Abstract
Chemical and physical properties of the environment control cell proliferation, differentiation, or apoptosis in the long term. However, to be able to move and migrate through a complex three-dimensional environment, cells must quickly adapt in the short term to the physical properties of their surroundings. Interactions with the extracellular matrix (ECM) occur through focal adhesions or hemidesmosomes via the engagement of integrins with fibrillar ECM proteins. Cells also interact with their neighbors, and this involves various types of intercellular adhesive structures such as tight junctions, cadherin-based adherens junctions, and desmosomes. Mechanobiology studies have shown that cell–ECM and cell–cell adhesions participate in mechanosensing to transduce mechanical cues into biochemical signals and conversely are responsible for the transmission of intracellular forces to the extracellular environment. As they migrate, cells use these adhesive structures to probe their surroundings, adapt their mechanical properties, and exert the appropriate forces required for their movements. The focus of this review is to give an overview of recent developments showing the bidirectional relationship between the physical properties of the environment and the cell mechanical responses during single and collective cell migration.
INTRODUCTION
Cells, tissues, and organs must constantly adapt to their surroundings. A cell’s interaction with its environment is crucial for physiological tissue organization and functions during development, as well as for homeostasis, regeneration, and aging. It is also involved in pathological conditions–for instance, during tumor progression or fibrosis. The cell microenvironment is composed of the extracellular matrix (ECM) neighboring cells and surrounding intercellular medium. The microenvironment varies in composition and organization, depending on the tissue or in vitro culture conditions. At the cellular level, when a cell touches a permissive surface, be it a substrate or another cell, it will form adhesive structures that allow it to sense and respond to the properties of its surrounding. Cells can sense two major types of information: chemical signals, such as small molecules and soluble factors, which are read through specific receptors, and physical properties, including substrate stiffness, topology, porosity, and elastic behavior, as well as compressive and traction forces (Figure 1). We focus here on the recent evidence pointing to substrate rigidity as a critical parameter controlling cell mechanical responses. However, it is important to keep in mind that other physical properties of the microenvironment are as likely to affect cell behavior. Each tissue has its own stiffness, which affects cell differentiation or behavior (Swift et al., 2013; Swift and Discher, 2014; Ivanovska et al., 2015). For example, axon elongation in Xenopus depends on a stiffness gradient that affects persistent growth and fasciculation of the retinal ganglion axon in the developing brain (Koser et al., 2016). Variations in tissue stiffness control cell proliferation or cell fate specification (Tse and Engler, 2011; Aleksandrova et al., 2015). Cardiac myocytes need a specific stiffness to become actively beating cells, and muscle cells need muscle-like stiffness to form myotubes; excessive stiffness will impede correct myofibril development and may lead to sclerosis and scars (Engler et al., 2004, 2008). In the case of mesenchymal stem cells, a stiffer environment induces bone-like development, whereas a soft one induces neuron-like behavior, and intermediate stiffness induces a muscle-like phenotype (Engler et al., 2006). Signaling cascades responsible for the control of gene expression in response to the physical properties of the environment are being deciphered. The protein Yes-associated protein (YAP) is involved, and its shuttling to the nucleus is controlled by mechanical cues (Aragona et al., 2013). In the nucleus, YAP interacts with TEAD transcription factors to induce specific gene transcription, promote proliferation, and inhibit differentiation (Dupont et al., 2011; Piccolo et al., 2014). Of interest, optogenetic control of contractility shows that the nuclear localization of YAP is a rapid process downstream of RhoA (Valon et al., 2017). The number of studies on the physics and mechanics of tumor tissues has steadily increased over the past decade, showing differences between normal and cancer cells. Most of these studies have been performed at the scale of the tumor or the tissue and shown that some tumors exhibit increased tissue stiffness (Egeblad et al., 2010). In addition, the mechanical properties of tumor cells could also contribute to the physical properties of tumor tissues (Baker et al., 2010). Tumor cells can also exploit stiffness to their advantage. Changes in tumor rigidity resulting from tumor cell activity or the physical remodeling of the ECM by surrounding stromal cells can promote tumor cell invasion, proliferation, and survival (Paszek et al., 2005; Kostic et al., 2009; Levental et al., 2009).
FIGURE 1:
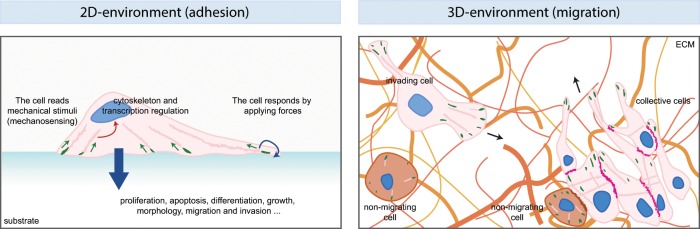
Mechanobiology and migration. Schematic of cells migrating on two-dimensional (2D) or 3D matrices. The 2D example shows the principles of mechanobiology by which a cell reads (green arrows) the mechanical properties of the ECM and converts them into a biochemical intracellular signal (red arrow) that affects the cytoskeleton, signaling, and transcription. Ultimately, the cell responds both by applying forces to the matrix itself (blue arrow) and undergoing processes such as proliferation, apoptosis, differentiation, and migration (large blue arrow). Mechanosensing occurs as the cells interact with the ECM through focal adhesions (green), which are linked to actin fibers (pink). The situation is more complicated in 3D migration, in which cells can move inside a matrix, here composed of fibers (different shades of orange) of different composition, structure, topology, and rigidity and other nonmigrating cells (pink). The drawing shows both a single invading cell (pink), moving in the direction of the arrow, and a group of migrating cells (pink) moving collectively and attached to one another by cell–cell junctions (magenta). Attached to the group of cells there can also be nonmigrating, nonpolarized cells (brown). In this complex situation, cells must integrate the signals transmitted by different types of focal adhesions and adherens junctions.
The interaction between the cell and its environment functions both ways. On one side, cells sense the signals from the environment (Figure 1). Cells read physical stimuli through the use of mechanical sensors. This can occur by opening a channel, stretching a protein, exposing cryptic binding sites, or inducing biochemical signaling pathways. Ultimately this information is integrated so that the cell can respond appropriately. Responses can occur quickly via cytoskeletal rearrangements and changes in cell shape and motility. For instance, endothelial cells realign their cytoskeleton in the direction of the flow when subject to shear stress (Takahashi et al., 1997). The reciprocal relationship between the mechanics of the cell and the physical properties of its surrounding is crucial during cell migration. During spreading and migration, cell adhesion to the substrate drives cytoskeletal rearrangements to promote membrane protrusion and cell spreading. To migrate, cells also use the adhesion sites located at the cell front as cortical anchors for the polymerizing actin meshwork that pushes against the plasma membrane and pulls on the substrate while contracting the cell body forward. As the cell forms new adhesions at the cell edge, it must test the mechanical resistance of the ECM to generate the appropriate amount of force to optimize migration. This mechanosensing process does not occur only at the single focal adhesion level: it is also integrated over the whole cell (Figure 1). After the establishment of myosin-based polarization (Raab et al., 2012), cells can follow a gradient of stiffness in the ECM in a process called durotaxis (Lo et al., 2000; Isenberg et al., 2009; Tse and Engler, 2011). Variations in the physical properties of the ECM may also trigger a particular type of invasion (Tozluoğlu et al., 2013). In cancer-associated fibroblasts (CAFs), the presence of a stiff matrix causes actomyosin contractility. This induces stress fiber formation and Src activation at focal adhesions. This in turn causes nuclear shuttling of YAP, which maintains the aggressive phenotype of CAFs, as it creates a feedback loop in which YAP transcription induces matrix stiffening (Calvo et al., 2013). In the case of tumors, mechanical changes, such as increased confinement during migration, can also induce changes in DNA organization that ultimately modulate the cell’s ability to migrate, participating in the invasive phenotype (Irianto et al., 2017). ECM remodeling by matrix metalloprotease secretion can generate paths or tunnels in which cells can migrate more easily. It can also mechanically facilitate migration by changing the orientation and the tension of ECM fibers (Hynes, 2009; Egeblad et al., 2010).
Collective migration is particularly important during development and in processes such as tissue shaping and wound healing. Collective migration corresponds to the coordinated movement of cell groups, sheets, or chains. It also plays a critical role in the progression of many tumors. Much as in the case of single-cell migration, collective cells are able to respond to mechanical cues. Different factors can affect collective migration, including crowding (cell density), cohesion (strength of adhesions), and constraints (boundary conditions imposed by the ECM; Doxzen et al., 2013). This implies that the cells must integrate information from the environment, which, in this case, also includes the neighboring migrating cells (Figure 1).
In this review, we focus first on the regulation of forces during single-cell migration and then in collectively migrating cells.
MECHANOCOUPLING BETWEEN SUBSTRATE RIGIDITY AND TRACTION FORCES DURING MIGRATION
Mechanosensing at focal adhesions
Single migrating cells must sense the physical properties of the ECM and, in response, apply the appropriate forces to generate movement. Adhesion and traction on the ECM mainly rely on integrins. Integrins are heterodimeric transmembrane receptors composed of an α and a β subunit, which bind specific ECM proteins. Their binding to ECM proteins is controlled by a conformational change that can be activated by outside-in signaling upon ECM binding and by inside-out signaling triggered by the association of partner proteins to the integrin cytoplasmic tail (Huttenlocher and Horwitz, 2011). As cells spread on the ECM, a growing number of integrins interact with the ECM proteins, which progressively form clusters, also called nascent adhesions or focal complexes. The initial steps leading to the formation of nascent adhesions occur before mechanosensing, at least during spreading, and are independent of myosin (Choi et al., 2008; Changede et al., 2015; Sun et al., 2016a). However, forces contribute to the maturation of nascent adhesions into focal adhesions (Riveline et al., 2001; Sun et al., 2016a).
The formation of focal adhesions is initiated when the cluster of ECM-engaged integrins is large enough. In NIH3T3 fibroblasts on fibronectin, clusters of integrins smaller than 0.11 μm2 are unstable and unable to exert forces because the cluster force cannot sustain the cytoskeletal force (Coyer et al., 2012). By developing the tension gauge tether method, Wang and Ha (2013) demonstrated that for CHO-K1 cells, a tension of 40 pN is necessary for integrins to form adhesive structures. During migration, two tension levels can be identified, corresponding to nonclustered integrins (40 pN) and clustered integrins (54 pN). The latter level depends on actomyosin and actin stress fibers that connect focal adhesions together and represents integrins under higher tension, which are found in motile focal adhesions (Wang et al., 2015).
In focal adhesions, integrin clusters can recruit up to 160 different proteins (Zaidel-Bar et al., 2007; Horton et al., 2015; for a review, see Li et al., 2016). These proteins form a physical bridge between integrins engaged with the ECM and the cell cytoskeleton, and more particularly to actomyosin contractile fibers (Figures 2 and 3B). A key article by the Waterman group defined focal adhesions structurally by three-dimensional (3D) super-resolution. Integrins and actin are separated by a 40-nm focal adhesion core subdivided into a lower group (integrin tails, paxillin, and focal adhesion kinase [FAK]), an intermediate, force-transduction group (talin and vinculin), and a higher, actin-regulatory group (zyxin, α-actinin, and vasodilator-stimulated phosphoprotein [VASP]; Kanchanawong et al., 2010). The intermediate proteins talin and vinculin are fundamental mechanosensors, as they can change conformation and signaling properties upon force-induced stretching (for a review, see Yan et al., 2015; Haining et al., 2016). Ultimately, talin and vinculin allow force transmission through β integrins, regulating migration and detection of stiffness (Austen et al., 2016; Nordenfelt et al., 2016).
FIGURE 2:
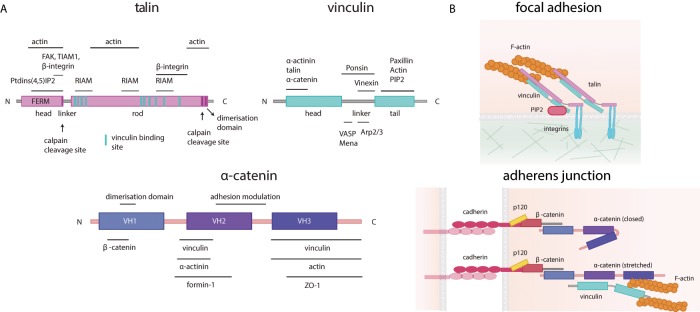
Tension-sensitive proteins are mechanical players of adhesion sites. (A) Schematic representation of the main tension-sensitive proteins involved in focal adhesions and adherens junctions: talin (purple), vinculin (light blue), and α-catenin (dark blue). The main protein interaction domains are shown, and the known interactors (with their binding sites) are indicated above or below each protein. (B) Top, components of focal adhesions and the structures of talin and vinculin when stretched (talin in pink, vinculin in light blue, PIP2 in purple, and F-actin in orange). Bottom, components of adherens junctions and the structure of the unstretched (closed, top) or stretched (bottom) α-catenin (cadherin in purple, p120 in yellow, β-catenin in coral, α-catenin in purple, vinculin in light blue, and actin in orange).
FIGURE 3:
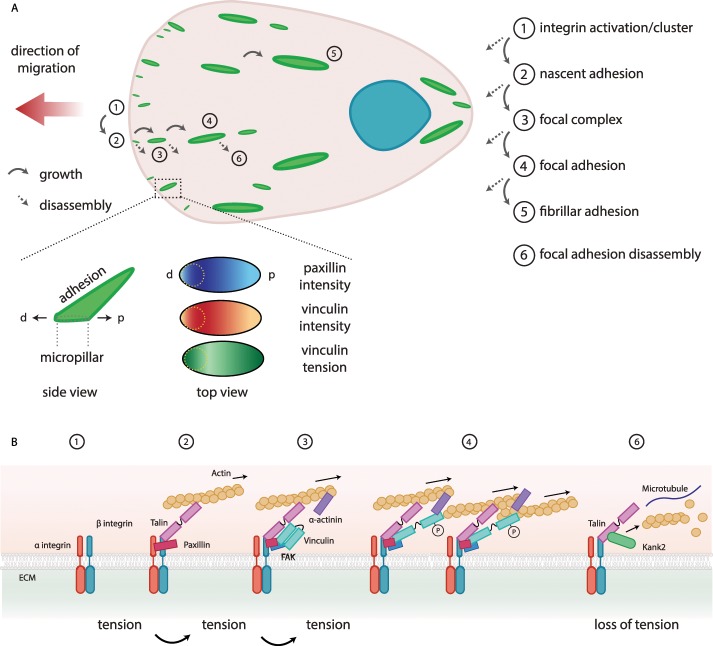
Turnover of focal adhesions. (A) Schematic representation of a migrating cell (pink; nucleus in blue; the arrow shows the direction of migration), highlighting focal adhesion (green) formation, maturation, and disassembly. Different maturation stages (labeled 1–6) can be observed, which can grow (full arrow) or disassemble (dotted arrow) at every step. Inset, summary of the work by Sarangi et al. (2016). The intensity of vinculin and paxillin is analyzed in parallel to vinculin tension (green, high; to white, low) on micropillars. The intensity of paxillin (blue, high; to white, low) and vinculin (red, high; to white, low) is higher in the region of the focal adhesion corresponding to the edge of the micropillar (yellow dotted lines), whereas the vinculin tension is higher at the distal (d) and proximal (p) sites in the adhesion. (B) Focal adhesions, from an integrin cluster to a mature focal adhesion that forms with tension. The disassembly occurs with loss of tension. The ECM (green), integrins (green and red), paxillin (purple), talin (pink), vinculin (light blue), FAK (blue), α-actinin (purple), actin (yellow), microtubules (blue line), and Kank2 (green).
Talin was one of the first proteins to be identified as an integrin partner (Horwitz et al., 1986; Figure 2A). In absence of talin, as in absence of integrins, focal adhesions cannot form properly (Zhang et al. 2008). Talin is recruited together with FAK to nascent adhesions (Lawson et al., 2012). Talin is a large protein of 270 kDa composed of an N-terminal head, a neck, and a C-terminal rod domain. It can adopt an autoinhibited, closed conformation and is activated upon release of this autoinhibition and opening into an extended form (Calderwood et al., 2013). Integrin binding to the rod domain activates talin, which reinforces the interaction (Himmel et al., 2009) and promotes the conformational change of the β integrin subunit. Talin binding to the integrin tail can be induced by inside-out signaling. Protein kinase Cα (PKCα), Rap1, Rap-1 GTP-interacting adaptor protein (RIAM), and phosphatidylinositol-4,5-biphosphate (PIP2) induce talin activation and promote integrin engagement with the ECM (Das et al., 2014). Talin-2 has a particularly high affinity for β integrins, which leads to higher traction forces and faster invasion (Qi et al., 2016). The whole rod can be stretched by a force in the range of 5–10 pN (Yao et al., 2016), allowing binding of many possible partners in response to force (Haining et al., 2016). In particular, stretching of the rod domain exposes more vinculin-binding sites (del Rio et al., 2009). A new tension sensor based on HP35 (a 35–amino acid–long villin head-piece peptide) flanked by two fluorophores has recently allowed the demonstration that talin experiences forces up to 7 pN, sometimes even reaching 10 pN when associated with vinculin and actin (Austen et al., 2016). Focal adhesion coupling to the actin retrograde flow is responsible for the generation of pulling forces (Case and Waterman, 2015; Comrie et al., 2015). However, actomyosin contraction induces even more conformational changes in both the rod and the linker domain of talin, exposing more vinculin-binding sites and promoting the formation of more stable focal adhesions (Calderwood et al., 2013). A recent study demonstrated by super-resolution microscopy that vinculin binds talin in a cooperative manner (the binding is optimal when talin is stretched to 180 nm; Hu et al., 2016). Multiple vinculin proteins actually bind the rod domain of talin when the latter is stretched as an antiparallel dimer.
Vinculin, a cytoplasmic 117-kDa protein, was initially identified as an actin-binding protein (Geiger et al., 1980) and later found to bind a high number of partners, including talin, α-actinin, Arp2/3, paxillin, VASP, catenins, and PIP2 (Carisey et al., 2013; for a review, see Peng et al., 2011; Goldmann, 2016; Figure 2A). Vinculin comprises an N-terminal globular head (Vh), which can bind talin, and a C-terminal rod tail (Vt), which can directly or indirectly interact with actin (Cavalheiro et al., 2017). These two major domains are separated by a short, flexible, proline-rich linker. Similar to talin, vinculin can be found in a closed, autoinhibited state in which Vh and Vt bind each other (Johnson and Craig, 1994, 1995a). Vinculin is recruited via talin to adhesion sites. After actin binding to the Vt domain, vinculin stretching dissociates the Vh and Vt (Bakolitsa et al., 2004; Izard et al., 2004; Cohen et al., 2005; Chen et al., 2006). At this point, vinculin can induce recruitment, activation, or release of other integrins, paxillin, focal adhesion proteins, and more actin, promoting the growth of the focal adhesion in a force-dependent manner (Humphries et al., 2007; Carisey et al., 2013). Owing to its activity and localization, vinculin is considered an optimal candidate in mechanotransduction (Atherton et al., 2016). Its loss induces small but dynamic focal adhesions and defects in locomotion (Coll et al., 1995; Saunders et al., 2006; Thievessen et al., 2013). Loss of vinculin is associated with cancer, as well as with developmental diseases such as cardiomyopathies (Olson et al., 2002; Goldmann et al., 2013). Depletion of the vinculin gene in mice leads to embryonic lethality by embryonic day 10, with defects consistent with problems in adhesion, such as neural tube defects and cardiac malformations. Mouse embryonic fibroblasts (MEFs) from vinculin knockout mice are faster but less adhesive, with disrupted focal adhesions (Xu and Baribault, 1998). A recent study showed that hyperactivation of vinculin also causes lethality and muscular defects in Drosophila due to the formation of cytoplasmic aggregates that resemble adhesion subcomplexes, which are bound to talin tail but not to integrins or actin (Maartens et al., 2016). In these complexes, vinculin can ectopically activate talin, mimicking the effect of force.
The recruitment of talin and vinculin to focal adhesions correlates with the mechanical force applied to the focal adhesion (Golji et al., 2011). The use of a vinculin fluorescence resonance energy transfer (FRET) tension sensor showed that vinculin is recruited to focal adhesions in a force-dependent manner (Grashoff et al., 2010). The Vt binding to actin induces actin fiber bundling to regulate migration and tractions (Johnson and Craig, 1995a; Janssen et al., 2006; Thompson et al., 2014; Jannie et al., 2015). Bundling is most likely mediated by the displacement of the first helix (H1) in Vt upon actin binding, partially unfolding vinculin (Ho Kim et al., 2016). Vinculin–actin interaction is also necessary for transmission of forces, mediating myosin contractility, which enhances forces (Dumbauld et al., 2010). The Vt domain is necessary to generate forces (Dumbauld et al., 2010), whereas the Vh domain probably enhances adhesion strength (Dumbauld et al., 2013). Vinculin also plays a role in engaging and stretching talin with the actomyosin system, locking it in an open conformation and stabilizing the talin–integrin complex (Dumbauld et al., 2013) and focal adhesion (Atherton et al., 2016).
Front-to-rear control of mechanotransduction
During migration, cells form, use, and dissociate focal adhesions. Much stronger traction forces are applied on mature focal adhesions than on nascent adhesions (Gardel et al., 2008). The newer adhesions at the cell front have a higher tension than the retracting ones at the cell rear (Grashoff et al., 2010), as talin tension is higher in peripheral focal adhesions than in older ones (Kumar et al., 2016). Vinculin appears incorporated in the proximal tip of new focal adhesions with minimal tension in the paxillin-rich lower layer of the focal adhesion. Vinculin then treadmills toward the distal end of the focal adhesion, binding actin and talin, opening and increasing its tension (Case and Waterman, 2015; Figure 3). Vinculin interaction with actin is necessary to regulate the actin retrograde flow, as it slows the flow and leads to higher traction forces (Humphries et al., 2007; Thievessen et al., 2013; Jannie et al., 2015). As tension increases, vinculin progressively detaches from the lower layer and is carried inward and upward by the actin retrograde flow as an open protein but without any tension (Case and Waterman, 2015).
Mechanotransduction through talin and vinculin is continuously influenced by the cytoskeletal dynamics and molecular signaling. As forces increase on new focal adhesions, p130Cas, a protein involved in integrin signaling, is stretched. In this case, the stretching renders phosphorylation sites accessible to the Src kinase (Sawada et al., 2006). The FAT domain of p130Cas appears essential in mechanosensing substrate rigidity and controlling cell speed (Bradbury et al., 2017). The following p130Cas phosphorylation increases integrin signaling to the small GTPase Rap1, which in turn can activate talin via RIAM and promote further integrin engagement. As the focal adhesions mature, vinculin competes with RIAM to bind talin and stabilizes the integrin-talin-actin complex independently of Rap1 (Lee et al., 2013). Vinculin phosphorylation by Src on Y100 and Y1065 promotes vinculin opening and increases adhesion and force transmission (Auernheimer et al., 2015). Phosphoinositide signaling affects both talin and vinculin activities. PIP2 activates autoinhibited talin (Ye et al., 2016). Vinculin also binds PIP2 (Johnson and Craig, 1995b; Izard and Brown, 2016), and vinculin stretching increases its binding to PIP2 (Dwivedi and Winter, 2016). A recent model resolved the structure for a short-chain PIP2 binding to vinculin tail and showed that vinculin dimerizes in the presence of PIP2 (Chinthalapudi et al., 2014, 2015). Vinculin mutants that cannot bind lipids are associated with altered focal adhesion turnover but are still able to reinforce cell stiffness upon mechanical deformation (Thompson et al., 2017).
Disassembly or sliding of focal adhesions can result from the negative regulation of talin (Figure 3). Kank2 was recently identified as a component of focal adhesions that forms a “belt” around more mature focal adhesions. In migrating cells, Kank2 concentrates around most mature focal adhesions and binds talin. This interaction displaces actin but maintains talin active and thereby uncouples integrins from actin fibers, reducing force transmission and promoting the sliding of the focal adhesions (Sun et al., 2016b). Ultimately, the loss of traction can also promote the disassembly of focal adhesions. A decrease in traction forces promotes the association of the clathrin adaptor Dab2 with integrin β3 while excluding talin and thereby promotes clathrin-mediated endocytosis of integrins (Yu et al., 2015).
Adaptation of forces to substrate rigidity
During migration, cells use focal adhesion to apply traction forces on the ECM. They respond in a linear manner to the substrate stiffness and change focal adhesion size accordingly (Pelham and Wang, 1997; Saez et al., 2005; Ghibaudo et al., 2008). To measure the traction exerted by the cell, different techniques have been developed over the past 25 years. Initially, cells were plated onto deformable silicon sheets, and the sheet wrinkling was an indirect measure of the cell’s traction onto the substrate (Harris et al., 1980). Toward the end of the 1990s, in a key article, Pelham and Wang (1997) reported that the deformation of a polyacrylamide gel could be used to infer the traction force field exerted by a cell, leading to the development of traction force microscopy (TFM; Dembo and Wang, 1999). TFM is based on the measurement of the displacement of fluorescent beads embedded into an inert hydrogel: the amplitude of displacement indicates how much traction the cell exerted. Other techniques, such as micropillars and atomic force microscopy, have also been developed to better study the cells from a mechanical point of view (Polacheck and Chen, 2016). Mechanotransduction at focal adhesions allows the cells to adapt the forces they exert to the physical properties of the substrate (Figure 1). Thus cells tend to exert higher forces on stiffer substrates and lower tractions on softer substrates (Saez et al., 2005; Ghibaudo et al., 2008). On soft gels, focal adhesions usually appear diffuse and dynamic; on stiffer gels (or glass), they are stabler and larger (Pelham and Wang, 1997; Ghibaudo et al., 2008). During branching on soft substrate, human mesenchymal stem cells show small patches of rapidly turning over focal adhesions but longer protrusions. The vinculin head–tail interaction is necessary for this response, as the mutation (T12) of vinculin that prevent this interaction stabilizes the talin–vinculin complex in focal adhesions in amounts that are not rigidity dependent (Liu et al., 2016). The size–force relationship in focal adhesions is not simple, and focal adhesions of the same size can exert different forces, depending on the substrate stiffness (Trichet et al., 2012). The nature of integrins associated with the substrate also influences the mechanical responses. β1 integrins induce Rac1 activation to assemble new small adhesions. αV integrins are involved in rigidity sensing and accumulate in areas of high tension to reinforce adhesions and actomyosin contractility by activating a RhoA-mDia pathway and the formation of additional actin bundles (Schiller et al., 2013). β1 and αV integrins cooperate to promote myosin II contractility and adapt the level of forces to the rigidity of the substrate.
A recent challenging study analyzed the relationship between focal adhesion internal forces and traction forces by combining micropillars of given stiffness and FRET tension sensors (Sarangi et al., 2016). The authors found that the tension inside focal adhesions correlates in space and time with the force exerted on the substrate, depending on the integrity of the stress fibers (Sarangi et al., 2016). Whereas a previous study found that the traction peak localizes distally a few micrometers from where paxillin is most abundant (Plotnikov et al., 2012), Sarangi et al. (2016) demonstrated that both paxillin and vinculin are concentrated at the distal end of the focal adhesions and are less abundant behind the central area (Figure 3A). Vinculin forces are higher in the region that directly contacts the substrate, where vinculin is not at its peak concentration (Sarangi et al., 2016). The coupling between integrins and actomyosin forces was initially explained in neurons by the “molecular clutch” hypothesis (Mitchison and Kirschner, 1988; Schwarz and Gardel, 2012). Actin rapidly polymerizes and pushes the lamellipodia forward, whereas its contraction through myosin II leads to net rearward flow of the actin network. When the retrograde actin flow is coupled to the ECM through integrins and focal adhesion proteins (in other words, when the clutch is engaged), the force of the actin polymerization at the leading edge is converted into a protrusion force pushing the leading edge forward. The force is transmitted to the ECM (rearward traction), allowing the cell to move forward (Swaminathan and Waterman, 2016).
The molecular clutch model allows a better understanding of how cells sense the environment. According to the Odde model, the molecular clutch behaves differently, depending on the substrate stiffness. On stiff matrices, the retrograde flow is fast, with low traction (“frictional slippage”), because the F-actin bundle is continuously disengaged from the clutch. On soft matrices, the retrograde flow is slower, tension can build up, and the clutch remains engaged for longer time, until the load reaches such high levels that some proteins in the clutch are lost and the whole clutch fails (“load-and-fail”; Chan and Odde, 2008). One limitation of the Odde model is that it predicts a biphasic force–rigidity relationship. However, in most conditions, a monotonic increase of traction forces is observed as a function of ECM stiffness. The Roca-Cusachs group (Elosegui-Artola et al., 2016) recently demonstrated that the biphasic curve can be masked by talin. Above a certain rigidity threshold, the force loading becomes fast enough to allow unfolding of talin before integrins disengage, leading to recruitment of vinculin and integrins, reinforcement of integrin binding, adhesion growth, and increase of force transmission. Moreover, when talin unfolds, YAP translocates to the nucleus, possibly through integrin clustering, signaling downstream of vinculin and talin and transmission of forces to the nucleus via actin stress fibers. Below this stiffness threshold, talin is not stretched rapidly enough, integrins disengage, and YAP is not shuttled to the nucleus (Elosegui-Artola et al., 2016). Vinculin, together with FAK and paxillin, is involved in sensing rigidity. Inhibition of FAK and paxillin reduces traction and decreases the rigidity threshold that promotes tugging on softer ECM, involving vinculin recruitment in strengthening the molecular clutch (Mierke et al., 2008; Plotnikov et al., 2012). In line with this, vinculin tends to adopt an inactive conformation on softer ECM and an active one on stiffer ECM, which is important for stiffness-dependent migration (Yamashita et al., 2014). Other proteins involved in the molecular clutch include α5β1 integrin (Schiller et al., 2013; Riaz et al., 2016) and α-actinin (Meacci et al., 2016), the loss of which causes aberrant rigidity sensing.
Adaptation to substrate rigidity is a very fast process (Mitrossilis et al., 2010). The cell must continuously sense and respond in a feedback loop to maintain the situation in a steady state (Roca-Cusachs et al., 2013). How the cell really measures rigidity is still debated; most probably the cell exerts submicrometer contractions and detects the local deformation of the substrate by “measuring” how much force/contraction it needs to generate such deformation. These contraction areas are similar to sarcomeres, as shown by the recruitment of α-actinin on the pillar tips and of myosin-II between pillars. The contraction units involve nanometer-size, myosin-dependent steps with a frequency of ∼2–3 steps/s; when the force reaches a threshold of ∼20 pN, a pause is triggered so that the adhesion can be reinforced by recruiting more α-actinin (Ghassemi et al., 2012; Wolfenson et al., 2015). A recent study on human skin fibroblasts highlighted the role of two receptor tyrosine kinases, AXL and ROR2, which regulate rigidity sensing by respectively modulating the strength or the duration of these contractions (Yang et al., 2016). In the case of MEFs on micropillars, cells measure displacements of 60 nm in early steps of adhesion, until the adhesion itself can grow and couples to the actin retrograde flow (Ghassemi et al., 2012). Moreover, the actin cytoskeleton responds through rheological changes, behaving like a fluid on soft substrates and a nematic solid on stiff ones (Gupta et al., 2015).
Mechanotransduction beyond actin
The role of other cytoskeletal networks in the cell mechanical responses is still understudied. However, evidence is accumulating suggesting a possible role of microtubules in the generation of adapted forces. First, microtubules are involved in the regulation of adhesion sites (Akhmanova et al., 2009; Etienne-Manneville, 2013), contractility (Kolodney and Elson, 1995; Rape et al., 2011), and RhoA signaling (Heck et al., 2012). In parallel, microtubules can modulate traction through FAK (Rape et al., 2011). Second, microtubule dynamics appear to be controlled by the rigidity of the substrate. In endothelial cells, both actin and microtubules participate in cell branching, and microtubule growth depends on substrate stiffness and myosin (Myers et al., 2011). Microtubules have also been shown to orient toward stiffer areas (Maiuri et al., 2015; Raab and Discher, 2016) and retract from the stiff area when contraction is locally inhibited (Kaverina et al., 2002). On stiff matrices, microtubules are important to regulate cell polarity, while protrusions are mainly generated by actin dynamics (Etienne-Manneville, 2013). However, in soft 3D matrices, microtubules are necessary for fibroblast dendritic-like extensions and endothelial cell migration (Rhee et al., 2007; Bouchet and Akhmanova, 2017). Kank family proteins were shown to regulate talin-mediated force transmission (Sun et al., 2016b). Kank1 was recently shown to bind talin, and this interaction is necessary to allow targeting of microtubules near focal adhesions (Bouchet et al., 2016); it will be interesting to see whether this mediates a mechanosensory response.
Detyrosination, a posttranslational modification of microtubules associated with their stability, participates in mechanotransduction of striated muscles. Reduced tubulin detyrosination is associated with decreased cytoskeletal stiffness and faster muscle contraction and relaxation, suggesting a role in mechanotransduction (Kerr et al., 2015). Oncogenes may increase tumor cell stiffness and invasion through HDAC6 (histone deacetylase 6), which inhibits microtubule acetylation and causes the reorganization of the vimentin intermediate filament network (Rathje et al., 2014). In different cell types, solubility of vimentin depends on ECM stiffness and correlates with cell ruffling. On softer substrates, the soluble pool is maintained by microtubules, whereas on stiffer substrates, it depends on contractility (Murray et al., 2014). Intermediate filaments have been shown to participate in and regulate cell migration (Gonzales et al., 2001; Bhattacharya et al., 2009; Mendez et al., 2010; Dupin et al., 2011; Weber et al., 2011; Sakamoto et al., 2013; Leduc and Etienne-Manneville, 2015; Liu et al., 2015a; Vincent et al., 2015). Because of this and their peculiar properties as highly elastic filaments and role in cell mechanics (Herrmann et al., 2007; Block et al., 2015), intermediate filaments are an ideal candidate to mediate mechanotransduction in cells.
THE MECHANICS OF COLLECTIVELY MIGRATING CELL GROUPS
Just as single cells, migrating cell groups are also clearly affected by the biochemical and physical properties of their environment (Figure 1B). However, migrating collectives cannot be simplified as a group of independent cells that happen to move at the same speed and direction. The collective behavior results in a more efficient migration and sometimes in the acquisition of specific features (Mayor and Etienne-Manneville, 2016). This relies on the communication between migrating cells, and the direction of each cell depends on its neighbors (Vicsek et al., 1995; Szabo et al., 2006). During collective migration, cells couple to one another mechanically and chemically through cell–cell contacts and the actin cytoskeleton (for a review, see Mayor and Etienne-Manneville, 2016). This allows the cells to influence the behavior of one another and modify the supracellular front–rear polarity. A hierarchy is established inside the group by selecting a population of leaders that sense the mechanical and chemical cues that induce migration. Leaders influence followers via mechanical coupling. Cells within the migrating group also influence each other. In the particular case of contact inhibition of locomotion, which will not be discussed here, cells can also repel each other as a mechanism of collective guidance (Theveneau et al., 2013; Scarpa et al., 2015; Zimmermann et al., 2016). How cells collectively adjust their forces and how they sense and transduce the mechanical properties of their neighbors is currently under intensive investigation.
Distribution of forces in migrating collectives
The first attempts to measure forces in collectively migrating Madin–Darby canine kidney (MDCK) cells on micropillars showed that tractions are mainly localized at the cell front and perpendicular to the monolayer edge, with an average force of 5 nN. This demonstrated that migration is due to the pulling of the leader cells onto the substrate and not to the pushing of follower cells (du Roure et al., 2005). The strongest tractions are applied at the leading edge, but, at least in epithelial cell monolayers, traction forces are generated up to several hundreds of micrometers inside the monolayer (Trepat et al., 2009; Tambe et al., 2011; Serra-Picamal et al., 2012; Figure 4B). However, recent measurements obtained with a new silicone wrinkling, temperature-sensitive substrate show that tractions are limited to the first row of MDCK leader cells (Yokoyama et al., 2016). In general, cells far from the leading edge exert smaller forces than leaders, but tractions in the monolayer are heterogeneous and change continuously over time, with small hot spots and fluctuations (Serra-Picamal et al. 2012). Although leader cells give biochemical and mechanical cues to followers, cells inside the monolayer can slow down, move in different directions (sometimes even opposite to the direction of the group), or form swirls (Petitjean et al., 2010; Vedula et al., 2012; Reffay et al., 2014). Thus, the distribution of forces across the monolayer is dynamic and fluctuating, with variations in both the adhesion to the substrate and contractility (Ng et al., 2014). These local variations are likely to induce cells to polarize and exert tractions in a direction that is not necessarily the same as that of the global movement during the entire duration of migration. Thus tractions must be regulated by velocity but also by other local parameters, such as cell polarity (Notbohm et al., 2016).
FIGURE 4:
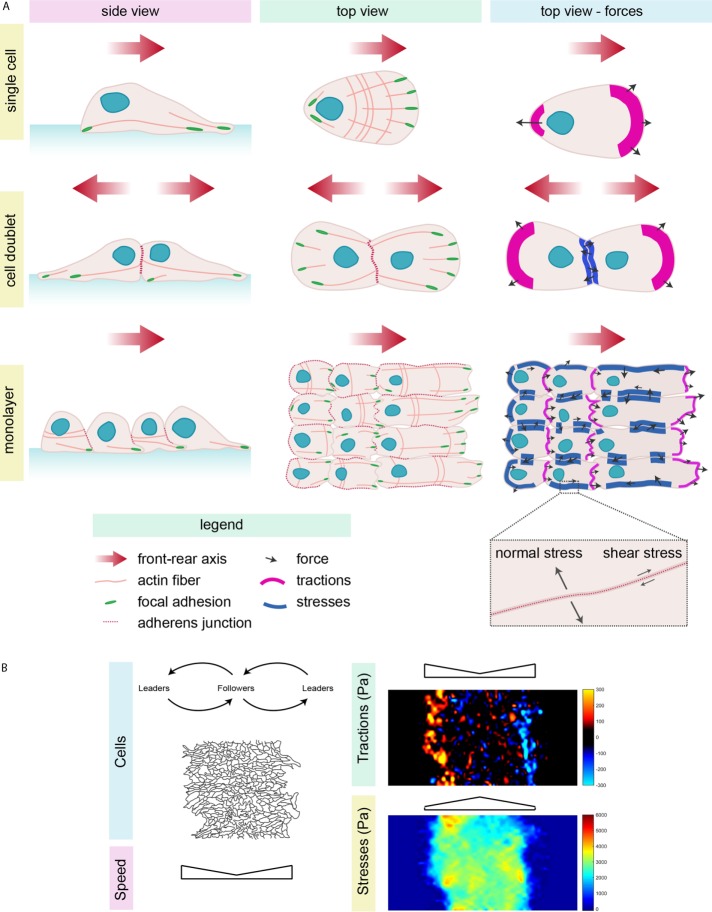
Cell migration and force transmission and their study in collective migration. (A) Single cell (top), doublets (middle), and a migrating monolayer (bottom) from the side and top views. Cells (light pink) show a polarized (red arrow, front–rear axis of migration) morphology, with the nucleus (blue) at the back, an asymmetric distribution of focal adhesions (green) and actin (pink lines), microtubule (not shown), and intermediate filament (not shown) networks. Cell–cell contacts (red dotted line) allow adhesion between cells. The third column shows representative forces (tractions on the substrate in magenta and intercellular stresses in blue); tractions are high at the cell front, whereas intercellular stresses concentrate at cell–cell contacts. The gray arrows represent possible forces and their directions. (B) Representative images of cells migrating on hydrogels (black lines show the cell edges). Collectively migrating cells are divided into leaders and followers, which influence one another. The speed of migration is higher at the edges of the monolayer, as are tractions. Tractions are calculated by TFM (bead displacements) and are higher where the color intensity is stronger—yellow and blue correspond to the maximal forces in opposite directions along the axis of migration. Stresses, calculated with MSM, are higher at the center of the migrating monolayer (strong intensity in red). TFM and MSM images were obtained on migrating astrocytes on a 9-kPa collagen-coated hydrogel by C.D.P. and C. Pérez González.
Owing to unequal distribution of tractions between the leaders and the followers, the cell sheet is under global tensile stress, and forces are transmitted at large scales (Trepat et al., 2009; Tambe et al., 2011; Serra-Picamal et al., 2012; Figure 4A). The leaders and the rest of the cells in the monolayer play a tug-of-war: the monolayer manages to migrate forward because the leader cells are stronger. However, leaders exert forces only up to ∼100 nN, which is not strong enough to pull the whole monolayer. They might nevertheless succeed because of a mechanical “X-wave” that propagates from the front to the back of the monolayer, communicating information on mechanics and polarity to the followers. This wave is continuously repeated so that the flow of information is maintained during the whole migration process (Serra-Picamal et al., 2012). The coordinated behavior of the followers may be simply explained by the fact that single cells in the group align their traction forces with the general velocity of the cell group (Basan et al., 2013). Development of monolayer stress microscopy (MSM) has allowed a better understanding of the distribution of stresses (σ) in the monolayer. Stresses are again heterogeneous, with large areas of tensile (positive) stresses alternating with regions of weak compressive (negative) stresses that vary over time. Stresses are defined by their components, shear stress (σxy and σyx) tangent to the surface and normal stress (σxx and σyy) perpendicular to the surface. In biological terms, these correspond respectively to the tangential and perpendicular forces exerted on cell–cell junctions by neighboring cells. During migration, cells in the monolayer migrate in the direction that maintains the shear stress minimum and the normal stress maximum (Figure 4A). The collective tendency of cells in a monolayer to migrate along the orientation of maximal principal stress is called plithotaxis (Tambe et al., 2011; Trepat and Fredberg, 2011). Leader cells induce traction forces on followers and shear stress on neighbors, transforming local forces into coordinated and polarized traction forces, ensuring plitothaxis (Zaritsky et al., 2015). Merlin, a tumor suppressor and regulator of the Hippo pathway, has been proposed to play a role in plithotaxis of epithelial cells through Rac1 modulation (Das et al., 2015). When leader cells start to migrate, Rac1 is activated toward the cell front. Leader cells pull the followers, which in turn release merlin from junctions to promote the polarized activation of Rac1 in the followers.
The transmission of forces within the cell group is mainly mediated by adherens junctions (Tambe et al., 2011; Trepat and Fredberg, 2011). To obtain direct information of the mechanical role of adherens junctions between neighboring cells migrating collectively on ECM, cells can be plated as doublets, the smallest possible group on adhesive micropatterns. Even if this is clearly not an example of collective cell migration per se, it still provides useful information. For example, a pair of endothelial cells sustains forces at cell–cell junctions of ∼100–120 nN perpendicular to the cell–cell contact (Liu et al., 2010; Maruthamuthu et al., 2011). However, the different adherens junction proteins can differently affect the physical parameters controlling either the monolayer kinematics or forces. For example, in the case of MCF10A cells, P-cadherin and E-cadherin show different responses to mechanical stress in magnetic tweezers experiments. E-cadherin allows the cells to adapt to an extracellular force by activating a mechanotransduction pathway via vinculin, whereas P-cadherin cannot reinforce junctions. However, the two proteins seem to compete for the same mechanotransduction pathway because P-cadherin can rescue the absence of E-cadherin (Bazellières et al., 2015). The specific role of each cadherin in mediating intercellular stresses during collective migration is still unclear. The expression of different levels of cadherins during epithelial-to-mesenchymal transition or tumor invasion might also help the cells migrate and invade collectively through a cadherin-dependent regulation of forces (Friedl and Mayor, 2017). In a carcinoma model, CAFs are able to pull the tumor cells to drive collective invasion. This is due to a heterophilic interaction between the N-cadherin of CAFs and the E-cadherin of the tumor cells, which actively responds to forces and allows polarization of CAFs (Labernadie et al., 2017).
Sensing and adjusting forces between adjacent migrating cells
Adherens junctions are the main cell–cell adhesion structures that mediate tissue mechanical integrity. Adherens junctions are typically composed of cadherins—transmembrane proteins that interact homotypically. Cadherins bind intracellularly to catenins (p120 catenin, α-catenin, and β-catenin) and can activate different signaling pathways to influence the cytoskeleton, differentiation, and the cell cycle (Gumbiner, 2005; Leckband and de Rooij, 2014). Cadherin cytoplasmic partners also associate with actin directly or indirectly via vinculin and zyxin. Thereby, they couple cell–cell interactions to the actin cytoskeleton.
In the Drosophila ovary, border cells migrate as a cohesive and coordinated group through the nurse cells that compress them. Migrating border cells express E-cadherin, which on one hand contributes to their migration—E-cadherin expressed by the immobile surrounding nurse cells being used as a substrate—and on the other hand mediates the communication between the leaders to follower cells of the moving cluster. To resist compression, the migrating border cell cluster activates cycles of myosin II contraction to promote cortical tension (Aranjuez et al., 2016). Moreover, the common direction of migration is controlled through E-cadherin and Rac. E-cadherin is under higher tension at the front of the border cells, where it activates Rac to increase E-cadherin tension (Cai et al., 2014).
Information on the forces exerted at the level of cell–cell junctions can be obtained from assays easier to interpret than whole migrating monolayers. As for TFM or micropillar experiments, where cells are plated on ECM substrates, cells can also be plated onto cadherin patterns. These experiments demonstrated that cell spreading and force transmission on N-cadherin–coated substrate is stiffness dependent. Cadherin adhesions are larger and stronger on stiff gels, whereas cells have smaller adhesions and a disorganized actin network on softer gels (Ladoux et al., 2010). Moreover, increasing forces on adherens junctions leads to a force-dependent reinforcement of their structure and of the associated actomyosin system (Lambert et al., 2007; le Duc et al., 2010). These observations imply that cadherin-mediated adhesions possess a mechanosensor and serve as a major site of mechanotransduction.
Cadherins form nanoclusters that associate with the actin cytoskeleton to mediate mechanotransduction (Changede and Sheetz, 2016; Cosgrove et al., 2016). The understanding of the role of cadherins in mechanical intercellular coupling has progressed significantly since the α-catenin/vinculin modulus has been involved in mechanotransduction. α-Catenin is a 102-kDa protein that possesses roles in different signaling pathways involved in proliferation and size, such as YAP, MAPK (mitogen-activated protein kinase), and Wnt (Figure 2A). It is also a key molecule in adherens junctions, where it is recruited through its association with β-catenin on one side and can bind actin filaments and vinculin on the other side to reinforce cell–cell junctions (Figure 2). Loss of α-catenin results in alterations of adherens junctions and loss of its connection to the actin cytoskeleton (Hirano et al., 1992; Vasioukhin et al., 2000). α-Catenin contains three main domains called vinculin homology domains (VH1, VH2, and VH3), which, as their names suggest, possess high homology with vinculin domains (27, 31, and 34%, respectively). VH1 is important for both β-catenin binding and α-catenin homodimerization. The VH2 domain contains binding sites for many of its partners, including vinculin and the actin-binding proteins α-actinin and formin-1. Part of the VH2 domain contains an adhesion modulation domain (M) of four α-helix bundles. The VH3 C-terminal domain binds actin (Kobielak and Fuchs, 2004). α-Catenin can be found in an autoinhibited conformation, where M1 and M2-3 domains interact. In a key study, Yonemura et al. (2010) showed that α-catenin is a mechanosensor. Stretching forces induce a change in α-catenin conformation that unmasks the vinculin-binding site. Disruption of the intramolecular inhibitory interaction requires only ∼5 pN and leads to an open catenin conformation (Yao et al., 2014). The interaction of α-catenin with vinculin reinforces the junction in a force-dependent manner by promoting actin recruitment (Yonemura et al., 2010). Vinculin binding stabilizes catenin in an intermediate conformation, which allows its activity as a mechanotransducer without excessively opening it. Forces >30 pN induce vinculin dissociation and junction disassembly (Ishiyama et al., 2013; Yao et al., 2014; Maki et al., 2016). A recent study described the nanoscale architecture of cadherin-based cell–cell junctions (Bertocchi et al., 2016). Similar to the structure of focal adhesions (Kanchanawong et al., 2010), cell–cell junctions are divided into compartments. The cytoplasmic tails of cadherins bound by catenins are separated from the actin and actin-regulatory protein compartment by vinculin, which bridges the two compartments and separates them by ∼30 nm. In this model, the conformation and position of vinculin depend on α-catenin and tension. Vinculin opening can also be induced by Abl kinase–mediated phosphorylation of Tyr-822, which can be dephosphorylated by protein tyrosine phosphatase 1B. Once open, vinculin recruits proteins such as VASP, probably promoting further actin polymerization and a feedback loop (Bertocchi et al., 2016; Figure 2).
Ultimately, α-catenin and vinculin cooperate to link cadherins and actin and allow a proper force response and junction reinforcement over time (le Duc et al., 2010; Borghi et al., 2012; Thomas et al., 2013; Figure 2). Defects in the connection to actin impair cell coordination and increase migration (Strale et al., 2015). This is also demonstrated by the fact that endothelial cells expressing a mutant α-catenin (ΔVBS) that cannot recruit vinculin show defects in junction reinforcement and mechanosensing (Twiss et al., 2012). The importance of the α-catenin–vinculin modulus in mechanotransduction has also been confirmed in vivo in Drosophila (Desai et al., 2013; Jurado et al., 2016) and zebrafish (Han et al., 2016). Although most reports focused on the α-catenin and vinculin modules in mechanotransduction, recent work in endothelial cells suggests a possible role for other proteins, such as zyxin, VASP, and testin, in the mechanical responses of adherens junctions (Oldenburg et al., 2015). In addition to actin, microtubules are probably involved in the strengthening of cadherin adhesions (Plestant et al., 2014) and could therefore influence mechanotransduction in adherens junctions. Moreover, the interaction of keratin intermediate filaments with desmosomal cadherin is also involved in mechanotransduction at cell–cell contacts (Weber et al., 2012). Finally, an elegant study in nontumorigenic breast epithelial cells (MCF10A) showed that many proteins involved in cell–cell junctions (not limited to adherens junctions) are important for force transmission (Bazellières et al., 2015). In the case of hepatocyte growth factor–stimulated MDCK cells plated on N-cadherin substrates, depletion of the cytoplasmic domain does not completely abolish tractions (Lee et al., 2016), suggesting that alternative mechanisms may also contribute.
Mechanical cross-talk between focal adhesions and cell–cell junctions
Focal adhesions and adherens junctions share similar structures and connection to the cytoskeleton, as well as similar mechanosensing mechanisms and mechanotransduction pathways (Han and de Rooij, 2016; Mui et al., 2016). It is thus tempting to speculate that these two major adhesive structures influence each other. Mui et al. (2016) addressed the most recent findings on adhesion cross-talk from the mechanical point of view. Several studies suggest that increasing forces in one compartment decreases them in the other; in other words, strong adhesion to the ECM decreases the strength of cell–cell junctions and vice versa (Guo et al., 2006; Wang et al., 2006). However, the relationship between the two structures is clearly more complex.
During development, mesenchymal cells have to adapt from a cell–cell adhesion-based system to one that relies more on cell–substrate interactions. A method has recently been developed to decouple the presentation of RGD (fibronectin) from that of HAVDI (N-cadherin) ligand peptides at different stiffnesses and assess mesenchymal stem cell mechanosensing (Cosgrove et al., 2016). On keeping RGD constant and presenting HAVDI, the cells read ECM stiffness as softer than it actually is. This is coupled to inhibition of Rac1, which reduces cell contractile forces and YAP nuclear localization and leads to errors in proliferation and differentiation (Cosgrove et al., 2016). A recent study showed that E-cadherin mediates force transmission by downstream activation of PI3K (phosphoinositide 3-kinase) in an epidermal growth factor receptor–dependent manner in epithelial cells, leading to integrin activation, probably by inside-out signaling, which in turn induces cell stiffening through ROCK and myosin II (Muhamed et al., 2016). The direct role of the cytoskeleton in the mechanical coupling between adherens junctions and focal adhesions is not entirely clear, and more complex biochemical signaling pathways are likely to be involved. In migrating astrocytes, which mainly express N-cadherin, loss of N-cadherin or alteration of its dynamics results in the faster and less-directed migration of the leader cells, which detach from their followers (Camand et al. 2012). Cadherin-mediated adherens junctions are necessary to regulate the lamellipodia activity, cell polarization, and the direction of migration (Borghi et al., 2010; Dupin et al., 2011). They control the position of focal adhesions and the recruitment of the β-PIX/Cdc42/Par6/aPKC pathway proteins that promote cell polarity and persistent migration (Dupin et al., 2009; Camand et al., 2012). In C2C12 myoblasts, expression of P-cadherin, but not other cadherins, induces efficient collective cell migration and polarization. In this system, activation of Cdc42 by the guanine nucleotide exchange factor β-PIX, recruited by P-cadherin, controls polarity and cadherin-dependent forces, leading to increased traction and intracellular stresses in the monolayer (Plutoni et al., 2016). The maintenance of adherens junctions between actively migrating cells is crucial for the collective behavior. In astrocytes, as well as in endothelial cells or fibroblasts, adherens junctions located on lateral contacts dynamically flow backward during collective migration (Peglion et al., 2014). This ensures that cells keep stable yet malleable interactions as they migrate through a complex environment. Given that lateral adherens junctions link the actin transverse arcs of adjacent cells, they also likely contribute to the coordination of the actin retrograde flow between cells migrating next to each other (Etienne-Manneville, 2014). It will be interesting to test whether the retrograde flow of adherens junctions is involved in the transmission of forces through the monolayer.
The cross-talk between focal adhesions and adherens junctions is bidirectional. The ECM also affects the localization and the forces exerted on cell–cell junctions. The physical proximity between ECM and junctions results in higher intercellular and intracellular forces, which control the position of the junctions (Tseng et al., 2012), supporting the fact that integrins and ECM regulate cell–cell adhesions (Marsden and DeSimone, 2003; De Rooij et al., 2005). This phenomenon may explain how durotaxis can be acquired during the collective migration of cells that do not normally durotax (Sunyer et al., 2016). Leader cells sense substrate rigidity and communicate mechanical information to their followers through actomyosin contractility. The efficiency of this mechanical signal decays over large distances in a stiffness-dependent manner (Ng et al., 2012). Changes in substrate rigidity modulate forces at the level of cell–cell junctions, demonstrating that tension at focal adhesions correlates with tension at junctions (Maruthamuthu et al., 2011). During convergent extension movements—for instance, during Xenopus development—β1 integrin modulates cell–cell adhesion. Blocking fibronectin or β1 integrins alters cadherin-mediated adhesion, aggregation, cell intercalation, and axial extension during gastrulation (Marsden and DeSimone, 2003).
CONCLUSIONS
Many open questions remain to be answered in the expanding field of mechanobiology and migration. First, reports have concentrated on proteins that act as mechanosensors by stretching and allowing the binding of other binding partners. It will be crucial to understand whether stretching of proteins can also induce enzymatic activity in addition to the unmasking of protein-binding sites. Stretching might not be limited to protein structure but could also be related to the deformation of a membrane (e.g., pulling forces, curved membranes, or protrusions), which is especially crucial during migration. A recent study describes cadherin fingers—polarized VE-cadherin–rich protrusions between leaders and followers—during endothelial migration (Hayer et al., 2016). These fingers are formed by convex curved membranes that recruit curvature-sensing proteins that might induce specific signaling pathways. The primary candidates are BAR proteins, which contain domains for reading membrane curvatures and are likely to be essential players in mechanosensing (McMahon and Boucrot, 2015). Much work still needs to be done to better understand the molecular mechanisms involved in mechanotransduction and the coupling between mechanotransduction sites. More attention will have to be devoted to understanding how the cytoskeleton—not only actin, but also microtubules, intermediate filaments, and septins—regulates mechanotransduction in migration. It is crucial to decipher which GTPases affect the cytoskeleton during mechanosensing. How exactly does a cell integrate information from both junctions and focal adhesions? Mechanically it is the same type of signal (a force), but biochemically, different signaling pathways are involved. From a biological point of view, adherens junctions and focal adhesions share similar molecular organization and common mechanosensing mechanisms but differ in their downstream signaling (Han and de Rooij, 2016). Key questions are whether and when one type of adhesion site predominates over the other, although it is probable that the two systems form a feedback loop. Finally, many different physical parameters other than substrate rigidity can affect migration. These include substrate topography, porosity, elasticity, and other physical constraints (Nelson and Tien, 2006; Liu et al., 2015b). There are influences of both nanoscale and microscale cues, although the microscale geometric cues tend to dominate (Nam et al., 2016). How these physical properties are sensed by cells and affect mechanotransduction to control cell migration needs to be further investigated. More generally, we do not understand how cells process the multiple physical inputs in a robust and coherent manner, clearly pointing to the need for a systems-level investigation of mechanobiology.
The majority of reports have given information on in vitro conditions. With the advent of new technologies and intravital imaging, studies will focus on the in vivo situation during morphogenesis and pathological conditions. Some studies have already started to describe what happens in vivo during migration (Koser et al., 2016) or fibrosis (Kai et al., 2016). A key question to answer is whether mechanobiological signals and cues are druggable. For example, during fibrosis or tumor growth and invasion, modifying the mechanobiological properties of the cell or its surrounding could be a valid treatment option that has not yet been approached.
Acknowledgments
We thank Shailaja Seetharaman and Jean-Baptiste Manneville for critical reading of the manuscript and Carlos Pérez González for analysis of TFM and MSM. C.D.P. is a scholar in the Pasteur–Paris University International PhD program and received a stipend from the Fondation pour la Recherche Médicale and Institut Carnot. This work was supported by La Ligue Contre le Cancer, the Institut Pasteur, and the Centre National de la Recherche Scientifique.
Abbreviations used:
- CAF
cancer-associated fibroblast
- ECM
extracellular matrix
- FAK
focal adhesion kinase
- FRET
fluorescence resonance energy transfer
- MDCK
Madin–Darby canine kidney
- MEF
mouse embryonic fibroblast
- MSM
monolayer stress microscopy
- PIP2
phosphatidylinositol-4,5-biphosphate
- PKC
protein kinase C
- RIAM
Rap-1 GTP-interacting adaptor protein
- TFM
traction force microscopy
- VASP
vasodilator-stimulated protein
- ΔVBS
Δ vinculin–binding site
- Vh
vinculin head
- VH1-2-3
vinculin homology domains 1-2-3
- Vt
vinculin tail
- YAP
Yes-associated protein.
Footnotes
REFERENCES
- Akhmanova A, Stehbens SJ, Yap AS. Touch, grasp, deliver and control: functional cross-talk between microtubules and cell adhesions. Traffic. 2009;10:268–274. doi: 10.1111/j.1600-0854.2008.00869.x. [DOI] [PubMed] [Google Scholar]
- Aleksandrova A, Czirok A, Kosa E, Galkin O, Cheuvront TJ, Rongish BJ. The endoderm and myocardium join forces to drive early heart tube assembly. Dev Biol. 2015;404:40–54. doi: 10.1016/j.ydbio.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragona M, Panciera T, Manfrin A, Giulitti S, Michielin F, Elvassore N, Dupont S, Piccolo S. A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell. 2013;154:1047–1059. doi: 10.1016/j.cell.2013.07.042. [DOI] [PubMed] [Google Scholar]
- Aranjuez G, Burtscher A, Sawant K, Majumder P, McDonald JA. Dynamic myosin activation promotes collective morphology and migration by locally balancing oppositional forces from surrounding tissue. Mol Biol Cell. 2016;27:1898–1910. doi: 10.1091/mbc.E15-10-0744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atherton P, Stutchbury B, Jethwa D, Ballestrem C. Mechanosensitive components of integrin adhesions: role of vinculin. Exp Cell Res. 2016;343:21–27. doi: 10.1016/j.yexcr.2015.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auernheimer V, Lautscham LA, Leidenberger M, Friedrich O, Kappes B, Fabry B, Goldmann WH. Vinculin phosphorylation at residues Y100 and Y1065 is required for cellular force transmission. J Cell Sci. 2015;128:3435–3443. doi: 10.1242/jcs.172031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austen K, Ringer P, Mehlich A, Chrostek-grashoff A, Kluger C, Klingner C, Sabass B, Zent R, Rief M, Grashoff C. Mechanical linkages. Nat Cell Biol. 2016;17:1597–1606. doi: 10.1038/ncb3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker EL, Lu J, Yu D, Bonnecaze RT, Zaman MH. Cancer cell stiffness: integrated roles of three-dimensional matrix stiffness and transforming potential. Biophys J. 2010;99:2048–2057. doi: 10.1016/j.bpj.2010.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakolitsa C, Cohen DM, Bankston LA, Bobkov AA, Cadwell GW, Jennings L, Critchley DR, Craig SW, Liddington RC. Structural basis for vinculin activation at sites of cell adhesion. Nature. 2004;430:583–586. doi: 10.1038/nature02610. [DOI] [PubMed] [Google Scholar]
- Basan M, Elgeti J, Hannezo E, Rappel W-J, Levine H. Alignment of cellular motility forces with tissue flow as a mechanism for efficient wound healing. Proc Natl Acad Sci USA. 2013;110:2452–2459. doi: 10.1073/pnas.1219937110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazellières E, Conte V, Elosegui-Artola A, Serra-Picamal X, Bintanel-Morcillo M, Roca-Cusachs P, Muñoz JJ, Sales-Pardo M, Guimerà R, Trepat X. Control of cell-cell forces and collective cell dynamics by the intercellular adhesome. Nat Cell Biol. 2015;17:409–420. doi: 10.1038/ncb3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertocchi C, Wang Y, Ravasio A, Hara Y, Wu Y, Sailov T, Baird MA, Davidson MW, Zaidel-Bar R, Toyama Y, et al. Nanoscale architecture of cadherin-based cell adhesions. Nat Cell Biol. 2016;19:28–37. doi: 10.1038/ncb3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya R, Gonzalez AM, Debiase PJ, Trejo HE, Goldman RD, Flitney FW, Jones JCR. Recruitment of vimentin to the cell surface by beta3 integrin and plectin mediates adhesion strength. J Cell Sci. 2009;122:1390–1400. doi: 10.1242/jcs.043042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block J, Schroeder V, Pawelzyk P, Willenbacher N, Köster S. Physical properties of cytoplasmic intermediate filaments. Biochim Biophys Acta. 2015;1853:3053–3064. doi: 10.1016/j.bbamcr.2015.05.009. [DOI] [PubMed] [Google Scholar]
- Borghi N, Lowndes M, Maruthamuthu V, Gardel ML, Nelson WJ. Regulation of cell motile behavior by crosstalk between cadherin- and integrin-mediated adhesions. Proc Natl Acad Sci USA. 2010;107:13324–13329. doi: 10.1073/pnas.1002662107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghi N, Sorokina M, Shcherbakova OG, Weis WI, Pruitt BL, Nelson WJ, Dunn AR. E-cadherin is under constitutive actomyosin-generated tension that is increased at cell-cell contacts upon externally applied stretch. Proc Natl Acad Sci USA. 2012;109:12568–12573. doi: 10.1073/pnas.1204390109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchet BP, Akhmanova A. Microtubules in 3D cell motility. J Cell Sci. 2017;130:39–50. doi: 10.1242/jcs.189431. [DOI] [PubMed] [Google Scholar]
- Bouchet BP, Gough RE, Ammon YC, van de Willige D, Post H, Jacquemet G, Maarten Altelaar AF, Heck AJR, Goult BT, Akhmanova A. Talin-KANK1 interaction controls the recruitment of cortical microtubule stabilizing complexes to focal adhesions. Elife. 2016;5:1–23. doi: 10.7554/eLife.18124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury PM, Turner K, Mitchell C, Griffin KR, Middlemiss S, Lau L, Dagg R, Taran E, Cooper-White J, Fabry B, et al. The focal adhesion targeting (FAT) domain of p130 Crk associated substrate (p130Cas) confers mechanosensing function. J Cell Sci. 2017;130:1263–1273. doi: 10.1242/jcs.192930. [DOI] [PubMed] [Google Scholar]
- Cai D, Chen SC, Prasad M, He L, Wang X, Choesmel-Cadamuro V, Sawyer JK, Danuser G, Montell DJ. Mechanical feedback through E-cadherin promotes direction sensing during collective cell migration. Cell. 2014;157:1146–1159. doi: 10.1016/j.cell.2014.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderwood D, Campbell ID, Critchley DR. Talins and kindlins: partners in integrin-mediated adhesion. Nat Rev Mol Cell Biol. 2013;14:503–517. doi: 10.1038/nrm3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo F, Ege N, Grande-Garcia A, Hooper S, Jenkins RP, Chaudhry SI, Harrington K, Williamson P, Moeendarbary E, Charras G, et al. Mechanotransduction and YAP-dependent matrix remodelling is required for the generation and maintenance of cancer-associated fibroblasts. Nat Cell Biol. 2013;15:637–646. doi: 10.1038/ncb2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camand E, Peglion F, Osmani N, Sanson M, Etienne-Manneville S. N-cadherin expression level modulates integrin-mediated polarity and strongly impacts on the speed and directionality of glial cell migration. J Cell Sci. 2012;125:844–857. doi: 10.1242/jcs.087668. [DOI] [PubMed] [Google Scholar]
- Carisey A, Tsang R, Greiner AM, Nijenhuis N, Heath N, Nazgiewicz A, Kemkemer R, Derby B, Spatz J, Ballestrem C. Vinculin regulates the recruitment and release of core focal adhesion proteins in a force-dependent manner. Curr Biol. 2013;23:271–281. doi: 10.1016/j.cub.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case LB, Waterman CM. Integration of actin dynamics and cell adhesion by a three-dimensional, mechanosensitive molecular clutch. Nat Cell Biol. 2015;17:955–963. doi: 10.1038/ncb3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalheiro RP, Lima MA, Jarrouge-Bouças TR, Viana GM, Lopes CC, Coulson-Thomas VJ, Dreyfuss JL, Yates EA, Tersariol ILS, Nader HB. Coupling of vinculin to F-actin demands Syndecan-4 proteoglycan. Matrix Biol. 2017 doi: 10.1016/j.matbio.2016.12.006. 2017(Jan 4), S0945-053X(16)30191-3. [DOI] [PubMed] [Google Scholar]
- Chan CE, Odde DJ. Traction dynamics of filopodia on compliant substrates. Science. 2008;322:1687–1691. doi: 10.1126/science.1163595. [DOI] [PubMed] [Google Scholar]
- Changede R, Sheetz M. Integrin and cadherin clusters: a robust way to organize adhesions for cell mechanics. BioEssays. 2016;39:1–12. doi: 10.1002/bies.201600123. [DOI] [PubMed] [Google Scholar]
- Changede R, Xu X, Margadant F, Sheetz MP. Nascent integrin adhesions form on all matrix rigidities after integrin activation. Dev Cell. 2015;35:614–621. doi: 10.1016/j.devcel.2015.11.001. [DOI] [PubMed] [Google Scholar]
- Chen H, Choudhury DM, Craig SW. Coincidence of actin filaments and talin is required to activate vinculin. J Biol Chem. 2006;281:40389–40398. doi: 10.1074/jbc.M607324200. [DOI] [PubMed] [Google Scholar]
- Chinthalapudi K, Patil DN, Rangarajan ES, Rader C, Izard T. Lipid-directed vinculin dimerization. Biochemistry. 2015;54:2758–2768. doi: 10.1021/acs.biochem.5b00015. [DOI] [PubMed] [Google Scholar]
- Chinthalapudi K, Rangarajan ES, Patil DN, George EM, Brown DT, Izard T. Lipid binding promotes oligomerization and focal adhesion activity of vinculin. J Cell Biol. 2014;207:643–656. doi: 10.1083/jcb.201404128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi CK, Vicente-Manzanares M, Zareno J, Whitmore LA, Mogilner A, Horwitz AR. Actin and alpha-actinin orchestrate the assembly and maturation of nascent adhesions in a myosin II motor-independent manner. Nat Cell Biol. 2008;10:1039–1050. doi: 10.1038/ncb1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen DM, Chen H, Johnson RP, Choudhury B, Craig SW. Two distinct head-tail interfaces cooperate to suppress activation of vinculin by talin. J Biol Chem. 2005;280:17109–17117. doi: 10.1074/jbc.M414704200. [DOI] [PubMed] [Google Scholar]
- Coll JL, Ben-Ze’ev A, Ezzell RM, Rodríguez Fernández JL, Baribault H, Oshima RG, Adamson ED. Targeted disruption of vinculin genes in F9 and embryonic stem cells changes cell morphology, adhesion, and locomotion. Proc Natl Acad Sci USA. 1995;92:9161–9165. doi: 10.1073/pnas.92.20.9161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comrie WA, Babich A, Burkhardt JK. F-actin flow drives affinity maturation and spatial organization of LFA-1 at the immunological synapse. J Cell Biol. 2015;208:475–491. doi: 10.1083/jcb.201406121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove BD, Mui KL, Driscoll TP, Caliari SR, Mehta KD, Assoian RK, Burdick JA, Mauck RL. N-cadherin adhesive interactions modulate matrix mechanosensing and fate commitment of mesenchymal stem cells. Nat Mater. 2016;15:1297–1306. doi: 10.1038/nmat4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyer SR, Singh A, Dumbauld DW, Calderwood DA, Craig SW, Delamarche E, Garcia AJ. Nanopatterning reveals an ECM area threshold for focal adhesion assembly and force transmission that is regulated by integrin activation and cytoskeleton tension. J Cell Sci. 2012;125:5110–5123. doi: 10.1242/jcs.108035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das M, Subbayya Ithychanda S, Qin J, Plow EF. Mechanisms of talin-dependent integrin signaling and crosstalk. Biochim. Biophys Acta. 2014;1838:579–588. doi: 10.1016/j.bbamem.2013.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das T, Safferling K, Rausch S, Grabe N, Boehm H, Spatz JP. A molecular mechanotransduction pathway regulates collective migration of epithelial cells. Nat Cell Biol. 2015;17:276–287. doi: 10.1038/ncb3115. [DOI] [PubMed] [Google Scholar]
- del Rio A, Perez-jimenez R, Liu R, Roca-cusachs P, Fernandez JM, Sheetz MP. Stretching single talin rod molecules activates vinculin binding. Science. 2009;323:638–641. doi: 10.1126/science.1162912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dembo M, Wang Y-L. Stresses at the cell-to-substrate interface during locomotion of fibroblasts. Biophys J. 1999;76:2307–2316. doi: 10.1016/S0006-3495(99)77386-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rooij J, Kerstens A, Danuser G, Schwartz MA, Waterman-Storer CM. Integrin-dependent actomyosin contraction regulates epithelial cell scattering. J Cell Biol. 2005;171:153–164. doi: 10.1083/jcb.200506152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai R, Sarpal R, Ishiyama N, Pellikka M, Ikura M, Tepass U. Monomeric α-catenin links cadherin to the actin cytoskeleton. Nat Cell Biol. 2013;15:261–273. doi: 10.1038/ncb2685. [DOI] [PubMed] [Google Scholar]
- Doxzen K, Vedula SRK, Leong MC, Hirata H, Gov NS, Kabla AJ, Ladoux B, Lim CT. Guidance of cell migration by substrate dimension. Biophys J. 2013;104:313–321. doi: 10.1016/j.bpj.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumbauld DW, Lee TT, Singh A, Scrimgeour J, Gersbach CA, Zamir EA, Fu J, Chen CS, Curtis JE, Craig SW, et al. How vinculin regulates force transmission. Proc Natl Acad Sci USA. 2013;110:9788–9793. doi: 10.1073/pnas.1216209110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumbauld DW, Shin H, Gallant ND, Michael KE, Radhakrishna H, García AJ. Contractility modulates cell adhesion strengthening through focal adhesion kinase and assembly of vinculin-containing focal adhesions. J Cell Physiol. 2010;223:746–756. doi: 10.1002/jcp.22084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupin I, Camand E, Etienne-Manneville S. Classical cadherins control nucleus and centrosome position and cell polarity. J Cell Biol. 2009;185:779–786. doi: 10.1083/jcb.200812034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupin I, Sakamoto Y, Etienne-Manneville S. Cytoplasmic intermediate filaments mediate actin-driven positioning of the nucleus. J Cell Sci. 2011;124:865–872. doi: 10.1242/jcs.076356. [DOI] [PubMed] [Google Scholar]
- Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- du Roure O, Saez A, Buguin A, Austin RH, Chavrier P, Silberzan P, Ladoux B. Force mapping in epithelial cell migration. Proc Natl Acad Sci USA. 2005;102:2390–2395. doi: 10.1073/pnas.0408482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi M, Winter R. Article binding of vinculin to lipid membranes in its inhibited and activated states. Biophys J. 2016;111:1444–1453. doi: 10.1016/j.bpj.2016.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egeblad M, Rasch MG, Weaver VM. Dynamic interplay between the collagen scaffold and tumor evolution. Curr Opin Cell Biol. 2010;22:697–706. doi: 10.1016/j.ceb.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elosegui-Artola A, Oria R, Chen Y, Kosmalska A, Pérez-González C, Castro N, Zhu C, Trepat X, Roca-Cusachs P. Mechanical regulation of a molecular clutch defines force transmission and transduction in response to matrix rigidity. Nat Cell Biol. 2016;18:540–548. doi: 10.1038/ncb3336. [DOI] [PubMed] [Google Scholar]
- Engler AJ, Carag-Krieger C, Johnson CP, Raab M, Tang H-Y, Speicher DW, Sanger JW, Sanger JM, Discher DE. Embryonic cardiomyocytes beat best on a matrix with heart-like elasticity: scar-like rigidity inhibits beating. J Cell Sci. 2008;121:3794–3802. doi: 10.1242/jcs.029678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler AJ, Griffin MA, Sen S, Bönnemann CG, Sweeney HL, Discher DE. Myotubes differentiate optimally on substrates with tissue-like stiffness: pathological implications for soft or stiff microenvironments. J Cell Biol. 2004;166:877–887. doi: 10.1083/jcb.200405004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S. Microtubules in cell migration. Annu Rev Cell Dev Biol. 2013;29:471–499. doi: 10.1146/annurev-cellbio-101011-155711. [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S. Neighborly relations during collective migration. Curr Opin Cell Biol. 2014;30:51–59. doi: 10.1016/j.ceb.2014.06.004. [DOI] [PubMed] [Google Scholar]
- Friedl P, Mayor R. Tuning collective cell migration by cell-cell junction regulation. Cold Spring Harb Perspect Biol. 2017;9:1–17. doi: 10.1101/cshperspect.a029199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardel ML, Sabass B, Ji L, Danuser G, Schwarz US, Waterman CM. Traction stress in focal adhesions correlates biphasically with actin retrograde flow speed. J Cell Biol. 2008;183:999–1005. doi: 10.1083/jcb.200810060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger B, Tokuyasu KT, Dutton AH, Singer SJ. Vinculin, an intracellular protein localized at specialized sites where microfilament bundles terminate at cell membranes. Proc Natl Acad Sci USA. 1980;77:4127–4131. doi: 10.1073/pnas.77.7.4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghassemi S, Meacci G, Liu S, Gondarenko AA, Mathur A, Roca-Cusachs P, Sheetz MP, Hone J. Cells test substrate rigidity by local contractions on submicrometer pillars. Proc Natl Acad Sci USA. 2012;109:5328–5333. doi: 10.1073/pnas.1119886109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghibaudo M, Saez A, Trichet L, Xayaphoummine A, Browaeys J, Silberzan P, Buguin A, Ladoux B. Traction forces and rigidity sensing regulate cell functions. Soft Matter. 2008;4:1836. [Google Scholar]
- Goldmann WH. Role of vinculin in cellular mechanotransduction. Cell Biol Int. 2016;40:241–256. doi: 10.1002/cbin.10563. [DOI] [PubMed] [Google Scholar]
- Goldmann WH, Auernheimer V, Thievessen I, Fabry B. Vinculin, cell mechanics and tumour cell invasion. Cell Biol Int. 2013;37:397–405. doi: 10.1002/cbin.10064. [DOI] [PubMed] [Google Scholar]
- Golji J, Lam J, Mofrad MRK. Vinculin activation is necessary for complete talin binding. Biophys J. 2011;100:332–340. doi: 10.1016/j.bpj.2010.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales M, Weksler B, Tsuruta D, Goldman RD, Yoon KJ, Hopkinson SB, Flitney FW, Jones JC. Structure and function of a vimentin-associated matrix adhesion in endothelial cells. Mol Biol Cell. 2001;12:85–100. doi: 10.1091/mbc.12.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grashoff C, Hoffman BD, Brenner MD, Zhou R, Parsons M, Yang MT, McLean MA, Sligar SG, Chen CS, Ha T, Schwartz MA. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature. 2010;466:263–266. doi: 10.1038/nature09198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumbiner BM. Regulation of cadherin-mediated adhesion in morphogenesis. Nat Cell Biol. 2005;14:1183–1197. doi: 10.1038/nrm1699. [DOI] [PubMed] [Google Scholar]
- Guo W, Frey MT, Burnham NA, Wang Y. Substrate rigidity regulates the formation and maintenance of tissues. Biophys J. 2006;90:2213–2220. doi: 10.1529/biophysj.105.070144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta M, Sarangi BR, Deschamps J, Nematbakhsh Y, Callan-Jones A, Margadant F, Mège R-M, Lim CT, Voituriez R, Ladoux B. Adaptive rheology and ordering of cell cytoskeleton govern matrix rigidity sensing. Nat Commun. 2015;6:7525. doi: 10.1038/ncomms8525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haining AWM, Lieberthal TJ, Del Río Hernández A. Talin: a mechanosensitive molecule in health and disease. FASEB J. 2016;30:2073–2085. doi: 10.1096/fj.201500080R. [DOI] [PubMed] [Google Scholar]
- Han MKL, de Rooij J. Converging and unique mechanisms of mechanotransduction at adhesion sites. Trends Cell Biol. 2016;26:612–623. doi: 10.1016/j.tcb.2016.03.005. [DOI] [PubMed] [Google Scholar]
- Han MKL, Hoijman E, Nöel E, Garric L, Bakkers J. αE-catenin-dependent mechanotransduction is essential for proper convergent extension in zebrafish. Biol Open. 2016;5:1461–1472. doi: 10.1242/bio.021378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris AK, Wild P, Stopak D. Silicone rubber substrata: a new wrinkle in the study of cell locomotion. Science. 1980;208:177–179. doi: 10.1126/science.6987736. [DOI] [PubMed] [Google Scholar]
- Hayer A, Shao L, Chung M, Joubert L-M, Yang HW, Tsai F-C, Bisaria A, Betzig E, Meyer T. Engulfed cadherin fingers are polarized junctional structures between collectively migrating endothelial cells. Nat Cell Biol. 2016;18:1311–1323. doi: 10.1038/ncb3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck JN, Ponik SM, Garcia-Mendoza MG, Pehlke CA, Inman DR, Eliceiri KW, Keely PJ. Microtubules regulate GEF-H1 in response to extracellular matrix stiffness. Mol Biol Cell. 2012;23:2583–2592. doi: 10.1091/mbc.E11-10-0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann H, Bär H, Kreplak L, Strelkov SV, Aebi U. Intermediate filaments: from cell architecture to nanomechanics. Mol Cell Biol. 2007;8:562–573. doi: 10.1038/nrm2197. [DOI] [PubMed] [Google Scholar]
- Himmel M, Ritter A, Rothemund S, Pauling BV, Rottner K, Gingras AR, Ziegler WH. Control of high affinity interactions in the talin C terminus: how talin domains coordinate protein dynamics in cell adhesions. J Biol Chem. 2009;284:13832–13842. doi: 10.1074/jbc.M900266200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano S, Kimoto N, Shimoyama Y, Hirohashi S, Takeichi M. Identification of a neural alpha-catenin as a key regulator of cadherin function and multicellular organization. Cell. 1992;70:293–301. doi: 10.1016/0092-8674(92)90103-j. [DOI] [PubMed] [Google Scholar]
- Ho Kim E, Sook Song H, Hoon Yoo S, Yoon M. Tumor treating fields inhibit glioblastoma cell migration, invasion and angiogenesis. Oncotarget. 2016;7:65125–65136. doi: 10.18632/oncotarget.11372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton ER, Byron A, Askari JA, Ng DH, Millon-Frémillon A, Robertson J, Koper EJ, Paul NR, Warwood S, Knight D, et al. Definition of a consensus integrin adhesome and its dynamics during adhesion complex assembly and disassembly. Nat Cell Biol. 2015;17:1577–1587. doi: 10.1038/ncb3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz A, Duggan K, Buck C, Beckerle MC, Burridge K. Interaction of plasma membrane fibronectin receptor with talin–a transmembrane linkage. Nature. 1986;320:531–533. doi: 10.1038/320531a0. [DOI] [PubMed] [Google Scholar]
- Hu X, Jing C, Xu X, Nakazawa N, Cornish VW, Margadant FM, Sheetz MP. Cooperative vinculin binding to talin mapped by time-resolved super resolution microscopy. Nano Lett. 2016;16:4062–4068. doi: 10.1021/acs.nanolett.6b00650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries JD, Wang P, Streuli C, Geiger B, Humphries MJ, Ballestrem C. Vinculin controls focal adhesion formation by direct interactions with talin and actin. J Cell Biol. 2007;179:1043–1057. doi: 10.1083/jcb.200703036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenlocher A, Horwitz A. Integrins in cell migration. Cold Spring Harb Perspect Biol. 2011;3:47–67. doi: 10.1101/cshperspect.a005074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes RO. Extracellular matrix: not just pretty fibrils. Science. 2009;326:1216–1219. doi: 10.1126/science.1176009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irianto J, Xia Y, Pfeifer CR, Athirasala A, Ji J, Alvey C, Tewari M, Bennett RR, Harding SM, Liu AJ, et al. DNA damage follows repair factor depletion and portends genome variation in cancer cells after article DNA damage follows repair factor depletion and portends genome variation in cancer cells after pore migration. Curr Biol. 2017;27:1–14. doi: 10.1016/j.cub.2016.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isenberg BC, DiMilla PA, Walker M, Kim S, Wong JY. Vascular smooth muscle cell durotaxis depends on substrate stiffness gradient strength. Biophys J. 2009;97:1313–1322. doi: 10.1016/j.bpj.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiyama N, Tanaka N, Abe K, Yang YJ, Abbas YM, Umitsu M, Nagar B, Bueler SA, Rubinstein JL, Takeichi M, et al. An autoinhibited structure of α-catenin and its implications for vinculin recruitment to adherens junctions. J Biol Chem. 2013;288:15913–15925. doi: 10.1074/jbc.M113.453928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanovska IL, Shin J-W, Swift J, Discher DE. Stem cell mechanobiology: diverse lessons from bone marrow. Trends Cell Biol. 2015;8:583–592. doi: 10.1016/j.tcb.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izard T, Brown DT. Mechanisms and functions of vinculin interactions with phospholipids at cell adhesion sites. J Biol Chem. 2016;291:2548–2555. doi: 10.1074/jbc.R115.686493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izard T, Evans G, Borgon RA, Rush CL, Bricogne G, Bois PRJ. Vinculin activation by talin through helical bundle conversion. Nature. 2004;427:171–175. doi: 10.1038/nature02281. [DOI] [PubMed] [Google Scholar]
- Jannie K, Ellerbroek S, Zhou D, Chen S, Crompton D, García A, DeMali K. Vinculin-dependent actin bundling regulates cell migration and traction forces. Biochem J. 2015;465:383–393. doi: 10.1042/BJ20140872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen MEW, Kim E, Liu H, Fujimoto LM, Bobkov A, Volkmann N, Hanein D. Three-dimensional structure of vinculin bound to actin filaments. Mol Cell. 2006;21:271–281. doi: 10.1016/j.molcel.2005.11.020. [DOI] [PubMed] [Google Scholar]
- Johnson RP, Craig SW. An intramolecular association between the head and tail domains of vinculin modulates talin binding. J Biol Chem. 1994;269:12611–12619. [PubMed] [Google Scholar]
- Johnson RP, Craig SW. F-actin binding site masked by the intramolecular association of vinculin head and tail domains. Nature. 1995a;373:261–264. doi: 10.1038/373261a0. [DOI] [PubMed] [Google Scholar]
- Johnson RP, Craig SW. The carboxy-terminal tail domain of vinculin contains a cryptic binding site for acidic phospholipids. Biochem Biophys Res Commun. 1995b;210:159–164. doi: 10.1006/bbrc.1995.1641. [DOI] [PubMed] [Google Scholar]
- Jurado J, de Navascués J, Gorfinkiel N. alpha-Catenin stabilises cadherin-catenin complexes and modulates actomyosin dynamics to allow pulsatile apical contraction. J Cell Sci. 2016;129:4496–4508. doi: 10.1242/jcs.193268. [DOI] [PubMed] [Google Scholar]
- Kai F, Laklai H, Weaver V. Force matters: biomechanical regulation of cell invasion and migration in disease. Trends Cell Biol. 2016;26:1–12. doi: 10.1016/j.tcb.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanchanawong P, Shtengel G, Pasapera AM, Ramko EB, Davidson MW, Hess HF, Waterman CM. Nanoscale architecture of integrin-based cell adhesions. Nature. 2010;468:580–584. doi: 10.1038/nature09621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaverina I, Krylyshkina O, Beningo K, Anderson K, Wang Y-L, Small JV. Tensile stress stimulates microtubule outgrowth in living cells. J Cell Sci. 2002;115:2283–2291. doi: 10.1242/jcs.115.11.2283. [DOI] [PubMed] [Google Scholar]
- Kerr JP, Robison P, Shi G, Bogush AI, Kempema AM, Hexum JK, Becerra N, Harki DA, Martin SS, Raiteri R, et al. Detyrosinated microtubules modulate mechanotransduction in heart and skeletal muscle. Nat Commun. 2015;6:8526. doi: 10.1038/ncomms9526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobielak A, Fuchs E. Alpha-catenin: at the junction of intercellular adhesion and actin dynamics. Nat Rev Mol Cell Biol. 2004;5:614–625. doi: 10.1038/nrm1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodney MS, Elson EL. Contraction due to microtubule disruption is associated with increased phosphorylation of myosin regulatory light chain. Proc Natl Acad Sci USA. 1995;92:10252–10256. doi: 10.1073/pnas.92.22.10252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koser DE, Thompson AJ, Foster SK, Dwivedy A, Pillai EK, Sheridan GK, Svoboda H, Viana M, Costa LD, Guck J, et al. Mechanosensing is critical for axon growth in the developing brain. Nat Neurosci. 2016;19:1592–1598. doi: 10.1038/nn.4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic A, Lynch CD, Sheetz MP. Differential matrix rigidity response in breast cancer cell lines correlates with the tissue tropism. PLoS One. 2009;4:e6361. doi: 10.1371/journal.pone.0006361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Ouyang M, Van den Dries K, McGhee EJ, Tanaka K, Anderson MD, Groisman A, Goult BT, Anderson KI, Schwartz MA. Talin tension sensor reveals novel features of focal adhesion force transmission and mechanosensitivity. J Cell Biol. 2016;213:371–383. doi: 10.1083/jcb.201510012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labernadie A, Kato T, Brugués A, Serra-Picamal X, Derzsi S, Arwert E, Weston A, González-Tarragó V, Elosegui-Artola A, Albertazzi L, et al. A mechanically active heterotypic E-cadherin/N-cadherin adhesion enables fibroblasts to drive cancer cell invasion. Nat Cell Biol. 2017;19:224–237. doi: 10.1038/ncb3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladoux B, Anon E, Lambert M, Rabodzey A, Hersen P, Buguin A, Silberzan P, Mège RM. Strength dependence of cadherin-mediated adhesions. Biophys J. 2010;98:534–542. doi: 10.1016/j.bpj.2009.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert M, Thoumine O, Brevier J, Choquet D, Riveline D, Mège RM. Nucleation and growth of cadherin adhesions. Exp Cell Res. 2007;313:4025–4040. doi: 10.1016/j.yexcr.2007.07.035. [DOI] [PubMed] [Google Scholar]
- Lawson C, Lim ST, Uryu S, Chen XL, Calderwood DA, Schlaepfer DD. FAK promotes recruitment of talin to nascent adhesions to control cell motility. J Cell Biol. 2012;196:223–232. doi: 10.1083/jcb.201108078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leckband DE, de Rooij J. Cadherin adhesion and mechanotransduction. Annu Rev Cell Dev Biol. 2014;30:291–315. doi: 10.1146/annurev-cellbio-100913-013212. [DOI] [PubMed] [Google Scholar]
- Leduc C, Etienne-Manneville S. Intermediate filaments in cell migration and invasion: the unusual suspects. Curr Opin Cell Biol. 2015;32:102–112. doi: 10.1016/j.ceb.2015.01.005. [DOI] [PubMed] [Google Scholar]
- le Duc Q, Shi Q, Blonk I, Sonnenberg A, Wang N, Leckband D, De Rooij J. Vinculin potentiates E-cadherin mechanosensing and is recruited to actin-anchored sites within adherens junctions in a myosin II-dependent manner. J Cell Biol. 2010;189:1107–1115. doi: 10.1083/jcb.201001149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E, Ewald ML, Sedarous M, Kim T, Weyers BW, Truong RH, Yamada S. Deletion of the cytoplasmic domain of N-cadherin reduces, but does not eliminate, traction force-transmission. Biochem Biophys Res Commun. 2016;478:1640–1646. doi: 10.1016/j.bbrc.2016.08.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HS, Anekal P, Lim CJ, Liu CC, Ginsberg MH. Two modes of integrin activation form a binary molecular switch in adhesion maturation. Mol Biol Cell. 2013;24:1354–1362. doi: 10.1091/mbc.E12-09-0695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levental KR, Yu H, Kass L, Lakins JN, Egeblad M, Erler JT, Fong SF, Csiszar K, Giaccia A, Weninger W, et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 2009;139:891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Lee H, Zhu C. Molecular mechanisms of mechanotransduction in integrin-mediated cell-matrix adhesion. Exp Cell Res. 2016;349:85–94. doi: 10.1016/j.yexcr.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Lin H, Tang M, Wang Y. Vimentin contributes to epithelial-mesenchymal transition cancer cell mechanics by mediating cytoskeletal organization and focal adhesion maturation. Oncotarget. 2015a;6:15966–15983. doi: 10.18632/oncotarget.3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YJ, Le Berre M, Lautenschlaeger F, Maiuri P, Callan-Jones A, Heuzé M, Takaki T, Voituriez R, Piel M. Confinement and low adhesion induce fast amoeboid migration of slow mesenchymal cells. Cell. 2015b;160:659–672. doi: 10.1016/j.cell.2015.01.007. [DOI] [PubMed] [Google Scholar]
- Liu Z, Bun P, Audugé N, Coppey-Moisan M, Borghi N. Vinculin head-tail interaction defines multiple early mechanisms for stem cell rigidity sensing. Integr Biol. 2016;8:693–703. doi: 10.1039/c5ib00307e. [DOI] [PubMed] [Google Scholar]
- Liu Z, Tan JL, Cohen DM, Yang MT, Sniadecki NJ, Ruiz SA, Nelson CM, Chen CS. Mechanical tugging force regulates the size of cell-cell junctions. Proc Natl Acad Sci USA. 2010;107:9944–9949. doi: 10.1073/pnas.0914547107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo C, Wang H-B, Dembo M, Wang Y. Cell movement is guided by the rigidity of the substrate. Biophys J. 2000;79:144–152. doi: 10.1016/S0006-3495(00)76279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maartens AP, Wellmann J, Wictome E, Klapholz B, Green H, Brown NH. Drosophila vinculin is more harmful when hyperactive than absent, and can circumvent integrin to form adhesion complexes. J Cell Sci. 2016;129:4354–4365. doi: 10.1242/jcs.189878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiuri P, Rupprecht JF, Wieser S, Ruprecht V, Bénichou O, Carpi N, Coppey M, De Beco S, Gov N, Heisenberg CP, et al. Actin flows mediate a universal coupling between cell speed and cell persistence. Cell. 2015;161:374–386. doi: 10.1016/j.cell.2015.01.056. [DOI] [PubMed] [Google Scholar]
- Maki K, Han S-W, Hirano Y, Yonemura S, Hakoshima T, Adachi T. Mechano-adaptive sensory mechanism of α-catenin under tension. Sci Rep. 2016;6:24878. doi: 10.1038/srep24878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsden M, DeSimone DW. Integrin-ECM interactions regulate cadherin-dependent cell adhesion and are required for convergent extension in Xenopus. Curr Biol. 2003;13:1182–1191. doi: 10.1016/s0960-9822(03)00433-0. [DOI] [PubMed] [Google Scholar]
- Maruthamuthu V, Sabass B, Schwarz US, Gardel ML. Cell-ECM traction force modulates endogenous tension at cell-cell contacts. Proc Natl Acad Sci USA. 2011;108:4708–4713. doi: 10.1073/pnas.1011123108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor R, Etienne-Manneville S. The front and rear of collective cell migration. Nat Rev Mol Cell Biol. 2016;17:97–109. doi: 10.1038/nrm.2015.14. [DOI] [PubMed] [Google Scholar]
- McMahon HT, Boucrot E. Membrane curvature at a glance. J Cell Sci. 2015;128:1065–1070. doi: 10.1242/jcs.114454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meacci G, Wolfenson H, Liu S, Stachowiak MR, Iskratsch T, Mathur A, Ghassemi S, Gauthier N, Tabdanov E, Lohner J, et al. α-actinin links ECM rigidity sensing contractile units with periodic cell edge. Mol Biol Cell. 2016;27:3471–3479. doi: 10.1091/mbc.E16-02-0107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez MG, Kojima S-I, Goldman RD. Vimentin induces changes in cell shape, motility, and adhesion during the epithelial to mesenchymal transition. FASEB J. 2010;24:1838–1851. doi: 10.1096/fj.09-151639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mierke CT, Kollmannsberger P, Zitterbart DP, Smith J, Fabry B, Goldmann WH. Mechano-coupling and regulation of contractility by the vinculin tail domain. Biophys J. 2008;94:661–670. doi: 10.1529/biophysj.107.108472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T, Kirschner M. Cytoskeletal dynamics and nerve growth. Neuron. 1988;1:761–772. doi: 10.1016/0896-6273(88)90124-9. [DOI] [PubMed] [Google Scholar]
- Mitrossilis D, Fouchard J, Pereira D, Postic F, Richert A, Saint-Jean M, Asnacios A. Real-time single-cell response to stiffness. Proc Natl Acad Sci USA. 2010;107:16518–16523. doi: 10.1073/pnas.1007940107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhamed I, Wu J, Sehgal P, Kong X, Tajik A, Wang N, Leckband DE. E-cadherin-mediated force transduction signals regulate global cell mechanics. J Cell Sci. 2016;129:1843–1854. doi: 10.1242/jcs.185447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mui KL, Chen CS, Assoian RK. The mechanical regulation of integrin-cadherin crosstalk organizes cells, signaling and forces. J Cell Sci. 2016;129:1093–1100. doi: 10.1242/jcs.183699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray ME, Mendez MG, Janmey PA. Substrate stiffness regulates solubility of cellular vimentin. Mol Biol Cell. 2014;25:87–94. doi: 10.1091/mbc.E13-06-0326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers KA, Applegate KT, Danuser G, Fischer RS, Waterman CM. Distinct ECM mechanosensing pathways regulate microtubule dynamics to control endothelial cell branching morphogenesis. J Cell Biol. 2011;192:321–334. doi: 10.1083/jcb.201006009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam K-H, Kim P, Wood DK, Kwon S, Provenzano PP, Kim D-H. Multiscale cues drive collective cell migration. Sci Rep. 2016;6:29749. doi: 10.1038/srep29749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson CM, Tien J. Microstructured extracellular matrices in tissue engineering and development. Curr Opin Biotechnol. 2006;17:518–523. doi: 10.1016/j.copbio.2006.08.011. [DOI] [PubMed] [Google Scholar]
- Ng MR, Besser A, Brugge JS, Danuser G. Mapping the dynamics of force transduction at cell-cell junctions of epithelial clusters. Elife. 2014;3:1–29. doi: 10.7554/eLife.03282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng MR, Besser A, Danuser G, Brugge JS. Substrate stiffness regulates cadherin-dependent collective migration through myosin-II contractility. J Cell Biol. 2012;199:545–563. doi: 10.1083/jcb.201207148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordenfelt P, Elliott HL, Springer TA. Coordinated integrin activation by actin-dependent force during T-cell migration. Nat Commun. 2016;7:13119. doi: 10.1038/ncomms13119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notbohm J, Banerjee S, Utuje KJC, Gweon B, Jang H, Park Y, Shin J, Butler JP, Fredberg JJ, Marchetti MC. Cellular contraction and polarization drive collective cellular motion. Biophys J. 2016;110:2729–2738. doi: 10.1016/j.bpj.2016.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenburg J, van der Krogt G, Twiss F, Bongaarts A, Habani Y, Slotman JA, Houtsmuller A, Huveneers S, de Rooij J. VASP, zyxin and TES are tension-dependent members of focal adherens junctions independent of the α-catenin-vinculin module. Sci Rep. 2015;5:17225. doi: 10.1038/srep17225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson TM, Illenberger S, Kishimoto NY, Huttelmaier S, Keating MT, Jockusch BM. Metavinculin mutations alter actin interaction in dilated cardiomyopathy. Circulation. 2002;105:431–437. doi: 10.1161/hc0402.102930. [DOI] [PubMed] [Google Scholar]
- Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A, Reinhart-King CA, Margulies SS, Dembo M, Boettiger D, et al. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8:241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Peglion F, Llense F, Etienne-Manneville S. Adherens junction treadmilling during collective migration. Nat Cell Biol. 2014;16:639–651. doi: 10.1038/ncb2985. [DOI] [PubMed] [Google Scholar]
- Pelham J, Wang Y-L. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc Natl Acad Sci USA. 1997;94:13661–13665. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X, Nelson ES, Maiers JL, DeMali KA. New insights into vinculin function and regulation. Int Rev Cell Mol Biol. 2011;287:191–231. doi: 10.1016/B978-0-12-386043-9.00005-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petitjean L, Reffay M, Grasland-Mongrain E, Poujade M, Ladoux B, Buguin A, Silberzan P. Velocity fields in a collectively migrating epithelium. Biophys J. 2010;98:1790–1800. doi: 10.1016/j.bpj.2010.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccolo S, Dupont S, Cordenonsi M. The biology of YAP/TAZ: hippo signaling and beyond. Physiol Rev. 2014;94:1287–1312. doi: 10.1152/physrev.00005.2014. [DOI] [PubMed] [Google Scholar]
- Plestant C, Strale P-O, Seddiki R, Nguyen E, Ladoux B, Mège R-M. Adhesive interactions of N-cadherin limit the recruitment of microtubules to cell-cell contacts through organization of actomyosin. J Cell Sci. 2014;127:1660–1671. doi: 10.1242/jcs.131284. [DOI] [PubMed] [Google Scholar]
- Plotnikov SV, Pasapera AM, Sabass B, Waterman CM. Force fluctuations within focal adhesions mediate ECM-rigidity sensing to guide directed cell migration. Cell. 2012;151:1513–1527. doi: 10.1016/j.cell.2012.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plutoni C, Bazellieres E, Le Borgne-Rochet M, Comunale F, Brugues A, Séveno M, Planchon D, Thuault S, Morin N, Bodin S, et al. P-cadherin promotes collective cell migration via a Cdc42-mediated increase in mechanical forces. J Cell Biol. 2016;212:199–217. doi: 10.1083/jcb.201505105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polacheck WJ, Chen CS. Measuring cell-generated forces: a guide to the available tools. Nat Methods. 2016;13:415–423. doi: 10.1038/nmeth.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L, Jafari N, Li X, Chen Z, Li L, Hytönen VP, Goult BT, Zhan C-G, Huang C. Talin2-mediated traction force drives matrix degradation and cell invasion. J Cell Sci. 2016;129:3661–3674. doi: 10.1242/jcs.185959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab M, Discher D. Matrix rigidity regulates the microtubule network polarization in migration. Cytoskeleton (Hoboken) 2016;74:114–124. doi: 10.1002/cm.21349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab M, Swift J, Dingal PCDP, Shah P, Shin JW, Discher DE. Crawling from soft to stiff matrix polarizes the cytoskeleton and phosphoregulates myosin-II heavy chain. J Cell Biol. 2012;199:669–683. doi: 10.1083/jcb.201205056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rape A, Guo W, Wang Y. Microtubule depolymerization induces traction force increase through two distinct pathways. J Cell Sci. 2011;124:4233–4240. doi: 10.1242/jcs.090563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathje L-SZ, Nordgren N, Pettersson T, Rönnlund D, Widengren J, Aspenström P, Gad AKB. Oncogenes induce a vimentin filament collapse mediated by HDAC6 that is linked to cell stiffness. Proc Natl Acad Sci USA. 2014;111:1515–1520. doi: 10.1073/pnas.1300238111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reffay M, Parrini MC, Cochet-Escartin O, Ladoux B, Buguin A, Coscoy S, Amblard F, Camonis J, Silberzan P. Interplay of RhoA and mechanical forces in collective cell migration driven by leader cells. Nat Cell Biol. 2014;16:217–223. doi: 10.1038/ncb2917. [DOI] [PubMed] [Google Scholar]
- Rhee S, Jiang H, Ho C-H, Grinnell F. Microtubule function in fibroblast spreading is modulated according to the tension state of cell-matrix interactions. Proc Natl Acad Sci USA. 2007;104:5425–5430. doi: 10.1073/pnas.0608030104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riaz M, Versaevel M, Mohammed D, Glinel K, Gabriele S. Persistence of fan-shaped keratocytes is a matrix-rigidity-dependent mechanism that requires α5β1 integrin engagement. Sci Rep. 2016;6:34141. doi: 10.1038/srep34141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riveline D, Zamir E, Balaban NQ, Schwarz US, Ishizaki T, Narumiya S, Kam Z, Geiger B, Bershadsky AD. Focal contacts as mechanosensors: externally applied local mechanical force induces growth of focal contacts by an mDia1-dependent and ROCK-independent mechanism. J Cell Biol. 2001;153:1175–1185. doi: 10.1083/jcb.153.6.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca-Cusachs P, Sunyer R, Trepat X. Mechanical guidance of cell migration: lessons from chemotaxis. Curr Opin Cell Biol. 2013;25:543–549. doi: 10.1016/j.ceb.2013.04.010. [DOI] [PubMed] [Google Scholar]
- Saez A, Buguin A, Silberzan P, Ladoux B. Is the mechanical activity of epithelial cells controlled by deformations or forces. Biophys J. 2005;89:L52–L54. doi: 10.1529/biophysj.105.071217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto Y, Boeda B, Etienne-Manneville S. APC binds intermediate filaments and is required for their reorganization during cell migration. J Cell Biol. 2013;200:249–258. doi: 10.1083/jcb.201206010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarangi BR, Gupta M, Doss BL, Tissot N, Lam F, Mege RM, Borghi N, Ladoux B. Coordination between intra- and extracellular forces regulates focal adhesion dynamics. Nano Lett. 2016;17:399–406. doi: 10.1021/acs.nanolett.6b04364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders RM, Holt MR, Jennings L, Sutton DH, Barsukov IL, Bobkov A, Liddington RC, Adamson EA, Dunn GA, Critchley DR. Role of vinculin in regulating focal adhesion turnover. Eur J Cell Biol. 2006;85:487–500. doi: 10.1016/j.ejcb.2006.01.014. [DOI] [PubMed] [Google Scholar]
- Sawada Y, Tamada M, Dubin-Thaler BJ, Cherniavskaya O, Sakai R, Tanaka S, Sheetz MP. Force sensing by mechanical extension of the Src family kinase substrate p130Cas. Cell. 2006;127:1015–1026. doi: 10.1016/j.cell.2006.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpa E, Szabo´ A, Bibonne A, Theveneau E, Parsons M, Mayor R. Cadherin switch during EMT in neural crest cells leads to contact inhibition of locomotion via repolarization of forces. Dev Cell. 2015;34:421–434. doi: 10.1016/j.devcel.2015.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller HB, Hermann MR, Polleux J, Vignaud T, Zanivan S, Friedel CC, Sun Z, Raducanu A, Gottschalk KE, Théry M, et al. beta1- and alphav-class integrins cooperate to regulate myosin II during rigidity sensing of fibronectin-based microenvironments. Nat Cell Biol. 2013;15:625–636. doi: 10.1038/ncb2747. [DOI] [PubMed] [Google Scholar]
- Schwarz US, Gardel ML. United we stand—integrating the actin cytoskeleton and cell-matrix adhesions in cellular mechanotransduction. J Cell Sci. 2012;125:3051–3060. doi: 10.1242/jcs.093716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra-Picamal X, Conte V, Vincent R, Anon E, Tambe DT, Bazellieres E, Butler JP, Fredberg JJ, Trepat X. Mechanical waves during tissue expansion. Nat Phys. 2012;8:628–634. [Google Scholar]
- Strale PO, Duchesne L, Peyret G, Montel L, Nguyen T, Png E, Tampé R, Troyanovsky S, Hénon S, Ladoux B, et al. The formation of ordered nanoclusters controls cadherin anchoring to actin and cell-cell contact fluidity. J Cell Biol. 2015;210:333–346. doi: 10.1083/jcb.201410111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Guo SS, Fässler R. Integrin-mediated mechanotransduction. J Cell Biol. 2016a;215:445–456. doi: 10.1083/jcb.201609037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Tseng HY, Tan S, Senger F, Kurzawa L, Dedden D, Mizuno N, Wasik AA, Thery M, Dunn AR, et al. Kank2 activates talin, reduces force transduction across integrins and induces central adhesion formation. Nat Cell Biol. 2016b;18:941–953. doi: 10.1038/ncb3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunyer R, Conte V, Escribano J, Elosegui-Artola A, Labernadie A, Valon L, Navajas D, García-Aznar JM, Muñoz JJ, Roca-Cusachs P, et al. Collective cell durotaxis emerges from long-range intercellular force transmission. Science. 2016;353:1157–1161. doi: 10.1126/science.aaf7119. [DOI] [PubMed] [Google Scholar]
- Swaminathan V, Waterman CM. The molecular clutch model for mechanotransduction evolves. Nat Cell Biol. 2016;18:459–461. doi: 10.1038/ncb3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift J, Discher DE. The nuclear lamina is mechano-responsive to ECM elasticity in mature tissue. J Cell Sci. 2014;127:3005–3015. doi: 10.1242/jcs.149203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift J, Ivanovska IL, Buxboim A, Harada T, Dingal PC, Pinter J, Pajerowski JD, Spinler KR, Shin JW, Tewari M, et al. Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science. 2013;341:1240104. doi: 10.1126/science.1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo B, Szollosi GJ, Gonci B, Juranyi Z, Selmeczi D, Vicsek T. Phase transition in the collective migration of tissue cells: experiment and model. Phys Rev E Stat Nonlin Soft Matter Phys. 2006;74:061908. doi: 10.1103/PhysRevE.74.061908. [DOI] [PubMed] [Google Scholar]
- Takahashi M, Ishida T, Traub O, Corson MA, Berk BC. Mechanotransduction in endothelial cells: temporal signaling events in response to shear stress. J Vasc Res. 1997;34:212–219. doi: 10.1159/000159225. [DOI] [PubMed] [Google Scholar]
- Tambe DT, Hardin CC, Angelini TE, Rajendran K, Park CY, Serra-Picamal X, Zhou EH, Zaman MH, Butler JP, Weitz DA, et al. Collective cell guidance by cooperative intercellular forces. Nat Mater. 2011;10:469–475. doi: 10.1038/nmat3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theveneau E, Steventon B, Scarpa E, Garcia S, Trepat X, Streit A, Mayor R. Chase-and-run between adjacent cell populations promotes directional collective migration. Nat Cell Biol. 2013;15:763–772. doi: 10.1038/ncb2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thievessen I, Thompson PM, Berlemont S, Plevock KM, Plotnikov SV, Zemljic-Harpf A, Ross RS, Davidson MW, Danuser G, Campbell SL, et al. Vinculin-actin interaction couples actin retrograde flow to focal adhesions, but is dispensable for focal adhesion growth. J Cell Biol. 2013;202:163–177. doi: 10.1083/jcb.201303129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas WA, Boscher C, Chu YS, Cuvelier D, Martinez-Rico C, Seddiki R, Heysch J, Ladoux B, Thiery JP, Mege RM, et al. alpha-Catenin and vinculin cooperate to promote high E-cadherin-based adhesion strength. J Biol Chem. 2013;288:4957–4969. doi: 10.1074/jbc.M112.403774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PM, Ramachandran S, Case LB, Tolbert CE, Tandon A, Pershad M, Dokholyan NV, Waterman CM, Campbell SL. A structural model for vinculin insertion into PIP2-containing membranes and the effect of insertion on vinculin activation and localization. Structure. 2017;25:1–12. doi: 10.1016/j.str.2016.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PM, Tolbert CE, Shen K, Kota P, Palmer SM, Plevock KM, Orlova A, Galkin VE, Burridge K, Egelman EH, et al. Identification of an actin binding surface on vinculin that mediates mechanical cell and focal adhesion properties. Structure. 2014;22:697–706. doi: 10.1016/j.str.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tozluoğlu M, Tournier AL, Jenkins RP, Hooper S, Bates PA, Sahai E. Matrix geometry determines optimal cancer cell migration strategy and modulates response to interventions. Nat Cell Biol. 2013;15:1–14. doi: 10.1038/ncb2775. [DOI] [PubMed] [Google Scholar]
- Trepat X, Fredberg JJ. Plithotaxis and emergent dynamics in collective cellular migration. Trends Cell Biol. 2011;21:638–646. doi: 10.1016/j.tcb.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trepat X, Wasserman MR, Angelini TE, Millet E, Weitz DA, Butler JP, Fredberg JJ. Physical forces during collective cell migration. Nat Phys. 2009;5:426–430. [Google Scholar]
- Trichet L, Le Digabel J, Hawkins RJ, Vedula SRK, Gupta M, Ribrault C, Hersen P, Voituriez R, Ladoux B. Evidence of a large-scale mechanosensing mechanism for cellular adaptation to substrate stiffness. Proc Natl Acad Sci USA. 2012;109:6933–6938. doi: 10.1073/pnas.1117810109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse JR, Engler AJ. Stiffness gradients mimicking in vivo tissue variation regulate mesenchymal stem cell fate. PLoS One. 2011;6:e15978. doi: 10.1371/journal.pone.0015978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng Q, Duchemin-Pelletier E, Deshiere A, Balland M, Guillou H, Filhol O, Théry M. Spatial organization of the extracellular matrix regulates cell-cell junction positioning. Proc Natl Acad Sci USA. 2012;109:1506–1511. doi: 10.1073/pnas.1106377109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twiss F, Le Duc Q, Van Der Horst S, Tabdili H, Van Der Krogt G, Wang N, Rehmann H, Huveneers S, Leckband DE, De Rooij J. Vinculin-dependent cadherin mechanosensing regulates efficient epithelial barrier formation. Biol Open. 2012;1:1128–1140. doi: 10.1242/bio.20122428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valon L, Marín-Llauradó A, Wyatt T, Charras G, Trepat X. Optogenetic control of cellular forces and mechanotransduction. Nat Commun. 2017;8:14396. doi: 10.1038/ncomms14396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasioukhin V, Bauer C, Yin M, Fuchs E. Directed actin polymerization is the driving force for epithelial cell-cell adhesion. Cell. 2000;100:209–219. doi: 10.1016/s0092-8674(00)81559-7. [DOI] [PubMed] [Google Scholar]
- Vedula SRK, Leong MC, Lai TL, Hersen P, Kabla AJ, Lim CT, Ladoux B. Emerging modes of collective cell migration induced by geometrical constraints. Proc Natl Acad Sci USA. 2012;109:12974–12979. doi: 10.1073/pnas.1119313109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicsek T, Czirok A, Ben-Jacob E, Cohen II, Schochet O. Novel type of phase transition in a system of self-driven particles. Phys Rev Lett. 1995;75:1226–1229. doi: 10.1103/PhysRevLett.75.1226. [DOI] [PubMed] [Google Scholar]
- Vincent R, Bazellières E, Pérez-González C, Uroz M, Serra-Picamal X, Trepat X. Active tensile modulus of an epithelial monolayer. Phys Rev Lett. 2015;115:1–5. doi: 10.1103/PhysRevLett.115.248103. [DOI] [PubMed] [Google Scholar]
- Wang X, Ha T. Defining single molecular forces required to activate integrin and notch signaling. Science. 2013;340:991–994. doi: 10.1126/science.1231041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Sun J, Xu Q, Chowdhury F, Roein-Peikar M, Wang Y, Ha T. Integrin molecular tension within motile focal adhesions. Biophys J. 2015;109:2259–2267. doi: 10.1016/j.bpj.2015.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Jin G, Miao H, Li JY-S, Usami S, Chien S. Integrins regulate VE-cadherin and catenins: dependence of this regulation on Src, but not on Ras. Proc Natl Acad Sci USA. 2006;103:1774–1779. doi: 10.1073/pnas.0510774103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber GF, Bjerke MA, DeSimone DW. Integrins and cadherins join forces to form adhesive networks. J Cell Sci. 2011;124:1183–1193. doi: 10.1242/jcs.064618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber GF, Bjerke MA, DeSimone DW. A mechanoresponsive cadherin-keratin complex directs polarized protrusive behavior and collective cell migration. Dev Cell. 2012;22:104–115. doi: 10.1016/j.devcel.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfenson H, Meacci G, Liu S, Stachowiak MR, Iskratsch T, Ghassemi S, Roca-Cusachs P, O’Shaughnessy B, Hone J, Sheetz MP. Tropomyosin controls sarcomere-like contractions for rigidity sensing and suppressing growth on soft matrices. Nat Cell Biol. 2015;18:33–42. doi: 10.1038/ncb3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Baribault H. Vinculin knockout results in heart and brain defects during embryonic development. Development. 1998;125:327–337. doi: 10.1242/dev.125.2.327. [DOI] [PubMed] [Google Scholar]
- Yamashita H, Ichikawa T, Matsuyama D, Kimura Y, Ueda K, Craig SW, Harada I, Kioka N. The role of the interaction of the vinculin proline-rich linker region with vinexin α in sensing the stiffness of the extracellular matrix. J Cell Sci. 2014;127:1875–1886. doi: 10.1242/jcs.133645. [DOI] [PubMed] [Google Scholar]
- Yan J, Yao M, Goult BT, Sheetz MP. Talin dependent mechanosensitivity of cell focal adhesions. Cell Mol Bioeng. 2015;8:151–159. doi: 10.1007/s12195-014-0364-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Lieu ZZ, Wolfenson H, Hameed FM, Bershadsky AD, Sheetz MP. Mechanosensing controlled directly by tyrosine kinases. Nano Lett. 2016;16:5951–5961. doi: 10.1021/acs.nanolett.6b02995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao M, Goult BT, Klapholz B, Hu X, Toseland CP, Guo Y, Cong P, Sheetz MP, Yan J. The mechanical response of talin. Nat Commun. 2016;7:11966. doi: 10.1038/ncomms11966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao M, Qiu W, Liu R, Efremov AK, Cong P, Seddiki R, Payre M, Lim CT, Ladoux B, Mège RM, et al. Force-dependent conformational switch of α-catenin controls vinculin binding. Nat Commun. 2014;5:4525. doi: 10.1038/ncomms5525. [DOI] [PubMed] [Google Scholar]
- Ye X, McLean MA, Sligar SG. Phosphatidylinositol 4,5-bisphosphate modulates the affinity of talin-1 for phospholipid bilayers and activates its autoinhibited form. Biochemistry. 2016;55:5038–5048. doi: 10.1021/acs.biochem.6b00497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama S, Matsui TS, Deguchi S. New wrinkling substrate assay reveals traction force fields of leader and follower cells undergoing collective migration. Biochem Biophys Res Commun. 2016;482:975–979. doi: 10.1016/j.bbrc.2016.11.142. [DOI] [PubMed] [Google Scholar]
- Yonemura S, Wada Y, Watanabe T, Nagafuchi A, Shibata M. alpha-Catenin as a tension transducer that induces adherens junction development. Nat Cell Biol. 2010;12:533–542. doi: 10.1038/ncb2055. [DOI] [PubMed] [Google Scholar]
- Yu CH, Rafiq NB, Cao F, Zhou Y, Krishnasamy A, Biswas KH, Ravasio A, Chen Z, Wang YH, Kawauchi K, et al. Integrin-beta3 clusters recruit clathrin-mediated endocytic machinery in the absence of traction force. Nat Commun. 2015;6:8672. doi: 10.1038/ncomms9672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidel-Bar R, Itzkovitz S, Ma’ayan A, Iyengar R, Geiger B. Functional atlas of the integrin adhesome. Nat Cell Biol. 2007;9:858–867. doi: 10.1038/ncb0807-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaritsky A, Welf ES, Tseng YY, Angeles Rabadan M, Serra-Picamal X, Trepat X, Danuser G. Seeds of locally aligned motion and stress coordinate a collective cell migration. Biophys J. 2015;109:2492–2500. doi: 10.1016/j.bpj.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Jiang G, Cai Y, Monkley SJ, Critchley DR, Sheetz MP. Talin depletion reveals independence of initial cell spreading from integrin activation and traction. Nat Cell Biol. 2008;10:1062–1068. doi: 10.1038/ncb1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann J, Camley BA, Rappel W-J, Levine H. Contact inhibition of locomotion determines cell-cell and cell-substrate forces in tissues. Proc Natl Acad Sci USA. 2016;113:2660–2665. doi: 10.1073/pnas.1522330113. [DOI] [PMC free article] [PubMed] [Google Scholar]