Micromanipulation force measurements of single isolated nuclei at physiological strains and strain rates reveal two distinct cell nuclear mechanical regimes differentially governed by chromatin and lamin A. Chromatin and its histone-modification compaction govern short extension, and the lamin A amount dictates long-extension strain stiffening.
Abstract
The cell nucleus must continually resist and respond to intercellular and intracellular mechanical forces to transduce mechanical signals and maintain proper genome organization and expression. Altered nuclear mechanics is associated with many human diseases, including heart disease, progeria, and cancer. Chromatin and nuclear envelope A-type lamin proteins are known to be key nuclear mechanical components perturbed in these diseases, but their distinct mechanical contributions are not known. Here we directly establish the separate roles of chromatin and lamin A/C and show that they determine two distinct mechanical regimes via micromanipulation of single isolated nuclei. Chromatin governs response to small extensions (<3 μm), and euchromatin/heterochromatin levels modulate the stiffness. In contrast, lamin A/C levels control nuclear strain stiffening at large extensions. These results can be understood through simulations of a polymeric shell and cross-linked polymer interior. Our results provide a framework for understanding the differential effects of chromatin and lamin A/C in cell nuclear mechanics and their alterations in disease.
INTRODUCTION
Nuclear mechanical response—the way in which the cell nucleus deforms and reacts to external forces—is essential to basic cell biological functions as diverse as migration, differentiation, and spatial ordering and regulation of genes (Butin-Israeli et al., 2012; Isermann and Lammerding, 2013). Consequently a number of major human diseases exhibit perturbed nuclear mechanics. Experiments have shown that a meshwork of intermediate filaments composed of lamin protein just inside the nuclear envelope is a crucial mechanical component of the nucleus (Dahl et al., 2004; Lammerding et al., 2006; Ho et al., 2013; Swift et al., 2013). Further experiments suggest that the chromatin that fills the nucleus acts as a viscoelastic component (Pajerowski et al., 2007; Mazumder et al., 2008; Furusawa et al., 2015; Schreiner et al., 2015). However, the distinction between the roles of lamins and chromatin in nuclear mechanical response has not been resolved. Because both lamins and chromatin are altered in diseases with nuclear disruption (Bank and Gruenbaum, 2011; Butin-Israeli et al., 2012), it is crucial to elucidate and disentangle their contributions to nuclear mechanics before their respective roles in disease processes can be understood.
A variety of human diseases are associated with aberrant cell nuclear mechanics and morphology. For instance, in laminopathies—diseases primarily caused by mutations in the LMNA gene (which codes lamin A and lamin C)—cell nuclei have altered mechanical strength and display nuclear blebs (Goldman et al., 2004; Dahl et al., 2006; Scaffidi and Misteli, 2006; Verstraeten et al., 2008; Swift et al., 2013; Zwerger et al., 2013; Booth et al., 2015). Lamin mutations also cause or parallel many major diseases and conditions, including aging, heart disease, and muscular dystrophy. Intriguingly, chromatin is also altered in these situations, as exhibited by elevated levels of decondensed euchromatin and/or loss of peripheral compact heterochromatin (Sullivan et al., 1999; Shumaker et al., 2006; Taimen et al., 2009; Butin-Israeli et al., 2012; Swift et al., 2013; Booth et al., 2015). Similarly, in many cancers, the levels of both lamins and euchromatin/heterochromatin are altered (Bank and Gruenbaum, 2011). However, because the roles of these two components have not been disentangled, the specific contributions of lamin A and chromatin to abnormal cell nuclear mechanics remain unclear.
Because the amount of lamin A correlates with both nuclear and tissue stiffness (Swift et al., 2013), experiments have primarily focused on lamin A as the main mechanical element. However, lamin A/C is not essential, and many cells have little or no lamin A/C (Butin-Israeli et al., 2012; Isermann and Lammerding, 2013). Furthermore, depletion of lamin A/C results in only an ∼50% decrease in nuclear stiffness, which suggests that other nuclear mechanical components account for the remaining strength (Lammerding et al., 2006; Pajerowski et al., 2007).
Early experiments, which used nonphysiological salt conditions to swell or compact chromatin, indirectly showed that chromatin is also involved in nuclear mechanical response (Dahl et al., 2005; Pajerowski et al., 2007; Mazumder et al., 2008). However, at physiological salt conditions, it was reported that chromatin merely flows when the nucleus is extended (Pajerowski et al., 2007). Thus chromatin is usually considered a secondary or minor viscous component. More recent experiments showed that disruption of LINC complexes connecting chromatin to the nuclear periphery in yeast (Schreiner et al., 2015) and overexpression of HMGN5, a high-mobility chromatin protein, in mammals (Furusawa et al., 2015) enhance nuclear deformations in vivo. However, the physiologically relevant contribution of chromatin and its histone modification–mediated compaction (euchromatin/heterochromatin) to whole nuclear mechanics has not been established quantitatively in a direct manner.
Previous experiments exploring cell nuclear mechanics did not separate the contributions of chromatin and lamins to whole nuclear response to physiological strains and strain rates. Atomic force microscopy measures a localized, compressive force (Schape et al., 2009), and micropipette aspiration experiments measure the mechanical response to a nonphysiological, sudden applied extensional stress to a small area. The latter, although widely used, typically produces large (>100%) and nonuniform strain (Vaziri and Mofrad, 2007), resulting in irreversible nuclear deformations (Pajerowski et al., 2007) in which static and dynamic (viscoelastic flow) effects are mixed. Magnetic beads attached to proteins in the nuclear envelope interrogate pathway-specific responses to localized small (∼300 nm) deformations (Guilluy et al., 2014). On the other hand, substrate-strain experiments stretch the entire nucleus, but force response measurements are confounded by the deformation and response of the whole cell (Caille et al., 1998).
Although these methods have successfully measured viscoelastic properties of the nucleus and identified lamin A/C and, indirectly, chromatin as important mechanical elements, they have not established a clear distinction between their contributions to mechanics. An alternative and complementary technique is micromanipulation, which can probe uniform physiological extensions over both short and long length scales and precisely controlled strain rates. In addition, micromanipulation excels at differentiating the roles of mechanical components of micrometer-sized structures because it allows for repeatable force measurements coupled with simultaneous biochemical treatments. This technique has been successfully used to separate the mechanical contributions of DNA and proteins in isolated mitotic chromosomes (Poirier et al., 2000; Poirier and Marko, 2002; Kawamura et al., 2010). Thus we tailored micromanipulation force measurement techniques to investigate the differential contributions of chromatin and lamin A/C in cell nuclear mechanics.
Here we report the use of our novel micromanipulation methods to isolate and gradually stretch individual cell nuclei at physiological speeds to physiological strains in a repeatable and reproducible manner to perform nuclear force-extension measurements. Our approach facilitates the study of the roles of specific molecules in nuclear mechanics. We identify for the first time the differential contributions of chromatin and lamin A/C to cell nuclear mechanical response. First, we find that the force response of the whole nucleus exhibits two deformation regimes. Then, by biochemically treating isolated nuclei with DNA-digesting enzymes, we demonstrate that chromatin is a major mechanical component that is required for maintaining nuclear morphology and resisting small deformations. Using physiologically relevant changes in histone modifications, we confirm that chromatin compaction determines the short-extension force response, whereas depletion of lamin A/C does not contribute significantly to this regime. Instead, we find that lamin A/C levels control the ability of cell nuclei to strain stiffen when subjected to large deformations. We thus propose a simple model for nuclear mechanical response in which chromatin and lamin A determine two different force response regimes of the nucleus, providing a framework for deducing their respective mechanical roles in cell functionality and human disease.
RESULTS
Individual microdissected nuclei stretched by micromanipulation exhibit strain stiffening
To measure the distinct contributions of chromatin and lamin A/C to nuclear mechanics, we developed a nucleus micromanipulation and force measurement technique. Our method was adapted from a microdissection and micromanipulation technique that has been used successfully to study the mechanics of mitotic chromosomes and the differential roles of proteins and DNA in these chromosomes (Poirier et al., 2000; Poirier and Marko, 2002; Kawamura et al., 2010; see Materials and Methods). Briefly, a single nucleus is isolated from a living mammalian cell in cell culture medium by cytoskeletal disruption and lysis (Figure 1A); this avoids harsh treatments, such as hypertonic swelling, shearing, and/or prolonged exposure to detergents used during bulk nuclei isolation procedures.
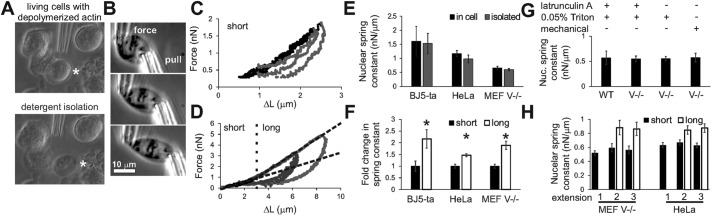
FIGURE 1:
Whole-nuclear force measurements of individual isolated nuclei are reproducible and exhibit strain stiffening. (A) HeLa nucleus isolated from a latrunculin A–treated cell (asterisk) via locally lysing with 0.05% Triton X-100. (B) Images of a representative HeLa experiment in which the “pull” pipette stretches the nucleus while the deflection of the calibrated “force” pipette measures force. Representative triplicate measurements for HeLa nuclei, showing (C) short and (D) long extensions and relaxations displaying two different linear regimes. Graphs of human BJ5-ta, HeLa, and mouse MEF vimentin-null (V−/−) cell nuclei (E) short-extension spring constants measured for in-cell (black) and isolated (gray) nuclei for short extensions of 3 µm and (F) relative change in the nuclear spring constants from short (<3 µm) to long (>3 µm) extensions of isolated nuclei. (G) MEF WT and V−/− short-extension nuclear spring constants after different isolation techniques. Note that V−/− nuclei can be isolated without actin depolymerization via latrunculin A. (H) Short- and long-extension nuclear spring constant averages for the first, second, and third successive extensions of the nucleus for both MEF V−/− and HeLa nuclei. Error bars denote SEM. n = 10–25. *p < 0.05.
Once suspended between pipettes, the whole nucleus is then stretched by moving a “pull” pipette attached to one end of the nucleus while force is measured by the deflection of a calibrated “force” pipette attached to the opposite end (Figure 1B). The nucleus is stretched and relaxed at a physiologically relevant speed of 50 nm/s, which is within the range of typical nuclear movement (Luxton et al., 2010) and cell migration (Harris et al., 2012). Similar to experiments with isolated chromosomes (Poirier et al., 2000; Poirier and Marko, 2002; Kawamura et al., 2010), this facilitates repeated extension and reproducible nuclear mechanics measurements with no detectable change in the chromatin or lamin content, as shown by fluorescence images (Figure 1,C, D, and H, and Supplemental Figure S1, G–I). Note that in this type of experiment, the strain is well controlled and nearly uniform across the whole nucleus, as opposed to aspiration experiments, in which there is large strain in the aspirated region and little strain in the nucleus region exterior to the aspirating pipette (Pajerowski et al., 2007; Vaziri and Mofrad, 2007). Furthermore, this allows measurement of mechanical response of an individual nucleus before and after biochemical treatment (see following sections).
Using this technique, we find that the nucleus exhibits a clear two-regime mechanical response to strain. First, we probed short nuclear extensions of <3 µm (∼30% strain), which correspond to the typical small strains experienced by nuclei in vivo (Kaminski et al., 2014). Nuclei extended short distances exhibit a linear force response (Figure 1C). We then extended nuclei greater distances (>3 µm) to probe the force response to the larger strains typically experienced in mechanically demanding tissue environments or during cell migration (Friedl et al., 2011; Wolf et al., 2013). As the nuclei are extended further, they stiffen under strain, as shown by the larger nuclear spring constant for larger deformations (stiffening for extensions >3 µm; Figure 1D). Strain stiffening was consistently observed as a 1.5- to 2.5-fold increase in the nuclear spring constant from short to long extensions for a variety of cells, indicating that this effect is not cell-type specific (human HeLa, human fibroblasts BJ5-ta, and mouse MEF vimentin null [MEF V−/−]; Figure 1F; also see later discussion of Figures 3 and 4).
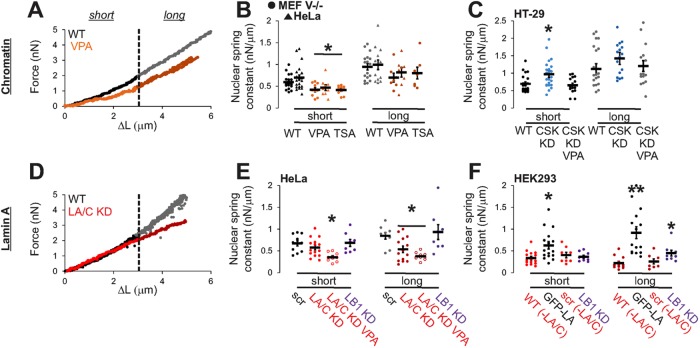
FIGURE 3:
Alterations to chromatin histone modifications and lamin A/C levels reveal differing contributions to nuclear mechanics. Nuclear spring constants for short (<3 µm) and long extensions (>3 µm) upon altering the histone modification state of chromatin or lamin protein levels. Representative force–extension plots for decreased (A) chromatin compaction and (D) lamin A/C. Nuclear spring constants upon modulating chromatin by (B) increasing euchromatin levels via pretreatment with VPA or TSA (orange) in MEF V−/− and HeLa cells or (C) increasing heterochromatin (HT-29 CSK KD; blue) and subsequently decreasing heterochromatin with VPA (CSK KD VPA). Nuclear spring constants upon modulating lamin levels in (E) HeLa cells with high lamin A/C levels and (F) HEK293 cells with low lamin A/C levels, denoted as –LA/C, via shRNA knockdown or overexpression. Red data represent nuclei with knockdown/low levels of lamin A/C protein; purple represents lamin B1 knockdown. For level change validations, see Supplemental Figures S3 and S4. Nucleus dimensions are similar for all nuclei except HEK293 WT and HeLa LB1 KD (p < 0.05); see Supplemental Table S1. Error bars denote SEM. n = 8–30. *,**p < 0.05, with different numbers of asterisks denoting statistically significant differences.
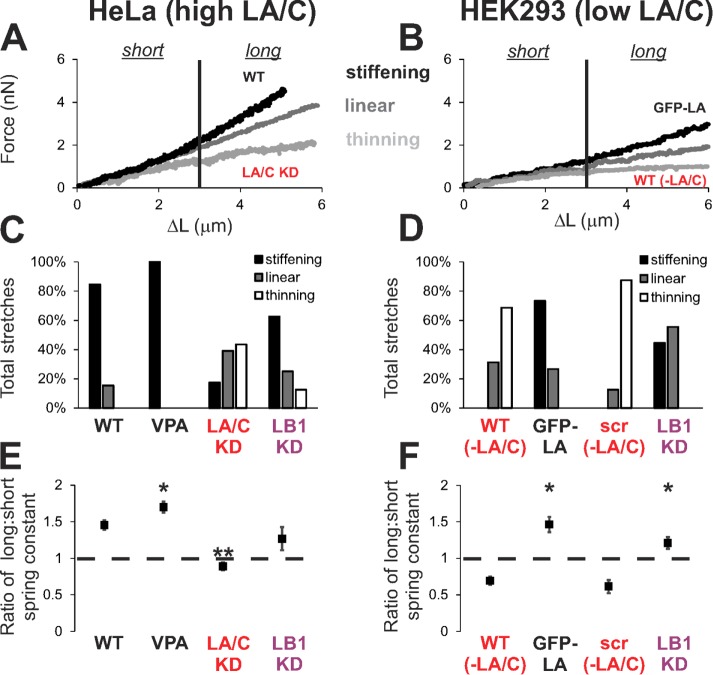
FIGURE 4:
Lamin A levels control strain-stiffening response, and loss of lamin A/C can lead to strain thinning. Representative force–extension plots displaying strain-stiffening (black), linear (gray), and strain-thinning (light gray) response for (A) HeLa nuclei with high lamin A/C levels and (B) HEK293 nuclei with low lamin A/C levels, denoted as –LA/C. Percentage of events displaying each behavior for (C) HeLa and (D) HEK293 for WT and different treatments (n = 8–25). The ratio of the nuclear spring constant for long extension (>3 µm) to that for short extension was used to identify strain-stiffening (fold change >1.2), linear (0.8–1.2), and strain-thinning (<0.8) response for each nucleus. Average ratios of long- to short-extension nuclear spring constants are shown for (E) HeLa and (F) HEK293. *,**p < 0.05, with different numbers of asterisks denoting statistically significant differences.
Typically, we observe nuclear spring constants on the order of nanonewtons/micrometer, consistent with atomic force microscopy (Schape et al., 2009) and micropipette aspiration experiments (Dahl et al., 2005). Although the nucleus exhibits hysteresis as it returns to rest length, after relaxation, successive stretches of the same nucleus do not alter nuclear force response to short and long extensions (Figure 1, C, D, and H). This differs from recent experiments showing adaptation of mechanical linkages (nesprin) in the nuclear envelope (Guilluy et al., 2014), but this is not surprising, considering the differences in scale of speeds (and time), lengths (and areas), and forces probed in our experiments. Finally, we note that our stretching experiments are quasistatic because we do not observe significant relaxation when the nucleus is held at maximal extension for several minutes (Supplemental Figure S1, A–F), and stretching the nucleus at the slower speed of 15 nm/s produces a similar force response (Supplemental Figure S1J). This suggests that “creep” effects observed in previous aspiration experiments are specific to that technique, which involves continuous hydrodynamic flow to distort nuclei (Rowat et al., 2006; Pajerowski et al., 2007; Vaziri and Mofrad, 2007). Thus, by microdissection and micromanipulation, we can reliably perform repeated mechanical measurements on individual isolated nuclei and find an intriguing two-regime force response.
Nucleus isolation does not perturb mechanical response
To assess the in vivo relevance of our technique, we compared force measurements of isolated nuclei to those of nuclei remaining in intact cells. Initial spring constants for nuclei in vivo and isolated nuclei are similar for BJ5-ta, HeLa, and MEF V−/− cells (Figure 1E), consistent with previous findings (Dahl et al., 2005; Rowat et al., 2005). Furthermore, comparison of the short-extension nuclear spring constant was similar for varied nucleus extractions methods in wild-type (WT) and V−/− MEFs, in which the latter does not require actin depolymerization for nucleus isolation (Figure 1G; see Materials and Methods). Although there may be active forces due to chromatin remodeling in vivo, the results suggest that these forces are secondary in mechanical response. We conclude that the mechanics of isolated nuclei are reasonably representative of the in vivo force response of the nucleus.
Chromatin dominates the small-strain force response regime
To determine the origins of the two regimes of nuclear mechanical response (to small and large deformations), we directly probed the role of chromatin by biochemically altering isolated nuclei from HeLa (human) or MEF V−/− (mouse) cells. We isolated and stretched nuclei up to ∼3 µm to obtain the nuclear spring constant for short extensions. After obtaining the native nuclear spring constant, we treated the nucleus with 10 mM MgCl2 via a “spray” micropipette. The nucleus visibly shrank and the nuclear spring constant increased approximately threefold upon compaction by MgCl2 (Figure 2, A, B, and E), consistent with previous experiments (Pajerowski et al., 2007). At 10-min posttreatment, after Mg2+ ions diffused away into the experimental well, the nucleus returned to its native shape and spring constant (Figure 2, B and E). To confirm directly the contribution of chromatin to this small strain response, we specifically induced chromatin stiffening via the DNA intercalator propidium iodide (PI; Vladescu et al., 2007; Banerjee et al., 2014). Treatment of the nucleus with PI similarly stiffened the initial mechanical response approximately fourfold (1 µg/ml PI; Figure 2, C–E). Pretreatment with the mild intercalator Hoechst also enhanced nuclear force resistance, although the effect was less than that of PI (Supplemental Figure S2, A and B). In both experiments, nuclei stretched >3 µm still exhibited WT-like strain stiffening (Supplemental Figure S2, A–C). Thus chromatin compaction or stiffening can enhance nuclear resistance to deformation, especially for small strains.
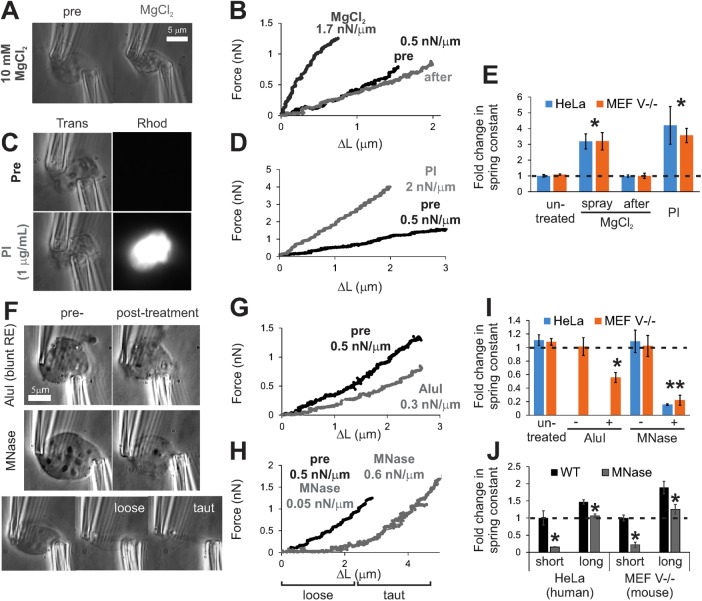
FIGURE 2:
Biochemical compaction or digestion of chromatin alters nuclear mechanical response. Images of isolated MEF V−/− nuclei before and after chromatin compaction by (A) MgCl2, (C) PI, or (F) digestion by restriction enzyme AluI or micrococcal nuclease (MNase). Visible chromatin digestion is noticeable upon MNase treatment. Example MEF V−/− nucleus force–extension plots for chromatin compaction via (B) MgCl2 or (D) PI and chromatin digestion by (G) AluI or (H) MNase. Graphs of fold change in short-extension nuclear spring constants for HeLa (human; blue) and MEF V−/− nuclei (mouse; orange) upon chromatin (E) compaction or (I) digestion. (J) Fold change for both short-extension (<3 µm) and long-extension (>3 µm) relative nuclear spring constants. See Supplemental Figure S2 for example of long-extension force extension plots for PI and AluI. Error bars denote SEM. n = 4–6. *p < 0.05.
Having found that chromatin can bolster nuclear mechanical response for short deformations, we sought to determine whether chromatin is the dominant component of the response. To address this question, we used AluI restriction endonuclease and micrococcal nuclease (MNase) to digest chromatin by cutting at low and high frequency along chromatin, respectively. AluI, a 4–base pair blunt endonuclease, cuts chromatin every few kilobases, primarily between nucleosomes (Polach et al., 2000). The nucleus is not visibly altered by 5 min of AluI treatment, but the nuclear spring constant decreases nearly 50% for small strains (Figure 2, F, G, and I). Intriguingly, the spring constant is nearly unaltered for larger extensions (Supplemental Figure S2D). On extensive chromatin digestion by 1 min of MNase treatment, nuclear morphology visibly changed and the ability of the nucleus to resist small deformations decreased drastically (Figure 2, F and H–J). Strikingly, the spring constant increased dramatically at large extensions, as in untreated nuclei, although the long-extension spring constant decreased compared with WT (Figure 2, H and J). This response, in the absence of chromatin, suggests that another remaining nuclear component, such as lamin A/C (Supplemental Figure S2, E–G), is responsible for nuclear resistance to large deformations. This suggests that the lamin shell is not a stiff cage but instead a flexible bag that is supported and shaped by the chromatin inside it. In turn, chromatin governs nuclear morphology and short-extension nuclear force response, with a moderate secondary role at long extensions.
Chromatin compaction and decompaction by alterations to histone modifications modulate nuclear mechanical response to short extensions
To decouple the mechanical contributions of chromatin from lamins in a physiologically relevant manner, we used drugs and knockdowns to control histone acetylation and methylation and thus chromatin compaction. Nuclei from MEF V −/− and HeLa cells were stretched 6 µm in order to measure both the short- and long-extension nuclear spring constants, with 3 µm (∼30% strain) providing a reliable transition between regimes (Figure 3, A and D). The histone deacetylase (HDAC) inhibitors valproic acid (VPA; Marchion et al., 2005) and trichostatin A (TSA; Yoshida et al., 1990) were used to increase euchromatin levels, thus decondensing chromatin (Supplemental Figure S3, A–C). Chromatin decompaction resulted in a decrease in mean nuclear stiffness at small extensions (from 0.60 to 0.41 nN/µm in MEF V−/− and from 0.70 to 0.47 nN/µm in HeLa) but no statistically significant change in mechanical response to larger deformations (Figure 3, A and B).
To contrast these results with the effects of increased heterochromatin and chromatin compaction, we used the HT-29 human colon cancer cell line in unmodified form (HT-29 WT) and a clinically more aggressive form with a c-Src tyrosine kinase (CSK) knockdown (HT-29 CSK KD), which is a well-characterized cell model of heterochromatinization (Kunte et al., 2005; Stypula-Cyrus et al., 2013). HT-29 CSK KD nuclei display overexpression of HDACs, increased levels of compact heterochromatin, and increased nuclear density relative to the HT-29 WT parental cell line (Supplemental Figure S3, D–F; Stypula-Cyrus et al., 2013). For these nuclei, the spring constant increased significantly for short extensions compared with HT-29 WT and moderately, but not significantly, at larger extensions (Figure 3C, blue dots). The treatment of HT-29 CSK KD with the HDAC inhibitor VPA reduces the high level of chromatin compaction (Stypula-Cyrus et al., 2013) and results in the recovery of lower, WT-level values of the short nuclear spring constant (Figure 3C, CSK KD VPA; Supplemental Figure S3, D–F). Although changes in the long-extension spring constant were not significant, the general trend suggests that chromatin has a secondary role in nuclear force resistance to large strains. These data show that, consistent with our prior biochemical studies, alterations to chromatin compaction through disease-mimicking alterations of histone modification levels determine mechanical response to small deformations and have a secondary role in the large-strain response.
Lamin A dominates the large-strain force response
Previous studies implicate lamin A/C as a major determinant of nuclear stiffness (Lammerding et al., 2006; Swift et al., 2013), but our experiments suggest that lamin A/C is not the sole generator of nuclear force resistance for small strains. We hypothesized that lamin A/C in fact is the major determinant of the force response to large deformations. To elucidate its specific role in whole nuclear mechanical response, we studied the mechanics of HeLa nuclei with altered levels of lamin A/C. We transfected HeLa cells with a short hairpin RNA (shRNA) lamin A/C knockdown (KD) construct with a green fluorescent protein (GFP) reporter (Supplemental Figure S4, A–C, ∼80% KD). Lamin A/C knockdown did not significantly affect nuclear stiffness at small strains compared with WT or the scrambled (scr) shRNA control (WT 0.70 ± 0.06 and scr 0.67 ± 0.06 vs. LA/C KD 0.58 ± 0.05 nN/µm, p > 0.05; Figure 3, D and E). However, depletion of lamin A/C reduced stiffness at large strains (from 0.85 to 0.54 nN/µm), resulting in a linear or even strain-thinning response, in contrast to the strain-stiffening response displayed by WT nuclei (Figures 3, D and E, and 4, A, C, and E). Although it is known that lamin A/C levels may perturb chromatin architecture (Bank and Gruenbaum, 2011), note that lamin A/C knockdown increased euchromatin by ∼20%, but this change is much smaller than the 100–200% increase upon HDACi treatment (Supplemental Figures S3, A–C, and S4B). Thus lamin A/C does not significantly contribute to short-extension force response but instead is a major contributor to resistance at long extensions.
To determine whether chromatin governs short-extension force response even upon loss of lamin A/C, we treated HeLa lamin A/C–knockdown cells with VPA. As expected, VPA treatment significantly decreased the short-extension nuclear spring constant in lamin A/C–depleted nuclei (Figure 3E). Consistent with lamin A/C depletion results in untreated cells, nuclei with lamin A/C knockdown and VPA treatment also displayed a decrease in long-extension nuclear spring constants, resulting in a loss of strain stiffening (Figure 3E). The separate and combined disruption of each mechanical component further confirms the differential mechanical roles of chromatin-governed short-extension and lamin A/C–governed long-extension force response.
To verify that strain stiffening is controlled by the level of lamin A/C, we measured force response in cells with inherently low levels of lamin A/C. Similar to HeLa lamin A/C knockdown, the low levels of lamin A/C in WT HEK293 nuclei result in strain thinning and plastic deformation upon stretching (Figures 3F and 4, B, D, and F, and Supplemental Figure S4, D–F and H). To verify further that lamin A modulates strain stiffening, we increased levels of lamin A in HEK293 cells via ectopic expression of GFP-lamin A (Supplemental Figure S4, D and E). HEK293 GFP-lamin A nuclei displayed strain stiffening and reproducible stretching along with a moderately increased nuclear spring constant for short extensions (Figures 3F and 4, B, D, and F, and Supplemental Figure S4F). These data demonstrate that lamin A/C is essential for strain stiffening and reproducible stretching, with a secondary role in determining the initial spring constant.
Lamin B1 depletion stiffens nuclei lacking lamin A/C
To determine whether lamin B1 contributes to nuclear mechanical response, we performed nuclear stretching experiments on nuclei depleted of lamin B1. Although lamin B1 is not regarded as a contributor to mechanical response (Lammerding et al., 2006), it has been reported that the stoichiometric ratio of lamin A:B determines nuclear rigidity (Swift et al., 2013). In our experiments, depletion of lamin B1 in HeLa cells, which have high levels of lamin A/C, did not induce a significant change in either the short or long nuclear spring constant (Figure 3E, purple dots), consistent with previous studies (Lammerding et al., 2006). However, depletion of lamin B1 in HEK293 cells, which have low levels of lamin A/C, resulted in an increased nuclear spring constant for long extensions (WT 0.22 ± 0.04 nN/µm vs. LB1 KD 0.45 ± 0.06 nN/µm, p < 0.05; Figure 3F). The increased long-extension spring constant in this case resulted in linear or strain-stiffening force response, a drastic change from the majority of low-level lamin A/C HEK293 WT nuclei, which exhibited strain thinning (Figure 4, B, D, and F). These results are consistent with micropipette aspiration data, which measured increased nuclear stiffness upon depletion of lamin B1 from proerythroblasts with low levels of lamin A/C (Shin et al., 2013). Thus we find that the long-extension force response, but not the short-extension response, is affected by lamin B1 levels, specifically for nuclei with low levels of lamin A.
A simple simulation model recapitulates the observations of separate chromatin- and lamin-A/C–dependent mechanical responses
To understand the mechanical response of stretched nuclei, we considered minimalistic models for cell nucleus mechanics. The nonlinear force response shown in Figures 1– 4 rules out a purely linear elastic model for the nucleus. Instead, our experiments demonstrate that cell nuclei exhibit a characteristic force response composed of two regimes (Figure 1D). For small deformations, extension is linear in applied force and the stiffness is primarily determined by chromatin (Figures 2 and 3, A–C). For highly stretched nuclei, the force response typically stiffens, depending on the level of lamin A/C (Figures 3, D–F, and 4).
Thus we developed a semiquantitative two-component simulation model composed of a polymeric shell representing the lamina (Shimi et al., 2015) and a cross-linked polymer to model the chromatin interior (Figure 5A, insets, green and blue, respectively). Because intermediate filaments have a low bending modulus and are elastically extensible (Panorchan et al., 2004; Koster et al., 2015; Mahamid et al., 2016), we connect nodes in the polymeric shell with extensible springs with zero bending modulus. To model chromatin, the shell is filled with a cross-linked polymer, which is also physically linked to the shell. This model for chromatin is based on experiments showing spatially correlated motions and interactions between chromatin domains (Fullwood et al., 2009; Lieberman-Aiden et al., 2009; Zidovska et al., 2013). To capture the results of the micropipette manipulation experiments with reasonable quantitative accuracy, we use the experimentally motivated parameters given in Table 1. Model details and parameters are described further in Methods and Materials.
FIGURE 5:
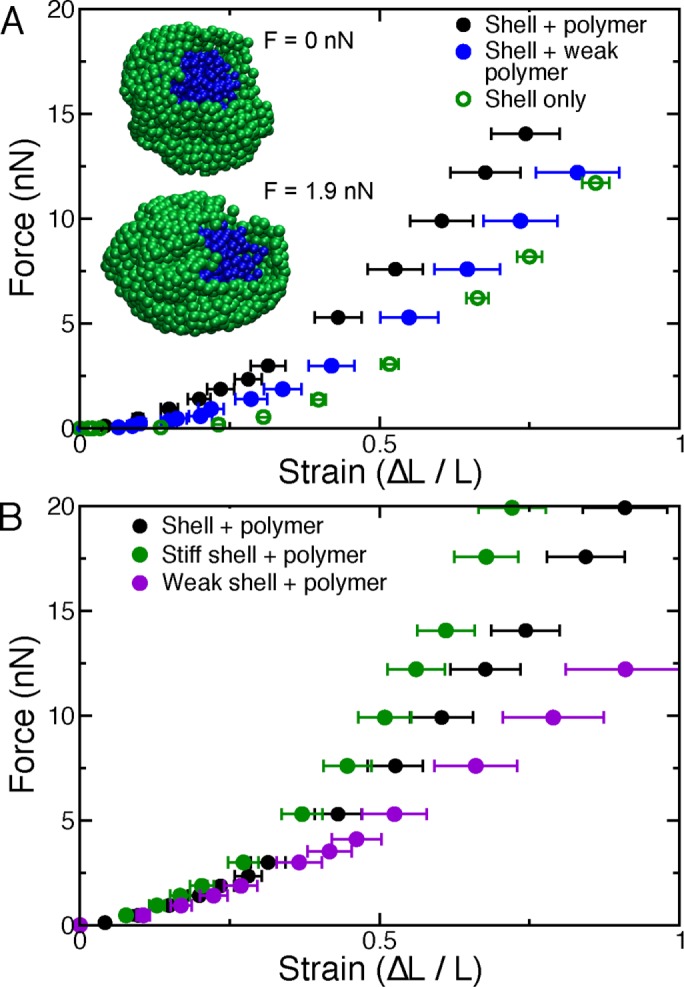
Polymeric shell with polymer interior model of the nucleus exhibits the experimentally observed two-regime mechanical response. (A) A model nucleus, composed of a polymeric shell (insets, green) and a cross-linked polymer interior (insets, blue), which is physically linked to the shell, stretches, and buckles when it is uniaxially stressed by forces exerted on its polar subunits. The force–extension relation for a typical model nucleus (solid black circles) exhibits two approximately linear force–response regimes and strain stiffening. Model nuclei lacking the interior polymer are highly extensible under small strains but stiffer for large deformations (open green circles). Model nuclei with a decondensed polymer filling, modeled by softer polymeric potentials (kp = 0.32 nN/μm), exhibit a weaker response to small deformations (ΔL/L < 0.3), but stiffness (slope of the force–extension curve) is not markedly altered for large strains (solid blue circles). (B) Compared to a typical model nucleus (solid black circles), simulated nuclei with shell monomers connected by stiff springs (k = 1.6 nN/μm; solid green circles) have a stiffer force response, most notably at large extensions. Model nuclei with a polymer filling but weaker shell springs (k = 0.32 nN/μm), deform more easily than typical model nuclei, especially for large strains (solid purple circles).
TABLE 1:
Values of parameters used and variables calculated in the simulation model.
| Parameter/variable | Standard simulation value | Approximate experimental value | Reference |
|---|---|---|---|
| System properties | |||
| Nuclear size, 2R (μm) | 10 | 10–15 | Present work |
| Viscosity, η (cP) | ∼1 | 1 | Present work |
| Duration of pulling, τ (s) | ∼30 | 100–200 | Present work |
| Maximum force, F (nN) | 15 | 7 | Present work |
| Lamina properties | |||
| Nodes in lamina, N | 1000 | 1000–3000 | Shimi et al. (2015) |
| Node connectivity, z | ∼4.5 | 4 | Shimi et al. (2015) |
| Lamin filament length, (4πR2/N)1/2 (μm) | 0.6 | 0.4 | Shimi et al. (2015) |
| Lamin A/C filament stiffness, k (nN/μm) | 0.8 | 6 | Mahamid et al. (2016) |
| Chromatin properties | Interpretation | |
|---|---|---|
| Number of subunits in polymer, Np | 552 | 1 subunit ≈ 1–10 Mbp |
| Subunit diameter, σp (μm) | 0.6 | Mbp domain size |
| Intersubunit spring constant, kp (nN/μm) | 1.6 | Mbp domain Young’s modulus ≈ 1 kPa (estimated from Guilak et al., 2000; de Vries et al., 2007; Pajerowski et al., 2007; present work) |
| Number of cross-links, Nc | 55 | 20% of domains physically linked to a distant domain |
| Number of links to shell, Ns | 40 | ∼50% of subunits in outer nucleus (near the shell) physically linked to the shell |
First, we study the properties of the shell in the absence of the interior cross-linked polymer. Owing to its weak entropic and bending elasticity, an empty shell stretches easily when subject to small forces (Figure 5A, green open circles). However, consistent with the behavior of lamin networks (Panorchan et al., 2004), the shell stiffens at large forces (Figure 5A, green open circles) as the bonds within the shell stretch. Note that because the model shell is composed of linear springs, stiffening arises due to the geometrical effects of axially stretching a sphere. The observed response qualitatively resembles the experimental observations of nuclei treated with the chromatin-digesting enzyme MNase, which stretch easily until the lamina becomes visibly taut (Figure 2, F and H).
To understand the mechanical role of chromatin, we next studied the properties of the model including the cross-linked polymer. Filling the shell with the cross-linked polymer increases the small-extension nuclear spring constant (by a factor of ∼10) without strongly affecting the large-extension response, as chromatin does for experimental nuclei (Figure 5A, black solid circles). Thus together the two components comprise a basic framework for understanding the different force response regimes observed experimentally.
The model provides qualitative predictions for how alterations to the two mechanical components may affect force response. For instance, a decondensed interior polymer, modeled by relaxing the physical constraints on the polymeric subunits, results in a weaker initial spring constant, similar to measurements of nuclei with chromatin decompaction via HDAC inhibitors (Figures 5A, blue solid circles, and 3, A and B). Note that the polymer must be cross-linked to itself and the polymeric shell in order to exhibit this interior-dependent behavior. This is consistent with experiments showing that chromatin tethering to the nuclear envelope is critical to robust nuclear mechanical response in yeast (Schreiner et al., 2015).
The large-strain response is dominated by the microscopic properties of the shell, as shown by simulations with weak and stiff shell springs (Figure 5B, purple and green solid circles, respectively). This is consistent with experiments manipulating lamin A levels (Figures 3, D–F, and 4). In addition, the transition between the different response regimes in the model occurs at ∼30% strain as in experimental measurements (∼10-µm nucleus transitions at 3 µm). This suggests that the geometry of axially deformed elastic shells is sufficient to explain lamin-dependent nuclear strain stiffening, although we do not rule out possible contributions from nonlinear material properties of chromatin and the lamina. Of interest, the model nucleus buckles and ruffles at large strains, similar to previous experimental observations (Dahl et al., 2005; Rowat et al., 2005; inset to Figure 5A). Thus, with just two basic components—the lamin network and the chromatin interior—we can understand typical nuclear force measurements (Figure 1D) and a variety of biological perturbations (Figures 2– 4).
DISCUSSION
The nucleus houses and protects the genome while directing mechanotransduction throughout the cell and dictating gene organization and expression. We developed and used a technique to apply mechanical stresses to individual cell nuclei via uniform stretching and differentiated between the contributions of different nuclear mechanical components. Our assay probes mechanical response at physiological forces (1–10 nN), length scales (0.1–10 μm), and strain rates (10–3–10–2 s–1; speeds of 15–50 nm/s); these are comparable to the physical conditions that a nucleus might experience during cellular processes such as nuclear migration (Luxton et al., 2010) and cell migration (Friedl et al., 2011; Wolf et al., 2013). Through our novel studies of whole-nucleus deformations under these conditions, we found that the chromatin interior and the peripheral lamina dictate different force response regimes. These experimental findings, along with our simple physical model, provide a basic quantitative framework with which we can begin to understand the functional roles of these two mechanical components of the nucleus.
Chromatin governs resistance to small nuclear deformations
Prior studies emphasized the importance of lamin A in nuclear mechanics. However, not all eukaryotes or cells express lamin A. Studies modulating chromatin indirectly through alterations in divalent ion concentrations suggested a limited mechanical role for chromatin in physiological contexts (Dahl et al., 2005; Pajerowski et al., 2007). These and other micropipette aspiration studies hypothesized, but did not test, that chromatin only contributes to severe deformations by resisting extreme chromatin compression (>70%) in the nucleus region aspirated into the pipette (Dahl et al., 2005; Rowat et al., 2006). This hypothesis, in part, was speculatively based on a simple comparison of the nuclear pressure–strain relation to the force–extension curve of a single chromatin fiber (Pajerowski et al., 2007).
In contrast, by using whole-nucleus deformations that mimic in vivo strains and strain rates, we find that chromatin is a major component of nuclear force response, responsible for resisting small deformations. Our evidence for this includes observations of dramatic mechanical and structural changes associated with the introduction of direct DNA modifications via intercalating dyes to stiffen the nucleus (Figure 2, C–E) and nucleases to dramatically weaken it (Figure 2, F–J). In addition, the loss of volume and stiffness resulting from extensive DNA digestion (Figure 2, F–J) casts strong doubt on the existence of a non–DNA-based “nuclear matrix” (Capco et al., 1982). Instead, this experiment indicates that chromatin itself is the major structural component of the nuclear interior, with the lamin network in the nuclear envelope acting as a flexible yet strong “bag” that is tensed and shaped by the chromatin within it. Combined with recent observations in yeast (Schreiner et al., 2015), these results suggest that chromatin comprises a primordial structural component of the nucleus.
Previous micropipette aspiration experiments reported lamin A/C as the main mechanical component of the nucleus (Pajerowski et al., 2007). However, we note that the aspiration of a small region of the nucleus into a pipette by the sudden application of 1- to 20-kPa pressure results in a nonuniform and high-strain measurement (strains of 1–5, or 100–500% of the initial rest length; Dahl et al., 2005; Pajerowski et al., 2007; Vaziri and Mofrad, 2007). Thus previous aspiration experiments likely have been primarily probing the same strain-stiffening, long-extension regime described here, with the mechanical signal from chromatin masked by the strong deformation of the region of the nucleus being aspirated. Although it is clear from other reports that chromatin indeed has a viscous component (Guilak et al., 2000; de Vries et al., 2007; Schreiner et al., 2015), our findings demonstrate that chromatin also comprises a major resistive element.
The dependence of small-strain nuclear elasticity on chromatin suggests that posttranslational modifications of histones might change overall nuclear mechanics by altering nucleosome–nucleosome interactions or chromatin higher-order structure. We observed precisely this effect, with hyperacetylation leading to softening (Figure 3, A and B) and increased heterochromatin leading to stiffening (Figure 3C). This indicates that chromatin modifications could play direct roles in modulating nuclear mechanics in vivo. The dominance of chromatin in the small-strain regime suggests that chromatin and its compaction state directly dictate nuclear mechanical response for many cells, which typically are subject to small strains in physiological scenarios.
Our findings raise further questions about the emergent biophysical properties of the genome and its organization. For instance, we find it likely that higher-order chromatin organization, such as chromosome domains (Lieberman-Aiden et al., 2009), lamin-associated domains (Guelen et al., 2008), and higher-order loops (Denker and de Laat, 2016), contribute to chromatin’s function as a structural and mechanical component of the nucleus (Bustin and Misteli, 2016). Thus major known chromatin proteins that establish and maintain the three-dimensional genome and/or determine the mechanical properties of chromosomes, such as CTCF, cohesin, condensin, and topoisomerase, could have similar roles in nuclear mechanical response. This hypothesis merits further investigation into the roles of these proteins in interphase nucleus mechanics.
Lamin A provides robust mechanical response to large nuclear deformations
Although chromatin provides the restoring force for small nuclear deformations, we find that lamin A/C is responsible for nuclear strain stiffening at larger deformations (Figures 3, D–F, and 4). This is consistent with its role in cell migration, in which nuclei incur large deformations (Rowat et al., 2013; Harada et al., 2014). Both experimental (MNase) and simulation (shell only) results suggest that this contribution to the long regime is largely due to geometry and delayed lamin engagement (Figures 2 and 5). Of importance, we find that lamins are fully engaged at ∼30% strain, so that the resulting stiffening helps protect nuclei from entering the very large strain regime in which they might suffer nuclear ruptures or other types of permanent damage (Le Berre et al., 2012; Harada et al., 2014; Denais et al., 2016). For example, it was recently observed that the lamina halts nuclear spreading once sufficiently stretched (Li et al., 2015).
Our results are also consistent with the increase in stoichiometric ratio of lamin A to lamin B in cells living in mechanically active or stiff environments (Swift et al., 2013). Previous measurement techniques reported a twofold increase in nuclear stiffness upon expression of lamin A/C (Lammerding et al., 2006; Pajerowski et al., 2007). This increase can be accounted for by the lamin-A–dependent strain stiffening we report (1.5- to 2.5-fold; Figures 1F and 4). Lamin B1 plays a secondary role in governing long-extension mechanics, in that its depletion does not alter the mechanical response of nuclei with high levels of lamin A/C (Figure 3E; Lammerding et al., 2006) but stiffens nuclei lacking lamin A/C (Figure 3F; Shin et al., 2013). Thus our findings are in line with past mechanics measurements and provide the additional insight that lamin A/C specifically controls the mechanics of the cell nucleus at large deformations.
Outlook for nuclear mechanical response experiments
Our mechanical measurements were mainly carried out in the extracellular medium, after removal of the nucleus from the cell. This change in environment does not appear to strongly perturb nuclear force response, as we observed similar mechanical response for nuclei inside living cells (Figure 1E). However, those in vivo experiments are experimentally far more challenging, requiring that most of our experiments, especially those at higher forces that probe nuclear envelope elasticity, be carried out outside the cell. It would be interesting to study the mechanics of isolated nuclei in an artificial in vivo–like medium, for example, made using cytoplasmic or nuclear extracts in order to examine in detail what aspects of nuclear force response depend on the in vivo environment.
From nuclear mechanics to nuclear function
The finding that chromatin and lamin A/C dictate different force response regimes provides insights into nuclear function and its aberrant behavior in disease. For example, increased incorporation of mutant lamin A/C in the laminopathy progeria alters the stability of the lamina (Butin-Israeli et al., 2012; Isermann and Lammerding, 2013). This mechanical alteration could play a role in the manifestations of laminopathies in muscle cells, which live in a mechanically demanding environment and likely undergo large deformations. Lamin perturbations are also relevant to some cancers, in which lamins are up- or down-regulated in a cell type– and cancer-dependent manner. This has been hypothesized to be linked to lamin A/C function. The evidence for this is that increased lamin A/C promotes survival, whereas decreased lamin A/C promotes motility (Rowat et al., 2013; Harada et al., 2014). Thus circulating tumor cells in the blood stream would benefit from higher levels of lamin A/C, which may help resist large shear stresses (Mitchell et al., 2015). On the other hand, prostate and ovarian cancers are more malignant and exhibit increased motility and invasion of surrounding tissue, which could be due to their decreased levels of lamin A/C (Gong et al., 2015; Saarinen et al., 2015). Our findings may thus clarify why different lamin A/C levels are associated with varying disease phenotypes.
Chromatin is also altered in laminopathies and cancers, and its role in determining short deformations (<3 µm) points specifically to a different set of mechanical and morphological phenomena. Nuclear distortions termed blebs occur in many laminopathies and have long been used as an indicator for cancer (Butin-Israeli et al., 2012). Despite their prevalence and diagnostic utility, it remains unclear how or why nuclear blebs form. Although these malformations are often attributed to mutant lamin A/C or altered lamin expression levels inducing the separation of the lamin A/C and B networks (Wren et al., 2012; Funkhouser et al., 2013), alternative hypotheses are that blebs form due to increased euchromatin (Shumaker et al., 2006), decreased chromatin tethering (Schreiner et al., 2015), and/or chromatin plasticity (Pajerowski et al., 2007). Indeed, our nuclear mechanics experiments revealing the dominance of chromatin in the small-deformation force response suggest that aberrant chromatin organization could perturb nuclear morphology. This is supported by recent results demonstrating that decondensation of chromatin by overexpression of HMGN5 induces nuclear blebbing (Furusawa et al., 2015). The finding that chromatin and lamin differentially regulate two regimes of mechanical response therefore sets the table for investigating how alterations to each individual component affect nuclear mechanics and influence important cellular behaviors in disease.
MATERIALS AND METHODS
Cell growth
HeLa, BJ-5ta, HEK293, and immortalized MEF WT vimentin-null (V−/−) cells were grown in 60-mm dishes (Corning) in DMEM (Corning) with phenol red, 0.1 mg/ml penicillin streptomycin (Corning), and 10% FBS (HyClone) at 37°C and 5% CO2. HT-29 cells were grown in McCoy’s medium (Life Technologies) at 37°C and 5% CO2. HT-29 CSK KD cells were maintained in McCoy’s medium supplemented with G418 (Corning) at 1 mg/ml. The ionic composition of this medium is ∼1–2 mM Ca2+, 1 mM Mg2+, 5 mM K+, and ∼150 mM Na+.
Nucleus extraction
Cells were passaged from 80–90% confluent 60-mm dishes and diluted 1:10 to 1:40 into microscope slide wells built of rubber rings coated in paraffin wax. Cells were grown in wells for 2–3 d before experiments. Cells were treated with 1 µg/ml latrunculin A (Enzo Life Sciences) for ∼45 min before single-nucleus isolation. The medium was removed and replaced with fresh prewarmed medium after treatment. MEF V−/− (Mendez et al., 2010) nuclei have a disrupted cytoskeleton, making it possible to isolate nuclei without latrunculin A treatment. MEF V−/− cells served as an important control allowing comparison of different isolation techniques (Figure 1G).
Nucleus isolation was adapted from previous techniques to isolate newt or human chromosomes (Poirier et al., 2000; Kawamura et al., 2010). Microscope coverslip wells were mounted onto the IX-70 Olympus wide-field microscope using a 60× oil 1.4 numerical aperture (NA) Olympus objective for phase imaging with a Pelco DSP black and white charge-coupled device (CCD) camera. Micropipettes were pulled by a Suter Instruments Flaming/Brown micropipette puller. Tapered pipettes were cut to create a diameter opening of 3–3.5 µm. Pipettes were loaded using a vacuum to fill the tip, and then the body was filled with a syringe. Pipettes were filled with either 0.05% Triton X-100 in phosphate-buffered saline (PBS; Lonza) for nucleus isolation or PBS only if the pipette was used to hold the nucleus. Pipettes were loaded into pipette holders and attached to micromanipulators (Sutter Instruments MP-285). An “isolation” pipette loaded with mild detergent, 0.05% Triton X-100 in PBS, was loaded opposite to the “pull” pipette. Gravity-well water sources attached to the pipette holders were used to expel or suck in liquid to the pipettes. Nuclei were isolated from living cells in the cell culture media in which the cells were grown at 24 or 37°C (see Supplemental Figure S1K). Cell membranes were lysed using a gentle expel of 0.05% Triton X-100 in PBS (Figure 1A). The “pull” pipette then captured the isolated nucleus and removed it from the cell by sucking in a small amount of liquid and 10–20% of the nucleus. Nonspecific binding occurred between the nucleus and the micropipette to create a seal. The spray pipette was then changed for a calibrated “force” pipette. The “force” pipette was attached to the opposite end of the nucleus and aligned in preparation for force measurement.
Nucleus force versus extension measurements
The method for making force measurements of single isolated nuclei by micromanipulation was adapted from the procedure developed for isolated chromosomes (Poirier et al., 2000). Force-measuring pipettes were precalibrated before use in experiments using the calibration method described in Poirier et al. (2000). These pipettes were made from thin-wall micropipettes with filament (World Precision Instruments) and pulled with a longer taper than other micropipettes. Force pipettes were cut to have an opening of 3–3.5 µm, which corresponds to spring constants of 1.4–2.1 nN/µm. This range of pipette diameters did not alter measured nuclear spring constants within the sensitivity of the measurement, whereas the use of smaller or larger pipettes significantly altered both the amount of nucleus captured by the pipette and the measured nuclear spring constant. Spring constants were measured by having a micropipette with a known spring constant push a micropipette with an unknown spring constant. The known pipette was set to push the unknown pipette a set distance (6 µm). The two pipettes moved the same distance while in contact (Xequal). At this point, the unknown pipette was removed; the known one then relaxed to a larger, zero-force position (Xtotal). The unknown force-pipette spring constant follows as Funknown = (Fknown/Xequal)(Xtotal – Xequal).
Extension and tracking of both the “pull” and the “force” pipettes was accomplished using a computer program written in LabView (National Instruments). The “pull” pipette was moved 5 µm and then returned in order to generate short nucleus extension of ∼3–4 µm. Long nucleus extensions were performed by moving the pull pipette 10 or 15 µm and then returning the same distance in order to generate 6- to 10-µm nucleus extension. Long nucleus extensions provided the ability to measure both a short-extension (<3 µm) and a long-extension (>3 µm) nuclear spring constant. Deflection of the calibrated force pipette accounts for the difference between the distance the pull pipette moved and the extension of the nucleus (pull pipette displacement = nucleus extension + force pipette defection). During extension of the nucleus, the computer program tracked and recorded the positions of both pipettes via contrast correlation. A data and beginning image file were saved for each experimental stretch. All nuclei were stretched at least three times.
Force measurements were analyzed with Excel (Microsoft). The position of the “force” pipette multiplied by its premeasured spring constant provided force data for each time point relative to nucleus extension, the latter being simply the distance between the two pipettes. Excel was used to graph force versus extension and then draw a best-fit linear slope to obtain the spring constant of the nucleus. Nuclear extension of 3 µm separated short-extension (<3 µm) and long-extension (>3 µm) spring constants.
Nucleus spray treatment
A third pipette was cut with a large opening of 5–7 µm and then loaded with a desired biochemical solution by vacuum filling the tip with ∼20–50 µl. A pen was used to mark the position of the loaded biochemical solution to track movement in the pipette. The rest of the micropipette was filled with water, with an air bubble used to separate the biochemical solution and the water, which aided expelling the solution onto a nucleus. The third micropipette was then loaded into a pipette holder and manual manipulator set 90° (perpendicular) to the micromechanically controlled micropipettes. The isolated nucleus was stretched twice before spray biochemical treatment. After treatment, the nucleus was stretched twice to provide posttreatment force measurements. The water gravity well was raised to expel the biochemical solution onto the isolated nucleus.
Treatments were as follows: 10 mM MgCl2 was sprayed continually during chromatin compaction measurements. After the spray was turned off, nuclei were measured every 5 min to determine whether the nucleus would return to prespray mechanical strength. Propidium iodide at 1 µg/ml was sprayed onto an isolated nucleus for 30 s and then stretched. The nucleus was imaged after force measurements to confirm PI binding. Restriction endonuclease AluI at 1 U/µl (New England Biolabs) was sprayed for 5 min to cut the DNA every few kilobases in between nucleosomes (Polach et al., 2000). MNase at 1 U/µL was sprayed for 30 s to 1 min to cause extensive cutting of the DNA in the nucleus.
HDACi treatment
Cells were treated with either 1 µM VPA or 100 nM TSA for 16–24 h to accumulate decondensed euchromatin. For validations, see Supplemental Figure S3.
Transfection/lamin modulation
HeLa cells were transiently transfected with a plasmid expressing a shRNA to silence lamin A/C expression (Kojima et al., 2004; Shimi et al., 2008) or a scrambled shRNA plasmid with a reporter GFP using HeLa Monster transfection reagent (Mirus). Three days after transfection, cells were trypsinized and then replated in microscope slide wells at a dilution of 1:20–1:40. Five days posttransfection, individual nuclei from cells expressing the reporter GFP were extracted and measured for mechanical response using micromanipulation. Lamin A/C levels were knocked down ∼80% as determined by immunofluorescence in nuclei of cells expressing GFP from the lamin A/C shRNA plasmid (Supplemental Figure S4B). Lamin B1 was depleted to ∼80% via a shRNA reporter plasmid in a similar manner (Supplemental Figure S4G). HEK293 WT parent and stable HEK293 cell lines overexpressing GFP-lamin A (Butin-Israeli et al., 2011) were grown and measured in parallel. HEK293 WT cells have effectively knockdown levels or decreased levels of lamin A/C compared with HeLa or HEK293 GFP-LA, which have similar levels of lamin A (Supplemental Figure S4, D and E).
Immunofluorescence
Cells were seeded on cover glasses in six-well plates to reach confluence in 3 d. Upon reaching 80–90% confluence, cells were fixed with 4% paraformaldehyde (Electron Microscopy Sciences) in PBS for 15 min at room temperature. Cells were then washed three times for 10 min each with PBS. Cells were then permeabilized with 0.1% Triton X-100 (US Biological) in PBS for 15 min. Cells were then washed with 0.06% Tween 20 (US Biological) in PBS for 5 min, followed by two more washes in PBS for 5 min each at room temperature. Cells were then blocked for 1 h at room temperature using 10% goat serum (Sigma-Aldrich) in PBS. Primary antibodies were diluted in blocking solution (10% goat serum in PBS) for lamin A/C 5G4 (Butin-Israeli et al., 2015) at 1:10,000 (Goldman lab), lamin A/C at 1:10,000 (Active Motif), lamin B1 at 1:1000 (ab16048, Abcam), H3K9me2-3 at 1:100 (6F12; Cell Signaling), and H3K9ac at 1:600 (C5B11; Cell Signaling). Primary antibodies were incubated with fixed cells overnight at 4°C in the dark. Cells were washed with PBS three times for 5 min each. Next cells were incubated with fluorescent secondary antibody anti-mouse or anti-rabbit Alexa 488 or 594 (2 mg/ml; Life Technologies) at 1:600 for 1 h at room temperature in the dark. Cells were first washed with 1 µg/ml Hoechst 33342 (Life Technologies) in PBS for 5 min and then washed three more times with PBS. Finally, cover slides were mounted onto microscope slides using ProLong Gold antifade reagent (Life Technologies) and allowed to dry for 2 h or overnight at room temperature.
Imaging and analysis
Immunofluorescence images were acquired with an IX-70 Olympus wide-field microscope using a 60× oil 1.4 NA Olympus objective with an Andor iXon3 electron-multiplying CCD camera using MetaMorph. Exposure times for 4′,6-diamidino-2-phenylindole, rhodamine, and fluorescein isothiocyanate were between 50 and 400 ms. Images were saved with MetaMorph and transferred to ImageJ for analysis. Nuclei were selected by ImageJ threshold or drawn by hand around Hoechst fluorescence if nuclei were too close. Background fluorescence was quantified by highlighting a 30 × 30 pixel area with no cells. Intensity values were acquired and moved into Excel. In Excel, all intensities were background subtracted. For shRNA-knockdown experiments and HEK293 GFP-LA ectopic expression, the fluorescence intensities of nuclei were normalized for each field of view to cells not expressing the GFP reporter or GFP-LA <1.25-fold average intensity compared with background. When comparing WT to mutants in HT-29 cells, the signal intensities for nuclei were normalized for Hoechst. Relative intensities were reported as fold intensity relative to WT or untreated nuclei.
Western blots
Cells were grown to 90% confluency on 10-cm dishes and then lysed using RIPA Lysis Buffer Complete (Santa Cruz Biotechnology) and tumbled for 30 min at 4C°. Protein extracts were separated from cell debris via centrifugation at 12,000 rpm for 15 min in a microfuge. Protein concentrations of cell extracts were measured using the Pierce 660-nm protein assay (Thermo Scientific) and recorded. Equal amounts of sample buffer were added to the protein extracts, followed by heating to 95°C for 5 min before storage at −20°C. A 30-μg protein extract of each condition was run in triplicate on a 4–20% gradient SDS–PAGE gel (Lonza) at 100 V. Protein samples were transferred to 0.2-µm-pore nitrocellulose (Life Sciences) at 100 V in a mini-PROTEAN apparatus (Bio-Rad) for 2 h in a 4°C room with additional ice cooling. The nitrocellulose membrane was washed in blocking buffer (5% bovine serum albumin [BSA] in Tris-buffered saline and Tween 20 [TBST; Fisher]) three times for 5 min and then incubated at room temperature for 1 h. The blot was probed with anti-H3K9ac (C5B11; Cell Signaling) at 1:1000 or lamin A/C (5G4; Goldman lab) at 1:5000 and anti–β-actin (926-42212; Li-Cor) diluted 1:4000 as a loading control overnight with agitation at 4°C in 5% BSA in TBST. The next day, the membrane was washed in blocking buffer three times for 5 min. Secondary horseradish peroxidase–linked antibodies (anti-mouse, 7076; Cell Signaling; anti-rabbit, 31464; Thermo Scientific) were diluted to 1:2000 in 5% BSA in TBST and incubated at room temperature for 1 h. The membrane was washed five times for 5 min each in TBST before imaging. Images were analyzed in ImageJ using Analyze Gels features. The H3K9ac signal was normalized to the β-actin loading control signal.
Simulations
Brownian dynamics simulations (Allen and Tildesley, 1989) were performed with custom-written C++ code. One thousand subunits (N = 1000) of diameter 0.7 μm were initially randomly placed on the surface of a sphere of radius R = 5 μm and linked to at least z = 4 of the nearest neighbors (<z> ≈ 4.5) by harmonic springs with stiffness k (k = 0.8 nN/μm in typical simulations) and rest length equal to the initial length (mean length (4πR2/N)1/2 ≈ 0.6 μm). Shell subunits repelled each other by excluded-volume interactions, modeled as a harmonic potential with spring constant kexc = k. A randomly configured polymer composed of Np = 552 subunits connected by springs with stiffness kp = 1.6 nN/μm (in typical simulations) was placed inside the shell (packing fraction ≈ 10%). All polymer subunits repelled each other by excluded-volume interactions, modeled by a harmonic spring potential with stiffness kexc,p = kp; this parameter was chosen to maintain excluded-volume interactions between subunits and does not quantitatively alter simulation results for kexc,p > 0.1 nN/μm and does not qualitatively alter simulation results for smaller values of kexc. Excluded-volume interactions between polymeric and shell subunits were modeled by a spring with constant kexc,inter = kexckexc,p/(kexc + kexc,p). A total of 2Nc = 110 (∼20%) subunits in the polymer were cross-linked to another subunit that resided at least 4 subunits away along the polymer contour by springs with stiffness kc = kp. Finally, Ns = 40 subunits in the polymer near the surface were linked to the shell by springs with stiffness ks = kp. Simulation parameters are listed in Table 1.
Subunits were subject to stochastic thermal forces, and the system was evolved using the coupled Langevin equations for the described interactions with time step Δt = 0.0005 for >107 steps. For stretching simulations, the closest 100 monomers to each of the two opposite poles were pulled outward along the axis at a constant force, F. The reported strains are averages after relaxation of at least eight different random configurations. The qualitative model behavior is robust to moderate parameter changes; for instance, the qualitative behavior described in the Results is present in each curve presented in Figure 5.
Supplementary Material
Acknowledgments
We thank fellow Northwestern University labs, the Backman lab for HT-29 cell lines and the Horvath lab for use of their equipment. We acknowledge helpful discussions with Aykut Erbaş. A.D.S. is supported by National Research Service Award Postdoctoral Fellowship F32GM112422 and a postdoctoral fellowship from the American Heart Association (14POST20490209; 7/1/14-2/29/16). A.D.S., E.J.B., and J.F.M. are supported by National Science Foundation Grants DMR-1206868 and MCB-1022117 and National Institutes of Health Grants GM105847 and CA193419, and by a subcontract to National Institutes of Health Grant DK107980. S.A.A. and R.D.G. are supported by National Institutes of Health Grants GM106023 and GM0969 and Progeria Research Foundation Grant PRF 2013-51. This research was supported in part through the computational resources and staff contributions provided for the Quest High Performance Computing Facility at Northwestern University, which is jointly supported by the Office of the Provost, the Office for Research, and Northwestern University Information Technology.
Abbreviations used:
- CSK
c-Src tyrosine kinase
- GFP
green fluorescent protein
- HDAC
histone deacetylase
- HDACi
HDAC inhibitor
- KD
knockdown
- LA/C
lamin A/C
- LB1
lamin B1
- MEF
mouse embryonic fibroblasts
- MEF V−/−
MEF vimentin null
- MNase
micrococcal nuclease
- PBS
phosphate-buffered saline
- PI
propidium iodide
- shRNA
short hairpin RNA
- TSA
trichostatin A
- VPA
valproic acid
- WT
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-09-0653) on January 5, 2017.
REFERENCES
- Allen MP, Tildesley DJ. Computer Simulation of Liquids. Oxford, UK: Clarendon Press; 1989. [Google Scholar]
- Banerjee A, Majumder P, Sanyal S, Singh J, Jana K, Das C, Dasgupta D. The DNA intercalators ethidium bromide and propidium iodide also bind to core histones. FEBS Open Bio. 2014;4:251–259. doi: 10.1016/j.fob.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bank EM, Gruenbaum Y. The nuclear lamina and heterochromatin: a complex relationship. Biochem Soc Trans. 2011;39:1705–1709. doi: 10.1042/BST20110603. [DOI] [PubMed] [Google Scholar]
- Booth EA, Spagnol ST, Alcoser TA, Dahl KN. Nuclear stiffening and chromatin softening with progerin expression leads to an attenuated nuclear response to force. Soft Matter. 2015;11:6412–6418. doi: 10.1039/c5sm00521c. [DOI] [PubMed] [Google Scholar]
- Bustin M, Misteli T. Nongenetic functions of the genome. Science. 2016;352:aad6933. doi: 10.1126/science.aad6933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butin-Israeli V, Adam SA, Goldman AE, Goldman RD. Nuclear lamin functions and disease. Trends Genet. 2012;28:464–471. doi: 10.1016/j.tig.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butin-Israeli V, Adam SA, Jain N, Otte GL, Neems D, Wiesmuller L, Berger SL, Goldman RD. Role of lamin b1 in chromatin instability. Mol Cell Biol. 2015;35:884–898. doi: 10.1128/MCB.01145-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butin-Israeli V, Ben-nun-Shaul O, Kopatz I, Adam SA, Shimi T, Goldman RD, Oppenheim A. Simian virus 40 induces lamin A/C fluctuations and nuclear envelope deformation during cell entry. Nucleus. 2011;2:320–330. doi: 10.4161/nucl.2.4.16371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caille N, Tardy Y, Meister JJ. Assessment of strain field in endothelial cells subjected to uniaxial deformation of their substrate. Ann Biomed Eng. 1998;26:409–416. doi: 10.1114/1.132. [DOI] [PubMed] [Google Scholar]
- Capco DG, Wan KM, Penman S. The nuclear matrix: three-dimensional architecture and protein composition. Cell. 1982;29:847–858. doi: 10.1016/0092-8674(82)90446-9. [DOI] [PubMed] [Google Scholar]
- Dahl KN, Engler AJ, Pajerowski JD, Discher DE. Power-law rheology of isolated nuclei with deformation mapping of nuclear substructures. Biophys J. 2005;89:2855–2864. doi: 10.1529/biophysj.105.062554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl KN, Kahn SM, Wilson KL, Discher DE. The nuclear envelope lamina network has elasticity and a compressibility limit suggestive of a molecular shock absorber. J Cell Sci. 2004;117:4779–4786. doi: 10.1242/jcs.01357. [DOI] [PubMed] [Google Scholar]
- Dahl KN, Scaffidi P, Islam MF, Yodh AG, Wilson KL, Misteli T. Distinct structural and mechanical properties of the nuclear lamina in Hutchinson-Gilford progeria syndrome. Proc Natl Acad Sci USA. 2006;103:10271–10276. doi: 10.1073/pnas.0601058103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denais CM, Gilbert RM, Isermann P, McGregor AL, te Lindert M, Weigelin B, Davidson PM, Friedl P, Wolf K, Lammerding J. Nuclear envelope rupture and repair during cancer cell migration. Science. 2016;352:353–358. doi: 10.1126/science.aad7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denker A, de Laat W. The second decade of 3C technologies: detailed insights into nuclear organization. Genes Dev. 2016;30:1357–1382. doi: 10.1101/gad.281964.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries AH, Krenn BE, van Driel R, Subramaniam V, Kanger JS. Direct observation of nanomechanical properties of chromatin in living cells. Nano Lett. 2007;7:1424–1427. doi: 10.1021/nl070603+. [DOI] [PubMed] [Google Scholar]
- Friedl P, Wolf K, Lammerding J. Nuclear mechanics during cell migration. Curr Opin Cell Biol. 2011;23:55–64. doi: 10.1016/j.ceb.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, Orlov YL, Velkov S, Ho A, Mei PH, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funkhouser CM, Sknepnek R, Shimi T, Goldman AE, Goldman RD, Olvera de la Cruz M. Mechanical model of blebbing in nuclear lamin meshworks. Proc Natl Acad Sci USA. 2013;110:3248–3253. doi: 10.1073/pnas.1300215110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusawa T, Rochman M, Taher L, Dimitriadis EK, Nagashima K, Anderson S, Bustin M. Chromatin decompaction by the nucleosomal binding protein HMGN5 impairs nuclear sturdiness. Nat Commun. 2015;6:6138. doi: 10.1038/ncomms7138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman RD, Shumaker DK, Erdos MR, Eriksson M, Goldman AE, Gordon LB, Gruenbaum Y, Khuon S, Mendez M, Varga R, Collins FS. Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson-Gilford progeria syndrome. Proc Natl Acad Sci USA. 2004;101:8963–8968. doi: 10.1073/pnas.0402943101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong G, Chen P, Li L, Tan H, Zhou J, Zhou Y, Yang X, Wu X. Loss of lamin A but not lamin C expression in epithelial ovarian cancer cells is associated with metastasis and poor prognosis. Pathol Res Pract. 2015;211:175–182. doi: 10.1016/j.prp.2014.11.008. [DOI] [PubMed] [Google Scholar]
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, van Steensel B. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- Guilak F, Tedrow JR, Burgkart R. Viscoelastic properties of the cell nucleus. Biochem Biophys Res Commun. 2000;269:781–786. doi: 10.1006/bbrc.2000.2360. [DOI] [PubMed] [Google Scholar]
- Guilluy C, Osborne LD, Van Landeghem L, Sharek L, Superfine R, Garcia-Mata R, Burridge K. Isolated nuclei adapt to force and reveal a mechanotransduction pathway in the nucleus. Nat Cell Biol. 2014;16:376–381. doi: 10.1038/ncb2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada T, Swift J, Irianto J, Shin JW, Spinler KR, Athirasala A, Diegmiller R, Dingal PC, Ivanovska IL, Discher DE. Nuclear lamin stiffness is a barrier to 3D migration, but softness can limit survival. J Cell Biol. 2014;204:669–682. doi: 10.1083/jcb.201308029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris TH, Banigan EJ, Christian DA, Konradt C, Tait Wojno ED, Norose K, Wilson EH, John B, Weninger W, Luster AD, et al. Generalized Levy walks and the role of chemokines in migration of effector CD8+ T cells. Nature. 2012;486:545–548. doi: 10.1038/nature11098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho CY, Jaalouk DE, Vartiainen MK, Lammerding J. Lamin A/C and emerin regulate MKL1-SRF activity by modulating actin dynamics. Nature. 2013;497:507–511. doi: 10.1038/nature12105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isermann P, Lammerding J. Nuclear mechanics and mechanotransduction in health and disease. Curr Biol. 2013;23:R1113–R1121. doi: 10.1016/j.cub.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski A, Fedorchak GR, Lammerding J. The cellular mastermind(?)—mechanotransduction and the nucleus. Prog Mol Biol Transl Sci. 2014;126:157–203. doi: 10.1016/B978-0-12-394624-9.00007-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura R, Pope LH, Christensen MO, Sun M, Terekhova K, Boege F, Mielke C, Andersen AH, Marko JF. Mitotic chromosomes are constrained by topoisomerase II-sensitive DNA entanglements. J Cell Biol. 2010;188:653–663. doi: 10.1083/jcb.200910085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S, Vignjevic D, Borisy GG. Improved silencing vector coexpressing GFP and small hairpin RNA. Biotechniques. 2004;36:74–79. doi: 10.2144/04361ST02. [DOI] [PubMed] [Google Scholar]
- Koster S, Weitz DA, Goldman RD, Aebi U, Herrmann H. Intermediate filament mechanics in vitro and in the cell: from coiled coils to filaments, fibers and networks. Curr Opin Cell Biol. 2015;32:82–91. doi: 10.1016/j.ceb.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunte DP, Wali RK, Koetsier JL, Hart J, Kostjukova MN, Kilimnik AY, Pyatkin IG, Strelnikova SR, Roy HK. Down-regulation of the tumor suppressor gene C-terminal Src kinase: an early event during premalignant colonic epithelial hyperproliferation. FEBS Lett. 2005;579:3497–3502. doi: 10.1016/j.febslet.2005.05.030. [DOI] [PubMed] [Google Scholar]
- Lammerding J, Fong LG, Ji JY, Reue K, Stewart CL, Young SG, Lee RT. Lamins A and C but not lamin B1 regulate nuclear mechanics. J Biol Chem. 2006;281:25768–25780. doi: 10.1074/jbc.M513511200. [DOI] [PubMed] [Google Scholar]
- Le Berre M, Aubertin J, Piel M. Fine control of nuclear confinement identifies a threshold deformation leading to lamina rupture and induction of specific genes. Integr Biol (Camb) 2012;4:1406–1414. doi: 10.1039/c2ib20056b. [DOI] [PubMed] [Google Scholar]
- Li Y, Lovett D, Zhang Q, Neelam S, Kuchibhotla RA, Zhu R, Gundersen GG, Lele TP, Dickinson RB. Moving cell boundaries drive nuclear shaping during cell spreading. Biophys J. 2015;109:670–686. doi: 10.1016/j.bpj.2015.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxton GW, Gomes ER, Folker ES, Vintinner E, Gundersen GG. Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear movement. Science. 2010;329:956–959. doi: 10.1126/science.1189072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahamid J, Pfeffer S, Schaffer M, Villa E, Danev R, Cuellar LK, Forster F, Hyman AA, Plitzko JM, Baumeister W. Visualizing the molecular sociology at the HeLa cell nuclear periphery. Science. 2016;351:969–972. doi: 10.1126/science.aad8857. [DOI] [PubMed] [Google Scholar]
- Marchion DC, Bicaku E, Daud AI, Sullivan DM, Munster PN. Valproic acid alters chromatin structure by regulation of chromatin modulation proteins. Cancer Res. 2005;65:3815–3822. doi: 10.1158/0008-5472.CAN-04-2478. [DOI] [PubMed] [Google Scholar]
- Mazumder A, Roopa T, Basu A, Mahadevan L, Shivashankar GV. Dynamics of chromatin decondensation reveals the structural integrity of a mechanically prestressed nucleus. Biophys J. 2008;95:3028–3035. doi: 10.1529/biophysj.108.132274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez MG, Kojima S, Goldman RD. Vimentin induces changes in cell shape, motility, and adhesion during the epithelial to mesenchymal transition. FASEB J. 2010;24:1838–1851. doi: 10.1096/fj.09-151639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell MJ, Denais C, Chan MF, Wang Z, Lammerding J, King MR. Lamin A/C deficiency reduces circulating tumor cell resistance to fluid shear stress. Am J Physiol Cell Physiol. 2015;309:C736–C746. doi: 10.1152/ajpcell.00050.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajerowski JD, Dahl KN, Zhong FL, Sammak PJ, Discher DE. Physical plasticity of the nucleus in stem cell differentiation. Proc Natl Acad Sci USA. 2007;104:15619–15624. doi: 10.1073/pnas.0702576104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panorchan P, Schafer BW, Wirtz D, Tseng Y. Nuclear envelope breakdown requires overcoming the mechanical integrity of the nuclear lamina. J Biol Chem. 2004;279:43462–43467. doi: 10.1074/jbc.M402474200. [DOI] [PubMed] [Google Scholar]
- Poirier M, Eroglu S, Chatenay D, Marko JF. Reversible and irreversible unfolding of mitotic newt chromosomes by applied force. Mol Biol Cell. 2000;11:269–276. doi: 10.1091/mbc.11.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier MG, Marko JF. Mitotic chromosomes are chromatin networks without a mechanically contiguous protein scaffold. Proc Natl Acad Sci USA. 2002;99:15393–15397. doi: 10.1073/pnas.232442599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polach KJ, Lowary PT, Widom J. Effects of core histone tail domains on the equilibrium constants for dynamic DNA site accessibility in nucleosomes. J Mol Biol. 2000;298:211–223. doi: 10.1006/jmbi.2000.3644. [DOI] [PubMed] [Google Scholar]
- Rowat AC, Foster LJ, Nielsen MM, Weiss M, Ipsen JH. Characterization of the elastic properties of the nuclear envelope. J R Soc Interface. 2005;2:63–69. doi: 10.1098/rsif.2004.0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowat AC, Jaalouk DE, Zwerger M, Ung WL, Eydelnant IA, Olins DE, Olins AL, Herrmann H, Weitz DA, Lammerding J. Nuclear envelope composition determines the ability of neutrophil-type cells to passage through micron-scale constrictions. J Biol Chem. 2013;288:8610–8618. doi: 10.1074/jbc.M112.441535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowat AC, Lammerding J, Ipsen JH. Mechanical properties of the cell nucleus and the effect of emerin deficiency. Biophys J. 2006;91:4649–4664. doi: 10.1529/biophysj.106.086454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saarinen I, Mirtti T, Seikkula H, Bostrom PJ, Taimen P. Differential predictive roles of A- and B-Type nuclear lamins in prostate cancer progression. PLoS One. 2015;10:e0140671. doi: 10.1371/journal.pone.0140671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaffidi P, Misteli T. Lamin A-dependent nuclear defects in human aging. Science. 2006;312:1059–1063. doi: 10.1126/science.1127168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schape J, Prausse S, Radmacher M, Stick R. Influence of lamin A on the mechanical properties of amphibian oocyte nuclei measured by atomic force microscopy. Biophys J. 2009;96:4319–4325. doi: 10.1016/j.bpj.2009.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiner SM, Koo PK, Zhao Y, Mochrie SG, King MC. The tethering of chromatin to the nuclear envelope supports nuclear mechanics. Nat Commun. 2015;6:7159. doi: 10.1038/ncomms8159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimi T, Kittisopikul M, Tran J, Goldman AE, Adam SA, Zheng Y, Jaqaman K, Goldman RD. Structural organization of nuclear lamins A, C, B1 and B2 revealed by super-resolution microscopy. Mol Biol Cell. 2015;26:4075–4086. doi: 10.1091/mbc.E15-07-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimi T, Pfleghaar K, Kojima S, Pack CG, Solovei I, Goldman AE, Adam SA, Shumaker DK, Kinjo M, Cremer T, Goldman RD. The A- and B-type nuclear lamin networks: microdomains involved in chromatin organization and transcription. Genes Dev. 2008;22:3409–3421. doi: 10.1101/gad.1735208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JW, Spinler KR, Swift J, Chasis JA, Mohandas N, Discher DE. Lamins regulate cell trafficking and lineage maturation of adult human hematopoietic cells. Proc Natl Acad Sci USA. 2013;110:18892–18897. doi: 10.1073/pnas.1304996110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumaker DK, Dechat T, Kohlmaier A, Adam SA, Bozovsky MR, Erdos MR, Eriksson M, Goldman AE, Khuon S, Collins FS, et al. Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proc Natl Acad Sci USA. 2006;103:8703–8708. doi: 10.1073/pnas.0602569103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stypula-Cyrus Y, Damania D, Kunte DP, Cruz MD, Subramanian H, Roy HK, Backman V. HDAC up-regulation in early colon field carcinogenesis is involved in cell tumorigenicity through regulation of chromatin structure. PLoS One. 2013;8:e64600. doi: 10.1371/journal.pone.0064600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan T, Escalante-Alcalde D, Bhatt H, Anver M, Bhat N, Nagashima K, Stewart CL, Burke B. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J Cell Biol. 1999;147:913–920. doi: 10.1083/jcb.147.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift J, Ivanovska IL, Buxboim A, Harada T, Dingal PC, Pinter J, Pajerowski JD, Spinler KR, Shin JW, Tewari M, et al. Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science. 2013;341:1240104. doi: 10.1126/science.1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taimen P, Pfleghaar K, Shimi T, Moller D, Ben-Harush K, Erdos MR, Adam SA, Herrmann H, Medalia O, Collins FS, et al. A progeria mutation reveals functions for lamin A in nuclear assembly, architecture, and chromosome organization. Proc Natl Acad Sci USA. 2009;106:20788–20793. doi: 10.1073/pnas.0911895106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaziri A, Mofrad MR. Mechanics and deformation of the nucleus in micropipette aspiration experiment. J Biomech. 2007;40:2053–2062. doi: 10.1016/j.jbiomech.2006.09.023. [DOI] [PubMed] [Google Scholar]
- Verstraeten VL, Ji JY, Cummings KS, Lee RT, Lammerding J. Increased mechanosensitivity and nuclear stiffness in Hutchinson-Gilford progeria cells: effects of farnesyltransferase inhibitors. Aging Cell. 2008;7:383–393. doi: 10.1111/j.1474-9726.2008.00382.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vladescu ID, McCauley MJ, Nunez ME, Rouzina I, Williams MC. Quantifying force-dependent and zero-force DNA intercalation by single-molecule stretching. Nat Methods. 2007;4:517–522. doi: 10.1038/nmeth1044. [DOI] [PubMed] [Google Scholar]
- Wolf K, Te Lindert M, Krause M, Alexander S, Te Riet J, Willis AL, Hoffman RM, Figdor CG, Weiss SJ, Friedl P. Physical limits of cell migration: control by ECM space and nuclear deformation and tuning by proteolysis and traction force. J Cell Biol. 2013;201:1069–1084. doi: 10.1083/jcb.201210152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wren NS, Zhong Z, Schwartz RS, Dahl KN. Modeling nuclear blebs in a nucleoskeleton of independent filament networks. Cell Mol Bioeng. 2012;5:73–81. doi: 10.1007/s12195-011-0196-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M, Kijima M, Akita M, Beppu T. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J Biol Chem. 1990;265:17174–17179. [PubMed] [Google Scholar]
- Zidovska A, Weitz DA, Mitchison TJ. Micron-scale coherence in interphase chromatin dynamics. Proc Natl Acad Sci USA. 2013;110:15555–15560. doi: 10.1073/pnas.1220313110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwerger M, Jaalouk DE, Lombardi ML, Isermann P, Mauermann M, Dialynas G, Herrmann H, Wallrath LL, Lammerding J. Myopathic lamin mutations impair nuclear stability in cells and tissue and disrupt nucleo-cytoskeletal coupling. Hum Mol Genet. 2013;22:2335–2349. doi: 10.1093/hmg/ddt079. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.