SUMMARY
The location of neurons early in development can be critical for their ability to differentiate and receive normal synaptic inputs. Indeed, disruptions in neuronal positioning lead to a variety of neurological disorders. Neurons have, however, shifted their positions across phylogeny, suggesting that changes in location do not always spell functional disaster. To investigate the functional consequences of abnormal positioning, we leveraged previously reported genetic perturbations to disrupt normal neuronal migration – and thus positioning – in a population of cranial motor neurons, the facial branchiomotor neurons (FBMNs). We used a combination of topographical, morphological, physiological, and behavioral analyses to determine if key functional features of FBMNs were still established in migration mutants, in spite of a dramatic rostrocaudal re-positioning of these neurons in hindbrain. We discovered that FBMNs seem remarkably resilient to a disruption in positioning, suggesting that they may not rely heavily on rostrocaudal positioning to guide their functional development. Thus, the role of positioning may vary across the developing nervous system, with some populations – like facial motor neurons – exhibiting greater resilience to abnormal positioning that permits them to shift location as a part of evolutionary change.
Keywords: zebrafish, hindbrain, motor neurons, neuronal migration, neural circuits, neural development, robustness
TOC image
The importance of proper positioning for the development of neuronal populations is still poorly understood. McArthur and Fetcho show that facial motor neurons in larval zebrafish exhibit a remarkable resilience to dramatic disruption in normal positioning, suggesting that hindbrain circuits may be robust to shifts in segmental organization.
INTRODUCTION
Spatial patterning supports neuronal differentiation and circuit formation in the developing nervous system. Neuronal subtypes arise and extend processes in specific locations, determined in part by extracellular signaling gradients [1]. Neurons then migrate from their birthplaces to particular locations, where they finish differentiating and extend processes to form synaptic connections [2]. Finally, neurons often arrange themselves into a specific topography within a given population – for example, according to relative age [3,4] – to further facilitate circuit development.
Somatic motor neurons provide a striking example of the relationship between spatial topography and circuit development. In mammalian spinal cord, discretely clustered motor pools are positioned in columns and columels according to the anatomical and functional arrangement of their target muscles [reviewed in 5]. Recent work has demonstrated the importance of this precise positioning for establishing appropriate synaptic connectivity. Disruption of motor pool clustering spares the integrity of the projection from motor neuron to muscle [6] but interferes with the formation of monosynaptic connections between muscle proprioceptors and limb motor neurons, with afferents targeting neurons in a specific region rather than recognizing neurons with a particular genetic identity [7]. These results imply that positioning, and not just molecular recognition cues, can be critical for establishing appropriate synaptic connectivity. Given that developing neurons are typically spatially structured, this seems a sensible strategy for guiding initial circuit formation.
The spatial structure of the developing nervous system has, however, evolved, with some neuronal populations exhibiting dramatic shifts in location across phylogeny. For example, some cranial motor nuclei are found in different rostrocaudal segments of the hindbrain in different taxa [8–12], attributed to inter-species differences in neuronal migration. Previous work has theorized that shifts in the final position of these motor nuclei correlate with changes in their dominant inputs [13,14], though the mechanisms driving this shift are not known. As these cranial nuclei shifted their positions across phylogeny, the motor neurons still needed to receive appropriate synaptic inputs to support the survival-critical behaviors mediated by this part of the brain. If position is critical for establishing circuit architecture, how did these shifting populations maintain or adapt their functional organization despite dramatic alterations to their segmental positions in the developing hindbrain?
To address this question, we investigated the functional development of facial branchiomotor neurons (FBMNs) in larval zebrafish. In wild type zebrafish, FBMNs arise and differentiate in one segment of the embryonic hindbrain, then migrate to settle in more caudal segments by two days post-fertilization (dpf) [15]. However, a number of genetic mutations have been identified that specifically disrupt this caudal migration while sparing the gross organization of the surrounding neuroepithelium [reviewed in 16], resulting in larval zebrafish with FBMNs located in the wrong hindbrain segment. We selected two of these mutant lines that disrupt caudal migration via distinct molecular mechanisms [17,18] and used cellular morphology, functional imaging, electrophysiology, and behavior to ask how the normal functional organization of FBMNs is affected by a disruption in caudal migration.
Because of the apparent importance of cell positioning in the developing nervous system, we expected that at least some functional properties of the facial motor nucleus would be grossly disrupted in migration mutants. Instead, we find that FBMNs in migration mutants still achieve similar intra-population motor pool and age topography, exhibit similar gross patterns of activity at the single cell and population level, and show similar rhythmic facial movements to their wild type counterparts. While we cannot conclude that abnormal migration has no impact on circuit architecture or function, the similarities in the properties of misplaced and correctly positioned neurons lead us to conclude that a shift in neuronal position need not result in a massive functional disruption. Instead, some neuronal populations may develop in a way that permits a degree of functional resilience in the face of positional shifts, perhaps conferring on that population the ability to maintain its participation in pre-existing circuits while opportunistically accessing novel synaptic inputs – thus providing a substrate for circuit evolution.
RESULTS
Functional Topography: Facial Motor Pools
In wild type zebrafish hindbrain, FBMNs migrate caudally from their birthplace in rhombomere 4 to settle in rhombomeres 5–7. In the llk(rw16) [17] and pk1b(fh122) [18] mutant lines, FBMNs fail to migrate caudally and remain instead in rhombomere 4 (Figure 1A–C). As a first step towards understanding the importance of rostrocaudal positioning for neural circuit development, we set out to determine whether the neuronal population in migration mutants could achieve a topographical organization similar to that in wild type. Because we already knew that spinal motor neurons cluster according to their muscle targets [5] and that this organization can be critical for establishing synaptic connectivity [7], we began by probing the topography of facial motor pools.
Figure 1. Facial motor neurons migrate caudally in wild type but not migration mutant larvae, and backfills from cranial muscles reveal the location of facial motor pools.
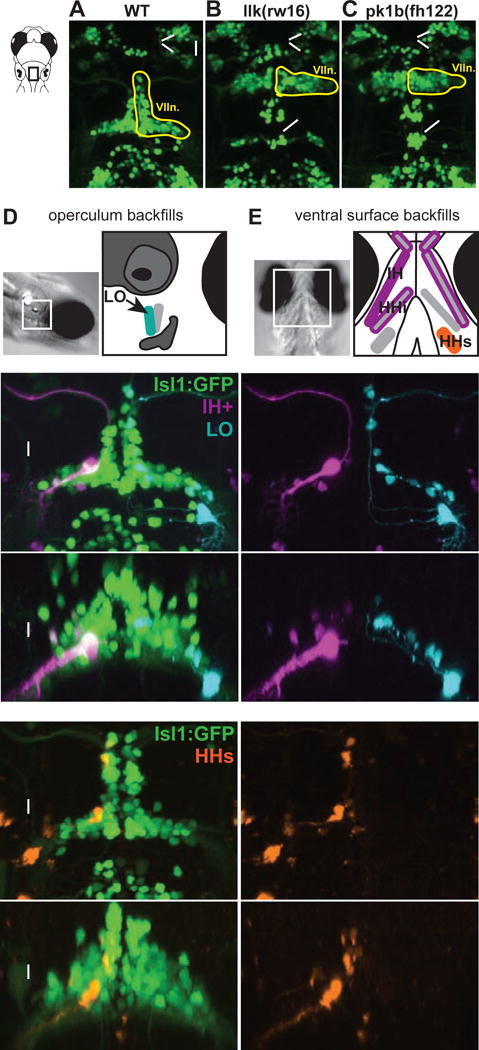
Top panels show the hindbrain of Tg(Islet1:GFP) larvae at 5 dpf. (A) In wild type larvae, facial motor neurons are born in rhombomere 4 (R4) and migrate caudally to settle in rhombomeres 5–7 by 48 hpf. In both llk(rw16) mutant larvae (B) and pk1b(fh122) mutant larvae (C), facial motor neurons fail to migrate caudally and remain in R4. Octavolateralis efferents (OLes) born in R4 and R6 also fail to migrate caudally. (D-E) Schematic representation of the cranial muscles targeted for backfills of the facial motor nerve. Filled muscles indicate the electroporation target, and outlined muscles correspond to other muscles labeled with dye after target electroporation. (F-G) Electroporation of fluorescent dyes into the IH (magenta) and LO (cyan) muscles on opposite sides of a single zebrafish larva (4dpf) backfills reveals a subset of each muscle’s facial motor pool, imaged at 5dpf. (H-I) Targeting the electroporation to the HHs (orange) backfills a subset of that muscle’s motor pool. Arrowheads indicate backfilled cell bodies. Vn. = trigeminal motor nuclei, VIIn. = facial motor nucleus, Xn. = vagal motor nucleus, IH+ = interhyoideus + hyohyoideus inferior, LO = levator operculi, HHs = hyohyoideus superior. See also Figure S1.
In larval zebrafish, the facial motor nerve innervates multiple muscles over the operculum (e.g. levator operculi, LO) and the ventral surface of the buccal cavity (interhyoideus, IH; hyohyoideus inferior, HHi; hyohyoideus superior, HHs) [19]. To determine the location of facial motor pools projecting to these muscles, we backfilled FBMNs contributing axons to specific branches of the facial motor nerve by electroporating fluorescent dye into the nerve over each muscle at 4dpf, and imaged the filled motor neuron cell bodies at 5dpf. Pilot experiments indicated that some FBMNs project to multiple muscles at this stage – confirmed by filling single cells with dye and visualizing the axon branching pattern – while other FBMNs project to only a single muscle (Figure S1). Based on the backfills, we set out to describe the spatial topography of three motor pools: one pool innervating the ipsilateral LO (Figure 1D); a second pool innervating the bilateral IH, ipsilateral HHi, and bilateral intermandibularis posterior (referred to as the IH+ pool; Figure 1E, magenta); and a third pool innervating the ipsilateral HHs (Figure 1E, orange). Each backfill labeled a subset of the FBMNs projecting to a given muscle in that animal (Figure 1F–I).
A summary of the results is shown in Figure 2. In wild type larvae, motor pools exhibit a significant amount of overlap, unlike the discrete clusters observed in mammalian spinal cord [5]. However, a consistent topographic arrangement is still apparent (Figure 2A). LO-projecting neurons are found throughout the dorsoventral and mediolateral extent of the facial nucleus, and make up most of the facial motor neurons in the ventrolateral arm of the facial nucleus. IH+-projecting neurons are found in the intermediate portion of the cross-section, while HHs-projecting neurons are found primarily in the most dorsomedial region. Though the shape of the facial nucleus itself differs a bit between wild type larvae and migration mutants, the spatial arrangement of the facial motor pools remains much the same, with a dorsomedial HHs pool, an intermediate IH+ pool, and an expansive LO pool in both llk(rw16) (Figure 2B) and pk1b(fh122) (Figure 2C) mutants. Thus, in a statistical comparison using a linear mixed effects model (Table S1), muscle target was a significant predictor of FBMN cross-sectional position (mediolateral: p < 0.001; dorsoventral: p < 0.001), but there was no significant interaction between muscle target and migration phenotype (mediolateral: p = 0.29; dorsoventral: p = 0.26) – consistent with a topographic arrangement of motor pools that is not significantly affected by the disruption of their caudal migration.
Figure 2. Facial motor pools exhibit a loose spatial topography that is maintained in caudal migration mutants.
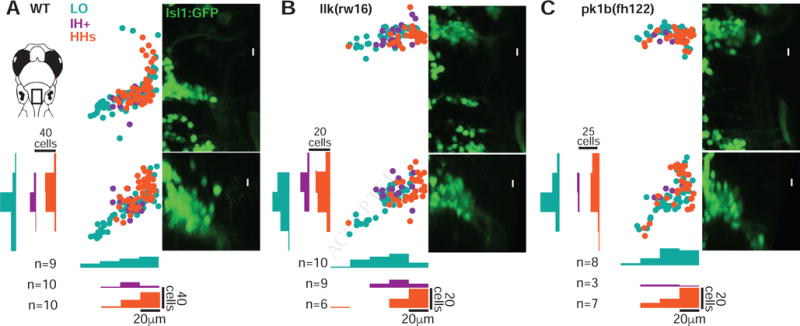
(A) Across wild type larvae, the LO motor pool (teal) extends throughout the entire facial motor nucleus, while the IH+ (purple) and HHs (orange) motor pools are found in more restricted but still overlapping regions – in the middle and dorsomedial portions of the nucleus, respectively. In llk(rw16) mutant larvae (B) and pk1b(fh122) mutant larvae (C), the cross-sectional topography of the motor pools is maintained (Table S1). All fish were backfilled at 4dpf and imaged at 5dpf. n = number of fish used in each condition. Histograms illustrate the distribution of cells along the mediolateral and dorsoventral axes, binned at 20μm intervals. LO = levator operculi, IH+ = interhyoideus + hyohyoideus inferior, HHs = hyohyoideus superior. See also Figure S1.
Functional Topography: Relative Age
Many neuronal populations in the developing nervous system are organized according to relative age. In larval zebrafish, spinal cord [20,21] and hindbrain [4,21–23] motor and interneurons are organized topographically by age along the dorsoventral axis, and the dorsoventral location of the soma correlates with key functional properties such as recruitment order and axon morphology [4,21,24]. This led us to use transgenic expression of the photo convertible protein Dendra2 to determine if facial motor neurons were organized according to relative age, and if this age topography was still established in caudal migration mutants.
Dendra2 was expressed under the zCREST1 promoter/enhancer element [25], causing FBMNs to start expressing green (unconverted) Dendra protein once they exited the cell cycle and began to differentiate as cranial efferent neurons. Exposure to UV light at 20, 24, or 30 hours post-fertilization (hpf) converted the green protein in differentiated neurons to red. Imaging at 3dpf then revealed which neurons were older (red) and which had differentiated after the conversion (green only).
In wild type fish, FBMNs are arranged by age along the dorsoventral axis, as shown in Figure 3A (n=17 fish across three conversion time points). For each conversion time point, the older neurons were found in the most ventral part of the facial nucleus, and younger neurons were found mostly dorsal to them. In migration mutants, FBMNs still established this dorsoventral age topography (llk(rw16): Figure 3B, n=13; pk1b(fh122): Figure S2, n=14). Although there was a modest tendency for the oldest neurons in pk1b(fh122) mutants to be located laterally, as compared to wild type and llk(rw16) mutants, there was still a clear dorsoventral arrangement by relative age in all three groups. Thus, while mutant FBMNs are located in the wrong rostrocaudal hindbrain segment, they still establish the cross-sectional organization seen in wild type, according to both muscle target and relative neuronal age.
Figure 3. Facial motor neurons are topographically organized according to age in both wild type and caudal migration mutants.
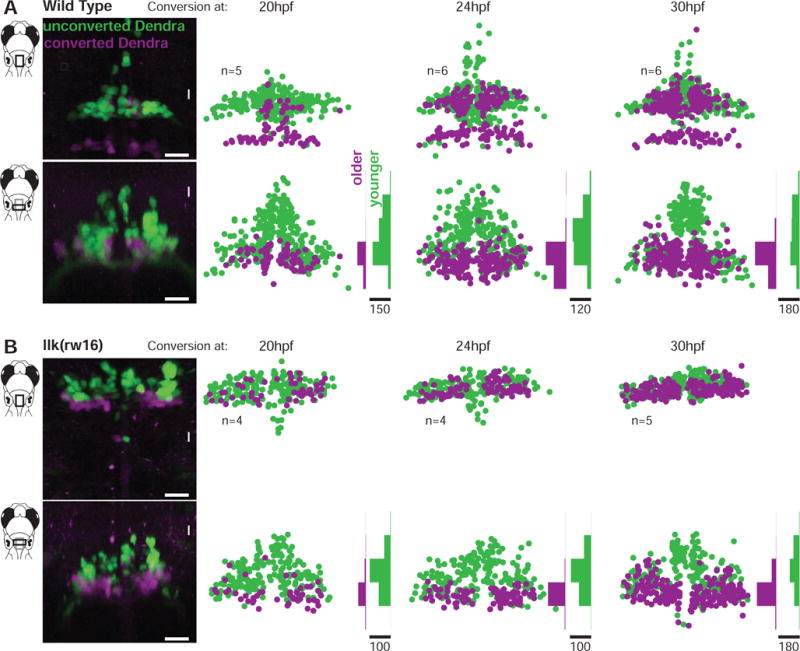
Zebrafish embryos expressed the photo convertible protein Dendra2 in cranial motor neurons and were exposed to ultraviolet light at one of three time points during facial motor neuron differentiation (20, 24 or 30 hpf), then imaged at 3dpf. Neurons older than the conversion time point contain converted (magenta/purple) Dendra, while younger neurons contain only unconverted (green) Dendra. In both wild type (A) and llk(rw16) mutant (B) larvae, older facial motor neurons are located in the ventral-most part of the facial motor nucleus. Examples in left panels of (A) and (B) have been color adjusted to increase visibility of the converted (magenta) Dendra signal. n = number of fish used in each condition. Histograms illustrate the distribution of cells along the dorsoventral axis binned at 20μm intervals, where the scale bars refer to the number of cells. See Figure S2 for data from pk1b(fh122) mutants.
Rostrocaudal Location of Dendrites
Although FBMN cell bodies are located in the wrong rostrocaudal segment in migration mutants, these neurons could still sample the same brain region for presynaptic inputs as wild type FBMNs if they extended their dendritic arbors into the same region of caudal neuropil that they occupy in wild type fish. To reveal FBMN dendritic arbors, we filled single FBMNs with fluorescent dye and traced their dendrites (Figure 4). We focused primarily on neurons from the IH+ motor pool, to facilitate a direct comparison between wild type and migration mutant neurons. In both wild type (n = 8; example shown in Figure 4A) and llk(rw16) migration mutant (n = 5; example shown in Figure 4B) larvae, FBMNs extended their dendrites primarily into the neuropil found lateral, ventral, and caudal to the cell body. Although there was some modest overlap, wild type FBMNs – which had migrated properly into rhombomere 6 – accessed much more caudal regions of hindbrain neuropil than their migration mutant counterparts located in rhombomere 4 (summarized in Figure 4C). Our data are, therefore, inconsistent with the hypothesis that mutant FBMNs simply extend their dendrites to reach wild type-innervated regions of neuropil – and instead suggest that the region of neuropil sampled by FBMN dendrites shifts along with the rostrocaudal position of their cell bodies.
Figure 4. The location of dendrites from facial motor neurons in wild type and migration mutants.
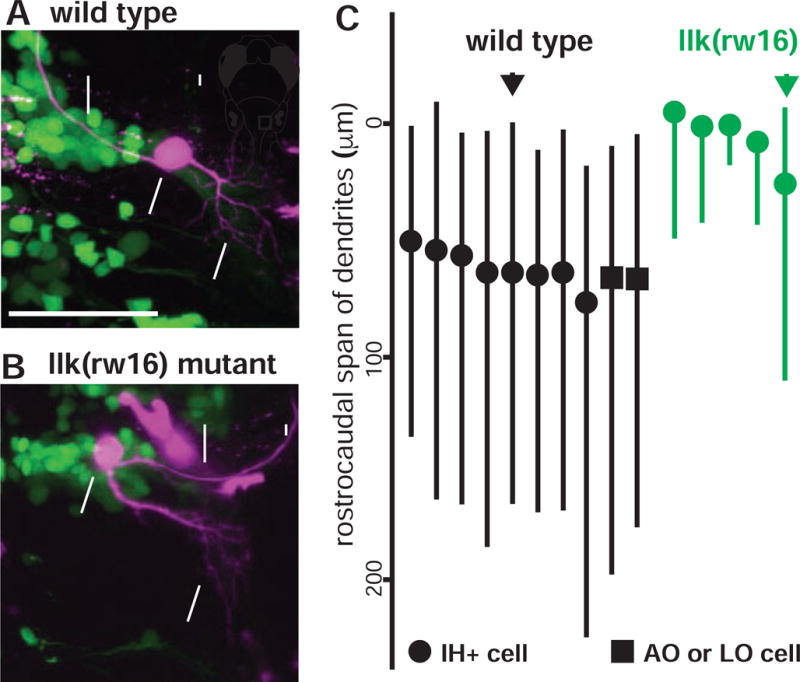
In both wild type (A) and llk(rw16) mutant (B) larvae, single-cell dye fills show that FBMNs extend their dendrites primarily into the neuropil caudal and lateral to the cell body. (C) However, a comparison of the rostrocaudal span of FBMN dendrites reveals that wild type FBMNs innervate more caudal regions of the neuropil than migration mutant FBMNs. Circles = IH+-projecting FBMNs; squares = operculum-projecting FBMNs. Black and green arrows correspond to the neurons used in (A) and (B), respectively. All cells were filled at 4dpf and imaged at 5dpf.
Patterns of Activity: Electrophysiology
The morphological data indicate that the internal spatial topography of FBMNs is resilient to a dramatic shift in segmental location, but both their cell bodies and dendrites are still in the wrong location in the developing brain. Does a segmental shift in neuronal position block FBMNs’ access to critical synaptic inputs, thereby disrupting patterns of activity, or can shifted FBMNs still achieve patterns of activity similar to wild type?
We began by using whole cell recordings to observe the spontaneous activity of wild type FBMNs in the ventrolateral arm of the facial nucleus in rhombomere 6 (Figure 5A), sometimes with simultaneous recording of ventral motor root activity in the tail (wild type: n=6/23 fish; llk(rw16) mutants: 3/8 fish). All FBMNs fired spontaneous bursts – depolarization hills (approximately 5–10mV in amplitude) typically topped by one or multiple small spikes. Some neurons exhibited only infrequent but strong bursting activity (n=8/23 cells; Figure 5B), which correlated with spontaneous motor activity in the tail when monitored. Others also exhibited bouts of shorter, rhythmic bursting (n=15/23 cells; Figure 5C) that were typically not accompanied by tail motor activity.
Figure 5. Whole cell recordings show that rhythmic and swim-related activity exhibited by wild type FBMNs is also present in migration mutant FBMNs.
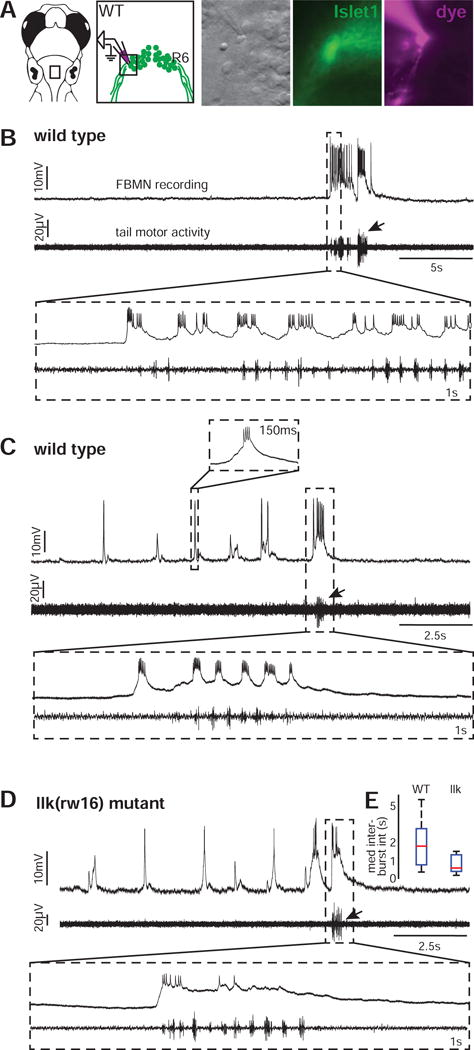
(A) Whole cell patch clamp recordings were made from the ventrolateral arm of the facial motor nucleus. (B) These recordings reveal a subset of wild type FBMNs (n=8/23 cells) that burst infrequently, correlated with tail motor activity. (C) Other wild type FBMNs also exhibit rhythmic bursting, typically in the absence of tail motor activity (n=15/23 cells). (D) Migration mutant llk(rw16) FBMNs can also respond during attempted tail movements (infrequent bursts only: n=2/8 cells) and in rhythmic bursts without large tail motor activity (n=6/8 cells). All recordings were done at 5dpf. Arrows indicate bursts of attempted tail movement activity. (E) Box-whisker plot showing the distribution of median rhythmic inter-burst intervals in wild type (n=15 cells) and llk(w16) mutant (n=6 cells) larvae.
Remarkably, when we recorded from FBMNs in the same ventrolateral region of the facial nucleus (now located in rhombomere 4) in llk(rw16) migration mutants, we found evidence that migration mutant FBMNs exhibit both types of activity observed in wild type: a minority of cells (n=2/8 cells) only fired infrequent bursts (correlated with strong tail motor activity when recorded simultaneously), and other FBMNs fired strongly during infrequent tail motor bouts but also exhibited rhythmic activity (n=6/8 cells; Figure 5D). These rhythmic bursts occur at overlapping frequency ranges (typically 0.25–2Hz) in wild type and mutant fish (Figure 5E), though there was more variability in the wild type sample and the mutants fell toward the bottom of the wild type range. Thus, electrophysiology shows us that the fundamental firing patterns of FBMNs are similar in wild type and mutants, but dissection might be disrupting sensory inputs to facial motor circuits and affecting firing frequency. Furthermore, recording a single cell per fish makes it difficult to draw conclusions about how activity patterns are distributed across the population. We therefore also monitored activity in a broader sample using functional imaging.
Patterns of Activity: Population Calcium Imaging
To explore how activity patterns were distributed across FBMNs in intact larvae, we imaged FBMNs expressing a nuclear localized calcium indicator (Tg(Elavl3:H2B-GCaMP6fast); [26]). When we imaged a series of slices through the dorsoventral extent of the wild type facial motor nucleus at 5dpf, we observed FBMN calcium signals consistent with activity patterns seen in whole cell recordings (Figure 6A). Some neurons only exhibited large, infrequent transients (typically one per minute), correlated in time with large body movements, while other neurons also exhibited bouts of rhythmically oscillating calcium activity. Consistent with the electrophysiological data, these two patterns were also observed in the llk(rw16) migration mutants (Figure 6B). In addition, when we targeted LO-projecting neurons by backfilling them prior to imaging, we found that both response patterns were exhibited by neurons within a single motor pool – and once again, this was true for both wild type and migration mutant larvae (raw data not shown; see summary data in Figure 6C).
Figure 6. Calcium imaging in intact larvae confirms patterns of FBMN activity, and reveals a robust functional spatial topography.
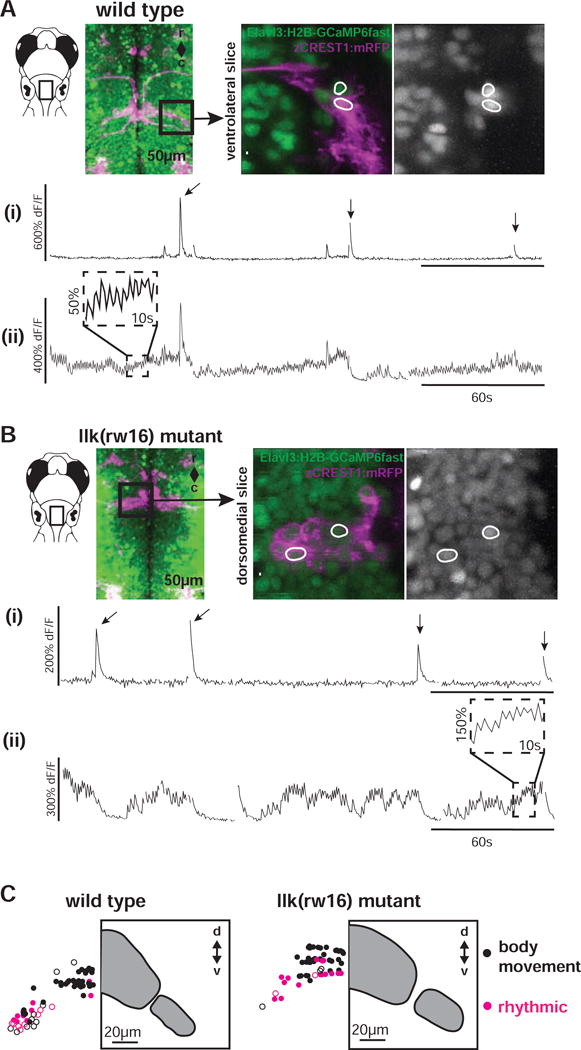
(A) In wild type larvae, some FBMNs exhibit only large, infrequent calcium transients during attempted body movements, while other FBMNs exhibit additional rhythmic activity. These two response types could be observed in neighboring FBMNs, as shown here. Outlined regions of interest (i and ii) correspond to the calcium traces shown in (i) and (ii). Sampling interval = 247ms. (B) FBMNs in llk(rw16) migration mutants also exhibit these two activity patterns. Sampling interval = 493ms. Arrows in (A) and (B) indicate large body movements. (C) In both wild type (left) and llk(rw16) mutant (right) larvae, there was significant overlap of neurons exhibiting infrequent-only (black symbols) and rhythmic (magenta symbols) activity, though rhythmic neurons were concentrated ventrolaterally (Table S2). Open symbols = neurons backfilled from the LO (operculum). Grey panels show the cross-sectional outline of the facial motor nucleus for each phenotype, based on backfill data shown in Figure 2. Number of fish used to assemble summary data: n=5 wild type (64 classified neurons in total), n=6 llk(rw16) mutant (60 classified neurons in total). All data shown here were obtained from unparalyzed fish at 5dpf.
To explore the topography of activity patterns, we categorized FBMN responses as rhythmic or non-rhythmic (with large infrequent responses only), then plotted their registered positions within the facial motor nucleus (Figure 6C). Across wild type larvae (n=7 fish; 2 fish without backfill, and 5 fish in which only LO-projecting neurons were sampled), FBMNs with different response patterns overlapped spatially, but rhythmically oscillating neurons were concentrated in the ventrolateral arm of the motor nucleus. In migration mutants (n=6 fish; 3 fish without backfill, and 3 fish in which only LO-projecting neurons were sampled), these rhythmic cells are still in the ventral-most portion of the sampled region. Indeed, when we fit a linear mixed effects model to the data (Table S2), we found that response type (rhythmic v. non-rhythmic) was a significant predictor of neurons’ dorsoventral position (p < 0.001). However, we failed to find statistical evidence for an interaction between migration phenotype and response type (p = 0.14) – indicating that the dorsoventral distribution of rhythmic neurons was similar in wild type and mutant larvae. This points to at least some preservation of not only similar activity patterns but a similar topographic distribution of those patterns in migration mutants.
What do these patterns of activity represent? At 5dpf, hypoxia evokes bouts of rhythmic contractions by buccal and opercular muscle, with infrequent whole-body movements [27,28]. These movements likely facilitate oxygen diffusion across the skin prior to gill maturation, and precede the continuous respiratory movements that develop a few days later [27,29]. Immobilization (by dissection or agarose embedding) likely evokes respiratory facial activity by decreasing fluid flow (compared to a freely swimming fish). Indeed, the frequency range of rhythmic FBMN bursts overlaps with the reported frequency range of hypoxia-induced facial behaviors [27]. Thus, the imaging and physiology indicate substantial preservation of the rhythmic respiratory control of facial motoneurons in the mutants, which we expected would lead to a preservation of the movements they drive.
Behavioral Patterns: Rhythmic Operculum Movements
To determine if rhythmic movements were, indeed, robust to abnormal FBMN migration, we imaged movements of the operculum, which rhythmically adducts and rotates (driven by the facially-innervated AO and LO muscles, respectively) in response to hypoxia [19,27] (Figure 7A). Wild type larvae restrained in agarose exhibit bouts of operculum movement, including smaller rhythmic movements and stronger movements during attempted whole-body motion (n = 8 fish; Figure 7B), as previously reported [27]. Consistent with our electrophysiological and calcium imaging results, migration mutant larvae also exhibit this pattern (n = 8 fish; Figure 7C). Focusing on the rhythmic component, we compared the distribution of inter-movement intervals for wild type and mutant larvae. Mutants exhibited a broader range of median movement frequencies across individuals (Figure 7D, left panel), perhaps due in part to the lengthening of the mutant interval histogram tail (Figure 7D, right panel) – corresponding to a modest increase in the number of slow operculum movements observed. However, the overall range and distribution of inter-movement intervals in wild type and mutant larvae were still quite similar (Figure 7D, right panel). Indeed, a statistical comparison using a linear mixed effects model failed to identify migration phenotype as a significant predictor of inter-movement interval (p = 0.613; Table S3) – consistent with statistically similar frequencies of opercular movements in wild type and mutant fish. Thus, while there are hints of potential subtle differences between wild type and mutant larval behavior, the topography of facial motor pools, the age topography of the motor nucleus, two major patterns of neuronal activity (and their topographic arrangement across the population), and the overall pattern and rhythmic frequency of operculum movements are still surprisingly robust to a radical re-positioning of facial motor neurons.
Figure 7. Wild type and migration mutant larvae exhibit rhythmic opercular movements with similar temporal properties.
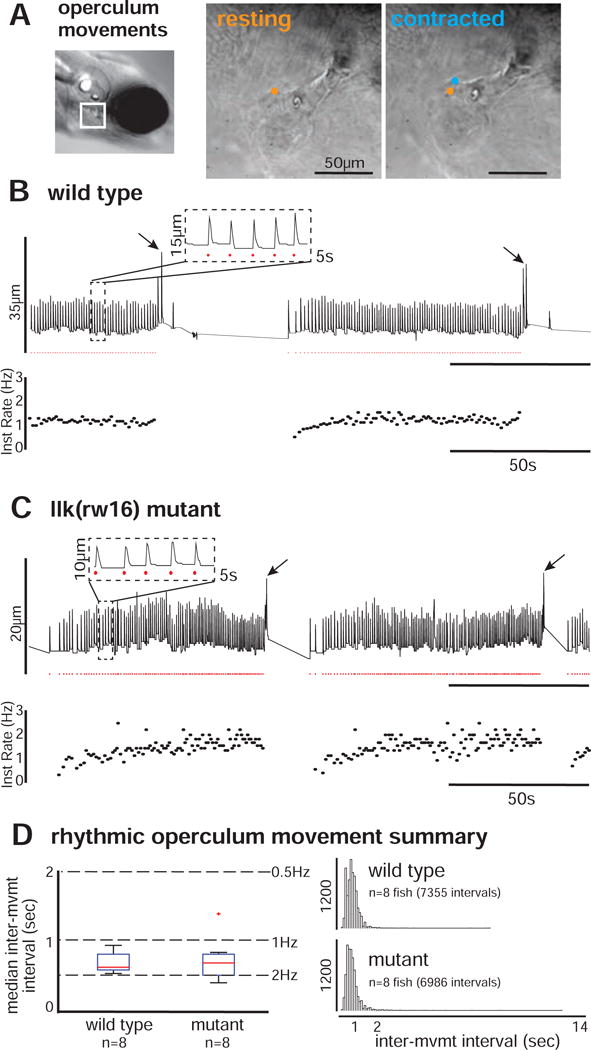
(A) Operculum rotations were manually recorded from restrained larvae, by tracking the position of landmarks over time across resting (orange) and contracted (blue) states (sampling frequency = 20Hz). For each example (B-C), vertical landmark position is plotted over time (top trace), with rhythmic operculum movement times indicated by red dots and instantaneous movement frequency plotted below. Both wild type (B) and llk(rw16) mutant (C) larvae exhibit bouts of rhythmic operculum rotation. (D) The box-whisker plot (left) shows the distribution of per-fish median inter-movement intervals in wild type (n=8) and mutant (n=8) larvae. The histograms (right) show the complete distribution of inter-movement intervals in wild type (top; n=7355 intervals) and mutant (bottom; n=6986 intervals) larvae. Migration phenotype was not a significant predictor of inter-movement interval (p = 0.613; see Table S3).
DISCUSSION
We set out to investigate the rules linking the position and function of a neuronal population in the developing brain. Genetic disruptions of the caudal migration of the facial motor nucleus in larval zebrafish allowed us to explore the effects of manipulating its segmental position in the developing hindbrain. A comparison of key features of wild type and migration mutant facial motor neurons revealed a striking similarity in both intra-population motor pool and age topography as well as spontaneous activity patterns, consistent with the similarity between wild type and mutant operculum behavior. This phenotypic resilience in the face of a dramatic positional shift has important implications for our understanding of the development and evolution of neural circuits.
Resilience to Abnormal Migration: Evidence and Caveats
After a change in the segmental position of FBMNs in migration mutants, their intra-population topography was maintained, as were their gross activity patterns – all of this despite the fact that their dendritic arbors, like their cell bodies, were in the wrong rostrocaudal location in the developing hindbrain. The similarity of the activity patterns is particularly interesting and important. The two firing patterns we observed correspond to two types of cranial respiratory activity in response to hypoxia, one rhythmic and one infrequent and correlated with large body movements [27]. Similar respiratory patterns have also been observed in lamprey [30]. The fact that FBMNs can dramatically shift their segmental position and still potentially receive inputs from the same (or functionally similar) respiratory pattern generators in caudal hindbrain [31] suggests that achieving a specific segmental position is not strictly necessary for the development of major features of facial motor output. If we had identified clear functional abnormalities in mutants (e.g. loss or severe disruption of rhythmic activity) – as we expected – then we would be able to confidently conclude that these neurons were not resilient to abnormal migration, consistent with a central role for segmental positioning in circuit development. Instead, we observed similar facial nucleus topography, similar facial motor neuron activity patterns, and similar facial motor behavior in wild type and migration mutant larvae. This leads us to conclude that facial motor circuits are, indeed, robust to abnormal migration – at least in terms of the spatial patterning and gross motor outputs that were studied herein.
Others have used genetic manipulation of hindbrain patterning genes to provide some evidence of flexibility and robustness in the developmental program, by studying the functional properties of ectopically induced neurons [32,33] and populations developing in the absence of normal segmental gene expression [34]. Indeed, a previous study in mice [35] provided indirect evidence that a mutation blocking FBMN caudal migration also extended the rostrocaudal range of the embryonic parafacial oscillator (e-pF) and spared specific features of respiratory connectivity. These results lend support to the conclusion that some circuits and associated behaviors can be robust to changes in neuronal locations.
Still, facial motor circuits might be sensitive to abnormal positioning in ways that we simply failed to detect in the current study. While mutant FBMNs still seem to incorporate into larval respiratory circuits, their connection to other sensorimotor circuits – such as those involved in suction feeding – might be more vulnerable to a positional shift. Further, normal positioning could be less critical for early circuitry and more critical for mature circuitry, particularly given the fact that the gills are not yet functional at the ages we studied. In addition, even when circuits achieve their normal function despite some developmental perturbation, they may be vulnerable to dysfunction in specific contexts. For example, Ma et al. [34] found that disruption of normal segmental gene expression in valentino zebrafish mutants resulted in sporadic, aberrant oculomotor behaviors (correlated with jaw and fin movements) layered on top of normal horizontal eye movements. The authors concluded that normal segmental signaling may protect developing circuits from aberrant connectivity. Studies like this one reinforce the importance of continuing to probe the boundaries of facial motor circuit resilience to abnormal positioning.
There may also be position-dependent reconfigurations of the underlying circuitry in mutants that preserve gross functional output. For example, there could be changes in the fidelity or strength of specific synaptic inputs, which might not be readily apparent from single-cell recordings or calcium imaging. Further, similar activity patterns could be achieved in distinct ways in wild type and migration mutant larvae if, for example, mutant FBMNs received rhythmic input from a different set of respiratory neurons. At the very least, given our observation that the dendritic arbors shift their position along with the cell body in migration mutants, premotor neurons likely send their axonal projections to a different region of the hindbrain in order to reach FBMN targets.
Development Rules Linking Position and Function
In many cases, normal positioning is clearly important for a neuron’s ability to incorporate into functional circuits. A particularly clear example can be seen in mammalian spinal cord, where disrupting the clustering and positioning of motor pools results in abnormal afferent-motoneuron synaptic connectivity [7]. Significant changes in cell morphology, physiology, and synaptic connectivity also follow abnormal neuronal migration in the olfactory bulb [36], hippocampus [37], and pre-cerebellar nuclei [38]. Indeed, abnormal neuronal migration in humans is associated with a variety of neurological problems, such as seizures and cognitive impairment [39]. It seems likely that different strategies for synaptic specification are used in different contexts, depending on the nature of the circuits they are used to construct. In cases where input-output mappings are rigid, we might predict that neurons should be more precisely positioned and that synaptic specification should rely more heavily on that positioning – as has been demonstrated for afferent-to-motoneuron synapses in mammalian spinal cord [7]. In the hindbrain, premotor circuits have a more flexible relationship to specific cranial motor nuclei – as the connectivity of these nuclei evolves in parallel with changes in peripheral musculoskeletal structure and function [8–11,31]. Thus, synaptic specification might rely less heavily on segmental position and more heavily on molecular recognition and activity-dependent refinement, with premotor axons (from respiratory central pattern generators, for example) simply extending along the rostrocaudal axis until they find their target motoneurons.
The resilience to rostrocaudal location does not entirely rule out a role for location in the connectivity of the facial motoneurons. Indeed, our evidence indicates that dorsoventral position (more so than segmental position) could be an important factor for normal functional connectivity. Migration mutant FBMNs still achieve wild type-like topography along the dorsoventral axis, according to relative age and muscle target. Others have shown that dorsoventral age topography in the surrounding hindbrain interneurons predicts those interneurons’ contributions to functional circuits [24]. If dorsoventral positioning is being used as an internal coordinate system for specifying synaptic connections within the facial motor nucleus, longitudinal premotor projections may reach the FBMNs at any rostrocaudal level and form appropriate synapses, so long as that intra-population topography remains intact – which it does in the case of migration mutant FBMNs.
Neuronal Migration and Evolution
If facial motor neurons can, indeed, shift position along the rostrocaudal axis and still receive functional inputs, why do they migrate at all? Though FBMNs arise in the same hindbrain segment across vertebrates, their final settling position along the rostrocaudal axis varies dramatically across species [12,14]. Others have noted that these phylogenetic shifts in position seem to correlate with changes in function: in species where FBMNs receive strong inputs from caudal hindbrain networks (like respiratory central pattern generators), FBMNs migrate and settle in more caudal positions [8–11]. Adjusting the extent of neuronal migration to shift FBMNs closer to the source of their predominant synaptic inputs could be adaptive, by minimizing the total length of projections – in accord with the notion of wiring minimization [40,41] – or by adjusting the probability or strength of synaptic connections without specifying them in a deterministic fashion (as suggested by [34]). These selective forces (and possibly others) may act in tandem to confer an adaptive advantage on one settling location over another in a particular species. Our discovery of the remarkable functional resilience of FBMNs to a shift in segmental position provides an opportunity to investigate how this resilience is achieved. This work will be important for our understanding of the fundamental ontogenetic processes that generate neuronal diversity and synaptic connectivity, and of how these processes facilitate or constrain the evolution of neural circuits.
STAR METHODS
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. Joseph Fetcho (jrf49@cornell.edu).
Experimental Model and Subject Details
Zebrafish (Danio rerio) larvae have not undergone sex specification at the ages used in these experiments (1–5 dpf). All zebrafish larvae were obtained from crosses between adult laboratory stocks. Zebrafish larvae were maintained in nursery tubes in our fish facility until selected for experiments and subjected to their first experimental procedure (i.e. photoconversion, electroporation of dye), after which they were maintained in dishes of 10% Hank’s solution (in mM: 13.7 NaCl, 0.54 KCl, 0.10 MgSO4, 0.044 KH2PO4, 0.025 Na2HPO4, 0.42 NaHCO3, 0.13 CaCl2 in purified water) in an incubator (28.5°C). Most fish were staged according to days post-fertilization (dpf); however, for photoconversion experiments, fish were initially staged according to hours post-fertilization (hpf) within +30 minutes. Electrophysiological experiments were performed with larvae lacking any mutation for skin pigmentation. Other experiments were performed using larvae with skin color mutations (either nacre or casper) [42,43] to increase visibility during confocal imaging. All fish were maintained on a 14:10 light:dark cycle. All experimental procedures conformed to the US National Institutes of Health guidelines regarding scientific use of animals and were approved by Cornell University’s Institutional Animal Care and Use Committee.
The transgenic zebrafish lines Tg(zCREST1:Dendra2) and Tg(zCREST1:GCaMP5) were generated via plasmid injection into one-cell stage embryos, as described in [44]. zCREST1:mRFP plasmid was generously donated by C. Moens. Transgenic zebrafish lines Tg(Islet1:GFP) [45], Tg(zCREST1:mRFP) [18], and Tg(Elavl3:H2B-GCaMP6fast) [26] were described previously, as were the mutant lines utilized. For our comparisons, we chose two previously described mutations that disrupt FBMN caudal migration. llk(rw16) is a point mutation in scribble1 (scrb1), which is widely transcribed in the developing neuroepithelium and codes for a protein involved in establishing planar cell polarity [17]. In homozygous llk(rw16) mutants, FBMNs are unable to migrate caudally at all – though the movement of migratory populations in other parts of the nervous system appears unaffected. pk1b(fh122) is a mutation in prickle1b (which is expressed by migrating FBMNs and a subset of nearby hindbrain neurons) that interferes with the protein’s ability to localize to the nucleus and interact with the transcription factor REST [18,46] and likely also participates in the planar cell polarity pathway [47]. In homozygous pk1b(fh122) mutants, FBMNs are believed to be unable to postpone terminal differentiation, causing them to stop migrating prematurely. Fish were classified as migration mutants according to the migration phenotype of the FBMNs. In general, a given data collection session included both wild type and mutant larvae from the same multi-pair cross.
Method Details
Backfills and Single-Cell Fills: Electroporation
Tg(Islet1:GFP) larvae (4dpf) were anesthetized (MS-222, tricaine methanesulfonate, Western Chemical, Inc.; 0.226mg/mL in 10% Hank’s solution) and immobilized in 2% low melting point agarose (Fisher Scientific). Voltage pulse trains were delivered with an Axoporator device (Molecular Devices) through a fine-tipped glass micropipette (same design used for whole cell recordings; see below). Backfills from muscle: Fish were mounted such that the target muscle was just underneath the agarose surface, and the agarose was covered with anesthetic solution. The electroporation pipette was filled with dye (Alexa Fluor 546 or 647, dextran conjugated, MW 10000, anionic, Molecular Probes; 20% in extracellular solution – see below) and lowered over the facial nerve where it branches over the target muscle. Voltage pulse trains were delivered to the tissue (20–30V, 120Hz, 1-ms pulses, 20s per train, 2–3 trains per site) to electroporate dye into the nerve. In some cases, two muscles in the same fish were targeted on opposite sides of the head, using different dyes. Single-cell fills: Fish were mounted with the dorsal surface of the head just underneath the agarose surface, and the agarose was covered with anesthetic solution. The electroporation pipette was filled with dye as described above and driven into the intact hindbrain behind the ear, until the tip of the pipette was just touching the cell body of a single facial motor neuron. Voltage pulse trains were delivered to the cell surface (3–5V, 50Hz, 1-ms pulses, 3–5s per train, one train per cell) to electroporate dye into a single neuron. After electroporations, fish were allowed to recover overnight, then imaged at 5dpf. For backfills, muscle target was confirmed by imaging the facial nerve and cranial muscles.
Photoconversion of Dendra
Tg(zCREST1:Dendra2) embryos were exposed in-chorion to ultraviolet light at 20, 24, or 30 hpf, using a mercury lamp (DAPI filtered) focused through a 10x objective. Four embryos at a time were suspended in a drop of 10% Hank’s solution near the glass bottom of a small specimen dish, then exposed to the ultraviolet light. Ultraviolet light power and exposure time were chosen in pilot experiments to maximize photo conversion of Dendra (from green to red emission) while minimizing tissue damage, then standardized across experimental subjects (2.5 min, 50% UV power). After photo conversion, fish were kept in a light-tight container in an incubator (28.5°C) until imaging at 3 dpf. Nearly all fish survived to imaging after photo conversion without apparent morphological abnormalities, suggesting that the length and power of ultraviolet light exposure did not have a detrimental effect on development.
Image Acquisition
Larvae were immobilized in 2% low-melting point agarose (Fisher Scientific) against the class coverslip of a small specimen dish. The agarose was covered with solution to prevent dessication, and imaging was done at room temperature. Images were collected using an inverted Zeiss LSM confocal microscope with a Zeiss C-Apochromat 40X water immersion objective. Green fluorescent proteins (Dendra2, GFP) were excited by a 488nm laser, and emission fluorescence was collected using a band pass (505–530nm) filter. Red fluorescent proteins (converted Dendra2, mRFP) and dyes (Alexa Fluor 564) were excited by a 543nm laser, and emission fluorescence was collected using a band pass (585–615nm) filter. Far red fluorescent dye (Alexa Fluor 647) was excited by a 633nm laser, and emission fluorescence was collected using a long pass (650nm) filter. Images were acquired and saved as .lsm files using Zen (Zeiss) software. For calcium imaging, larvae (5dpf) were either paralyzed with ɑ-bungarotoxin or immobilized in agarose without drug, and time series were collected from selected z-planes within the facial motor nucleus (typically several minutes per plane, sampling intervals 240 – 500 ms), and image stacks through the facial motor nucleus were collected at the conclusion of the imaging session. Fish health was assessed between time series by visualization of tissue clarity and blood flow, and only healthy fish were included in subsequent analyses.
Electrophysiology
Whole cell recordings were performed in 5dpf Tg(Islet1:GFP) larvae, specifically targeting the ventrolateral arm of the facial motor nucleus. Larvae were paralyzed with acute (3–5min) exposure to α-bungarotoxin (Biotoxins, Inc.; 1 mg/mL in purified water) and secured to a Sylgard-coated glass-bottomed recording chamber with two etched tungsten pins inserted through the notochord. Larvae were then submerged in extracellular recording solution (in mM: 134 NaCl, 2.9 KCl, 1.2 MgCl2, 10 HEPES, 10 glucose, 2.1 CaCl2; adjusted to pH 7.8 with NaOH). The brain was exposed by dissection of the surrounding head and gut tissue with a sharp tungsten probe. The exposed brain was then secured to the dish with etched tungsten pins through the rostral spinal cord and midbrain-forebrain junction. Facial motor neurons were visualized with an Olympus BX51WI inverted microscope equipped with infrared DIC optics, epifluorescence, and a 40X immersion objective, and were viewed with a CCD camera controlled by Q Capture Pro 6.0 software (QImaging). Each recorded neuron was filled with fluorescent dye during recording (see below) and imaged after recording to confirm neuronal identity. All experiments were performed at room temperature (23–25°C).
Micropipettes for whole cell recording were pulled from thin-walled filamented capillary glass (AM Systems) on a Flaming-Brown puller (Sutter Instruments), to a tip diameter of 1–3 μm and resistance of 8–15M . Micropipettes were backfilled with intracellular recording solution (in mM: 125 K-gluconate, 2 MgCl2, 10 HEPES, 10 EGTA, 4 Na2-ATP; adjusted to pH 7.2 with KOH) containing 0.1% Alexa Fluor 546 hydrazide (Molecular Probes) or 0.1% sulforhodamine B (Sigma). Micropipettes were advanced into the hindbrain using a motorized micro manipulator (MP-225, Sutter Instruments), under positive tip pressure (60–65 mmHg) maintained by a pneumatic transducer (DPM-1B, Bio-Tek Instruments). Once the tip was near the cell body, positive pressure was released to form a gigaohm seal. A holding voltage of −50mV was applied, and a sharp suction pulse ruptured the cell membrane to initiate whole cell recording. All recordings were performed in current-clamp mode, and standard corrections were made for bridge and pipette capacitance.
Electrophysiological data were acquired with a MultiClamp 700A amplifier (Molecular Devices) and a Digidata 1322A digitizer (Molecular Devices), recorded with Clampex 8.2 software (Molecular Devices), and analyzed offline with Clampfit (Molecular Devices) and custom scripts in Matlab (Mathworks). Electrical signals from facial motor neurons were filtered at 30 kHz and digitized at 100 kHz (Rf = 500M). Access resistance was usually monitored before and after recording, and was typically under 120M . Although action potential heights were generally quite small (riding atop depolarizing hills), this was the case for neurons with both higher and lower (40–60M) access resistance values. All recorded neurons had stable resting membrane potentials between −40 and −70 mV. Since we were concerned primarily with changes in membrane potential rather than absolute values, we have reported values as recorded without applying a correction for junction potential, which was estimated to be 18.5 mV at 23°C.
In a subset of whole cell recordings (wild type: n=6/23 fish; llk(rw16) mutants: n=5/8 fish), we simultaneously recorded extracellular activity from the ventral motor root innervating locomotor muscle in the tail – as a means of recording fictive attempted swimming bouts. A section of skin covering the tail was removed, exposing the ventral muscle, and a wide-tipped glass pipette filled with extracellular recording solution was lowered onto the muscle surface. Electrical signals from the motor nerve were recorded (AC: 300 Hz), filtered at 1 kHz and digitized at 100 kHz (Rf = 50M).
Behavior
Larvae (5dpf) were immobilized side-down in low-melt agarose (2% in 10% Hank’s) at the bottom of a glass-bottomed specimen dish, covered with 10% Hank’s solution. After 5 minutes (to allow the agarose to cool to room temperature, and the fish to acclimate to the dish), larvae were transferred to an Olympus BX51WI inverted microscope equipped with a 40X immersion objective. Movements of the operculum were imaged at 20Hz using a high-speed camera (Fastec Imaging). Three sessions were recorded per fish, and each session lasted approximately 5 minutes (5770 frames). Two or three of these sessions were analyzed per fish – sessions were discarded for poor image quality or excessive movement of the entire fish within the imaging frame (both of which were rare). Experiments were done at room temperature (23–25°C).
Quantification and Statistical Analyses
Spot Analysis and Registration
To determine neuronal locations, we first used spots functions in Imaris (Bitplane) to semi-automatically place spots defining single neurons and extract each cell’s native coordinates in the frame of the original image, then registered neurons with a common coordinate system in the following way. Wild type models and standardized coordinates: We selected a representative wild type animal to serve as the model. In the model animal, we used measure points in Imaris (Bitplane) to locate the native coordinates of several anatomical landmarks within the facial motor nucleus, then used these landmarks to specify new standardized coordinate axes and a new origin. In this standardized system, mediolateral coordinates fall along the axis connecting the inflection points (found in rhombomere 4) in the left and right facial nerves, and rostrocaudal coordinates fall along the axis running midway between the left and right facial nerves (as they run in parallel through rhombomere 5). The origin is specified by the intersection of these two axes, and dorsoventral coordinates fall along the axis that is mutually perpendicular to the other two axes. The cell positions for the wild type model were re-specified within this standardized coordinate system, then used to register other wild type data. Wild type data registration: The positions of trigeminal and facial nucleus neurons (rhombomeres 2–6) were obtained for each wild type fish. These position data were treated as point clouds and registered to a corresponding (standardized coordinate) point cloud from the wild type model using an iterative closest point algorithm implemented in Matlab (Mathworks; code obtained from [48]). Mutant data registration: We selected a representative mutant animal for each mutation type. To obtain a mutant model in the standardized coordinate system, we registered each mutant model to the wild type model using only the positions of neurons in the trigeminal nuclei (rhombomeres 2 and 3), then used the resulting registration transform to obtain registered positions for the mutant facial nucleus neurons as well. Other mutant animal data were then registered to the mutant model using the iterative closest point algorithm described above, using position data from the mutant trigeminal and facial nuclei (rhombomeres 2–4).
Analysis of Dendra Photoconversion
In Tg(zCREST1:Dendra2) embryos, the photo convertible protein Dendra is expressed in facial motor neurons once they differentiate as motor neurons – since zCREST1 is a motor neuron enhancer from the Islet1 promoter [25]. Thus, neurons that have already begun to differentiate at a given photo conversion time point (20, 24, or 30 hpf) will contain green Dendra at that time point, which will be converted to red upon exposure to ultraviolet light. Those neurons will continue to express green Dendra after the photo conversion time point, so neurons older than the conversion time point will contain both green and red Dendra. Younger neurons that begin to differentiate after the conversion time point will only contain green Dendra. To determine which neurons contain red (converted) Dendra, we set a minimum threshold for mean absolute red intensity of a 1μm spot at the center of the neuron, obtained using the spots function in Imaris (Bitplane). Since expression levels and imaging conditions varied across subjects, we chose this threshold separately for each fish, by viewing the red intensity histogram for all spots and the red channel image itself. Although the red Dendra was often dim, it was clear both within and across individual fish (upon viewing the raw images) that the red-containing cells were in the ventral part of the facial nuclei.
Dendrite Tracing and Analysis
Image stacks were collected from Tg(Islet1:GFP) larvae at 5dpf, in which single-cell electroporation at 4dpf had been used to fill the processes of one FBMN with fluorescent dye. In some cases, additional non-FBMN cells were also filled with dye, likely due to the accidental electroporation of cellular processes while targeting a specific FBMN cell body. Typically, the dendritic arbor of the targeted FBMN could still be clearly distinguished and cleanly traced; if not, the ambiguous dye fill was discarded from the analysis. The dendritic arbor was traced semi-manually using filament functions in Imaris (Bitplane), and the muscle target of the backfilled cell was confirmed by imaging the cranial muscles and the facial nerve. The position of the filled FBMN was registered, using the same method described above (Spot Analysis and Registration), to permit comparison across fish. A spot was placed at the rostral-most and caudal-most points on the traced dendritic arbor, and the registered position of these points was used to mark the rostrocaudal extent of the arbor.
Analysis of Calcium Imaging Data
We initially generated a transgenic line expressing calcium indicator in the cytosol (Tg(zCREST1:GCaMP5)). With this line, we confirmed that FBMNs in intact wild type larvae exhibit the two gross patterns of activity that we had previously observed in whole cell recordings (data not shown). However, because most FBMNs are tightly clustered together, the cytosolic indicator made it difficult to extract the calcium transients from a single FBMN, without contamination of the signal from adjacent neurons. To solve this problem, we used Tg(Elavl3:H2B-GCaMP6fast) larvae, in which calcium indicator is restricted to the nucleus, to simplify identification of single-cell regions of interest [26]. The two key calcium signal patterns were observed in both paralyzed and unparalyzed larvae, of both migration phenotypes; however, in paralyzed larvae, the larger, less frequent bursts were extended in time, which obscured the distinction between the two patterns in a subset of fish. Thus, we focused our analysis on the unparalyzed fish, where the two patterns of response could be readily distinguished in most cases.
Time series were converted from .lsm files to tiff stacks in ImageJ (NIH) and manually inspected to determine if image registration was necessary. When necessary, tiff stacks were registered using the native imregister function in Matlab (Mathworks), then manually inspected for suitable registration. Images that were improperly registered or unable to be registered (e.g. tissue distortion during attempted movements) were excluded from further analysis. We found the maximum intensity projection (MIP) of the registered images in Imaris (Bitplane), then manually drew regions-of-interest (ROIs) using the MIP. In Tg(Elavl3:H2B-GCaMP6fast) fish, ROIs corresponding to FBMNs were chosen based on Tg(zCREST1:mRFP) expression and location in the hindbrain.
For each time series, we extracted the mean intensity of GCaMP in each putative FBMN (at each time point) and subtracted the mean intensity of a background ROI from the time series, yielding the background-corrected trace. Then, for each ROI, we computed the baseline calcium signals, by dividing the trial into four equal parts, computing the minimum background-corrected intensity of the ROI for each trial segment, and taking the mean of these values. We subtracted this baseline value from the ROI intensity series, then divided by the baseline (% dF/F). Only those neurons with clear responses (maximum % dF/F > 50%), for which we could classify the response type, were included in our analyses; neurons with flat or noisy activity were not analyzed further. Some neurons were also discarded because that slice’s time series contained too many tissue movement artefacts – which was primarily a concern in the dorsal-most portion of the facial nucleus. Further, the region of the facial motor nucleus also contains lateral line efferent neurons, which also show red membrane expression in the (Tg(zCREST1:mRFP)) line. In whole cell recordings, we could exclude lateral line efferents from our analysis using cell morphology, as lateral line efferents (but not FBMNs) have contralateral dendrites [49]. However, with imaging calcium, we were unable to exclude lateral line efferents from the analysis, except in cases where we backfilled neurons projecting to a specific cranial muscle. Thus, it is also possible that some of the unresponsive neurons were lateral line efferents and not FBMNs. Note that those neurons expressing nuclear localized calcium indicator that exhibit additional rhythmic activity tend to have a mean value that builds over time – that is, calcium transients do fall, but not to baseline. We believe this to be the result of how these neurons fire – in bursts of spikes riding on hills of depolarization, which may cause an accumulation of nuclear calcium.
Analysis of Electrophysiology Data
Recorded neurons were filled with fluorescent dye from the recording pipette and inspected post-recording. The facial motor nucleus also contains octavolateralis efferent neurons (OLes); however, unlike FBMNs, OLes have both ipsilateral and contralateral dendrites [49]. Thus, any recorded neurons with crossing processes post-recording were excluded from analysis.
Neurons were classified into two groups, based on their spontaneous responses. Some neurons responded with infrequent (typically 1–2 per minute or less) and irregular large bursts, which correlated with strong tail motor activity (40–100Hz beat frequency, 5–10 beats or more) when available. Some neurons also exhibited regular, rhythmic (0.25–3Hz) hills of depolarization, typically accompanied by a burst of small spikes. In a few fish, this rhythmic firing was accompanied by very brief (1–2 beats) tail motor activity; in other fish, this rhythmic firing occurred in the absence of any measurable ventral root activity.
To calculate each rhythmic neuron’s median inter-burst interval, burst times were manually identified (placed on the rising slope of the depolarizing hill) for a single 10–30 second period of rhythmic bursting during that cell’s recording session, in the absence of hyper- or de-polarizing current injection.
Analysis of Behavioral Data
Images were analyzed using manual tracking in FIJI (NIH). For each fish, a landmark on the operculum was manually tracked across frames, and the vertical pixel positions were exported into Matlab (Mathworks). Operculum movements were quantified during rhythmic bouts -excluding between-bout intervals and large movements made during attempted whole-body movements. Movement times were detected using the findpeaks function and manually spot-checked for accuracy. Summary plots and statistical comparisons were made using the inter-peak intervals. For display, instantaneous movement frequency was calculated as the inverse of the previous inter-peak interval. Box-whisker plots were constructed using the median inter-peak interval for each fish, as the median gives a better representation of central tendency given the skewed nature of the distributions.
Statistics & Plots
For each quantitative analysis, information about the value and meaning of n can be found in the text and figure legend, and details of statistical test output can be found in the supplemental tables. All linear mixed effects models were evaluated with the maximum likelihood method, using the fitlme function in Matlab (Mathworks). The assumption of homogeneous variances was evaluated by plotting residual variances as a function of fitted value, and the assumption of normally distributed residuals was evaluated with a quartile-quartile plot. In general, model assumptions were reasonably well met; see below for details of each test. When assumptions were not strictly met, this was generally due to a modest deviation from normally distributed residuals (specifically at very high and very low ends of the range). When p-values were unambiguous (very large or very high) and assumptions were reasonably well met, the results of the linear model were used in the text – as linear mixed model outputs are robust to modest deviations from the assumption of normal residuals, particularly for large values of n. All plots were created in Matlab (Mathworks) and edited in Illustrator (Adobe). All figures were created in Illustrator (Adobe).
To determine if caudal migration phenotype had a significant effect on motor pool topography (Table S1), we fit each component of the registered position data (mediolateral, dorsoventral, and rostrocaudal coordinates) with a linear mixed-effects model, with fixed effects of migration phenotype and muscle target and a random effect of experimental subject. In this way, we tested for the significance of the fixed effects (phenotype, muscle target) and the interaction between the two. Aside from a slight decrease in variance at higher fitted values for the mediolateral coordinate, all three coordinate models met the assumption of homogeneous variance. Residuals were close to normally distributed for all three models.
To determine if caudal migration phenotype had a significant effect on response type topography (Table S2), we fit the dorsoventral component of the registered position data obtained for neurons categorized by calcium imaging as rhythmic and non-rhythmic with a linear mixed-effects model, with fixed effects of migration phenotype and response type and a random effect of experimental subject. For this statistical analysis, we only included data from fish in which we had sampled the entire dorsoventral extent of the facial nucleus, without backfill (WT: n = 2; llk(rw16) mutant: n = 3). Variances were homogeneous, and residuals were close to normally distributed. However, because the p-value for the interaction effect was not large (p = 0.14), we fit models to the wild type and mutant data separately, with a fixed effect of response type and a random effect of experimental subject – to confirm that there was a significant effect of response type in both migration phenotypes. These p-values were very low and highly significant.
To determine if caudal migration phenotype had a significant effect on rhythmic operculum movements (Table S3), we fit inter-movement interval data with a linear mixed-effects model, with a fixed effect of migration phenotype and a random effect of experimental subject. Once again, the variances were homogeneous, and the residuals distribution was near-normal – but the sample size was high, and the p-value was highly significant. We also fit a model to the data when we removed the first three response intervals from each movement bout, as those intervals tend to be longer (as operculum movements typically begin slow). However, removing those data points did not alter our conclusion, so we have taken a conservative approach and utilized the model fit to the entire dataset here.
KEY RESOURCES TABLE
The table highlights the genetically modified organisms and strains, cell lines, reagents, software, and source data essential to reproduce results presented in the manuscript. Depending on the nature of the study, this may include standard laboratory materials (i.e., food chow for metabolism studies), but the Table is not meant to be comprehensive list of all materials and resources used (e.g., essential chemicals such as SDS, sucrose, or standard culture media don’t need to be listed in the Table). Items in the Table must also be reported in the Method Details section within the context of their use. The number of primers and RNA sequences that may be listed in the Table is restricted to no more than ten each. If there are more than ten primers or RNA sequences to report, please provide this information as a supplementary document and reference this file (e.g., See Table S1 for XX) in the Key Resources Table.
Please note that ALL references cited in the Key Resources Table must be included in the References list. Please report the information as follows:
REAGENT or RESOURCE: Provide full descriptive name of the item so that it can be identified and linked with its description in the manuscript (e.g., provide version number for software, host source for antibody, strain name). In the Experimental Models section, please include all models used in the paper and describe each line/strain as: model organism: name used for strain/line in paper: genotype. (i.e., Mouse: OXTRfl/fl: B6.129(SJL)-Oxtrtm1.1Wsy/J). In the Biological Samples section, please list all samples obtained from commercial sources or biological repositories. Please note that software mentioned in the Methods Details or Data and Software Availability section needs to be also included in the table. See the sample Table at the end of this document for examples of how to report reagents.
SOURCE: Report the company, manufacturer, or individual that provided the item or where the item can obtained (e.g., stock center or repository). For materials distributed by Addgene, please cite the article describing the plasmid and include “Addgene” as part of the identifier. If an item is from another lab, please include the name of the principal investigator and a citation if it has been previously published. If the material is being reported for the first time in the current paper, please indicate as “this paper.” For software, please provide the company name if it is commercially available or cite the paper in which it has been initially described.
IDENTIFIER: Include catalog numbers (entered in the column as “Cat#” followed by the number, e.g., Cat#3879S). Where available, please include unique entities such as RRIDs, Model Organism Database numbers, accession numbers, and PDB or CAS IDs. For antibodies, if applicable and available, please also include the lot number or clone identity. For software or data resources, please include the URL where the resource can be downloaded. Please ensure accuracy of the identifiers, as they are essential for generation of hyperlinks to external sources when available. Please see the Elsevier list of Data Repositories with automated bidirectional linking for details. When listing more than one identifier for the same item, use semicolons to separate them (e.g. Cat#3879S; RRID: AB_2255011). If an identifier is not available, please enter “N/A” in the column.
o A NOTE ABOUT RRIDs: We highly recommend using RRIDs as the identifier (in particular for antibodies and organisms, but also for software tools and databases). For more details on how to obtain or generate an RRID for existing or newly generated resources, please visit the RII or search for RRIDs.
Please see the sample Table at the end of this document for examples of how reagents should be cited. To see how the typeset table will appear in the PDF and online, please refer to any of the research articles published in Cell in the August 25, 2016 issue and beyond.
Please use the empty table that follows to organize the information in the sections defined by the subheading, skipping sections not relevant to your study. Please do not add subheadings. To add a row, place the cursor at the end of the row above where you would like to add the row, just outside the right border of the table. Then press the ENTER key to add the row. You do not need to delete empty rows. Each entry must be on a separate row; do not list multiple items in a single table cell.
TABLE FOR AUTHOR TO COMPLETE
Please upload the completed table as a separate document. Please do not add subheadings to the Key Resources Table. If you wish to make an entry that does not fall into one of the subheadings below, please contact your handling editor.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Bacterial and Virus Strains | ||
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| MS-222 (tricaine methanesulfonate) | Western Chemical | |
| Alexa Fluor 546 (dextran-conjugated, MW 10000, anionic) | Molecular Probes | |
| Alexa Fluor 546 hydrazide | Molecular Probes | |
| Alexa Fluor 647 (dextran-conjugated, MW 10000, anionic) | Molecular Probes | |
| Sulforhodamine B | Sigma | |
| α-bungarotoxin | Biotoxins, Inc. | |
| Critical Commercial Assays | ||
| Deposited Data | ||
| Experimental Models: Cell Lines | ||
| Experimental Models: Organisms/Strains | ||
| Zebrafish: Tg(zCREST1:Dendra2) | Current publication | |
| Zebrafish: Tg(zCREST1:GCaMP5) | Current publication | |
| Zebrafish: Tg(Islet1:GFP) | [45] | |
| Zebrafish: Tg(Elavl3:H2B-GCaMP6fast) | [26] | |
| Zebrafish: llk(rw16) | [17] | ZFIN: ZDB-FISH- 150901-11117 |
| Zebrafish: pk1b(fh122) | [18] | ZFIN: ZDB-ALT- 110819-4 |
| Oligonucleotides | ||
| Recombinant DNA | ||
| Software and Algorithms | ||
| Fiji | http://fiji.sc | |
| Matlab (R2015a) | Mathworks | |
| Imaris (8.2.1) | Bitplane | |
| Matlab: Finite Iterative Closest Point | https://www.mathworks.com/matlabcentral/fileexchange/24301-finite-iterative-closest-point | |
| Illustrator | Adobe | |
| Other | ||
TABLE WITH EXAMPLES FOR AUTHOR REFERENCE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-Snail | Cell Signaling Technology | Cat#3879S; RRID: AB 2255011 |
| Mouse monoclonal anti-Tubulin (clone DM1A) | Sigma-Aldrich | Cat#T9026; RRID: AB 477593 |
| Rabbit polyclonal anti-BMALI | This paper | N/A |
| Bacterial and Virus Strains | ||
| pAAV-hSyn-DIO-hM3D(Gq)-mCherry | Krashes et al., 2011 | Addgene AAV5; 44361-AAV5 |
| AAV5-EF1a-DIO-hChR2(H134R)-EYFP | Hope Center Viral Vectors Core | N/A |
| Cowpox virus Brighton Red | BEI Resources | NR-88 |
| Zika-SMGC-1, GENBANK: KX266255 | Isolated from patient (Wang et al., 2016) | N/A |
| Staphylococcus aureus | ATCC | ATCC 29213 |
| Streptococcus pyogenes: M1 serotype strain: strain SF370; M1 GAS | ATCC | ATCC 700294 |
| Biological Samples | ||
| Healthy adult BA9 brain tissue | University of Maryland Brain & Tissue Bank; http://medschool.umaryland.edu/btbank/ | Cat#UMB1455 |
| Human hippocampal brain blocks | New York Brain Bank | http://nybb.hs.columbia.edu/ |
| Patient-derived xenografts (PDX) | Children’s Oncology Group Cell Culture and Xenograft Repository | http://cogcell.org/ |
| Chemicals, Peptides, and Recombinant Proteins | ||
| MK-2206 AKT inhibitor | Selleck Chemicals | S1078; CAS: 1032350-13-2 |
| SB-505124 | Sigma-Aldrich | S4696; CAS: 694433-59-5 (free base) |
| Picrotoxin | Sigma-Aldrich | P1675; CAS: 12487-8 |
| Human TGF-ß | R&D | 240-B; GenPept: P01137 |
| Activated S6K1 | Millipore | Cat#14-486 |
| GST-BMAL1 | Novus | Cat#H00000406-P01 |
| Critical Commercial Assays | ||
| EasyTag EXPRESS 35S Protein Labeling Kit | Perkin-Elmer | NEG772014MC |
| CaspaseGlo 3/7 | Promega | G8090 |
| TruSeq ChIP Sample Prep Kit | Illumina | IP-202-1012 |
| Deposited Data | ||
| Raw and analyzed data | This paper | GEO: GSE63473 |
| B-RAF RBD (apo) structure | This paper | PDB: 5J17 |
| Human reference genome NCBI build 37, GRCh37 | Genome Reference Consortium | http://www.ncbi.nlm.nih.gov/projects/genome/assembly/grc/human/ |
| Nanog STILT inference | This paper; Mendeley Data | http://dx.doi.org/10.17632/wx6s4mj7s8.2 |
| Affinity-based mass spectrometry performed with 57 genes | This paper; and Mendeley Data | Table S8; http://dx.doi.org/10.17632/5hvpvspw82.1 |
| Experimental Models: Cell Lines | ||
| Hamster: CHO cells | ATCC | CRL-11268 |
| D. melanogaster: Cell line S2: S2-DRSC | Laboratory of Norbert Perrimon | FlyBase: FBtc0000181 |
| Human: Passage 40 H9 ES cells | MSKCC stem cell core facility | N/A |
| Human: HUES 8 hESC line (NIH approval number NIHhESC-09-0021) | HSCI iPS Core | hES Cell Line: HUES-8 |
| Experimental Models: Organisms/Strains | ||
| C. elegans: Strain BC4011: srl-1(s2500) II; dpy- 18(e364) III; unc-46(e177)rol-3(s1040) V. | Caenorhabditis Genetics Center | WB Strain: BC4011; WormBase: WBVar00241916 |
| D. melanogaster: RNAi of Sxl: y[1] sc[*] v[1 ]; P{TRiP.HMS00609}attP2 | Bloomington Drosophila Stock Center | BDSC:34393; FlyBase: FBtp0064874 |
| S. cerevisiae: Strain background: W303 | ATCC | ATTC: 208353 |
| Mouse: R6/2: B6CBA-Tg(HDexon1)62Gpb/3J | The Jackson Laboratory | JAX: 006494 |
| Mouse: OXTRfl/fl: B6.129(SJL)-Oxtrtm11Wsy/J | The Jackson Laboratory | RRID: IMSR JAX:008471 |
| Zebrafish: Tg(Shha:GFP)t10: t10Tg | Neumann and Nuesslein- Volhard, 2000 | ZFIN: ZDB-GENO- 060207-1 |
| Arabidopsis: 35S::PIF4-YFP, BZR1-CFP | Wang et al., 2012 | N/A |
| Arabidopsis: JYB1021.2: pS24(AT5G58010)::cS24:GFP(-G):NOS #1 | NASC | NASC ID: N70450 |
| Oligonucleotides | ||
| siRNA targeting sequence: PIP5K I alpha #1: ACACAGUACUCAGUUGAUA | This paper | N/A |
| Primers for XX, see Table SX | This paper | N/A |
| Primer: GFP/YFP/CFP Forward: GCACGACTTCTTCAAGTCCGCCATGCC | This paper | N/A |
| Morpholino: MO-pax2a GGTCTG CTTTG CAGTGAATATCCAT | Gene Tools | ZFIN: ZDB- MRPHLNO-061106- 5 |
| ACTB (hs01060665_g1) | Life Technologies | Cat#4331182 |
| RNA sequence: hnRNPAI ligand: UAGGGACUUAGGGUUCUCUCUAGGGACUUAGGGUUCUCUCUAGGGA | This paper | N/A |
| Recombinant DNA | ||
| pLVX-Tight-Puro (TetOn) | Clonetech | Cat#632162 |
| Plasmid: GFP-Nito | This paper | N/A |
| cDNA GH111110 | Drosophila Genomics Resource Center | DGRC:5666; FlyBase:FBcl0130415 |
| AAV2/1-hsyn-GCaMP6- WPRE | Chen et al., 2013 | N/A |
| Mouse raptor: pLKO mouse shRNA 1 raptor | Thoreen et al., 2009 | Addgene Plasmid #21339 |
| Software and Algorithms | ||
| Bowtie2 | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| Samtools | Li et al., 2009 | http://samtools.sourceforge.net/ |
| Weighted Maximal Information Component Analysis v0.9 | Rau et al., 2013 | https://github.com/ChristophRau/wMICA |
| ICS algorithm | This paper; Mendeley Data | http://dx.doi.org/10.17632/5hvpvspw82.1 |
| Other | ||
| Sequence data, analyses, and resources related to the ultra-deep sequencing of the AML31 tumor, relapse, and matched normal. | This paper | http://aml31.genome.wustl.edu |
| Resource website for the AML31 publication | This paper | https://github.com/chrisamiller/aml31SuppSite |
HIGHLIGHTS.
Abnormal migration of facial motor neurons mis-positions cell bodies and dendrites.
Abnormal migration spares the internal topography of the facial nucleus.
Abnormal migration spares gross patterns of neuronal and behavioral activity.
Development of facial motor neurons seems resilient to abnormal migration.
Acknowledgments
The authors would like to thank M. Ahrens, S. Higashijima, C. Moens, H. Okamoto, and V. Prince for generously sharing zebrafish lines and reagents, and B. Miller and N. Gilbert for expert technical assistance. This work was supported in part by grants from the National Institutes of Health (NINDS: F32-NS083099 and R01-NS26539).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
K.L.M. conducted the experiments and analyzed the data. K.L.M. and J.R.F. designed the experiments and wrote the paper.
References
- 1.Tripodi M, Arber S. Regulation of motor circuit assembly by spatial and temporal mechanisms. Curr Opin Neurobiol. 2012;22:615–623. doi: 10.1016/j.conb.2012.02.011. [DOI] [PubMed] [Google Scholar]
- 2.Cooper JA. Mechanisms of cell migration in the nervous system. J Cell Biol. 2013;202:725–734. doi: 10.1083/jcb.201305021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gilmore EC, Herrup K. Cortical development: layers of complexity. Curr Biol. 1997;7:R231–R234. doi: 10.1016/s0960-9822(06)00108-4. [DOI] [PubMed] [Google Scholar]
- 4.Kinkhabwala A, Riley M, Koyama M, Monen J, Satou C, Kimura Y, Higashijima S, Fetcho J. A structural and functional ground plan for neurons in the hindbrain of zebrafish. Proc Natl Acad Sci USA. 2011;108:1164–1169. doi: 10.1073/pnas.1012185108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jessell TM, Sürmeli G, Kelly JS. Motor neurons and the sense of place. Neuron. 2011;72:419–424. doi: 10.1016/j.neuron.2011.10.021. [DOI] [PubMed] [Google Scholar]
- 6.Demireva EY, Shapiro LS, Jessell TM, Zampieri N. Motor neuron position and topographic order imposed by β- and γ-catenin activities. Cell. 2011;3:641–652. doi: 10.1016/j.cell.2011.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sürmeli G, Akay T, Ippolito GC, Tucker PW, Jessell TM. Patterns of spinal sensory-motor connectivity prescribed by a dorsoventral positional template. Cell. 2011;147:653–665. doi: 10.1016/j.cell.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Black D. The motor nuclei of the cerebral nerves in phylogeny: a study of the phenomena of neurobiotaxis. I Cyclostomi and pisces. J Comp Neurol. 1917;27:467–564. [Google Scholar]
- 9.Black D. The motor nuclei of the cerebral nerves in phylogeny: a study of the phenomena of neurobiotaxis. II Amphibia. J Comp Neurol. 1917;28:379–427. [Google Scholar]
- 10.Black D. The motor nuclei of the cerebral nerves in phylogeny: a study of the phenomena of neurobiotaxis. III Reptilia. J Comp Neurol. 1920;32:61–98. [Google Scholar]
- 11.Black D. The motor nuclei of the cerebral nerves in phylogeny: a study of the phenomena of neurobiotaxis. IV Aves. J Comp Neurol. 1922;34:233–275. [Google Scholar]
- 12.Gilland E, Baker R. Evolutionary patterns of cranial nerve efferent nuclei in vertebrates. Brain Behav Evol. 2005;66:234–254. doi: 10.1159/000088128. [DOI] [PubMed] [Google Scholar]
- 13.Ariëns Kappers CU. On neurobiotaxis: a physical law in the structure of the nervous system. Acta Psych Scand. 1927;2:118–145. [Google Scholar]
- 14.Straka H, Baker R, Gilland E. Preservation of segmental hindbrain organization in adult frogs. J Comp Neurol. 2006;494:228–245. doi: 10.1002/cne.20801. [DOI] [PubMed] [Google Scholar]
- 15.Chandrasekhar A. Turning heads: development of vertebrate branchiomotor neurons. Dev Dyn. 2004;229:143–161. doi: 10.1002/dvdy.10444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wanner SJ, Saeger I, Guthrie S, Prince VE. Facial motor neuron migration advances. Curr Opin Neurobiol. 2013;23:943–950. doi: 10.1016/j.conb.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wada H, Iwasaki M, Sato T, Masai I, Nishiwaki Y, Tanaka H, Sato A, Nojima Y, Okamoto H. Dual roles of zygotic and maternal Scribble1 in neural migration and convergent extension movements in zebrafish embryos. Development. 2005;132:2273–2285. doi: 10.1242/dev.01810. [DOI] [PubMed] [Google Scholar]
- 18.Mapp OM, Walsh GS, Moens CB, Tada M, Prince VE. Zebrafish Prickle1b mediates facial branchiomotor neuron migration via a farnesylation-dependent nuclear activity. Development. 2011;138:2121–2132. doi: 10.1242/dev.060442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diogo R, Hinits Y, Hughes SM. Development of mandibular, hyoid and hypobranchial muscles in the zebrafish: homologies and evolution of these muscles within bony fishes and tetrapods. BMC Dev Biol. 2008;8:24. doi: 10.1186/1471-213X-8-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beattie CE, Hatta K, Halpern ME, Liu H, Eisen JS, Kimmel CB. Temporal separation in the specification of primary and secondary motoneurons in zebrafish. Dev Biol. 1997;187:171–182. doi: 10.1006/dbio.1997.8604. [DOI] [PubMed] [Google Scholar]
- 21.McLean DL, Fetcho JR. Spinal interneurons differentiate sequentially from those driving the fastest swimming movements in larval zebrafish to those driving the slowest ones. J Neurosci. 2009;29:13566–13577. doi: 10.1523/JNEUROSCI.3277-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caron SJC, Prober D, Choy M, Schier AF. In vivo birthdating by BAPTISM reveals that trigeminal sensory neuron diversity depends on early neurogenesis. Development. 2008;135:3259–3269. doi: 10.1242/dev.023200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greaney MR, Privorotskiy AE, D’Elia KP, Schoppik D. Extraocular motoneuron pools develop along a dorsoventral axis in zebrafish, Danio rerio. J Comp Neurol. 2017;525:65–78. doi: 10.1002/cne.24042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koyama M, Kinkhabwala A, Satou C, Higashijima S, Fetcho J. Mapping a sensory-motor network onto a structural and functional ground plan in the hindbrain. Proc Natl Acad Sci USA. 2011;108:1170–1175. doi: 10.1073/pnas.1012189108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uemura O, Okaya Y, Ando H, Guedj M, Higashijima S, Shimazaki T, Chino N, Okano H, Okamoto H. Comparative functional genomics revealed conservation and diversification of three enhancers of the isl1 gene for motor and sensory neuron-specific expression. Dev Biol. 2005;278:587–606. doi: 10.1016/j.ydbio.2004.11.031. [DOI] [PubMed] [Google Scholar]
- 26.Dunn TW, Mu Y, Narayan S, Randlett O, Naumann EA, Yang C, Schier AF, Freeman J, Engert F, Ahrens MB. Brain-wide mapping of neural activity controlling zebrafish exploratory locomotion. eLife. 2016;5:e12741. doi: 10.7554/eLife.12741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jonz MG, Nurse CA. Development of oxygen sensing in the gills of zebrafish. J Exp Biol. 2005;208:1537–1549. doi: 10.1242/jeb.01564. [DOI] [PubMed] [Google Scholar]
- 28.Erickstad M, Hale LA, Chalasani SH, Groisman A. A microfluidic system for studying the behavior of zebrafish larvae under acute hypoxia. Lab Chip. 2015;15:857–866. doi: 10.1039/c4lc00717d. [DOI] [PubMed] [Google Scholar]
- 29.Hale ME. Developmental change in the function of movement systems: transition of the pectoral fins between respiratory and locomotor roles in zebrafish. Int Comp Biol. 2014;54:238–249. doi: 10.1093/icb/icu014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Missaghi K, Le Gal JP, Gray PA, Dubuc R. The neural control of respiration in lampreys. Resp Physiol Neurobiol. 2016;234:14–25. doi: 10.1016/j.resp.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 31.Bass AH, Baker R. Phenotypic specification of hindbrain rhombomeres and the origins of rhythmic circuits in vertebrates. Brain Behav Evol 50 Suppl. 1997;1:3–16. doi: 10.1159/000113351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.del Toro ED, Borday V, Davenne M, Neun R, Rijli FM, Champagnat J. Generation of a novel functional neuronal circuit in Hoxa1 mutant mice. J Neurosci. 2001;21:5637–5642. doi: 10.1523/JNEUROSCI.21-15-05637.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hale ME, Kheirbek MA, Schriefer JE, Prince VE. Hox gene misexpression and cell-specific lesions reveal functionality of homeotically transformed neurons. J Neurosci. 2004;24:3070–3076. doi: 10.1523/JNEUROSCI.5624-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma LH, Grove CL, Baker R. Development of oculomotor circuitry independent of hox3 genes. Nat Comm. 2014;5:4221. doi: 10.1038/ncomms5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thoby-Brisson M, Bouvier J, Glasco DM, Stewart ME, Dean C, Murdoch JN, Champagnat J, Fortin G, Chandrasekhar A. Brainstem respiratory oscillators develop independently of neuronal migration defects in the Wnt/PCP mouse mutant looptail. PLoS One. 2012;7:e31140. doi: 10.1371/journal.pone.0031140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Belvindrah R, Nissant A, Lledo PM. Abnormal neuronal migration changes the fate of developing neurons in the postnatal olfactory bulb. J Neurosci. 2011;31:7551–7562. doi: 10.1523/JNEUROSCI.6716-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Belvindrah R, Nosten-Bertrand M, Francis F. Neuronal migration and its disorders affecting the CA3 region. Front Cell Neurosci. 2014;8:1–15. doi: 10.3389/fncel.2014.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Di Meglio T, Kratochwil CF, Vilain N, Loche A, Vitobello A, Yonehara K, Hrycaj SM, Roska B, Peters AHFM, Eichmann A, Wellik D, Ducret S, Rijli FM. Ezh2 orchestrates topographic migration and connectivity of mouse precerebellar neurons. Science. 2013;339:204–207. doi: 10.1126/science.1229326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moffat JJ, Minhan K, Jung EM, Kim WY. Genes and brain malformations associated with abnormal neuron positioning. Molec Brain. 2015;8:72. doi: 10.1186/s13041-015-0164-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen BL, Hall DH, Chklovskii DB. Wiring optimization can relate neuronal structure and function. Proc Natl Acad Sci USA. 2006;103:4723–4728. doi: 10.1073/pnas.0506806103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gushchin A, Tang A. Total wiring length minimization of C. elegans neural network: a constrained optimization approach. PLoS One. 2015;10:e0145029. doi: 10.1371/journal.pone.0145029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lister JA, Robertston CP, Lepage T, Johnson SL, Raible DW. nacre encodes a zebrafish microphthalmia-related protein that regulates neural-crest-derived pigment cell fate. Development. 1999;126:3757–3767. doi: 10.1242/dev.126.17.3757. [DOI] [PubMed] [Google Scholar]
- 43.White RM, Sessa A, Burke C, Bowman T, LeBlance J, Ceol C, Bourque C, Dovey M, Goessling W, Burnes CE, Zon LI. Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell. 2008;2:183–189. doi: 10.1016/j.stem.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clark KJ, Urban MD, Skuster KJ, Ekker SC. Transgenic zebrafish using transposable elements. Methods Cell Biol. 2011;104:137–149. doi: 10.1016/B978-0-12-374814-0.00008-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Higashijima S, Hotta Y, Okamoto H. Visualization of cranial motor neurons in live transgenic zebrafish expressing green fluorescent protein under the control of the islet-1 promoter/enhancer. J Neurosci. 2000;20:206–218. doi: 10.1523/JNEUROSCI.20-01-00206.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rohrschneider MR, Elsen GE, Prince VE. Zebrafish Hoxb1a regulates multiple downstream genes including prickle1b. Dev Biol. 2007;309:358–372. doi: 10.1016/j.ydbio.2007.06.012. [DOI] [PubMed] [Google Scholar]
- 47.Love CE, Prince VE. Rest represses maturation within migration facial branchiomotor neurons. Dev Biol. 2015;401:220–235. doi: 10.1016/j.ydbio.2015.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kroon DJ. Finite iterative closest point (https://www.mathworks.com/matlabcentral/fileexchange/24301-finite-iterative-closest-point) Matlab Central File Exchange. 2009 Retrieved September 29, 2015. [Google Scholar]
- 49.Suli A, Mortimer N, Shepher I, Chien CB. Netrin/DCC signaling controls contralateral dendrites of octavolateralis efferent neurons. J Neurosci. 2006;26:13328–13337. doi: 10.1523/JNEUROSCI.2858-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


