Main Text
The Hippo pathway controls organ size, and tissue homeostasis with deregulation leading to cancer. The core hippo components in mammals are composed of upstream serine/threonine kinases Mst1/2, MAPK4Ks, and Lats1/2. Inactivation of these upstream kinases leads to dephosphorylation, stabilization, nuclear translocation, and thus activation of the major functional transducers of the Hippo pathway, YAP and its paralog TAZ1, 2. YAP/TAZ are transcription co-activators that regulate gene expression primarily through interaction with TEA domain DNA-binding family of transcription factors (TEAD)3. The current paradigm for regulation of this pathway centers on phosphorylation-dependent nucleocytoplasmic shuttling of YAP/TAZ through a complex network of upstream components2. However, unlike other transcription factors, such as SMAD, NF-κB, NFAT, and STAT, the regulation of TEAD nucleocytoplasmic shuttling has been largely overlooked. In the present study, we show that environmental stress promotes TEAD cytoplasmic translocation via p38 MAPK in a Hippo-independent manner. Importantly, stress-induced TEAD inhibition predominates YAP activating signals and selectively suppresses YAP-driven cancer cell growth. Our data reveal a mechanism governing TEAD nucleocytoplasmic shuttling and show that TEAD localization is the final determinant of Hippo signaling output.
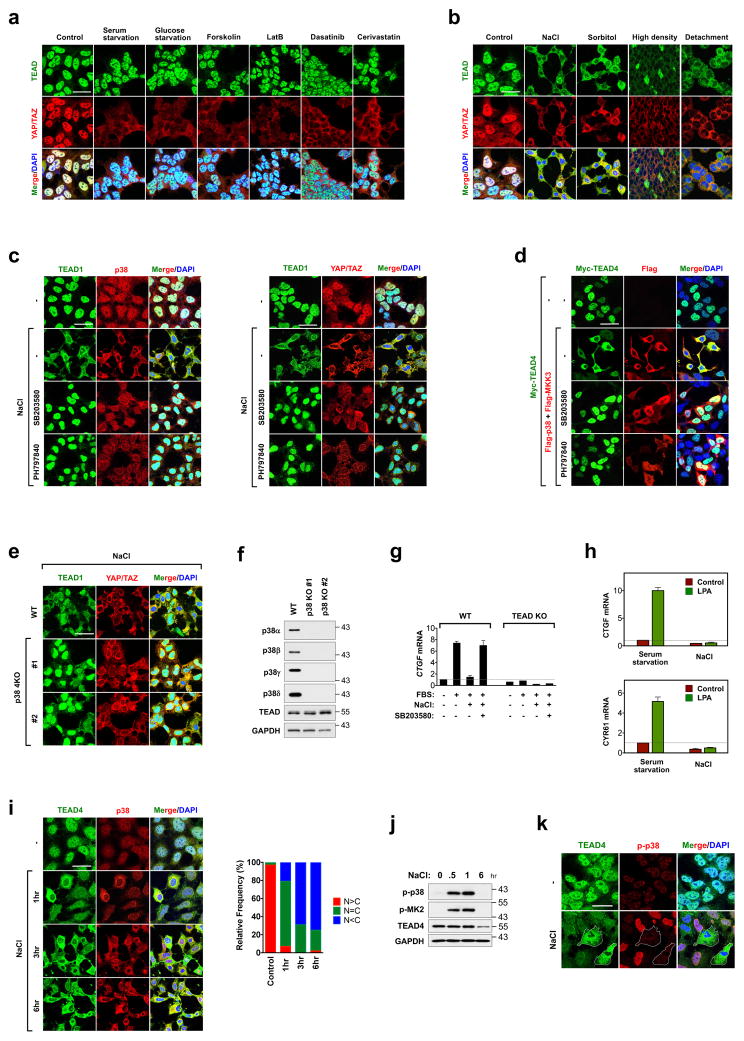
We set out to identify signals that may regulate TEAD subcellular localization by focusing on conditions known to inhibit YAP/TAZ such as serum starvation4, energy stress by glucose starvation5, 6, PKA activation by forskolin4, disruption of the actin cytoskeleton by latrunculin B7, 8, Src inhibition by dasatinib9, and inhibition of mevalonate synthesis by cerivastatin10. These well-known YAP/TAZ inhibitory stimuli indeed induced YAP/TAZ cytoplasmic localization, but failed to alter TEAD subcellular localization (Fig. 1a). In contrast, environmental stresses such as osmotic stress, high cell density and cell detachment induced cytoplasmic translocation of TEAD and YAP/TAZ (Fig. 1b and Supplementary Fig. 1a, b), demonstrating that only a subset of signals that induce YAP/TAZ cytoplasmic localization are capable of driving TEAD cytoplasmic localization.
Figure 1. p38-mediates stress-induced TEAD cytoplasmic translocation.
a, Immunofluorescence staining of TEAD and YAP/TAZ in HEK293A cells treated with YAP-inhibiting signals. b, Immunofluorescence detects TEAD cytoplasmic translocation by environmental stress. c, Effect of p38 inhibitors on osmotic stress induced-TEAD cytoplasmic translocation. HEK293A cells were pretreated with p38 inhibitors, and stimulated with NaCl and stained for immunofluorescence. d, Ectopic expression of MKK3/p38 promotes TEAD4 cytoplasmic translocation. 24 hr after transfection, cells were treated with p38 inhibitors for 8hr and stained for immunofluorescence. e, Immunofluorescence staining shows deletion of p38 impairs TEAD nucleocytoplasmic shuttling by osmotic stress. Data for two independent p38 4KO clones are shown. f, Western blotting of p38 isoforms in p38 4KO cells. g, p38 mediates inhibition of YAP-TEAD target gene expression by stress. WT and TEAD KO cells were pretreated with NaCl and SB203580 as indicated, and then stimulated with 10% serum. CTGF mRNA expression was measured by qRT-PCR. Data are presented as mean ± s.e.m. from n=3 independent experiments. h, Osmotic stress inhibits YAP-TEAD target gene expression. Cells were subject to serum starvation or NaCl, and then LPA-induced CTGF and CYR61 mRNA expression was measured by qRT-PCR. Data are presented as mean ± s.e.m. from n=3 independent experiments. i, Correlation between stress-induced cytoplasmic translocation of TEAD and p38. Cells were stimulated with NaCl for the indicated times and then subjected to immunofluorescence to detect p38. Quantification of TEAD nuclear localization (N) and cytoplasmic localization (C) is provided. Random views (~100 cells) were selected for quantification. j, Time course of p38 activation by NaCl. Western blotting of phospho-p38 and its substrate phospho-MK2 upon NaCl treatment. k, Inverse correlation between stress-induced cytoplasmic translocation of TEAD and phospho-p38. Cells were stimulated with NaCl for 1 hr and then subjected to immunofluorescence using a phospho-p38 antibody. Scale bars in a–e, i, and k are 20μm. Statistics source data are shown in Supplementary Table 1. Unprocessed scans of blots are shown in Supplementary Figure 5.
The p38 MAP kinase is activated by stress, including hyperosmotic conditions; therefore, we examined whether p38 plays a role in regulation of TEAD during stress. Treatment with p38 inhibitors (SB203580 or PH797840) blocked osmotic stress-induced, but not high density-induced, TEAD cytoplasmic localization, indicating that p38 is specifically involved in TEAD cytoplasmic translocation upon osmotic stress (Fig. 1c and Supplementary Fig. 1c). Activation of p38 by ectopic expression of p38 and its upstream kinase MKK3 also induced cytoplasmic translocation of TEAD and this effect was blocked by p38 inhibitor treatment (Fig. 1d and Supplementary Fig. 1d). We predicted that all four isoforms of p38 may play compensatory roles as ablating TEAD translocation required concentrations of p38 inhibitor which were sufficient for inhibiting all p38 isoforms (Supplementary Fig. 1e). Deletion of p38α/β (p38 2KO) resulted in p38γ/δ upregulation and did not impede TEAD cytoplasmic translocation (Supplementary Fig. 1f, g), further supporting the pharmacological evidence that all four isoforms of p38 play a role in TEAD regulation. When all four p38 genes were deleted in the p38α/β/γ/δ knockout (KO) (p38 4KO) cells, TEAD localization was insensitive to osmotic stress and largely retained in the nucleus (Fig. 1e, f and Supplementary Fig. 1f, h). Under basal conditions, deletion of p38 had no effect on TEAD localization and marginally increased YAP-TEAD activity, indicating that p38 plays a role in regulation of TEAD mainly under conditions of cellular stress. (Supplementary Fig. 1i, j). The specific role of p38 in osmotic stress is further supported by the result that p38 inhibition or knockout had no effect on density-induced TEAD cytoplasmic localization (Supplementary Fig. 1c). Collectively, the above observations demonstrate a critical role of p38 in stress-induced TEAD nucleocytoplasmic translocation. Consequently, TEAD cytoplasmic translocation by osmotic stress suppressed YAP/TAZ target gene expression induced by YAP-activating signals, such as lysophosphatidic acid (LPA) and serum4, which was rescued by p38 inhibition (Fig. 1g, h). The NaCl-induced cytoplasmic localization of TEAD was slower than p38 phosphorylation and activation (Fig. 1i, j) but occurred concurrently with p38 dephosphorylation and cytoplasmic translocation (Fig. 1k). It is well established that post osmotic stress, p38 undergoes dephosphorylation and cytoplasmic translocation, indicating that TEAD cytoplasmic translocation occurs during the adaptation phase of stress response11.
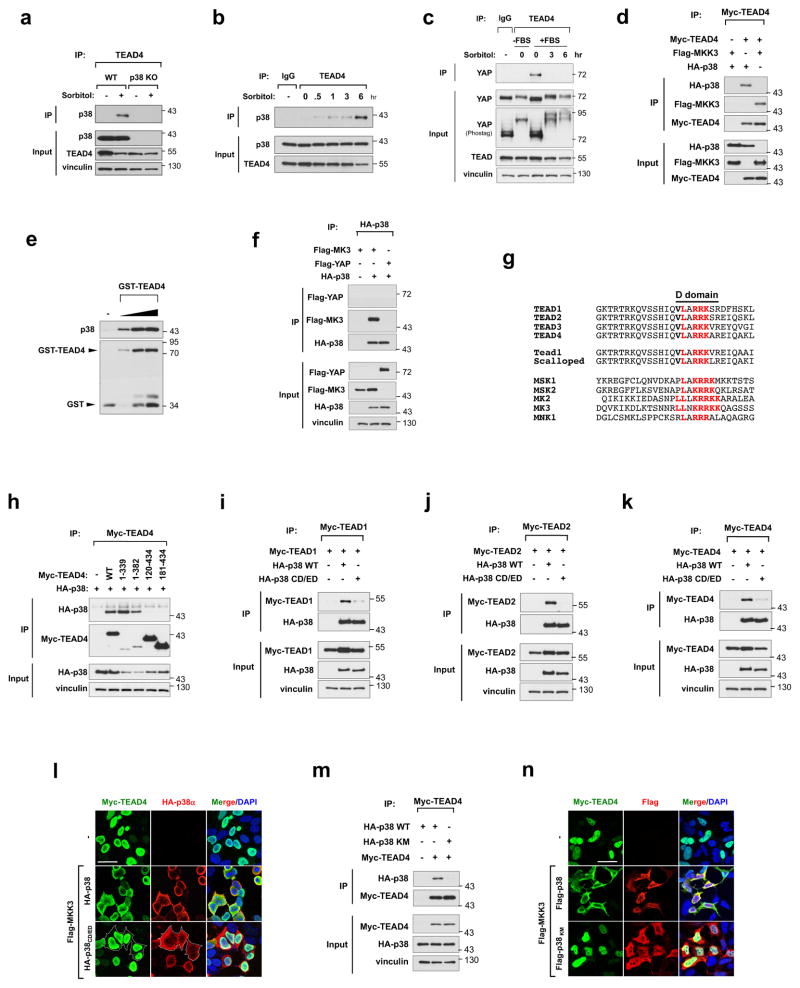
To further gain mechanistic insight into TEAD regulation by p38, we investigated the role of p38 protein-protein interaction and kinase activity, both of which are critically involved in p38 signal transduction12. We sought to determine whether p38 directly interacts with TEAD to promote cytoplasmic translocation. Interestingly, osmotic stress induced endogenous TEAD-p38 interaction, whereas serum-induced TEAD-YAP interaction was abolished by osmotic stress (Fig. 2a–c). In addition, exogenous p38 and MKK3 both showed interaction with TEAD (Fig. 2d). Using bacterially-purified proteins in an in vitro binding assay, we show TEAD can interact directly with p38 without scaffold proteins (Fig. 2e). Furthermore, p38 does not bind to YAP and thus regulates TEAD independently of YAP (Fig. 2f). The D domain, found in p38 binding partners, serves as a docking site for p38 protein-protein interactions12. We identified a putative D domain that is highly conserved within the TEAD family (Fig. 2g). Deletion of the D domain in TEAD abolished TEAD-p38 interaction (Fig. 2h). Consistently, p38 CD/ED, a p38 mutant that has lost its ability to interact with D domain-containing substrates13, 14, significantly dampened TEAD-p38 interaction and was unable to induce TEAD cytoplasmic translocation (Fig. 2i–l). Ectopic expression of kinase deficient mutant, p38 KM, was also insufficient in binding to TEAD and driving cytoplasmic translocation (Fig. 2m, n). To determine whether TEAD cytoplasmic translocation is due to p38-mediated phosphorylation, we constructed TEAD4-4SP, in which the four putative p38 phosphorylation sites were mutated to alanine (Supplementary Fig. 2a). Using an in vitro kinase assay, we found TEAD4 to be a poor substrate for p38 phosphorylation with complete ablation of phosphorylation in the TEAD4-4SP mutant, indicating the absence of alternative phosphorylation sites (Supplementary Fig. 2b–d). Additionally, TEAD4-4SP displayed normal cytoplasmic translocation upon osmotic stress (Supplementary Fig. 2e), suggesting that p38 kinase activity is required for TEAD interaction but does not directly phosphorylate TEAD to regulate its subcellular localization. Disruption of a putative TEAD nuclear export signal, as well as inhibition of Chromosomal Maintenance 1 (CRM1) using Leptomycin B (LMB), largely ablated TEAD translocation, indicating that TEAD cytoplasmic translocation is an active, CRM1-mediated process (Supplementary Fig. 2f–h). In contrast to TEAD cytoplasmic localization, osmotic stress stimulates nuclear translocation of the transcription factor Nuclear Factor of Activated T-cells 5 (NFAT5)15, thus TEAD cytoplasmic translocation is a specific cellular response upon osmotic stress (Supplementary Fig. 2i).
Figure 2. p38 mediates stress-induced TEAD cytoplasmic translocation via protein-protein interaction.
a, Detection of osmotic stress-induced TEAD4 and p38 interaction by immunoprecipitation (IP) assay. IP and WB denote immunoprecipitation and Western blot, respectively. b, Immunoprecipitation showing time course of sorbitol-induced TEAD-p38 interaction. c, TEAD immunoprecipitation shows osmotic stress ablates TEAD-YAP interaction. d, TEAD interacts with p38 and MKK3 as shown by immunoprecipitation. e, In vitro binding assay using bacterially-purified proteins shows a direct interaction between p38 and TEAD. f, p38 does not interact with YAP. g, Sequence alignment of TEAD with canonical p38 substrates. Putative D domain (red) is conserved in N-terminus of all TEAD isoforms. h, The TEAD D domain is required for interaction with p38. p38 binds TEAD4 C-terminal truncation constructs (1–339 and 1–382), but not N-terminal truncations (120–434 or 181–434) in an immunoprecipitation assay. i–k, The p38 CD/ED docking motif is required for interaction with TEAD. p38 WT, but not p38 CD/ED mutant co-immunoprecipitates with TEAD1 (g), TEAD2 (h), and TEAD4 (i). l, TEAD-p38 interaction mediates TEAD cytoplasmic translocation. Immunofluorescence staining shows TEAD cytoplasmic translocation occurs in cells transfected with p38 WT, but not CD/ED mutant. m, Effect of p38 kinase activity on p38-TEAD binding. Immunoprecipitation assay shows TEAD binds p38 WT but not kinase-dead mutant, p38KM. n, Effect of p38 kinase activity on TEAD cytoplasmic translocation. Immunofluorescence shows ectopic expression of p38 WT, but not kinase-dead mutant p38 KM, induces TEAD4 cytoplasmic translocation. Scale bars in l and n are 20μm. Unprocessed scans of blots are shown in Supplementary Figure 5.
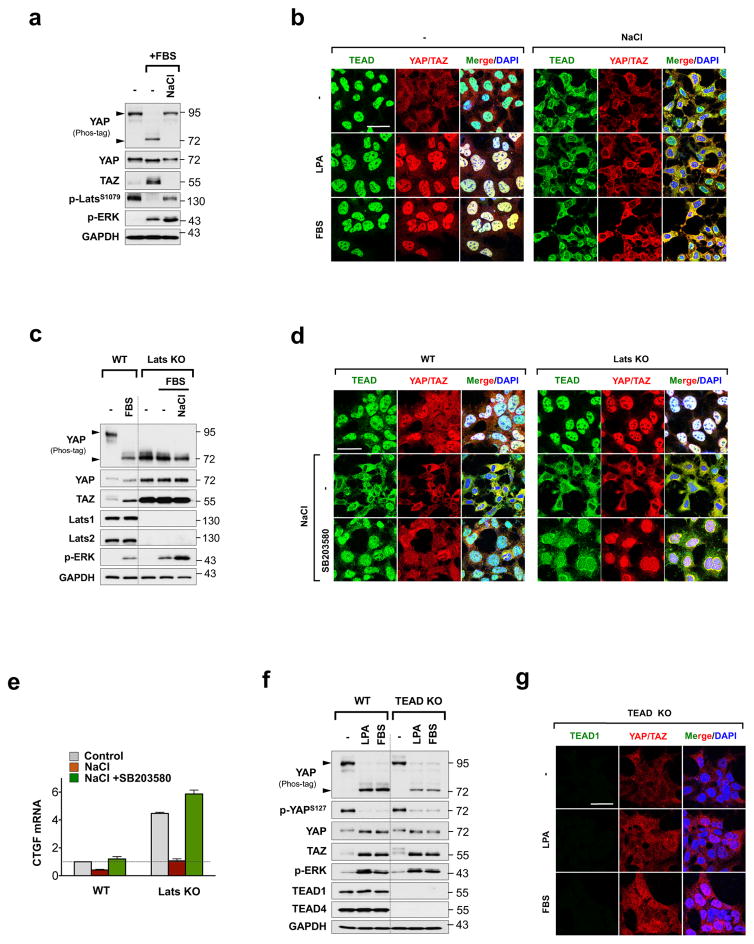
Next, we tested the effect of stress-induced TEAD cytoplasmic sequestration on YAP activation. Under osmotic stress, YAP activating signals, such as serum and LPA, failed to induce YAP dephosphorylation and nuclear accumulation (Fig. 3a, b). ERK phosphorylation, however, was not affected, suggesting specificity of osmotic stress on YAP inhibition. Unexpectedly, stress evoked TEAD and YAP/TAZ cytoplasmic translocation in MAP4K 4/6/7, Mst1/2 and Lats1/2 KO cells, despite constitutively dephosphorylated YAP (Fig. 3c, d and Supplementary Fig. 3a–g). These results indicate that stress-induced TEAD and YAP/TAZ cytoplasmic retention is a Hippo-independent process and uncouples YAP dephosphorylation from its nuclear localization. Consistently, p38 inhibition restored TEAD in the nucleus in Lats1/2 KO cells (Fig. 3d). Compared to WT cells, p38 inhibition enhanced YAP/TAZ nuclear accumulation and target gene expression in the absence of Lats (Fig. 3d, e and Supplementary Fig. 3g). To test whether nuclear TEAD is required for YAP nuclear translocation upon activating signals, we generated TEAD1/2/4 KO cells. YAP-activating signals promoted normal YAP dephosphorylation in the TEAD KO cells, but failed to elicit nuclear YAP/TAZ accumulation (Fig. 3f, g), suggesting that nuclear localization of TEAD is a prerequisite for proper YAP nuclear localization. No interaction was detected between YAP and p38, indicating that YAP cytoplasmic translocation is a consequence of TEAD regulation by p38 (Fig. 2f). Our data suggests that YAP nuclear localization is contingent upon two conditions, dephosphorylation and nuclear localization of TEAD.
Figure 3. TEAD cytoplasmic translocation prevents YAP activation.
a, b, Effect of osmotic stress on serum-, and LPA-induced YAP activation. Osmotic stress blocks serum and LPA-induced YAP dephosphorylation as shown by Western blot (a, lower arrow), and nuclear translocation as shown by immunofluorescence staining (b). c, d, Stress-induced TEAD and YAP cytoplasmic translocation is p38-dependent, but Hippo-independent. YAP is constitutively dephosphorylated in Lats KO cells as indicated by YAP phostag gel (c). Immunofluorescence shows stress induces YAP and TEAD cytoplasmic translocation in the Lats KO cells, which is blocked by SB203580 treatment (d). e, Quantification of CTGF mRNA by qRT-PCR in WT and Lats KO cells stimulated with osmotic stress with or without SB203580 treatment. Data are presented as mean ± s.e.m. from n=3 independent experiments. f, g, Detection of YAP/TAZ localization by immunofluorescence staining in TEAD KO cells stimulated with LPA or serum. Western blotting indicates YAP dephosphorylation by LPA or serum stimulation is intact in TEAD KO cells (f), whereas immunofluorescence shows YAP nuclear accumulation is impaired (g). Scale bars in b, d, and g are 20μm. Statistics source data are shown in Supplementary Table 1. Unprocessed scans of blots are shown in Supplementary Figure 5.
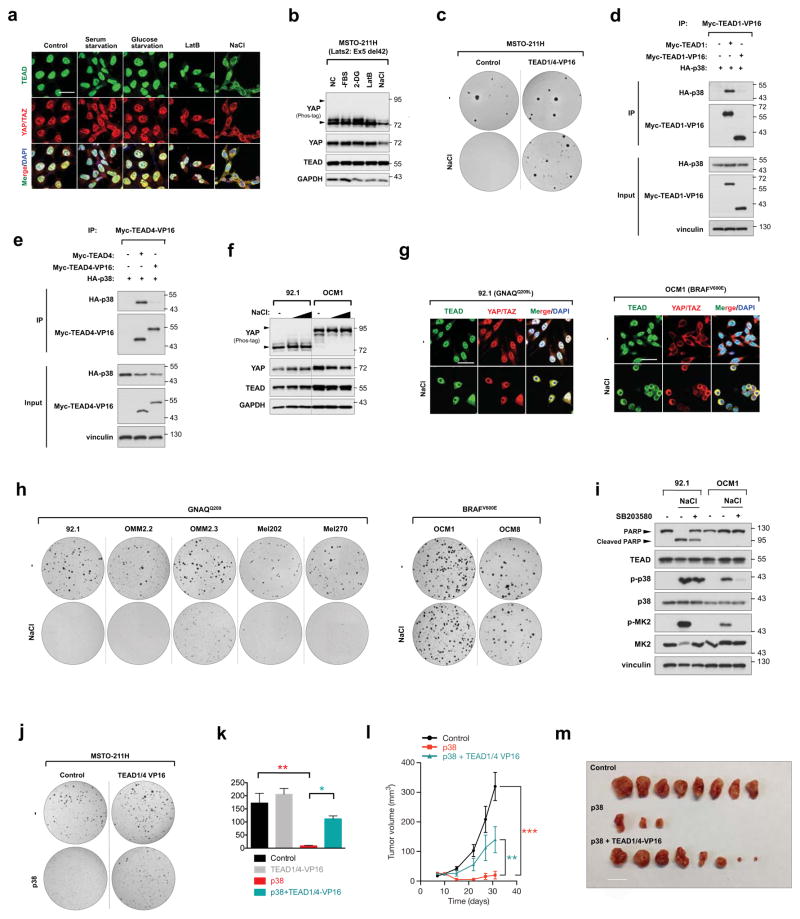
YAP is highly active in many cancers, particularly in uveal melanoma (UM) and mesothelioma, due to mutations in upstream components of the Hippo pathway16–18. YAP was constitutively hypophosphorylated and nuclear in mesothelioma cells MSTO-211H (Lats2 mutation) and H2373 (NF2 mutation), even under YAP-inhibitory conditions (Fig. 4a, b and Supplementary Fig. 4a, b). However, osmotic stress promoted cytoplasmic translocation of both TEAD and YAP/TAZ, and consequently suppressed anchorage-independent growth (Fig. 4a–c and Supplementary Fig. 4a, b). Importantly, ectopic expression of a fusion of the TEAD DNA binding domain to the VP16 transactivation domain, which is constitutively active and p38 binding deficient, restored colony forming ability of mesothelioma cells (Fig. 4c–e Supplementary Fig. 4c), suggesting that osmotic stress-induced growth arrest is due to TEAD inhibition. To further examine whether stress-induced TEAD inhibition selectively suppresses YAP-driven cancer cell growth, we compared a series of UM cell lines with mutations in either GNAQ or BRAF. The GNAQ-mutant UM cells are YAP-dependent, while the BRAF-mutant UM cells are YAP-independent (Fig. 4f and Supplementary Fig. 4d, e)17. We observed that osmotic stress evoked p38-dependent cytoplasmic translocation of TEAD and YAP/TAZ in both 92.1 (GNAQQ209L) and OCM1 (BRAFV600E) cells (Fig. 4g and Supplementary Fig. 4d–g). However, TEAD inhibition by osmotic stress or stable expression of p38 preferentially suppressed anchorage-independent growth of GNAQ-mutant UM cell lines, 92.1, OMM2.2, OMM2.3, Mel202, Mel270, but not BRAF mutant UM cell lines, OCM1 and OCM8 (Fig. 4h and Supplementary Fig. 4h). Consistently, TEAD translocation induced apoptosis specifically in the YAP-driven 92.1 cells but not the YAP-independent OCM1 cells (Fig. 4i). Furthermore, promotion of anchorage independent growth by YAP-5SA transformation of MCF10A cells was also stunted by TEAD inhibition (Supplementary Fig. 4i). These results indicate that YAP-driven cancer cells are highly susceptible to stress-induced TEAD inhibition. To further elucidate the role of TEAD inhibition on YAP-driven cancers, MSTO-211H cells were used as an isogenic model. Growth inhibitory effects resulting from stable expression of p38 were rescued by expression of constitutively active TEAD in vitro and in vivo. (Fig. 4j–m and Supplementary Fig. 4j). Under physiological osmotic stress, TEAD was cytoplasmic in tubule cells of normal kidney tissue, but was nuclear in malignant renal clear cell carcinoma as well as other normal tissues not exposed to osmotic stress (Supplementary Fig. 4k–m). Taken together, our results suggest that regulation of TEAD is important for modulating cancer growth and indicate TEAD as a potential therapeutic target.
Figure 4. TEAD inhibition restricts YAP-driven cancer cell growth.
a, b, Effect of osmotic stress on TEAD and YAP cytoplasmic sequestration in MSTO-211H mesothelioma cells. Note that unlike HEK293A (Fig. 1a), serum starvation, glucose starvation, and latruculin B did not induce YAP cytoplasmic localization because of Lats2 mutation in MSTO-211H cells. Only NaCl treatment elicited TEAD and YAP cytoplasmic translocation as detected by immunofluorescence (a) despite constitutive YAP dephosphorylation as detected by Western blot (b). c, Stress-induced TEAD inhibition suppresses anchorage independent growth. Osmotic stress inhibited colony formation in control, but not TEAD1/4-VP16 expressing MSTO-211H cells. d, e, Immunoprecipitation shows p38 cannot bind TEAD1-VP16 (d) or TEAD4-VP16 (e). f, Western blotting of YAP phosphorylation status in UM cell lines 92.1 and OCM1. g, Immunostaining of TEAD and YAP/TAZ in UM cell lines 92.1 and OCM1 upon osmotic stress. Note that YAP displays cytoplasmic staining under normal condition in OCM1 cells. Scale bars in a and g are 20μm. h, Differential effect of TEAD inhibition on anchorage-independent growth of GNAQ-mutant and BRAF-mutant UM cells. Stress-induced TEAD inhibition ablated colony formation in all GNAQ-mutant cell lines, whereas BRAF-mutant cell growth was insensitive. i, Western blot for PARP cleavage in 92.1 and OCM1 cells. NaCl stimulation induces apoptosis in 92.1 but not OCM1. Pretreatment with p38 inhibitor rescues cells from osmotic stress-induced apoptosis. j, TEAD1/4 -VP16 rescues p38-induced inhibition of colony formation of MSTO-211H cells. k, Quantification of (j). n=3 biological replicates. Data are presented as mean ± s.e.m. *p < 0.05; **p < 0.01; p values were determined using one-way ANOVA followed by Tukey’s multiple comparison test. l, TEAD1/4-VP16 rescues p38-induced inhibition of MSTO-211H in vivo tumor xenograft growth. Nude mice were injected with control, p38, or p38 + TEAD1/4-VP16 expressing MSTO-211H cells and tumor growth was measured at the indicated times. Data are presented as mean ± s.e.m. n = 4 mice per group. **p < 0.01; ***p < 0.001; p values were determined using two-way ANOVA. m, Nude mice were injected with control, p38, or p38 + TEAD1/4-VP16 expressing MSTO-211H cells and tumors were harvested after 4 weeks. Only three tumors developed in the p38 group. Scale bar, 10mm.
In the present study, we report that the Hippo pathway transcription factor TEAD is regulated through nucleocytoplasmic shuttling. TEAD is regulated by different upstream signals with distinct mechanisms as compared to YAP/TAZ. Many signals, such as serum and energy status, which modulate the localization of YAP, have no effect on TEAD localization. We identified certain environmental stresses that can induce cytoplasmic TEAD translocation. In the case of osmotic stress, TEAD cytoplasmic translocation is mediated by p38 MAPK and independent of the Hippo core kinases. It should be noted that osmotic stress initially induces acute nuclear accumulation of YAP 19, 20, then at later stages, induces cytoplasmic translocation of TEAD, and consequently YAP, as an adaptive response to stress. Mechanistically, osmotic stress-induced cytoplasmic TEAD translocation occurs via direct protein-protein interaction with p38, independent of Hippo. Disruption of TEAD-p38 interaction abolishes TEAD cytoplasmic translocation, resulting in nuclear retention of transcriptionally active TEAD. Cytoplasmic localization of TEAD is also observed in different cellular contexts, such as cell density that is p38-independent, as well as developmental contexts 21, 22. Importantly, stress-induced TEAD inhibition predominates YAP-activating signals by preventing YAP nuclear accumulation, regardless of the phosphorylation status of YAP. Thus, inhibition of TEAD presents a Hippo pathway independent avenue of regulating YAP activity, thereby providing a mechanism of controlling its functional output without targeting Hippo core components Mst and Lats. Moreover, stress-induced TEAD nucleocytoplasmic shuttling is intact in cancer cells that harbor mutations in Hippo pathway upstream components and renders YAP-driven cancer cells highly susceptible to stress-induced growth inhibition. Therefore, pharmacological agents that promote TEAD cytoplasmic localization may be a viable therapeutic strategy for treatment of cancers, especially those with high YAP activity.
Methods
Cell culture
All cell lines were maintained at 37°C with 5% CO2. HEK293A cells were cultured in DMEM (Invitrogen, 11965118) and uveal melanoma and mesothelioma cell lines were cultured in RPMI (Invitrogen, 11875119) containing 10% FBS (Gibco, 10437028) and 50 μg/ml penicillin/streptomycin (Invitrogen, 15140122). MCF10A cells were cultured in DMEM-F12 supplemented with 5% horse serum (Invitrogen, 26050088), 20 ng/ml EGF (Peprotech, AF-100-15), 0.5 μg/ml hydrocortisone (Sigma, H4001-25G), 100 ng/ml cholera toxin (Sigma, C8052-2MG), and 10 μg/ml insulin (Sigma, I1882-100MG). YAP inhibitory signals and environmental stresses included the following: serum starvation (16hr), glucose starvation (2-DG, 25mM, 2hr), PKA activation (forskolin, 10μM, 1hr), disruption of F-actin (latrunculin B, 0.1μg/ml, 1hr), Src inhibition (dasatinib, 5μM, 6hr), inhibition of the mevalonate synthesis (cerivastatin, 2μM, 6hr), NaCl (200mM, 6hr), sorbitol (0.5M, 6hr), high cell density (2 day post-confluent), and cell detachment (1hr). No cell lines used in this study were found in the database of commonly misidentified cell lines that is maintained by ICLAC and NCBI Biosample. The cell lines were not authenticated. Cells lines were tested and confirmed to be free of mycoplasma.
Induction of osmotic stress and p38 inhibitor treatment
Cells were treated with either 200mM NaCl or 0.5M sorbitol for 6 hr. p38 inhibitors SB203580 (S1076) (40μM) and PH-797840 (S2726) (30μM) were purchased from Selleckchem and cells were treated 2 hr prior to osmotic stress exposure.
Transfection and viral infection
Cells were transfected with plasmid DNA using PolyJet Reagent (Signagen Laboratories) according to manufacturer’s protocol. Cells were transfected with (pCDNA3) Flag-p38 or (pCDNA3) HA-p38, (pCDNA3) Flag-MKK3, and (pRK5) Myc-TEAD4.
92.1, OCM1, and MSTO-211H cells stably expressing empty vector; p38 and MKK3; and p38, MKK3, and TEAD1/4-VP16 were generated by retroviral and lentiviral infection. HEK293T packaging cells were transfected with empty vector, (pHIV puro) p38, (pQCXIH) MKK3, and (pHIV GFP) TEAD1/4-VP16 constructs. 48 hr after transfection, retroviral and lentiviral supernatant was filtered through 0.45 μm filter, supplemented with 8 μg/ml polybrene, and used for infection. 48 hr after infection, cells were selected with puromycin (2μg/mL) and hygromycin (200μg/mL) and FACS sorted for GFP expression.
Animal Work
NU/J (nude mice) were purchased from Jackson Laboratory (Bar Harbor, ME, USA). For tumor xenograft models, MSTO-211H cells (5×106) were injected subcutaneously into both flanks of 8–12 week old female nude mice. Four mice were assigned to each group. The investigators were not blinded to allocation during experiments and outcome assessment. Tumor height and width were measured with a caliper every 2–3 days to calculate tumor volume ( = width2 × height × 0.523). Mice were sacrificed 4 weeks post engraftment. All animal experiments were approved by the University of California, San Diego, Institutional Animal Care and Use Committee.
Antibodies
The following antibodies were purchased from Cell Signaling and used at the indicated dilution for western blot analysis, immunohistochemistry, and immunofluorescence: pan-TEAD (13295, 1:1000), p38 MAPK (8690, 1:1000), phospho-p38 MAPK (4511, 1:1000), YAP (14074, 1:1000), TAZ (4883, 1:1000), Lats1 (3477, 1:1000), p-MK2 (3007, 1:1000), p-ERK (4370, 1:1000), DYKDDDDK tag (2368, 1:1000), Myc tag (2276, 1:1000), p38α (2371, 1:1000), p38β (2339, 1:1000), p38γ (2307, 1:1000), and p38δ (2308, 1:1000). The following antibodies were purchased from Santa Cruz Biotechnology and used at the indicated dilution for western blot analysis and immunofluorescence: YAP (sc-101199, 1:1000), HA (sc-7392, 1:5000), Myc (sc-40, 1:5000), GAPDH (sc-25778, 1:1000). TEAD4 (ab58310, 1:1000) was purchased from Abcam, Flag (A8592, 1:10,000) and vinculin (V9131, 1:5000) was purchased from Sigma, TEAD1 (610923, 1:1000) was purchased from BD Biosciences, Lats2 (A300-479A, 1:1000) was purchased from Bethyl Laboratories, and NFAT5 (bs-9473R OWL 1:1000) was purchased from One World Lab.
Generation of knockout cells and mutagenesis
pSpCas9(BB)-2A-Puro (PX459) was a gift from Dr. Feng Zhang (Addgene plasmid #48139)23. The nucleotide guide sequences were designed using the CRISPR design tool at http://www.genome-engineering.org/crispr. Single-guide RNAs (sgRNAs) were cloned into PX459 expression vector. HEK293A cells were transfected using PolyJet DNA in vitro Transfection Reagent according to the manufacturer’s instructions. 24 hr post transfection, cells were selected with puromycin for 2–3 days. Following removal of puromycin, cells were allowed to recover in regular growth media for 24 hr before being single-cell sorted by FACs (UCSD; Human Embryonic Stem Cell Core, BDInflux) into a 96-well plate format. Single clones were expanded and screened by protein immunoblotting, genomic sequencing, and functional assays. Lats KO, Mst KO, and MAP4K KO cells were generated as previously described 24, 25.
Guide sequences:
p38α: 5′-3′ AGCTCCTGCCGGTAGAACGT
p38β: 5′-3′ CCACGCGCGCAGAACGTACC
p38γ: 5′-3′ GGACGGCCGCACCGGCGCTA
p38δ: 5′-3′ TCCCCGACGCACGTCGGCAG
TEAD1: 5′-3′ TGGCAGTGGCCGAGACGATC
TEAD2: 5′-3′ AGATAGGTGGGACGCCGGCG
TEAD4: 5′-3′ CTCAAGGATCTCTTCGAACG
p38 and TEAD site-directed mutagenesis was carried out using Q5 Hot Start High Fidelity DNA Polymerase from New England Biolabs (M0494) per manufacturer protocol.
RNA extraction, cDNA synthesis, and quantitative real-time PCR analysis
Cells were harvested for RNA extraction using RNeasy Plus mini kit (QIAGEN, 74136). RNA samples were reverse-transcribed to complementary DNA (cDNA) using iScript reverse transcriptase (Bio-Rad, 1708891). qRT-PCR was performed using KAPA SYBR FAST qPCR kit (Kapa Biosystems, KK4605) and the 7300 real-time PCR system (Applied Biosystems).
Immunofluorescent microscopy
Cells were seeded in 12 well plates on coverslips 2 days prior to experimentation. Coverslips were pretreated with Poly-L-ornithine solution (Sigma, P4957) diluted 1:20 at 37°C for 15 mins with a quick phosphate-buffered saline (PBS) wash prior to cell seeding. Cells were fixed in 4% paraformaldehyde (Electron Microscopy Sciences, 2280) for 15 min followed by permeabilization with 0.1% Triton-X for 5 mins. Cells were blocked in 3% BSA for 1 hr and incubated overnight at 4°C in primary antibodies diluted in 3% BSA. Secondary antibodies were diluted in 3% BSA and incubated for 1 hr. Slides were mounted with prolong gold antifade reagent with DAPI (Invitrogen, P36931). Each image is a single Z section at the same cellular level. Images were captured with a Nikon Eclipse Ti confocal microscope. Images depicted in figures were exported from NIS elements imaging software.
Immunohistochemistry
Kidney tissue arrays were purchased from U.S. Biomax Inc. Tissues were subject to heat induced antigen retrieval using 10mM sodium citrate buffer followed by 3% H2O2 for 30 min to quench endogenous peroxidase activity. Sections were incubated overnight at 4°C with pan-TEAD antibody and detected using Vectastain elite ABC kit and DAB Peroxidase Substrate kit (Vector Laboratories) as per manufacturer protocol.
Western blot and immunoprecipitation
Immunoblotting was performed using a standard protocol. Phos-tag reagents were purchased from Wako Chemicals, gels containing phos-tag were prepared according to manufacturer’s instructions. For immunoprecipitations, cells were rinsed twice with ice-cold PBS and lysed in ice-cold lysis buffer (0.15M NaCl, 0.05M Tris-HCl, 0.5% Triton X-100, and one tablet each of EDTA-free protease inhibitors (Roche, 11873580001) and phosphatase inhibitor (Thermo Fisher, 88667) per 50 ml). For immunoprecipitations, primary antibodies were added to the lysates and incubated with rotation overnight at 4°C. 10 μl magnetic protein A/G beads (Thermo Fisher, 88802) were added and incubated for an additional 2 hr. Immunoprecipitates were washed three times with lysis buffer. Immunoprecipitated proteins were denatured by the addition of sample buffer and boiling for 5 mins, resolved by 9% SDS-PAGE, and analyzed via Western blot analysis.
In vitro kinase assay
To analyze p38 kinase activity, HEK293A cells were transfected with WT or kinase mutant p38α. p38γ and p38δ were purchased from SignalChem. Cells were collected and p38 was immunoprecipitated as described above. Immunoprecipitates were washed with kinase assay buffer (25mM HEPES pH 7.4, 25mM MgCl2, and 2mM DTT) and subjected to a kinase assay in kinase assay buffer along with 500μM ATP-γ-S. GST–ATF2, GST-TEAD4, or GST-TEAD4-4SP fusion proteins were used as substrates. Reactions were incubated for 30 min at 30°C. p-Nitrobenzyl mesylate (Sigma, A1388) was added to the kinase reactions and incubated for 1 hr to alkylate the thiophosphorylation sites on the substrates. Reactions were terminated with sample buffer and resolved on 9% SDS-PAGE. A thiophosphate ester antibody (Abcam, ab92570) was used to detect substrate phosphorylation.
Soft agar colony growth
Each 6-well plate was coated with 1.5 ml of bottom agar (DMEM containing 10% FBS and 0.5% Difco agar noble). Various cells (5 × 103) were suspended in 1.5 ml of top agar (DMEM containing 10% FBS and 0.35% Difco agar noble) into each well. Cells were incubated for approximately three weeks and replaced with fresh medium containing 50 mM NaCl every three days. Colonies were stained using 0.005% crystal violet.
Statistics and reproducibility
The experiments shown in Fig. 2b, 2f, 4j, and Supplementary Fig. 1h, 2b–d, 3f, 4h–j, 4l–m are representative of 2 independent experiments performed with similar results. All other experiments are representative of at least 3 independent repeats. Data are presented as mean ± s.e.m. p values were determined using one-way ANOVA followed by Tukey’s multiple comparison test or two-way ANOVA as noted in the figure legends.
Data Availability
Source data for Fig. 1g–h, 3e and Supplementary Fig. 1j, 4j have been provided as Supplementary Table 1. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health (CA196878, DE15964, and GM51586) to K.-L.G., the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIP) (2014R1A5A2009936) to H.-S.J. and by a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant number: HI17C1560), and the National Research Foundation of Korea(NRF) grant funded by the Korea government (MOE) (2017R1D1A1B0303479) to H.W.P. K.C.L. and S.W.P. was supported in part by the University of California, San Diego (UCSD) Graduate Training Program in Cellular and Molecular Pharmacology (T32 GM007752).
Footnotes
Author contributions:
K.C.L. and H.W.P. designed experiments, performed research, analyzed data and wrote the manuscript. T.M., Z.M., H.-S.J. and S.W.P. designed experiments, performed research, analyzed data and reviewed the manuscript. Y.S. and J.H. provided reagents and reviewed the manuscript. K.-L.G. designed experiments, analyzed data and wrote the manuscript.
Competing financial interests
K.-L.G. is a co-founder and has an equity interest in Vivace Therapeutics, Inc. The terms of this arrangement have been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies.
References and Notes
- 1.Johnson R, Halder G. The two faces of Hippo: targeting the Hippo pathway for regenerative medicine and cancer treatment. Nat Rev Drug Discov. 2014;13:63–79. doi: 10.1038/nrd4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yu FX, Zhao B, Guan KL. Hippo Pathway in Organ Size Control, Tissue Homeostasis, and Cancer. Cell. 2015;163:811–828. doi: 10.1016/j.cell.2015.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao B, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu FX, et al. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell. 2012;150:780–791. doi: 10.1016/j.cell.2012.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mo JS, et al. Cellular energy stress induces AMPK-mediated regulation of YAP and the Hippo pathway. Nat Cell Biol. 2015;17:500–510. doi: 10.1038/ncb3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang W, et al. AMPK modulates Hippo pathway activity to regulate energy homeostasis. Nat Cell Biol. 2015;17:490–499. doi: 10.1038/ncb3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dupont S, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 8.Zhao B, et al. Cell detachment activates the Hippo pathway via cytoskeleton reorganization to induce anoikis. Genes Dev. 2012;26:54–68. doi: 10.1101/gad.173435.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim NG, Gumbiner BM. Adhesion to fibronectin regulates Hippo signaling via the FAK-Src-PI3K pathway. J Cell Biol. 2015;210:503–515. doi: 10.1083/jcb.201501025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sorrentino G, et al. Metabolic control of YAP and TAZ by the mevalonate pathway. Nat Cell Biol. 2014;16:357–366. doi: 10.1038/ncb2936. [DOI] [PubMed] [Google Scholar]
- 11.de Nadal E, Ammerer G, Posas F. Controlling gene expression in response to stress. Nat Rev Genet. 2011;12:833–845. doi: 10.1038/nrg3055. [DOI] [PubMed] [Google Scholar]
- 12.Cargnello M, Roux PP. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol Mol Biol Rev. 2011;75:50–83. doi: 10.1128/MMBR.00031-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanoue T, Adachi M, Moriguchi T, Nishida E. A conserved docking motif in MAP kinases common to substrates, activators and regulators. Nat Cell Biol. 2000;2:110–116. doi: 10.1038/35000065. [DOI] [PubMed] [Google Scholar]
- 14.Tanoue T, Maeda R, Adachi M, Nishida E. Identification of a docking groove on ERK and p38 MAP kinases that regulates the specificity of docking interactions. Embo j. 2001;20:466–479. doi: 10.1093/emboj/20.3.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Estrada-Gelonch A, Aramburu J, Lopez-Rodriguez C. Exclusion of NFAT5 from mitotic chromatin resets its nucleo-cytoplasmic distribution in interphase. PLoS One. 2009;4:e7036. doi: 10.1371/journal.pone.0007036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moroishi T, Hansen CG, Guan KL. The emerging roles of YAP and TAZ in cancer. Nat Rev Cancer. 2015;15:73–79. doi: 10.1038/nrc3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu FX, et al. Mutant Gq/11 promote uveal melanoma tumorigenesis by activating YAP. Cancer Cell. 2014;25:822–830. doi: 10.1016/j.ccr.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murakami H, et al. LATS2 is a tumor suppressor gene of malignant mesothelioma. Cancer Res. 2011;71:873–883. doi: 10.1158/0008-5472.CAN-10-2164. [DOI] [PubMed] [Google Scholar]
- 19.Hong AW, et al. Osmotic stress-induced phosphorylation by NLK at Ser128 activates YAP. EMBO Rep. 2017;18:72–86. doi: 10.15252/embr.201642681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moon S, et al. Phosphorylation by NLK inhibits YAP-14-3-3-interactions and induces its nuclear localization. EMBO Rep. 2017;18:61–71. doi: 10.15252/embr.201642683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Home P, et al. Altered subcellular localization of transcription factor TEAD4 regulates first mammalian cell lineage commitment. Proc Natl Acad Sci U S A. 2012;109:7362–7367. doi: 10.1073/pnas.1201595109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cagliero J, Forget A, Daldello E, Silber J, Zider A. The Hippo kinase promotes Scalloped cytoplasmic localization independently of Warts in a CRM1/Exportin1-dependent manner in Drosophila. Faseb j. 2013;27:1330–1341. doi: 10.1096/fj.12-216424. [DOI] [PubMed] [Google Scholar]
- 23.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park HW, et al. Alternative Wnt Signaling Activates YAP/TAZ. Cell. 2015;162:780–794. doi: 10.1016/j.cell.2015.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meng Z, et al. MAP4K family kinases act in parallel to MST1/2 to activate LATS1/2 in the Hippo pathway. Nat Commun. 2015;6:8357. doi: 10.1038/ncomms9357. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data for Fig. 1g–h, 3e and Supplementary Fig. 1j, 4j have been provided as Supplementary Table 1. All other data supporting the findings of this study are available from the corresponding author on reasonable request.