The contractile myoepithelial cells of the Caenorhabditis elegans somatic gonad are stretched by oocyte entry and subsequently contract to expel the fertilized embryo into the uterus. Formation of aligned, parallel actomyosin bundles during the first ovulation is triggered by oocyte entry and regulated by myosin contractility.
Abstract
Stress fibers—contractile actomyosin bundles—are important for cellular force production and adaptation to physical stress and have been well studied within the context of cell migration. However, less is known about actomyosin bundle formation and organization in vivo and in specialized contractile cells, such as smooth muscle and myoepithelial cells. The Caenorhabditis elegans spermatheca is a bag-like organ of 24 myoepithelial cells that houses the sperm and is the site of fertilization. During ovulation, spermathecal cells are stretched by oocyte entry and then coordinately contract to expel the fertilized embryo into the uterus. Here we use four-dimensional confocal microscopy of live animals to observe changes to spermathecal actomyosin network organization during cell stretch and contraction. Oocyte entry is required to trigger cell contraction and concomitant production of parallel actomyosin bundles. Actomyosin bundle size, connectivity, spacing, and orientation are regulated by myosin activity. We conclude that myosin drives actomyosin bundle production and that myosin activity is tightly regulated during ovulation to produce an optimally organized actomyosin network in C. elegans spermathecae.
INTRODUCTION
Actin is one of the most highly conserved proteins across eukaryotes and plays a central role in cellular adaptation to and generation of force (Gunning et al., 2015). This is required for numerous biological processes, including cell-cycle progression (Provenzano and Keely, 2011), cell migration (Li et al., 2005), tissue morphogenesis (Wozniak and Chen, 2009; Nelson and Gleghorn, 2012), wound closure (Brugues et al., 2014), and cell/tissue adaptation to physical stress (Davis et al., 2001; Burridge and Wittchen, 2013). The actin cytoskeleton is able to produce the different structures required to meet diverse cellular needs through complex and tightly regulated protein networks (dos Remedios et al., 2003; Dominguez, 2004; Zaidel-Bar et al., 2015). Stress fibers—contractile actomyosin bundles—are common among cells exposed to physical stress (Tojkander et al., 2012) and have been well studied within the context of cell migration (Pollard et al., 2000; Pellegrin and Mellor, 2007; Naumanen et al., 2008; Burridge and Wittchen, 2013). However, contractile stress fiber–like structures are found in many specialized contractile cells, such as contractile epithelial, contractile endothelial, smooth muscle, and myoepithelial cells, and much less is known about the regulation of actomyosin contractility in these contexts (Pellegrin and Mellor, 2007; Pollard and Cooper, 2009). Alterations in actomyosin contractility underlie the pathophysiology of numerous conditions, including cardiac disease, hypertension, vasospasm, asthma, erectile dysfunction, and preterm labor (Uehata et al., 1997; Wettschureck and Offermanns, 2002; Somlyo and Somlyo, 2003; Seguchi et al., 2007; Lavoie et al., 2009). There are few in vivo systems that allow for visualization of the actomyosin cytoskeleton in cells under dynamic mechanical conditions. Most of the in vivo work comes from elegant studies of cell movements and cell contractility during Drosophila elongation, gastrulation, egg chamber rotation, and tracheal tube development (Mason et al., 2013; Cetera et al., 2014; Kasza et al., 2014; Hannezo et al., 2015; Hosono et al., 2015). Although cell shape changes and large perturbations to the localization of the actomyosin contractile apparatus can be observed in these cell types, their small size makes imaging of dynamic reorganization of the actin cytoskeleton a challenge. The somatic gonad of the transparent, hermaphroditic nematode Caenorhabditis elegans is composed of a single layer of contractile myoepithelial cells (Mccarter et al., 1999; Michaux et al., 2001). Because the cells of the gonad are large and clearly visible in the intact animal and the gonad naturally stretches and contracts during the ovulation process, this system is ideal for studying how cells in an intact tissue modulate their actomyosin cytoskeletons in response to stretch and contraction.
The C. elegans somatic gonad contains two symmetrical, u-shaped gonad arms connected to a common uterus. Sheath cells surround the developing oocytes, and the spermatheca—a contractile, bag-like organ of 24 myoepithelial cells—houses the sperm and is the site of fertilization (Hirsh et al., 1976; Hubbard and Greenstein, 2000). During ovulation, sheath cell contractions increase and the distal spermathecal neck opens to allow entry of the proximal oocyte to the spermatheca, where it is immediately fertilized (Hirsh et al., 1976; Ward and Carrel, 1979; Hubbard and Greenstein, 2000). After a regulated period of time, spermathecal cells coordinately contract, the spermathecal-uterine (SP-UT) valve opens, and the embryo is expelled into the uterus. This process occurs ∼150 times per gonad arm, requiring robust regulation of somatic tissue contractility to successfully propel the oocyte from the sheath through the spermatheca and into the uterus (Mccarter et al., 1999; Yamamoto et al., 2006).
Contractions of both sheath and spermathecal cells are regulated by the conserved Ca2+ and RhoA signaling pathways (Clandinin et al., 1998; Wissmann et al., 1999; Bui and Sternberg, 2002; Kariya et al., 2004; Yin et al., 2004; McMullan and Nurrish, 2011; Kovacevic et al., 2013; Meighan et al., 2015; Ono and Ono, 2016), which cooperate to increase phosphorylation of the myosin regulatory light chain (p-MRLC) and actomyosin contractility in smooth muscle and nonmuscle cells (Somlyo and Somlyo, 2000). In the C. elegans gonad, two phospholipase C isoforms, Cγ (PLC-3) and Cε (PLC-1), cleave phosphatidyl inositol to produce inositol 1,4,5-triphosphate (IP3), which triggers Ca2+ release from the endoplasmic reticulum (Clandinin et al., 1998; Bui and Sternberg, 2002; Kariya et al., 2004; Yin et al., 2004; Kovacevic et al., 2013). This likely increases myosin activity by activating a Ca2+-calmodulin–dependent myosin light chain kinase (Adelstein, 1982; Somlyo and Somlyo, 2000). In both tissues, RHO-1/RhoA signaling activates Rho-associated kinase, LET-502/ROCK, which phosphorylates and inactivates the myosin-associated phosphatase regulatory subunit MEL-11 (referred to here as myosin phosphatase), leading to increased p-MRLC (Wissmann et al., 1999; McMullan and Nurrish, 2011; Meighan et al., 2015; Ono and Ono, 2016). However, unlike sheath cells, spermathecal cells lack the troponin-tropomyosin complex (Ono and Ono, 2004; Ono et al., 2007) responsible for Ca2+ regulation of actomyosin contraction in muscle cells (Ebashi, 1984). Sheath and spermathecal cells also have distinct cytoskeletal organizations (Strome, 1986). Sheath cells exhibit long, longitudinal actin bundles that contain both nonmuscle and muscle isoforms of myosin II heavy chain (Ono and Ono, 2016), whereas the spermatheca has stress fiber–like, parallel actin bundles oriented along the long axis of each cell (Figure 1; Strome, 1986) and lacks muscle isoforms of myosin II (Ardizzi and Epstein, 1987; Ono et al., 2007). This difference in actomyosin organization may reflect differences in the contractile properties of each cell type. The sheath cells, wrapped around the maturing oocytes, experience a relatively constant degree of stretch and produce multiple contractions that increase in intensity and frequency during ovulation, whereas spermathecal cells are dramatically stretched by the incoming oocyte and produce a single smooth contraction as the oocyte is expelled into the uterus (Ward and Carrel, 1979; Hubbard and Greenstein, 2000).
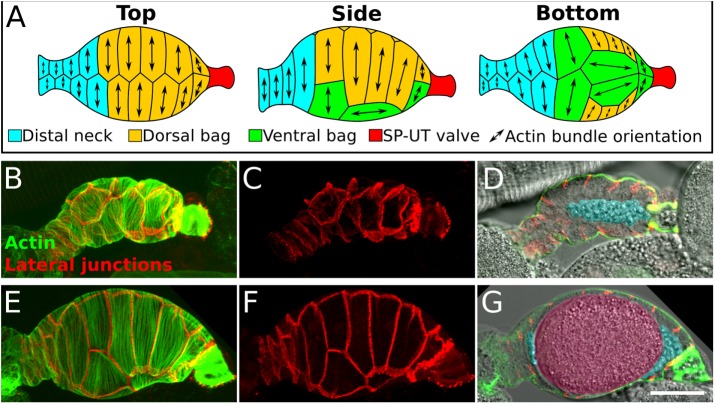
FIGURE 1:
Anatomy of the C. elegans spermatheca. (A) Schematic diagram showing actin bundle orientation in spermathecal cells. (B–G) Confocal images of two fixed and stained spermathecae, one that is unoccupied, sperm only (B–D), and one that is occupied, sperm and oocyte present (E–G). (B, C, E, F) Confocal maximum intensity projections of spermathecae expressing INX-12::mApple to label lateral junctions (red) stained with phalloidin to label F-actin (green). Note the difference in cell stretch in an unoccupied (C) and an occupied (F) spermatheca. (D, G) A central sagittal z-slice showing a cross section of the spermatheca with basal actin bundles (green), lateral junctions (red), and bright-field image (grayscale). Sperm and oocyte are false colored in blue and pink, respectively. Scale bar, 20 μm. In all images, the spermatheca is oriented distal to proximal.
We previously observed that spermathecal cells before the first ovulation have tortuous, randomly oriented actin bundles and that the mature actin cytoskeleton is not present until after the first ovulation (Kovacevic and Cram, 2010). Here we use four-dimensional (4D) confocal microscopy to observe changes to spermathecal actomyosin network organization during cell stretch and contraction. We show that 1) maturation of the actin cytoskeleton requires and is proportionate to the degree of spermathecal cell stretch during the first ovulation; 2) actin network maturation coincides with the onset of cell contraction and requires Ca2+ signaling to trigger myosin II activity; and 3) both reduced and increased myosin II activity cause changes in actomyosin network connectivity, tortuosity, spacing, and orientation. We conclude that tight spatiotemporal regulation of myosin II activity during cell stretch and contraction is required for optimal actomyosin network organization and tissue contractility in C. elegans spermathecae.
RESULTS
Parallel actomyosin bundle formation requires oocyte entry to the spermatheca
The most prominent features of the mature actin cytoskeleton in the C. elegans spermatheca are basal stress fiber–like actin bundles oriented along the long axis of each cell (Figures 1 and 2; Strome, 1986). We previously reported that these parallel actin bundles are absent in spermathecae of young animals before the first ovulation (Kovacevic and Cram, 2010). To investigate what drives production of parallel actin bundles in mature adults and determine whether these actin structures are actomyosin bundles, we used phalloidin staining to visualize filamentous-actin (F-actin) and functional green fluorescent protein–labeled nonmuscle myosin II (GFP::NMY-1) to visualize myosin (Supplemental Figure S1). In late-L4 animals, parallel basal bundles are absent, and the majority of F-actin is located at lateral junctions and the apical cell surface. GFP-labeled myosin II appears diffusely throughout the cytosol and, similarly to F-actin, accumulates at lateral junctions and the apical cell surface, forming small punctae. In some cases, a few thin actomyosin bundles are visible at the basal surface (Figure 2A). By young adulthood, F-actin and myosin II colocalize into basal actomyosin bundles that differ in their organization from those seen in mature adults. Before the first ovulation, the basal actomyosin bundles are tortuous, branching, and randomly oriented (Figure 2B). After the first ovulation, branching and tortuosity decrease, and prominent, parallel actomyosin bundles aligned along the long axis of each cell are apparent (Figure 2C). These actomyosin bundles appear to result primarily from reorganization of existing F-actin. However, using DNase I to specifically bind globular actin (G-actin; Cramer et al., 2002), we did observe a slight but significant increase in the ratio of F-actin to G-actin in spermathecae from animals after the first ovulation, suggesting that de novo actin polymerization may also be involved (Supplemental Figure S2). Once parallel actin bundles develop, they are maintained throughout successive rounds of ovulation and are consistently seen in both occupied (oocyte present) and empty (only sperm present) spermathecae. These results demonstrate that the actin structures observed in adults (Strome, 1986) are actomyosin bundles and suggested to us that oocyte entry might be required for development of the mature actomyosin network during the first ovulation.
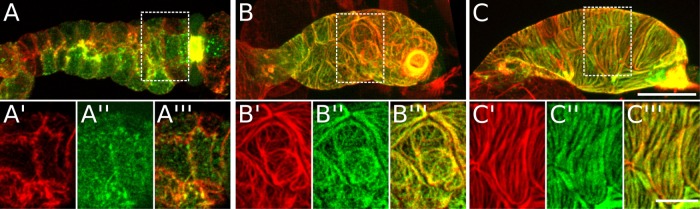
FIGURE 2:
Myosin and actin colocalize into actomyosin bundles in spermathecal cells. Confocal maximum intensity projections of excised spermathecae at different developmental stages expressing GFP-labeled myosin (GFP::NMY-1; green) and stained with phalloidin to label F-actin (red). Yellow indicates actin and myosin colocalization. (A) Late L4 animal, (B) young adult prior to the first ovulation, and (C) mature adult after the first ovulation. The spermatheca in C is unoccupied (after oocyte exit). White boxes indicate section magnified in insets below. All insets are a single confocal z-slice showing only the most basal surface. (A’, C’) Actin, (A’’, C’’) myosin, (A’’’, C’’’) merge. Scale bar: 20 μm (A–C), 10 μm (insets).
To determine whether animal age rather than oocyte entry per se initiates actin bundle alignment, we used RNA interference (RNAi) to block entry of the oocyte to the spermatheca and observed the cytoskeleton in larval stage 1 (L1) arrest–synchronized populations after 45 h. At this time point, wild-type (WT) animals have undergone approximately five ovulations. First, we abolished oocyte production by knocking down prp-17, a germline-expressed gene involved in regulating the sperm-to-oocyte switch (Kerins et al., 2010). Most prp-17(RNAi) animals produce only sperm (Kerins et al., 2010; Cecchetelli et al., 2016). In these animals, the immature actomyosin network is maintained, and parallel bundles do not develop. In some cases, prp-17(RNAi) animals produce and ovulate a few abnormal oocytes (Kerins et al., 2010; Cecchetelli et al., 2016). When these oocytes enter the spermatheca, the actomyosin network forms aligned bundles. Quantification of network anisotropy—a measurement of alignment—in individual cells using FibrilTool (Boudaoud et al., 2014) reveals a significant increase in anisotropy in prp-17(RNAi) animals, which produced abnormal oocytes, compared with control animals after the first ovulation. Because the prp-17(RNAi) oocytes do not form an eggshell and remain deformable, this apparent increase in anisotropy over WT is probably the result of increased flattening of the spermatheca during imaging. These results suggest that knockdown of prp-17 influences spermathecal actin organization through its effect on oocyte production.
We next explored the role of oocyte entry in actomyosin maturation by inhibiting genes required for sheath cell contraction. RNAi knockdown of the phospholipase Cγ, plc-3 (Yin et al., 2004), or the ER calcium sensor STIM1, stim-1 (Yan et al., 2006), disrupts Ca2+ signaling and sheath contractions, and knockdown of scribble homologue, let-413 (Pilipiuk et al., 2009), or the atypical protein kinase C (PKC), pkc-3 (Aono et al., 2004), disrupts dilation of the distal spermathecal neck. Knockdown of each gene prevents entry of the oocyte to the spermatheca, trapping oocytes in the oviduct, where they undergo endomitotic (Emo) DNA replication and produce abnormally large nuclei (Iwasaki et al., 1996; Mccarter et al., 1999). Although the mechanism of ovulation disruption differs for each gene, the effect on spermathecal actomyosin bundle organization is similar. In all cases, animals with an Emo phenotype have basal, poorly aligned actomyosin bundles that resemble those in control animals before the first ovulation (Figure 3). Quantification of actomyosin organization in individual cells with FibrilTool shows that bundle anisotropy in animals treated with plc-3 or let-413 RNAi does not differ significantly from that in control animals before the first ovulation. RNAi knockdown of stim-1 results in a modest decrease in anisotropy compared with the preovulation control, whereas RNAi knockdown of pkc-3 results in a more significant reduction in anisotropy (Figure 3). Because pkc-3 is involved in establishing apicobasal polarity (Aono et al., 2004), PKC-3 may be required for wild-type morphology of preovulation actomyosin bundles. Of importance, in all cases in which oocyte entry to the spermatheca was blocked, the actomyosin network failed to mature into the parallel bundles. These findings indicate that oocyte entry is required to initiate actomyosin network maturation.
FIGURE 3:
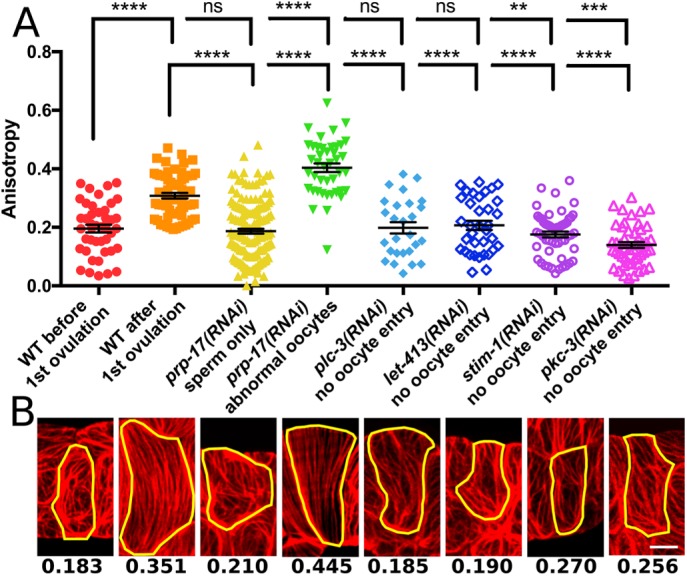
Oocyte entry during ovulation is required for alignment of spermathecal actin bundles. (A) Quantification of actin bundle anisotropy (degree of alignment) using FibrilTool. Each point represents a single cell, and no more than three cells were measured from the same animal. For each condition, from left to right, 44, 63, 152, 42, 27, 35, 54, and 49 cells. (B) Representative phalloidin staining results for each condition in A. Yellow line indicates a single cell selected for measurement with FibrilTool, and numbers indicate anisotropy measured for each cell. Error bars represent SEM. Unpaired t test: ns, p > 0.05; **p ≤ 0.01; ***p ≤ 0.001; ****p ≤ 0.0001. Scale bar, 5 μm.
Parallel actin bundle formation coincides with cell contraction triggered by and proportional to cell stretch during ovulation
Cell stretch (Thoumine et al., 1995; Sears and Kaunas, 2016) and actomyosin-driven contraction (Peterson et al., 2004; Sears and Kaunas, 2016) have been shown to influence actomyosin network organization. To determine whether cell stretch, contraction, or both are required for parallel actomyosin bundle production, we used confocal microscopy to capture 4D ovulation movies of live whole animals. Spermathecal actin was visualized using three different fluorescent actin reporters: GFP-labeled actin, GFP::ACT-1; GFP fused to the actin-binding calponin homology domain from utrophin, GFP::UtrCH (Burkel et al., 2007); and the moesin actin-binding domain fused to mCherry, moeABD::mCherry (Edwards et al., 1997). All spermathecal actin reporter lines recapitulate the F-actin organization observed using phalloidin staining and are capable of ovulating normally, suggesting that the actin markers accurately label endogenous actin with minimal disruption of the actin cytoskeleton (Supplemental Figure S3). Fortuitously, we found that during image acquisition, the SP-UT valve in the GFP::ACT-1 animals remains partially closed, prolonging oocyte dwell time in the spermatheca. This allows us to monitor changes in actin bundle organization during cell stretch caused by oocyte entry and subsequent cell contraction. When the embryo exits, the spermatheca returns to a compressed conformation, making it difficult to image cells during contraction (Supplemental Figure S3). For this reason, we present here data obtained using the GFP::ACT-1 line. However, similar results were obtained with the GFP::UtrCH and moeABD::mCherry lines (Supplemental Figure S3).
As previously discussed, oocyte entry stretches the cells of the spermatheca (Figure 1), and this cell stretch precedes and is required for parallel actin bundle production (Figure 2). To determine whether the degree of stretch correlates with the degree of actomyosin bundle alignment, we observed actin dynamics in animals in which the distal spermathecal neck had closed prematurely, permitting only a fraction of the oocyte to enter the spermatheca (Supplemental Movie S1). In contrast to the actomyosin bundle production and alignment evident in normally stretched cells, entry of small oocyte fragments results in minimal change to bundle organization (Figure 4 and Supplemental Movie S1). This suggests that stretch is required for bundle alignment. To determine whether actomyosin bundle alignment coincides temporally with cell stretch upon oocyte entry or instead with onset of contraction, we simultaneously tracked changes in cell length—an indirect measurement of cell stretch and contraction—and actomyosin bundle anisotropy. Immediately after oocyte entry, stretched spermathecal cells exhibit the preovulation actin network organization. Production of prominent, parallel actomyosin bundles does not begin until ∼500 s after the initiation of cell stretch, coincident with the onset of cell contraction (Figure 5 and Supplemental Movie S2). This suggests that active actomyosin contraction drives bundle formation and alignment and that this process is activated by and proportionate to cell stretch.
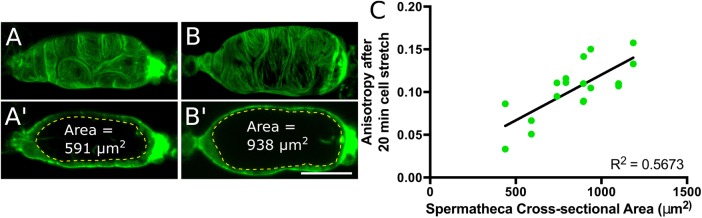
FIGURE 4:
Actin bundle alignment is proportional to cell stretch. (A, B) Confocal maximum intensity projections showing spermathecal actin labeled with GFP (GFP::ACT-1) in a slightly stretched spermatheca containing an oocyte fragment (A) and normally stretched spermatheca containing the entire oocyte (B) at ∼20 min after oocyte entry. (A’, B’) A central, sagittal z-slice from A and B. The yellow dashed line indicates measurement of spermathecal cross-sectional area. (C) Comparison of spermathecal cross-sectional area and actin network anisotropy (degree of alignment) achieved ∼20 min after oocyte entry, depicting a positive relationship between spermathecal cell stretch and actin network anisotropy. Two cells were measured from nine animals (18 cells). Black line: best-fit linear regression. Scale bar, 20 μm.
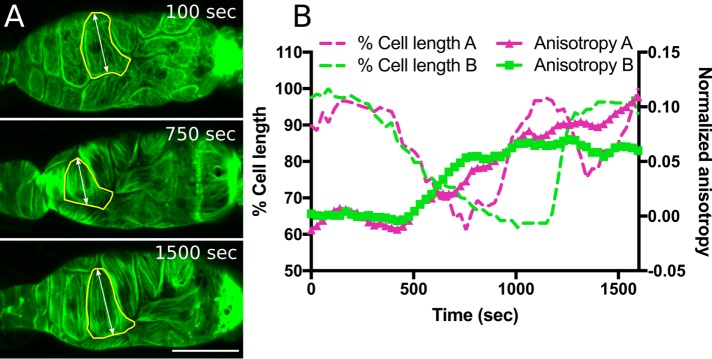
FIGURE 5:
Actin bundle alignment coincides with cell contraction during ovulation. (A) Maximum intensity projections from a 4D confocal ovulation movie of spermathecal cells expressing GFP-labeled actin (GFP::ACT-1). At 100 s after oocyte entry, spermathecal cells are stretched, yet display the webby preovulation actomyosin network. At 750 s after oocyte entry, contracting cells show development of parallel actomyosin bundles. At 1500 s after oocyte entry, spermathecal cells exhibit parallel actomyosin bundles oriented along the long axis of each cell. (B) Quantification of cell contraction and actomyosin bundle anisotropy (degree of alignment) in two cells from independent ovulation movies. FibrilTool was used to quantify actin anisotropy in individual cells (yellow outline in A). Cell shortening coincides with a rapid increase in actin anisotropy ∼500 s after oocyte entry. Scale bar, 20 μm.
To determine whether increased contractility is able to overcome the requirement of cell stretch to induce contraction and produce aligned actin bundles, we used RNAi to deplete myosin phosphatase, mel-11, previously shown to regulate spermathecal contractility (Piekny and Mains, 2002). RNAi depletion of mel-11 produces hypercontractile spermathecae that are capable of contracting without oocyte entry (Supplemental Figure S4). However, in these spermathecae, the cells become compact, and actin structures are difficult to visualize (Supplemental Figure S4). This shows that although enhanced cell contractility can overcome the requirement of oocyte entry to trigger contraction, we were not able to observe the effect on the actin cytoskeleton under these conditions.
Myosin activity drives parallel actomyosin bundle formation
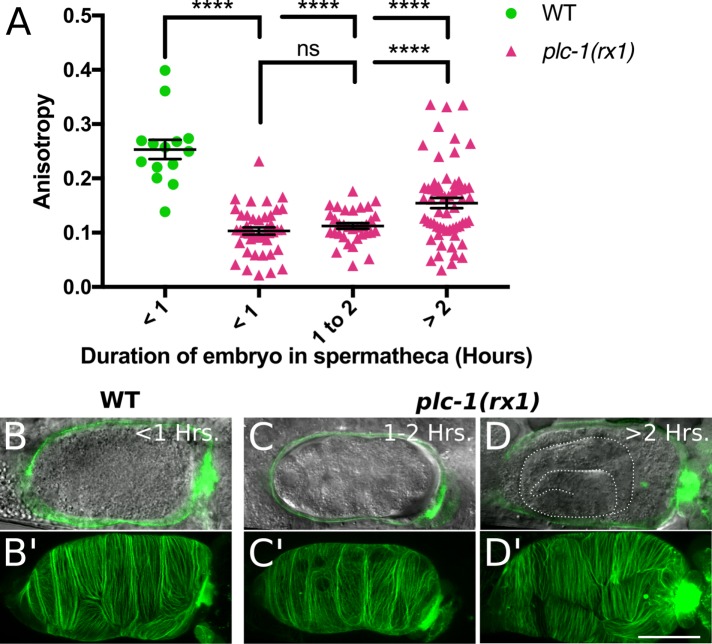
Because formation of the parallel actomyosin bundles coincides with contraction, we next investigated whether phospholipase-induced cell contraction is required. Phospholipase-stimulated Ca2+ release is predicted to lead to actomyosin contraction through the activation of myosin light chain kinase (MLCK) and the phosphorylation of MRLC. Although the MLCK active in the spermatheca has not been identified, the connection between phospholipases, IP3 production, Ca2+ release, and contraction in C. elegans is well established (Clandinin et al., 1998; Bui and Sternberg, 2002; Kariya et al., 2004; Yin et al., 2004; Kovacevic et al., 2013). We showed previously that intracellular Ca2+ levels peak during spermathecal contraction and that loss of the phospholipase Cε, PLC-1, abolishes Ca2+ signaling and tissue contraction (Kovacevic et al., 2013). If contraction is required for actin network maturation, plc-1–null animals should retain the immature, webby actomyosin network after spermathecal cell stretch. To investigate the effect of plc-1 knockout on actomyosin organization during ovulation, we generated animals homozygous for the putative null allele, plc-1(rx1), expressing GFP-labeled actin in the spermatheca. In these animals, the oocyte enters the spermatheca normally, is fertilized, and then becomes trapped as spermatheca cells fail to contract. This phenotype allowed us to use embryonic development to roughly determine the duration of the embryo in the spermatheca and spermathecal cell stretch. We find that actomyosin network maturation is severely delayed in plc-1(rx1) animals. In wild-type animals, actin organization rapidly increases after ∼500 s of cell stretch (Figure 5 and Supplemental Movie S2). In contrast, plc-1(rx1) animals require at least 2 h to form quantifiable parallel bundles (Figure 6). This suggests that cell contraction, lost in plc-1(rx1) animals, is required to drive timely actomyosin organization.
FIGURE 6:
In phospholipase Cε–null animals, actin bundle alignment is delayed. (A) Actin bundle alignment requires at least 2 h of spermathecal cell stretch in phospholipase Cε–null, plc-1(rx1), animals. FibrilTool was used to measure anisotropy (degree of alignment) in individual cells. Each point represents a single cell from 14 WT cells and 43, 33, and 60 plc-1(rx1) cells for <1, 1–2, and >2 h, respectively. Embryos between 2 and 8 h of age are lumped into the >2-h category. No more than three cells were measured from the same animal. Embryonic developmental stages were used to estimate embryo duration in the spermatheca. (B–D) A central, sagittal z-slice of spermathecae expressing GFP-labeled actin (GFP::ACT-1; green). Bright-field image shows development of the embryo trapped within the spermatheca. Note the advanced embryonic development indicating lengthy spermathecal cell stretch of at least 8 h (D). (B’–D’) Confocal maximum intensity projections of the spermathecae shown in B–D. Note the lack of prominent, parallel actin bundles in C’ only. Error bars represent SEM. Unpaired t test: ns, p > 0.05, ****p ≤ 0.0001. Scale bar, 20 μm.
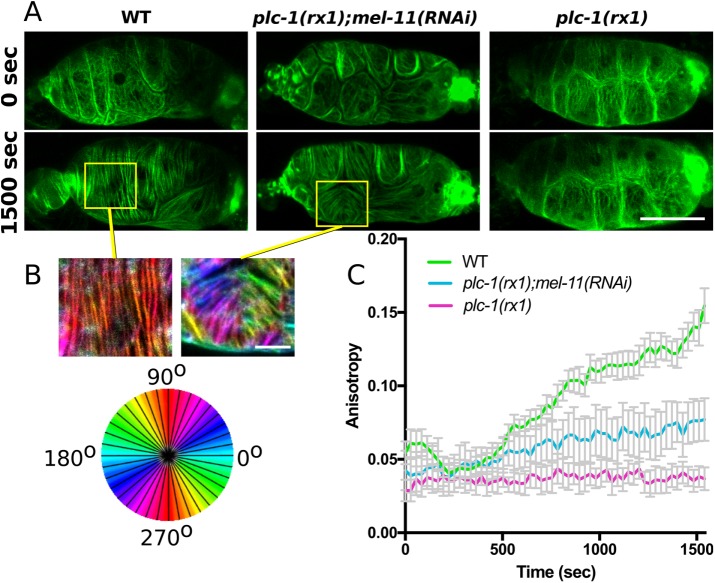
To determine whether delayed actomyosin network maturation seen in plc-1(rx1) animals can be rescued by increasing myosin activity, we used 4D confocal microscopy to capture ovulation movies of WT, plc-1(rx1), and plc-1(rx1) animals treated with RNAi against the myosin phosphatase, mel-11. If the noncontractile plc-1(rx1) phenotype is primarily the result of decreased p-MRLC levels and reduced myosin activity, then loss of the phosphatase mel-11 should rescue timely actomyosin bundle production and alignment. In WT animals, actomyosin network maturation begins during cell contraction, ∼500 s after the start of cell stretch, and mature, parallel bundles are well developed after 1000–1500 s (Figures 5 and 7 and Supplemental Movie S2). During this time, plc-1(rx1) spermathecal cells do not contract and show no significant change in actomyosin bundle organization (Figure 7 and Supplemental Movie S3). Treatment of plc-1(rx1) animals with RNAi against mel-11 partially rescues timely actomyosin network maturation, and prominent bundles, similar to those in wild-type cells, are apparent after ∼1500 s of cell stretch (Figure 7 and Supplemental Movie S4). Although mel-11 RNAi treatment rescues the production of prominent actomyosin bundles in plc-1(rx1) animals, these bundles are frequently misaligned with respect to the long axis of the cell, and populations of differentially oriented bundles are seen within the same cell (Figure 7). This phenotype is distinct from WT cells, which consistently develop parallel actomyosin bundles oriented along the long axis of each cell, and likely contributes to the reduced anisotropy measured in plc-1(rx1);mel-11(RNAi) animals. Overall these results suggest that the delayed actomyosin network maturation in plc-1(rx1) animals is the result of insufficient activation of myosin and suggest that precise regulation of myosin activity during cell contraction is required to achieve wild-type actin network morphology.
FIGURE 7:
Depletion of myosin phosphatase rescues timely actin bundle formation but not orientation in phospholipase Cε–null spermathecae. (A) Confocal maximum intensity projections from frames of 4D ovulation movies of spermathecae from WT, phospholipase Cε–null animals (plc-1(rx1)) and plc-1(rx1) animals treated with RNAi against myosin phosphatase (plc-1(rx1);mel-11(RNAi)) expressing actin labeled with GFP (GFP::ACT-1). Times indicate time after oocyte entry. (B) OrientationJ was used to false-color actin bundles according to their orientation in selected cells indicated by yellow boxes in A. Note the production of populations of differently oriented actin bundles within a single plc-1(rx1);mel-11(RNAi) cell. (C) Quantification of anisotropy (degree of alignment) in individual spermathecal cells during ovulation. FibrilTool was used to measure anisotropy in individual cells at 28-s intervals during ovulation. No more than two cells were measured from the same animal. For WT, n = 12 cells (6 animals), for plc-1(rx1);mel-11(RNAi), n = 7 cells (4 animals), and for plc-1(rx1) n = 8 cells (4 animals). Error bars represent SEM. Scale bar, 20 μm.
Both increased and decreased myosin activity alter actin network organization
To probe more directly the role of myosin activity in actin bundle formation, we used RNAi of nmy-1, the major nonmuscle myosin II heavy chain expressed in the spermatheca (Kovacevic et al., 2013), to reduce the level of myosin, and RNAi of myosin phosphatase, mel-11, to elevate myosin activity. Spermathecal actin was visualized in whole live animals using the actin reporter lines described earlier and in N2 animals using phalloidin staining. Similar results were obtained using all techniques (Supplemental Figures S5–S7). As expected, RNAi depletion of nmy-1 reduces spermathecal cell contraction, producing the flaccid phenotype described previously (Kovacevic et al., 2013) characterized by a distended distal neck and SP-UT valve (Supplemental Figure S5). Surprisingly, we find that nmy-1(RNAi) spermathecae have basal actin bundles indistinguishable from WT animals before the first ovulation (Supplemental Figure S8). We obtained similar results with the null allele, nmy-1(sb115), indicating RNAi against nmy-1 is sufficient to produce a null phenotype (Supplemental Figure S5). Knockdown of mel-11 also has no noticeable effect on preovulation spermathecal actin organization (Supplemental Figure S5), suggesting that myosin is dispensable for organization of preovulation actin bundles.
After ovulation, both loss of myosin and elevated myosin activity disrupt spermathecal actin organization. Animals treated with nmy-1 RNAi maintain the preovulation actin network characterized by thin, interconnected, tortuous actin bundles (Figure 8 and Supplemental Figures S5–S7). Similar to plc-1(rx1) spermathecae, spermathecae of nmy-1(RNAi) animals fail to contract after oocyte entry, resulting in trapped embryos in the spermatheca. However, unlike plc-1(rx1) spermathecae, nmy-1(RNAi) spermathecae never develop the prominent, parallel actin bundles seen in mature, postovulation, WT animals, regardless of the duration of cell stretch. Loss of mel-11 produces the opposite phenotype. After oocyte entry, the spermatheca hyperconstricts, the SP-UT valve remains tightly closed, blocking oocyte exit, and contraction continues until mounting force culminates in rupture of cell–cell contacts. Before tissue rupture, mel-11(RNAi) spermathecae develop very prominent actin bundles. These bundles are frequently thicker and spaced farther apart than bundles in WT cells and appear to result from neighboring bundles being pulled together (Figure 9 and Supplemental Figures S6 and S7). In addition, mel-11(RNAi) cells frequently contain populations of actin bundles oriented in different directions (Figure 9 and Supplemental Figure S5). Overall these results show that myosin is required for actin network maturation and suggest that myosin activity is regulated during contraction to produce WT parallel actin bundles.
FIGURE 8:
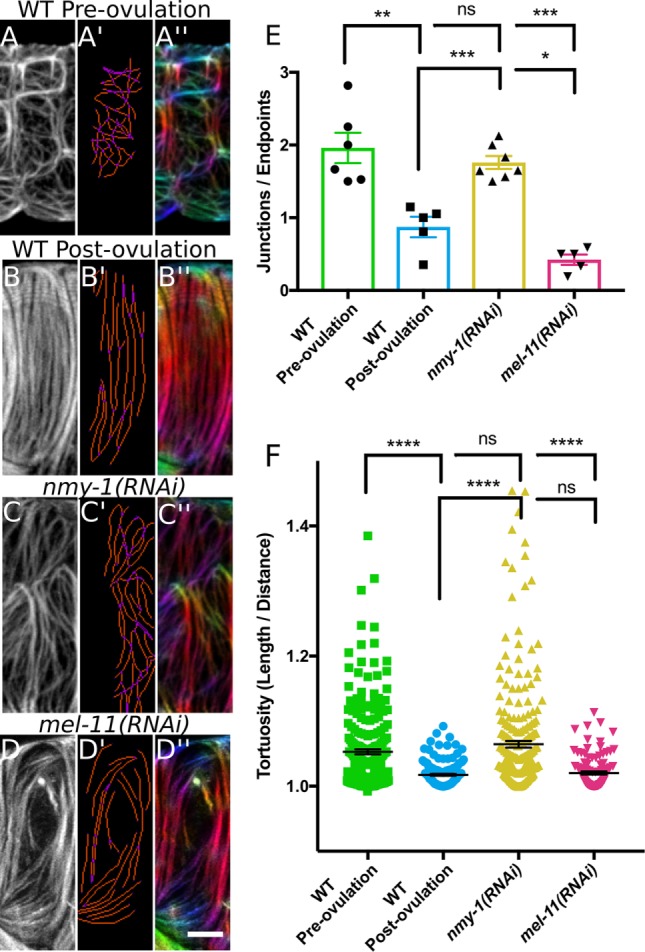
Myosin activity influences actin bundle development and organization. (A–D) A single confocal z-slice of a spermathecal cell stained with phalloidin–Texas Red to label F-actin. (A’–D’) AnalyzeSkeleton analysis of bundles. Blue, endpoint (less than two neighbors); purple, junction (more than two neighbors); orange, slab (exactly two neighbors). Analysis indicates high actin bundle branching and/or intersection in a preovulation, WT cell (A’) and a nmy-1(RNAi) cell (C’); note increased purple pixels in A’ and C’ compared with B’ and D’. (A’’–D’’) OrientationJ was used to false-color actin bundles according to their orientation. Note that a postovulation, WT cell has parallel actin bundles oriented along the long cell axis. In a preovulation, WT cell (A’’) and a nmy-1(RNAi) cell (C’’), actin bundles are randomly oriented, as illustrated by many different colors. In a mell-11(RNAi) cell, some bundles are oriented correctly parallel to the long cell axis, but subpopulations are misaligned. (E) Quantification of actin bundle connectivity based on AnalyzeSkeleton results. The number of junctions (purple pixels) was divided by the number of endpoints (blue pixels). Each point represents analysis of a single cell with no more than two cells from the same animal and six, five, seven, and five cells for WT preovulation, WT postovulation, nmy-1(RNAi), and mel-11(RNAi), respectively. A value >1 indicates actin bundles more frequently intersected with or branched from neighboring bundles. A value <1 indicates actin bundles more frequently extended the length of the cell without contacting neighboring bundles. Note the increased actin bundle intersection and/or branching in preovulation, WT and nmy-1(RNAi) cells. (F) Quantification of actin bundle tortuosity (total length/linear distance between ends). Note increased actin bundle tortuosity in preovulation, WT cells and nmy-1(RNAi) cells. Each point represents an individual bundle measurement from five to seven different cells and 241, 145, 256, and 110 bundles for WT preovulation, WT postovulation, nmy-1(RNAi), and mel-11(RNAi), respectively. Error bars represent SEM. Unpaired t test (E) or Mann–Whitney test (F): ns, p > 0.05; *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001; ****p ≤ 0.0001. Scale bar, 5 μm.
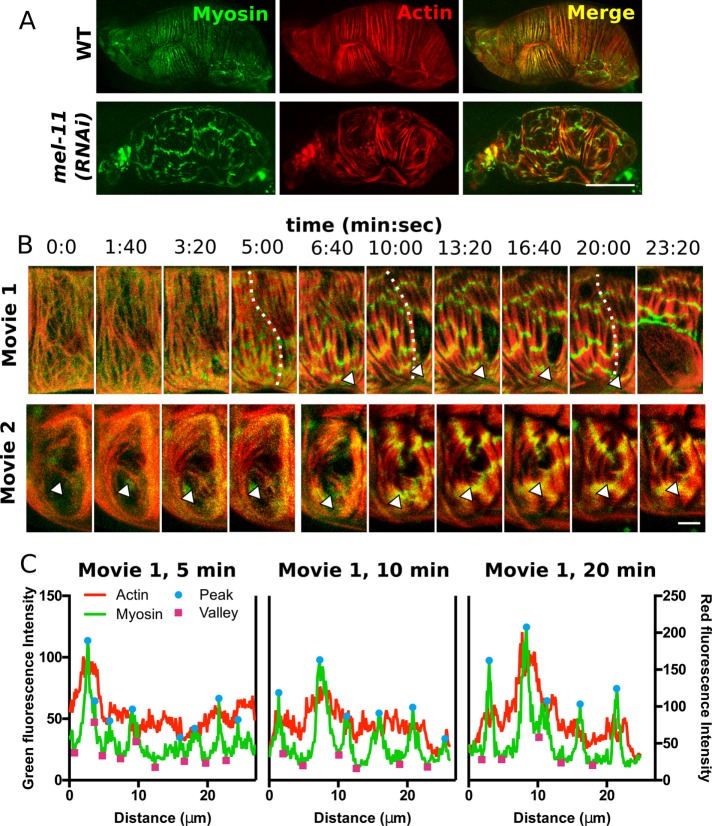
FIGURE 9:
Elevated myosin activity alters myosin organization and interaction with actomyosin bundles. (A) Confocal maximum intensity projections of fixed spermathecae from a WT animal and an animal treated with RNAi against myosin phosphatase, mel-11(RNAi), expressing moeABD::mCherry to label actin (red) and GFP::NMY-1 to label myosin (green). Note the homogeneous distribution of myosin in actomyosin bundles in WT and laterally associated myosin clusters that create transverse bands across several actomyosin bundles in a mel-11(RNAi) spermatheca. (B) Confocal maximum intensity projections from two 4D ovulation movies showing progression of mel-11(RNAi) phenotypes in selected cells. In movie 1, the actomyosin bundles rupture. The arrowhead indicates the site of bundle detachment. At 6:40, bundles are attached, and they retract by 10:00. By 23:20, the basal cell layer pulls back (last frame). In movie 2, misaligned actin bundles appear to be stabilized and reinforced (arrowhead). (C) Line-scan analysis along a single actomyosin bundle in movie 1 (indicated by a dashed line) at 5, 10, and 20 min after oocyte entry reveals that myosin clusters increase in intensity and grow by fusion of smaller clusters along a single bundle. In addition, actin fluorescence intensity is increased under prominent myosin peaks. Scale bar, 20 μm (A), 5 μm (B).
Increased myosin activity leads to clustering of myosin within highly contractile actomyosin bundles
To better understand how altered myosin activity influences actin network organization, we used 4D confocal microscopy to visualize both actin and myosin during cell stretch and contraction. Myosin and actin were coimaged in a line expressing moeABD:: mCherry and GFP::NMY-1 to label actin and myosin, respectively. To confirm that GFP does not interfere with myosin activity, we expressed GFP::NMY-1 in the nmy-1(sb115) null background. GFP::NMY-1 rescues the brood size defect of nmy-1(sb115), producing 142 ± 8.737 (mean ± SEM, n = 3) live offspring per animal, compared with only 21 ± 4.509 (mean ± SEM, n = 3) produced by nmy-1(sb115) (Supplemental Figure S1). This indicates that the GFP-myosin is functional and likely reports localization of endogenous myosin.
During ovulation in wild-type animals, myosin is highly dynamic. Immediately after oocyte entry, GFP-myosin becomes diffuse throughout the cytosol. This is followed by rapid recruitment of GFP-myosin into actomyosin bundles and cell contraction as the oocyte is expelled from the spermatheca. Because oocytes are not retained in the spermatheca in this line, visualizing actomyosin organization during contraction is difficult as structures become obscured in the highly compact, contracted cells (Supplemental Figure S3 and Supplemental Movie S5). Imaging fixed whole animals mid ovulation using slow acquisition settings (see Materials and Methods) more clearly shows that GFP-myosin is incorporated into and homogeneously distributed throughout actomyosin bundles (Figure 9A).
In mel-11(RNAi) spermathecae, myosin was not distributed throughout actomyosin bundles and instead accumulated in large, laterally associated clusters, creating transverse bands across several actin bundles (Figure 9). In mel-11(RNAi) spermathecae, immediately after oocyte entry, actomyosin organization appears similar to WT. However, 3–5 min later, myosin is recruited to actin bundles, producing small clusters of myosin along actin bundles. These foci rapidly increase in intensity and appear to grow by fusion with neighboring clusters along a single actin bundle until reaching a steady state, ∼10 min after initiation of cell stretch, when they increase in intensity but maintain a distance of 5–10 μm along the actin bundles (Figure 9 and Supplemental Figure S9 and Supplemental Movie S6). We also observe changes in actomyosin bundle behavior, including rupture of bundles and maintenance of misaligned bundles present in the immature network (Figure 9B). Eventually, sufficient force builds up to rupture cell contacts. Rupture occurs 27 ± 2.1 min (mean ± SEM, n = 3) after oocyte entry and always at horizontal cell junctions, whereas vertical junctions remain intact. Furthermore, loss of cell–cell adhesion begins at basal cell–cell contacts. Tissue integrity is presumably maintained by apical adherens junctions until cells completely pull apart (Figure 9B and Supplemental Movie S6). Tissue rupture indicates that these actomyosin bundles are highly contractile. Together these results show elevated myosin activity influences myosin organization within actomyosin bundles and generates sufficient force to alter bundle behavior and rupture bundles and cell–cell contacts.
DISCUSSION
We previously observed that spermathecal basal actomyosin bundles mature from a webby, isotropic network to one with prominent, evenly spaced, and aligned bundles. Here we show that this maturation requires and is proportionate to spermathecal cell stretch, suggesting that spermathecal cells are mechanoreceptive. One recently identified mechanotransducer in the spermatheca is the RhoA GAP SPV-1, which contains an F-BAR domain capable of perceiving membrane curvature (Tan and Zaidel-Bar, 2015). During cell stretch, SPV-1 is displaced from the membrane, releasing inhibition of RHO-1/RhoA (Tan and Zaidel-Bar, 2015) and triggering actomyosin contraction by LET-502/ROCK inhibition of the myosin phosphatase, MEL-11 (Wissmann et al., 1999). This is likely part of the mechanism by which cells that experience greater stretch have higher p-MRLC and active myosin than minimally stretched cells. We also show here that cell stretch alone is not sufficient to align actin bundles. Immediately after oocyte entry, spermathecal cells are fully stretched, yet retain the webby, immature, actomyosin network. Actomyosin network maturation instead coincides with cell contraction ∼500 s after the initiation of cell stretch. This is contrary to some cell culture work, in which actin bundle alignment is proportional to the degree of uniaxial stretch (Sears and Kaunas, 2016) but still occurs in stretched cells when actomyosin contractility is disrupted by inhibition of ROCK (Kaunas et al., 2005; Lee et al., 2010) or MLCK (Lee et al., 2010). However, complete loss of myosin activity by simultaneous inhibition of ROCK and MLCK does prevent actin bundle formation in cultured cells (Lee et al., 2010). Similarly, we find myosin activity is required for spermathecal actomyosin bundle production and alignment. We propose that increase in active myosin during cell contraction, triggered by stretch, drives reorganization of the actomyosin network into parallel bundles. This may occur by the network contraction behavior of myosin (Verkhovsky and Borisy, 1993; Fenix et al., 2016) or poorly understood attractive forces between distant myosin bipolar filaments capable of merging actin bundles (Hu et al., 2017).
In smooth muscle and nonmuscle cells, an increase in cytosolic Ca2+ causes cell contraction primarily through activation of the Ca2+-calmodulin–dependent myosin light chain kinase (Adelstein, 1982). Unlike muscle myosin, nonmuscle myosin Mg2+-ATPase activity is not activated by actin alone but requires phosphorylation of the MRLC (Sellars et al., 1981). We showed previously that intracellular Ca2+ levels peak during spermathecal contraction and that loss of phospholipase Cε, plc-1, abolishes measurable Ca2+ transients and tissue contraction (Kovacevic et al., 2013). Here we show that loss of plc-1 delays the rate of actomyosin bundle maturation and alignment, extending a process that normally occurs in minutes over several hours. However, Ca2+ and the ubiquitous Ca2+-sensing protein calmodulin have numerous downstream effects, ranging from activation of protein kinases to regulation of ion channels (Clapham, 2007). We show that treatment of plc-1 null animals with mel-11 RNAi rescues timely actomyosin bundle production. This suggests that the delayed production of actomyosin bundles formed in plc-1–null animals is driven by a plc-1–independent pool of p-MRLC due to either undetectably low levels of Ca2+ or an alternative mechanism for phosphorylating MRLC, such as phosphorylation by ROCK (Amano et al., 1996; Gally et al., 2009; Beach et al., 2017), and that knockdown of mel-11 is able to unmask this pool, resulting in timely actomyosin bundle formation. Of interest, these bundles were frequently misaligned with respect to the long axis of the cell, and differently oriented populations of bundles were seen within the same cell. This suggests that precise regulation of myosin activity during cell contraction is required to achieve WT actomyosin network morphology. It is also possible that the observed disruption of actomyosin bundle orientation is due to dysregulation of a myosin phosphatase substrate other than MRLC. For example, in Madin–Darby canine kidney cells, phosphorylation of moesin, a protein involved in actin network organization (Tsukita and Yonemura, 1999; Haynes et al., 2011), is regulated by myosin phosphatase (Fukata et al., 1998).
To probe more directly the influence of myosin activity on actomyosin network organization, we knocked down the primary nonmuscle myosin II heavy chain isoform expressed in the spermatheca, nmy-1 (Kovacevic et al., 2013), to eliminate myosin activity or mel-11, to increase myosin activity. Knockdown of either nmy-1 or mel-11 had no effect on preovulation actomyosin bundles. This was unexpected, given that myosin and actin appear to colocalize in these bundles. Perhaps the presence of other actin cross-linking proteins such as filamin (Kovacevic and Cram, 2010) is sufficient to produce these thin, webby actin bundles independently of myosin. However, as is seen in other contractile actomyosin structures, such as stress fibers (Lamb et al., 1988; Totsukawa et al., 2000) and the cytokinetic contractile ring (Murthy and Wadsworth, 2005; Takaine et al., 2015), regulated myosin activity is required for development of the WT mature actin network. Without nmy-1, the webby immature actin network persists and, unlike plc-1–null animals, parallel bundles never develop even after prolonged cell stretch. Similarly, the requirement of mel-11 for WT actomyosin morphology is apparent once cell contraction is stimulated by cell stretch during oocyte entry. Depletion of mel-11 produces large, irregularly spaced, misaligned bundles that appear to result from merging of neighboring bundles.
During contraction in WT cells, myosin is recruited to and homogeneously distributed throughout actomyosin bundles. Increased myosin activity by mel-11 RNAi results in rapid accumulation of myosin into actin bundles and the production of myosin foci that grow by fusion with neighboring clusters along a single bundle. Myosin foci also appear to interact with myosin on neighboring bundles, producing laterally associated bands of myosin that span several actin bundles and seem to pull bundles together. Similar transverse myosin bands have been observed in contractile structures of cultured cells, including stress fibers and the cytokinetic ring (Verkhovsky and Borisy, 1993; Verkhovsky et al., 1995; Fenix et al., 2016; Hu et al., 2017). These bands appeared to result from interactions between adjacent bipolar filaments, producing an interconnected myosin network (Verkhovsky and Borisy, 1993; Fenix et al., 2016). In addition, it has also been observed that alignment of myosin bands across neighboring actomyosin bundles occurs via long-range interactions (Hu et al., 2017) such as mechanical communication through an elastic deformable substrate (Friedrich et al., 2011). Our observations suggest that myosin clusters on adjacent actomyosin bundles physically interact either directly or through an unknown binding partner. This idea is supported by the observation that myosin clusters appear to pull neighboring bundles together and hold on to ruptured bundles, preventing further retraction. We also observe precocious stabilization and reinforcement of misaligned bundles present in the immature actomyosin network. This inappropriate stabilization of nascent actomyosin bundles may explain the observation that mel-11 RNAi in a plc-1–null background frequently produces misaligned actin bundles but not the large bundles seen in mel-11 RNAi in a WT background.
Conclusion
Although actin nucleators (Hegsted et al., 2016) and cross-linkers (Kovacevic and Cram, 2010) clearly play an important role in spermathecal actin organization, we find that myosin activity is the dominant factor driving actomyosin network maturation and alignment during cell contraction. This is consistent with work in Drosophila tracheal tubes, where properties of the circumferential actomyosin bundles, including bundle production (Hannezo et al., 2015) and orientation (Hosono et al., 2015), are regulated by myosin activity. In vitro–reconstituted systems provide mechanistic insight into how modulation of myosin activity alone can alter actomyosin network properties. In vitro, actin and myosin are able to self-organize into contractile bundles (Thoresen and Gardel, 2011; Thoresen et al., 2013), and myosin is capable of cross-linking actin filaments (Haviv et al., 2008; Thoresen and Gardel, 2011), fusing actin bundles (Miyazaki et al., 2015), and producing forces sufficient to buckle (Molloy et al., 1995; Lenz et al., 2012) and depolymerize actin filaments (Haviv et al., 2008). Furthermore, in vitro phosphorylation of the MRLC is cooperative, allowing a small increase in kinase activity to cause a large increase in active myosin (Persechini and Hartshorne, 1981). This suggests that tight regulation of p-MRLC and actin–myosin interactions is required for establishment and maintenance of functional actomyosin networks in vivo. Now that we have characterized the role of myosin activity in spermathecal actomyosin network development, we see this as an ideal system for investigating how myosin activity is regulated to maintain optimal tissue contractility in a mechanically stressed tissue. The spermatheca must undergo >100 successive rounds of stretch and contraction during ovulation. Identification of protein networks that regulate myosin activity throughout this process, such as MRLC kinases and proteins that modulate the actin–myosin interaction (Zaidel-Bar et al., 2015), may help us better understand how actomyosin networks are formed and maintained in vivo and address the pathophysiology of contractile tissue diseases such as asthma (Lavoie et al., 2009) and hypertension (Uehata et al., 1997).
MATERIALS AND METHODS
C. elegans strains and culture
All nematode strains were maintained on NGM (0.107 M NaCl, 0.25% [wt/vol] Peptone [Fischer Science Education], 1.7% [wt/vol] BD Bacto-Agar [Fisher Scientific], 0.5% nystatin [Sigma-Aldrich], 0.1 mM CaCl2, 0.1 mM MgSO4, 0.5% [wt/vol] cholesterol, and 2.5 mM KPO4) agar plates seeded with OP50 Escherichia coli at 23°C (Hope, 1999). Generation of nematode strains for this study was done by standard microinjection (Mello et al., 1991). All constructs were injected at a concentration between 5 and 10 ng/µl with injection markers rol-6 and ttx3p::RFP injected at 40 ng/µl. Carrier DNA, sheared, denatured salmon sperm DNA (Clontech, Mountain View, CA) was added to bring the final DNA concentration to 100 ng/µl for each injection mix. Extrachromosomal arrays were integrated by ultraviolet irradiation essentially as described in Mariol et al. (2013). Nematode observations and manipulations were performed at 23°C unless otherwise noted. The C. elegans nonmuscle myosin II allele HR1184 nmy-1(sb115) and phospholipase Cε allele PS4112 plc-1(rx1) were obtained from the Caenorhabditis Genetics Center and outcrossed at least four times with N2 (wild-type strain from Bristol). Transgene qyIs198[inft-1p::moeABD::mCherry, unc-119(+)] was obtained from strain NK1069, a kind gift from the Sherwood lab (Duke University, Durham, NC). For a list of strains used in this study, see Supplemental Table S1.
RNA interference
The RNAi feeding protocol was performed essentially as described in Timmons et al. (2001). To prepare seeded NGM–isopropylthio-β-galactoside (IPTG) plates, HT115(DE3) bacteria transformed with the double-strand RNA (dsRNA) construct of interest were grown overnight at 37°C in Luria broth (LB) supplemented with 40 µg/ml ampicillin. The next day, 150 µl of the culture was seeded on NGM-IPTG agar (NGM supplemented with 25 µg/ml carbenicillin and 1 mM IPTG) and incubated at room temperature for 24–72 h to induce dsRNA expression. Age-synchronized animals, prepared as described next, were then transferred to these plates.
Partially synchronized populations were obtained by alkaline lysis procedure (“egg prep”), and tightly synchronized populations were obtained by L1 arrest. For egg prep, starved dauer nematodes were allowed to recover for 48 h on NGM plates newly seeded with OP50. This produces young gravid adults for egg collection. Eggs were released using an alkaline hypochlorite solution as described in Hope (1999) and washed three times with filter-sterilized M9 buffer (22 mM KH2PO4, 42 mM NaHPO4, 86 mM NaCl, and 1 mM MgSO4; Hope, 1999). Clean eggs were then transferred to seeded NGM-IPTG plates. The L1 arrest protocol was adapted from Hope (1999). For L1 arrest, the same alkaline lysis procedure was performed, except that clean eggs were transferred onto NGM without bacteria. Eggs were allowed to hatch and develop until L1 at 16°C overnight before being transferred to seeded NGM-IPTG plates. Populations produced by egg prep are ∼10 h behind L1 arrest populations developmentally. All RNAi experiments were performed at 23°C. Strains used in each RNAi experiment are indicated.
RNAi constructs were obtained from the ORFeome-RNAi v1.1 library or were constructed by PCR amplification of WT cDNA and cloned into pPD129.36 (Fire Vector Kit). Empty pPD129.36 vector was used as a negative control in RNAi experiments. All primer sequences and cloning details are available upon request.
Histochemistry
For observations of N2 and UN1534 (GFP::NMY-1), partially synchronized populations were obtained by egg prep and grown at 23°C for 50–55 h. For RNAi experiments, tightly synchronized populations were obtained by L1 arrest and grown at 23°C for 45 h. At this time point, N2 control animals have undergone approximately five ovulations, and the Emo phenotype is obvious in RNAi treatments that disrupt oocyte entry. F-actin staining was adapted from Ono et al. (2007). Briefly, animals were dissected using a 25-gauge hypodermic needle in phosphate-buffered saline (PBS), and dissected gonads were fixed in 1.85% formaldehyde in PBS for 25 min at room temperature. After fixation, gonads were washed twice with PBS, permeabilized for 15 min in PBST (PBS + 0.1% Triton X-100), and then incubated with 0.4 U/ml Texas Red-X–phalloidin in PBS (Invitrogen, Carlsbad, CA) overnight at 4°C or 4 h at room temperature. For identification of the Emo phenotype, the nuclear stain 4′,6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich, St. Louis, MO) was added at 100 ng/ml and incubated for at least 20 min at room temperature. Labeled samples were washed twice with PBS and mounted on 2% agarose pads for observation.
Confocal microscopy
For ovulation movies and observations of live or fixed whole animals, partially synchronized populations were obtained by egg prep, and animals were grown at 23°C for ∼50 h, around the time of the first ovulation. Live animals were immobilized with 0.01% tetramisole and 0.1% tricaine in M9 buffer (Kirby et al., 1990; Mccarter et al., 1997) and mounted on 2% agarose pads or with 0.05-μm Polybead Microspheres (Polysciences, Warrington, PA) diluted 1:2 in water and mounted on 5% agarose pads (adapted from Wang and Audhya, 2014). In this imaging preparation, WT transits occur in ∼500 s. Although Mccarter et al. (1999) show more rapid oocyte transit times (∼340 s), our results agreed with earlier work from our laboratory and others (Ward and Carrel, 1979; Kovacevic et al., 2013). To prevent alteration of GFP::NMY-1 localization as was occasionally observed in excised gonads, live whole animals were fixed by immersion in 1.25% formaldehyde in PBS for 25 min at room temperature. Fixed animals were washed three times with PBS and mounted on 2% agarose pads. Confocal microscopy was performed on an LSM 710 confocal microscope (Zeiss) equipped with Zen software (Zeiss) using the Plan-Apochromat 63x/1.40 Oil DIC M27 objective. The 405-nm laser was used to excite DAPI, the 488-nm laser was used for GFP, and the 561-nm laser was used for mCherry and Texas Red. For movies, 40 z-slices were acquired at 14-s intervals for imaging a single channel or 30 z-slices at 20-s intervals for imaging two channels. Illumination of the spermathecae from animals expressing actin labeled with GFP (GFP::ACT-1) with the 488-nm laser for ∼5 min before oocyte entry frequently caused the valve to remain partially closed during ovulation, increasing oocyte dwell time. For movies of all lines, live animals were imaged for ∼30 min total. For still images of live and fixed animals and tissue, z-slices were acquired at 0.38-μm intervals with each slice representing the average of two or four scans for slow acquisition settings.
Image analysis
ImageJ software was used for all image analysis. For consistency, all analysis was performed on cells of the main spermathecal bag. Cells of the distal neck and cells most proximal to the valve were not used. FibrilTool (Boudaoud et al., 2014) was used for quantification of actin anisotropy in selected cells. For images of fixed and stained tissue, FibrilTool was used on a single z-slice, capturing only the basal surface of the cell. For movie analysis, maximum intensity projections were generated, and FibrilTool was used on individual cells at 28-s intervals (every other frame). In Figure 5, anisotropy measurements were normalized by taking the average anisotropy of the first 10 frames measured and subtracting this number from each measurement. The anisotropy curve in Figure 5 was smoothed using GraphPad Prism software set to average each point according to the five nearest points on each side of it.
To quantify actin network tortuosity (length of the bundle/linear distance between bundle ends) and connectivity in images of fixed tissue, ImageJ was used to enhance image contrast of the entire image, and NeuronJ (Meijering et al., 2004) was used for computer-guided tracing of individual bundles to allow for tortuosity measurements and to generate a skeletonized image. Network connectivity of skeletonized images was measured using AnalyzeSkeleton (Arganda-Carreras et al., 2010) to determine the number of junction (more than two neighbors), endpoint (less than two neighbors), and slab (exactly two neighbors) pixels per image. Connectivity was calculated by dividing the number of junction pixels by the number of endpoint pixels for each skeletonized image of a single cell. OrientationJ (Rezakhaniha et al., 2012), configured using a Gaussian fit with a pixel size of 2, was used to measure the orientation distribution of actin bundles in individual cells and generate color-coded images, where color indicates orientation, hue indicates coherency, and brightness is the brightness of the original image. Unless otherwise indicated, line-scan measurements of fluorescence intensity were conducted using line pixel width of 5. All measurements were conducted on raw images except when NeuronJ was used. Contrast is enhanced in images to clarify structures of interest. All statistical analysis was performed with GraphPad Prism software. Unpaired t test was used to determine whether the difference between the means of two data sets was significant when data had a normal distribution, and Welch's correction was included if the different treatments were expected to have different SDs. The Mann–Whitney test was used when data did not have a normal distribution. In all cases, the statistical test used and resulting p values are noted in the figure captions. ns, p > 0.05; *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001; ****p ≤ 0.0001.
Supplementary Material
Acknowledgments
We thank Ronen Zaidel-Bar for many helpful discussions. C. elegans strain NK1069 was generously provided by the Sherwood lab. Some C. elegans strains used in this study were provided by the Caenorhabditis Genetics Center, which is funded by the National Center for Research Resources, National Institutes of Health. This work was supported by a grant from the National Institutes of Health/National Institute of General Medical Sciences (GM110268) to E.J.C.
Abbreviations used:
- Emo
endomitotic
- F-actin
filamentous-actin
- G-actin
globular actin
- GFP
green fluorescent protein
- MRLC
myosin regulatory light chain
- p-MRLC
phosphorylated MRLC
- RNAi
RNA interference
- ROCK
Rho-associated kinase
- SP-UT
spermatheca-uterine
- WT
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-01-0029) on March 22, 2017.
REFERENCES
- Adelstein RS. Calmodulin and the regulation of the actin-myosin interaction in smooth muscle and nonmuscle cells. Cell. 1982;30:349–350. doi: 10.1016/0092-8674(82)90232-x. [DOI] [PubMed] [Google Scholar]
- Amano M, Ito M, Fukata Y, Chihara K, Nakano T, Matsuura Y, Kaibuchi K. Phosphorylation and activation of myosin by Rho-associated kinase (Rho-kinase) J Biol Chem. 1996;271:20246–20249. doi: 10.1074/jbc.271.34.20246. [DOI] [PubMed] [Google Scholar]
- Aono S, Legouis R, Hoose WA, Kemphues KJ. PAR-3 is required for epithelial cell polarity in the distal spermatheca of C. elegans. Development. 2004;131:2865–2874. doi: 10.1242/dev.01146. [DOI] [PubMed] [Google Scholar]
- Ardizzi JP, Epstein HF. Immunochemical localization of myosin heavy chain isoforms and paramyosin in developmentally and structurally diverse muscle cell types of the nematode Caenorhabditis elegans. J Cell Biol. 1987;105:2763–2770. doi: 10.1083/jcb.105.6.2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arganda-Carreras I, Fernández-González R, Muñoz-Barrutia A, Ortiz-De-Solorzano C. 3D reconstruction of histological sections: application to mammary gland tissue. Microsc Res Tech. 2010;73:1019–1029. doi: 10.1002/jemt.20829. [DOI] [PubMed] [Google Scholar]
- Beach JR, Bruun KS, Shao L, Li D, Swider Z, Remmert K, Zhang Y, Conti MA, Adelstein RS, Rusan NM, et al. Actin dynamics and competition for myosin monomer govern the sequential amplification of myosin filaments. Nat Cell Biol. 2017;19:85–93. doi: 10.1038/ncb3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudaoud A, Burian A, Borowska-Wykre D, Uyttewaal M, Wrzalik R, Kwiatkowska D, Hamant O. FibrilTool, an ImageJ plug-in to quantify fibrillar structures in raw microscopy images. Nat Protoc. 2014;9:457–463. doi: 10.1038/nprot.2014.024. [DOI] [PubMed] [Google Scholar]
- Brugues A, Anon E, Conte V, Veldhuis JH, Gupta M, Colombelli J, Munoz JJ, Brodland GW, Ladoux B, Trepat X. Forces driving epithelial wound healing. Nat Phys. 2014;10:683–690. doi: 10.1038/nphys3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui YK, Sternberg PW. Caenorhabditis elegans inositol 5-phosphatase homolog negatively regulates inositol 1,4,5-triphosphate signaling in ovulation. Mol Biol Cell. 2002;13:1641–1651. doi: 10.1091/mbc.02-01-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkel BM, Dassow G, von Bement WM. Versatile fluorescent probes for actin filaments based on the actin-binding domain of utrophin. Cell Motil Cytoskeleton. 2007;64:822–832. doi: 10.1002/cm.20226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge K, Wittchen ES. The tension mounts: stress fibers as force-generating mechanotransducers. J Cell Biol. 2013;200:9–19. doi: 10.1083/jcb.201210090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecchetelli AD, Hugunin J, Tannoury H, Cram EJ. CACN-1 is required in the Caenorhabditis elegans somatic gonad for proper oocyte development. Dev Biol. 2016;414:58–71. doi: 10.1016/j.ydbio.2016.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cetera M, Ramirez-San Juan GR, Oakes PW, Lewellyn L, Fairchild MJ, Tanentzapf G, Gardel ML, Horne-Badovinac S. Epithelial rotation promotes the global alignment of contractile actin bundles during Drosophila egg chamber elongation. Nat Commun. 2014;5:5511. doi: 10.1038/ncomms6511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clandinin TR, DeModena Ja, Sternberg PW. Inositol trisphosphate mediates a RAS-independent response to LET-23 receptor tyrosine kinase activation in C. elegans. Cell. 1998;92:523–533. doi: 10.1016/s0092-8674(00)80945-9. [DOI] [PubMed] [Google Scholar]
- Clapham DE. Calcium signaling. Cell. 2007;131:1047–1058. doi: 10.1016/j.cell.2007.11.028. [DOI] [PubMed] [Google Scholar]
- Cramer LP, Briggs LJ, Dawe HR. Use of fluorescently labelled deoxyribonuclease I to spatially measure G-actin levels in migrating and non-migrating cells. Cell Motil Cytoskeleton. 2002;38:27–38. doi: 10.1002/cm.10013. [DOI] [PubMed] [Google Scholar]
- Davis MJ, Wu XIN, Nurkiewicz TR, Kawasaki J, Davis GE, Hill MA, Meininger GA. Integrins and mechanotransduction of the vascular myogenic response. Am J Physiol Heart Circ Physiol. 2001;280:1427–1433. doi: 10.1152/ajpheart.2001.280.4.H1427. [DOI] [PubMed] [Google Scholar]
- Dominguez R. Actin-binding proteins—a unifying hypothesis. Trends Biochem Sci. 2004;29:572–578. doi: 10.1016/j.tibs.2004.09.004. [DOI] [PubMed] [Google Scholar]
- dos Remedios CG, Chhabra D, Kekic M, Dedova IV, Tsubakihara M, DA Berry, Nosworthy NJ. Actin binding proteins: regulation of cytoskeletal microfilaments. Physiol Rev. 2003;83:433–473. doi: 10.1152/physrev.00026.2002. [DOI] [PubMed] [Google Scholar]
- Ebashi S. Ca2+ and the contractile proteins. J Mol Cell Cardiol. 1984;16:129–136. doi: 10.1016/s0022-2828(84)80701-4. [DOI] [PubMed] [Google Scholar]
- Edwards KA, Demsky M, Montague RA, Weymouth N, Kiehart DP. GFP-moesin illuminates actin cytoskeleton dynamics in living tissue and demonstrates cell shape changes during morphogenesis in Drosophila. Dev Biol. 1997;191:103–117. doi: 10.1006/dbio.1997.8707. [DOI] [PubMed] [Google Scholar]
- Fenix AM, Taneja N, Buttler CA, Lewis J, Van Engelenburg SB, Ohi R, Brunette DT. Expansion and concatenation of nonmuscle myosin IIA filaments drive cellular contractile system formation during interphase and mitosis. Mol Biol Cell. 2016;27:1465–1478. doi: 10.1091/mbc.E15-10-0725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich BM, Buxboim A, Discher DE, Safran SA. Striated acto-myosin fibers can reorganize and register in response to elastic interactions with the matrix. Biophys J. 2011;100:2706–2715. doi: 10.1016/j.bpj.2011.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukata Y, Kimura K, Oshiro N, Saya H, Matsuura Y, Kaibuchi K. Association of the myosin-binding subunit of myosin phosphatase and moesin: dual regulation of moesin phosphorylation by Rho-associated kinase and myosin phosphatase. J Cell Biol. 1998;141:409–418. doi: 10.1083/jcb.141.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gally C, Wissler F, Zahreddine H, Quintin S, Landmann F, Labouesse M. Myosin II regulation during C. elegans embryonic elongation: LET-502/ROCK, MRCK-1 and PAK-1, three kinases with different roles. Development. 2009;136:3109–3119. doi: 10.1242/dev.039412. [DOI] [PubMed] [Google Scholar]
- Gunning PW, Ghoshdastider U, Whitaker S, Popp D, Robinson RC. The evolution of compositionally and functionally distinct actin filaments. J Cell Sci. 2015;128:2009–2019. doi: 10.1242/jcs.165563. [DOI] [PubMed] [Google Scholar]
- Hannezo E, Dong B, Recho P, Joanny J-F, Hayashi S. Cortical instability drives periodic supracellular actin pattern formation in epithelial tubes. Proc Natl Acad Sci USA. 2015;112:201504762. doi: 10.1073/pnas.1504762112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haviv L, Gillo D, Backouche F, Bernheim-Groswasser A. A cytoskeletal demolition worker: myosin II acts as an actin depolymerization agent. J Mol Biol. 2008;375:325–330. doi: 10.1016/j.jmb.2007.09.066. [DOI] [PubMed] [Google Scholar]
- Haynes J, Srivastava J, Madson N, Wittmann T, Barber DL. Dynamic actin remodeling during epithelial-mesenchymal transition depends on increased moesin expression. Mol Biol Cell. 2011;22:4750–4764. doi: 10.1091/mbc.E11-02-0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegsted A, Wright FA, Votra S, Pruyne D. INF2- and FHOD-Related formins promote ovulation in the somatic gonad of C. elegans. Cytoskeleton. 2016;73:712–728. doi: 10.1002/cm.21341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsh D, Oppenheim D, Klass M. Development of the reproductive system of Caenorhabditis elegans. Dev Biol. 1976;49:200–219. doi: 10.1016/0012-1606(76)90267-0. [DOI] [PubMed] [Google Scholar]
- Hope IA. C. elegans—A Practical Approach. Oxford, UK: Oxford University Press; 1999. [Google Scholar]
- Hosono C, Matsuda R, Adryan B, Samakovlis C. Transient junction anisotropies orient annular cell polarization in the Drosophila airway tubes. Nat Cell Biol. 2015;17:1569–1576. doi: 10.1038/ncb3267. [DOI] [PubMed] [Google Scholar]
- Hu S, Dasbiswas K, Guo Z, Tee Y, Thiagarajan V, Hersen P, Chew T, Safran SA, Zaidel-bar R, Bershadsky AD. Long-range self-organization of cytoskeletal myosin II filament stacks. Nat Cell Biol. 2017;19:133–141. doi: 10.1038/ncb3466. [DOI] [PubMed] [Google Scholar]
- Hubbard EJ, Greenstein D. The Caenorhabditis elegans gonad: a test tube for cell and developmental biology. Dev Dyn. 2000;218:2–22. doi: 10.1002/(SICI)1097-0177(200005)218:1<2::AID-DVDY2>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Iwasaki K, McCarter J, Francis R, Schedl T. emo-1, a Caenorhabditis elegans Sec 61p gamma homologue, is required for oocyte development and ovulation. J Cell Biol. 1996;134:699 LP-714. doi: 10.1083/jcb.134.3.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariya KI, Kim Bui Y, Gao X, Sternberg PW, Kataoka T. Phospholipase Cε regulates ovulation in Caenorhabditis elegans. Dev Biol. 2004;274:201–210. doi: 10.1016/j.ydbio.2004.06.024. [DOI] [PubMed] [Google Scholar]
- Kasza KE, Farrell DL, Zallen JA. Spatiotemporal control of epithelial remodeling by regulated myosin phosphorylation. Proc Natl Acad Sci USA. 2014;111:11732–11737. doi: 10.1073/pnas.1400520111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaunas R, Nguyen P, Usami S, Chien S. Cooperative effects of Rho and mechanical stretch on stress fiber organization. Proc Natl Acad Sci USA. 2005;102:15895–15900. doi: 10.1073/pnas.0506041102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerins JA, Hanazawa M, Dorsett M, Schedl T. PRP-17 and the pre-mRNA splicing pathway are preferentially required for the proliferation versus meiotic development decision and germline sex determination in Caenorhabditis elegans. Dev Dyn. 2010;239:1555–1572. doi: 10.1002/dvdy.22274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby C, Kusch M, Kemphues K. Mutations in the par genes of Caenorhabditis elegans affect cytoplasmic reorganization during the first cell cycle. Dev Biol. 1990;142:203–215. doi: 10.1016/0012-1606(90)90164-e. [DOI] [PubMed] [Google Scholar]
- Kovacevic I, Cram EJ. FLN-1/Filamin is required for maintenance of actin and exit of fertilized oocytes from the spermatheca in C. elegans. Dev Biol. 2010;347:247–257. doi: 10.1016/j.ydbio.2010.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacevic I, Orozco JM, Cram EJ. Filamin and phospholipase C-ε are required for calcium signaling in the Caenorhabditis elegans spermatheca. PLoS Genet. 2013;9:e1003510. doi: 10.1371/journal.pgen.1003510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb NJC, Fernandez A, Conti MA, Adelstein R, Glass DB, Welch WJ, Feramisco JR. Regulation of actin microfilament integrity in living nonmuscle cells by the cAMP-dependent protein kinase and the myosin light chain kinase. J Cell Biol. 1988;106:1955–1971. doi: 10.1083/jcb.106.6.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie TL, Dowell ML, Lakser OJ, Gerthoffer WT, Fredberg JJ, Seow CY, Mitchell RW, Solway J. Disrupting actin-myosin-actin connectivity in airway smooth muscle as a treatment for asthma? Proc Am Thorac Soc. 2009;6:295–300. doi: 10.1513/pats.200808-078RM. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Haase C, Deguchi S, Kaunas R. Cyclic stretch-induced stress fiber dynamics – dependence on strain rate, Rho-kinase and MLCK. Biochem Biophys Res Commun. 2010;401:344–349. doi: 10.1016/j.bbrc.2010.09.046. [DOI] [PubMed] [Google Scholar]
- Lenz M, Gardel ML, Dinner AR. Requirements for contractility in disordered cytoskeletal bundles. New J Phys. 2012:14, 033037. doi: 10.1088/1367-2630/14/3/033037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Huang NF, Hsu S. Mechanotransduction in endothelial cell migration. J Cell Biochem. 2005;96:1110–1126. doi: 10.1002/jcb.20614. [DOI] [PubMed] [Google Scholar]
- Mariol M, Walter L, Bellemin S, Gieseler K. A rapid protocol for integrating extrachromosomal arrays with high transmission rate into the C. elegans genome. J Vis Exp. 2013;82:e50773. doi: 10.3791/50773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason FM, Tworoger M, Martin AC. Apical domain polarization localizes actin-myosin activity to drive ratchet-like apical constriction. Nat Cell Biol. 2013;15:926–936. doi: 10.1038/ncb2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mccarter J, Bartlett B, Dang T, Schedl T. On the control of oocyte meiotic maturation and ovulation in Caenorhabditis elegans. Dev Biol. 1999;205:111–128. doi: 10.1006/dbio.1998.9109. [DOI] [PubMed] [Google Scholar]
- Mccarter J, Bartlett B, Thanh D, Schedl T. Soma–germ cell interactions in Caenorhabditis elegans: multiple events of hermaphrodite germline development require the somatic sheath and spermathecal lineages. Dev Biol. 1997;143:121–143. doi: 10.1006/dbio.1996.8429. [DOI] [PubMed] [Google Scholar]
- McMullan R, Nurrish SJ. The RHO-1 RhoGTPase modulates fertility and multiple behaviors in adult C. elegans. PLoS One. 2011;6:e17265. doi: 10.1371/journal.pone.0017265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meighan CM, Kelly VE, Krahe EC, Gaeta AJ. α Integrin cytoplasmic tails can rescue the loss of Rho-family GTPase signaling in the C. elegans somatic gonad. Mech Dev. 2015;136:111–122. doi: 10.1016/j.mod.2014.12.006. [DOI] [PubMed] [Google Scholar]
- Meijering E, Jacob M, Sarria JCF, Steiner P, Hirling H, Unser M. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry A. 2004;58:167–176. doi: 10.1002/cyto.a.20022. [DOI] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: extrachromosoal maintenance and integration of transforming sequences. EMBO. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaux G, Legouis R, Labouesse M. Epithelial biology: lessons from Caenorhabditis elegans. Gene. 2001;277:83–100. doi: 10.1016/s0378-1119(01)00700-4. [DOI] [PubMed] [Google Scholar]
- Miyazaki M, Chiba M, Eguchi H, Ohki T, Ishiwata S. Cell-sized spherical confinement induces the spontaneous formation of contractile actomyosin rings in vitro. Nat Cell Biol. 2015;17:480–489. doi: 10.1038/ncb3142. [DOI] [PubMed] [Google Scholar]
- Molloy JE, Burns JE, Kendrick-Jones J, Tregear RT, White DC. Movement and force produced by a single myosin head. Nature. 1995;378:209–212. doi: 10.1038/378209a0. [DOI] [PubMed] [Google Scholar]
- Murthy K, Wadsworth P. Myosin-II-dependent localization and dynamics of F-actin during cytokinesis. Curr Biol. 2005;15:724–731. doi: 10.1016/j.cub.2005.02.055. [DOI] [PubMed] [Google Scholar]
- Naumanen P, Lappalainen P, Hotulainen P. Mechanisms of actin stress fibre assembly. J Microsc. 2008;231:446–454. doi: 10.1111/j.1365-2818.2008.02057.x. [DOI] [PubMed] [Google Scholar]
- Nelson CM, Gleghorn JP. Sculpting organs: mechanical regulation of tissue development. Annu Rev Biomed Eng. 2012;14:129–154. doi: 10.1146/annurev-bioeng-071811-150043. [DOI] [PubMed] [Google Scholar]
- Ono K, Ono S. Tropomyosin and troponin are required for ovarian contraction in the Caenorhabditis elegans reproductive system. Mol Biol Cell. 2004;15:2782–2793. doi: 10.1091/mbc.E04-03-0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono K, Ono S. Two distinct myosin II populations coordinate ovulatory contraction of the myoepithelial sheath in the Caenorhabditis elegans somatic gonad. Mol Biol Cell. 2016;27:1131–1142. doi: 10.1091/mbc.E15-09-0648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono K, Yu R, Ono S. Structural components of the nonstriated contractile apparatuses in the Caenorhabditis elegans gonadal myoepithelial sheath and their essential roles for ovulation. Dev Dyn. 2007;236:1093–1105. doi: 10.1002/dvdy.21091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrin S, Mellor H. Actin stress fibres. J Cell Sci. 2007;120:3491–3499. doi: 10.1242/jcs.018473. [DOI] [PubMed] [Google Scholar]
- Persechini A, Hartshorne DJ. Phosphorylation of smooth muscle myosin: evidence for cooperativity between the myosin heads. Science. 1981;213:1383–1385. doi: 10.1126/science.6455737. [DOI] [PubMed] [Google Scholar]
- Peterson LJ, Rajfur Z, Maddox AS, Freel CD, Chen Y, Edlund M, Otey C, Burridge K. Simultaneous stretching and contraction of stress fibers in vivo. Mol Biol Cell. 2004;15:3751–3737. doi: 10.1091/mbc.E03-09-0696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piekny AJAJ, Mains PEPE. Rho-binding kinase (LET-502) and myosin phosphatase (MEL-11) regulate cytokinesis in the early Caenorhabditis elegans embryo. J Cell Sci. 2002;115:2271–2282. doi: 10.1242/jcs.115.11.2271. [DOI] [PubMed] [Google Scholar]
- Pilipiuk J, Lefebvre C, Wiesenfahrt T, Legouis R, Bossinger O. Increased IP3/Ca2+ signaling compensates depletion of LET-413/DLG-1 in C. elegans epithelial junction assembly. Dev Biol. 2009;327:34–47. doi: 10.1016/j.ydbio.2008.11.025. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Blanchoin L, Mullins RD. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu Rev Biophys Biomol Struct. 2000;29:545–576. doi: 10.1146/annurev.biophys.29.1.545. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Cooper JA. Actin, a central player in cell shape and movement. Science. 2009;326:1208–1212. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provenzano PP, Keely PJ. Mechanical signaling through the cytoskeleton regulates cell proliferation by coordinated focal adhesion and Rho GTPase signaling. J Cell Sci. 2011;124:1195–1205. doi: 10.1242/jcs.067009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezakhaniha R, Agianniotis A, Schrauwen JTC, Griffa A, Sage D, Bouten CVC, van de Vosse FN, Unser M, Stergiopulos N. Experimental investigation of collagen waviness and orientation in the arterial adventitia using confocal laser scanning microscopy. Biomech Model Mechanobiol. 2012;11:461–473. doi: 10.1007/s10237-011-0325-z. [DOI] [PubMed] [Google Scholar]
- Sears C, Kaunas R. The many ways adherent cells respond to applied stretch. J Biomech. 2016;49:1347–1354. doi: 10.1016/j.jbiomech.2015.10.014. [DOI] [PubMed] [Google Scholar]
- Seguchi O, Takashima S, Yamazaki S, Asakura M, Asano Y, Shintani Y, Wakeno M, Minamino T, Kondo H, Furukawa H, et al. A cardiac myosin light chain kinase regulates sarcomere assembly in the vertebrate heart. J Clin Invest. 2007;117:2812–2824. doi: 10.1172/JCI30804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellars JR, Pato MD, Adelstein RS. Reversible phosphorylation of smooth muscle myosin, heavy meromyosin, and platelet myosin. J Biol Chem. 1981;256:13137–13142. [PubMed] [Google Scholar]
- Somlyo AP, Somlyo AV. Signal transduction by G-proteins, rho-kinase and protein phosphatase to smooth muscle and non-muscle myosin II. J Physiol. 2000;522:177–185. doi: 10.1111/j.1469-7793.2000.t01-2-00177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somlyo AP, Somlyo AV. Ca2+ sensitivity of smooth muscle and nonmuscle myosin II: modulated by G proteins, kinases, and myosin phosphatase. Physiol Rev. 2003;83:1325–1358. doi: 10.1152/physrev.00023.2003. [DOI] [PubMed] [Google Scholar]
- Strome S. Fluorescence visualization of the distribution of microfilaments in gonads and early embryos of the nematode Caenorhabditis elegans. J Cell Biol. 1986;103:2241–2252. doi: 10.1083/jcb.103.6.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaine M, Numata O, Nakano K. An actin-myosin-II interaction is involved in maintaining the contractile ring in fission yeast. J Cell Sci. 2015;128:2903–2918. doi: 10.1242/jcs.171264. [DOI] [PubMed] [Google Scholar]
- Tan PY, Zaidel-Bar R. Transient membrane localization of SPV-1 drives cyclical actomyosin contractions in the C. elegans spermatheca. Curr Biol. 2015;25:141–151. doi: 10.1016/j.cub.2014.11.033. [DOI] [PubMed] [Google Scholar]
- Thoresen T, Gardel M. Reconstitution of actomyosin bundles. Biophys J. 2011;100:446a. doi: 10.1016/j.bpj.2011.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoresen T, Lenz M, Gardel ML. Thick filament length and isoform composition determine self-organized contractile units in actomyosin bundles. Biophys J. 2013;104:655–665. doi: 10.1016/j.bpj.2012.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoumine O, Ziegler T, Girard PR, Nerem RM. Elongation of confluent endothelial cells in culture: the importance of fields of force in the associated alterations of their cytoskeletal structure. Exp Cell Res. 1995;219:427–441. doi: 10.1006/excr.1995.1249. [DOI] [PubMed] [Google Scholar]
- Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- Tojkander S, Gateva G, Lappalainen P. Actin stress fibers—assembly, dynamics and biological roles. J Cell Sci. 2012;125:1855–1864. doi: 10.1242/jcs.098087. [DOI] [PubMed] [Google Scholar]
- Totsukawa G, Yamakita Y, Yamashiro S, Hartshorne DJ, Sasaki Y, Matsumura F. Distinct roles of ROCK (Rho-kinase) and MLCK in spatial regulation of MLC phosphorylation for assembly of stress fibers and focal adhesions in 3T3 fibroblasts. J Cell Biol. 2000;150:797–806. doi: 10.1083/jcb.150.4.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukita S, Yonemura S. Cortical actin organization: lessons from ERM (ezrin/radixin/moesin) proteins. J Biol Chem. 1999;274:34507–34510. doi: 10.1074/jbc.274.49.34507. [DOI] [PubMed] [Google Scholar]
- Uehata M, Ishizaki T, Satoh H, Ono T, Kawahara T, Morishita T, Tamakawa H, Yamagami K, Inui J, Maekawa M, et al. Calcium sensitization of smooth muscle mediated by a Rho-associated protein kinase in hypertension. Nature. 1997;389:990–994. doi: 10.1038/40187. [DOI] [PubMed] [Google Scholar]
- Verkhovsky AB, Borisy GG. Non-sarcomeric mode of myosin II organization in the fibroblast lamellum. J Cell Biol. 1993;123:637–652. doi: 10.1083/jcb.123.3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkhovsky AB, Svitkina TM, Borisy GG. Myosin II filament assemblies in the active lamella of fibroblasts: their morphogenesis and role in the formation of actin filament bundles. J Cell Biol. 1995;131:989–1002. doi: 10.1083/jcb.131.4.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Audhya A. In vivo imaging of C. elegans endocytosis. Methods. 2014;68:518–528. doi: 10.1016/j.ymeth.2014.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward S, Carrel JS. Fertilization and sperm competition in the nematode Caenorhabditis elegans. Dev Biol. 1979;73:304–321. doi: 10.1016/0012-1606(79)90069-1. [DOI] [PubMed] [Google Scholar]
- Wettschureck N, Offermanns S. Rho/Rho-kinase mediated signaling in physiology and pathophysiology. J Mol Med. 2002;80:629–638. doi: 10.1007/s00109-002-0370-2. [DOI] [PubMed] [Google Scholar]
- Wissmann A, Ingles J, Mains PE. The Caenorhabditis elegans mel-11 myosin phosphatase regulatory subunit affects tissue contraction in the somatic gonad and the embryonic epidermis and genetically interacts with the Rac signaling pathway. Dev Biol. 1999;209:111–127. doi: 10.1006/dbio.1999.9242. [DOI] [PubMed] [Google Scholar]
- Wozniak MA, Chen CS. Mechanotransduction in development: a growing role for contractility. Nat Rev Mol Cell Biol. 2009;10:34–43. doi: 10.1038/nrm2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto I, Kosinski ME, Greenstein D. Start me up: cell signaling and the journey from oocyte to embryo in C. elegans. Dev Dyn. 2006;235:571–585. doi: 10.1002/dvdy.20662. [DOI] [PubMed] [Google Scholar]
- Yan X, Xing J, Lorin-Nebel C, Estevez AY, Nehrke K, Lamitina T, Strange K. Function of a STIM1 homologue in C. elegans: evidence that store-operated Ca2+ entry is not essential for oscillatory Ca2+ signaling and ER Ca2+ homeostasis. J Gen Physiol. 2006;128:443–459. doi: 10.1085/jgp.200609611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X, Gower NJD, Baylis HA, Strange K. Inositol 1,4,5-trisphosphate signaling regulates rhythmic contractile activity of myoepithelial sheath cells in Caenorhabditis elegans. Mol Biol Cell. 2004;15:3938–3949. doi: 10.1091/mbc.E04-03-0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidel-Bar R, Zhenhuan G, Luxenburg C. The contractome—a systems view of actomyosin contractility in non-muscle cells. J Cell Sci. 2015;128:2209–2217. doi: 10.1242/jcs.170068. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.