Abstract
Monitoring of single cell signal transduction in leukemic cellular subsets has been proposed to provide deeper understanding of disease biology and prognosis, but has so far not been tested in a clinical trial of targeted therapy. We developed a complete mass cytometry analysis pipeline for characterization of intracellular signal transduction patterns in the major leukocyte subsets of chronic phase chronic myeloid leukemia. Changes in phosphorylated Bcr-Abl1 and the signaling pathways involved were readily identifiable in peripheral blood single cells already within three hours of the patient receiving oral nilotinib. The signal transduction profiles of healthy donors were clearly distinct from those of the patients at diagnosis. Furthermore, using principal component analysis, we could show that phosphorylated transcription factors STAT3 (Y705) and CREB (S133) within seven days reflected BCR-ABL1IS at three and six months. Analyses of peripheral blood cells longitudinally collected from patients in the ENEST1st clinical trial showed that single cell mass cytometry appears to be highly suitable for future investigations addressing tyrosine kinase inhibitor dosing and effect. (clinicaltrials.gov identifier: 01061177)
Introduction
Chronic myeloid leukemia (CML) is a hematologic stem cell disorder driven by transcription of the fusion protein Bcr-Abl1, a constitutively active tyrosine kinase.1 The deregulated kinase activity is sufficient to maintain the CML phenotype2,3 and leads to activation of several pathways, including JAK/STAT,4 PI3K/Akt5 and Ras/MAPK.6 Accordingly, specific tyrosine kinase inhibitors (TKIs) targeting Bcr-Abl1 have revolutionized CML treatment. The majority of patients display excellent cytogenetic and molecular responses and remain free of disease progression.7–9 However, for many CML patients, a cure will probably not be achieved by TKIs alone because of the persistence of leukemic stem cells that may cause relapse.10,11 Furthermore, a significant number of patients are switched from one TKI to another because of insufficient efficacy or adverse effects.12 Early identification of suboptimal responders to TKI could facilitate optimal dosing and choice of TKI thereby limiting adverse events and securing optimal molecular responses.13 This could be accomplished by the direct assessment of Bcr-Abl1 kinase activity in the leukemic cells early after start of TKI therapy.
Pharmacodynamic analysis of TKI effect on Bcr-Abl1 tyrosine kinase activity and subsequent downstream signal transduction in CML patients has been technically challenging and thus rarely exploited in clinical trials.14,15 However, recent advances in single cell analysis of intracellular signal transduction show promise both in diagnostics and in therapeutic leukemia monitoring.16–18 Cytometry by Time of Flight (CyTOF) allows single cell characterization of immunophenotypes and intracellular signaling status in healthy individuals19,20 and patients with hematologic malignancies.21 The technique enables measurement of more than 40 parameters per cell, thereby dramatically increasing the dimensionality of acquired data compared to conventional flow cytometry. This is achieved by exploiting the resolution of mass spectrometry and stable rare-earth isotope-conjugated antibodies to perform highly multiplexed assays on single cells. This has permitted the dynamic signal transduction events within the hematologic hierarchy in acute myeloid leukemia to be evaluated.22–24
A panel of antibodies targeting more than 30 cell-surface and intracellular phospho-specific epitopes covering the major signaling molecules in Bcr-Abl1 oncogenic signaling (pBCR Y177, pAbl Y245, pCRKL Y207, pSTAT1 Y701, pSTAT3 Y705, pSTAT5 Y694, pCREB S133, pERK 1/2 T202/Y204, pS6 ribosomal protein S235/36 and S240/44) were developed. By analyzing longitudinally collected samples we show that both TKI-mediated changes in signal transduction and immunophenotypes is measurable, and propose that a 1-tube-based analysis by mass cytometry is a feasible biomarker strategy in clinical trials of leukemia.
Methods
Patients
Patients were eligible for the present study if they were enrolled in the international phase IIIb ENEST1st study.25 Patients were enrolled when they had newly diagnosed chronic phase CML, were 18 years of age or older, and had a World Health Organization (WHO) performance status no higher than 2. The demographic and clinical parameters of all patients included in this study (n=17) are shown in Online Supplementary Tables S1 and S2. Treatment consisted of nilotinib 300 mg BID. A peripheral blood (PB) sample was collected before the first nilotinib dose, and three hours, seven days and 28 days after the start of treatment. The study was performed in accordance with the Declaration of Helsinki, and all patients provided written informed consent. The study was approved by the local institutional review boards of all participating centers and is registered at clinicaltrials.gov identifier: 01061177. PB and bone marrow (BM) were collected from healthy individuals after written informed consent (University of Bergen, Norway, local ethical committee approval 2012/1045).
Materials
The primary material was fixed and erythrocytes lysed by Lyse/Fix buffer (BD Phosflow) in local hospital laboratories and immediately frozen in saline for long-term storage and shipping at −80°C. The longitudinal PB samples from each patient (before first dose of nilotinib, and after 3 hours, 7 days, and 28 days) were barcoded for identification, pooled and stained with the antibody panels (Online Supplementary Table S3) according to the manufacturer’s recommendation (MaxPar Phospho-Protein Staining Protocol). In the preliminary dataset #1, a protocol for barcoding was adapted from Zunder et al.26 (Online Supplementary Figure S1) and barcoded reagents were a kind gift from Prof. Bernd Bodenmiller, University of Zurich, Switzerland. The acquisition of samples was performed on the CyTOF2 mass cytometer (Fluidigm) at the Mass Cytometry Facility, University of Zurich, Switzerland. In dataset #2, a commercially available barcoding kit was used (Fluidigm). Acquisition of this dataset was performed using the Helios mass cytometer (Fluidigm) at the Flow Cytometry Core Facility, University of Bergen, Norway.
Statistical analysis
Friedman non-parametric test with Dunn’s multiple comparison was used to find statistically significant changes in phosphorylation of intracellular targets (before, and after 3 hours and 7 days of treatment). Wilcoxon matched-pairs signed rank test was used to identify statistically significant changes in cell abundances before and after initiation of nilotinib therapy. Prism software (GraphPad Software Inc.) was used to calculate P-values and plot the graphs. P≤0.05 was considered statistically significant.
Results
The immunophenotype and signal transduction in CD34+cells of CML patients
It has been proposed that both number of hematopoietic stem and progenitor cells (CD34+CD38low and CD34+CD38high, respectively), and their signaling response patterns to TKIs are important for response prediction in CML.27 Therefore, we performed an initial analysis of the PB CD34+ compartment before and after receiving the first dose (3 hours and 7 days) of nilotinib in 17 patients enrolled in the ENEST1st study. The 3 samples from each patient were barcoded, pooled and stained with a validated panel of antibodies (Online Supplementary Figures S2 and S3). The high dimensional single-cell data for each patient were clustered using the SPADE algorithm,28 where manual annotation identified several major hematologic cell subsets, including the CD34+ cells. This population was further gated manually into CD34+CD38low and CD34+CD38high cells. Due to inter-patient variation in CD38 expression range, the gating was tailored to each longitudinal patient sample set. Only patients for whom a minimum of 100 cells could be identified as CD34+CD38low and CD34+CD38high for all samples were included in the final analysis (n=10). To characterize the debulking of the tumor load at the single cell level and over time, all CD34+ cells were analyzed using viSNE algorithm.21 This allowed single cells from different samples to be graphed in a unifying 2-dimensional plot, where its (x,y) position contains information about cell surface marker expression (see Figure 1A for 2 representative patients’ sample sets). CD34+CD38low cells resided in the lower part of the viSNE plot, while CD34+CD38high cells resided in the middle/top right. It has been suggested that CD25 can be used to identify leukemic stem cells in CML.29 In this pool of CD34+ cells collected from PB, CD34+CD25+ cells are strongly aggregated towards the very bottom of the viSNE plot, in the CD34+CD38low region. During seven days of TKI treatment, we saw a statistically significant (P≤0.01) depletion of CD34+ cells from PB leukocytes (Figure 1B). Furthermore, after gating of CD34+CD25+ double positive cells, we saw a statistically significant depletion of CD34+CD25+ cells compared to CD34+ cells at seven days of therapy (Figure 1C). The 85th percentile metal intensity of pAbl Y245, pCRKL Y207, pSTAT3 Y705 and pSTAT5 Y694 was calculated for the manually gated CD34+CD38low and CD34+CD38high single cell populations. Comparing the changes of phosphorylation level across patients over time, we found a statistically significant increase in pSTAT3 Y705 in the CD34+CD38high population (P≤0.05) between samples collected before and after seven days of TKI therapy (Figure 1D). Based on these observations in the CML progenitor compartment, we decided to examine more highly differentiated CML cells based on a more extensive panel of antibodies characterizing the myeloid and lymphoid lineages in greater detail (Online Supplementary Figures S4 and S5)
Figure 1.
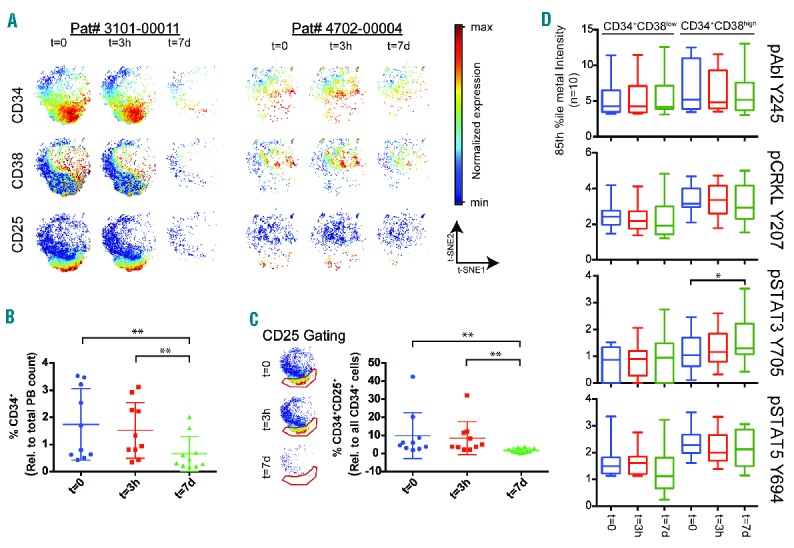
Nilotinib dosing altered signal transduction of the CD34+ cell population in chronic phase chronic myeloid leukemia (CML) patients analyzed by mass cytometry. Patient peripheral blood (PB) was collected at trial inclusion (t=0; before dosing), and after three hours (t=3h) and at day 7 (t=7d) of nilotinib (300 mg BID). (A) The CD34+ subset identified by the SPADE algorithm was further analyzed by the viSNE algorithm.21 The CD34+ cells from 2 representative patients at the three time points are shown. For each patient, the range of expression of each cell surface marker (CD34, CD38 and CD25) is color-coded from minimum to maximum expression of the three longitudinal samples. (B) The ratio of CD34+ cells and the total PB counts was calculated for all patients (n=10) in all three longitudinal samples. We found a statistically significant decrease of CD34+ cells in the PB between t=0 and day 7, and between after three hours and day 7 (Friedman non-parametric with Dunn’s multiple comparison; **P≤0.01). (C) For all patients, CD34+CD25+ cells were identified on the viSNE plots (see gating scheme). The ratio between CD34+CD25+ and all CD34+ cells was calculated showing a statistically significant decrease in CD34+CD25+ cells between diagnosis and day 7, and between after three hours and day 7 (Friedman non-parametric with Dunn’s multiple comparison; **P≤0.01). (D) Intracellular signaling transduction targets of Bcr-Abl1. The 85th percentile metal intensities of pAbl Y245, pCRKL Y207, pSTAT3 Y705, pSTAT5 Y694 and pCREB S133 were calculated and data of the patient cohort (n=10) for the CD34+CD38low and CD34+CD38high populations box plotted as a function of time. We observed a statistically significant change of pSTAT3 Y705 in the CD34+CD38high population (Friedman non-parametric with Dunn’s multiple comparison, *P≤0.05) between samples collected at diagnosis and day 7.
Mass cytometry identified hematologic remission in clinical trial samples
To validate our methodological approach, we analyzed a second cohort of patients (n=8) enrolled in the ENEST1st clinical trial. From each patient, 3 longitudinally collected samples (before, and after 3 hours and day 7 of TKI treatment) were barcoded, along with 4 healthy PB and 4 healthy BM samples, using the commercially available 20-plex metal barcoding kit (Fluidigm). Each longitudinal set of 3 samples from a patient, and healthy PB and BM, was analyzed using the SPADE clustering algorithm,28 and the major leukocyte subsets were manually identified and annotated (Figure 2A). The relative abundance of a selection of cellular subsets quantified in longitudinal samples collected before and after seven days of TKI therapy is shown in Figure 2B and C. The abundance of the corresponding healthy cell subset is shown in a lighter shade, identified either from the healthy PB or BM samples (n=4 each). Similar to previous results (Online Supplementary Figure S3), already at day 7 of TKI therapy we observed a statistically significant change in the relative abundance of several cellular subsets. We measured a statistically significant reduction of myelocytes and progenitor cells (CD34+CD38low), and an expansion of mature neutrophils, CD8+ T cells, and activated T cells, (Wilcoxon matched pairs signed rank test P>0.05 for all).
Figure 2.
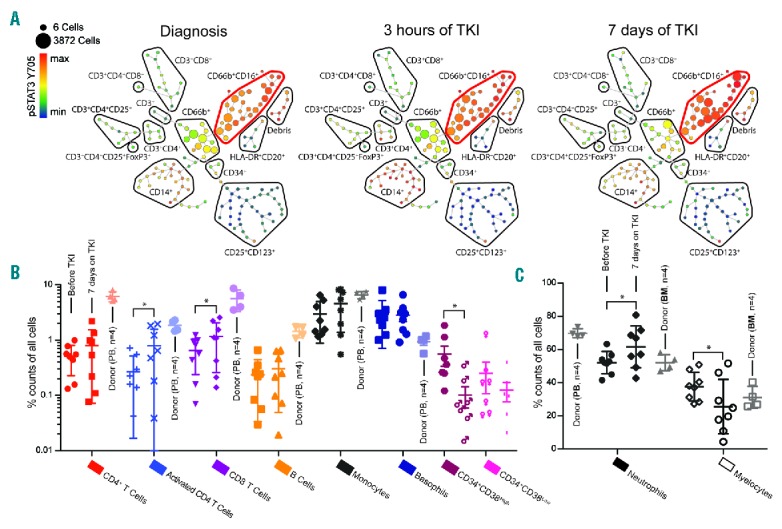
High-resolution single cell immune profiles of healthy and patient leukocytes. Longitudinally collected samples (before, after 3 hours and day 7 on nilotinib) from 8 patients in the ENEST1st trial, together with 4 healthy peripheral blood (PB) and bone marrow (BM) samples, were barcoded using the 20-plex metal barcoding kit (Fluidigm). (A) Data from the longitudinal samples from each patient, and the 4 healthy PB and BM samples, were pooled and clustered using the SPADE algorithm28 and manually annotated to identify cellular subsets. The SPADE tree analysis of patient 4702_0004 is shown. The size of each node represents the number of cells clustered and the expression of pSTAT3 Y705 is color-coded. The red bubble highlights the mature neutrophil population. (B and C) The relative abundance of the major PB populations identified in samples collected at diagnosis and after seven days of tyrosine kinase inhibitor (TKI) therapy is shown for the patient cohort (n=8, error bars showing standard error of mean, SEM), with the appropriate subpopulation in the healthy samples shown in a lighter shade. CD34+ cells could only be identified in 7 out of 8 patients. Wilcoxon matched-pairs rank test was used to identify statistically significant changes from before and after seven days of TKI therapy, where P≤0.05 was considered statistically significant.
The direct measurement of phosphorylated Bcr-Abl1 in single cells from patients treated with nilotinib
The phosphorylation level of several key signaling molecules in the Bcr-Abl1 signal transduction network was characterized in each patient, as a function of time and nilotinib treatment (before, and after 3 hours and 7 days of TKI therapy; n=8), and in the healthy PB and BM (n=4 each) (Figure 3A). In general, these data support our initial findings (Online Supplementary Figure 3C), reproducing the clearly distinct signaling status observed in healthy PB and CML at diagnosis, including the attenuated level of pSTAT3 Y694 phosphorylation in the CML neutrophils. In the larger patient cohort (n=8), we are able to measure a statistically significant decrease in pCRKL Y207 as a function of Bcr-Abl1 inhibition in the basophils (between before and after 7 days on TKI; P≤0.05), and an increase in pSTAT5 Y694 in the CD4 T helper cells (between diagnosis and after 3 hours of TKI; P≤0.01). We also measured a clear trend of pAbl Y245 downregulation in the CD34+CD38− population as a function of TKI-therapy (simple Wilcoxon test after 7 days of TKI P=0.0313). The seemingly ubiquitous regulation of pCREB S133, initial decrease and subsequent increase, reached statistical significance in the neutrophils (between before and after 7 days, and between diagnosis and after 3 hours of TKI; P≤0.05 for both) but was observed in several cellular subsets. In the hematopoietic stem and progenitor compartment (Figure 3B), we reproduced a statistically significant increase in pSTAT3 Y694 (between after 3 hours of TKI and after 7 days of TKI; P≤0.05) similar to what can be seen in the CD34+CD38high progenitor cell population (Figure 1D).
Figure 3.
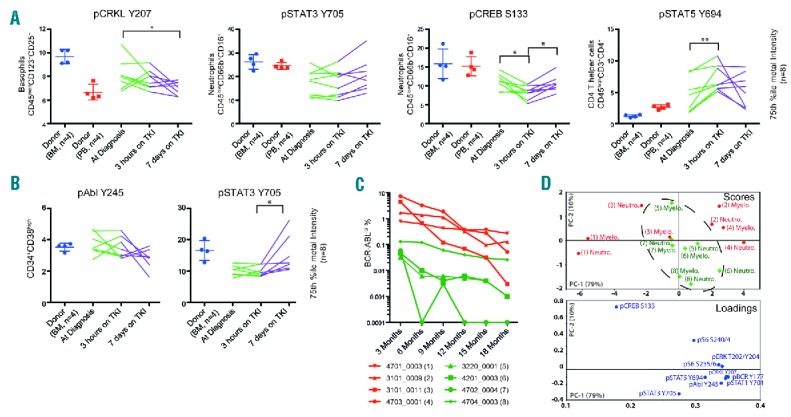
Single cell signaling profiles correlate to BCR-ABL1IS molecular response. The phosphorylation level (75th percentile of metal intensity) of the intracellular Bcr-Abl1 signaling network (n=10) was measured in the longitudinal samples (before, after 3 hours and after 7 days of nilotinib) in the patient cohort (n=8), together with 4 healthy peripheral blood (PB) and bone marrow (BM) samples. (A) We observed distinctly different states of signal transduction in the patient samples compared to the healthy controls. (B) The Lin-CD34+CD38low and Lin−CD34+CD38high cell populations were manually gated from the CD34+ population identified by SPADE, and the intracellular signal transduction measured. CD34+ cells could only be identified in 7 out of 8 patients. The same strategy was followed for healthy CD34+ cells. For both (A and B) statistical significance was determined using Friedman non-parametric with Dunn’s multiple comparison test, where P≤0.05 was considered statistically significant (*P≤0.05, **P≤0.01). (C) The BCR-ABL1IS of each patient during tyrosine kinase inhibitor (TKI) therapy. The patient cohort was divided into two groups based on the BCR-ABL1IS at three and six months (red and green). (D) An unsupervised PCA analysis of the signal transduction arcsinh fold change compared samples before and after seven days of nilotinib treatment in the mature neutrophils (Neutro) and myelocytes (Myelo) was performed using Unscramble software (CAMO Software). The categorical color-coding from (C) was superimposed on the resulting PCA plot, and the dashed line was drawn manually.
TKI-mediated changes in single cell signal transduction reflects BCR-ABL1IS at 3 and 6 months
Finally, we examined whether the observed TKI-induced changes in signal transduction networks could provide prognostic information. We split the patients according to their BCR-ABL1IS at three and six months into two response groups (Figure 3C, low in green and high in red). The arcsinh fold change of each signal transduction epitope (n=10) between diagnosis and after seven days of TKI treatment was calculated for the neutrophils and myelocytes for all patients (n=8). We performed an unsupervised multivariate principle component analysis (PCA) of the fold change data using Unscrambler X (CAMO Software), and then manually overlaid the response group affiliation (Figure 3D). The PCA analysis revealed underlying differences in signaling status between the response groups, driven mainly by the rates of downregulation of pCREB S133 and upregulation of pSTAT3 Y705.
Discussion
Mass cytometry enabled the measurement of more than 30 intracellular and cell surface markers on each single cell, with an additional six channels reserved for metal barcoding for sample multiplexing.30 The convenient single tube labeling of multiplexed samples combined with semiautomatic analysis makes the technique highly efficient. We were able to recapitulate the expected hematologic response, and could also demonstrate the debulking of leukemic CD34+ cells in the PB by immunophenotyping as early as one week after start of therapy. Importantly, by simultaneously probing key intracellular phosphorylation targets of the Bcr-Abl1 signaling network,2,3 we monitored changes in signal transduction of individual cell types for each patient undergoing TKI therapy. Unsupervised principle component analysis of these early changes in signal transduction allowed patients to be identified according to their BCR-ABL1IS, indicating a possible future prognostic impact of this approach.
The proportion of leukemic stem cells (as determined by FISH) in the BM of chronic phase CML have been shown to have a prognostic significance.26 In the PB of our patients, we were able to monitor the therapy-dependent debulking of CD34+ cells (Figure 1B), and both CD34+CD38low and CD34+CD38high cells (data not shown), at day 7 of therapy. Interestingly, at day 7 we saw a statistically significant depletion of CD34+CD25+, a putative CML stem cell subset27 compared to all CD34+ cells (Figure 1C). This could indicate an increased sensitivity to TKI or alternatively an increased bone marrow homing of this cell subset.29 Early relative increase of lymphocytes in the PB of CML patients treated with TKI has also previously been shown to have prognostic importance.31 Our immune profiling of patient leukocytes allowed a high-resolution picture of PB subpopulations, enabling the simultaneous monitoring of the size of all major hematopoietic lineages. Here, the analysis of healthy PB and BM side-by-side with primary material clearly delineates cell populations dominating in CML compared to healthy individuals (Figure 2). Interestingly, we were able to measure statistically significant changes in abundances for several cellular subpopulations, including an increase in PB CD8+ cytotoxic T cells already after seven days of TKI therapy.
We believe that a particular strength of our single cell analysis is the direct monitoring of nilotinib-induced changes in signal transduction. This includes the primary target kinase for nilotinib, Abl1, as well as its immediate substrates like pCRKL Y20732 and pivotal Abl1-affected signal transduction pathways in myeloid neoplasias, e.g. the STAT3/5 pathway.16,17,33
We established a metal barcoded pool of stimulated cell lines for functional validation of our panel of phospho-specific antibodies (Online Supplementary Figure S5). Indeed, we could measure a statistically significant reduction of pCRKL Y207 in the basophils as a function of nilotinib therapy in our patents (n=8) (Figure 3A). Compared to healthy controls, we could measure a clearly attenuated pSTAT3 Y705 in mature neutrophils (Figure 3A and Online Supplementary Figure S3C) and in the CD34+CD38high progenitor compartment (Figures 1C and 3C) collected before treatment initiation, which subsequently increased during TKI treatment. A similar increase in pSTAT3 Y705 has previously been reported in both mature neutrophils and CD34+ cells.34,35 In BCR-ABL1 oncogene-addicted CML cells, it has been shown that growth factor receptor signaling is dampened through MEK/ERK-dependent negative feedback of the BCR-ABL1 signal.36 This may explain the attenuated basal level of phosphorylation of pSTAT3. Finally, we observe a decrease of pCREB S133 as early as three hours after first TKI dosing in several cellular subsets followed by an increase after seven days. Interestingly, we see that, compared to non-CML cell lines, K562 cells express high levels of pCREB S133 at baseline that was reduced by TKI treatment (Online Supplementary Figure S5C). Using a principle component analysis to analyze changes in signal transduction between diagnosis and after seven days of TKI treatment, we could discriminate patient groups according to their BCR-ABL1IS at three and six months (Figure 3D). The PCA analysis was driven mainly by mutual regulation of pCREB S133 and pSTAT3 Y705, and underscores a potential prognostic application for this assay.
In summary, here we demonstrate the first human study of signal transduction in cancer cells from nilotinib-treated CML patients in a prospective clinical trial, demonstrating significant phosphorylation of CREB and STAT3 in chronic phase CML patients early after start on therapy. We propose that mass cytometry, enabling simultaneous measurement of more than 30 parameters on single cells, may provide a highly detailed characterization of CML that allows comprehensive 1-tube-based monitoring of the disease in patients during TKI therapy. The method outlined here will allow early changes in individual patient’s immunophenotype and signal transduction as a function of TKI therapy to be monitored in clinical trials. Furthermore, this approach may shed light on the relation between the kinetics of TKI-modulated signal transduction and clinical response. We anticipate this technology to have high clinical impact by providing cancer cell signal transduction profiles in patients close to real time after start of TKI therapy. Additional work on standardization of antibody panels is a prerequisite if mass cytometry is to be established as a future tool to guide signal transduction-targeted therapy.
Supplementary Material
Acknowledgments
The authors would like to thank personnel at the following ENEST1st study centers for participating in this sub-study: VU University Medical Center, Amsterdam, the Netherlands; Cliniques Universitaires St Luc, Brussels, Belgium; University of Helsinki and Comprehensive Cancer Center, Helsinki University Hospital, Finland; Cliniques Universitaires St Luc, Brussels, Belgium; University Hospital Bonn (UKB), Bonn, Germany; Wels-Grieskirchen Hospital, Wels, Austria; Hospital Feldkirch, Austria; University Hospital, Aarhus, Denmark; Oslo University Hospital, Oslo, Norway; St Olavs Hospital Trondheim, Norway; Stavanger University Hospital, Norway; Haukeland University Hospital, Bergen; Norway; MTZ Clinical Research, Warsaw, Poland; University Hospital Leuven, Belgium; University Hospital Center Rebro, Zagreb, Croatia; Karolinska University Hospital, Stockholm, Sweden; Medical University of Gdańsk, Poland; University Hospital Sofia, Bulgaria.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/102/8/1361
Funding
This study was supported by The Research Council of Norway (Petromaks program grant #220759), Helse Vest health trust and the Norwegian Cancer Society with Solveig & Ole Lunds Legacy. Novartis is acknowledged for financially supporting data and sample collection for this sub-study.
References
- 1.Nowell PC, Hungerford DA. A minute chromosome in human granulocytic leukemia. Science. 1960;32:1497–1501. [Google Scholar]
- 2.Deininger MW, Goldman JM, Melo JV. The molecular biology of chronic myeloid leukemia. Blood. 2000;96(10):3343–3356. [PubMed] [Google Scholar]
- 3.Cilloni D, Saglio G. Molecular pathways: BCR-ABL. Clin Cancer Res. 2012;18(4): 930–937. [DOI] [PubMed] [Google Scholar]
- 4.Chai SK, Nichols GL, Rothman P. Constitutive activation of JAKs and STATs in BCR-Abl-expressing cell lines and peripheral blood cells derived from leukemic patients. J Immunol. 1997;159(10):4720–4728. [PubMed] [Google Scholar]
- 5.Ly C, Arechiga AF, Melo JV, Walsh CM, Ong ST. Bcr-Abl kinase modulates the translation regulators ribosomal protein S6 and 4E-BP1 in chronic myelogenous leukemia cells via the mammalian target of rapamycin. Cancer Res. 2003;63(18):5716–5722. [PubMed] [Google Scholar]
- 6.Cortez D, Reuther G, Pendergast AM. The Bcr-Abl tyrosine kinase activates mitogenic signaling pathways and stimulates G1-to-S phase transition in hematopoietic cells. Oncogene. 1997;15(19):2333–2342. [DOI] [PubMed] [Google Scholar]
- 7.Hughes TP, Hochhaus A, Branford S, et al. Long-term prognostic significance of early molecular response to imatinib in newly diagnosed chronic myeloid leukemia: an analysis from the International Randomized Study of Interferon and STI571 (IRIS). Blood. 2010;116(19):3758–3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosti G, Palandri F, Castagnetti F, et al. Nilotinib for the frontline treatment of Ph(+) chronic myeloid leukemia. Blood. 2009;114(24):4933–4938. [DOI] [PubMed] [Google Scholar]
- 9.Cortes JE, Jones D, O’Brien S, et al. Results of dasatinib therapy in patients with early chronic-phase chronic myeloid leukemia. J Clin Oncol. 2010;28(3):398–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graham SM, Jørgensen HG, Allan E, et al. Primitive, quiescent, Philadelphia-positive stem cells from patients with chronic myeloid leukemia are insensitive to STI571 in vitro. Blood. 2002;99(1):319–325. [DOI] [PubMed] [Google Scholar]
- 11.Jorgensen HG, Allan EK, Jordanides NE, Mountford JC, Holyoake TL. Nilotinib exerts equipotent antiproliferative effects to imatinib and does not induce apoptosis in CD34+ CML cells. Blood. 2007; 109(9):4016–4019. [DOI] [PubMed] [Google Scholar]
- 12.Hochhaus A, O’Brien SG, Guilhot F, et al. Six-year follow-up of patients receiving imatinib for the first-line treatment of chronic myeloid leukemia. Leukemia. 2009;23(6):1054–1061. [DOI] [PubMed] [Google Scholar]
- 13.Quintás-Cardama A, Kantarjian H, Jones D, et al. Delayed achievement of cytogenetic and molecular response is associated with increased risk of progression among patients with chronic myeloid leukemia in early chronic phase receiving high-dose or standard-dose imatinib therapy. Blood. 2009;113(25):6315–6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iliuk AB, Tao WA. Is phosphoproteomics ready for clinical research¿ Clin Chim Acta. 2013;420:23–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baca Q, Cosma A, Nolan G, Gaudilliere B. The road ahead: Implementing mass cytometry in clinical studies, one cell at a time. Cytometry B Clin Cytom. 2017; 92(1):10–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skavland J, Jørgensen KM, Hadziavdic K, et al. Specific cellular signal-transduction responses to in vivo combination therapy with ATRA, valproic acid and theophylline in acute myeloid leukemia. Blood Cancer J. 2011;1(2):e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kotecha N, Flores NJ, Irish JM, et al. Single-cell profiling identifies aberrant STAT5 activation in myeloid malignancies with specific clinical and biologic correlates. Cancer Cell. 2008;14(4):335–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jalkanen SE, Lahesmaa-Korpinen AM, Heckman CA, et al. Phosphoprotein profiling predicts response to tyrosine kinase inhibitor therapy in chronic myeloid leukemia patients. Exp Hematol. 2012;40(9):705–714.e03. [DOI] [PubMed] [Google Scholar]
- 19.Bendall SC, Simonds EF, Qiu P, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011; 332(6030):687–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bodenmiller B, Zunder ER, Finck R, et al. Multiplexed mass cytometry profiling of cellular states perturbed by small-molecule regulators. Nat Biotechnol. 2012;30(9):858–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amir el-AD, Davis KL, Tadmor MD, et al. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol. 2013;31(6):545–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levine JH, Simonds EF, Bendall SC, et al. Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis. Cell. 2015;162(1):184–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Behbehani GK, Samusik N, Bjornson ZB, Fantl WJ, Medeiros BC, Nolan GP. Mass Cytometric Functional Profiling of Acute Myeloid Leukemia Defines Cell-Cycle and Immunophenotypic Properties That Correlate with Known Responses to Therapy. Cancer Discov. 2015;5(9):988–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han L, Qiu P, Zeng Z, et al. Single-cell mass cytometry reveals intracellular survival/proliferative signaling in FLT3-ITD-mutated AML stem/progenitor cells. Cytometry A. 2015;87(4):346–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hochhaus A, Rosti G, Cross NC, et al. Frontline nilotinib in patients with chronic myeloid leukemia in chronic phase: results from the European ENEST1st study. Leukemia. 2016;30(1):57–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zunder ER, Finck R, Behbehani GK, et al. Palladium-based mass tag cell barcoding with a doublet-filtering scheme and single-cell deconvolution algorithm. Nat Protoc. 2015;10(2):316–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mustjoki S, Richter J, Barbany G, et al. Impact of malignant stem cell burden on therapy outcome in newly diagnosed chronic myeloid leukemia patients. Leukemia. 2013;27(7):1520–1526. [DOI] [PubMed] [Google Scholar]
- 28.Qiu P, Simonds EF, Bendall SC, et al. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat Biotechnol. 2011;29(10):886–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kobayashi CI, Takubo K, Kobayashi H, et al. The IL-2/CD25 axis maintains distinct subsets of chronic myeloid leukemia-initiating cells. Blood. 2014;123(16):2540–2549. [DOI] [PubMed] [Google Scholar]
- 30.Gavasso S, Gullaksen SE, Skavland J, Gjertsen BT. Single-cell proteomics: potential implications for cancer diagnostics. Expert Rev Mol Diagn. 2016;16(5):579–589. [DOI] [PubMed] [Google Scholar]
- 31.Kumagai T, Matsuki E, Inokuchi K, et al. Relative increase in lymphocytes from as early as 1 month predicts improved response to dasatinib in chronic-phase chronic myelogenous leukemia. Int J Hematol. 2014;99(1):41–52. [DOI] [PubMed] [Google Scholar]
- 32.Nichols GL, Raines MA, Vera JC, Lacomis L, Tempst P, Golde DW. Identification of CRKL as the constitutively phosphorylated 39-kD tyrosine phosphoprotein in chronic myelogenous leukemia cells. Blood. 1994; 84(9):2912–2918. [PubMed] [Google Scholar]
- 33.Irish JM, Hovland R, Krutzik PO, et al. Single cell profiling of potentiated phospho-protein networks in cancer cells. Cell. 2004;118(2):217–228. [DOI] [PubMed] [Google Scholar]
- 34.Jalkanen SE, Vakkila J, Kreutzman A, Nieminen JK, Porkka K, Mustjoki S. Poor cytokine-induced phosphorylation in chronic myeloid leukemia patients at diagnosis is effectively reversed by tyrosine kinase inhibitor therapy. Exp Hematol. 2011;39(1):102–113.e1. [DOI] [PubMed] [Google Scholar]
- 35.Eiring AM, Page BD, Kraft IL, et al. Combined STAT3 and BCR-ABL1 inhibition induces synthetic lethality in therapy-resistant chronic myeloid leukemia. Leukemia. 2015;29(3):586–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Asmussen J, Lasater EA, Tajon C, et al. MEK-dependent negative feedback underlies BCR-ABL-mediated oncogene addiction. Cancer Discov. 2014;4(2):200–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


