Splenic marginal zone lymphoma (SMZL) is a small B-cell lymphoma distinguished by frequent 7q loss and the biased use of IGHV01-02. Massive sequencing techniques have demonstrated NOTCH2-and KLF2-gene mutations to be the most frequent mutations in SMZL, with NOTCH2-mutations occurring in 10–25% of SMZL cases,1–3 and transcription factor KLF2 mutations present in 12–44% of SMZLs.1,4,5 The clinical impact of NOTCH2-alterations is controversial, with different studies reaching very distinct conclusions.3 Herein, we have analysed NOTCH2-and KLF2-mutations in a large series of SMZLs and examined the associations of these alterations with clinicopathological features. Our results confirm the association of the NOTCH2-mutation with shorter median treatment-free survival and suggest the possible usefulness of the identification of these changes for the diagnosis of SMZL.
The study panel comprised a series of 84 SMZL patients, and, for comparison purposes, 76 non-SMZL B-cell lymphomas, including 12 splenic diffuse red pulp B-cell lymphoma (SDRPL), 13 chronic lymphocytic leukemia (CLL), 14 mantle cell lymphoma (MCL), 14 follicular lymphoma (FL), 1 hairy cell leukemia (HCL) and 18 lymphoplasmacytic lymphoma (LPL). Four cases of monoclonal B-cell lymphocytosis (MBL) with non-CLL phenotype were also included. The diagnosis of all the lymphoma types included in the series was performed according to WHO criteria and Matutes et al.6,7 MBL cases had a B-cell monoclonal population without lymphadenopathy or splenomegaly.
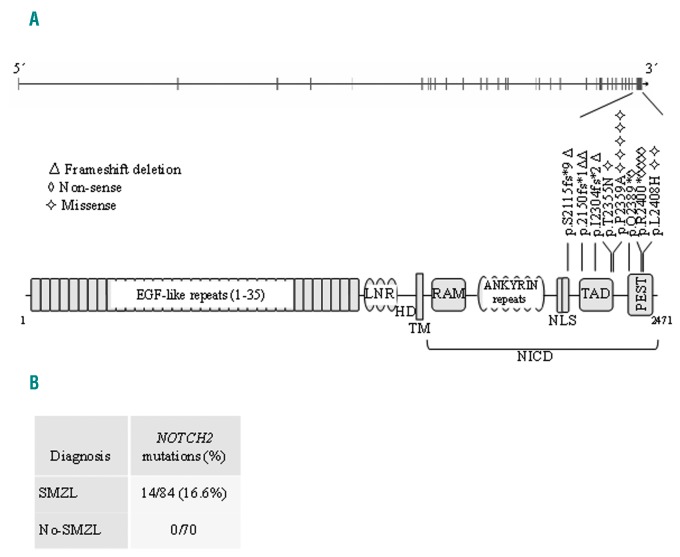
The NOTCH2-C-terminal coding exon 34 and the KLF2 exons 1–3 were sequenced by Sanger sequencing for all tumor samples in the study herein (Online Supplementary Table S1). We identified 17 NOTCH2-mutations in 14 of the 84 (16.6%) cases of SMZL, noting double mutations in 3 cases. Their somatic nature was confirmed in 5 cases for which non-tumor DNA was available. Of the 17 NOTCH2-mutations identified in SMZL, 9 were truncating (frameshift or nonsense) mutations. These deleterious mutations were localized in the nuclear localization signal (NLS), transactivation domain (TAD) and PEST domains (Figure 1), where they are predicted to alter the correct transport of the notch intracellular domain (NICD) to the nucleus or to eliminate degradation signals in the PEST domain, increasing the stability of the NICD. The remaining 8 mutations seen in SMZL were missense variants localized in the TAD and PEST domains. Two of them were predicted to be deleterious by the PolyPhen-2 program8 (Online Supplementary Table S2). No NOTCH2-abnormality was seen in the other B-cell lymphomas examined: in SDRPL (0/9), CLL (0/13), MCL (0/14), FL (0/14), HCL (0/1), LPL (0/15) and MBL (0/4). Consistent with our findings, this alteration has been reported only occasionally in other B-cell lymphomas, such as extranodal marginal zone lymphoma (1.5%), in diffuse large B-cell lymphoma (3.7–4.3%),3,9 MCL (5.2%)10 and FL (1.94%).11 It is of note that NOTCH-mutated FLs showed significantly more frequent splenic involvement than wild-type cases.11 The analysis of the relationship of NOTCH2-mutations with other alterations involved in SMZL pathogenesis has revealed an association with the deletion of 7q31–32,3 IgHV1-2 overrepresentation5 and higher promoter-methylation status.12
Figure 1.
NOTCH2-mutation distribution in SMZL. A. Schematic representation of the human NOTCH2-gene (top) and the protein (bottom), with its key functional domains. B. Number of cases of mutation for diagnosis. Symbols indicate the type and position of the mutation. EGF: epithelial growth factor; LNR: NOTCH repeats; HD: heterodimerization; TM: transmembrane; RAM: regulation of amino acid metabolism; NLS: nuclear localization signal; TAD: transactivation domain; PEST: pro-line (P), glutamic acid (E), serine (S) and threonine (T) peptide sequence; SMZL: splenic marginal zone lymphoma; NICD: notch intracellular domain.
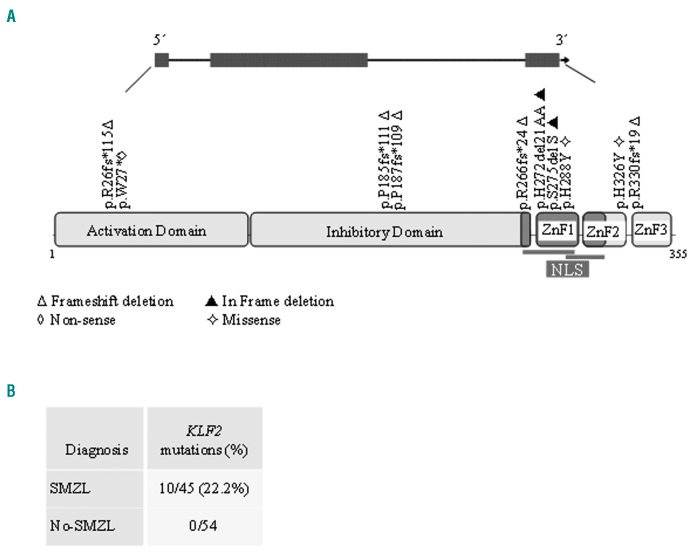
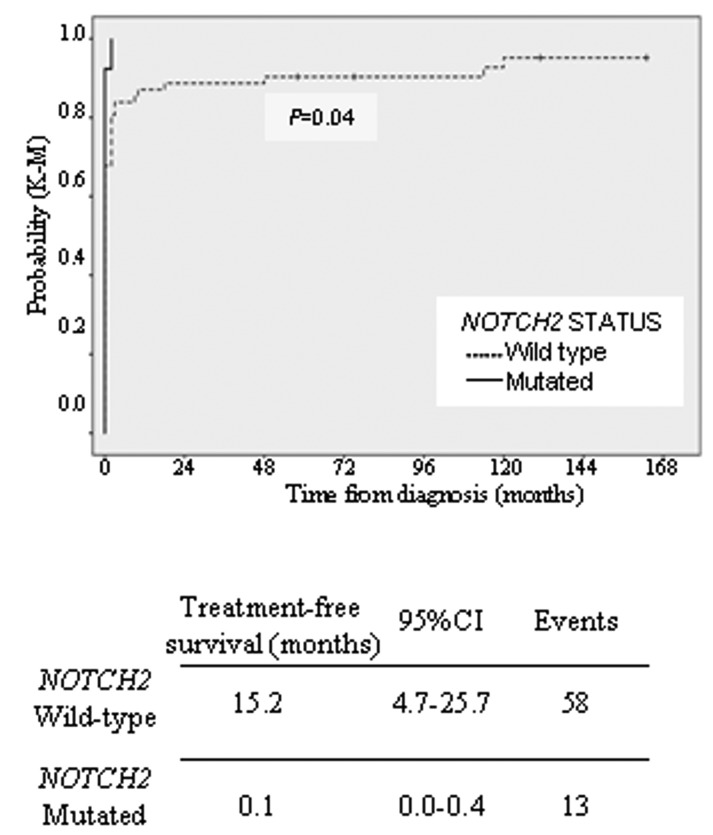
KLF2-is also frequently targeted by mutation in SMZL.4,5 We investigated KLF2-mutations (exon 1–3) in this SMZL series with available material (45/84). The KLF2 protein comprises activating and inhibitory domains, 2 NLS and 3 zinc finger motifs (C2H2 type 1, 2 and 3). In our study, we identified 10 mutations in 10 of the 45 (22.2%) cases of SMZL, similar to the frequencies reported by other authors. Two of the cases with KLF2-mutations also had a mutation in NOTCH2. Their somatic nature was confirmed in 1 case for which non-tumor DNA was available. KLF2-mutations included 5 frameshift deletions and 1 nonsense mutation. The c.76 del 1 base pair (bp) frameshift occurs in the splice site inside exon 2. Based on their distribution in KLF2, these truncating mutations are predicted either to disrupt the entire protein or to remove the zinc-finger domains of KLF2, including the putative NLS and part of the inhibitory domain (Figure 2). Two inframe deletions were localized in the first zinc-finger domain that also include part of NLS sequence. Two missense mutations according to the PolyPhen-2 program8 were predicted to be deleterious (Online Supplementary Table S2). KLF2-was investigated across the clinicopathological spectrum of other B-cell tumors (n=54). In contrast to other reports, the KLF2-mutation was not found in SDRPL (0/5), CLL (0/13), MCL (0/9), FL (0/13), HCL (0/1), LPL (0/9) or MBL (0/4). KLF2-lesions are known to be enriched in SMZL cases with 7q deletion,4,5 NOTCH2-mutations4,5 and IGHV1-02 usage.4,5 In our series, we found a significant association between NOTCH2-and KLF2-mutations (P<0.005), but there are insufficient cases with molecular data about IGHV1-02 and 7q deletion which precludes the obtainment of statistical results. The clinical findings of SMZL patients are summarized in the Online Supplementary Table S3. The median age of patients was 64 years (range, 32–98 years), and there was a strong predominance of females (M:F, 1:2). The median follow up was 70 months (range, 2–209 months). Around 60% (46/76) of patients received some type of treatment (splenectomy and/or chemotherapy) at the time of diagnosis. In 25 patients, the mean time from diagnosis to first treatment was 31 months (range, 2–148 months). Four patients had not received any treatment by the last followup date, and were managed following solely a “wait-and-see” policy. Splenectomy was performed in 70/78 (89.7%) patients. Chemotherapy was administered in 30/60 patients. Clinical progression, large B-cell transformation or death attributable to the tumor was observed in 29/67 (43.2%) patients. Adopting the clinical scoring system proposed by Montalban et al.,13 we distinguished 3 risk groups with significantly different overall survival (OS). We defined OS as the time from the date of diagnosis to death from any cause. Event-free survival (EFS) was considered as the time from diagnosis to clinical progression, large B-cell transformation or death attributable to the tumor. In our series, neither NOTCH2-nor KLF2-status was correlated with OS or EFS. We found a relationship between NOTCH2-status and treatment-free survival (TFS), defined as the time between date of diagnosis and the date of initiation of first treatment or date of last follow up at which the patient was known to be untreated. Wild-type and mutant patients exhibited a median TFS of 15.2 and 0.1 months, respectively (P=0.04) (Figure 3). Data on NOTCH2-and KLF2-mutations and their clinical relevance from more than 100 SMZL cases have been published.1–3,5 The lack of an association between the mutation status of these genes and OS or EFS is the most frequent result, including in our study. Given that SMZL is a low-grade lymphoma, it seems most appropriate to examine other parameters to assess the impact of these changes. Thus, Parry et al. identified NOTCH2-mutations as a marker of reduced TFS.1 In our series, although a high percentage of patients received treatment in the first year following diagnosis, and the number of splenectomized patients was higher than that in others series, we also found a significant relationship between NOTCH2-mutations and TFS.
Figure 2.
KLF2-mutation distribution in SMZL. A. Schematic representation of the human KLF2-gene (top) and the protein (bottom), with its key functional domain (Activating and Inhibitory domain; NLS and ZnF, zinc finger domain). The symbols denote the type and position of the mutation. B. Number of cases of mutation for diagnosis. NLS: nuclear localization signal; SMZL: splenic marginal zone lymphoma.
Figure 3.
NOTCH2-status are associated with TFS. Kaplan–Meier plots of time to first treatment for patients with NOTCH2-mutations.
In other subtypes of lymphomas with NOTCH2-mutations, including large B-cell lymphoma (LBCL), CLL and FL, no significant differences in OS were observed between cases with and without NOTCH2-mutations,11 although a significant association of NOTCH-mutation with LBCL transformation in FL has been reported. However, NOTCH2-mutation is associated with aggressive behavior in MCL.10 It is of note that the NOTCH pathway is affected in approximately 25% of cases of LBCL that develop in Hepatitis C virus (HCV)-positive patients, including 20% of NOTCH2-and 4% of NOTCH1,14 in contrast to the low frequency (1.6%) in HCV-negative patients. Furthermore, the cases with NOTCH-mutations were more often associated with the low grade component, and had an adverse prognostic impact.
In summary, we confirmed the relevance of NOTCH2-and KLF2-mutations in the clinical outcome of SMZL, and the probable usefulness of that alteration in the diagnosis of SMZL. Additional studies are required to confirm our findings and to determine the pathogenic consequences of these alterations.
Supplementary Material
Acknowledgements
We acknowledge the Tumor Bank of the Hospital Virgen de la Salud (BioB-HVS, Toledo, Spain) for providing tumor samples.
Footnotes
Funding: his study was supported by Grants from the Ministerio de Ciencia e Innovación (RETICS, FIS PI12/1682, PT13/0010/0007, RTICC, CIBER de Cancer, PI16/01294) and the Asociación Española Contra el Cancer (AECC).
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Parry M, Rose-Zerilli MJ, Ljungström V, et al. Genetics and prognostication in splenic marginal zone lymphoma: revelations from deep sequencing. Clin Cancer Res. 2015;21(18):4174–4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kiel MJ, Velusamy T, Betz BL, et al. Whole-genome sequencing identifies recurrent somatic NOTCH2 mutations in splenic marginal zone lymphoma. J Exp Med. 2012;209(9):1553–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rossi D, Trifonov V, Fangazio M, et al. The coding genome of splenic marginal zone lymphoma: activation of NOTCH2 and other pathways regulating marginal zone development. J Exp Med. 2012;209(9):1537–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Piva R, Deaglio S, Famà R, et al. The Krüppel-like factor 2 transcription factor gene is recurrently mutated in splenic marginal zone lymphoma. Leukemia. 2015;29(2):503–507. [DOI] [PubMed] [Google Scholar]
- 5.Clipson A, Wang M, de Leval L, et al. KLF2 mutation is the most frequent somatic change in splenic marginal zone lymphoma and identifies a subset with distinct genotype. Leukemia. 2015;29(5):1177–1185. [DOI] [PubMed] [Google Scholar]
- 6.Swerdlow SH, Campo E, Harris NL, et al. WHO classification of tumours of haematopoietic and lymphoid tissues. Press I, editor. Lyon: 2008. [Google Scholar]
- 7.Matutes E, Oscier D, Montalban C, et al. Splenic marginal zone lymphoma proposals for a revision of diagnostic, staging and therapeutic criteria. Leukemia. 2008;22(3):487–495. [DOI] [PubMed] [Google Scholar]
- 8.Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang X, Shi Y, Weng Y, et al. The truncate mutation of Notch2 enhances cell proliferation through activating the NF-κB signal pathway in the diffuse large B-cell lymphomas. PLoS One. 2014;9(10):e108747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beà S, Valdés-Mas R, Navarro A, et al. Landscape of somatic mutations and clonal evolution in mantle cell lymphoma. Proc Natl Acad Sci U S A. 2013;110(45):18250–18255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karube K, Martínez D, Royo C, et al. Recurrent mutations of NOTCH genes in follicular lymphoma identify a distinctive subset of tumours. J Pathol. 2014;234(3):423–430. [DOI] [PubMed] [Google Scholar]
- 12.Arribas AJ, Rinaldi A, Mensah AA, et al. DNA methylation-profiling identifies two splenic marginal zone lymphoma subgroups with different clinical and genetic features. Blood. 2015;125(12):1922–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Montalban C, Abraira V, Arcaini L, et al. Simplification of risk stratification for splenic marginal zone lymphoma: a point-based score for practical use. Leuk Lymphoma. 2014;55(4):929–931. [DOI] [PubMed] [Google Scholar]
- 14.Arcaini L, Rossi D, Lucioni M, et al. The NOTCH pathway is recurrently mutated in diffuse large B cell lymphoma associated with hepatitis C virus infection. Haematologica. 2014;100(2):246–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.