Summary
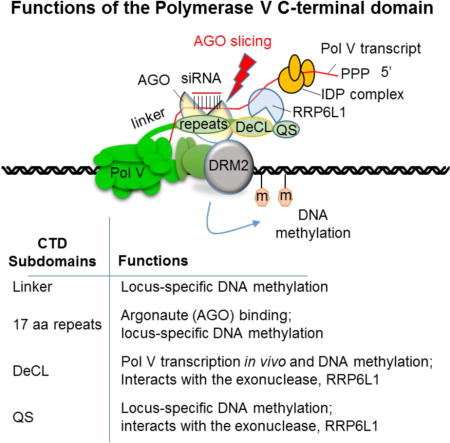
Plant multisubunit RNA Polymerase V transcription recruits Argonaute-siRNA complexes that specify sites of RNA-directed DNA methylation (RdDM) for gene silencing. Pol V’s largest subunit, NRPE1, evolved from the largest subunit of Pol II but has a distinctive carboxyl-terminal domain (CTD). We show that the Pol V CTD is dispensable for catalytic activity in vitro, yet essential in vivo. One CTD subdomain (DeCL) is required for Pol V function at virtually all loci. Other CTD subdomains have locus-specific effects. In a yeast two-hybrid screen, the 3′–>5′ exoribonuclease, RRP6L1 was identified as an interactor with the DeCL and glutamine-serine-rich (QS) subdomains located downstream from an Argonaute-binding subdomain. Experimental evidence indicates that RRP6L1 trims the 3′ ends of Pol V transcripts sliced by ARGONAUTE 4 (AGO4), suggesting a model whereby the CTD enables the spatial and temporal coordination of AGO4 and RRP6L1 RNA processing activities.
Keywords: gene silencing, cytosine methylation, chromatin modification, Pol V transcription, noncoding RNA, siRNA
eTOC Blurb
In plants, transcription by RNA Polymerase V specifies sites of RNA-directed DNA methylation (RdDM) and gene silencing. Wendte et al. define locus-specific functions for subdomains of the Pol V largest subunit’s carboxyl-terminal domain. The exonuclease, RRP6L1, is shown to interact with the CTD and trim the 3′ ends of Pol V-transcripts.
Introduction
NUCLEAR RNA POLYMERASE V (Pol V) is a plant-specific multisubunit RNA polymerase important for siRNA-directed DNA methylation (RdDM) and transcriptional gene silencing, primarily of transposable elements (reviewed in: Haag and Pikaard, 2011; Ream et al., 2013; Zhou and Law, 2015). In the major RdDM pathway (reviewed in: Matzke and Mosher, 2014; Wendte and Pikaard, 2017), NUCLEAR RNA POLYMERASE IV (Pol IV) and RNA-DEPENDENT RNA POLYMERASE 2 (RDR2) generate short double-stranded RNAs that are then diced into 24 nt short interfering RNAs (siRNAs) (Blevins et al., 2015; Li et al., 2015; Zhai et al., 2015). These siRNAs associate with an Argonaute family protein, primarily AGO4 or AGO6, and guide cytosine methylation through basepairing interactions with nascent Pol V transcripts (Wierzbicki et al., 2009).
Pol V is composed of twelve subunits, like Pol II (Haag et al., 2014; Ream et al., 2013; Ream et al., 2009). Approximately half of the Pol V subunits are encoded by the same genes as Pol II subunits (Ream et al., 2013; Ream et al., 2009). Remaining subunits, including the catalytic subunits (the two largest subunits), are encoded by genes that arose through duplication and sub-functionalization of ancestral Pol II subunit genes (Huang et al., 2015; Luo and Hall, 2007; Ream et al., 2009; Tucker et al., 2010; Wang and Ma, 2015). A major distinguishing feature of Pol V is the largest subunit, NRPE1, which is characterized by a long C-terminal domain (CTD), spanning ~700 amino acids. At the very C-terminus is a subdomain rich in repeated glutamine-serine (QS) motifs. The QS subdomain is preceded by a DeCL subdomain, which shares sequence similarity with a chloroplast protein implicated in plastid ribosomal RNA processing, DEFECTIVE CHLOROPLASTS AND LEAVES (abbreviated DeCL) (Bellaoui and Gruissem, 2004; Bellaoui et al., 2003; Haag and Pikaard, 2011; Huang et al., 2015; Keddie et al., 1996). N-terminal to the DeCL domain is a subdomain consisting of ten, 17 amino-acid repeats rich in glycine-tryptophan (GW) or WG dipeptide “ago-hooks” that bind to Argonaute family proteins, including AGO4 and AGO6 (El-Shami et al., 2007; Pontier et al., 2012). A fourth subdomain, termed the linker, is located between the 17aa repeats and the conserved sequences (domains A–H) that are characteristic of all multisubunit RNA polymerase largest subunits (see Figure 1A).
Figure 1.
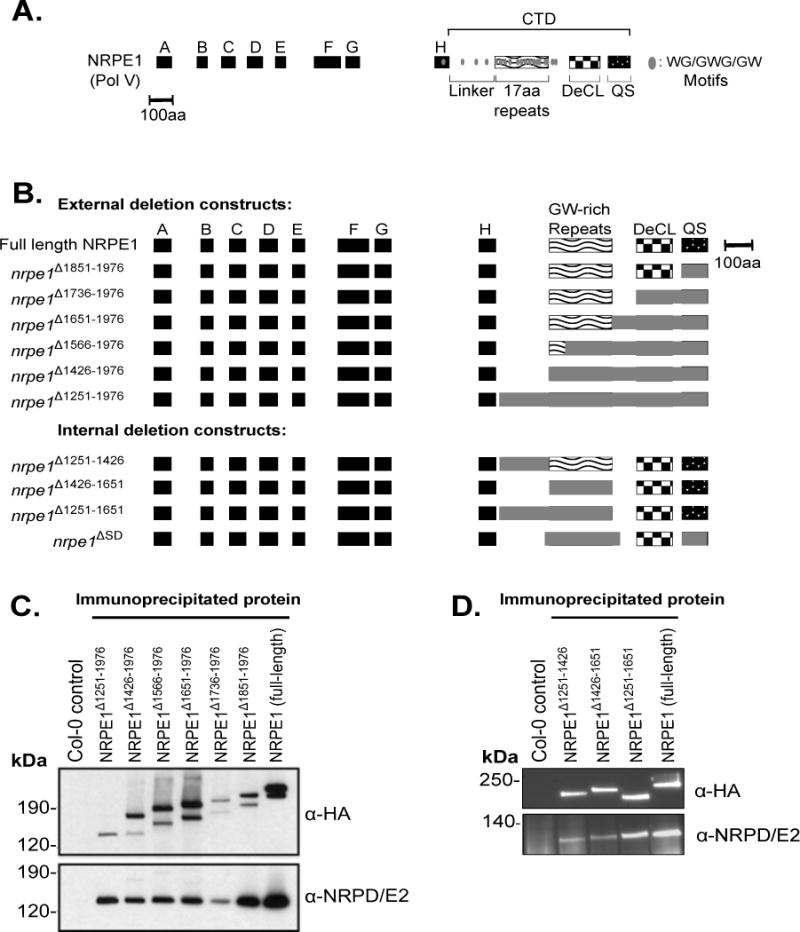
Engineering and expression of Pol V largest subunit (NRPE1) proteins with deletions in the C-terminal domain (CTD).
A. Diagram of the NRPE1 protein of A. thaliana ecotype Col-0, showing the relative positions of domains A–H, conserved among multisubunit RNA polymerase largest subunits, and subdomains of the CTD.
B. Diagrams of recombinant NRPE1 constructs tested in the study. All constructs were engineered to have a C-terminal HA epitope tag, except ΔSD, described previously (El-Shami et al., 2007). Numbering denotes amino acid positions measured from the N-terminus.
C–D. Immunoblot assays following anti-HA immunoprecipitation of C-terminal (C) or internal (D) CTD deletion constructs, with non-transgenic Col-0 serving as a control. Blots were probed using antibodies recognizing the HA epitope tag or the second-largest subunit of Pol V, NRP(D/E)2. See also Figure S1.
Despite originating from a duplication of the Pol II largest subunit, NRPB1, the CTD of the Pol V largest subunit shares no sequence similarity with the CTD of NRPB1 (Haag and Pikaard, 2011; Huang et al., 2015; Pontier et al., 2005; Trujillo et al., 2016). Pol II largest subunit CTDs consist of repeats of the heptad consensus sequence, YSPTSPS. These repeats mediate numerous aspects of Pol II function, including polymerase recruitment, transcriptional activation, elongation, termination, and RNA processing (reviewed in: Zaborowska et al., 2016). Whether the NRPE1 CTD mediates analogous functions is unclear.
To functionally dissect the NRPE1 CTD, we engineered a series of NRPE1 transgenes bearing targeted CTD deletions and then tested their function. We show that the CTD is dispensable for Pol V transcription in vitro but is critical for Pol V transcript abundance and cytosine methylation in vivo. The biological activity of the CTD is explained primarily by the DeCL subdomain. However, the QS, 17aa repeat and linker subdomains are needed at loci that tend to be highly methylated and dependent on proteins known to interact with Pol V or its transcripts. We show that the exonuclease, RRP6L1 interacts with the QS and DeCL subdomains. In vivo, our evidence indicates that RRP6L1 trims the 3′ ends of Pol V transcripts after slicing by AGO4, without completely degrading the RNAs, consistent with a prior study indicating that RRP6L1 stabilizes Pol V transcript association with chromatin (Zhang et al., 2014). We propose a model whereby the Pol V CTD coordinates AGO4 slicing of Pol V transcripts and subsequent enzymatic engagement by RRP6L1, which might increase the dwell time of Pol V transcripts at target sites, promoting RNA-directed DNA methylation.
Results
The NRPE1 CTD is required for Pol V-dependent RdDM
We engineered deletions affecting four CTD subdomains of the NRPE1 protein of A. thaliana ecotype Col-0: the linker subdomain (amino acid (aa) positions 1251–1426); the repeat subdomain (aa 1426–1651), containing ten complete, and two degenerate, repeats of a 17 aa consensus sequence, DKKNSETESGPAAWGSW; the DeCL subdomain (aa 1736–1851); and the QS-rich subdomain (aa 1851–1976) (Figure 1A). Within the CTD are seventeen WG (Tryptophan-Glycine), one GWG, and one GW motif(s). Twelve of these Ago-hook motifs occur within the 17 aa repeat subdomain (aa 1426–1651) and mediate interactions with AGO4 (El-Shami et al., 2007).
Transgenes expressing the deleted NRPE1 proteins were tested for their ability to rescue an nrpe1-11 null mutant, compared to a full-length NRPE1 transgene. One set of recombinant NRPE1 proteins was sequentially deleted from the C-terminus (Δ1851–1976, Δ1736–1976, Δ1651–1976, Δ1566–1976, Δ1426–1976, and Δ1251–1976). Three additional transgenes had internal CTD deletions: Δ1251–1426 (deleting the linker subdomain), Δ1426–1651 (deleting all 17aa repeats), and Δ1251–1651 (deleting the linker and repeats). We also tested NRPE1ΔSD, engineered by the Lagrange lab (El-Shami et al., 2007), which has both the repeat and QS-rich subdomains deleted (Δ1411–1707 and Δ1875–1976) (Figure 1B). The transgenes were expressed from the native NRPE1 promoter and each recombinant protein (except NRPE1ΔSD) was engineered to have a C-terminal HA epitope tag. Immunoprecipitation using anti-HA resin showed that the recombinant proteins are expressed at similar levels (Figure 1C–D). The Pol V second-largest subunit, NRP(D/E)2, co-immunoprecipitates with all of the recombinant NRPE1 proteins, indicating that polymerase assembly is not disrupted by the CTD deletions (Figure 1C–D). Moreover, full length NRPE1 and NRPE1 Δ1251–1976 (full CTD deletion) both localize to the nucleus and yield signals similar to native NRPE1 (Figure S1) (Pontes et al., 2006). However, NRPE1 Δ1251–1976 shows a slightly higher proportion of nuclei with smaller, dispersed foci of staining compared to full length NRPE1 (Figure S1)
To test the effects of CTD deletions, we first examined DNA methylation at restriction endonuclease sites within known Pol V target loci, using Chop-PCR (Figure 2A). In this method, genomic DNA is digested using a methylation-sensitive restriction enzyme prior to conducting PCR with primers flanking the endonuclease recognition site. If the site is methylated, it is not cut, and PCR amplification occurs. If the site is unmethylated, the DNA is cut and PCR fails. All loci yielded PCR products using DNA of wild-type Col-0 but not using DNA of the nrpe1-11 mutant, in which RdDM is lost (Figure 2A, S2A). The full-length NRPE1 transgene, and NRPE1 missing only the QS-rich domain (Δ1851–1976), both restored methylation at nine loci analyzed (Figure 2A, S2A). However, deletion of the DeCL domain (constructs Δ1736–1976, Δ1651–1976) impaired restoration of DNA methylation at P6, IGN5A and 5S rRNA gene repeats (Figure 2A, S2A), and prevented restoration of AtSN1 transposon silencing (Figure S2B) or siRNA levels at AtCopia or 45S rRNA gene loci (Figure S2C). For other loci, including P9, IGN5B, IGN23, IGN26, and AtSN1, deletion of additional CTD subdomains was needed to reduce Chop-PCR signals to nrpe1-11 mutant levels (Figure 2A). At the soloLTR locus, all constructs restored methylation at the restriction site assayed (Figure 2A).
Figure 2.
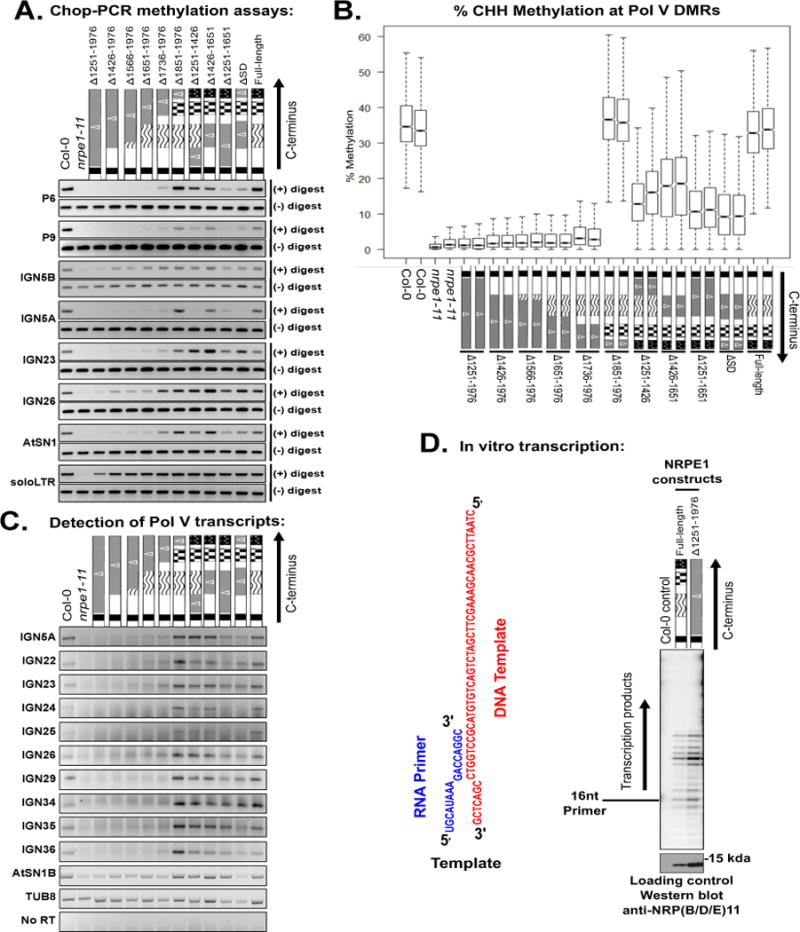
Effects of NRPE1 CTD deletions on CHH cytosine methylation and Pol V transcript abundance.
A. Chop-PCR assays conducted using AluI or HaeIII restriction endonucleases (see text for details). nrpe1-11 mutants expressing the indicated transgenes are compared to wild-type (Col-0) and nrpe1-11 controls. IGN5A and IGN5B adjacent regions, with A being intergenic and B corresponding to sequences internal to a LINE element targeted by RdDM (Wierzbicki et al., 2008).
B. Box plots displaying overall CHH cytosine methylation levels, determined by whole-genome bisulfite sequencing, within Pol V-dependent differentially methylated regions (DMRs).
C. RT-PCR detection of Pol V transcriptional products. TUB8 (Tubulin), transcribed by Pol II, serves as a control.
D. In vitro transcription activities of immunoprecipitated full length NRPE1 and NRPE1 missing the entire CTD on the indicated DNA template hybridized to an RNA primer. An immunoprecipitation fraction of non-transgenic Col-0 serves as a negative control. Relative protein input levels were compared by immunoblotting for the 11th subunit using anti-NRP(B/D/E)11. Transcription products are labeled by virtue of incorporation of 32P-CTP and visualized by phosphorimaging (Haag et al., 2012). See also Figures S2–S4, and Tables S1–S2.
NRPE1 proteins lacking the linker domain (Δ1251–1426) or the 17 aa repeat domain (Δ1426–1651) showed partially reduced DNA methylation, silencing and siRNA levels (Figure 2A, Figure S2). NRPE1 Δ1251–1426 failed to restore methylation at P6 and IGN5A and NRPE1 Δ1426–1651 failed to restore methylation at P6 (Figure 2A). NRPE1 proteins missing two CTD subdomains, namely Δ1251–1651 (linker and repeat domains deleted) and ΔSD (repeat and QS domains deleted), showed a stronger defect, failing to restore methylation at P6, P9, IGN5A, or 5S rRNA genes (Figure 2A, Figure S2).
The DeCL subdomain accounts for most CTD activity
To further assess the impacts of CTD deletions genome-wide, including regional changes in DNA methylation not detectable by Chop-PCR, we conducted whole genome bisulfite sequencing (Table S1).
In nrpe1-11 plants, compared to wild-type, CHH methylation was lost from 2,259 differentially methylated regions (DMRs; see Methods) (Figure 2B, Table S2). Expressing full-length NRPE1, or NRPE1 missing only the QS domain (Δ1851–1976), restored CHH methylation at ~99% of these DMRs (Figure 2B, Figure S3A, S3G, Table S2). In contrast, in lines expressing NRPE1 missing, at a minimum, the DeCL and QS domains (Δ1736–1976, Δ1651–1976, Δ1566–1976, Δ1426–1976, and Δ1251–1976), nearly 90% (~2,000) of the DMRs failed to significantly regain methylation (Figure 2B, Figure S3 B–F, Table S2). NRPE1 proteins missing the linker or 17 aa repeat CTD subdomains restored methylation at 40–60 % of the DMRs (Figure 2B). Specifically, 922, 685, 1,246, and 1,421 DMRs remained demethylated in plants expressing NRPE1 missing the linker subdomain (greatly reduced, or eliminated. Deletion of the linker and 17 aa repeat domains affected Pol V1251–1426), the 17 aa repeat subdomain (Δ1426–1651), the linker and repeat subdomains (Δ1251–1651), or the repeat plus QS domain (ΔSD), respectively (Figure S3H–I, Table S2).
CG and CHG methylation can be established by RdDM, but the majority of CG and CHG methylation results from maintenance methylation (Law and Jacobsen, 2010). Approximately 100 CG DMRs were identified in each of the CTD test lines, but little overlap exists among the DMRs in the different lines, suggesting that these are spontaneous losses in methylation (Becker et al., 2011; Schmitz et al., 2011) rather than losses attributable to NRPE1 or its CTD (Figure S4).
For CHG methylation, ~600 regions showed Pol V-dependent methylation (Figure S4). Full length NRPE1 and NRPE1 missing only the QS domain both fully restored methylation at these DMRs. Deleting both the DeCL and QS domains prevented restoration of methylation at ~50–65% of the regions, whereas deleting domains other than the DeCL had a lesser effect, but still impaired methylation restoration at ~20–40% of the CHG DMRs (Figure S4). A milder effect of CTD deletions on CHG vs CHH methylation is consistent with maintenance methylation pathways contributing to CHG methylation, decreasing the negative consequences of CTD deletions impairing Pol V activity and RdDM.
The NRPE1 CTD is required for Pol V transcript detection in vivo but not enzymatic activity in vitro
We tested whether methylation defects of CTD mutants correlate with defects in Pol V transcript levels in vivo using RT-PCR (Figure 2C). CTD deletion mutants missing the DeCL domain, in which CHH methylation is most reduced, have Pol V transcript levels that are also greatly reduced, or eliminated. Deletion of the linker and 17 aa repeat domains affected Pol V transcript levels in a locus-dependent manner, whereas deleting the QS domain was inconsequential at all loci (Figure 2C).
The severe loss of Pol V transcripts observed in vivo when the full CTD is deleted led us to test whether the Pol V CTD is required for the intrinsic activity of Pol V, which we tested using in vitro transcription assays (Haag et al., 2012). We found no difference in activity comparing full-length NRPE1 to NRPE1 missing the entire CTD (Δ1251–1976) (Figure 2D), indicating that the CTD is not required for Pol V’s core catalytic activity.
The full length NRPE1 CTD is required for RdDM at a subset of highly methylated loci
We examined the influence of CTD subdomains on CHH methylation of individual Pol V-dependent DMR loci using clustering analysis. Plants expressing full-length NRPE1, or NRPE1 missing just the QS domain (Δ1851–1976), display methylation profiles similar to wild-type plants (Figure 3A). In contrast, plants expressing NRPE1 lacking the DeCL domain (Δ1736–1976, Δ1651–1976, Δ1566–1976, Δ1426–1976, and Δ1251–1976) have methylation profiles similar to the nrpe1-11 null mutant (Figure 3A). Profiles for NRPE1 proteins lacking subdomains other than the DeCL domain (Δ1251–1426, Δ1426–1651, Δ1251–1651, and ΔSD) showed intermediate CHH methylation levels, and affected overlapping subsets of loci (Figure 3A).
Figure 3.
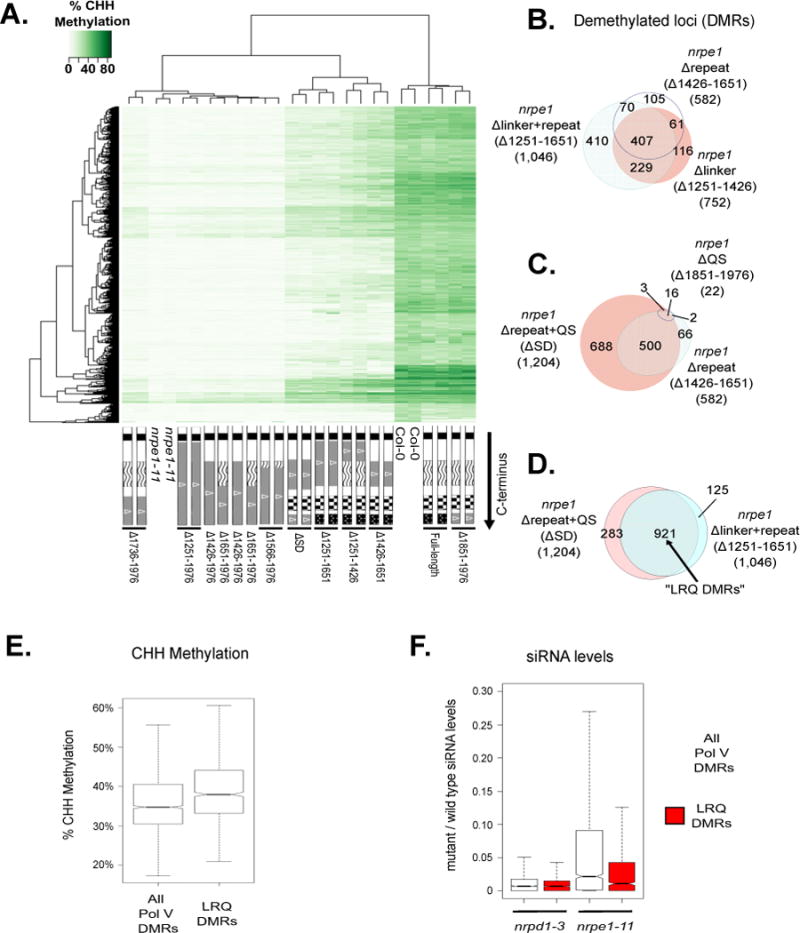
Functional relationships among Pol V CTD subdomains.
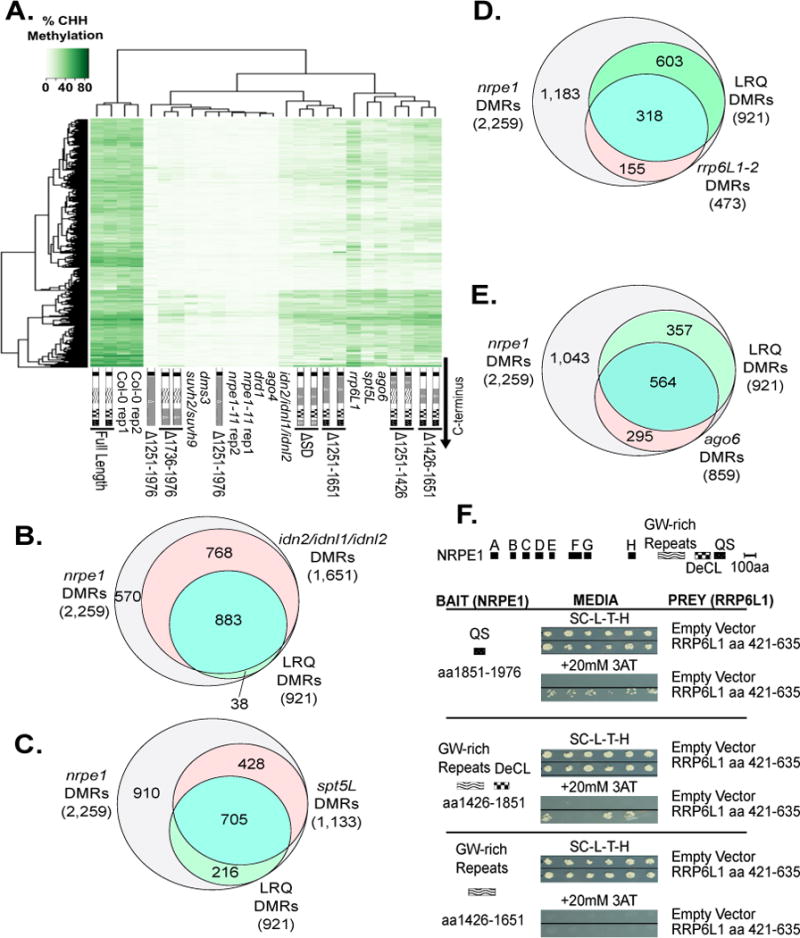
A. Heatmap clustering of % CHH methylation levels within Pol V DMRs for CTD deletion
mutants, wild-type (Col-0) and nrpe1-11 controls.
B–D. Venn diagrams showing subsets of the 2,259 total Pol V CHH DMRs that remain significantly hypo-methylated when the indicated CTD mutants are expressed in the nrpe1 mutant background. nrpe1 Δ1251–1651 and nrpe1 ΔSD in (D) collectively represent deletions in the linker, repeat, and QS subdomains, thus the subset of DMRs un-rescued by both constructs are referred to as LRQ DMRs.
E. Box plot of % CHH methylation in wild type plants within all Pol V CHH DMRs, compared to the 921 LRQ DMRs defined in panel D.
F. Box plot showing the ratio of siRNA levels (in reads per million; RPM) in nrpd1-3 and nrpe1- 11 mutants relative to wild-type Col-0. Total Pol V DMRs (white) are compared to the 921 LRQ DMRs defined in panel D (red). See also Table S2 and S3.
Comparing Pol V-dependent DMRs that fail to regain methylation when NRPE1 is missing either the linker subdomain (Δ1251–1426; 752 DMRs) or 17 aa repeat subdomain (Δ1426–1651; 582 DMRs) revealed an overlap of 468 DMRs (Figure 3B). If the linker and 17 aa repeat subdomains are both deleted (Δ1251–1651), the effect is more severe, such that nearly half of all Pol V-dependent DMRs (1,046 out of 2,259) remain significantly hypo-methylated. This includes 410 DMRs that were not affected by deletion of either subdomain alone (Figure 3B), suggesting partially redundant functions for these subdomains.
We next compared the overlap between DMRs that remain significantly hypo-methylated in lines in which NRPE1 lacks the QS domain (Δ1851–1976) or the repeat domain (Δ1426–1651) (Figure 3C). Eighteen of the 22 DMRs identified in Δ1851–1976 are among the 582 DMRs remaining in Δ1426–1651 (Figure 3C). Interestingly, deleting both the QS and 17 aa repeat subdomains (ΔSD) is substantially more deleterious that deleting either domain alone, such that 1,204 DMRs are not rescued, suggesting a functional interaction between these domains (Figure 3C).
We compared the effect of deleting the linker plus 17 aa repeat subdomains (Δ1251–1651) to ΔSD, in which the 17aa repeat and QS subdomains are deleted. 921 hypo-methylated regions overlap among the 1,204 (ΔSD) and 1,046 (Δ1251–1651)-affected DMRs, accounting for approximately half of the 2,259 Pol V- dependent DMRs (Figure 3D). Because the Δ1251–1651 and ΔSD forms of NRPE1 both have the critical DeCL subdomain, but are deleted for the linker (L), repeat (R), and QS (Q) subdomains, we refer to the 921 DMRs affected by both constructs as LRQ DMRs for the remainder of the manuscript (Table S2). LRQ DMRs have at least two notable characteristics. One is that their methylation level in wild-type Col-0 is higher, on average, than total Pol V-dependent DMRs (Figure 3E). Second, they differ with respect to siRNA levels. Pol IV is required for the biogenesis of virtually all 24nt siRNAs, whereas Pol V affects siRNA production at only a subset of loci (Mosher et al., 2008; Pontier et al., 2005; Zhang et al., 2007). Dividing the number of 24nt siRNAs detected in Pol IV (nrpd1-3) mutants by the number in wild type (Col-0) yields an average value near zero at both LRQ DMRs and total Pol V DMRs (Figure 3F, Table S3). However, at LRQ DMRs, siRNA levels in nrpe1-11 mutants are substantially lower than for Pol V DMRs as a whole, indicating that siRNA levels at LRQ DMRs have a greater dependency on Pol V (Figure 3F, Table S3).
LRQ DMRs correlate with loci whose methylation is similarly dependent on Pol V transcript binding proteins
Comparing CHH methylation profiles of CTD mutants and mutants acting in Pol V-dependent steps of the RdDM pathway revealed intriguing relationships (Figure 4A). Mutants with severe methylation defects, clustering with nrpe1-11, include NRPE1 with the full CTD deleted (Δ1251–1976), NRPE1 missing the DeCL and QS subdomains (Δ1736–1976) and several mutants defective for proteins implicated in Pol V recruitment to target sites, including drd1, dms3, and suvh2 suvh9 (Jing et al., 2016; Johnson et al., 2014; Liu et al., 2014; Wierzbicki et al., 2008; Wierzbicki et al., 2009; Zhong et al., 2012) (Figure 4A, Table S2). Mutants with less severe effects on methylation, resembling CTD deletion mutants lacking the linker, 17 aa repeat, or QS subdomains, include mutants defective for proteins that interact with Pol V transcripts and/or Pol V transcription elongation complexes, including the IDN2-IDP complex (idn2 idnl1 idnl2), spt5L (also known as ktf1), and rrp6L1, (Ausin et al., 2009; Bies-Etheve et al., 2009; He et al., 2009; Kollen et al., 2015; Rowley et al., 2011; Zhang et al., 2012) (Figure 4A, Table S2). The IDN2-IDP complex binds Pol V transcripts prior to recruitment of the de novo DNA methyltransferase, DRM2 (Ausin et al., 2012; Ausin et al., 2009; Bohmdorfer et al., 2014). SPT5L is a paralog of the Pol II transcription elongation factor, SPT5, and can bind Pol V, Pol V transcripts, and help recruit Argonaute (Bies-Etheve et al., 2009; He et al., 2009; Huang et al., 2009; Rowley et al., 2011). RRP6L1 binds Pol V transcripts and has been implicated in maintaining these RNAs in chromatin (Zhang et al., 2014). Comparing statistically significant hypo-DMRs in each mutant line to the LRQ DMRs, revealed a high degree of overlap among affected loci (Figure 4B–D, Table S2).
Figure 4.
Relationship of methylation defects in Pol V CTD mutants to other RdDM mutants.
A. Heatmap clustering of % CHH methylation at Pol V DMRs, comparing CTD mutants, other RdDM pathway mutants (suvh2 suvh9, dms3, drd1, ago4, ago6, idn2 idnl1 idnl2, rrp6L1, spt5), and wild-type (Col-0) and nrpe1-11 controls.
B–E. Venn diagrams comparing total Pol V CHH DMRs, and LRQ DMRs in combination with DMRs dependent on idn2/idnl1/idnl2 (B), spt5L (C), rrp6L1 (D), or ago6 (E).
F. Yeast two hybrid (Y2H) interaction tests for Pol V CTD subdomains (“Bait”) and RRP6L1 aa 426–635 (“Prey”). Each bait was also tested against empty vector (pDEST22) controls. SC-L-T-H refers to media lacking leucine, tryptophan and histidine (top panel). Bait-prey interaction allows growth in the presence of 20 mM E-Amino-1, 2, 4 Triazol (3-AT), a HIS3 inhibitor (bottom panel). See also Figure S5 and Tables S1 and S2.
AGO4 and AGO6 interact with Pol V transcripts as well as AGO-hook motifs of the CTD 17 aa repeat subdomain (aa 1426–1651) (El-Shami et al., 2007; Pontier et al., 2012; Rowley et al., 2011; Wierzbicki et al., 2009). Thus, one might predict that methylation defects resulting from targeted deletion of the 17 aa repeat subdomain might be phenocopied by ago4 or ago6 mutants. Interestingly, DMRs affected by ago6 and the 17 aa repeat subdomain fit this expectation and show substantial overlap (Figure 4A, E). However, ago4 mutants exhibit a much more severe loss of CHH methylation (Stroud et al., 2013), clustering with the nrpe1-11, full CTD, or DeCL subdomain deletions (Figure 4A). This finding is consistent with evidence that AGO4 recruitment entails interactions with Pol V transcripts and SPT5L, not just the NRPE1 CTD (El-Shami et al., 2007; Lahmy et al., 2016; Pontier et al., 2012; Wierzbicki et al., 2009).
The exonuclease, RRP6L1, interacts with the CTD and trims Pol V transcripts
We conducted a yeast two hybrid (Y2H) screen for A. thaliana proteins that interact with the Pol V CTD. We tested individual subdomains, including constructs expressing the repeat domain (NRPE1 aa 1426–1651), the repeat plus DeCL domain (aa 1426–1851), and the QS subdomain (aa 1851–1976) as bait. Only one confirmed interactor was identified in the initial screen using the QS subdomain (aa 1851–1976), namely RRP6L1, previously identified by the Zhu lab (Zhang et al., 2014) as a protein affecting RdDM at a subset of loci and stabilizing Pol V transcripts in the context of chromatin (Figure 4F). Additional, targeted Y2H tests showed that RRP6L1 also interacts with the 17 aa repeat plus DeCL subdomain (aa 1426–1851), but the interaction is weaker than with the QS domain, which likely explains why the interaction was not detected in the initial screen (Figure 4F). In contrast, no interaction was detected with the 17 aa repeat subdomain alone (aa 1426–1651). Collectively, these results suggest that RRP6L1 can interact with both the DeCL and QS subdomains of the Pol V CTD (Figure 4F). Confirming the Y2H results, full-length RRP6L1 expressed in insect cells physically interacts with a recombinant QS subdomain polypeptide in vitro (Figure S5A–B). These physical interactions are consistent with RRP6L1 and the interacting CTD domains affecting an overlapping set of target loci (Figure 4D), as well as RRP6L1 binding to Pol V transcripts (Zhang et al., 2014). However, we were unable to confirm RRP6L1-NRPE1 interactions in vivo in co-immunoprecipitation assays. No firm conclusions can be drawn from a negative result, but because rrp6L1 mutants affect only ~20% of RdDM target loci (Figure 4D), it is plausible that RRP6L1- Pol V interactions are transient or involve only a fraction of the Pol V pool, making detection difficult.
If RRP6L1 degrades Pol V transcripts, as one might expect of a predicted exonuclease, Pol V transcript levels should increase in rrp6L1 mutants, but this is not the case (Figure 5A). This led us to test whether RRP6L1 expressed in insect cells (Figure S5A) exhibits exoribonuclease activity. Similar to its homolog, yeast Rrp6p, RRP6L1 does, indeed, have exonuclease activity, acting on single-stranded RNA that has a 3′ hydroxyl group, but not single-stranded RNA with a 3′ phosphate, single-stranded DNA, single-stranded RNA with secondary structure, or double-stranded RNA (Figure 5B).
Figure 5.
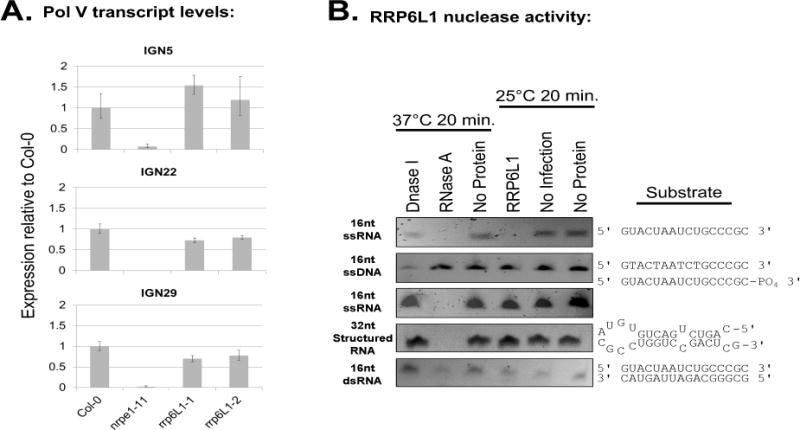
The CTD-interactor, RRP6L1 is a 3′ to 5′ exonuclease
A. Quantitative RT-PCR of Pol V-transcript levels in the wild-type (Col-0), nrpe1-11 or rrp6L1 mutants at three IGN loci. Histograms show ratios of ΔCt (Ct − CtActin2) values in the indicated genotypes relative to Col-0. Error bars represent the propagated standard error of the mean for 3 technical replicates.
B. SYBR gold-stained polyacrylamide gels of RNA or DNA substrates following incubations with DNase I, RNase A, rRRP6L1 expressed in insect cells, or buffer (no protein). No infection controls are extracts of insect cells not infected with baculovirus but otherwise treated the same as for RRP6L1 extracts. See also Figure S5.
Our experimental evidence shows that RRP6L1 is a functional exonuclease that interacts with the Pol V CTD, yet the Zhu lab has shown that RRP6L1 interacts with Pol V transcripts to stabilize the RNAs in the context of chromatin (Zhang et al., 2014). A potential solution to this paradox might be that RRP6L1 does not completely degrade Pol V transcripts, but merely trims their 3′ ends, analogous to yeast Rrp6p’s trimming of several nuclear RNA species (Fox and Mosley, 2016). To test this hypothesis, we conducted 3′ RACE at Pol V-transcribed loci (Table S4). At the IGN5A and IGN25 loci, where CHH methylation is RRP6L1-dependent, the median length of transcript 3′ ends increased in rrp6L1-1 and rrp6L1-2 mutants, relative to wild-type (Figures 6A and 6B). In contrast, at IGN17, IGN23, IGN29 and IGN35, where RRP6L1 has no effect on CHH methylation, transcript 3′ end lengths were unaffected, or slightly shorter, in rrp6L1 mutants (Figure 6C–D, Figure S6). Overall, these results suggest that at loci where CHH methylation is dependent on RRP6L1, the exonuclease trims the 3′ ends of Pol V transcripts.
Figure 6.
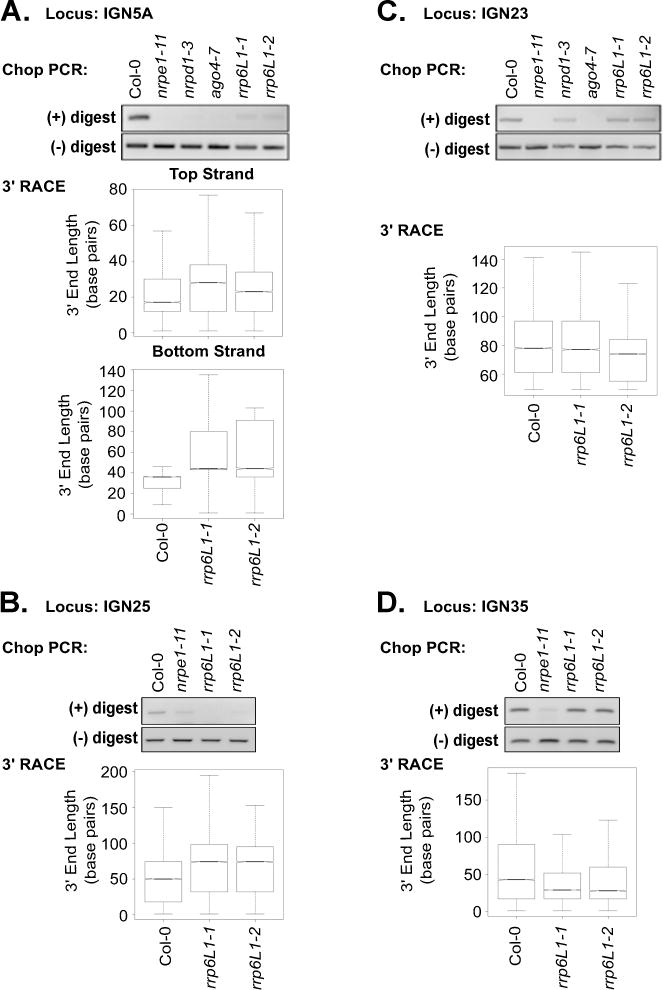
Evidence for RRP6L1 trimming of Pol V transcript 3′ ends. Chop-PCR and 3′ RACE results for Pol V transcribed loci: A. IGN5A, B. IGN25, C. IGN23, D. IGN35. Chop PCR was conducted with the enzymes HaeIII or AluI, both of which are inhibited by CHH methylation. Box plots show 3′ end lengths in basepairs, measured from the internal gene specific primer used in the 3′ RACE reactions to the ends of the RNAs. See also Figures S6–S7 and Table S4.
Interestingly, the 3′ ends at all loci examined are distributed within an ~100–200 basepair region, both at loci that undergo trimming and loci that do not (Figure S7). These results are reminiscent of the dispersed distribution of Pol V transcript 5′ ends (Wierzbicki et al., 2008), and suggests that Pol V initiates and terminates in a probabilistic manner within RdDM targeted regions.
As an independent test for longer RNAs at RRP6L1-dependent loci, we examined the IGN5 locus, where Pol V-dependent transcripts can be readily detected by RT-PCR using a primer pair defining what is termed amplicon 1 in Figure S6C. Moving one PCR primer an additional 58 bp in the 3′ direction defines a larger amplicon, amplicon 2, for which corresponding RNAs are not detected in wild-type. However, amplicon 2 transcripts are detected above background levels in rrp6L1 mutants, but lost in rrp6L1 nrpe1 double mutants, indicating that Pol V transcripts with extended 3′ ends accumulate if not processed by RRP6L1. Longer amplicon 2 transcripts are also detected nrpd1 (defective for siRNA biogenesis) and ago4 mutants, but do not increase in ago4 rrp6L1 double mutants (Figure S6C). Collectively, we interpret these results as evidence that RRP6L1 trims Pol V transcript 3′ ends generated by siRNA-directed AGO4 slicing.
Discussion
The CTD of NRPE1 is essential for Pol V function in vivo, but not for Pol V subunit assembly, nuclear localization or RNA polymerase activity in vitro. Thus, the reduced or undetectable levels of Pol V transcripts in full CTD or DeCL subdomain deletion mutants suggests an important role for the CTD in transcript production, possibly by affecting Pol V recruitment to target sites, initiation, elongation, or transcript stability. Consistent with this interpretation, Pol V transcripts and RdDM are similarly reduced in mutants for DRD1or DMS3 (Wierzbicki et al., 2008; Wierzbicki et al., 2009), which are needed for Pol V to be detected at target loci by chromatin immunoprecipitation (Law et al., 2010; Zhong et al., 2012). Likewise, the methylcytosine binding proteins SUVH2 and SUVH9 are also critical for Pol V recruitment to methylated target loci (Johnson et al., 2008; Liu et al., 2014), and the methylation profiles for suvh2 suvh9 double mutants, drd1, dms3, and nrpe1 mutants lacking the full CTD or DeCL subdomain are similar (see Figure 4A).
Deleting the DeCL subdomain has nearly the same effect as deleting the entire CTD. Other CTD subdomains are not as critical as the DeCL, but they significantly affect methylation at subsets of loci. Interestingly, methylated loci dependent on the linker, 17aa repeat, or QS subdomains are similarly dependent on proteins that interact with Pol V or its transcripts, including SPT5L, the IDP complex (IDN2, IDNL1, IDNL2), AGO6, or RRP6L1, implicating these CTD subdomains in diverse co-transcriptional steps of the RdDM process. In support of the possibility that the CTD subdomains may serve multiple roles, a study was published while this manuscript was in preparation that demonstrated restoring the 17 aa repeats into the NRPE1ΔSD construct rescues DNA methylation at affected loci, even upon changing the WG motifs to AG motifs, suggesting an unknown function for the repeats other than AGO binding (Lahmy et al., 2016).
The QS subdomain, located at the extreme C-terminus of NRPE1, is the least conserved feature of plant NRPE1 proteins, being present in A. thaliana but absent in other genera of the Brassicaceae family, including the closely related species, A. lyrata. Although the QS subdomain can be deleted without apparent consequence for Pol V function, its deletion has a synergistic effect when in combination with deletion of the 17 aa repeat subdomain. The QS and DeCL subdomains each interact with RRP6L1, suggesting that the QS subdomain may serve a recently evolved function that is at least partially redundant with the functions of other CTD subdomains.
Arabidopsis RRP6L1 was identified previously in a genetic screen for mutants disrupted in RdDM and found to stabilize Pol V transcript associations with chromatin, despite being a predicted exonuclease (Zhang et al., 2014). Our results provide several new insights, summarized in the model of Figure 7. First, our biochemical evidence indicates that RRP6L1 is an exonuclease that requires RNA with a free 3′ hydroxyl group, which could be accessed co-transcriptionally upon cleavage internally by an endonuclease, such as AGO4. The fact that Pol V transcripts have similarly extended 3′ ends in ago4 and rrp6L1 mutants, or mutants defective for siRNA biogenesis, is consistent with this hypothesis. Furthermore, Pol V transcripts exist in cells as a mixed population of species possessing either a 5′ triphosphate or 5′ monophosphate, indicative of both primary and sliced products (Wendte and Pikaard, 2017; Wierzbicki et al., 2008). It is noteworthy that AGO4 and RRP6L1-interacting subdomains of the Pol V CTD are adjacent to one another, such that siRNA-guided AGO4 slicing (Qi et al., 2006) may be coupled to RRP6L1 engagement of resulting RNA 3′ ends. Pausing of RRP6L1 trimming could promote retention of Pol V transcripts at chromatin, increasing the chances for engagement of the Pol V transcript-binding IDP complex and subsequent recruitment of the DNA methyltransferase, DRM2 (Figure 7).
Figure 7.
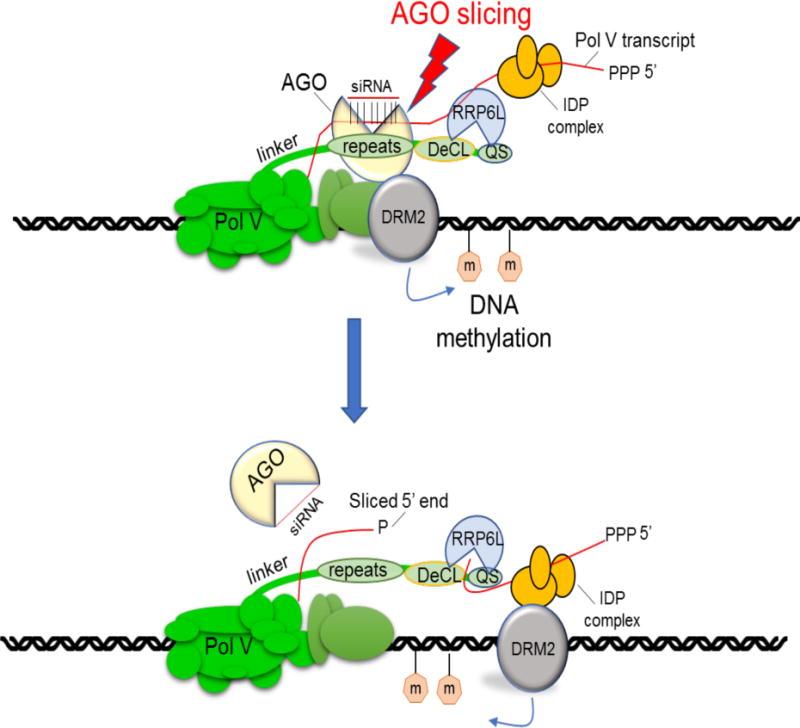
Model for CTD-mediated coordination of AGO4 transcript cleavage and RRP6L1 engagement of cleaved RNA 3′ ends. AGO4 and RRP6L1 bind adjacent subdomains of the NRPE1 CTD, such that co-transcriptional slicing of Pol V transcripts by AGO4, guided by basepaired 24 nt siRNAs, may be coupled to RRP6L1 engagement of cleaved RNA 3′ ends. RRP6L1’s trimming of Pol V transcripts, with pausing at sites of secondary structure, may facilitate RNA retention, allowing Pol V transcript-binding proteins, such as the IDP complex, to recruit the de novo cytosine methyltransferase, DRM2 to cleaved RNAs while Pol V transcription of nascent RNA continues.
Instead of serving as interaction sites for proteins important for mRNA synthesis and processing, like the Pol II CTD, the diverged Pol V CTD subdomains have evolved to mediate a potentially equally diverse repertoire of processes specific to RNA-directed silencing, including Argonaute and RRP6L interactions. Further understanding of the molecular details of CTD-mediated processes will likely help illuminate the full spectrum of Pol V functions.
Materials and Methods
Plant material
A. thaliana mutant lines, nrpe1-11 (Salk 029919), nrpd1-3 (Salk 128428), rrp6L1-1 (Salk 004432), rrp6L1-2 (Gabi 344G09) have been described previously (Onodera et al., 2005; Pontier et al., 2005; Zhang et al., 2014). The ago4-1 mutant is described in (Wierzbicki et al., 2009). Plants were grown in soil in long day conditions (16 hours light, 8 hours dark).
Targeted CTD deletion construct generation and plant transformation
The pENTR-NRPE1 genomic sequence with its endogenous promoter (Pontes et al., 2006) was recombined into pEarleyGate301 (Earley et al., 2006) to add a C-terminal HA tag. C-terminal domain deletions were obtained by using the pENTR-NRPE1 genomic clone as a DNA template with reverse primers that truncated the 3′ end (see Supplemental Experimental Procedures for primer sequences). Internal C-terminal domain deletions, nrpe1 Δ1251–1426 and nrpe1 Δ1251–1651, were obtained by the SLIM method (Chiu et al., 2004), and nrpe1 Δ1426–1651 was obtained using Stratagene cloning (now StrataClone from Agilent), using appropriate primers (Supplemental Experimental Procedures). pEarleyGate plasmids were transformed into Agrobacterium tumefaciens which was used to transform nrpe1-11 plants using the floral dip method (Bechtold and Pelletier, 1998; Clough and Bent, 1998).
Immunoprecipitation and western blot analysis
Protein for immunoprecipitation was extracted from frozen leaf tissue (4.0 g) as described in (Pontes et al., 2006). Antibodies were diluted in TBST + 5% (w/v) nonfat dried milk as follows: 1:500 NRPD/E2, 1:500 anti-NRPB/D/E11, and 1:3,000 anti-HA-HRP. Anti-rabbit-HRP (Amersham or Santa Cruz Biotechnology) diluted 1:5,000 was used as secondary antibody for native antibodies. Rabbit antibodies to NRPB11/NRPD11/NRPE11 (AT3G52090) were generated by Sigma and purified using immobilized NRPB/D/E11 protein.
Chop PCR
Chop PCR methylation analyses described in Figure 2A and Figure 6 were conducted using DNA extracted from 2.5 week old above ground plant tissues using the CTAB method (Murray and Thompson, 1980). ~350 ng of DNA was double digested using the enzymes AluI and HaeIII (NEB) at 37°C for 3 hours. PCR of regions of interest was conducted using GoTAQ Green (Promega) and primers described in Supplemental Experimental Procedures.
RT-PCR
RNA for RT-PCR was extracted from ~2.5 week old above ground plant tissues. Semi-quantitative RT-PCR analyses shown in Figure 2C and S6C were conducted as described in (Wierzbicki et al., 2008).Quantitative real time PCR shown in Figure 5A was conducted as described in (Rowley et al., 2013). All primer sequences are in Supplemental Experimental Procedures.
In vitro transcription assays
In vitro transcription assays were conducted as described in (Haag et al., 2012) with minor modifications described in the Supplemental Experimental Procedures.
Bisulfite sequencing
100 ng of DNA extracted from ~2.5 week old above ground plant tissues was prepared for Illumina sequencing using the TruSeq DNA methylation library prep kit. Mapping was completed using Bismark version 0.16.1 (Krueger and Andrews, 2011) and differentially methylated regions (DMRs) were identified using methylKit version 0.9.5 (Akalin et al., 2012) based on a 300 basepair sliding window. Significant hypo-DMRs were calculated based on two biologic replicates of each line using a logistic regression with a minimum cutoff of 25% (CHH and CHG context) or 40% (CG context) decrease relative to Col-0 and a q value less than or equal to 0.01. P-values were corrected for multiple testing using the SLIM method (Akalin et al., 2012). Additional details for mapping and analysis are in the Supplemental Experimental Procedures.
Small RNA sequencing
Small RNA sequencing for nrpe1-11 was completed as described in (Blevins et al., 2015). Procedures for mapping and analysis of sRNA sequencing are in the Supplemental Experimental Procedures.
Yeast two hybrid analyses
Yeast two hybrid (Y2H) was performed by the Indiana University Yeast Two Hybrid Facility. Briefly, an uncut custom cDNA Library for A. thaliana in the entry vector, pENTR222 (Invitrogen) was cloned into the prey vector, pDEST22 using LR Clonase (Invitrogen). NRPE1 cDNA fragments corresponding to amino acids 1426–1851 (repeat + DeCL domains), 1426–1651 (repeat domain), and 1851–1976 (QS) were cloned into the bait vector, pDEST32, and screening against the A. thaliana cDNA library was conducted in yeast strain MaV203 (Invitrogen). Screening for interactions using lacZ assays, ura- media, and his- media containing 20mM or 100mM of the HIS3 inhibitor, E-Amino-1, 2, 4 Triazol (3-AT). Follow-up Y2H assays, shown in Figure 4, assessed interactions by detecting growth in the presence of 20mM 3-AT on his-media. Primers are in Supplemental Experimental Procedures.
In vitro nuclease activity assays
50 ng rRRP6L1 was added to a 50 μl reaction containing 0.1 mM nucleic acid substrate and 1X Turbo DNase buffer (Ambion). Reactions were placed at 25°C for 30 minutes and inactivated by cleaning with a Zymogen Oligo Clean and Concentrate Kit. Samples were visualized on 15% denaturing polyacrylamide gels stained with Sybr Gold (Invitrogen). Negative controls included a no protein control and the addition of an equal volume of nickel column purified protein extract from uninfected insect cells. For positive controls, nucleic acid substrates were also digested with commercially available Turbo DNase (2.5 U/reaction) (Ambion) or RNase A (100 ng/reaction) (Thermo Scientific) at 37°C for 30 minutes.
3′ RACE
3′RACE was conducted using a modified version of the protocol provided in the 3′ RACE System by Invitrogen (Cat. # 18373019). Procedures for 3′ RACE, sequencing, and mapping and analysis are described in the Supplemental Experimental Procedures.
Supplementary Material
Highlights.
The NRPE1 CTD is essential for Pol V function in vivo
The DeCL subdomain is critical; three other subdomains affect specific loci
CTD subdomains and other silencing pathway proteins affect similar loci
RRP6L1 is a CTD-interacting exonuclease that trims Pol V transcripts
Acknowledgments
The authors thank James Ford of the Indiana University Center for Genomics and Bioinformatics for help with library preparation and sequencing, Vibhor Mishra for help with RRP6L1 expression in insect cells, Laurel Bender and Michael Alley of the Indiana University Yeast Two Hybrid Facility, Todd Blevins for sequencing of nrpe1-11 small RNAs, and Thierry Lagrange for providing seed for ΔSD plants. This work was supported by funds to CSP as an Investigator of the Howard Hughes Medical Institute and by a grant from the Gordon and Betty Moore Foundation and grant GM077590 from the National Institutes of Health. J.M.W. was supported by NIH training Grant, T32GM007757, and predoctoral fellowship Award F31GM116346. The content of this work is solely the responsibility of the authors and does not necessarily represent the official views of our sponsors.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no conflicts of interests.
Author contributions
J.R.H., J.M.W., and C.S.P. designed the experiments, analyzed the data, and wrote the paper. J.S. conducted in vitro transcription assays. O.P. and A.M. conducted immunolocalization assays. J.R.H. and J.M.W. conducted all other experiments.
Accession numbers
The deep sequencing data have been deposited in NCBI’s Gene Expression Omnibus (Edgar et al., 2002) with the accession number GSE93360.
References
- Akalin A, Kormaksson M, Li S, Garrett-Bakelman FE, Figueroa ME, Melnick A, Mason CE. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 2012;13:R87. doi: 10.1186/gb-2012-13-10-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausin I, Greenberg MV, Simanshu DK, Hale CJ, Vashisht AA, Simon SA, Lee TF, Feng S, Espanola SD, Meyers BC, et al. INVOLVED IN DE NOVO 2-containing complex involved in RNA-directed DNA methylation in Arabidopsis. Proc Natl Acad Sci U S A. 2012;109:8374–8381. doi: 10.1073/pnas.1206638109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausin I, Mockler TC, Chory J, Jacobsen SE. IDN1 and IDN2 are required for de novo DNA methylation in Arabidopsis thaliana. Nat Struct Mol Biol. 2009;16:1325–1327. doi: 10.1038/nsmb.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold N, Pelletier G. In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol Biol. 1998;82:259–266. doi: 10.1385/0-89603-391-0:259. [DOI] [PubMed] [Google Scholar]
- Becker C, Hagmann J, Muller J, Koenig D, Stegle O, Borgwardt K, Weigel D. Spontaneous epigenetic variation in the Arabidopsis thaliana methylome. Nature. 2011;480:245–249. doi: 10.1038/nature10555. [DOI] [PubMed] [Google Scholar]
- Bellaoui M, Gruissem W. Altered expression of the Arabidopsis ortholog of DCL affects normal plant development. Planta. 2004;219:819–826. doi: 10.1007/s00425-004-1295-5. [DOI] [PubMed] [Google Scholar]
- Bellaoui M, Keddie JS, Gruissem W. DCL is a plant-specific protein required for plastid ribosomal RNA processing and embryo development. Plant molecular biology. 2003;53:531–543. doi: 10.1023/B:PLAN.0000019061.79773.06. [DOI] [PubMed] [Google Scholar]
- Bies-Etheve N, Pontier D, Lahmy S, Picart C, Vega D, Cooke R, Lagrange T. RNA-directed DNA methylation requires an AGO4-interacting member of the SPT5 elongation factor family. EMBO Rep. 2009;10:649–654. doi: 10.1038/embor.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blevins T, Podicheti R, Mishra V, Marasco M, Tang H, Pikaard CS. Identification of Pol IV and RDR2-dependent precursors of 24 nt siRNAs guiding de novo DNA methylation in Arabidopsis. Elife. 2015;4:e09591. doi: 10.7554/eLife.09591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohmdorfer G, Rowley MJ, Kucinski J, Zhu Y, Amies I, Wierzbicki AT. RNA-directed DNA methylation requires stepwise binding of silencing factors to long non-coding RNA. Plant J. 2014;79:181–191. doi: 10.1111/tpj.12563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu J, March PE, Lee R, Tillett D. Site-directed, Ligase-Independent Mutagenesis (SLIM): a single-tube methodology approaching 100% efficiency in 4 h. Nucleic Acids Res. 2004;32:e174. doi: 10.1093/nar/gnh172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 2006;45:616–629. doi: 10.1111/j.1365-313X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Shami M, Pontier D, Lahmy S, Braun L, Picart C, Vega D, Hakimi MA, Jacobsen SE, Cooke R, Lagrange T. Reiterated WG/GW motifs form functionally and evolutionarily conserved ARGONAUTE-binding platforms in RNAi-related components. Genes Dev. 2007;21:2539–2544. doi: 10.1101/gad.451207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MJ, Mosley AL. Rrp6: Integrated roles in nuclear RNA metabolism and transcription termination. Wiley Interdiscip Rev RNA. 2016;7:91–104. doi: 10.1002/wrna.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag JR, Brower-Toland B, Krieger EK, Sidorenko L, Nicora CD, Norbeck AD, Irsigler A, LaRue H, Brzeski J, McGinnis K, et al. Functional Diversification of Maize RNA Polymerase IV and V Subtypes via Alternative Catalytic Subunits. Cell reports. 2014;9:378–390. doi: 10.1016/j.celrep.2014.08.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag JR, Pikaard CS. Multisubunit RNA polymerases IV and V: purveyors of non-coding RNA for plant gene silencing. Nat Rev Mol Cell Biol. 2011;12:483–492. doi: 10.1038/nrm3152. [DOI] [PubMed] [Google Scholar]
- Haag JR, Ream TS, Marasco M, Nicora CD, Norbeck AD, Pasa-Tolic L, Pikaard CS. In vitro transcription activities of Pol IV, Pol V, and RDR2 reveal coupling of Pol IV and RDR2 for dsRNA synthesis in plant RNA silencing. Molecular cell. 2012;48:811–818. doi: 10.1016/j.molcel.2012.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He XJ, Hsu YF, Zhu S, Wierzbicki AT, Pontes O, Pikaard CS, Liu HL, Wang CS, Jin H, Zhu JK. An effector of RNA-directed DNA methylation in arabidopsis is an ARGONAUTE 4- and RNA-binding protein. Cell. 2009;137:498–508. doi: 10.1016/j.cell.2009.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Jones AM, Searle I, Patel K, Vogler H, Hubner NC, Baulcombe DC. An atypical RNA polymerase involved in RNA silencing shares small subunits with RNA polymerase II. Nat Struct Mol Biol. 2009;16:91–93. doi: 10.1038/nsmb.1539. [DOI] [PubMed] [Google Scholar]
- Huang Y, Kendall T, Forsythe ES, Dorantes-Acosta A, Li S, Caballero-Perez J, Chen X, Arteaga-Vazquez M, Beilstein MA, Mosher RA. Ancient Origin and Recent Innovations of RNA Polymerase IV and V. Mol Biol Evol. 2015;32:1788–1799. doi: 10.1093/molbev/msv060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing Y, Sun H, Yuan W, Wang Y, Li Q, Liu Y, Li Y, Qian W. SUVH2 and SUVH9 Couple Two Essential Steps for Transcriptional Gene Silencing in Arabidopsis. Mol Plant. 2016;9:1156–1167. doi: 10.1016/j.molp.2016.05.006. [DOI] [PubMed] [Google Scholar]
- Johnson LM, Du J, Hale CJ, Bischof S, Feng S, Chodavarapu RK, Zhong X, Marson G, Pellegrini M, Segal DJ, et al. SRA- and SET-domain-containing proteins link RNA polymerase V occupancy to DNA methylation. Nature. 2014;507:124–128. doi: 10.1038/nature12931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LM, Law JA, Khattar A, Henderson IR, Jacobsen SE. SRA-domain proteins required for DRM2-mediated de novo DNA methylation. PLoS genetics. 2008;4:e1000280. doi: 10.1371/journal.pgen.1000280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keddie JS, Carroll B, Jones JD, Gruissem W. The DCL gene of tomato is required for chloroplast development and palisade cell morphogenesis in leaves. Embo J. 1996;15:4208–4217. [PMC free article] [PubMed] [Google Scholar]
- Kollen K, Dietz L, Bies-Etheve N, Lagrange T, Grasser M, Grasser KD. The zinc-finger protein SPT4 interacts with SPT5L/KTF1 and modulates transcriptional silencing in Arabidopsis. FEBS Lett. 2015;589:3254–3257. doi: 10.1016/j.febslet.2015.09.017. [DOI] [PubMed] [Google Scholar]
- Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahmy S, Pontier D, Bies-Etheve N, Laudie M, Feng S, Jobet E, Hale CJ, Cooke R, Hakimi MA, Angelov D, et al. Evidence for ARGONAUTE4-DNA interactions in RNA-directed DNA methylation in plants. Genes Dev. 2016;30:2565–2570. doi: 10.1101/gad.289553.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Ausin I, Johnson LM, Vashisht AA, Zhu JK, Wohlschlegel JA, Jacobsen SE. A protein complex required for polymerase V transcripts and RNA- directed DNA methylation in Arabidopsis. Curr Biol. 2010;20:951–956. doi: 10.1016/j.cub.2010.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Vandivier LE, Tu B, Gao L, Won SY, Li S, Zheng B, Gregory BD, Chen X. Detection of Pol IV/RDR2-dependent transcripts at the genomic scale in Arabidopsis reveals features and regulation of siRNA biogenesis. Genome Res. 2015;25:235–245. doi: 10.1101/gr.182238.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ZW, Shao CR, Zhang CJ, Zhou JX, Zhang SW, Li L, Chen S, Huang HW, Cai T, He XJ. The SET domain proteins SUVH2 and SUVH9 are required for Pol V occupancy at RNA-directed DNA methylation loci. PLoS genetics. 2014;10:e1003948. doi: 10.1371/journal.pgen.1003948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Hall BD. A multistep process gave rise to RNA polymerase IV of land plants. J Mol Evol. 2007;64:101–112. doi: 10.1007/s00239-006-0093-z. [DOI] [PubMed] [Google Scholar]
- Matzke MA, Mosher RA. RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat Rev Genet. 2014;15:394–408. doi: 10.1038/nrg3683. [DOI] [PubMed] [Google Scholar]
- Mosher RA, Schwach F, Studholme D, Baulcombe DC. PolIVb influences RNA-directed DNA methylation independently of its role in siRNA biogenesis. Proc Natl Acad Sci U S A. 2008;105:3145–3150. doi: 10.1073/pnas.0709632105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera Y, Haag JR, Ream T, Costa Nunes P, Pontes O, Pikaard CS. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell. 2005;120:613–622. doi: 10.1016/j.cell.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Pontes O, Li CF, Costa Nunes P, Haag J, Ream T, Vitins A, Jacobsen SE, Pikaard CS. The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center. Cell. 2006;126:79–92. doi: 10.1016/j.cell.2006.05.031. [DOI] [PubMed] [Google Scholar]
- Pontier D, Picart C, Roudier F, Garcia D, Lahmy S, Azevedo J, Alart E, Laudie M, Karlowski WM, Cooke R, et al. NERD, a plant-specific GW protein, defines an additional RNAi-dependent chromatin-based pathway in Arabidopsis. Molecular cell. 2012;48:121–132. doi: 10.1016/j.molcel.2012.07.027. [DOI] [PubMed] [Google Scholar]
- Pontier D, Yahubyan G, Vega D, Bulski A, Saez-Vasquez J, Hakimi MA, Lerbs-Mache S, Colot V, Lagrange T. Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes Dev. 2005;19:2030–2040. doi: 10.1101/gad.348405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, He X, Wang XJ, Kohany O, Jurka J, Hannon GJ. Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature. 2006;443:1008–1012. doi: 10.1038/nature05198. [DOI] [PubMed] [Google Scholar]
- Ream T, Haag J, Pikaard CS. In: Plant multisubunit RNA polymerases IV and V In Nucleic Acid Polymerases. Murakami K, Trakselis M, editors. Berlin Heidelberg: Springer-Verlag; 2013. pp. 289–308. [Google Scholar]
- Ream TS, Haag JR, Wierzbicki AT, Nicora CD, Norbeck AD, Zhu JK, Hagen G, Guilfoyle TJ, Pasa-Tolic L, Pikaard CS. Subunit compositions of the RNA-silencing enzymes Pol IV and Pol V reveal their origins as specialized forms of RNA polymerase II. Molecular cell. 2009;33:192–203. doi: 10.1016/j.molcel.2008.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley MJ, Avrutsky MI, Sifuentes CJ, Pereira L, Wierzbicki AT. Independent chromatin binding of ARGONAUTE4 and SPT5L/KTF1 mediates transcriptional gene silencing. PLoS genetics. 2011;7:e1002120. doi: 10.1371/journal.pgen.1002120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley MJ, Bohmdorfer G, Wierzbicki AT. Analysis of long non-coding RNAs produced by a specialized RNA polymerase in Arabidopsis thaliana. Methods. 2013;63:160–169. doi: 10.1016/j.ymeth.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz RJ, Schultz MD, Lewsey MG, O’Malley RC, Urich MA, Libiger O, Schork NJ, Ecker JR. Transgenerational epigenetic instability is a source of novel methylation variants. Science. 2011;334:369–373. doi: 10.1126/science.1212959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H, Greenberg MV, Feng S, Bernatavichute YV, Jacobsen SE. Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell. 2013;152:352–364. doi: 10.1016/j.cell.2012.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo JT, Beilstein MA, Mosher RA. The Argonaute-binding platform of NRPE1 evolves through modulation of intrinsically disordered repeats. New Phytol. 2016;212:1094–1105. doi: 10.1111/nph.14089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker SL, Reece J, Ream TS, Pikaard CS. Evolutionary history of plant multisubunit RNA polymerases IV and V: subunit origins via genome-wide and segmental gene duplications, retrotransposition, and lineage-specific subfunctionalization. Cold Spring Harbor symposia on quantitative biology. 2010;75:285–297. doi: 10.1101/sqb.2010.75.037. [DOI] [PubMed] [Google Scholar]
- Wang Y, Ma H. Step-wise and lineage-specific diversification of plant RNA polymerase genes and origin of the largest plant-specific subunits. New Phytol. 2015;207:1198–1212. doi: 10.1111/nph.13432. [DOI] [PubMed] [Google Scholar]
- Wendte JM, Pikaard CS. The RNAs of RNA-directed DNA methylation. Biochim Biophys Acta. 2017;1860:140–148. doi: 10.1016/j.bbagrm.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicki AT, Haag JR, Pikaard CS. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell. 2008;135:635–648. doi: 10.1016/j.cell.2008.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicki AT, Ream TS, Haag JR, Pikaard CS. RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat Genet. 2009;41:630–634. doi: 10.1038/ng.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaborowska J, Egloff S, Murphy S. The pol II CTD: new twists in the tail. Nat Struct Mol Biol. 2016;23:771–777. doi: 10.1038/nsmb.3285. [DOI] [PubMed] [Google Scholar]
- Zhai J, Bischof S, Wang H, Feng S, Lee TF, Teng C, Chen X, Park SY, Liu L, Gallego-Bartolome J, et al. A One Precursor One siRNA Model for Pol IV-Dependent siRNA Biogenesis. Cell. 2015;163:445–455. doi: 10.1016/j.cell.2015.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CJ, Ning YQ, Zhang SW, Chen Q, Shao CR, Guo YW, Zhou JX, Li L, Chen S, He XJ. IDN2 and its paralogs form a complex required for RNA-directed DNA methylation. PLoS genetics. 2012;8:e1002693. doi: 10.1371/journal.pgen.1002693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Tang K, Qian W, Duan CG, Wang B, Zhang H, Wang P, Zhu X, Lang Z, Yang Y, et al. An Rrp6-like protein positively regulates noncoding RNA levels and DNA methylation in Arabidopsis. Molecular cell. 2014;54:418–430. doi: 10.1016/j.molcel.2014.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Henderson IR, Lu C, Green PJ, Jacobsen SE. Role of RNA polymerase IV in plant small RNA metabolism. Proc Natl Acad Sci U S A. 2007;104:4536–4541. doi: 10.1073/pnas.0611456104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X, Hale CJ, Law JA, Johnson LM, Feng S, Tu A, Jacobsen SE. DDR complex facilitates global association of RNA polymerase V to promoters and evolutionarily young transposons. Nat Struct Mol Biol. 2012;19:870–875. doi: 10.1038/nsmb.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Law JA. RNA Pol IV and V in gene silencing: Rebel polymerases evolving away from Pol II’s rules. Curr Opin Plant Biol. 2015;27:154–164. doi: 10.1016/j.pbi.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


