Abstract
Antiphospholipid antibody syndrome is an autoimmune disease characterized by the presence of so-called antiphospholipid antibodies and clinical manifestations such as recurrent thromboembolic or pregnancy complications. Although the main antigenic determinant for antiphospholipid antibodies has been identified as the β-2-glycoprotein 1 (β2GP1), the precise epitope recognized by antiphospholipid antibodies still remains largely unknown. In the study herein, we wanted to identify a sequence in domain I of β2GP1 able to induce the proliferation of CD4+ T cells isolated from antiphospholipid antibody syndrome patients, but not from healthy donors, and to interact with antiphospholipid antibodies. We have characterized a sequence in domain I of β2GP1 that triggers CD4+ T-cell proliferation. A comparison of this sequence with the previously reported binding of antiphospholipid antibodies to discontinuous epitope R39-R43 reveals the presence of an indeterminate motif in β2GP1, in which the polarity determines the characteristics and specificity of antiphospholipid antibodies-interacting motifs. Using point mutations, we characterized the main antiphospholipid antibodies-interacting motif as ϕϕϕζζFxC, but also established ϕϕϕζζFxϕ-related motifs as potential antiphospholipid antibodies epitopes, in which ϕ represents nonpolar residues and ζ polar residues, with charges of the residues not being involved. Of specific importance, these different motifs are present at least once in all antiphospholipid antibodies-related receptors described so far. We have further demonstrated, in vitro, that peptides and domains of β2GP1 containing these motifs were able to interact with antiphospholipid antibodies and inhibit their monocyte activating activity. These results established that the antiphospholipid antibodies-interacting motifs are determined by the polarity, but not by the sequence or charge, of amino acids. These data could also contribute to the future development of more sensitive and specific diagnostic tools for antiphospholipid antibody syndrome determination and potential peptide- or β2GP1 domain-based clinical therapies.
Introduction
Antiphospholipid syndrome (APS) is described as a common risk factor for recurrent thromboembolic events and/or pregnancy complications resulting from circulating antiphospholipid antibodies (aPLA).1 It is now widely accepted that the plasma phospholipid binding protein β-2-Glycoprotein 1 (β2GP1) is the main antigenic target for aPLA.2 β2GP1 is a protein of 43 kDa composed of 5 short consensus repeat domains called “sushi” domains. Two different conformations exist for β2GP1: a circular plasma conformation and a fishhook conformation.3 Epitopes within domains I and V are involved in maintaining a circular conformation, whereas binding of domain V to anionic surfaces induces a fishhook conformation and exposure of a cryptic epitope in domain I.3,4 This cryptic epitope is described as being located around residues 39 and 43; however, Iverson et al. have identified additional residues involved in the recognition of residues by pathogenic anti-β2GP1 antibodies in domain I.5,6 Ioannou et al. have also studied mutations including residues R39 to R43 describing complex, and probably discontinuous, epitopes.7 Their data suggest that the epitope(s) are not “classical” and that several epitopes are present in domain I and could potentially be present elsewhere in β2GP1.
Humoral immunophysiology studies of APS and the treatment of APS patients with an anti-CD20 monoclonal antibody (rituximab) have aroused interest in B cells as therapeutic targets. Anti-CD20-treated APS patients have a normal distribution of anti-β2GP1, anti-cardiolipin (aCL) and Lupus anticoagulant (LAC) antibody titers and improved clinical manifestations.8 The isotype of anti-β2GP1 antibody is mainly immunoglobulin G (IgG), suggesting that the production of these antibodies requires antigen-specific CD4+ T helper cells.9 Hattori et al. have shown that β2GP1 induce an in vitro proliferative response of T cells from APS patients. These β2GP1-specific CD4+ T cells are able to induce the production of anti-β2GP1 antibodies by autologous peripheral blood B cells through human leukocyte antigen-D related (HLA-DR) interactions.10,11 The identity of the principal T-cell epitopes on β2GP1 has not been established as of yet. It seems that all 5 β2GP1 domains are able to induce a T-cell proliferative response depending on the APS patient.11 Moreover, analysis of T-cell responses to a β2GP1-derived peptide library have shown that CD4+ T cells are reactive to different peptides independently of HLA.12 In the study herein, we have investigated and identified an immunodominant β2GP1-specific CD4+ T-cell epitope using a peptide-associated major histocompatibility complex (pMHC) II tetramer-based assay. We have shown that the immunodominant β2GP1-specific CD4+ T-cell epitope shares a common peptide motif, which is present in the β2GP1 peptide sequence R39-R43. We have further determined that the characteristic ϕϕϕζζFxC motif, in which ϕ represents non-polar residues (AVILMFWCPG) and ζ polar residues (YTSHKREDQN), as well as motifs closely related to ϕϕϕζζFxC are not only present several times in β2GP1 but also in every receptor described for aPLA.13
Methods
Ethics statement
Buffy coats of blood from healthy donors were provided by the Geneva Hospital Blood Transfusion Center. In accordance with the ethical committee of the Geneva Hospital and with the Declaration of Helsinki, the blood bank obtained informed consent from the donors, who were informed that part of their blood would be used for research purposes.
Patient characteristics
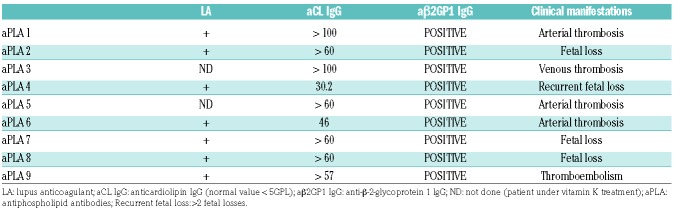
All patients had an APS, as defined by the revised Sapporo criteria.2 Control antibodies and peripheral blood mononuclear cells (PBMC) were isolated from the blood plasma of healthy volunteers. The characteristics of the patients used in this study are listed in Table 1.
Table 1.
Clinical and laboratory profiles of the 9 patients providing the aPLA.
Recombinant β2GP1 fusion proteins and peptide libraries
Recombinant fusion proteins, corresponding to sushi domains I and II, and domains III, IV and V, respectively, of β2GP1 were generated. Briefly, a series of β2GP1 complementary DNA (cDNA) constructs, encoding these domains, were inserted into the vector pcDNA3.1_mycHis_A_A130 between cloning sites BamHI / XhoI. The plasmids were prepared by Life Technology (Carlsbad, CA, USA) and transfected into HEK293 cells. The fusion proteins were purified using nickel resin affinity chromatography (GE Healthcare) and dialysed with Amicon® Ultra (Millipore, Billerica, MA, USA). The concentration was then adjusted to 5mg/ml in peripheral blood smear (PBS). The peptides libraries were generated by Mimotopes (Clayton, VIC, Australia). Lyophilized non water-soluble peptides were reconstituted in 50% dimethyl sulfoxide (DMSO) and 7.5% acetic acid before dilution in PBS. All peptides had 95% purity as assessed by analytical reverse phase high performance liquid chromatography (RP-HPLC). Native β2GP1 was purified from human plasma with 96% purity (Prospecbio, NJ, USA).
T-cell proliferation assays
Peripheral blood mononuclear cells (PBMC) were isolated by density gradient centrifugation over Lymphoprep™ (Axis-Shield PoC) according to the manufacturer’s instructions. T-cell proliferation was evaluated by 5,6-carboxyfluorescein diacetate succinimidyl ester (CFSE) dilution assays (eBioscience). PBMC were stained with 0.1 mM CFSE (eBioscience), according to the manufacturer’s instructions. Cells were cultured in the presence of antigens (10μg/ml) for 10 days. T-cell proliferation was assessed by flow cytometry evaluation of CFSE dilution. Proliferation was expressed as the cell division index (defined as the number of CFSElow T cells cultured with antigen/number of CFSElow T cells without antigen). In all cases, the culture medium consisted of X-VIVO 15 (Lonza, Walkersville, MD, USA).
Flow cytometry
Single-cell suspensions stained with CFSE (eBioscience) were incubated with anti-CD32 (Biolegend) to prevent nonspecific antibody binding, then stained with antibodies against AF488-CD3, AF647-CD4, (BD Biosciences) and major histocompatibility complex (MHC) class II tetramer (Benaroya Research Institute). Staining was assessed with ACCURI C6 flow cytometer (BD Biosciences).
ELISA epitope mapping assay
MaxiSorp™ 96 well plates (Nunc) were coated with 10 μg/ml recombinant domains or peptides of β2GP1 prior to incubation with aPLA or control IgG. Secondary anti-human antibodies conjugated to IR800CW (Rockland) were used. Protein- or peptide-bound antibodies were detected and quantified by the Odyssey system (Li-Cor Biosciences).
Statistical analysis
When required, the significance of differences between groups was assessed using the nonparametric Mann-Whitney U test. *P≤0.05; **P≤0.005; ***P≤0.0005. All data are represented as mean ± SEM of at least 3 independent experiments.
Results
Identification of β2GP1 domains recognized by T cells
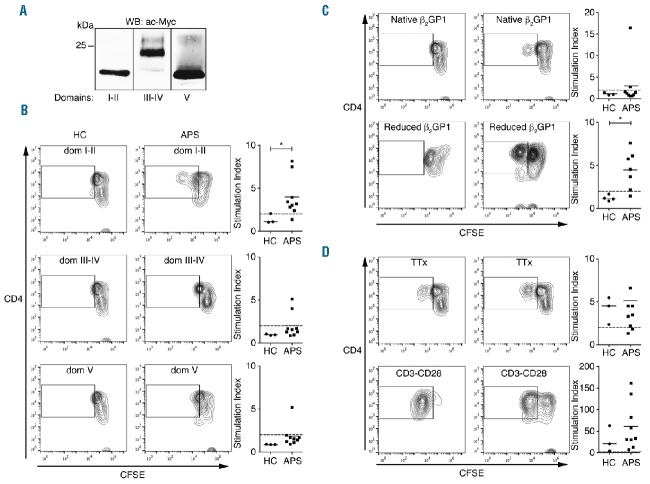
It was previously demonstrated that β2GP1-specific T cells are present in APS patients, although the specific domain(s) and peptide(s) inducing T-cell proliferation were not well defined.10,12 To identify β2GP1 domain-specific T cells in APS patients, PBMCs were isolated from 9 APS patients (Table 1), and 3 healthy controls (HC) were stimulated with native β2GP1, reduced β2GP1 and recombinant β2GP1 domain I–II, domain III–IV, and domain V (Figure 1A). We tested the proliferation of PBMC by the CFSE dilution assay. In contrast with HC, we detected CD4+ T-cell proliferative responses in primary culture to β2GP1 domain I–II and reduced β2GP1 from APS patients (Figure 1B,C). While the cryptic epitope exposed by reduced β2GP1 was recognized by β2GP1 reactive T cells in the normal T-cell repertoire of APS patients, native β2GP1 was unable to significantly stimulate T-cell proliferation (Figure 1C). The proliferation controls performed with tetanus toxoid (TTx) and CD3-CD28-beads, i.e., specific and unspecific T-cell proliferation stimuli, showed that the proliferative ability of T cells from HC and APS were unaltered (Figure 1D). These results suggest that the core sequence(s) of the β2GP1 peptide(s) inducing T-cell proliferation were present in β2GP1 domain I–II.
Figure 1.
Domain I–II and reduced β2GP1 induce proliferation of T cells from APS patients. (A) β2GP1 recombinant domains are purified and analyzed by Western blot. Data are representative of 3 independent experiments. (B–D) Flow cytometry analysis of the proliferation assay performed on PBMC of APS patients in the presence of domains I–II, II–IV, V β2GP1, reduced β2GP1 and tetanus toxin (TTx). CD3-CD28 is positive proliferation control. CD4+CFSElow population is gated on CD3+ cells. Data are mean ± SEM of 9 different donors. Statistical significance was determined by Mann-Whitney U analysis. HC: healthy controls; APS: Antiphospholipid syndrome; WB: western blot; ac-MYC: anti-c-Myc; CFSE: carboxyfluorescein diacetate succinimidyl ester.
Identification of a core peptide fragment recognized by T cells
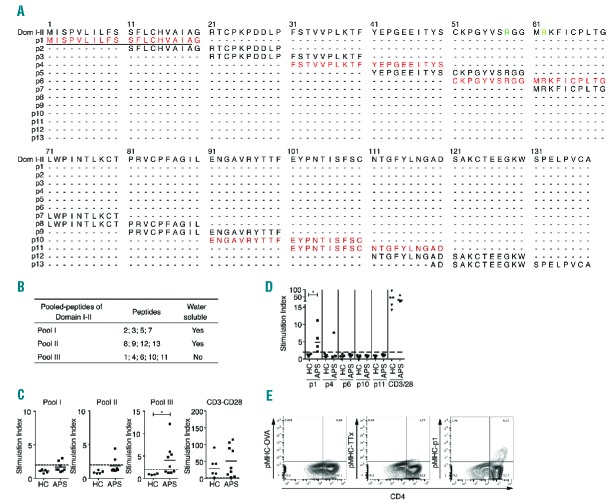
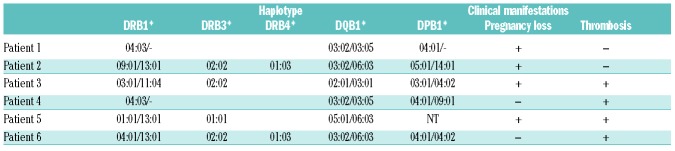
Having identified candidate regions of β2GP1 containing T-cell determinants, we then tested proliferative responses of APS patients against a pool of β2GP1 peptides. For this purpose, we tested the proliferation of PBMC from APS patients and HC to the pools of peptides containing 4 to 5 β2GP1 domain I–II peptides from a library of 13 synthetic overlapping 20-mer peptides present in the 138 amino-acid sequence of β2GP1 domain I–II (Figure 2A,B). By CFSE dilution assay, we identified proliferative responses in primary cultures to β2GP1 domain III peptide pool III (Figure 2C). T-cell proliferations of HC and APC triggered by CD3-CD28 have shown similar responses. β2GP1 domain I–II peptide pool III was composed of 5 peptides consistently insoluble in water with the cryptic character of epitope recognized by β2GP1 reactive T cells. Interestingly, this pool of peptides contained the region targeted by aPLA, i.e., peptide n°6 (p6).14 We then examined the responses to individual β2GP1 peptides contained in pool III. T-cell proliferation response against leader peptide n°1 (p1) was significantly increased (but not against peptides n°4, 6, 10 or 11), while T-cell proliferations of HC showed a similar response to CD3-CD28 (Figure 2D). To confirm the presence of circulating T cells specific for p1, we defined the HLA class II haplotype profiles of APS patients used previously (Table 2). We then used pMHCII-p1 tetramer DRB3* 02:02 staining to detect specific circulating T cells in β2GP1 domain I-II pulsed T-cell population from APS patients. Although, as expected, no population was observed with negative control pMHC-OVA (tetramer with a peptide of ovalbumin), a pMHC-p1 positive population was strongly present in β2GP1 domain I–II specific pulsed T-cell population (Figure 2E). pMHC-TTx staining was used as a positive control (Figure 2E). These results demonstrated that the immunodominant β2GP1-specific CD4+ T-cells’ epitope in patients with APS is present in a sequence of amino acids corresponding to MISPVLILFSSFLCHVAIAG, which correlates with the signal peptide of β2GP1.
Figure 2.
Peptide 1 of β2GP1 carries antigenic determinant for CD4+ T-cell proliferation. (A) Representation of the peptide library of domain I–II. Peptides in red are peptides contained in Pool III. Residues in green represent R39-R43 epitopes. Underlining represents a signal peptide. (B) Table of peptide distribution for proliferation assay. (C) Flow cytometry analysis of proliferation assay performed on PBMC of APS patients in the presence of pool I, II or III. CD3-CD28 shows positive proliferation control. (D) Flow cytometry analysis of proliferation assay performed on the PBMC of APS patients in the presence of p1, p4, p6, p10 and p11. CD3-CD28 is positive proliferation control. CD4+CFSElow population is gated on CD3+ cells. Data are mean ± SEM of 9 different donors. Statistical significance was determined by Mann-Whitney U analysis. (E) Flow cytometry analysis of pMHC class II tetramer staining loaded with p1 on PBMS of APS patients pulsed with recombinant domain I–II. pMHC-OVA (ovalbumin) and pMHC-TTx (Tetanus toxin) are negative controls. Populations are gated on CD3+ cells. HC: healthy controls; APS: antiphospholipid syndrome.
Table 2.
Haplotype profiles of 6 patients from Table 1.
Characterization of common motif shared by p1 and p6 of domain I–II
To identify a potential consensus motif between p1 and p6, which was previously reported as containing the aPLA binding discontinuous epitope R39-R43, we aligned their sequences and observed a common motif present around a phenylalanine (Figure 3A). This motif seemed to be determined by intrinsic physical properties (polar and nonpolar) and not by amino acid sequences. To investigate the ability of p1 to interact with patient-derived anti-β2GP1 antibodies and to identify critical residues in the motif, we generated an alanine scanning library of p1 and p6 including the common motif (Figure 3B) to perform an ELISA epitope mapping assay. As shown in Figure 3B, aPLA are able to bind p1 as potently as p6. The mutations I7A, L8A F9A, S11A, F12A and C14A of p1 significantly decreased the binding of aPLA. Similarly, mutations R58A (R39), G59A (G40), G60A (G41), M61A (M42), R62A (R43), F64A and C66A were involved in the binding of aPLA to p6 (Figure 3B) (numbering corresponds to the numbering in the mature β2GP1 protein, i.e., without the signal peptide). aPLA interacted similarly with both peptides, although the sequences of amino acids present in the motif were not the same. It seems that the aPLA interacting motif, ϕϕϕζζFxC, is dependent on polar or nonpolar properties more than hydropathy classes or charge of residues. Alanine is nonpolar, but it decreased the ability of aPLA to bind I7A, L8A, F9A, and C14A p1-mutated peptides as well as R58A, G59A, G60A, M61A, and C66A p6-mutated peptides. These data suggest that the volume of amino acids and the ability to form electrostatic or hydrogen bonds may also contribute to interactions between aPLA and the motif.
Figure 3.
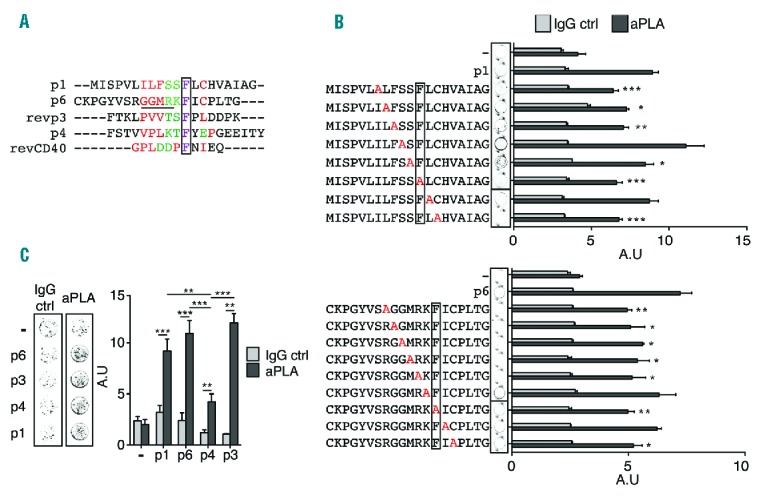
aPLA epitope contains ϕϕϕζζFxf motifs. (A) Alignment of p1, p6, p4, anti-sense p3 and CD40 consensus sequence. Red color shows nonpolar residues while green color shows polar residues in the motif. Underlining represents R39-43 epitopes and a frame represents conserved residue. (B) Binding of aPLA to microplate wells coated with p1 and p6 with consecutive point mutations. Substituted residues are in red. Frame represents conserved residue. Data are mean ± SEM of 9 different donors. (C) Representative picture of IgG ctrl (control) or aPLA binding to peptide-coated well. Quantification of aPLA binding to p1-, p6-, p4- and p3-coated wells. Data are mean ± SEM of 9 different donors. Statistical significance was determined by Mann-Whitney U analysis. aPLA: antiphospholipid antibodies; IgG: immunoglobulin G; A.U: arbitrary unit.
We then performed bioinformatical analysis using Prosite resource (SIB Swiss Institute of Bioinformatics) to find not only the ϕϕϕζζFxC motif (N- to C-term) but also the ϕxFζζϕϕϕ motif which is the reverse motif (C- to N-term) in the human proteome. Four sequences were present in domain I of β2GP1; 2 ϕϕϕζζFxC motifs and 2 ϕxFζζϕϕϕ motifs (Table 3). ϕxFζζϕϕϕ motifs were present in p1 and p3 (Figure 3A). We thus assessed the ability of aPLA to interact with p3. We also included a slightly different motif that was present in p4 to investigate whether a degenerated motif was able to interact with aPLA. As expected, aPLA bound to p3 with a similar efficiency compared to p1 and p6 (Figure 3C). We further observed that p4, although less potent in interactions with aPLA, bound significantly to the aPLA (Figure 3C). Altogether, these data demonstrated that aPLA interact with a ϕϕϕζζFxC motif, and more generally with a ϕϕϕζζFxϕ motif, in which the polarity of the residues was more important than the hydropathy classes. This motif could also interact with aPLA even if the motif was slightly modified by insertion or deletion of an amino acid before or after the key phenylalanine residue. Finally, aPLA were able to bind motifs in sense or anti-senses.
Table 3.
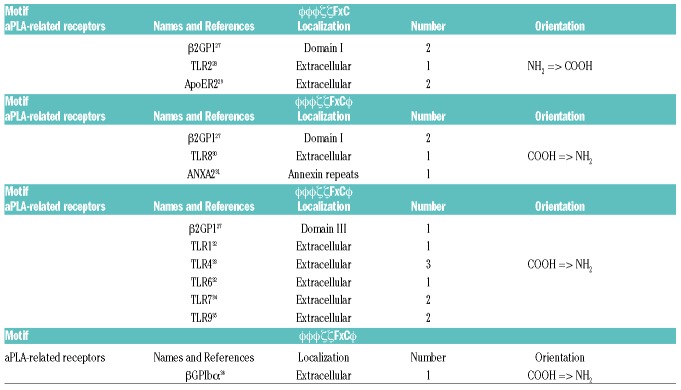
Different motifs present in antiphospholipid antibodies (aPLA)-related receptors and proteins.
aPLA-related receptors described possess closely related motifs
Vlachoyiannopoulos et al.15 identified a consensus motif between CD40 and p3 capable of interacting with aPLA. Although this sequence had one additional nonpolar amino acid between the polar and phenylalanine (ϕxFfζζϕϕϕ) amino acids, it was strongly related to the initial motif (Figure 3A). In addition, it seemed that both polar amino acids present in the ϕϕϕζζFxϕ motif were not fully required (Figure 3B). Thus we performed additional bioinformatical analysis relative to ϕxFζϕϕϕ and ϕxFfζζϕϕϕ motifs. Among the proteins containing these motifs, we identified β2GP1 domain I, toll-like receptor (TLR)1, TLR4, TLR6, TLR7, TLR8, TLR9 and GPIbα (Table 3). In addition to identified proteins containing the motifs ϕϕϕζζFxϕ and ϕxFζζϕϕϕ, all potential aPLA receptors described in the literature possess at least 1 of these 2 motifs. These results suggest that ϕϕϕζζFxC and other closely related motifs constitute the potential links between controversial data obtained by different research groups working on aPLA.
Functional characterization of recombinant domains of β2GP1, peptides 1 and 6
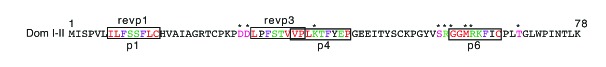
To examine the functional ability of recombinant domains of β2GP1, p1, p4 and p6 to inhibit the activity of aPLA isolated from APS patients studied in our previous experiments (Table 1), we evaluated tumor necrosis factor (TNF) production in human monocytes activated with aPLA which had previously been treated with peptides or recombinant domains. While incubation of human monocytes with aPLA pretreated with β2GP1 and reduced β2GP1 have no effect on aPLA activity, incubation of the aPLA with domain I–II, domain III–IV and domain V significantly decreased the production of TNF induced by aPLA (Figure 4A). This inhibition of TNF production involved the binding of aPLA to β2GP1, and reduced β2GP1, domain I–II and domain III, but not domain V (Figure 4B). Similarly, pre-incubation with peptides n°1, 4 and 6 (p1, p4 and p6) diminished the production of TNF induced by aPLA, whereas p10 and p11 had no effect (Figure 4C). This inhibition showed that p1, p4 and p6, but not p10 or p11, interact with aPLA (Figure 4D). We observed that p1 and p6 had similar aPLA binding activity, while p4 was significantly less potent in its interaction with aPLA (Figure 4D). These results were consistent with the presence of 4 aPLA interaction motifs in domain I–II, while p1 and p6 had only 1 motif (considering that p1 and revp1 were not simultaneously available) (Figure 5). As shown in Figure 3C, the interaction of p4 with aPLA was less effective due to a slightly degenerated motif (Figure 4D and 3A). Taken together, our results clearly revealed that the motifs recognized by aPLA, which were present in p1, p4 and p6 (Figure 3A), but not in p10 or p11 (Figure 2A), were able to functionally decrease the activity of aPLA. They further demonstrated that the aPLA interacting motif (ϕxFζϕϕϕ) present in domain III–IV (Table 3) efficiently interacted with aPLA and inhibited its activity, although to a lesser extent than domain I–II (Figure 4B), confirming the potential targets exposed in Table 3.
Figure 4.
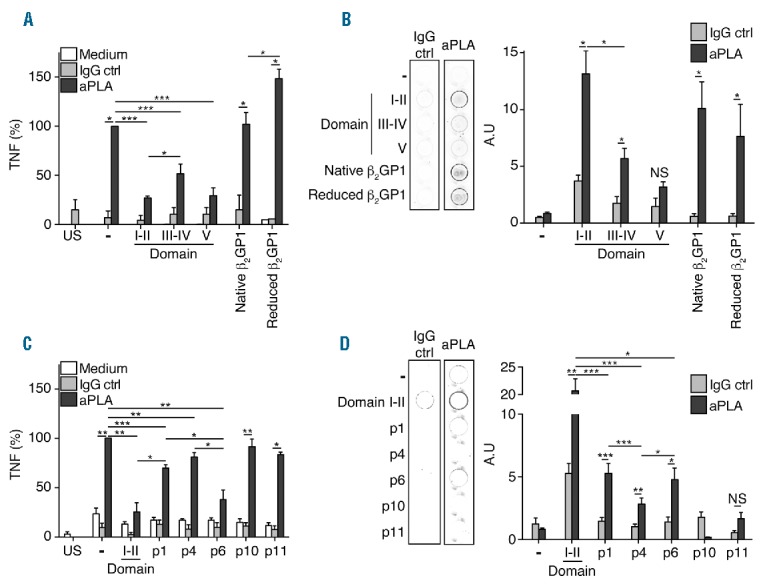
Motif containing domains and peptides inhibits aPLA activity. (A) Inhibition of tumor necrosis factor (TNF) production induced by aPLA by domains I–II, III–IV, V, β2GP1 of reduced β2GP1. (B) Representative picture of IgG ctrl (control) or aPLA binding to domains I–II, III–IV, V and β2GP1-coated well. Quantification of aPLA binding to domains I–II, III–IV, V or β2GP1-coated well. aPLA to well coated with domains I–II, III–IV, V, β2GP1 of reduced β2GP1. (C) Inhibition by domain I–II, p1, p4, p6, p10 and p11 of TNF production induced by aPLA. (D) Representative picture of IgG ctrl or aPLA binding to domains I–II-, p1-, p4-, p6-, p10- and p11-coated well. Quantification of aPLA binding to domains I–II, p1, p4, p6, p10 and p11-coated well. aPLA to well coated with domains I–II, III–IV, V, β2GP1 of reduced β2GP1. Data are mean ± SEM of 9 different donors. Statistical significance was determined by Mann-Whitney U analysis. aPLA: antiphospholipid antibodies; IgG: immunoglobulin G; US: unstimulated; NS: non significant.
Figure 5.
Representation of domain I–II. Frames represent motifs containing sequence. Red color shows nonpolar residues while green color shows polar residues in the motif. Violet color shows phenylalanine present in these motifs. Pink color and asterisks show point mutations already described. Dom: domain.
Discussion
The present study was undertaken to investigate the T-cell response associated with the production of autoantibodies against β2GP1 and their related epitopes in APS patients. We established that the β2GP1 signal peptide (p1) induced the proliferation of CD4+ T cells from APS patients but not from healthy donors. Furthermore, autoreactive CD4+ T cells were detected in APS patient blood samples and bound HLA-DRB3* 02:02 tetramer loaded with p1, thus endorsing the fact that specific HLA-DR alleles are associated with susceptibility to APS.10,16,17 We further observed that the signal peptide contained a motif that was similar to that of the R39-R43 epitope. We thus identified the aPLA interacting motif (ϕϕϕζζFxϕ) to be dependent on polar or nonpolar properties more than on hydropathy classes of amino acids. The recombinant domains of β2GP1, as well as individual peptides containing this motif, or closely related motifs, were able to inhibit the activity of aPLA on human monocytes. This study provides an appealing explanation for the controversial results obtained on potential aPLA-associated receptors. ϕϕϕζζFxC and ϕϕϕζζFxϕ-related motifs are indeed present in all aPLA-associated receptors described in the literature13 (Table 3). Bioinformatics analyses have revealed, however, that the ϕϕϕζζFxC motif is present and accessible in 16 different proteins within the whole human proteome (Online Supplementary Table S1).
Although it is well accepted that the β2GP1 domain I is the target of aPLA, the exact sequence of the epitope remains largely unknown. Herein, we have identified ϕϕϕζζFxϕ as being a common aPLA interacting motif. While the signal peptide (p1), containing an aPLA interacting motif, was not present in the mature β2GP1, CD4+ T cells were able to recognize the p1 sequence. This suggests that a signal peptide could trigger autoimmune responses. Consistently, it was shown that a polymorphism in the signal peptide of the cytotoxic T-lymphocyte antigen-4 (CTLA-4) leads to autoimmune predispositions.18 Interestingly, the aPLA interacting motif present in p1 was also present in revp1, corresponding to a palindromic form of p1 (Figure 5). Furthermore, APS patients autoreactive T-cell proliferation was triggered by p1 (Figure 2D and 2E). Smith et al.19 suggested that complementary sequences of proteins may have a function in the induction of autoimmunity. This theory was confirmed by the discovery that autoimmunity can be initiated through an immune response against a peptide that is antisense or complementary to the autoantigen, which then induced anti-idiotypic antibodies (autoantibodies) that cross-reacted with the autoantigen.20 Furthermore, as a potential origin of APS pathogenicity, the palindromic sequence of p1 could explain the aPLA interactions with p3 and domain III, in which the binding sequences were antisense. Six sense and antisense epitopes are present in β2GP1. Five of these are found in domain I (Table 3 and Figure 5) and could be the reason for its identification as the main interacting protein of aPLA.
To address the question of aPLA binding epitopes, several research groups performed epitope mapping using point mutations of domain I.6,7 They identified D8, D9, K19, S38, R39, G40, M42, R43 and T50 as residues participating in, or contributing to, aPLA domain I interactions. While 44% of these mutations were present in β2GP1 domain I motifs, the proportion increased to 100% if the mutations directly adjacent to motifs were included (Figure 5). Furthermore the adjacent mutations were at the same position as the motifs (Figure 5).6,7 While additional investigation is required in order to define in further detail the exact sequence required to attain the highest affinity for aPLA, these parallels suggest that the motifs proposed could be slightly larger than those exposed in the study herein. Zager et al. have further shown, through phage display techniques based on an affinity for anti-β2GP1, that epitopes recognized by aPLA contained the ϕϕϕζζ part of the motifs.21 They suggested consistently that polarity of the residues mainly mediate the interaction between epitope and aPLA.21 Another research group highlighted a sequence homology between residues 239–245 of CD40 and residues 26–32 of β2GP1.15 Residues 26–32 of β2GP1 corresponded to p3 and belonged to the ϕϕϕζζ part of the ϕϕϕζζϕFxϕ motif (Figure 3 and Figure 5). Moreover, the hexapeptide TLRVYK, described by Blank et al. as an aPLA binding epitope also corresponded to the ϕϕϕζζ part of the motifs.22 Mice immunized with a panel of microbial preparations have been shown to produce high titers of anti-β2GP1 antibodies. This panel of bacteria is composed of 6 different preparations, which include Streptococcus pneumonia, Shigella dysenteriae and haemophilus influenzae.22 Employing bioinformatical analysis (Prosite ressource, SIB), we found that all bacteria strains used in different preparations carry the ϕϕϕζζFxC motif. In fact, not only the bacteria strains but also the virus strains carry the ϕϕϕζζFxC motif. Several cases or clinical studies including patients with established APS or APS with thromboembolic phenomena have revealed a correlation between increased aPLA levels and HIV, Epstein-Barr virus, hepatitis C virus or herpesvirus-6 infection.23–26 Our bioinformatical analysis has revealed that all these virus strains carry the ϕϕϕζζFxC motif. Taken together these data demonstrate that the aPLA interaction motif ϕϕϕζζFxC, and by extension ϕϕϕζζFxϕ-related motifs characterized in the present study were strongly related to other previously depicted epitopes, thereby placing emphasis on polarity properties of residues and not on particular sequences.
The characterization of the aPLA epitope performed herein leads to a better understanding of the surprising number of candidate receptors described for aPLA.13 We have indeed shown that all TLRs suggested as aPLA receptors carry at least 1 motif in their extracellular region. Thus, TLR2 has 1 ϕϕϕζζ FxC motif and TLR4 3 ϕϕϕζFxϕ motifs. As presented in Figure 4D, the ϕϕϕζζ FxC motif may be more potent than the ϕϕϕζFxϕ motif in binding aPLA, but the presence of up to 3 ϕϕϕζFxϕ motifs on TLR4 may explain why TLR2 and TLR4 were proposed as aPLA receptors. Annexin A2, apolipoprotein E receptor 2 (ApoER2) and GPIbα also have at least 1 motif accessible by aPLA (Table 3). The most described aPLA interacting protein, β2GP1, has no less than 5 motifs which are mainly present in domain I, but also in domain III. Several other targets carrying ϕϕϕζζFxϕ motifs are important for APS, such as proteins involved in coagulation processes. Thus, thrombin, proteinase-activated receptor 2 (PAR-2 and 3) and the complements C5, C4a and C4b are some of the aPLA-targeted proteins carrying ϕϕϕζζFxϕ motifs (data not shown).
Although additional developments are necessary in order to find the exact association of residues needed to obtain the highest aPLA affinity, the study herein offers the opportunity to provide an accurate tool for the detection of β2GP1 antibodies for diagnostic purposes. Furthermore, aPLA interacting motifs present in peptides have the ability to inhibit aPLA activity and represent a prevention strategy for APS as an alternative to the use of anticoagulants. Finally, p1-tetramers associated with inducers of cell death could be used to specifically disrupt autoreactive T cells in APS patients, thus representing a potential therapeutic approach.
Supplementary Material
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/102/8/1324
Funding
This work was supported by two grants: the Swiss National Funds (n°310030–127639) and an unrestricted grant from the ISTH2007 Presidential Fund.
References
- 1.Giannakopoulos B, Krilis SA. The pathogenesis of the antiphospholipid syndrome. N Engl J Med. 2013;368(11):1033–1044. [DOI] [PubMed] [Google Scholar]
- 2.Miyakis S, Lockshin MD, Atsumi T, et al. International consensus statement on an update of the classification criteria for definite antiphospholipid syndrome (APS). J Thromb Haemost. 2006;4(2):295–306. [DOI] [PubMed] [Google Scholar]
- 3.Agar C, van Os GM, Morgelin M, et al. Beta2-glycoprotein I can exist in 2 conformations: implications for our understanding of the antiphospholipid syndrome. Blood. 2010;116(8):1336–1343. [DOI] [PubMed] [Google Scholar]
- 4.de Laat B, van Berkel M, Urbanus RT, et al. Immune responses against domain I of beta(2)-glycoprotein I are driven by conformational changes: domain I of beta(2)-glycoprotein I harbors a cryptic immunogenic epitope. Arthritis Rheum. 2011; 63(12):3960–3968. [DOI] [PubMed] [Google Scholar]
- 5.de Laat B, Derksen RH, van Lummel M, Pennings MT, de Groot PG. Pathogenic anti-beta2-glycoprotein I antibodies recognize domain I of beta2-glycoprotein I only after a conformational change. Blood. 2006; 107(5):1916–1924. [DOI] [PubMed] [Google Scholar]
- 6.Iverson GM, Reddel S, Victoria EJ, et al. Use of single point mutations in domain I of beta(2)-glycoprotein I to determine fine antigenic specificity of antiphospholipid autoantibodies. J Immunol. 2002; 169(12):7097–7103. [DOI] [PubMed] [Google Scholar]
- 7.Ioannou Y, Pericleous C, Giles I, et al. Binding of antiphospholipid antibodies to discontinuous epitopes on domain I of human beta(2)-glycoprotein I: mutation studies including residues R39 to R43. Arthritis Rheum. 2007;56(1):280–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khattri S, Zandman-Goddard G, Peeva E. B-cell directed therapies in antiphospholipid antibody syndrome–new directions based on murine and human data. Autoimmun Rev. 2012;11(10):717–722. [DOI] [PubMed] [Google Scholar]
- 9.Cousins L, Pericleous C, Khamashta M, et al. Antibodies to domain I of beta-2-glycoprotein I and IgA antiphospholipid antibodies in patients with ‘seronegative’ antiphospholipid syndrome. Ann Rheum Dis. 2015;74(1):317–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hattori N, Kuwana M, Kaburaki J, et al. T cells that are autoreactive to beta(2)-glycoprotein I in patients with antiphospholipid syndrome and healthy individuals. Arthritis Rheum. 2000;43(1):65–75. [DOI] [PubMed] [Google Scholar]
- 11.Arai T, Yoshida K, Kaburaki J, et al. Autoreactive CD4(+) T-cell clones to beta2-glycoprotein I in patients with antiphospholipid syndrome: preferential recognition of the major phospholipid-binding site. Blood. 2001;98(6):1889–1896. [DOI] [PubMed] [Google Scholar]
- 12.Ito H, Matsushita S, Tokano Y, et al. Analysis of T cell responses to the β2-glycoprotein I-derived peptide library in patients with anti-β2-glycoprotein I antibody-associated autoimmunity. Hum Immunol. 2000;61(4):366–377. [DOI] [PubMed] [Google Scholar]
- 13.Brandt KJ, Kruithof EKO, de Moerloose P. Receptors involved in cell activation by antiphospholipid antibodies. Thromb Res. 2013;132(4):408–413. [DOI] [PubMed] [Google Scholar]
- 14.de Laat B, de Groot PG. Autoantibodies directed against domain I of beta2-glycoprotein I. Curr Rheumatol Rep. 2011;13(1):70–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vlachoyiannopoulos PG, Mavragani CP, Bourazopoulou E, Balitsari AV, Routsias JG. Anti-CD40 antibodies in antiphospholipid syndrome and systemic lupus erythematosus. Thromb Haemost. 2004;92(6):1303–1311. [DOI] [PubMed] [Google Scholar]
- 16.Tanimura K, Jin H, Suenaga T, et al. β2-Glycoprotein I/HLA class II complexes are novel autoantigens in antiphospholipid syndrome. Blood. 2015;125(18):2835–2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Domenico Sebastiani G, Minisola G, Galeazzi M. HLA class II alleles and genetic predisposition to the antiphospholipid syndrome. Autoimmun Rev. 2003;2(6):387–394. [DOI] [PubMed] [Google Scholar]
- 18.Anjos S, Nguyen A, Ounissi-Benkalha H, Tessier MC, Polychronakos C. A common autoimmunity predisposing signal peptide variant of the cytotoxic T-lymphocyte antigen 4 results in inefficient glycosylation of the susceptibility allele. J Biol Chem. 2002;277(48):46478–46486. [DOI] [PubMed] [Google Scholar]
- 19.Smith LR, Bost KL, Blalock JE. Generation of idiotypic and anti-idiotypic antibodies by immunization with peptides encoded by complementary RNA: a possible molecular basis for the network theory. J Immunol. 1987;138(1):7–9. [PubMed] [Google Scholar]
- 20.Pendergraft WF, Preston GA, Shah RR, et al. Autoimmunity is triggered by cPR-3(105–201), a protein complementary to human autoantigen proteinase-3. Nat Med. 2004; 10(1):72–79. [DOI] [PubMed] [Google Scholar]
- 21.Žager U, Lunder M, Hodnik V, et al. Significance of K(L/V)WX(I/L/V)P Epitope of the B2Gpi in Its (Patho)Physiologic Function. EJIFCC. 2011;22(4):118–124. [PMC free article] [PubMed] [Google Scholar]
- 22.Blank M, Krouse I, Fridkin M, et al. Bacterial induction of autoantibodies to b2-glycoprotein-1 accounts for the infectious etiology of antiphospholipid syndrome. J Clin Invest. 2002;109:797–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toyoshima M, Maegaki Y, Yotsumata K, Takei S, Kawano Y. Antiphospholipid syndrome associated with human herpesvirus-6 infection. Pediatr Neurol. 2007;37(6):449–451. [DOI] [PubMed] [Google Scholar]
- 24.Grunewald T, Burmester GR, Schuler-Maue W, Hiepe F, Buttgereit F. Anti-phospholipid antibodies and CD5+ B cells in HIV infection. Clin Exp Immunol. 1999;115(3):464–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abdel-Wahab N, Lopez-Olivo MA, Pinto-Patarroyo GP, Suarez-Almazor ME. Systematic review of case reports of antiphospholipid syndrome following infection. Lupus. 2016;25(14):1520–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Durkin ML, Marchese D, Robinson MD, Ramgopal M. Catastrophic antiphospholipid syndrome (CAPS) induced by influenza A virus subtype H1N1. BMJ Case Rep. 2013;2013:bcr2013200474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McNeil HP, Simpson RJ, Chesterman CN, Krilis SA. Antiphospholipid antibodies are directed against a complex antigen includes a lipid-binding inhibitor of coagulation: B2-glycoprotein I (apolipoprotein H). Proc Natl Acad Sci USA. 1990;87:4120–4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Satta N, Dunoyer-Geindre S, Reber G, et al. The role of TLR2 in the inflammatory activation of mouse fibroblasts by human antiphospholipid antibodies. Blood. 2007; 109(4):1507–1514. [DOI] [PubMed] [Google Scholar]
- 29.Lutters BC, Derksen RH, Tekelenburg WL, et al. Dimers of beta 2-glycoprotein I increase platelet deposition to collagen via interaction with phospholipids and the apolipoprotein E receptor 2′. J Biol Chem. 2003;278(36):33831–33838. [DOI] [PubMed] [Google Scholar]
- 30.Döring Y, Hurst J, Lorenz M, et al. Human antiphospholipid antibodies induce TNFα in monocytes via Toll-like receptor 8. Immunobiology. 2010;215(3):230–241. [DOI] [PubMed] [Google Scholar]
- 31.Romay-Penabad Z, Montiel-Manzano MG, Shilagard T, et al. Annexin A2 is involved in antiphospholipid antibody-mediated pathogenic effects in vitro and in vivo. Blood. 2009;114:3074–3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brandt KJ, Fickentscher C, Boehlen F, Kruithof EK, de Moerloose P. NF-kappaB is activated from endosomal compartments in antiphospholipid antibodies-treated human monocytes. J Thromb Haemost. 2014;12(5): 779–791. [DOI] [PubMed] [Google Scholar]
- 33.Pierangeli SS, Vega-Ostertag ME, Raschi E, et al. Toll-like receptor and antiphospholipid mediated thrombosis: in vivo studies. Ann Rheum Dis. 2007;66(10):1327–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hurst J, Prinz N, Lorenz M, et al. TLR7 and TLR8 ligands and antiphospholipid antibodies show synergistic effects on the induction of IL-1β and caspase-1 in monocytes and dendritic cells. Immunobiology. 2009;214(8):683–691. [DOI] [PubMed] [Google Scholar]
- 35.Aguilar-Valenzuela R, Nickerson K, Romay-Penabad Z, et al. Involvement of TLR7 and TLR9 in the Production of Antiphospholipid Antibodies. Arthritis Rheum. 2011;63(10): S281–S282. [Google Scholar]
- 36.Shi T, Giannakopoulos B, Yan X, et al. Anti-β2-glycoprotein I antibodies in complex with β2-glycoprotein I can activate platelets in a dysregulated manner via glycoprotein Ib-IX-V. Arthritis Rheum. 2006;54(8):2558–2567. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.