Abstract
Myostatin is a transforming growth factor-β family member that acts as a negative regulator of skeletal muscle mass. To identify possible myostatin inhibitors that may have applications for promoting muscle growth, we investigated the regulation of myostatin signaling. Myostatin protein purified from mammalian cells consisted of a noncovalently held complex of the N-terminal propeptide and a disulfide-linked dimer of C-terminal fragments. The purified C-terminal myostatin dimer was capable of binding the activin type II receptors, Act RIIB and, to a lesser extent, Act RIIA. Binding of myostatin to Act RIIB could be inhibited by the activin-binding protein follistatin and, at higher concentrations, by the myostatin propeptide. To determine the functional significance of these interactions in vivo, we generated transgenic mice expressing high levels of the propeptide, follistatin, or a dominant-negative form of Act RIIB by using a skeletal muscle-specific promoter. Independent transgenic mouse lines for each construct exhibited dramatic increases in muscle mass comparable to those seen in myostatin knockout mice. Our findings suggest that the propeptide, follistatin, or other molecules that block signaling through this pathway may be useful agents for enhancing muscle growth for both human therapeutic and agricultural applications.
Myostatin is a transforming growth factor-β (TGF-β) family member that plays an essential role in regulating skeletal muscle growth (1). Myostatin is expressed initially in the myotome compartment of developing somites and continues to be expressed in the myogenic lineage throughout development and in adult animals. Mice carrying a targeted deletion of the myostatin gene have a dramatic and widespread increase in skeletal muscle mass. Individual muscles of myostatin null mice weigh approximately twice as much as those of wild-type mice as a result of a combination of muscle fiber hyperplasia and hypertrophy. The myostatin sequence has been highly conserved through evolution (2). Remarkably, the human, rat, murine, porcine, turkey, and chicken myostatin sequences are identical in the biologically active C-terminal portion of the molecule following the proteolytic processing site. The function of myostatin also appears to be conserved across species, as mutations in the myostatin gene have been shown to result in the double muscling phenotype in cattle (2–5).
These findings have raised the possibility that pharmacological agents capable of blocking myostatin activity may have applications for promoting muscle growth in human disease settings as well as in livestock animals. To identify novel strategies for blocking myostatin activity, we investigated the regulation of myostatin signaling. Here, we present evidence that myostatin, like TGF-β, may normally exist in vivo in a latent complex with the propeptide (the portion of the precursor protein upstream of the proteolytic processing site) and that on activation, myostatin may signal by binding to activin type II receptors.
Materials and Methods
Purification of Myostatin.
A Chinese hamster ovary cell line carrying amplified copies of a myostatin expression construct (1) was transfected with an expression construct for the furin protease PACE (kindly provided by Monique Davies, Genetics Institute, Cambridge, MA) to improve processing of the precursor protein. Conditioned medium (prepared by Cell Trends, Middletown, MD) was passed successively over hydroxylapatite (eluted with 200 mM sodium phosphate, pH 7.2), lentil lectin Sepharose (eluted with 50 mM Tris, pH 7.4/500 mM NaCl/500 mM methyl mannose), DEAE agarose (collected flowthrough), and heparin Sepharose (eluted with 50 mM Tris, pH 7.4/200 mM NaCl). The heparin eluate was then bound to a reverse-phase C4 HPLC column and eluted with an acetonitrile gradient in 0.1% trifluoroacetic acid. Antibodies directed against the mature C-terminal protein have been described (1). To raise antibodies against the propeptide, the portion of the human myostatin protein spanning amino acids 122–261 was expressed in bacteria by using the RSET vector (Invitrogen) and purified by nickel chelate chromatography. Immunization of rabbits was carried out by Spring Valley Labs (Woodbine, MD).
Receptor Binding.
Purified myostatin was radioiodinated by using the chloramine T method (6). COS-7 cells grown in 6- or 12-well plates were transfected with 1–2 μg pCMV5 or pCMV5/receptor construct by using lipofectamine (GIBCO). Crosslinking experiments were carried out as described (7). For quantitative receptor-binding assays, cell monolayers were incubated with labeled myostatin (in PBS containing 1 mg/ml of BSA) in the presence or absence of unlabeled myostatin, propeptide, or follistatin at 4°C. Cells were then washed, lysed in 0.5 M NaOH, and counted in a gamma counter. Specific binding was calculated as the difference in bound myostatin between cells transfected with Act RIIB and cells transfected with vector. This method of calculating specific binding was especially important in assessing the effect of the propeptide as the addition of the propeptide also reduced nonspecific binding in a concentration-dependent manner. Recombinant human follistatin was obtained through the National Hormone and Pituitary Program, the National Institute of Diabetes and Digestive and Kidney Diseases, and A. F. Parlow (Harbor–University of California, Los Angeles, Medical Center, Torrance, CA).
Transgenic Mice.
DNAs encoding a truncated form of murine Act RIIB (amino acids 1–174), the murine myostatin propeptide (amino acids 1–267), and the human follistatin short form were cloned into the MDAF2 vector containing the myosin light chain promoter and 1/3 enhancer and simian virus 40 processing sites (8). All microinjections and embryo transfers were carried out by the Johns Hopkins School of Medicine Transgenic Core Facility. Transgenic founders in a hybrid SJL/C57BL/6 background were mated to wild-type C57BL/6 mice, and all studies were carried out by using F1 offspring. For analysis of muscle weights, individual muscles were dissected from both sides of nearly all animals, and the average of the left and right muscle weights was used. Analysis of fiber numbers and sizes was carried out as described (1). RNA isolation and Northern analysis were carried out as described (9).
Results
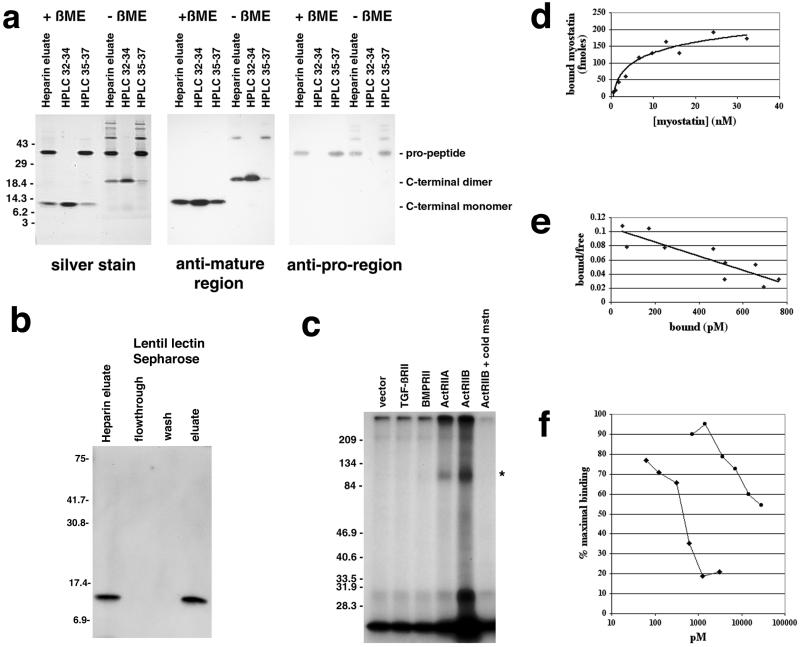
To overproduce myostatin protein, we generated a Chinese hamster ovary cell line carrying amplified copies of a myostatin expression construct. We purified myostatin protein from the conditioned medium of this cell line by successive fractionation on hydroxylapatite, lentil lectin Sepharose, DEAE agarose, and heparin Sepharose. Silver stain analysis of the purified protein preparation revealed the presence of two protein species of 36 and 12.5 kDa (Fig. 1a). A variety of data suggested that this purified protein consisted of a noncovalent complex of two propeptide molecules bound to a disulfide-linked C-terminal dimer. First, by Western analysis, the 36- and 12.5-kDa species were immunoreactive with antibodies raised against bacterially expressed fragments of myostatin spanning the propeptide and C-terminal mature region, respectively. Second, in the absence of reducing agents, the C-terminal region had an electrophoretic mobility consistent with that of a dimer. Third, the two species were present in a molar ratio of ≈1:1. And fourth, when the purified protein preparation was passed over a lentil lectin column, the C-terminal dimer was retained on the column and could be eluted with methyl mannose (Fig. 1b), even though this portion of the protein contains no potential N-linked glycosylation sites; the simplest interpretation of these data is that the C-terminal region bound the lectin indirectly by being present in a tight complex with the propeptide, which does have a glycosylation signal.
Figure 1.
Binding of myostatin to activin type II receptors. (a) Analysis of purified myostatin protein. Myostatin protein preparation following the heparin column (heparin eluate) or following reverse-phase HPLC (fractions 32–34 or 35–37 containing the C-terminal region or propeptide, respectively) was electrophoresed under reducing (+βME) or nonreducing (−βME) conditions and either silver stained or subjected to Western analysis. (b) Binding of myostatin to lentil lectin. The heparin eluate was bound to lentil lectin Sepharose and eluted with methyl mannose. Samples were electrophoresed under reducing conditions, blotted, and probed with antibodies directed against the C-terminal region. Similar analysis by using antibodies directed against the pro region showed that the pro region was also retained on the column and eluted with methyl mannose (data not shown). (c) Crosslinking experiments. COS-7 cells transfected with expression constructs for the indicated receptors were incubated with 125I-myostatin followed by the crosslinking agent disuccinimidyl suberate. Crosslinked complexes were analyzed by SDS/PAGE. Asterisk denotes predicted size for myostatin bound to an activin type II receptor. In the rightmost lane, excess unlabeled myostatin was included in the binding reaction. (d) Binding of myostatin to ActRIIB. All points represent the average of triplicate samples. (e) Scatchard analysis of the data shown in d. (f) Inhibition of myostatin binding to ActRIIB by follistatin (diamonds) and the propeptide (circles). Each experiment was carried out in triplicate, and each curve represents the average of three independent experiments.
Because the C-terminal dimer is known to be the biologically active molecule for other TGF-β family members, we further purified the C-terminal dimer of myostatin away from its propeptide by reverse-phase HPLC. As shown in Fig. 1a, the fractions containing the purified C-terminal dimer (fractions 32–34) appeared to be homogeneous. However, the fractions most enriched for the propeptide (fractions 35–37) were contaminated with small amounts of C-terminal dimer (see rightmost lane, Fig. 1a Center) and with high molecular weight complexes that most likely represented misfolded proteins.
Most members of the TGF-β superfamily have been shown to signal by binding serine/threonine kinase receptors followed by activation of Smad proteins (for review, see refs. 10 and 11). The initial event in triggering the signaling pathway is the binding of the ligand to a type II receptor. To determine whether myostatin is capable of binding any of the known type II receptors for related ligands, we carried out crosslinking studies with radioiodinated myostatin C-terminal dimer on COS-7 cells transfected with expression constructs for either TGF-β, BMP, or activin type II receptors. As shown in Fig. 1c, crosslinked complexes of the predicted size (full length receptor bound to myostatin) were detected for cells expressing either Act RIIA or Act RIIB. Because we consistently observed higher levels of binding to Act RIIB than to Act RIIA in both crosslinking and standard receptor-binding assays (data not shown), we focused our receptor-binding studies on Act RIIB. Binding of myostatin to Act RIIB was specific (binding could be competed by excess unlabeled myostatin; Fig. 1c) and saturable (Fig. 1d), and assuming that all of the myostatin protein was bioactive, we estimated the dissociation constant by Scatchard analysis to be ≈10 nM (Fig. 1e). Although this dissociation constant is higher than that reported for other TGF-β family members and their cognate receptors, it is possible that the binding affinity of myostatin for Act RIIB measured in our experiments may not accurately reflect that in vivo. For example, it is known in the case of TGF-β that the affinity for the type II receptor is significantly higher in the presence of the appropriate type I receptor (12) and that other molecules are involved in presenting the ligand to the receptor (13, 14).
To determine whether activin type II receptors may be involved in myostatin signaling in vivo, we investigated the effect of expressing a dominant-negative form of Act RIIB in mice. For this purpose, we generated a construct in which a truncated form of Act RIIB lacking the kinase domain was placed downstream of a skeletal muscle-specific myosin light chain promoter/enhancer. From pronuclear injections of this construct, we obtained a total of seven founder animals positive for the transgene. All seven showed significant increases in skeletal muscle mass with individual muscles weighing up to 125% more than those of control nontransgenic animals derived from similar injections (Table 1).
Table 1.
Muscle weights, mg
| Transgenic animals | Pectoralis | Triceps | Quadriceps | Gastrocnemius/ plantaris |
|---|---|---|---|---|
| Male controls (7 mo, n = 10) | 100.8 ± 5.4 | 115.6 ± 5.5 | 243.8 ± 12.5 | 168.1 ± 7.6 |
| Dominant-negative Act RIIB (7 mo) | ||||
| C5 male founder | 148 | 155 | 318 | 252 |
| C11 male founder | 227 | 250 | 454 | 338 |
| C33 male founder | 158 | 176 | 352 | 244 |
| C42 male founder | 196 | 212 | 309 | 269 |
| Female controls (7 mo, n = 10) | 68.9 ± 2.7 | 96.9 ± 3.5 | 208.3 ± 7.1 | 140.3 ± 4.3 |
| Dominant-negative Act RIIB (7 mo) | ||||
| C2 female founder | 104 | 163 | 352 | 263 |
| C4 female founder | 103 | 139 | 303 | 194 |
| C27 female founder | 135 | 117 | 181 | 256 |
| Male controls (4 mo, n = 12) | 98.3 ± 3.3 | 110.9 ± 2.9 | 251.7 ± 8.5 | 169.3 ± 4.7 |
| Follistatin (4 mo) | ||||
| F3 male founder | 296 | 494 | 736 | 568 |
| F66 male founder | 169 | 263 | 421 | 409 |
All animals (including controls) represent hybrid SJL/C57BL/6 F0 mice born from injected embryos.
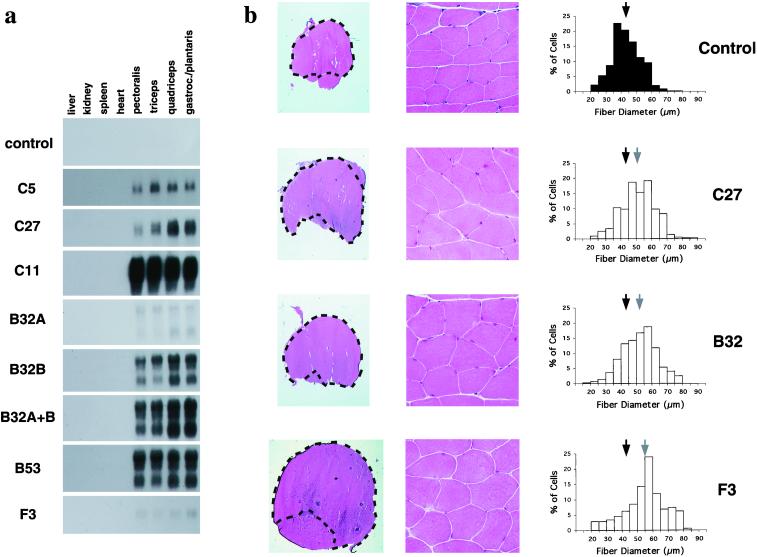
Three lines of evidence suggested that the increases in muscle weights in these founder animals resulted from the expression of the transgene. First, analysis of offspring derived from matings of three founder animals (the other four did not generate sufficient numbers of offspring for analysis) with wild-type C57BL/6 mice showed that the increases in muscle weights correlated with the presence of the transgenes (Table 2 and Fig. 2). Second, although muscle weights varied among the different transgenic lines, the magnitude of the increase was highly consistent among animals in any given line for all muscles examined and for both males and females (Table 2). For example, all muscles of both male and female mice from the C5 line weighed ≈30–60% more than those of control animals, whereas all muscles from C11 mice weighed ≈110–180% more. Third, Northern analysis of RNA samples prepared from transgenic animals showed that the expression of the transgene was restricted to skeletal muscle and that the relative levels of transgene expression (Fig. 3a) correlated with the relative magnitude of the increase in muscle weights (Table 2). For example, animals from the C11 line, which had the greatest increases in muscle weights, also had the highest levels of transgene expression.
Table 2.
Muscle weights, mg
| Transgenic line | Pectoralis | Triceps | Quadriceps | Gastrocnemius/plantaris |
|---|---|---|---|---|
| Males | ||||
| Controls (n = 50) | 104.6 ± 1.5 | 113.9 ± 1.6 | 246.2 ± 3.0 | 167.7 ± 2.1 |
| Dominant-negative Act RIIB | ||||
| C5 (n = 11) | 153.7 ± 6.0** | 177.5 ± 6.0** | 322.7 ± 9.3** | 247.1 ± 8.1** |
| C27 (n = 5) | 190.4 ± 7.1** | 230.8 ± 13.0** | 406.8 ± 11.6** | 283.8 ± 6.9** |
| C11 (n = 2) | 278.0 ± 18.4* | 244.5 ± 4.9** | 515.5 ± 7.8** | 366.0 ± 21.2* |
| Propeptide | ||||
| B32A (n = 8) | 139.9 ± 7.1** | 160.6 ± 7.8** | 322.5 ± 10.3** | 222.6 ± 7.1** |
| B32B (n = 4) | 214.0 ± 19.9** | 206.5 ± 6.7** | 435.8 ± 15.0** | 289.5 ± 8.6** |
| B32A + B (n = 8) | 212.4 ± 8.4** | 220.3 ± 6.4** | 429.1 ± 11.1** | 288.3 ± 8.5** |
| B53 (n = 8) | 215.1 ± 6.4** | 229.6 ± 8.1** | 413.3 ± 13.2** | 293.5 ± 10.5** |
| Females | ||||
| Controls (n = 50) | 64.7 ± 1.4 | 75.7 ± 1.1 | 164.5 ± 2.0 | 109.6 ± 1.4 |
| Dominant-negative Act RIIB | ||||
| C5 (n = 15) | 89.7 ± 2.8** | 115.9 ± 4.0** | 229.3 ± 6.5** | 161.8 ± 4.7** |
| C27 (n = 5) | 117.6 ± 10.9** | 138.6 ± 12.3** | 314.0 ± 27.7** | 207.6 ± 18.3** |
| C11 (n = 3) | 180.3 ± 38.9 | 208.7 ± 45.7 | 430.3 ± 72.2* | 291.7 ± 48.8* |
| Propeptide | ||||
| B32A (n = 9) | 78.8 ± 2.9** | 100.1 ± 3.7** | 206.0 ± 2.7** | 138.9 ± 3.1** |
| B32B (n = 2) | 131.0 ± 18.4 | 151.5 ± 23.3 | 315.5 ± 58.7 | 199.5 ± 24.7 |
| B32A + B (n = 4) | 109.3 ± 9.5* | 132.8 ± 6.0** | 270.8 ± 6.9** | 177.0 ± 2.4** |
| B53 (n = 6) | 134.7 ± 7.7** | 148.2 ± 12.1** | 303.8 ± 18.5** | 212.8 ± 12.9** |
All animals (including controls) represent 4-month-old offspring of transgenic founders (SJL/C57BL/6) mated with wild-type C57BL/6 mice.
, P < 0.05;
, P < 0.01;
, P < 0.001.
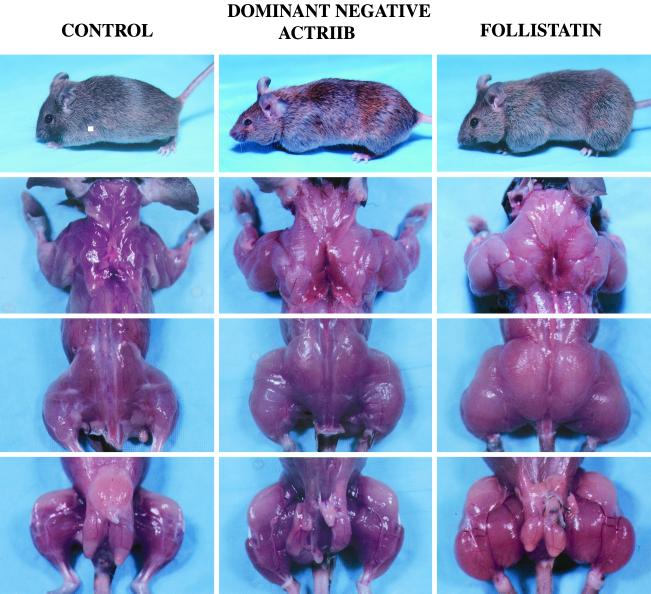
Figure 2.
Increased muscling in mice overexpressing a dominant-negative form of ActRIIB or full length follistatin. A control male nontransgenic mouse, a male transgenic mouse from the C11 line (dominant-negative ActRIIB), and the F3 male founder mouse (follistatin) are shown. Pictures of live mice are shown in the top row, and pictures of animals that had been killed and skinned are shown in the bottom three rows.
Figure 3.
Analysis of transgenic mice. (a) Specific expression of transgenes in skeletal muscle. Northern analysis of RNA samples prepared from female mice was carried out by using simian virus 40 sequences as a probe. On longer exposures of these blots, no expression of the transgenes was observed in liver, kidney, spleen, or heart in any of these lines. (b) Muscle analysis. Sections were stained with hematoxylin and eosin. The gastrocnemius and plantaris muscles are outlined (Left). Center shows higher magnifications of representative areas from the gastrocnemius muscle. Right shows distribution of fiber diameters. Each graph represents the composite of 450 fiber measurements from 3 animals (150 per animal), except for the F3 graph, which represents 175 measurements from the F3 founder animal. Standard deviations of fiber sizes were 9, 11, 11, and 13 μm for control, B32, C27, and F3 animals, respectively. Black and gray arrows show the mean fiber diameters for control and transgenic animals, respectively. Note that in each case, the mean fiber diameter was increased in the transgenic animals (P < 0.001).
These data showed that expression of a dominant-negative form of Act RIIB can cause increases in muscle mass similar to those seen in myostatin knockout mice. In myostatin knockout mice, the increase in muscle mass has been shown to result from increases in both fiber number and fiber size (1). To determine whether expression of dominant-negative Act RIIB also causes both hyperplasia and hypertrophy, we analyzed sections of the gastrocnemius and plantaris muscles of animals from the C27 line. Compared with control muscles, the muscles of the C27 animals showed a clear increase in overall cross-sectional area (Fig. 3b). This increase in area resulted partially from an increase in fiber number. At the widest point, the gastrocnemius and plantaris muscles had a total of 10,015 ± 1,143 fibers in animals from the C27 line (n = 3) compared with 7,871 ± 364 fibers in control animals (n = 3). However, muscle fiber hypertrophy also contributed to the increase in total area. As shown in Fig. 3b, the mean fiber diameter was 51 μm in animals of the C27 line compared with 43 μm in control animals. Hence, the increase in muscle mass appeared to result from an ≈27% increase in the number of fibers and 19% increase in fiber diameter (assuming the fibers to be roughly cylindrical, this increase in diameter would result in an ≈40% increase in cross-sectional area). Except for the increase in fiber number and size, however, the muscles from the transgenic animals looked grossly normal. In particular, there were no obvious signs of degeneration, such as widely varying fiber sizes (note that the standard deviation of fiber sizes was similar between control and transgenic animals) or extensive fibrosis or fat infiltration.
We also used these approaches to explore other possible strategies for inhibiting myostatin. First, we investigated the effect of the myostatin propeptide. In the case of TGF-β, it is known that the C-terminal dimer is held in an inactive latent complex with other proteins, including its propeptide (15), and that the propeptide of TGF-β can have an inhibitory effect on TGF-β activity both in vitro (16) and in vivo (17). Our observation that the myostatin C-terminal dimer and propeptide copurified raised the possibility that myostatin may normally exist in a similar latent complex and that the myostatin propeptide may have inhibitory activity. Second, we examined the effect of follistatin, which has been shown to be capable of binding and inhibiting the activity of several TGF-β family members. In particular, follistatin can block the activity of GDF-11 (18), which is highly related to myostatin (1, 18–20), and follistatin knockout mice have been shown to have reduced muscle mass at birth (21), which would be consistent with overactivity of myostatin.
We first examined the effect of the propeptide and follistatin in vitro. As shown in Fig. 1f, both the myostatin propeptide and follistatin were capable of blocking the binding of the C-terminal dimer to Act RIIB. We estimated the Ki of follistatin to be ≈470 pM and that of the propeptide to be at least 50-fold higher. The calculation of the Ki for the propeptide, however, assumes that all of the protein in the final preparation represented biologically active propeptide and therefore is likely to be an overestimate. As discussed above, our propeptide preparation was contaminated both with small amounts of C-terminal dimer and with misfolded high molecular weight species.
To determine whether these molecules are also capable of blocking myostatin activity in vivo, we generated transgenic mice in which the myosin light chain promoter/enhancer was used to drive expression of either the myostatin propeptide or follistatin. From pronuclear injections of the propeptide construct, we obtained three transgenic mouse lines (two of these, B32A and B32B, represented independently segregating transgene insertion sites in one original founder animal) that showed increased muscling. As shown in Table 2, muscle weights of animals from each line were increased by ≈20–110% compared with those of nontransgenic control animals. Northern analysis of RNA samples prepared from representative animals of each of these lines showed that the expression levels of the transgene correlated with the magnitude of the increase in muscle weights (Fig. 3a). Specifically, animals from the B32A line, which had only an ≈20–40% increase in muscle mass, had the lowest levels of transgene expression, and animals from the B32B and B53 lines, which had an ≈70–110% increase in muscle mass, had the highest levels of transgene expression. Perhaps significantly, muscle weights in animals that were doubly transgenic for the B32A and B32B insertion sites were similar to those observed in animals transgenic only for the B32B insertion site (Table 2), despite the fact that the doubly transgenic animals appeared to have higher levels of transgene expression (Fig. 3a). These findings suggest that the effects seen in the B32B line (and B53 line) were the maximal achievable from overexpressing the propeptide. As in the case of animals expressing the dominant-negative form of Act RIIB, animals expressing the propeptide showed increases in both muscle fiber number and size. Analysis of the gastrocnemius and plantaris muscles from two animals that were doubly transgenic for the B32A and B32B insertion sites showed that fiber numbers were increased by ≈40% (the two animals had 11,940 and 10,420 fibers), and fiber diameters were increased by ≈21% (to 52 μm) compared with control animals.
The most dramatic effects on skeletal muscle were obtained by using the follistatin construct. We obtained two founder animals (F3 and F66) that showed increased muscling (Table 1, Figs. 2 and 3b). In one of these animals (F3), muscle weights were increased by 194–327% relative to control animals, resulting from a combination of hyperplasia (66% increase in fiber number to 13,051 in the gastrocnemius/plantaris) and hypertrophy (28% increase in fiber diameter to 55 μm). Although we have not analyzed muscle weights of myostatin knockout mice in a hybrid SJL/C57BL/6 background, the increases in muscle mass observed in the F3 founder animal were significantly greater than the increases we have seen in myostatin null animals in other genetic backgrounds (unpublished results; see also ref. 1). These results suggest that at least part of the effect of follistatin may result from inhibition of another ligand besides myostatin. Clearly, analysis of additional follistatin transgenic lines will be essential in determining whether other ligands may also be involved in negatively regulating muscle growth.
Discussion
On the basis of the in vitro and transgenic mouse data presented here, we propose the following working model for the regulation of myostatin activity. After proteolytic processing, the myostatin C-terminal dimer is maintained in a latent complex with its propeptide and perhaps other proteins as well. Myostatin is also negatively regulated by follistatin, which binds the C-terminal dimer and inhibits its ability to bind to receptors. Release of the C-terminal dimer from these inhibitory proteins by unknown mechanisms allows myostatin to signal through activin type II receptors. By analogy with other family members, we presume that activation of these receptors then leads to activation of a type I receptor and Smad proteins.
Our overall model for myostatin regulation and signaling is consistent not only with the data presented here but also with other genetic data. As discussed earlier, follistatin knockout mice have been shown to have reduced muscle mass at birth (21), which is what one might expect for uninhibited myostatin activity. A similar muscle phenotype has been reported for mice lacking ski (22), which has been shown to inhibit the activity of Smad2 and 3 (23–27), and the opposite phenotype, namely excess skeletal muscle, has been observed in mice overexpressing ski (28). On the basis of our findings, an appealing hypothesis is that these observed phenotypes reflect the overactivity and underactivity, respectively, of myostatin in these mice.
Although all of the in vitro and genetic data are consistent with the overall model that we have put forth here, these data would also be consistent with alternative models involving other receptors and ligands. For example, we do not know the mechanism by which the truncated form of Act RIIB enhances muscle growth in our transgenic mice. It is possible that the truncated receptor is not acting to block signaling in the target cell but is rather merely acting as a sink to deplete extracellular concentrations of myostatin. It is also possible that the truncated receptor is blocking signaling of other ligands besides myostatin. In other species, it has been shown that dominant-negative forms of type II activin receptors can block signaling of a variety of different TGF-β-related ligands (29–32). Similarly, our data do not show definitively that follistatin is blocking myostatin activity in vivo to promote muscle growth. In this regard, the extraordinary degree of muscling seen in one of the follistatin-expressing founder animals suggests that other follistatin-sensitive ligands may be involved in regulating muscle growth. One obvious candidate is GDF-11, which is highly related to myostatin (1, 18–20) and also expressed in skeletal muscle (unpublished results). Moreover, it is known that GDF-11 activity in Xenopus can be blocked by follistatin (18). Other candidate ligands would also include the activins, which have been shown to be capable of inhibiting muscle cell differentiation in vitro (33).
To date, however, myostatin is the only secreted protein that has been demonstrated to play a negative role in regulating muscle mass in vivo. Although additional experiments will be required to prove aspects of this overall model and to identify the other signaling components, our data suggest that myostatin antagonists, such as follistatin and the myostatin propeptide, or activin type II receptor antagonists may be effective muscle-enhancing agents for both human and agricultural applications.
Acknowledgments
We thank Neil Wolfman for assistance with the HPLC, Christine Moss for assistance with the manuscript, and Joan Massague, Peter ten Dijke, Carl-Henrik Heldin, Jeffrey Wrana, Liliana Attisano, Wylie Vale, Sonia Pearson-White, Martin Matzuk, and Monique Davies for providing clones. This work was supported by National Institutes of Health Grants R01HD35887 and R01CA88866 (to S.J.-L.). Myostatin was licensed by the Johns Hopkins University to MetaMorphix (MMI) and sublicensed to American Home Products and Cape Aquaculture Technologies (CAT). The authors are entitled to a share of sales royalty received by the University from sales of this factor. The authors and the University own MMI stock, and the authors also own CAT stock, which are subject to certain restrictions under University policy. S.-J.L. is a consultant to MMI and CAT, and A.C.M. is a consultant to CAT. The terms of these arrangements are being managed by the University in accordance with its conflict of interest policies.
Abbreviations
- TGF-β
transforming growth factor-β
- BMP
bone morphogenetic protein
- GDF-11
growth/differentiation factor-11
References
- 1.McPherron A C, Lawler A M, Lee S-J. Nature (London) 1997;387:83–90. doi: 10.1038/387083a0. [DOI] [PubMed] [Google Scholar]
- 2.McPherron A C, Lee S-J. Proc Natl Acad Sci USA. 1997;94:12457–12461. doi: 10.1073/pnas.94.23.12457. . (First Published October 17, 2000; 10.1073/pnas.220421797) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grobet L, Martin L J R, Poncelet D, Pirottin D, Brouwers B, Riquet J, Schoeberlein A, Dunner S, Ménissier F, Massabanda J, et al. Nat Genet. 1997;17:71–74. doi: 10.1038/ng0997-71. [DOI] [PubMed] [Google Scholar]
- 4.Kambadur R, Sharma M, Smith T P L, Bass J J. Genome Res. 1997;7:910–916. doi: 10.1101/gr.7.9.910. [DOI] [PubMed] [Google Scholar]
- 5.Grobet L, Poncelet D, Royo L J, Brouwers B, Pirottin D, Michaux C, Ménissier F, Zanotti M, Dunner S, Georges M. Mamm Genome. 1998;9:210–213. doi: 10.1007/s003359900727. [DOI] [PubMed] [Google Scholar]
- 6.Frolik C A, Wakefield L M, Smith D M, Sporn M B. J Biol Chem. 1984;259:10995–11000. [PubMed] [Google Scholar]
- 7.Franzén P, ten Dijke P, Ichijo H, Yamashita H, Schultz P, Heldin C-H, Miyazono K. Cell. 1993;75:681–692. doi: 10.1016/0092-8674(93)90489-d. [DOI] [PubMed] [Google Scholar]
- 8.Donoghue M, Merlie J, Rosenthal N, Sanes J. Proc Natl Acad Sci USA. 1991;88:5847–5851. doi: 10.1073/pnas.88.13.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McPherron A C, Lee S-J. J Biol Chem. 1993;268:3444–3449. [PubMed] [Google Scholar]
- 10.Heldin C-H, Miyazono K, ten Dijke P. Nature (London) 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- 11.Massagué J, Blain S W, Lo R S. Cell. 2000;103:295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- 12.Attisano L, Cárcamo J, Ventura F, Weis F M B, Massagué J, Wrana J. Cell. 1993;75:671–680. doi: 10.1016/0092-8674(93)90488-c. [DOI] [PubMed] [Google Scholar]
- 13.Massagué J. Cell. 1992;69:1067–1070. doi: 10.1016/0092-8674(92)90627-o. [DOI] [PubMed] [Google Scholar]
- 14.Wang X-F, Lin H, Ng-Eaton E, Downward J, Lodish H, Weinberg R. Cell. 1991;67:797–805. doi: 10.1016/0092-8674(91)90074-9. [DOI] [PubMed] [Google Scholar]
- 15.Miyazono K, Hellman U, Wernstedt C, Heldin C-H. J Biol Chem. 1988;263:6407–6415. [PubMed] [Google Scholar]
- 16.Gentry L E, Nash B W. Biochemistry. 1990;29:6851–6857. doi: 10.1021/bi00481a014. [DOI] [PubMed] [Google Scholar]
- 17.Böttinger E P, Factor V M, Tsang M L-S, Weatherbee J A, Kopp J B, Qian S W, Wakefield L M, Roberts A B, Thorgeirsson S S, Sporn M B. Proc Natl Acad Sci USA. 1996;93:5877–5882. doi: 10.1073/pnas.93.12.5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gamer L, Wolfman N, Celeste A, Hattersley G, Hewick R, Rosen V. Dev Biol. 1999;208:222–232. doi: 10.1006/dbio.1998.9191. [DOI] [PubMed] [Google Scholar]
- 19.Nakashima M, Toyono T, Akamine A, Joyner A. Mech Dev. 1999;80:185–189. doi: 10.1016/s0925-4773(98)00205-6. [DOI] [PubMed] [Google Scholar]
- 20.McPherron A C, Lawler A M, Lee S-J. Nat Genet. 1999;22:260–264. doi: 10.1038/10320. [DOI] [PubMed] [Google Scholar]
- 21.Matzuk M M, Lu N, Vogel H, Sellheyer K, Roop D R, Bradley A. Nature (London) 1995;374:360–363. doi: 10.1038/374360a0. [DOI] [PubMed] [Google Scholar]
- 22.Berk M, Desai S, Heyman H, Colmenares C. Genes Dev. 1997;11:2029–2039. doi: 10.1101/gad.11.16.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo K, Stroschein S, Wang W, Chen D, Martens E, Zhou S, Zhou Q. Genes Dev. 1999;13:2196–2206. doi: 10.1101/gad.13.17.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stroschein S, Wang W, Zhou S, Zhou Q, Luo K. Science. 1999;286:771–774. doi: 10.1126/science.286.5440.771. [DOI] [PubMed] [Google Scholar]
- 25.Sun Y, Liu X, Ng-Eaton E, Lodish H, Weinberg R. Proc Natl Acad Sci USA. 1999;96:12442–12447. doi: 10.1073/pnas.96.22.12442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun Y, Liu X, Ng-Eaton E, Lane W, Lodish H, Weinberg R. Mol Cell. 1999;4:499–509. doi: 10.1016/s1097-2765(00)80201-4. [DOI] [PubMed] [Google Scholar]
- 27.Akiyoshi S, Inoue H, Hanai J-i, Kusanagi K, Nemoto N, Miyazono K, Kawabata M. J Biol Chem. 1999;274:35269–35277. doi: 10.1074/jbc.274.49.35269. [DOI] [PubMed] [Google Scholar]
- 28.Sutrave P, Kelly A, Hughes S. Genes Dev. 1990;4:1462–1472. doi: 10.1101/gad.4.9.1462. [DOI] [PubMed] [Google Scholar]
- 29.Hemmati-Brivanlou A, Melton D A. Nature (London) 1992;359:609–614. doi: 10.1038/359609a0. [DOI] [PubMed] [Google Scholar]
- 30.Schulte-Merker S, Smith J C, Dale L. EMBO J. 1994;13:3533–3541. doi: 10.1002/j.1460-2075.1994.tb06660.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hemmati-Brivanlou A H, Thomsen G H. Dev Genet (Amsterdam) 1995;17:78–89. doi: 10.1002/dvg.1020170109. [DOI] [PubMed] [Google Scholar]
- 32.Meno C, Gritsman K, Ohishi S, Ohfuji Y, Heckscher E, Mochida K, Shimono A, Kondoh H, Talbot W S, Robertson E J, et al. Mol Cell. 1999;4:287–298. doi: 10.1016/s1097-2765(00)80331-7. [DOI] [PubMed] [Google Scholar]
- 33.Link B, Nishi R. Exp Cell Res. 1997;233:350–362. doi: 10.1006/excr.1997.3575. [DOI] [PubMed] [Google Scholar]