9400 variable mode imager (GE Healthcare). Total protein staining was performed using G250 Coomassie Brilliant Blue (29).
MS analysis of HIF-1α phosphorylation
Purified GST-HIF-1α proteins were incubated with active recombinant Ca in the presence of ATP (as described above) for 1 h. Samples were reduced in 5 mM DTT at 60 °C for 1 h, alkylated in 15 mM iodoacetamide for 15 min at room temperature protected from light, and digested at 37 °C overnight with either endoproteinase Lys-C (Promega) for GST-HIF-1α1-80 or trypsin (Promega) for all other phosphorylated GST-HIF-1α proteins. GST-HIF-1α531-826 was also digested with Glu-C (Promega) at 37 °C overnight.
LC-MS/MS analyses were performed in the Johns Hopkins Mass Spectrometry and Proteomics Core on house-packed columns connected to an LTQ Velos Orbitrap (Thermo Scientific) equipped with a nanospray electron source. A portion of the peptides were trapped for 5 min on a house-packed YMC trap precolumn (10 µm, 120 Å; YMC Co. Ltd.) for 5 min at 5 ml/min and then separated on an analytical column (75 µm × 15 cm) packed with C18H reverse-phase resin (5 µm, 120 Å; ProntoSIL) connected to a 10 µm PicoTIP emitter (New Objective) at a flow rate of 300 nl/min using a linear gradient of 10–45% Buffer B over 90 min. Eluted peptides were injected directly into the Velos Orbitrap at a spray voltage of 2.2 kV. MS1 scans were acquired from 350–1800 m/z with up to 8 peptide masses individually isolated with a 1.9 Da window and fragmented (MS2) using a stepped collision energy of 34, 30 s dynamic exclusion. MS1 and MS2 scans were acquired at a resolution of 30,000 and 15,000 FWHM, respectively.
Raw MS/MS data files were searched against a custom HIF-1α fragment database using PEAKS algorithm version 7.0 (Bioinformatics Solution Inc.), set at the following search parameters: variable modifications of oxidation (M), carbamidomethylation (C), deamidation (NQ), phosphorylation (STY), acetylation (K, N-term); enzyme trypsin [D/P], with 4 maximum missed cleavages and non-specific cleavage at both ends; parent tolerance: 25.0 ppm; fragment tolerance: 0.03 Da peptide and phosphorylation false discovery rates (FDRs) were set at 1%, equivalent to a -10lgP value ≥ 30.7. The phosphorylated Thr63 and Ser692 peptides each had FDRs < 0.1% with -10lgP values of 58.32 and 76.86, respectively.
Pull-down, immunoprecipitation, and immunoblot assays
As previously described (28, 29), 2 µg of GST protein bound to glutathione beads was incubated with 2.5 mg of lysate overnight at 4°C, washed 3 times, fractionated by SDS-PAGE, and subjected to immunoblot assays. Immunoprecipitation of FLAG-HIF-1α was performed using anti-FLAG(M2) beads (Sigma-Aldrich). For other antibodies used for immunoprecipitation and immunoblot assays, see Table S2.
Luciferase reporter assays
HIF-dependent (p2.1) and Gal4-HIF-1α fusion protein luciferase reporter assays were performed as previously described (10, 39). HeLa cells (60,000 per well) were seeded onto 24-well plates, grown for 20 h, and cotransfected with 20 ng of pSV-Renilla, 25 ng of expression vector (where applicable), and 80 ng of p2.1, or 50 ng each of pG5-E1b-Luc and pGal4-HIF-1α. After 6 h, cells were grown for 20 h in fresh media, treated for 16 h for pharmacological studies, or grown for an additional 24 h for overexpression studies, at 20% or 1% O2, and subjected to the Dual Luciferase Assay (Promega) (10).
RT-qPCR assays
RNA extraction, RT, and qPCR were performed as previously described (39). See Table S1 for primer sequences. All treatments were performed as described for luciferase assays above.
Statistical analyses
Densitometry software (ImageJ, NIH) was used to quantify immunoblot band pixel intensities. Statistical software (JMP12, SAS Institute) was used to determine whether two sample sets were statistically different from each other with a confidence interval of 95% (p < 0.05). Total data sets amalgamated from independent experiments for each respective graph was plotted in a frequency histogram with equal class intervals and subjected to the Goodness-of-Fit Shapiro-Wilk W Test to determine if the data was normally distributed. All data exhibited non-normal distribution, thus the nonparametric Mann-Whitney U test was performed to assess statistical significance. All quantified data are expressed as means ± standard deviations (SD).
Supplementary Material
Data File 1. Summary of MS/MS data for R1a Peptide and Protein Group identification.
Table S1. List of primer and shRNA nucleotide sequences.
Table S2. Antibodies used for immunoprecipitation and immunoblot assays.
Acknowledgments
We thank Jin Zhang (Johns Hopkins University) for providing rsTagRFP and rsTagRFP-Ca vector and Karen Padgett (Novus Biologicals) for providing antibodies against p300, CREB, FIH-1, CD73, PKA-R1a and PKA-Ca.
Funding: This work was supported by Public Health Service contract HHS-N268201000032C, National Research Service Award F32-GM116326, NHLBI HL-119012 and NHLBI HL-107153. G.L.S. is the C. Michael Armstrong Professor at the Johns Hopkins University School of Medicine.
Footnotes
Author contributions: JWB and GLS designed the study, analyzed data, and wrote the paper. IT, RJH, VV and JVE performed MS/MS analyses of HIF-1α-interacting proteins. LD and RNC performed MS/MS analyses of phosphorylated HIF-1α. FW and DLK provided primary neonatal rat cardiomyocytes. All authors reviewed the results and approved the final version of the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: The HIF-1a interacting protein and phosphorylated HIF-1α residues mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE (40) partner repository with the dataset identifiers PXD003792 and PXD003795, respectively.
REFERENCES AND NOTES
- 1.Semenza GL. Oxygen sensing hypoxia-inducible factors and disease pathophysiology. Annu. Rev. Pathol. Mech. Dis. 2014;9:47–71. doi: 10.1146/annurev-pathol-012513-104720. [DOI] [PubMed] [Google Scholar]
- 2.Yang J, Liu Y, Fan X, Li Z, Cheng Y Y. A pathway and network review on β-adrenoreceptor signaling and β-blockers in cardiac remodeling. Heart Fail. Rev. 2014;19:799–814. doi: 10.1007/s10741-013-9417-4. [DOI] [PubMed] [Google Scholar]
- 3.Poth JM, Brodsky K, Ehrentraut H, Grenz A, Eltzschig HK. Transcriptional control of adenosine signaling by hypoxia-inducible factors during ischemic or inflammatory disease. J. Mol. Med. 2013;91:183–193. doi: 10.1007/s00109-012-0988-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tang J, Li Z, Lu L, Cho CH. β-Adrenergic system a backstage manipulator regulating tumor progression and drug target in cancer therapy. Semin. Cancer Biol. 2013;23:533–542. doi: 10.1016/j.semcancer.2013.08.009. [DOI] [PubMed] [Google Scholar]
- 5.Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl. Acad. Sci. U.S.A. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taylor SS, Ilouz R, Zhang P, Kornev AP. Assembly of allosteric macromolecular switches: lessons from PKA. Nat. Rev. Mol. Cell Biol. 2012;13:646–658. doi: 10.1038/nrm3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scott JD. Cyclic nucleotide-dependent protein kinases. Pharmacol. Ther. 1991;50:123–145. doi: 10.1016/0163-7258(91)90075-w. [DOI] [PubMed] [Google Scholar]
- 8.Soni S, Scholten A, Vos MA, van Veen TA. Anchored protein kinase A signaling in cardiac cellular electrophysiology. J. Cell Mol. Med. 2014;20:1–12. doi: 10.1111/jcmm.12365. [DOI] [PMC free article] [PubMed] [Google Scholar]; Semenza GL, Jiang BH, Leung SW, Passantino R, Concordet JP, Maire P, Giallongo A. Hypoxia response elements in the aldolase A, enolase and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J. Biol. Chem. 1996;271:32529–32537. doi: 10.1074/jbc.271.51.32529. [DOI] [PubMed] [Google Scholar]
- 9.Jiang BH, Zheng JZ, Leung SW, Roe R, Semenza GL. Transactivation and inhibitory domains of hypoxia-inducible factor 1α: modulation of transcriptional activity by oxygen tension. J. Biol. Chem. 1997;272:19253–19260. doi: 10.1074/jbc.272.31.19253. [DOI] [PubMed] [Google Scholar]
- 10.Mahon PC, Hirota K, Semenza GL. FIH-1: a novel protein that interacts with HIF-1α and VHL to mediate repression of HIF-1 transcriptional activity. Genes Dev. 2001;15:2675–2686. doi: 10.1101/gad.924501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sitkovsky M, Ohta A. Targeting the hypoxia-adenosinergic signaling pathway to improve the adoptive immunotherapy of cancer. J. Mol. Med. 2013;91:147–155. doi: 10.1007/s00109-013-1001-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Toffoli S, Feron O, Raes M, Michiels C. Intermittent hypoxia changes HIF-1α phosphorylation pattern in endothelial cells: unravelling of a new PKA-dependent regulation of HIF-1alpha. Biochim. Biophys. Acta. 2007;1773:1558–1571. doi: 10.1016/j.bbamcr.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 13.Liu YV, Baek JH, Zhang H, Diez R, Cole RN, Semenza GL. RACK1 competes with HSP90 for binding to HIF-1α and is required for O2-independent and HSP90 inhibitor-induced degradation of HIF-1α. Mol. Cell. 2007;25:207–217. doi: 10.1016/j.molcel.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amir S, Wang R, Simons JW, Mabjeesh NJ. SEPT9_v1 up-regulates hypoxia-inducible factor 1 by preventing its RACK1-mediated degradation. J. Biol. Chem. 2009;284:11142–11151. doi: 10.1074/jbc.M808348200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trisciuoglio D, Gabellini C, Desideri M, Ziparo E, Zupi G, Del Bufalo D. Bcl-2 regulates HIF-1α protein stabilization in hypoxic melanoma cells via the molecular chaperone HSP90. PLoS One. 5:e11772. doi: 10.1371/journal.pone.0011772. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Cam H, Easton JB, High A, Houghton PJ. mTORC1 signaling under hypoxic conditions is controlled by ATM-dependent phosphorylation of HIF-1α. Mol. Cell. 2010;40:509–520. doi: 10.1016/j.molcel.2010.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Warfel NA, Dolloff NG, Dicker DT, Malysz J, El-Deiry WS. CDK1 stabilizes HIF-1α via direct phosphorylation of Ser668 to promote tumor growth. Cell Cycle. 2013;12:3680–3701. doi: 10.4161/cc.26930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flügel D, Görlach A, Michiels C, Kietzmann T. Glycogen synthase kinase 3 phosphorylates hypoxia-inducible factor 1α and mediates its destabilization in a VHL-independent manner. Mol. Cell Biol. 2007;27:3253–3265. doi: 10.1128/MCB.00015-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu D, Yao Y, Lu L, Costa M, Dai W. Plk3 functions as an essential component of the hypoxia regulatory pathway by direct phosphorylation of HIF-1α. J. Biol. Chem. 2010;285:38944–38950. doi: 10.1074/jbc.M110.160325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carrero P, Okamoto K, Coumailleau P, O’Brien S, Tanaka H, Poellinger L. Redox-regulated recruitment of the transcriptional coactivators CREB-binding protein and SRC-1 to hypoxia-inducible factor 1α. Mol. Cell Biol. 2000;20:402–415. doi: 10.1128/mcb.20.1.402-415.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuan G, Nanduri J, Bhasker CR, Semenza GL, Prabhakar NR. Ca2+/calmodulin kinase-dependent activation of hypoxia-inducible factor 1 transcriptional activity in cells subjected to intermittent hypoxia. J. Biol. Chem. 2005;280:4321–4328. doi: 10.1074/jbc.M407706200. [DOI] [PubMed] [Google Scholar]
- 22.Shaikh D, Zhou Q, Chen T, Ibe JC, Raj JU, Zhou G. cAMP-dependent protein kinase is essential for hypoxia-mediated epithelial-mesenchymal transition migration and invasion in lung cancer cells. Cell. Signal. 2012;24:2396–2406. doi: 10.1016/j.cellsig.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 23.Sano M, Minamino T, Toko H, Miyauchi H, Orimo M, Qin Y, Akazawa H, Tateno K, Kayama Y, Harada M, Shimizu I, Asahara T, Hamada H, Tomita S, Molkentin JD, Zou Y, Komuro I. p53-induced inhibition of Hif-1 causes cardiac dysfunction during pressure overload. Nature. 2007;446:444–448. doi: 10.1038/nature05602. [DOI] [PubMed] [Google Scholar]
- 24.Eltzschig HK, Bratton DL, Colgan SP. Targeting hypoxia signaling for treatment of ischemic and inflammatory diseases. Nat. Rev. Drug Discov. 2014;13:852–869. doi: 10.1038/nrd4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Semenza GL. Hypoxia-inducible factors in medicine and physiology. Cell. 2012;148:399–408. doi: 10.1016/j.cell.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee DI, Zhu G, Sasaki T, Cho GS, Hamdani N, Holewinski R, Jo SH, Danner T, Zhang M, Rainer PP, Bedja D, Kirk JA, Ranek MJ, Dostmann WR, Kwon C, Margulies KB, Van Eyk JE, Paulus WJ, Takimoto E, Kass DA. Phosphodiesterase 9A controls nitric-oxide-independent cGMP and hypertrophic heart disease. Nature. 2015;519:472–476. doi: 10.1038/nature14332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo W, Zhong J, Chang R, Hu H, Pandey A, Semenza GL. Hsp70 and CHIP selectively mediate ubiquitination and degradation of hypoxia-inducible factor (HIF)-1α but not HIF-2α. J. Biol. Chem. 2010;285:3651–3663. doi: 10.1074/jbc.M109.068577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bullen JW, Balsbaugh JL, Chanda D, Shabanowitz J, Neumann D, Hunt DF, Hart GW. Cross-talk between two essential nutrient-sensitive enzymes:. O-GlcNAc transferase and AMP-activated protein kinase. J. Biol. Chem. 2014;289:10592–10606. doi: 10.1074/jbc.M113.523068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo W, Chang R, Zhong J, Pandey A, Semenza GL. Histone demethylase JMJD2C is a coactivator for hypoxia-inducible factor 1 that is required for breast cancer progression. Proc. Natl. Acad. Sci. U.S.A. 2012;109:E3367–E3376. doi: 10.1073/pnas.1217394109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amode D, Ruderman DL, Neumann S, Gatto L, Fischer B, Pratt B, Egertson J, Hoff K, Kessner D, Tasman N, Shulman N, Frewen B, Baker TA, Brusniak MY, Paulse C, Creasy D, Flashner L, Kani K, Moulding C, Seymour SL, Nuwaysir LM, Lefebvre B, Kuhlmann F, Roark J, Rainer P, Detlev S, Hemenway T, Huhmer A, Langridge J, Connolly B, Chadick T, Holly K, Eckels J, Deutsch EW, Moritz RL, Katz JE, Agus DB, MacCoss M, Tabb DL, Mallick P. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012;30:918–920. doi: 10.1038/nbt.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Craig R, Beavis R. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 20:1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 32.Geer LY, Markey SP, Kowalak JA, Wagner L, Xu M, Maynard DM, Yang X, Shi W, Bryant SH. Open mass spectrometry search algorithm. J. Proteome Res. 2004;3:958–964. doi: 10.1021/pr0499491. [DOI] [PubMed] [Google Scholar]
- 33.Apweiler R, Bairoch A, Wu C, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, Martin MJ, Natale DA, O’Donovan C, Redaschi N, Yeh LS. Uniprot: the Universal Protein knowledgebase. Nucleic Acids Res. 2004;32:D115–D119. doi: 10.1093/nar/gkh131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keller A, Eng J, Zhang N, Li XJ, Aebersold R. A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Mol. Syst. Biol. 2005;1:2005.0017. doi: 10.1038/msb4100024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 36.Shteynberg D, Deutsch EW, Lam H, Eng JK, Sun Z, Tasman N, Mendoza L, Moritz RL, Aebersold R, Nesvizhskii AI. iProphet: multi-level integrative analysis of shotgun proteomic data improves peptide and protein identification rates and error estimates. Mol. Cell Proteomics. 2011;10 doi: 10.1074/mcp.M111.007690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 38.Hubbi ME, Gilkes DM, Baek JH, Semenza GL. Four-and-a-half LIM domain proteins inhibit transactivation by hypoxia-inducible factor 1. J. Biol. Chem. 2012;287:6139–6149. doi: 10.1074/jbc.M111.278630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vizcaíno JA, Csordas A, del-Toro N, Dianes JA, Griss J, Lavidas I, Mayer G, Perez-Riverol Y, Reisinger F, Ternent T, Xu QW, Wang R, Hermjakob H. 2016 update of the PRIDE database and related tools. Nucleic Acids Res. 2016;44:D447–D456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data File 1. Summary of MS/MS data for R1a Peptide and Protein Group identification.
Table S1. List of primer and shRNA nucleotide sequences.
Table S2. Antibodies used for immunoprecipitation and immunoblot assays.