Abstract
Superoxide dismutase (SOD) proteins are widely present in the plant kingdom and play important roles in different biological processes. However, little is known about the SOD genes in cucumber. In this study, night SOD genes were identified from cucumber (Cucumis sativus) using bioinformatics-based methods, including 5 Cu/ZnSODs, 3 FeSODs, and 1 MnSOD. Gene structure and motif analysis indicated that most of the SOD genes have relatively conserved exon/intron arrangement and motif composition. Phylogenetic analyses with SODs from cucumber and several other species revealed that these SOD proteins can be traced back to two ancestral SODs before the divergence of monocot and dicot plants. Many cis-elements related to stress responses and plant hormones were found in the promoter sequence of each CsSOD gene. Gene expression analysis revealed that most of the CsSOD genes are expressed in almost all the tested tissues. qRT-PCR analysis of 8 selected CsSOD genes showed that these genes could respond to heat, cold, osmotic, and salt stresses. Our results provide a basis for further functional research on SOD gene family in cucumber and facilitate their potential applications in the genetic improvement of cucumber.
1. Introduction
Under natural conditions, plants are frequently exposed to various stresses such as drought, salt, extreme temperature, and heavy metals, which may seriously affect their growth and development [1]. These stresses are surely accompanied by the generation of reactive oxygen species (ROS), including superoxide anion radicals (O2−), hydrogen peroxide (H2O2), hydroxyl radical (OH−), peroxyl radical (HOO−), and singlet oxygen (1O2), causing peroxidation and degradation of macromolecules and damage to cell membranes and ultimately leading to the death of cells [2–4]. In plant cells, ROS production is strictly controlled by enzymatic and nonenzymatic antioxidant defense systems including superoxide dismutase (SOD), catalase (CAT), ascorbate peroxidase (APX), monodehydroascorbate reductase (MDHAR), dehydroascorbate reductase (DHAR), thioredoxin, and glutathione. Among the antioxidant enzymes, superoxide dismutase (SOD, EC 1.15.1.1) can serve as an efficient scavenger of ROS by catalyzing the decomposition of superoxide anion radicals (O2−) to hydrogen peroxide (H2O2), which is further converted to nontoxic water and oxygen.
According to the binding metal cofactor(s) that interact with the active site, SODs can be classified into four types, including iron (FeSOD), manganese (MnSOD), copper/zinc (Cu/ZnSOD), and nickel (NiSOD) [4–6]. Different SOD isoforms have similar functions, but have different metal cofactors and amino acid sequences, crystal structures, and subcellular localizations, and exhibit different sensitivities to H2O2 in vitro [5, 7]. For example, KCN and H2O2 can inactivate Cu/ZnSODs and FeSODs irreversibly, but MnSOD is not sensitive to either of the chemicals [8]. Cu/ZnSODs, which are mainly located in chloroplasts, cytoplasm, and/or the extracellular space, are present in some bacteria and all eukaryotic species [9, 10], whereas MnSODs are mainly found in plant mitochondria [11]. At least one copy of MnSODs present in plant genomes plays a role in the scavenging of ROS in mitochondria [12]. FeSODs are distributed in prokaryotes, protozoans, and chloroplasts and cytoplasm of plants [13], while NiSODs were shown to be present in Streptomyces species, were predicted to exist in some cyanobacteria [14, 15], and have not been discovered in plants yet.
Many studies have shown that SOD gene transcripts are induced in various plant species under different forms of stress such as heat, cold, drought, and salt as well as osmotic and oxidative stresses and hormone signaling [5, 6, 9, 10, 16]. Additionally, different types of SOD genes exhibit different expression patterns under diverse stresses. In tomato, SlSOD1 was the only gene that displayed markedly upregulated expression among the nine SlSOD genes, while the expression of four SlSOD genes (SlSOD2, SlSOD5, SlSOD6, and SlSOD8) was downregulated under salt treatment [6]. However, the four SlSOD genes (SlSOD2, SlSOD5, SlSOD6, and SlSOD8) showed increased expression levels under drought treatment [6]. Moreover, the same type of SOD genes do not share the same expression patterns under stresses. In banana, the transcript of MaMSD1A was increased under heat, drought, and salt stresses, and the expression of MaMSD1B was induced by heat and drought stresses, while the transcription of MaMSD1C was only induced by heat stress and MaMSD1D was not responsive to any stress [10]. Furthermore, some studies have revealed that the expression of some SOD genes can be regulated by alternative splicing [5, 10, 17, 18] and miRNAs [19, 20]. Certain genes were identified as positive regulators of abiotic stresses and can be one of the most important factors for plant's defense against various stresses. For example, transgenic plants overexpressing SOD genes displayed increased tolerance to a wide range of abiotic stresses including drought and salinity [21–26], oxidative stress [27], and stresses caused by the application of insecticides [28, 29].
Cucumber (Cucumis sativus L.), which belongs to the Cucurbitaceae family, is an economically and nutritionally important vegetable crop consumed worldwide [30]. SOD genes exist as a superfamily; however, no genome-wide information is available for SOD gene family in cucumber. In this study, we performed genome-wide identification of SOD gene family in cucumber and comprehensively analyzed their phylogenetic relationships, genome distribution, gene structure arrangement, conserved motifs, expression profiles in different tissues, and their expression patterns in response to various abiotic stresses, aiming to lay the foundation for functional characterization of them under various stresses.
2. Materials and Methods
2.1. Identification and Annotation of SOD Genes in Cucumber
A cucumber Cu/ZnSOD (GenBank number: XP_011648784.1) was used as a query sequence for tBLASTn against the cucumber genome from Cucumber Genome Initiative (CuGI, http://cucumber.genomics.org.cn/), which was released by the Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences (IVF-CAAS). A name search for superoxide dismutase also helped to identify SOD genes in the NCBI database. The candidate SOD members were aligned to each other to guarantee that no member was analyzed for multiple times. To further confirm the accuracy of these SOD sequences, the cDNA sequences were translated into amino acid sequences, which were submitted to NCBI for domain prediction to eliminate the sequences without any SOD superfamily conserved domains. Subsequently, the conserved domains of these amino acid sequences were searched using the Simple Modular Architecture Research Tool (SMART) (http://smart.embl-heidelberg.de/) and Pfam (http://www.sanger.ac.uk/software/pfam/) online tools. The molecular weight and isoelectric point (pI) of each SOD protein were analyzed using the online ProtParam program (http://web.expasy.org/protparam/).
2.2. Subcellular Localization and Conserved Motif Prediction
Subcellular localization and signal peptide of SOD proteins were predicted using ProtComp 9.0 (http://linux1.softberry.com/berry.phtml/) and SignalP 4.1 (http://www.cbs.dtu.dk/services/SignalP/), respectively. Conserved motifs of SOD proteins were detected using the MEME program (http://meme-suite.org/tools/meme).
2.3. Multiple Sequence Alignments and Phylogenetic Analyses
Multiple sequence alignments of the deduced amino acid sequences of SOD proteins were performed using the ClustalW software with default parameters and then redrawn with the software Geneious Pro 4.8.3 (http://www.geneious.com/). Neighbor-joining (NJ) phylogenetic tree was constructed with the aligned SOD protein sequences using MEGA 5.1 software with the following parameters: poisson correction, pairwise deletion, and bootstrap (1000 replicates).
2.4. Intron/Exon Structure and Genome Distribution
The genomic sequences of SOD genes and their coding sequences were downloaded from CuGI (http://cucumber.genomics.org.cn/). The intron distribution pattern and splicing phase of each CsSOD gene were analyzed using Gene Structure Display Server (GSDS, http://gsds.cbi.pku.edu.cn/). To obtain the chromosomal locations of the CsSOD genes, mapping of the distribution of CsSOD genes throughout cucumber genome was performed with the MapInspect tool as previously described [31].
2.5. Promoter Region Analysis
The 1.0 kb DNA sequences upstream of the start codons of each CsSOD gene were retrieved from CuGI. Subsequently, the cis-elements in the promoters of each CsSOD gene were predicted using the PlantCARE tool (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
2.6. Plant Materials and Treatments
Cucumber (Cucumis sativus L. cv. Chinese long 9930) was used in this research. The cucumber plants were planted in the field of Jiangxi Agricultural University (Nanchang, China). To examine the transcript levels of CsSOD members in different tissues, root, stem, leaf, flower, and fruit were collected from cucumber and immediately frozen in liquid nitrogen for storage at −80°C until use.
To analyze CsSOD members under various abiotic stresses, 2-week-old cucumber seedlings were adequately watered and grown in a greenhouse under the conditions of 16 h light/8 h dark at 24/18°C before the induction of different stresses. For heat treatment, cucumber seedlings were transferred to 50°C growth chambers under normal light conditions. For drought treatment, cucumber seedlings were grown in liquid Murashige and Skoog (MS) medium containing 300 mM PEG-6000. Cold and salt experiments for cucumber seedlings were conducted as previously described [31]. Leaves were collected at 0, 3, 6, 12, and 24 h after treatments and immediately frozen in liquid nitrogen for storage at −80°C until use.
2.7. RNA Extraction, RT-PCR, and Quantitative Real-Time PCR (qRT-PCR)
Total RNA was extracted with Trizol reagent (Tiangen Biotech Co., Ltd. Beijing, China) according to the manufacturer's instructions. Synthesis of first-strand cDNA and RT-PCR was performed using the methods described previously [31]. qRT-PCR was conducted as previously described [32]. Three replicates were performed for the analysis of each gene. The cucumber Actin gene was used as an internal control. The primers used in gene expression analysis are listed in Table S1 available online at https://doi.org/10.1155/2017/7243973.
3. Results
3.1. Genome-Wide Characterization of CsSODs in Cucumber
A total of nine cucumber SOD genes were identified from CuGI and NCBI database. The candidate genes were translated into amino acid sequences and analyzed for the presence of a SOD domain by searching SMART and Pfam database. The results showed that the cucumber genome contained five Cu/ZnSODs (CsCSD1–5), three FeSODs (CsFSD1–3), and one MnSOD (CsMSD), respectively (Table 1). All the three putative CsFSDs contained the C-terminal iron/manganese SOD domain (PF02777), and CsFSD2 and CsFSD3 had the N-terminal iron/manganese SOD alpha-hairpin domain (PF00081) (Table S2). All CsCSDs contained the copper-zinc domain (PF00080), which is a typical characteristic of Cu/ZnSOD. CsCSD3 contained an additional heavy-metal-associated domain (PF00403). Both of the N-terminal iron/manganese SOD alpha-hairpin domain (PF00081) and C-terminal iron/manganese SOD domain (PF02777) were present in CsMSD. In addition, CsFSD2 contained a glyoxalase domain (PF00903), which belongs to dioxygenase superfamily (Table S2). The three domains (PF00080, PF00081, and PF02777) were predicted to possess molecular function of oxidation-reduction process (GO:0055114) and superoxide metabolic process (GO:0006801) according to Gene Ontology [33], indicating that the putative CsSOD members belong to the SOD family.
Table 1.
A complete list of 9 CsSODs identified in this study.
| Isoforms | Gene ID | Chromosamal location | CuGI (5′–3′) | gDNA size (bp) | Intron number | Length (aa) | Theoretical Mw (kDa) | pI | ProtComp |
|---|---|---|---|---|---|---|---|---|---|
| CsCSD1 | Csa001740 | 1 | 17723177–17725232 | 2056 | 6 | 152 | 15.3 | 5.44 | Cytoplasm |
| CsCSD2 | Csa006777 | 2 | 1729871–1727288 | 2584 | 6 | 152 | 15.4 | 4.97 | Cytoplasm |
| CsCSD3 | Csa018002 | 3 | 2262114–2259007 | 3108 | 5 | 328 | 34.8 | 5.16 | Cytoplasm |
| CsCSD4 | Csa017461 | 4 | 6531097–6533698 | 2602 | 7 | 220 | 23.3 | 8.77 | Cytoplasm |
| CsCSD5 | Csa015219 | 6 | 8963271–8966782 | 3512 | 7 | 216 | 21.9 | 5.87 | Chloroplast |
| CsMSD | Csa004428 | 1 | 2960859–2958014 | 2846 | 6 | 243 | 27.0 | 8.54 | Mitochondrion |
| CsFSD1 | Csa001483 | 1 | 17559641–17556948 | 2694 | 6 | 203 | 23.4 | 8.29 | Chloroplast |
| CsFSD2 | Csa016620 | 4 | 9018790–9014683 | 4108 | 8 | 338 | 38.6 | 5.75 | Chloroplast |
| CsFSD3 | Csa022092 | Scaffold000274 | 12020–15398 | 3379 | 3 | 377 | 42.5 | 5.16 | Extracellular |
These CsSODs were chosen for further analyses, and their characteristics are listed in Table 1. The gDNA lengths of CsSODs from the start to stop codons varied between 2056 and 4108 bp. Physicochemical analysis revealed that the lengths of putative CsSOD proteins varied between 152 and 377 amino acids. Besides, the putative CsSODs had a calculated molecular mass ranging from 15.3 to 42.5 kDa and a theoretical pI from 4.97 to 8.77 (Table 1). According to previous studies, all Cu/ZnSODs are acidic in character, while Fe-MnSODs are basic or acidic in character [6, 34–36]. In the present study, most of the CsSODs were acidic in character, except for three CsSODs (CsFSD1, CsMSD, and CsCSD4), which were basic in character.
Subcellular localizations of SOD proteins were predicted using ProtComp 9.0. The results showed that most of the CsCSDs (CsCSD1, CsCSD2, CsCSD3, and CsCSD4) were localized in the cytoplasm, while CsCSD5 was localized in the chloroplast (Table 1). In addition, two members (CsFSD1 and CsFSD2) and one member (CsFSD3) of FeSODs were located in the chloroplast and extracellular space, respectively. It seemed that SOD proteins with close phylogenetic relationships tended to have the same patterns of subcellular localization. The only member in group III, CsMSD, which is an MnSOD, was predicted to be localized in the mitochondrion. The prediction of the signal peptide by SignalP 4.1 indicated that only CsFSD3 had a signal peptide sequence with 16 amino acid residues that belongs to a secretory protein, confirming that CsFSD3 is an extracellular protein (Figure S1).
3.2. Conserved Motifs and Clustering Analysis of CsSODs
To investigate the evolutionary relationship of CsSODs in cucumber, an unrooted phylogenetic tree was created by aligning the 9 CsSOD protein sequences. CsSODs could be clustered into four groups (I, II, III, and IV), which was highly consistent with the types of their metal cofactors (Figure 1(a)).
Figure 1.
Unrooted neighbor-joining phylogenetic tree and conserved motif analysis of CsSOD proteins. (a) Phylogenetic tree was generated based on the protein sequences of CsSOD proteins. (b) Conserved motif analysis of CsSOD proteins. Different color boxes represent different types of motifs.
We also conducted prediction of conserved motifs for CsSODs by MEME server. As shown in Figure 1(b), 7 motifs (motif 1 to motif 7) were identified. As expected, the SOD members in the same group of phylogenetic tree demonstrated very similar patterns of conserved motif distribution. Among them, the copper-zinc SOD domain (motifs 1, 2, and 7) was widely present in CSDs (group I: CsCSD1, CsCSD2, CsCSD4, and CsCSD5), except for CsCSD3 (group II), which included motifs 1 and 7. Three CsFSDs (CsFSD1, CsFSD2, and CsFSD3) composed group IV, which shared the iron/manganese SOD domain (motifs 3, 5, and 6) with CsMSD (group III). Motif 4 was related to iron/manganese SOD alpha-hairpin domain, which was located in group III (CsMSD) and group IV (CsFSD2 and CsFSD3, except for CsFSD1).
3.3. Multiple Sequence Alignment of CsSOD Proteins
To further explore the evolutionary relationships among cucumber SOD proteins, pairwise identities were calculated (Table S3). The results revealed that the similarity between CsSOD proteins was notably variable, and CsCSD proteins shared 27–72% identity at amino acid levels, while the identity of CsMSD and CsFSD proteins was 32–43%, which was also in accordance with the types of their metal cofactors.
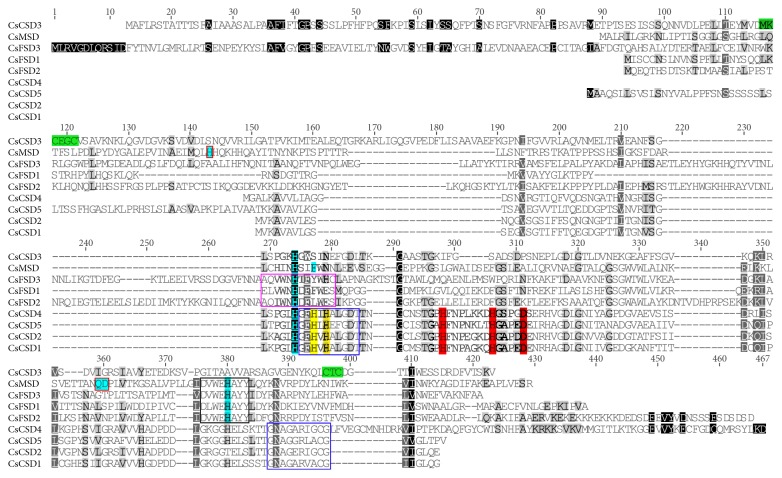
Multiple sequence alignment of the 9 CsSOD amino acid sequences was performed using the ClustalW software. The results showed that nearly all of the CsCSDs (except for CsCSD3) contained the Cu/ZnSOD signatures (GFH[VLI]H[EA][FL]GDTT and GNAG[GAE]R[VLI]ACG) and the metal-binding sites for Cu2+ and Zn2+ (Figure 2()). CsMSD along with CsFSDs had the conserved metal-binding domain “DVWEHAYY” with a conserved His residue found in almost all CsSODs. Three residues (His, Gln, and Asp) were present in CsMSD but absent in CsFSDs [10, 22], suggesting that these residues are specific for Mn2+. In addition, the signature ([AE][QL][VI]WNH[TD]F[YFL]W[EH][CS]) was responsible for the recognition of iron ion by FeSOD, which was also reported in previous studies [10, 37]. CsCSD3 was predicted to be located in extracellular space, suggesting that it participates in the secretory pathway. A blastp search in the NCBI database showed that CsCSD3 was 74.0% and 70.1% identical to copper chaperone for superoxide dismutase (CCS) proteins from rice (CuZn-SOD-CCh) [38] and banana (MaCCS) [39], respectively, implying that it belongs to the CCS subfamily. A conserved metal-binding motif (MxCxxC) and another conserved metal-binding motif (CxC) were near the N-terminus and in the C-terminus, respectively (Figure 2()).
Figure 2.
Multiple sequence alignment of deduced amino acid sequences of CsSOD proteins. Cu/ZnSOD signatures (GFH[VLI]H[EA][FL]GDTT and GNAG[GAE]R[VLI]ACG) are boxed in blue. The metal-binding sites for Cu2+ and Zn2+ are highlighted with yellow and red, respectively. The conserved metal-binding domain (DVWEHAYY) for Fe-MnSOD is in black box. The signature ([AE][QL][VI]WNH[TD]F[YFL]W[EH][CS]) responsible for the recognition of iron ion by FeSODs is boxed in pink. The conserved metal-binding motifs (MxCxxC and CxC) of CsCSD3 are highlighted in green. Six metal-binding sites in MnSOD are marked with cyan, and three residues (His, Gln, and Asp) that are present only in MnSOD and absent in FeSODs are marked with cyan in red box.
3.4. Chromosomal Location and Gene Structure of CsSODs
The chromosomal distributions of cucumber SOD genes are shown in Figure 3(). Accordingly, the 9 CsSODs were distributed on 5 chromosomes in Cucumis sativus, except for CsFSD3, which was found on an unassembled sequence scaffold000274 (Figure 3()). The largest number of SOD genes was determined on Chr1 with 3 genes, while Chr4 possessed two SOD genes and all the others contained only one.
Figure 3.
Chromosomal locations of 9 CsSOD genes on chromosomes of cucumber. The chromosome numbers are indicated at the top of chromosomes, and the size of chromosome is represented with a vertical scale.
To further examine the structural features of CsSODs, a comparison of the ORF sequences with the corresponding gDNA sequences was performed by GSDS server. As shown in Figure 4(), all CsSOD genes contained introns in their genomic sequences in cucumber, and the intron numbers in the CsSOD genes ranged from 3 to 7. Four CsSOD members (CsCSD4, CsCSD5, CsMSD, and CsFSD2) contained the largest number of introns (7 introns), while the smallest number was found in CsFSD3 (3 introns). It is worth noting that two pairs of CsSOD genes (CsCSD1 and CsCSD2, CsFSD1 and CsFSD2) exhibited similar intron/exon organization patterns, respectively.
Figure 4.
Exon-intron structures of CsSOD genes based on the phylogenetic relationship. Exons and introns are shown as green boxes and thin lines, respectively. Intron phases 0, 1, and 2 are indicated by numbers 0, 1, and 2.
3.5. Phylogenetic Analysis of SOD Proteins in Plants
To further investigate the phylogenetic relationships of SOD proteins between cucumber and other plant species, an unrooted neighbor-joining phylogenetic tree was generated using the amino acid sequences of the SOD proteins from Arabidopsis thaliana, Brachypodium distachyon, Solanum lycopersicum, Oryza sativa, Sorghum bicolor, Gossypium raimondii, and Setaria italica. The phylogenetic tree revealed that these SOD proteins could be classified into two major groups: Cu/ZnSODs and Fe-MnSODs. Cu/ZnSODs included three subgroups (a–c), while Fe-MnSODs were divided into two subgroups (d and e) (Figure 5), implying that FeSODs and MnSODs originated from a common ancestor [11].
Figure 5.
Phylogenetic tree of SOD proteins from cucumber and other plants. Deduced protein sequences of SOD proteins from different plant species were analyzed by ClustalW and then redrawn with the Geneious Pro software. Five colors (yellow, orange, cyan, blue, and green) represent five subgroups (a, b, c, d, and e), respectively. CsSOD proteins were highlighted in red. The amino acid sequences used to build the phylogenic tree were listed in Table S4.
Like in other plants, CsSODs were present in all the five subgroups. Three CsCSDs (CsCSD1, CsCSD2, and CsCSD4) along with SbSOD1, SlSOD1, and other cytoplasmic Cu/ZnSODs were clustered into subgroup a, while CsCSD5 together with SbSOD4, SlSOD3, and other chloroplastic Cu/ZnSODs was clustered in another cluster (subgroup b). The remaining CsCSDs (CsCSD3), along with SlSOD4, SbSOD3, GrCSD4, and other CSDs, were clustered in subgroup c, which was like a junction between Cu/ZnSODs and Mn-FeSODs clusters. Notably, all of the three CsFSDs were clustered in subgroup d with other plant FeSODs (Figure 5()). Among them, CsFSD1 and CsFSD2 were clustered with other plant chloroplastic FeSODs, while CsFSD3, which was found on an unassembled sequence and predicted to be located in extracellular space (Table 1; Figure 3()), was separated from other plant FeSODs. In addition, CsCSDs and CsFSDs (except for CsFSD3) were clustered into Cu/ZnSODs and FeSODs of dicot-specific branches, respectively, revealing the presence of two ancestral SODs before the divergence of monocot and dicot plants. Moreover, CsMSD together with other plant mitochondrial MSDs were clustered in subgroup e (Figure 5()).
3.6. Analysis of cis-Acting Regulatory Elements in the Promoter of CsSODs
To understand the gene function and regulation patterns of CsSODs, sequences of 1000 bp upstream regions from the translation start site of each CsSOD were identified from CuGI and were subsequently determined using PlantCARE. The results showed that 59 types of cis-acting regulatory elements were found in the promoter region of CsSODs and all CsSOD promoters contained typical TATA and CAAT boxes, which are essential elements of the promoters (Table S5). In addition, several cis-elements related to developmental processes such as circadian control (circadian), flavonoid biosynthetic gene regulation (MBSII), meristem expression (CAT-box) and zein metabolism regulation (O2-site), and tissue-specific expression, including shoot-specific element (as-2-box), meristem specific activation element (CCGTCC-box), and endosperm-specific elements (GCN4 and Skn-1 motif), were present in CsSOD promoters. Among them, the light-responsive elements included 23 different types and were the largest group of elements. Each CsSOD promoter contained 3–7 types of light-responsive elements, implying that CsSODs might be differentially regulated by light. The type numbers of the remaining cis-acting regulatory elements, including stress-related and hormone-related elements, were shown in Table 2. The cis-acting regulatory elements involved in abiotic stress, such as heat stress-responsive element (HSE), drought-responsive element (MBS), wound-responsive element (WUN motif), defense and stress-responsive element (TC-rich repeats), anaerobic induction element (ARE), and elicitor-responsive elements (EIRE and Box-W1), were also detected in a series of members. Moreover, 8 types of hormone-responsive cis-elements were identified, and all of the CsSOD promoter sequences contained at least one type of cis-elements involved in plant hormone response, including abscisic acid- (ABA-) responsive element (ABRE), methyl jasmonate- (MeJA-) responsive element (CGTCA-motif), ethylene-responsive element (ERE), gibberellin-responsive elements (GARE-motif, P-box, and TATC-box), salicylic acid-responsive elements (TCA-element), and auxin-responsive elements (TGA-element). Numerous cis-elements related to light, stress, or hormone responses were present in the promoter of CsSODs, suggesting that CsSODs are related to the responses to various stresses and hormones.
Table 2.
Kinds and numbers of known stress-related and hormone-related cis-elements in the promoter regions of CsSOD genes.
| Stress-related cis-element | Hormone-related cis-element | |
|---|---|---|
| CsCSD1 | TC-rich repeats (1), ARE (2) | ABRE (3), TCA-element (1), TGA-element (1) |
| CsCSD2 | MBS (3), WUN-motif (1), TC-rich repeats (1), ARE (1) | GARE-motif (2), TCA-element (1) |
| CsCSD3 | HSE (3), TC-rich repeats (3), ARE (1) | ABRE (1), CGTCA-motif (1), GARE-motif (2), TATC-box (1), TCA-element (1) |
| CsCSD4 | HSE (3), MBS (3), ARE (1) | ABRE (1), CGTCA-motif (1), ERE (3), TCA-element (1) |
| CsCSD5 | MBS (3), TC-rich repeats (1), ARE (1) | TATC-box (1), TGA-element (1) |
| CsMSD | HSE (1), MBS (1), TC-rich repeats (2), ARE (1), Box-W1 (1) | ABRE (1), ERE (1), GARE-motif (1), TGA-element (1) |
| CsFSD1 | HSE (2), MBS (2), TC-rich repeats (1) | GARE-motif (1) |
| CsFSD2 | HSE (1) | ABRE (3), P-box (2), TCA-element (2) |
| CsFSD3 | HSE (2), MBS (1), EIRE (1) | ABRE (1), CGTCA-motif (1) |
The numbers in parentheses indicate the number of the cis-element in the promoter of CsSOD.
3.7. Expression Profiles of CsSODs in Different Tissues
To investigate the functions of CsSODs, their expression profiles in different tissues (root, stem, leaf, flower, and fruit) were determined by RT-PCR. The results showed that most of the CsSODs were expressed in almost all 5 tissues, except for CsMSD, which was not expressed in fruit, and CsFSD3, whose transcript was not detected in all the tested tissues (Figure 6). In addition, CsCSD1 was consistently highly expressed in all 5 tissues, whereas CsCSD4 displayed an extremely low expression level. CsCSD1 was more highly expressed in leaf and flower than in other tested tissues. The expression of CsCSD2 in root and leaf was significantly higher than that in other tested tissues, while that of CsCSD5 in root was significantly lower than that in other tested tissues. Unlike other CsCSDs, CsCSD3 displayed a distinctly tissue-specific expression pattern and had the highest expression level in flower. CsFSD1 and CsFSD2, both of which were FeSODs (Table 1), showed similar expression patterns (Figure 6). CsFSD1 and CsFSD2 showed varied expression levels across tissues and were abundantly expressed in leaf and flower, but moderately expressed in other tissues (Figure 6). The expression of CsMSD was relatively lower in leaf, extremely low in flower and stem, and even not detected in root and fruit.
Figure 6.
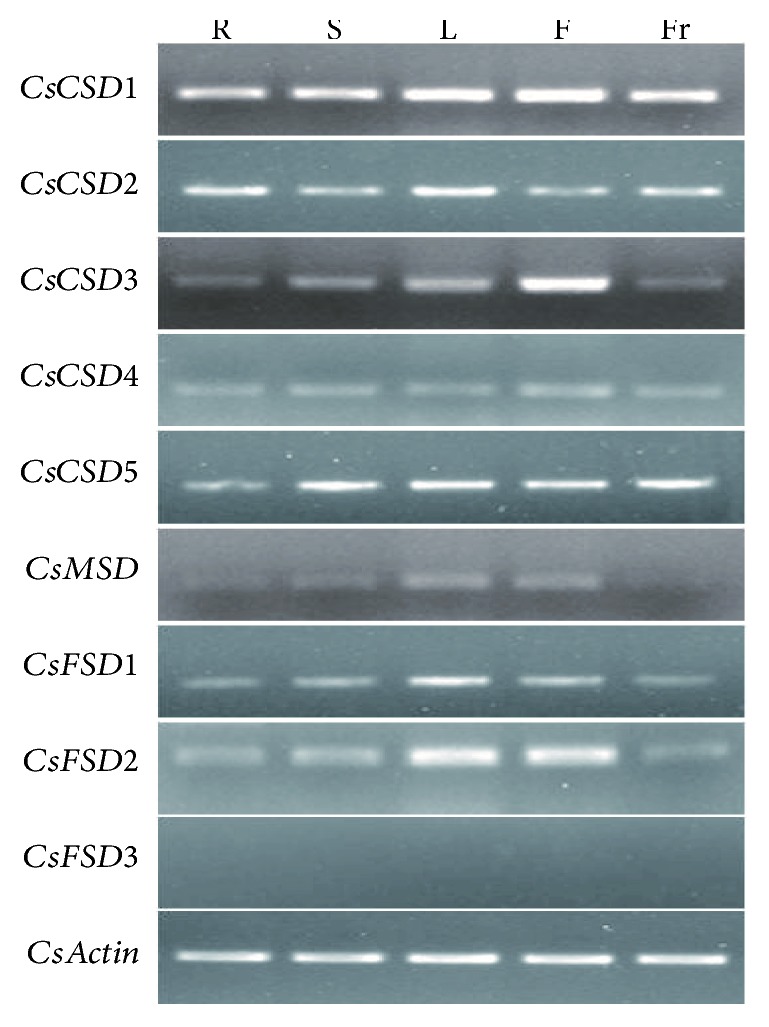
RT-PCR analysis of cucumber SOD genes in different tissues. RNA was isolated from different tissues, including root (R), stem (S), leaf (L), flower (F), and fruit (Fr).
3.8. Expression of CsSODs in Response to Various Abiotic Stresses
To investigate the roles of CsSOD genes in response to various abiotic stresses, we analyzed the expression patterns of CsSOD genes under heat, cold, drought, and NaCl treatments by qRT-PCR. As shown in Figure 7, the expression of CsSOD genes was noticeably different under different abiotic stress treatments, and their expression patterns were complex. Exceptionally, no expression of CsFSD3 was detected in response to these stresses. In addition, the expression of CsFSD3 was not detected in root, stem, leaf, flower, and fruit (Figure 6). These results implied that CsFSD3 is a pseudogene.
Figure 7.

qRT-PCR analysis of cucumber SOD genes in response to various abiotic stresses including heat (a), cold (b), PEG (c), and NaCl (d). Error bars indicate SD based on three biological replicates.
All of the remaining 8 CsSOD genes were strongly induced by heat treatment and exhibited more than 3- to 24-fold increase in expression. Among them, 1 gene (CsFSD1), 2 genes (CsCSD4, CsCSD5), 1 gene (CsCSD1), and 4 genes (CsCSD2, CsCSD3, CsMSD, and CsFSD2) reached the highest expression levels at 3 h, 6 h, 12 h, and 24 h, respectively. Interestingly, the expression of a pair of CSDs, CsCSD3 and CsCSD4, whose transcripts were induced gradually until 6 h, subsequently decreased at 12 h and finally reached a higher level at 24 h (Figure 7(a)). In addition, the expression of CsFSD1 was increased sharply, reached the highest level at 3 h, then dropped dramatically at 6 h, and subsequently increased gradually as the treatment continued (Figure 7(a)). Under cold treatment, the expression levels of most CsSOD genes decreased at 3 h (CsCSD3 remained constant, and CsFSD1 was upregulated slightly) and then increased markedly at 6 h (Figure 7(b)). After 6 h, all of the CsCSD genes were downregulated sharply at 12 h and continued to decline at 24 h (except for CsCSD2, whose expression was increased at 24 h). After 6 h, the expression of CsFSD1 and CsFSD2 was induced gradually, and the induction peaked at 12 h and 24 h, respectively. During PEG treatment, nearly all CsCSD genes were strongly induced, their expression peaked at 3 h, and then the transcript levels were decreased, whereas CsCSD1 was slightly downregulated at 3 h and its transcript remained constant (Figure 7(c)). Under NaCl treatment, the expression of most CsSOD genes was induced gradually until 6 h, then declined at 12 h, and ultimately increased at 24 h (Figure 7(d)). It was worth noting that the transcript of CsCSD2 reached the highest level (38.6-fold) at 24 h, which was obviously higher than that of other CsSOD genes. The expression of CsCSD5 increased gradually until 6 h, then declined gradually after 12 h of treatment, and finally decreased dramatically at 24 h (Figure 7(d)).
4. Discussion
4.1. SOD Genes in Cucumber and Their Evolution
SOD plays important roles in multiple processes of plant growth and resistance against environment stresses, and SOD gene family has been reported to be widely distributed in different plant species, such as Arabidopsis [40], longan [41], rice [34, 42], poplar [22], banana [10], sorghum [35], tomato [6], and cotton [36, 43]. However, no comprehensive analysis of this gene family has been reported in cucumber. In the present study, a total of 9 SOD genes were identified in cucumber, which cover all three major types of plant SOD genes, including 5 Cu/ZnSODs, 3 FeSODs, and 1 MnSOD (Table 1). Similar results were obtained in other plant species. For example, previous studies revealed that the numbers of SOD genes in Arabidopsis, rice, sorghum, and tomato are 8 (3 Cu/ZnSODs, 2 MnSODs, and 3 FeSODs), 8 (5 Cu/ZnSODs, 1 MnSOD, and 2 FeSODs), 8 (5 Cu/ZnSODs, 1 MnSOD, and 2 FeSODs), and 9 (4 Cu/ZnSODs, 1 MnSOD, and 4 FeSODs), respectively. There are large differences in genome size, and the number of SOD genes varies among these plant species, but does not vary proportionally along with the changes in genome size. The discrepancy in the number of SOD genes in different plant species may be attributed to gene duplication, which consists of tandem and segmental duplications and plays a crucial role in the expansion of SOD genes for diversification. Gene duplication of SOD genes was also found in different plant species [35, 36, 43]. In our study, no segmental duplication event was observed between these genes according to the criteria of a previous study [35]. Therefore, tandem duplication most likely plays an important role in the expansion of CsSOD genes, just like the two adjacent genes CsFSD1 and CsCSD1 in Chr1 (Figure 3()).
Gene structure analysis revealed that the intron numbers of the 9 cucumber SOD genes were 3–7 (Figure 4()). A previous research reported that plant SOD genes have highly conserved intron patterns, and most cytosolic and chloroplastic SODs harbor 7 introns [44]. In our study, four members (CsCSD4, CsCSD5, CsMSD, and CsFSD2) were predicted to contain 7 introns (Figure 4()). The divergence of CsSOD gene structure may be due to the mechanisms including exon/intron gain/loss, exonization/pseudoexonization, and insertion/deletion according to a previous study [45], and the SOD members in the same clade of phylogenetic tree displayed similar exon-intron organization patterns (such as CsCSD1 and CsCSD2, CsFSD1 and CsFSD2), suggesting that they may have similar functions related to various abiotic stresses.
Phylogenetic analysis of SOD proteins between cucumber and 7 other plant species showed that the SODs could be divided into two groups of Cu/ZnSODs and Fe-MnSODs, and these two groups could be further divided into 5 subgroups with robust statistical support, which is consistent with the results of previous reports [6, 10, 11, 35]. Most of the subcellular localization data of SODs supported the phylogenetic data. For example, several pairs of Cu/ZnSODs, including CsCSD1 and CsCSD2, SbSOD1 and SbSOD2 [35], SlSOD1 and SlSOD2 [6], cCuZn-SOD-1 and cCuZn-SOD-2 [42], GrCSD1 and GrCSD2 [36], were predicted to be located in the cytoplasm in subgroup a (Figure 5()). Chloroplastic Cu/ZnSODs, chloroplastic FeSODs, and mitochondrial MnSODs were clustered into subgroups b, d, and e, respectively. CsCSD3, along with CuZn-SOD-CCh and other chloroplastic CSDs, was observed in subgroup c, suggesting that it is a member of plant CCS family. In addition, almost all CsSODs were phylogenetically clustered with at least one member from all the considered plant species (Figure 5()), suggesting that CsSODs probably have the same functions as SODs in other plant species. However, CsFSD3, which was found on an unassembled sequence and predicted to be located in extracellular space (Figures 1() and 3()), was separated from other plant FeSODs. Besides, the prediction of the signal peptide revealed that CsFSD3 has a signal peptide sequence belonging to a secretory protein (Figure S1), implying that CsFSD3 is an extracellular protein.
4.2. Expression Patterns of CsSODs in Different Tissues
Comprehensive gene expression analyses of SOD family genes have demonstrated that SODs have diverse expression patterns in different tissues of plants, such as Arabidopsis [40], rice [34, 42], poplar [22], banana [10], tomato [6], and cotton [36, 43]. In this study, the results of RT-PCR analysis showed that most of the CsSOD genes are ubiquitously expressed and a few genes display tissue-specific expression patterns except for CsFSD3, whose transcript was not detected in any tested tissues (Figure 6). For instance, CsCSD1 showed high expression in all the tested tissues, whereas CsCSD4 displayed extremely low expression level (Figure 6), suggesting that they have differential expression patterns in various tissues. In addition, many CsSOD genes showed the highest expression in leaves, which is consistent with previous studies of other species [9, 36]. Notably, all of the CsSOD genes except for CsMSD were expressed in fruit (Figure 6) and shared a common cis-acting regulatory element Skn-1_motif (Table 2), which is required for endosperm expression, suggesting that these genes may be involved in fruit development. CsCSD3 exhibited high expression in flower, indicating its specific roles required in flower development.
4.3. Functions of CsSODs in Response to Various Abiotic Stresses
Analysis of the cis-elements in CsSOD gene promoters resulted in the detection of three major types of cis-elements associated with light, abiotic stress, and hormone response as well as cis-elements related to developmental processes and tissue-specific expression (Table S5). A relatively large number of light-responsive cis-elements were detected in CsSOD gene promoters, suggesting that CsSODs might participate in light response. Some studies have shown that SOD genes are involved in the response to light in different plants including Arabidopsis thaliana [19, 40], Nicotiana tabacum [46], and Nicotiana plumbaginifolia [47]. Transgenic plants overexpressing a pea chloroplastic Cu/ZnSOD showed increased resistance to high light [48]. In addition, the cis-acting regulatory element involved in circadian control was found in six CsSODs except for CsCSD4, CsCSD5, and CsMSD (Table S5). Therefore, CsSODs, especially chloroplastic SODs, may be responsible for light response and function in the efficient removal of the superoxide formed during photosynthetic electron transport and under various stress conditions in the light.
In addition, a series of cis-elements related to abiotic stress responses were identified in CsSOD gene promoters, such as HSE, MBS, WUN-motif, TC-rich repeats, ARE, EIRE, and Box-W1 (Table 2), which may regulate gene expression under various stresses. Most of the SOD genes in Arabidopsis, tomato, and banana can be induced in response to various abiotic stresses such as heat, cold, drought, and salinity [6, 10, 40] and are considered to participate in the elimination of ROS caused by various abiotic stresses [11, 40]. In the present study, the expression of CsFSD3 was not detected under various abiotic stresses (Figure 7), and its transcripts were also undetectable in different tissues (Figure 6), indicating that it is a pseudogene. The expression levels of other 8 CsSOD genes were largely altered under stress conditions, suggesting that these genes play important regulatory roles in stress response and that there may be functional redundancy. For example, in this study, all of the detected CsSOD genes were upregulated under heat treatment, and some of them displayed similar expression patterns (Figure 7(a)). Under cold treatment, the expression levels of all CsSOD genes were obviously altered and their expression patterns were distinct from each other (Figure 7(b)), implying that CsSOD genes may have different functions in response to cold stress. Among them, the expression of CsCSD1 was decreased gradually, with an expression pattern similar to that of MaCSD1B and MaCSD2B [10]. For PEG treatment, nearly all of the CsSOD genes had similar expression patterns, which reached the highest levels at 3 h and then decreased (Figure 7(c)), implying that the functions of CsSOD genes are related to osmotic stress. During NaCl treatment, although the transcripts of most CsSOD genes were changed, CsCSD2 was the only gene that showed a remarkable increase among the 8 CsSOD genes (Figure 7(d)), demonstrating that CsCSD2 might play a predominant antioxidant role under salt stress. This phenomenon was previously observed for SlSOD1 in tomato [6]. Both SlSOD1 and CsCSD2 were clustered in subgroup a and showed a relatively high amino acid sequence identity (72.8%) (Figure 5). Taken together, we propose that the CsSOD genes have different functions in ROS elimination caused by different abiotic stresses, and may play vital roles in adversity adaptation of plants. In addition, some CsSOD genes were found to respond to various abiotic stresses with different expression patterns. For example, CsCSD1 was downregulated by cold treatment and upregulated by heat and NaCl treatments, while its expression remained constant without a significant change under PEG stress (Figure 7), implying a divergence in the functions of CsCSD1 in different signal pathways. Further studies are needed to reveal the exact functions of CsSOD genes during various abiotic stresses in cucumber.
5. Conclusion
In the present study, we identified 9 putative SOD genes from cucumber and analyzed their classification, genome organization, gene structure, conserved motif, and phylogenetic relationships. Subsequently, RT-PCR and qRT-PCR were carried out to analyze the expression profiles of CsSOD genes in different tissues and under various abiotic stresses including heat, cold, PEG, and NaCl. It could be suggested that CsSOD genes play vital roles in almost all tissues at the stage of growth and development in cucumber. This study provides a comprehensive understanding and facilitates further functional characterization of the SOD gene family in cucumber and lays a foundation for elucidating the molecular mechanisms of SOD genes during stress response and development of plants.
Supplementary Material
Fig. S1. The signal peptide prediction of the CsSOD proteins. The purple line indicate the discrimination score which is used to discriminate signal peptides from non-signal peptides. Table S1. Primers used in this study. Table S2. A list of predicted domains in 9 CsSODs in our study. Table S3. Pairwise amino acid sequence comparison of CsSOD proteins. Table S4. SOD sequences used for phylogenetic tree analysis. Table S5. The predicted promoter elements of CsSOD genes.
Acknowledgments
This work was funded by the National Natural Science Foundation of China (Grants nos. 31460522 and 31660578), the Key Project of Youth Science Foundation of Jiangxi Province (Grant no. 20171ACB21025), and the Doctoral Scientific Research Foundation of Jiangxi Agricultural University (Grant no. 9232305179). The authors are grateful to Professor Zuoxiong Liu (Huazhong Agricultural University) for critical reading of the manuscript.
Conflicts of Interest
The authors declare that they have no conflict of interest.
Authors' Contributions
Yong Zhou and Lifang Hu contributed equally to this work.
References
- 1.Cramer G. R., Urano K., Delrot S., Pezzotti M., Shinozaki K. Effects of abiotic stress on plants: a systems biology perspective. BMC Plant Biology. 2011;11:p. 163. doi: 10.1186/1471-2229-11-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.You J., Chan Z. ROS regulation during abiotic stress responses in crop plants. Frontiers in Plant Science. 2015;6:p. 1092. doi: 10.3389/fpls.2015.01092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Del Rio L. A. ROS and RNS in plant physiology: an overview. Journal of Experimental Botany. 2015;66:2827–2837. doi: 10.1093/jxb/erv099. [DOI] [PubMed] [Google Scholar]
- 4.Mittler R., Vanderauwera S., Gollery M., Van Breusegem F. Reactive oxygen gene network of plants. Trends in Plant Science. 2004;9:490–498. doi: 10.1016/j.tplants.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 5.Yan J. J., Zhang L., Wang R. Q., et al. The sequence characteristics and expression models reveal superoxide dismutase involved in cold response and fruiting body development in Volvariella volvacea. International Journal of Molecular Sciences. 2016;17:34–46. doi: 10.3390/ijms17010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng K., Yu J., Cheng Y., et al. The SOD gene family in tomato: identification, phylogenetic relationships, and expression patterns. Frontiers in Plant Science. 2016;7:p. 1279. doi: 10.3389/fpls.2016.01279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abreu I. A., Cabelli D. E. Superoxide dismutases-a review of the metal-associated mechanistic variations. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics. 2010;1804:263–274. doi: 10.1016/j.bbapap.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 8.Asada K., Yoshikawa K., Takahashi M., Maeda Y., Enmanji K. Superoxide dismutases from a blue-green alga, Plectonema boryanum. Journal of Biological Chemistry. 1975;250:2801–2807. [PubMed] [Google Scholar]
- 9.Pilon M., Ravet K., Tapken W. The biogenesis and physiological function of chloroplast superoxide dismutases. Biochimica et Biophysica Acta (BBA) - Bioenergetics. 2011;1807:989–998. doi: 10.1016/j.bbabio.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 10.Feng X., Lai Z., Lin Y., Lai G., Lian C. Genome-wide identification and characterization of the superoxide dismutase gene family in Musa acuminata cv. Tianbaojiao (AAA group) BMC Genomics. 2015;16:p. 823. doi: 10.1186/s12864-015-2046-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alscher R. G., Erturk N., Heath L. S. Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. Journal of Experimental Botany. 2002;53:1331–1341. doi: 10.1093/jxb/53.372.1331. [DOI] [PubMed] [Google Scholar]
- 12.Moller I. M. Plant mitochondria and oxidative stress: electron transport, NADPH turnover, and metabolism of reactive oxygen species. Annual Review of Plant Physiology and Plant Molecular Biolog. 2001;52:561–591. doi: 10.1146/annurev.arplant.52.1.561. [DOI] [PubMed] [Google Scholar]
- 13.Miller A. F. Superoxide dismutases: ancient enzymes and new insights. FEBS Letters. 2012;586:585–595. doi: 10.1016/j.febslet.2011.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wuerges J., Lee J. W., Yim Y. I., Yim H. S., Kang S. O., Djinovic Carugo K. Crystal structure of nickel-containing superoxide dismutase reveals another type of active site. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:8569–8574. doi: 10.1073/pnas.0308514101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Youn H. D., Kim E. J., Roe J. H., Hah Y. C., Kang S. O. A novel nickel-containing superoxide dismutase from Streptomyces spp. Biochemical Journal. 1996;318(Part 3):889–896. doi: 10.1042/bj3180889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang B., Luttge U., Ratajczak R. Specific regulation of SOD isoforms by NaCl and osmotic stress in leaves of the C3 halophyte Suaeda salsa L. Journal of Plant Physiology. 2004;161:285–293. doi: 10.1078/0176-1617-01123. [DOI] [PubMed] [Google Scholar]
- 17.Srivastava V., Srivastava M. K., Chibani K., et al. Alternative splicing studies of the reactive oxygen species gene network in Populus reveal two isoforms of high-isoelectric-point superoxide dismutase. Plant Physiology. 2009;149:1848–1859. doi: 10.1104/pp.108.133371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng W., Hongbin W., Bing L., Jinfa W. Cloning and characterization of a novel splicing isoform of the iron-superoxide dismutase gene in rice (Oryza sativa L.) Plant Cell Reports. 2006;24:734–742. doi: 10.1007/s00299-005-0030-4. [DOI] [PubMed] [Google Scholar]
- 19.Sunkar R., Kapoor A., Zhu J. K. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell. 2006;18:2051–2065. doi: 10.1105/tpc.106.041673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dugas D. V., Bartel B. Sucrose induction of Arabidopsis miR398 represses two Cu/Zn superoxide dismutases. Plant Molecular Biology. 2008;67:403–417. doi: 10.1007/s11103-008-9329-1. [DOI] [PubMed] [Google Scholar]
- 21.Lee S. H., Ahsan N., Lee K. W., et al. Simultaneous overexpression of both CuZn superoxide dismutase and ascorbate peroxidase in transgenic tall fescue plants confers increased tolerance to a wide range of abiotic stresses. Journal of Plant Physiology. 2007;164:1626–1638. doi: 10.1016/j.jplph.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 22.Molina-Rueda J. J., Tsai C. J., Kirby E. G. The Populus superoxide dismutase gene family and its responses to drought stress in transgenic poplar overexpressing a pine cytosolic glutamine synthetase (GS1a) PLoS One. 2013;8, article e56421 doi: 10.1371/journal.pone.0056421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang F. Z., Wang Q. B., Kwon S. Y., Kwak S. S., Su W. A. Enhanced drought tolerance of transgenic rice plants expressing a pea manganese superoxide dismutase. Journal of Plant Physiology. 2005;162:465–472. doi: 10.1016/j.jplph.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 24.Wang M., Zhao X., Xiao Z., Yin X., Xing T., Xia G. A wheat superoxide dismutase gene TaSOD2 enhances salt resistance through modulating redox homeostasis by promoting NADPH oxidase activity. Plant Molecular Biology. 2016;91:115–130. doi: 10.1007/s11103-016-0446-y. [DOI] [PubMed] [Google Scholar]
- 25.Jing X., Hou P., Lu Y., et al. Overexpression of copper/zinc superoxide dismutase from mangrove Kandelia candel in tobacco enhances salinity tolerance by the reduction of reactive oxygen species in chloroplast. Frontiers in Plant Science. 2015;6:p. 23. doi: 10.3389/fpls.2015.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Negi N. P., Shrivastava D. C., Sharma V., Sarin N. B. Overexpression of CuZnSOD from Arachis hypogaea alleviates salinity and drought stress in tobacco. Plant Cell Reports. 2015;34:1109–1126. doi: 10.1007/s00299-015-1770-4. [DOI] [PubMed] [Google Scholar]
- 27.Kim Y. H., Lim S., Han S. H., et al. Expression of both CuZnSOD and APX in chloroplasts enhances tolerance to sulfur dioxide in transgenic sweet potato plants. Comptes Rendus Biologies. 2015;338:307–313. doi: 10.1016/j.crvi.2015.03.012. [DOI] [PubMed] [Google Scholar]
- 28.Perl A., Perl-Treves R., Galili S., et al. Enhanced oxidative-stress defense in transgenic potato expressing tomato Cu, Zn superoxide dismutases. Theoretical and Applied Genetics. 1993;85:568–576. doi: 10.1007/BF00220915. [DOI] [PubMed] [Google Scholar]
- 29.Luo X., Wu J., Li Y., et al. Synergistic effects of GhSOD1 and GhCAT1 overexpression in cotton chloroplasts on enhancing tolerance to methyl viologen and salt stresses. PLoS One. 2013;8, article e54002 doi: 10.1371/journal.pone.0054002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu L., Liu S. Genome-wide analysis of the MADS-box gene family in cucumber. Genome. 2012;55:245–256. doi: 10.1139/g2012-009. [DOI] [PubMed] [Google Scholar]
- 31.Hu L., Yang Y., Jiang L., Liu S. The catalase gene family in cucumber: genome-wide identification and organization. Genetics and Molecular Biology. 2016;39:408–415. doi: 10.1590/1678-4685-GMB-2015-0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou Y., Liu L., Huang W., et al. Overexpression of OsSWEET5 in rice causes growth retardation and precocious senescence. PLoS One. 2014;9, article e94210 doi: 10.1371/journal.pone.0094210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gotz S., Garcia-Gomez J. M., Terol J., et al. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Research. 2008;36:3420–3435. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dehury B., Sarma K., Sarmah R., et al. In silico analyses of superoxide dismutases (SODs) of rice (Oryza sativa L.) Journal of Plant Biochemistry and Biotechnology. 2013;22:150–156. doi: 10.1007/s13562-012-0121-6. [DOI] [Google Scholar]
- 35.Filiz E., Tombuloğlu H. Genome-wide distribution of superoxide dismutase (SOD) gene families in Sorghum bicolor. Turkish Journal of Biology. 2015;39:49–59. doi: 10.3906/biy-1403-9. [DOI] [Google Scholar]
- 36.Zhang J., Li B., Yang Y., et al. Genome-wide characterization and expression profiles of the superoxide dismutase gene family in Gossypium. International Journal of Genomics. 2016;2016:11. doi: 10.1155/2016/8740901.8740901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parker M. W., Blake C. C. F. Iron- and manganese-containing superoxide dismutases can be distinguished by analysis of their primary structures. FEBS Letters. 1988;229:377–382. doi: 10.1016/0014-5793(88)81160-8. [DOI] [PubMed] [Google Scholar]
- 38.Huang C. H., Kuo W. Y., Weiss C., Jinn T. L. Copper chaperone-dependent and -independent activation of three copper-zinc superoxide dismutase homologs localized in different cellular compartments in Arabidopsis. Plant Physiology. 2012;158:737–746. doi: 10.1104/pp.111.190223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feng X., Chen F., Liu W., et al. Molecular characterization of MaCCS, a novel copper chaperone gene involved in abiotic and hormonal stress responses in Musa acuminata cv. Tianbaojiao. International Journal of Molecular Sciences. 2016;17:p. 441. doi: 10.3390/ijms17040441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kliebenstein D. J., Monde R. A., Last R. L. Superoxide dismutase in Arabidopsis: an eclectic enzyme family with disparate regulation and protein localization. Plant Physiology. 1998;118:637–650. doi: 10.1104/pp.118.2.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin Y. L., Lai Z. X. Superoxide dismutase multigene family in longan somatic embryos: a comparison of CuZn-SOD, Fe-SOD, and Mn-SOD gene structure, splicing, phylogeny, and expression. Molecular Breeding. 2013;32:595–615. doi: 10.1007/s11032-013-9892-2. [DOI] [Google Scholar]
- 42.Nath K., Kumar S., Poudyal R. S., et al. Developmental stage-dependent differential gene expression of superoxide dismutase isoenzymes and their localization and physical interaction network in rice (Oryza sativa L.) Genes & Genomics. 2014;36:45–55. doi: 10.1007/s13258-013-0138-9. [DOI] [Google Scholar]
- 43.Wang W., Xia M., Chen J., et al. Genome-wide analysis of superoxide dismutase gene family in Gossypium raimondii and G. arboreum. Plant Gene. 2016;6:18–29. doi: 10.1016/j.plgene.2016.02.002. [DOI] [Google Scholar]
- 44.Fink R. C., Scandalios J. G. Molecular evolution and structure―function relationships of the superoxide dismutase gene families in angiosperms and their relationship to other eukaryotic and prokaryotic superoxide dismutases. Archives of Biochemistry and Biophysics. 2002;399:19–36. doi: 10.1006/abbi.2001.2739. [DOI] [PubMed] [Google Scholar]
- 45.Xu G., Guo C., Shan H., Kong H. Divergence of duplicate genes in exon-intron structure. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:1187–1192. doi: 10.1073/pnas.1109047109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kurepa J., Montagu M. V., Inzé D. Expression of sodCp and sodB genes in Nicotiana tabacum: effects of light and copper excess. Journal of Experimental Botany. 1997;48:2007–2014. doi: 10.1093/jxb/48.12.2007. [DOI] [Google Scholar]
- 47.Tsang E. W., Bowler C., Herouart D., et al. Differential regulation of superoxide dismutases in plants exposed to environmental stress. Plant Cell. 1991;3:783–792. doi: 10.1105/tpc.3.8.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gupta A. S., Heinen J. L., Holaday A. S., Burke J. J., Allen R. D. Increased resistance to oxidative stress in transgenic plants that overexpress chloroplastic Cu/Zn superoxide dismutase. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:1629–1633. doi: 10.1073/pnas.90.4.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. The signal peptide prediction of the CsSOD proteins. The purple line indicate the discrimination score which is used to discriminate signal peptides from non-signal peptides. Table S1. Primers used in this study. Table S2. A list of predicted domains in 9 CsSODs in our study. Table S3. Pairwise amino acid sequence comparison of CsSOD proteins. Table S4. SOD sequences used for phylogenetic tree analysis. Table S5. The predicted promoter elements of CsSOD genes.