Abstract
ARID1A , encoding a subunit of the SWI/SNF chromatin-remodelling complex, is the most frequently mutated epigenetic regulator across all human cancers. ARID1A and TP53 mutations are typically mutually exclusive. Therapeutic approaches that correlate with this genetic characteristic remain to be explored. Here, we show that HDAC6 activity is essential in ARID1A-mutated ovarian cancers. Inhibition of HDAC6 activity using a clinically applicable small molecule inhibitor significantly improved the survival of mice bearing ARID1A-mutated tumours. This correlated with the suppression of growth and dissemination of ARID1A-mutated, but not wildtype, tumours. The dependence on HDAC6 activity in ARID1A-mutated cells correlated with a direct transcriptional repression of HDAC6 by ARID1A. HDAC6 inhibition selectively promoted apoptosis of ARID1A-mutated cells. HDAC6 directly deacetylates Lys-120 of p53, a pro-apoptotic post-translational modification. Thus, ARID1A mutation inactivates p53’s apoptosis-promoting function by upregulating HDAC6. Together, these results indicate that pharmacological inhibition of HDAC6 is a therapeutic strategy for ARID1A-mutated cancers.
Introduction
SWI/SNF chromatin remodelling complexes regulate gene transcription by changing chromatin structure through hydrolysing ATP 1. Cancer genome sequencing found that mutations in genes encoding for the subunits of the SWI/SNF complexes collectively occur in ~20% of all human cancers 2. For example, saturation analysis of The Cancer Genome Atlas (TCGA) cancer mutational profile reveals that the ARID1A subunit of the SWI/SNF complex shows one of the highest mutation rates among epigenetic regulators 3. Notably, ARID1A is mutated in over 50% of ovarian clear cell carcinomas and 30% of ovarian endometrioid carcinomas 4, 5. ARID1A mutation is a known genetic driver of ovarian cancer 6–8. ARID1A and TP53 mutations are typically mutually exclusive in ovarian cancer 9. Consistently, ARID1A mutated ovarian cancers often lack genomic instability 5. However, therapeutic approaches harnessing this genetic characteristic of ARID1A-mutated cancers remain to be explored.
Over 90% of the ARID1A mutations observed in ovarian cancer are frame-shift or nonsense mutations that result in loss of ARID1A protein expression 4, 5, 10. Loss of ARID1A correlates with late-stage disease and predicts early recurrence of ovarian clear cell carcinoma 11. Ovarian clear cell carcinoma ranks second as the cause of death from epithelial ovarian cancer 12 and is associated with the worst prognosis amongst the major ovarian cancer subtypes when diagnosed at advanced stages 13, 14. Additionally, for advanced stage disease, there is currently no effective therapy. Notably, in Japan, its prevalence is higher than in western countries, with an estimated incidence of ~25% of epithelial ovarian cancer 15.
Histone deacetylase 6 (HDAC6) belongs to class IIb HDACs 16. Unlike other HDACs, HDAC6 primarily functions in the cytoplasm 16. HDAC6 deacetylates various substrates to regulate protein trafficking and degradation, cell shape and migration 16. HDAC6 expression is increased in several cancer types including ovarian cancer 17. Specific small molecule HDAC6 inhibitors have been developed and are in clinical trials for human hematopoietic malignancies 18. Here we show that inhibition of HDAC6 activity is selective against ARID1A-mutated ovarian cancer. Our findings provide scientific rationale for targeting ARID1A-mutation in ovarian cancer using pharmacological inhibition of HDAC6 activity.
Results
ARID1A-inactivated cells are selectively sensitive to HDAC6 inhibition
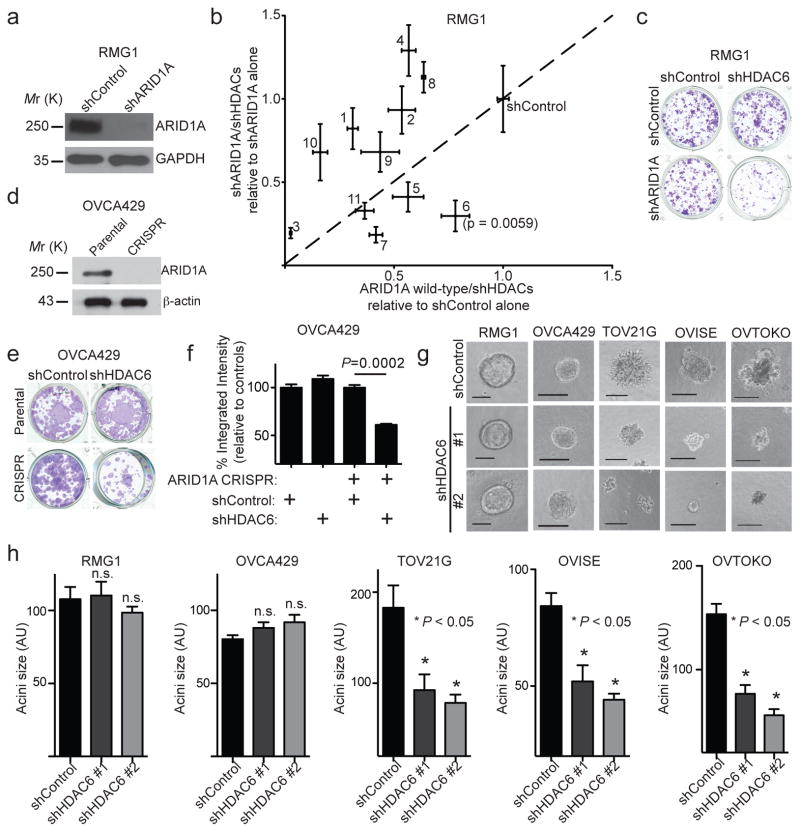
To examine the role of specific HDACs in the context of ARID1A-mutated ovarian cancers, we performed an unbiased short hairpin RNA (shRNA) knockdown-based evaluation against eleven histone deacetylase genes. This was done in the context of ARID1A wildtype ovarian clear cell RMG1 cells with or without ARID1A knockdown (Fig. 1a). ARID1A knockdown allows us to mimic loss of ARID1A protein expression caused by >90% of ARID1A mutations in ovarian cancer 5 and ensure the same genetic background for the unbiased evaluation. We transiently transduced pooled shRNAs for each of the 11 individual HDACs in ARID1A wildtype RMG1 cells with or without ARID1A knockdown (Supplementary Table 1). We confirmed knockdown of all the HDACs by qRT-PCR and found a similar degree of knockdown of the HDACs regardless of ARID1A expression (Supplementary Fig. 1a). To measure changes in cell viability, these cells were subjected to a colony formation assay. Similar to previous reports 19, we observed no significant difference between ARID1A wildtype RMG1 cells with or without ARID1A knockdown (Fig. 1b–c). HDAC6 knockdown showed the highest selectivity against ARID1A knockdown with the least growth inhibitory effects on controls (Fig. 1b and Supplementary Table 2). Likewise, HDAC6 knockdown was selective against ARID1A knockout in ARID1A wildtype OVCA429 cells (Fig. 1d–f and Supplementary Fig. 1b). Consistently, HDAC6 knockdown was selective against ARID1A-mutated ovarian clear cell and endometrioid cancer cell lines in the Project Achilles synthetic lethality screen database (Supplementary Fig. 1c) 20.
Figure 1. ARID1A-inactivated cells are selectively sensitive to HDAC6 knockdown.
(a–c) ARID1A and a loading control GAPDH protein expression in ARID1A wildtype RMG1 cells with or without ARID1A knockdown (a). Cells were transduced with lentivirus encoding shRNA to each of the 11 individual HDACs (HDAC1-11) and subjected to colony formation assay. Scatterplot of the integrated density normalized to control. The x-axis indicates changes in cell growth induced by individual shHDACs in control ARID1A wildtype treated cells, while the y-axis indicates changes in cell growth induced by the same shHDACs in shARID1A-expressing cells. Individual HDACs are indicated with their respective numbers (b). n=4 independent experiments. Colony formation by the indicated cells (c). (d–f) ARID1A protein expression in parental and ARID1A CRIPSR OVCA429 cells (d). Colony formation assay using the indicated OVCA429 cells with or without HDAC6 knockdown (e), which was quantified (f). n=3 independent experiments. (g–h) A panel of cell lines with known ARID1A mutational status with or without HDAC6 knockdown were grown in 3D using Matrigel. Shown are acini formed by the indicated cells (g). Scale bar = 75 AU in NIH Image J software. The diameters of acini (n=50; representative of three biological repeats) were quantified (h). n.s. indicates not significant. Error bars represent mean with S.E.M. P-value calculated via two-tailed t-test. Statistical source data are provided in Supplementary Table 6. Unprocessed original scans of all blots with size marker are shown in Supplementary Fig. 9.
ARID1A status correlates with response to HDAC6 inhibition
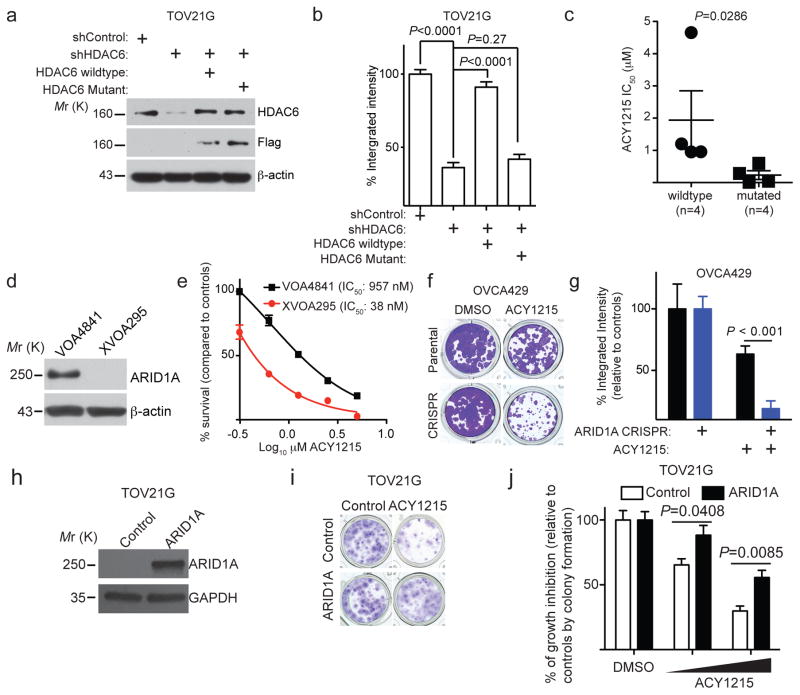
We next validated the initial findings in a panel of clear cell ovarian cancer cell lines in 3 dimensional (3D) cultures using Matrigel extracellular matrix that more closely mimics the tumour microenvironment. HDAC6 knockdown had no appreciable effect on the growth of ARID1A wildtype cells but significantly suppressed the growth of ARID1A-mutated cells (Fig. 1g–h and Supplementary Fig. 1d). The observed growth inhibition depends on the enzymatic activity of HDAC6 because the growth inhibition was rescued by a wildtype HDAC6 but not a catalytically inactive H216/611A mutant 21 (Fig. 2a–b and Supplementary Fig. 2a–b). Notably, pan-HDAC inhibitor Vorinostat that inhibits all other class I and class II HDACs 22 was not selectively against ARID1A knockdown in ARID1A wildtype RMG1 cells (Supplementary Fig. 2c). Given that class I and class II HDACs are not selective against ARID1A knockdown (e.g. Fig. 1b), the non-selective nature of pan-HDAC inhibitor Vorinostat against both class I and class II HDACs may have masked its inhibitory effects on HDAC6. Selective and specific HDAC6 inhibitors have been developed. We tested the HDAC6 inhibitor ACY1215 (Rocilinostat) 23 in a panel of cell lines with or without ARID1A mutation because it was safe in clinical trials 18. Compared with ARID1A wildtype cells, the IC50 of ACY1215 was significantly lower in ARID1A-mutated cells (Fig. 2c and Supplementary Table 3). Primary clear cell ovarian tumour cultures without ARID1A expression are more sensitive to ACY1215 compared to those with ARID1A expression (Fig. 2c–d). The IC50 values of ACY1215 in primary cells are comparable to those observed in cell lines (Fig. 2c–e and Supplementary Table 3). ARID1A knockout significantly increased the sensitivity of ARID1A wildtype OVCA429 cells to ACY1215 (Fig. 2f–g). Conversely, restoration of wildtype ARID1A in ARID1A-mutated TOV21G cells reduced the sensitivity of these cells to ACY1215 (Fig. 2h–j). Interestingly, knockdown of other SWI/SNF subunits such as BRG1 1 did not increase ACY1215 sensitivity (Supplementary Fig. 2d–f). This correlates with a compensation of BRG1 loss by the mutually exclusive catalytic subunit BRM (Supplementary Fig. 2g–i). We conclude that ARID1A-inactivated cells are selectively sensitive to HDAC6 inhibition.
Figure 2. The selectivity against ARID1A mutation depends on the enzymatic activity of HDAC6.
(a–b) Expression of HDAC6, FLAG and a loading control β-actin in ARID1A-mutated TOV21G cells expressing a shHDAC and concurrent expression of FLAG-tagged shRNA resistant wildtype HDAC6 or a catalytically inactive H216/611A mutant (a). The indicated cells were subjected to colony formation assay and integrated density was measured (b). n=4 independent experiments. (c) IC50 of HDAC6 inhibitor ACY1215 is significantly higher in ARID1A wildtype (n=4 cell lines) than mutated (n=4 cell lines) cells. (d) Expression of ARID1A and a loading control β-actin in the indicated primary cultures of human ovarian clear cell carcinomas determined by immunoblot. (e) HDAC6 inhibitor ACY1215 dose response curves of primary clear cell ovarian tumour cultures with (VOA4841) and without (XVOA295) ARID1A expression. n=3 independent experiments. (f) Control and ARID1A CRISPR OVCA429 cells were treated with or without 1.25 μM ACY1215 in a colony formation assay. (g) Quantification of (f). n=4 independent experiments. (h–j) Immunoblots of the indicated proteins in ARID1A-mutated TOV21G cells with or without wild-type ARID1A restoration (h). The indicated cells treated with or without ACY1215 were plated in 24-well plates in quadruplicates and subjected to colony formation assay for 12 days, after which they were stained with 0.05% crystal violet (shown are cells treated with 625 nM ACY1215). Note that ARID1A restoration inhibits the growth of ARID1A-mutated cells 19 9. To limit the potential bias in colony formation, the number of cells used for ARID1A restored cells were 2-fold of the control ARID1A-mutated cells (i). Integrated density was measured with NIH Image J software as a surrogate for cell growth (j). The concentrations of ACY1215 were 312 nM and 625 nM, respectively. n=4 independent experiments. Error bars represent mean with S.E.M. P-value calculated via two-tailed t-test. Statistical source data are provided in Supplementary Table 6. Unprocessed original scans of all blots with size marker are shown in Supplementary Fig. 9.
HDAC6 inhibition triggers apoptosis in ARID1A-inactivated cells
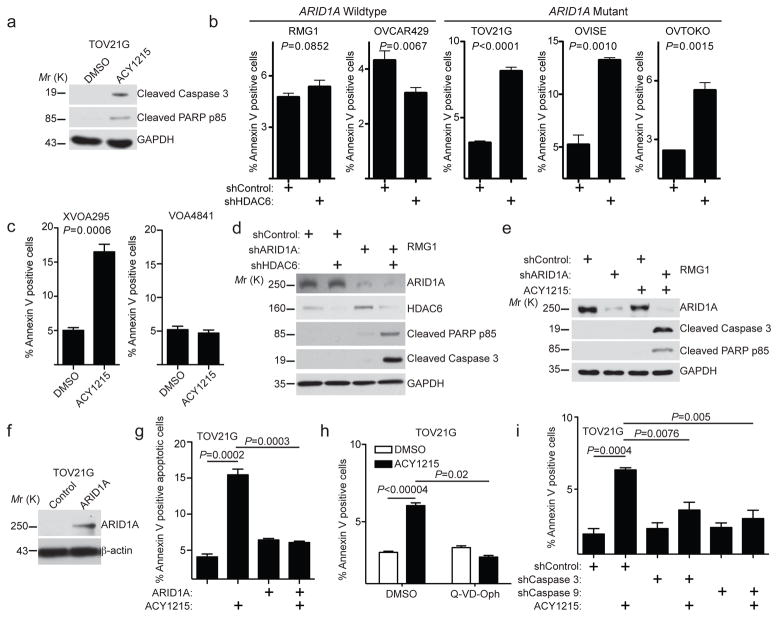
We next determined the mechanism whereby HDAC6 inhibition suppresses the growth of ARID1A-inactivated cells. HDAC6 inhibitor ACY1215 treatment induced apoptosis of ARID1A-inactivated cells as shown by an increase in Annexin V positive cells and upregulation of cleaved caspase 3 and cleaved PARP p85 (Fig. 3a–c and Supplementary Fig. 3a). Consistent with the observed selectivity of HDAC6 inhibition in cells with ARID1A inactivation (Fig. 1), ACY1215 did not induce a significant increase in apoptosis in ARID1A wildtype cells (Fig. 3b–e), and wildtype ARID1A restoration suppressed ACY1215 induced apoptosis in ARID1A-mutated TOV21G cells (Fig. 3f–g). Compared with ARID1A wildtype controls, the HDAC6 inhibitor ACY1215 or knockdown of HDAC6 increased markers of apoptosis in ARID1A knockdown cells (Fig. 3d–e). Notably, a pan-caspase inhibitor Q-VD-Oph or knockdown of intrinsic apoptotic pathway initiator caspase 9 or effector caspase 3 24 significantly suppressed the apoptosis induced by ACY1215 (Fig. 3h–i and Supplementary Fig. 3b–c). In contrast, knockdown of Caspase 8, the caspase of the extrinsic apoptotic pathway 24, did not affect the apoptosis induced by ACY1215 (Supplementary Fig. 3d–e). We conclude that HDAC6 inhibition promotes apoptosis in ARID1A-inactivated cells.
Figure 3. HDAC6 inhibition induces apoptosis in ARID1A inactivated cells.
(a) ARID1A-mutated TOV21G cells treated with 1.25 μM ACY1215 or DMSO control were examined for expression of markers of apoptosis cleaved caspase 3, cleaved PARP p85 or a loading control GAPDH by immunoblot. (b) Percent apoptosis in the indicated cell lines was quantified by FACS based on Annexin V staining. (c) Percent apoptosis of in the indicated primary clear cell ovarian tumour cultures was quantified by FACS based on Annexin V staining. (d) ARID1A wildtype RMG1 cells with the indicated knockdown of ARID1A, HDAC6 or a combination was examined for expression of apoptotic markers cleaved caspase 3 and cleaved PARP p85 and a loading control GAPDH by immunoblot. (e) ARID1A wildtype RMG1 cells with or without ARID1A knockdown were treated with 1.25 μM ACY1215. Expression of apoptotic markers cleaved caspase 3 and cleaved PARP p85 and a loading control GAPDH determined by immunoblot. (f) Expression of ARID1A and a loading control β-actin in TOV21G cells with or without wildtype ARID1A restoration. (g) Percent apoptosis based on Annexin V staining in ARID1A-mutated TOV21G cells with or without wildtype ARID1A restoration and treated with 1.25 μM ACY1215 or DMSO controls for 96 hrs. (h) Percent apoptosis based on Annexin V staining in ARID1A-mutated TOV21G cells treated with 1.25 μM ACY1215, 20 μM pan-caspase inhibitor Q-VD-Oph or a combination for 48 hrs. (i) Percent apoptosis based on Annexin V staining in ARID1A-mutated TOV21G cells treated with 1.25 μM ACY1215 for 48 hrs with or without knockdown of caspase 3 or caspase 9. For quantifications in b, c, g, h and i, n=3 independent experiments. Error bars represent mean with S.E.M. P-value calculated via two-tailed t-test. Statistical source data are provided in Supplementary Table 6. Unprocessed original scans of all blots with size marker are shown in Supplementary Fig. 9.
ARID1A directly represses HDAC6 gene transcription
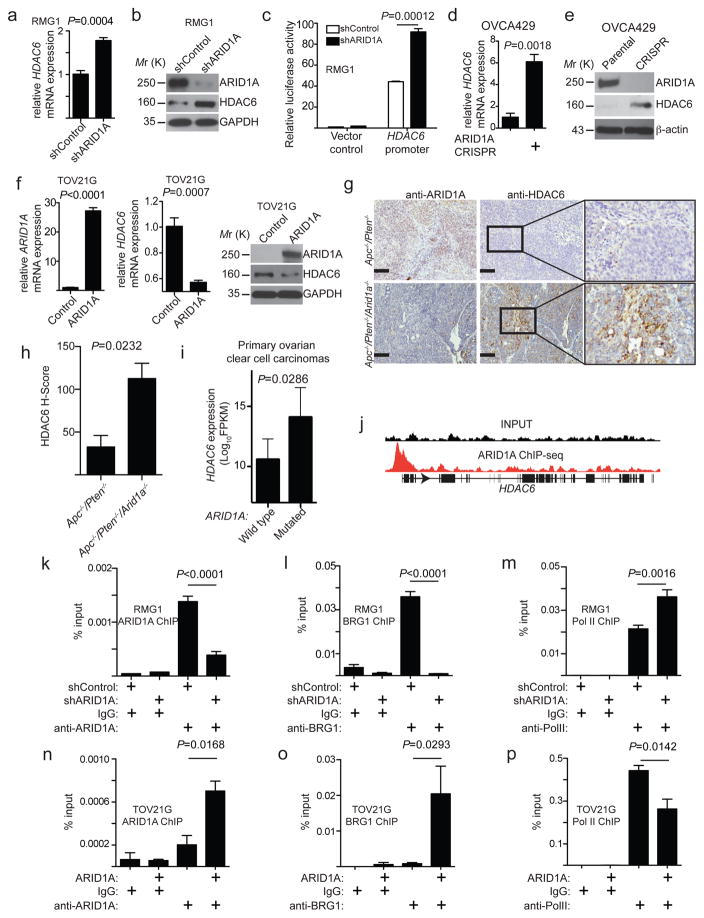
We next determined whether ARID1A affects HDAC6 expression levels. We observed a significant increase in HDAC6 mRNA and protein expression in an ARID1A-wildtype cells upon ARID1A knockdown (Fig. 4a–b), which correlates with an increase in HDAC6 promoter activity (Fig. 4c). Similarly, HDAC6 was expressed at a higher level in ARID1A knockout cells compared with parental ARID1A wildtype cells (Fig. 4d–e). Conversely, HDAC6 expression was significantly repressed when wildtype ARID1A was restored in ARID1A-mutated cells (Fig. 4f). BRG1 knockdown did not affect repression of HDAC6 by wildtype ARID1A restoration in ARID1A-mutated cells (Supplementary Fig. 4a), which is consistent with the observation that BRG1 knockdown did not affect HDAC6 expression in ARID1A wildtype cells (Supplementary Fig. 2f–g). Notably, HDAC6 is the only HDAC that is upregulated by ARID1A knockdown in ARID1A wildtype RMG1 cells and downregulated by wildtype ARID1A restoration in ARID1A-mutated TOV21G cells (Supplementary Fig. 4b). We next determined whether ARID1A regulates HDAC6 in vivo. We compared the HDAC6 expression in genetic mouse models of ovarian carcinomas developed from conditional Apc−/−/Pten−/− and Apc−/−/Pten−/−/Arid1a−/− mice as previously reported 8. These two mouse ovarian carcinoma models allowed us to examine Arid1a-dependent changes on a comparable genetic background. We examined HDAC6 expression by immunohistochemical (IHC) staining. Indeed, compared with ovarian tumours developed from Apc−/−/Pten−/− mice, HDAC6 was expressed at a significantly higher level in tumours developed from Apc−/−/Pten−/−/Arid1a−/− mice (Fig. 4g–h). Consistently, cells derived from Apc−/−/Pten−/−/Arid1a−/− tumours are more sensitive to ACY1215 compared with those derived from Apc−/−/Pten−/− tumours (Supplementary Fig. 4c). HDAC6 was the only class II HDAC that is expressed at significantly higher levels in ARID1A-mutated compared to wildtype primary human clear cell ovarian carcinomas (Fig. 4i and Supplementary Fig. 4d). In addition, ARID1A expression negatively correlates with HDAC6 expression in both clear cell and endometrioid ovarian cancer cell lines and laser capture microdissected specimens based on database mining (Supplementary Fig. 4e–f) 25, 26. We conclude that ARID1A represses HDAC6 expression, and ARID1A inactivation upregulates HDAC6 expression.
Figure 4. ARID1A represses HDAC6 expression.
(a–b) ARID1A wildtype RMG1 cells with or without ARID1A knockdown were determined for HDAC6 mRNA expression (a) n=4 independent experiments; or ARID1A, HDAC6 and GAPDH protein expression (b). (c) The human HDAC6 gene promoter activity in ARID1A wildtype RMG1 cells with or without ARID1A knockdown. n=3 independent experiments. (d–e) ARID1A wildtype and knockout OVCA429 cells were examined for expression of HDAC6 mRNA (d), n=3 independent experiments; or ARID1A, HDAC6 and β-actin protein expression (e). (f) ARID1A-mutated TOV21G cells with or without wildtype ARID1A restoration were examined for ARID1A and HDAC6 mRNA and protein expression. GAPDH was used as a loading control. n=3 for ARID1A and 4 for HDAC6 independent experiments. (g) Representative images of immunohistochemical staining of ARID1A and HDAC6 on consecutive sections of endometrioid tumours developed from Apc−/−/Pten−/− or Apc−/−/Pten−/−/Arid1a−/− conditional genetic mouse models. Scale bar = 100 μm. (h) Quantification of (g). Histological score (H-score) was quantified (n=3 independent tumours) for the indicated groups. (i) Relative HDAC6 mRNA expression in ARID1A wildtype (n=12) and mutated (n=7) human ovarian clear cell carcinoma specimens. Mann-Whitney test was used to compare the two groups and generate the P value. (j) ARID1A ChIP-seq and input tracks at the human HDAC6 gene promoter based on a ChIP-seq dataset 27. (k–m) ARID1A wildtype RMG1 cells with or without ARID1A knockdown were subjected to ChIP analysis for the HDAC6 gene promoter using antibodies against ARID1A (k), n=6 independent experiments; BRG1 (l), n=4 independent experiments; or Pol II (m), n=7 independent experiments. An isotype matched IgG was used as a control. (n–p) ARID1A-mutated TOV21G cells with or without wildtype ARID1A restoration were subjected to ChIP analysis for the HDAC6 gene promoter using antibodies against ARID1A (n), n=3 independent experiments; anti-BRG1 (o), n=4 independent experiments; or Pol II (p), n=4 independent experiments. An isotype matched IgG was used as a control. Error bars represent mean with S.E.M. P-value calculated via two-tailed t-test unless otherwise specified. Statistical source data are provided in Supplementary Table 6. Unprocessed original scans of all blots with size marker are shown in Supplementary Fig. 9.
SWI/SNF complexes contribute to both gene activation and repression in a context-dependent manner 1. We determined whether ARID1A directly represses HDAC6 expression based on published data from chromatin immunoprecipitation followed by next-generation sequencing (ChIP-seq) of ARID1A 27. Indeed, there is a significant enrichment of ARID1A at HDAC6 promoter regions (Fig. 4j). Validating these findings, we observed a significant association of ARID1A with the HDAC6 gene promoter in ARID1A wildtype cells (Fig. 4k). Supporting the notion that ARID1A directly suppresses HDAC6 transcription, ARID1A knockdown reduced its association with the HDAC6 gene promoter (Fig. 4k and Supplementary Fig. 4g–h). This correlated with a decrease in BRG1, an increase in RNA polymerase II (Pol II) and an increase in acetylated histone H3’s association with the HDAC6 gene promoter (Fig. 4l–m and Supplementary Fig. 4i–j). Conversely, wildtype ARID1A restoration in ARID1A-mutated TOV21G cells correlated with an increase in ARID1A and BRG1 and a decrease in Pol II’s association with the HDAC6 gene promoter (Fig. 4n–p). Thus, we identified ARID1A as a direct repressor of HDAC6 gene transcription.
p53 acetylated at Lys-120 (p53K120Ac) is a direct substrate for HDAC6-mediated deacetylation
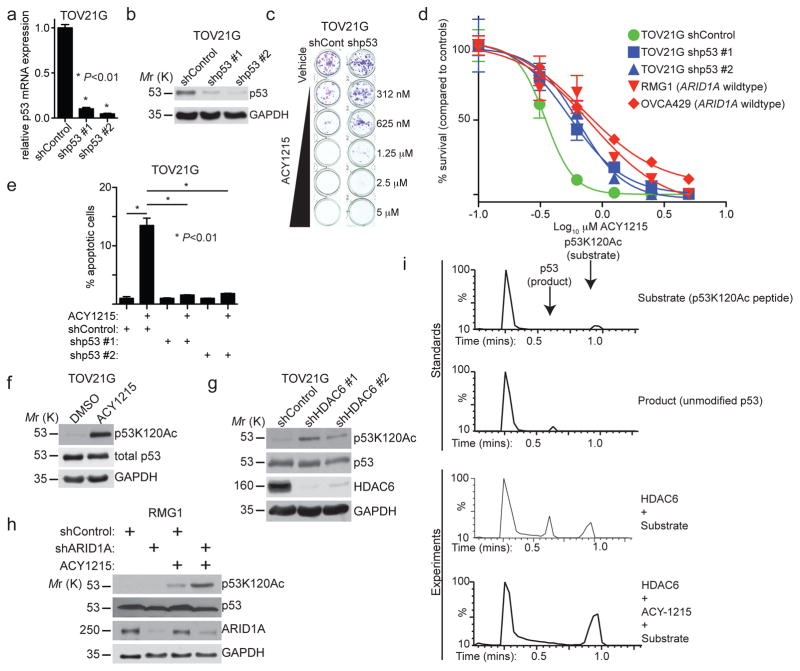
Next-generation sequencing revealed that ARID1A and TP53 mutation are typically mutually exclusive in the TCGA database (Supplementary Table 4). Indeed, ARID1A and TP53 mutations also show a mutually exclusive pattern in ovarian carcinomas 9. Since HDAC6 inhibition induces apoptosis in ARID1A inactivated cells and p53 is a key regulator of apoptosis, we determined whether p53 is necessary for the observed growth inhibition and apoptosis. Notably, knockdown of p53 expression significantly impaired the apoptosis and growth inhibition induced by the HDAC6 inhibitor ACY1215 in ARID1A-mutated TOV21G (Fig. 5a–e) and OVISE cells (Supplementary Fig. 5a–c). Similar results were also obtained for another HDAC6 inhibitor CAY10603 (Supplementary Fig. 5d). However, HDAC6 inhibition did not affect p53 expression levels (Fig. 5f), indicating that HDAC6 may regulate p53 post-translationally. Since HDAC6 is a deacetylase, we evaluated the changes of p53 acetylation status on the lysine residues that are known to regulate apoptosis such as lysine 120, 373, and 382. These residues were evaluated in ARID1A-mutated TOV21G cells following treatment with the HDAC6 inhibitor ACY1215. p53K120Ac was upregulated by ACY1215 treatment (Fig. 5f), while the acetylation status of lysine 373 or 382 residue was unchanged (Supplementary Fig. 5e). To confirm the increase in p53K120Ac was specific for HDAC6 inhibition, we knocked down HDAC6 with two individual shRNAs. We observed a strong concordance of the level of HDAC6 knockdown and the observed increase of p53K120Ac (Fig. 5g). We also evaluated p53K120Ac in ARID1A wildtype RMG1 cells with or without ARID1A knockdown after ACY1215 treatment. Indeed, we observed a significant increase in p53K120Ac in the ARID1A knockdown cells compared with controls (Fig. 5h). Thus, we conclude that HDAC6 inhibition increases p53K120Ac.
Figure 5. The selectivity of HDAC6 inhibition against ARID1A inactivation depends on p53 and HDAC6 deacetylase lysine 120 residues on p53.
(a–d) ARID1A-mutated TOV21G cells with or without p53 knockdown were examined for TP53 mRNA expression (a), n=3 independent experiments; p53 and GAPDH protein expression (b); or treated with the indicated doses of the HDAC6 inhibitor ACY1215. Shown are representative images of colonies formed (c) and dose responsive curves of the indicated cells (d). ARID1A wildtype RMG1 and OVCA429 cells were used as controls for comparison. n=4 independent experiments. Error bars represent S.E.M. (e) ARID1A-mutated TOV21G with or without p53 knockdown treated with ACY1215 (1.25 μM) or DMSO controls for 48 hours. Percent apoptosis was quantified by FACS analysis based on Annexin V staining. n=3 independent experiments. P-value calculated via two-tailed t-test. (f) ARID1A-mutated TOV21G treated with vehicle DMSO control or the HDAC6 inhibitor ACY1215 (1.25 μM). Expression of the indicated proteins was determined. (g) ARID1A-mutated TOV21G cells without or with shHDAC6 knockdown were examined for expression of p53K120Ac, total p53, HDAC6 and GAPDH by immunoblot. (h) ARID1A wildtype RMG1 with or without ARID1A knockdown were treated with vehicle DMSO control or the HDAC6 inhibitor ACY1215 (1.25 μM). Expression of the indicated proteins was examined. (i) In vitro deacetylase assay using a recombinant human HDAC6 construct and the p53K120Ac-based peptide substrate Ac-Leu-His-Ser-Gly-Thr-Ala-Lys(Ac)-Ser-Val-Thr-COOH. Deacetylation substrate and product were detected and quantified using a discontinuous liquid chromatography-mass spectrometry (LC-MS) assay. The negative control assay was run in the presence of the HDAC6 inhibitor ACY-1215. All assays were performed in triplicate independent experimental trials and yielded a specific activity of 3.4 ± 0.4 nmol product·nmol enzyme−1·min−1. Statistical source data are provided in Supplementary Table 6. Unprocessed original scans of all blots with size marker are shown in Supplementary Fig. 9.
We next determined whether HDAC6 is capable of directly catalysing the deacetylation of p53K120Ac. We utilized an in vitro deacetylation biochemical assay using a recombinant human HDAC6 expressed and purified from Escherichia coli 28 and a synthetic p53-based peptide containing the K120Ac modification, Ac-Leu-His-Ser-Gly-Thr-Ala-Lys(Ac)-Ser-Val-Thr. Indeed, this substrate was efficiently deacetylated by the purified HDAC6. The specific activity of HDAC6 with this substrate was 3.4 ± 0.4 nmol product·nmol enzyme-1·min-1, which was comparable to that of 10.5 ± 0.5 nmol product·nmol enzyme-1·min-1 measured for the standard assay substrate Ala-Lys(Ac)-Ala-NH2 28. Moreover, addition of the HDAC6 inhibitor ACY1215 blocked this activity (Fig. 5i). We conclude that p53K120Ac is a direct substrate of HDAC6’s deacetylase activity.
HDAC6 inhibition promotes p53-transcription-independent apoptosis
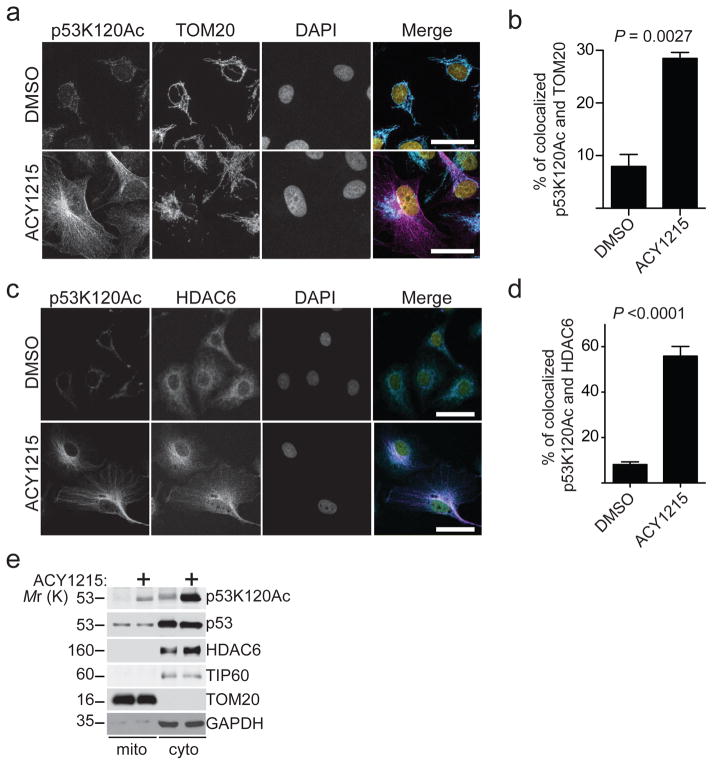
Apoptosis induced by HDAC6 inhibition in ARID1A inactivated cells is p53 dependent and correlates with upregulation of p53K120Ac (Fig. 5). Notably, p53K120Ac promotes apoptosis in both a transcription-dependent manner through upregulating p53 target apoptosis-promoting genes such as BAX and PUMA and transcription-independent mechanisms through its cytoplasmic localization in mitochondria 29–32. Thus, we evaluated transcriptional changes by RNA-seq after HDAC6 inhibition using two different HDAC6 inhibitors (namely ACY1215 or CAY10603 33) or HDAC6 knockdown. Gene expression profiling did not reveal a canonical p53-dependent apoptotic pathway by HDAC6 inhibition (GEO Accession Number: GSE84405). For example, known p53K120Ac target genes such as BAX and PUMA were not significantly upregulated by HDAC6 inhibition (Supplementary Fig. 6a and Table 5). This indicates that p53 may regulate apoptosis induced by HDAC6 inhibition in a transcription-independent manner. p53K120Ac can also promote apoptosis through its mitochondrial localization 30. We therefore measured the localization of p53K120Ac to the mitochondria following HDAC6 inhibition in ARID1A-mutated cells. Immunofluorescence analysis revealed that HDAC6 inhibition induced a significant increase in co-localization of p53K120Ac and the mitochondrial marker TOM20 or HDAC6 (Fig. 6a–d). Notably, NU9056, an inhibitor of TIP60 that acetylates p53K120 32, suppressed apoptosis induced by ACY1215, which correlated with the reduction of the p53K120Ac levels (Supplementary Fig. 6b–c). Cellular fractionation showed an increase in p53K120Ac in the mitochondrial fraction in HDAC6 inhibitor ACY1215 treated cells compared to controls (Fig. 6e).
Figure 6. Apoptosis induced by HDAC6 inhibition in ARID1A-mutated cells correlates with mitochondrial localization of p53K120Ac.
(a) ARID1A-mutated TOV21G cells treated with vehicle DMSO control or the HDAC6 inhibitor ACY1215 (1.25 μM) were fixed and subjected to immunofluorescence staining using antibodies against p53K120Ac (Magenta), TOM20 (Turquoise, a mitochondrial marker) and DAPI (Yellow, nuclei). Bar=25μm. (b) Confocal images were processed and co-localization between p53K120Ac and TOM20 was quantified using Leica Application Suite X (LASX) software. Error bars represent S.E.M. (n=6 independent experiments). (c) ARID1A-mutated TOV21G cells treated with vehicle DMSO control or the HDAC6 inhibitor ACY1215 (1.25 μM) were fixed and subjected to immunofluorescence staining using antibodies against p53K120Ac (Magenta), HDAC6 (Turquoise) and DAPI (Yellow, nuclei). Images were captured using confocal microscopy. Bar=25μm. (d) Confocal images were processed, and co-localization between p53K120Ac and HDAC6 was quantified using Leica Application Suite X (LASX) software. Error bars represent S.E.M. (n=6 independent experiments). P-value calculated via two-tailed t-test. (e) ARID1A-mutated TOV21G cells were treated with vehicle DMSO control or the HDAC6 inhibitor ACY1215 (1.25 μM) were fractionated to isolate mitochondria and cytosol. Expression of p53K120Ac, total p53, HDAC6, mitochondrial marker TOM20, and TIP60 that is known to acetylate p53K120 residue 32 in the indicated fractions was examined by immunoblot. GAPDH expression was used as a loading control. Statistical source data are provided in Supplementary Table 6. Unprocessed original scans of all blots with size marker are shown in Supplementary Fig. 9.
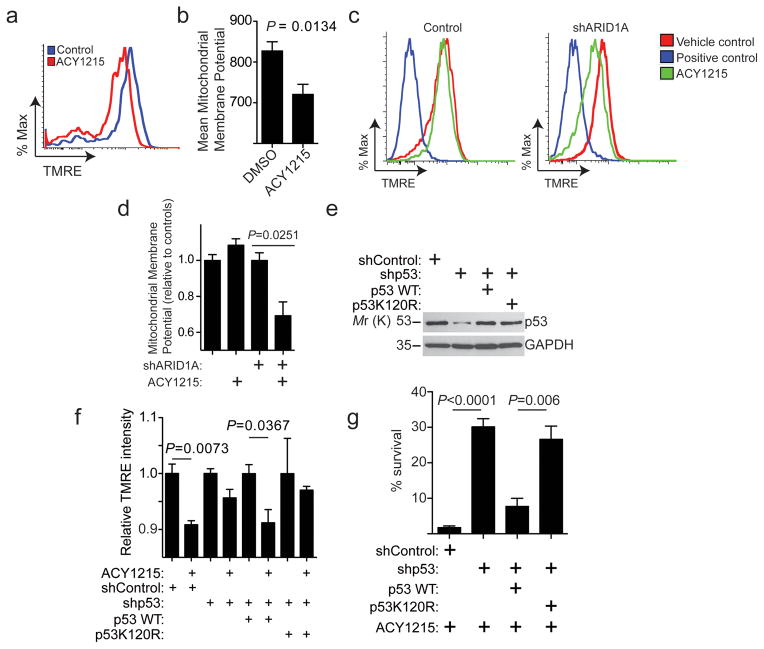
Mitochondrial p53K120Ac promotes apoptosis through decreasing mitochondrial membrane potential 34. Indeed, ACY1215 significantly decreased the mitochondrial membrane potential in ARID1A-mutated cells (Fig. 7a–b and Supplementary Fig. 6d). Consistent with the observed selectivity against ARID1A inactivation by HDAC6 inhibition (Fig. 1), ARID1A knockdown in ARID1A-wildtype cells significantly deceased mitochondrial membrane potential in cells treated with ACY1215 compared with controls (Fig. 7c–d). Mitochondrial membrane potential decrease by ACY1215 was both p53 and p53K120Ac dependent, because knockdown of p53 suppressed the observed decrease in mitochondrial membrane potential and this was rescued by wildtype p53 but not a p53K120R mutant (Fig. 7d–f). Indeed, wildtype p53 but not the p53K120R mutant rescued the p53 knockdown-mediated impairment of ACY1215-induced growth inhibition (Fig. 7g). We conclude that HDAC6 inhibition promotes transcription-independent apoptosis that correlates with p53K120Ac mitochondrial localization (Supplementary Fig. 7).
Figure 7. Apoptosis induced by HDAC6 inhibition in ARID1A-mutated cells correlates with a decrease in mitochondrial membrane potential.
(a) ARID1A-mutated TOV21G cells were treated with vehicle DMSO control or the HDAC6 inhibitor ACY1215 (1.25 μM) were examined for mitochondrial membrane potential by FACS. (b) Quantification of (a). n=5 independent experiments. P-value calculated via two-tailed t-test. (c) ARID1A wildtype RMG1 cells with or without ARID1A knockdown were treated with vehicle control, FCCP (positive control) or ACY1215 (1.25 μM). The mitochondrial membrane potential was quantified by TMRE using FACS analysis. Data was collected via FACS and is representative of 3 independent experiments. (d) Quantification of (c). n=3 independent experiments. (e–g) ARID1A-mutated TOV21G cells with or without endogenous p53 knockdown by a shp53 that targets the 3′ UTR region of the human TP53 gene together with a lentivirus encoding a control, wildtype p53 or a p53K120R mutant. Immunoblotting of p53 and GAPDH in the indicated cells (e); the indicated cells were treated with or without ACY1215 and examined for mitochondrial membrane potential by FACS (f), n=3 independent experiments; and the percentage of surviving cells of the indicated cells treated with ACY1215 was determined by colony formation assay (g). n=4 independent experiments. Error bars represent mean with S.E.M. P-value calculated via two-tailed t-test. Statistical source data are provided in Supplementary Table 6. Unprocessed original scans of all blots with size marker are shown in Supplementary Fig. 9.
HDAC6 inhibition by ACY1215 improves the survival of mice bearing ARID1A-mutated ovarian tumours
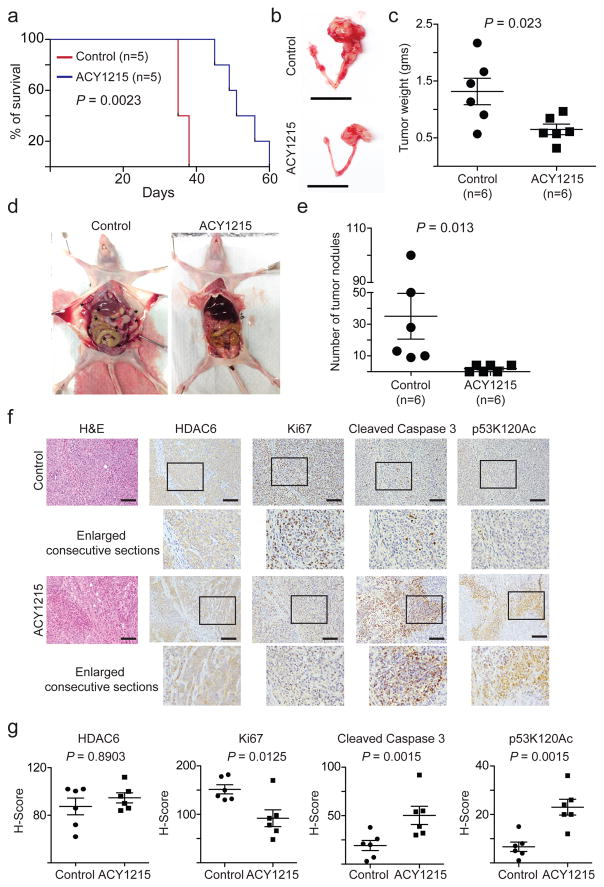
Clinical studies show that the HDAC6 inhibitor ACY1215 is well-tolerated without a dose-limiting toxicity 18. To determine the effects of HDAC6 inhibition in vivo on the growth of ARID1A-mutated tumours, we orthotopically transplanted luciferase-expressing ARID1A-mutated TOV21G cells into the bursa-sac covering the ovary of immunocompromised nude mice to mimic the tumour microenvironment. The injected ARID1A wildtype or mutant cells were allowed to grow for 2 weeks to establish the orthotopic tumours. Mice were then randomized and treated daily with vehicle control or ACY1215 (50 mg/kg) by intraperitoneal (i.p.) injection, the same dose as previously reported 35. Indeed, ACY1215 treatment significantly inhibited the growth of ARID1A-mutated tumours (Supplementary Fig. 8a–b). We next followed the survival of the treated mice after discontinuing the treatment regimens. Importantly, ACY1215 significantly improved the survival of mice bearing the orthotopically-transplanted ARID1A-mutated tumours compared with controls (Fig. 8a). Specifically, the median survival was improved from 35 days in the vehicle control group to 51 days in the ACY1215 treated group. Thus, we conclude that the HDAC6 inhibitor ACY1215 significantly improves the survival of mice bearing ARID1A-mutated tumours.
Figure 8. HDAC6 inhibition improves the survival of mice bearing ARID1A-mutated ovarian tumours.
ARID1A-mutated TOV21G cells were orthotopically transplanted into the ovarian bursa sac of SCID/nude female mice. Tumours were allowed to establish for 2 weeks before the mice were randomized into two different treatment groups (n=5 mice/group). Mice were treated with vehicle control or the HDAC6 inhibitor ACY1215 (50 mg/kg) daily for an additional 3 weeks. After stopping the treatment, the mice from the indicated groups were followed for survival. Shown is the Kaplan Meier survival curves for ACY1215 or vehicle control treated mice (a). P-value was calculated by log-rank test. At the end of treatment, the mice were euthanized (n=6 mice/group). Shown are representative images of reproductive tracts with tumours from control or ACY1215 treated mice (b). Scale bar = 2 cm. Tumour weight was measured as a surrogate for tumour burden from the control and ACY1215 treated mice (c) or examined for disseminated tumour nodules in the peritoneal cavity. Representative images of disseminated tumour nodules in control and ACY1215 treated mice (d). Asterisks (*) indicate the disseminated tumour nodules in peritoneal cavity. The number of disseminated tumour nodules in peritoneal cavity was quantified (e). The consecutive sections of tumours dissected from the indicated treatment groups were subjected to immunohistochemical staining for HDAC6, Ki67, cleaved caspase 3 and p53K120Ac (f). Scale bar = 100 μm. Histological score (H-score) was calculated for 5 separate fields from 6 tumours from 6 individual mice from each of the indicated groups (g). Error bars represent mean with S.E.M. P-value calculated via two-tailed t-test. Statistical source data are provided in Supplementary Table 6.
We next directly examined the effects of the HDAC6 inhibitor ACY1215 on tumour burden of the transplanted ARID1A-mutated or wildtypes cells. Indeed, using tumour weight as a surrogate for tumour burden, we found that ACY1215 treatment significantly reduced the burden of ARID1A-mutated orthotopically xenografted tumours (Fig. 8b–c). Likewise, ACY1215 significantly suppressed the tumour growth in the conditional Arid1a−/−/Pik3caH1047R genetic clear cell ovarian tumour mouse model 6 (Supplementary Fig. 8c). Ovarian cancer often progresses by disseminating to the intraperitoneal cavity 36. Thus, we quantified the number of grossly visible tumour nodules in the peritoneal cavity following treatment with vehicle control or ACY1215 in the pre-established ARID1A-mutated tumours. There was a significant decrease in the number of tumour nodules in ACY1215 treated mice bearing ARID1A-mutated tumours compared to controls (Fig. 8d–e). As a control, luciferase-expressing ARID1A wildtype RMG1 cells were orthotopically transplanted in parallel. In contrast to what we observed in ARID1A-mutated tumours, ACY1215 treatment did not significantly affect the growth, tumour burden or dissemination of ARID1A wildtype tumours (Supplementary Fig. 8d–g).
Finally, we sought to correlate the observed improvement of survival, suppression of tumour growth and reduction in tumour burden in vivo with the molecular pathways we have revealed for the observed dependence of ARID1A-mutated cells on HDAC6 activity in vitro. To do so, we performed IHC analysis for markers of cell proliferation (Ki67), apoptosis (cleaved caspase 3), HDAC6 and p53K120Ac in dissected ARID1A-mutated tumours treated with ACY1215 or controls. ACY1215 significantly decreased the cell proliferation marker Ki67 and increased the apoptotic marker cleaved caspase 3 (Fig. 8f–g). As a control, HDAC6 expression was not affected by ACY1215 (Fig. 8f–g). Furthermore, p53K120Ac staining was significantly increased by ACY1215 treatment (Fig. 8h–i). In contrast, ACY1215 did not affect the expression of Ki67, cleaved caspase 3 or p53K120Ac in ARID1A wildtype tumours (Supplementary Fig. 8h–i). This is consistent with the finding that ACY1215 did not affect the growth of ARID1A wildtype tumours in vivo (Supplementary Fig. 8d–f). Together, we conclude that the HDAC6 inhibitor ACY1215 selectively suppresses the growth and dissemination of ARID1A-mutated ovarian tumours and improves the survival of ARID1A-mutated tumour bearing mice. This correlates with a decrease in cell proliferation, an increase in apoptosis and an accumulation of apoptosis-promoting p53K120Ac in the treated ARID1A-mutated tumours.
Discussion
Our data demonstrate a dependence of ARID1A-mutated cells on HDAC6 activity. This was due to the direct suppression of HDAC6 transcription by ARID1A. Consequently, ARID1A inactivation upregulates HDAC6 expression. Although the SWI/SNF complex mostly promotes the transcription of its target genes, it can also repress gene transcription 1. Previous reports established that ARID1A inactivation correlates with silencing of tumour suppressive genes such as PIK3IP1 19. Here we showed that HDAC6 is a direct target of ARID1A-mediated transcriptional repression. ARID1A inactivation leads to upregulation of HDAC6; therefore, HDAC6 inhibition is selective against ARID1A inactivation. This suggests that both transcriptional repression of oncogenic genes and transcriptional activation of tumour suppressor genes contribute to the tumour suppressive activity of ARID1A.
Here we show that ARID1A inactivation upregulates HDAC6, and HDAC6 directly deacetylates the apoptosis-promoting p53K120Ac post-translational modification. Our biochemical experiments show that p53K120Ac is a substrate of HDAC6 and thus identifies a deacetylase for p53 post-translational modification. This suggests that ARID1A mutation functionally inactivates p53 to suppress apoptosis. This, at least in part, resolves the typical mutual exclusivity of mutations between ARID1A and TP53 in human cancers 9. Previous studies showed that p53K120Ac selectively regulates apoptosis, while it does not affect the expression of cell cycle regulatory p53 target genes such as CDKN1A 29–32. Thus, ARID1A mutation contributes to inactivation of p53’s apoptosis-promoting function by suppressing apoptosis-promoting p53K120Ac.
In summary, our studies demonstrate that targeting HDAC6 activity using HDAC6 inhibitors in ARID1A-mutated cells represents a therapeutic strategy. This approach shows great promise an example of precision medicine at work since it is based on ARID1A mutational status and the mutual exclusivity of ARID1A and TP53 mutations. Notably, HDAC6 inhibitors such as ACY1215 are well-tolerated and show minimal toxicity in clinical trials 18. Thus, our studies provide scientific rationale for potential translation of these findings by repurposing clinically applicable HDAC6 inhibitors for ARID1A-mutated ovarian cancers, for which no effective therapies currently exist. Given that ARID1A shows one of the highest mutation rates among epigenetic regulators 3 and loss of expression of ARID1A also occurs in many cancer types 2, our findings may have far-reaching implications for improving therapy for a wide array of cancer types.
Methods
Cell lines and 3D culture conditions
The protocol for using primary cultures of human ovarian clear cell tumour cells was approved by the University of British Columbia Institutional Review Board. Informed consent was obtained from human subjects. All relevant ethical regulations have been complied. The primary tumour cells were cultured in RPMI 1640 supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. Ovarian clear cell carcinoma cell lines (TOV21G, OVTOKO, OVISE and RMG1) were purchased from JCRB. TOV21G, OVTOKO, and OVISE cells were cultured in RPMI 1640 supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. RMG1 cells were cultured in 1:1 Dulbecco’s modified Eagle’s medium (DMEM):F12 supplemented with 10% FBS. Viral packaging cells were cultured in DMEM supplemented with 10% FBS at 37°C supplied with 5% CO2. Cells lines are authenticated at The Wistar Institute’s Genomics Facility using short tandem repeat DNA profiling. Regular Mycoplasma testing was performed using LookOut Mycoplasma PCR detection (Sigma). 3D culture was adapted from previously published methods 37 using growth factor reduced-Matrigel (GFR-Matrigel; BD Biosciences). Briefly, a single cell suspension was plated in 8-well chambers covered with Matrigel. Matrigel media with either vehicle control (DMSO) or drug was changed every 4 days and cells were grown for 12 days. Each of the experiments was performed in duplicate in three independent experimental repeats.
Reagents and antibodies
ACY1215, Q-VD-Oph and CAY10603 were obtained from Selleckchem. NU9056 was purchased from Tocris. The following antibodies were obtained from the indicated suppliers: mouse anti-acetylated-p53 K120 (Abcam, Cat. No. ab78316, 1:1000 for western blot and 1:100 for IHC), rabbit anti-acetylated-p53 K373 (Abcam, Cat. No. ab62376, 1:1000 for western blot), rabbit anti-acetylated-p53 K382 (Abcam, Cat. No. ab75754, 1:1000 for western blot), mouse anti-ARID1A (Santa Cruz, Cat. No. sc-32761, 1:1000 for western blot and 10μg per sample for ChIP), mouse anti-p53 (Millipore, Cat. No. OP43, 1:1000 for western blot), mouse anti-GAPDH (Millipore, Cat. No. MAB374, 1:10000 for western blot), rabbit anti-cleaved PARP p85 (Promega, Cat. No. 7344, 1:1000 for western blot), mouse anti-Ki67 (Cell Signaling, Cat. No. 9449, 1:1000 for IHC), rabbit anti-cleaved caspase 3 (Cell Signaling, Cat. No. 9661, 1:1000 for western blot and 1:50 for IHC), rabbit anti-HDAC6 (Cell Signaling, Cat. No. 7612, 1:1000 for western blot and Santa Cruz, Cat. No. sc-11420, 1:100 for IHC), rabbit anti-RNA polymerase II (Santa Cruz, Cat. No. sc-899 X, 2 μg per sample for ChIP), rabbit anti-TOM20 (Santa Cruz, Cat. No. sc-11415, 1:1000 for western blot), rabbit anti-H3Ac (Active Motif, Cat. No. 39139, 5 μl/immunoprecipitation for ChIP), rabbit anti-TIP60 (Santa Cruz, Cat. No. sc-166323, 1:1000 for western blot), mouse anti-BRG1 (Santa Cruz, Cat. No. sc-17796, 1:1000 for western blot and 2 μg/immunoprecipitation for ChIP) and rabbit anti-BRM (Cell Signaling, Cat. No. 11966, 1:1000 for western blot and 3 μg/immunoprecipitation for ChIP). Growth factor reduced Matrigel was purchased from Corning.
Immunoblotting
Protein was isolated as previously described 19. Briefly, protein was extracted with RIPA buffer (150mM NaCl, 1% NP40, 0.5% sodium deoxycholate, 0.1% SDS, 50mM Tris pH 8.0, and 1mM PMSF). Protein was separated on a SDS-PAGE and transferred to PVDF membrane. For immunoblot of p53 post-translation modifications, cells were treated with a proteasome inhibitor MG132 (10 μM) to stabilize p53 protein.
HDAC6 promoter reporter assay
Human HDAC6 gene promoter (genomic position: chrX: 48658920-48660419) was cloned into pGL2 basic reporter plasmid with firefly luciferase activity (Promega). pGL2-HDAC6 promoter was transfected into RMG1 cells expressing shARID1A or controls. pRL-SV40 reporter plasmid with Renilla luciferase activity (Promega) was used to normalize the transfection efficiency. The firefly and Renilla luciferase activity was measured by Dual-Luciferase Reporter Assay Kit (Promega) 24-hour post-transfection.
Generation of ARID1A CRISPR OVCA429 cells
OVCA429 cells were transfected with CRISPR-ARID1A (pSpCas9(BB)-2A-Puro (PX459)). The ARID1A gRNA sequence is: 5′-CGGGTTGCCCAGGCTGCTGGcgg-3′. The plasmid was a generous gift from Dr. Cigall Kadoch. Fugene6 transfection reagent (Promega) was used as per manufacturer’s specifications. Clonal populations for the loss of ARID1A expression were screened through immunoblot.
Retrovirus and Lentivirus infection
Retrovirus production and transduction were performed as described previously 19, 38. Phoenix cells were used to package the viruses (a gift of Dr. Gary Nolan, Stanford University). Lentivirus was packaged using the Virapower Kit from Life Technologies (Carlsbad, CA) following the manufacturer’s instructions as described previously 19, 38. pLKO.1-shARID1As (TRCN0000059090), pLKO.1-shp53 (TRCN0000010814 and TRCN0000003755), pLKO.1-shHDAC6 (TRCN0000004839 and TRCN0000004841), pLKO.1-shCaspase 3 (TRCN0000003550), shCaspase 9 (TRCN0000003583), shCaspase 8 (TRCN0000003577 and TRCN0000003579) and shBRG1 (TRCN0000015549 and TRCN0000015552) were obtained from Open Biosystems. A shRNA to luciferase was used as a control. HDAC6 wildtype and a catalytically inactive H216/611A mutant 21 were obtained from Addgene (Cat. No. 30482 and 30483) and subcloned into lentivirus plasmid pLVX-Puro (Promega) by XbaI and AgeI sites using standard molecular cloning protocols. Cells infected with viruses encoding the puromycin resistance gene were selected in 1 μg/ml puromycin.
Reverse-transcriptase quantitative PCR (RT-qPCR)
RNA was isolated from cells with RNeasy Mini Kit followed by on-column DNAse digest (Qiagen). mRNA expression for HDAC1-11, ARID1A, and TP53 was determined using SYBR green 1-step iScript (Bio-Rad) with Life Technologies QuantStudio 3. β-2-microglobulin (B2M) was used as an internal control. All primer sequences are listed in Supplementary Table 1.
Annexin V and Mitochondria Membrane Potential
Phosphatidylserine externalization was detected using an Annexin V FITC and PI kit (Thermo Fisher, Cat. No. V13242) following the manufacturer’s instructions. Briefly, cells were washed with cold PBS and resuspended in Annexin V binding buffer and stained with Annexin V and PI at room temperature and then analyzed immediately. To measure change in mitochondria membrane potential cells were treated with 200nM of tetramethylrhodamine, ethyl ester (TMRE; Abcam, Cat. No. ab113852) for 15min at 37 °C. Annexin V and TMRE-positive cells were detected using the Becton-Dickinson LSR18 machine and analysed with FlowJo version 7 software module.
Colony formation assay
Cell lines were infected with lentivirus pLKO.1-shRNAs or pLKO.1-control with puromycin selection marker. Infected cells were selected with 1 μg/mL of puromycin for 72 hours and the selected cells were seeded in 12-well or 24-well plates. Cell medium was changed every three days with appropriate drug doses for 12 days. Colonies were washed twice with PBS and fixed with 10% methanol and 10% acetic acid in distilled water. Fixed colonies were stained with 0.005% crystal violet. Integrated density was measured using NIH ImageJ software.
In vitro biochemical HDAC6 deacetylation assay
The catalytic activity of a recombinant human HDAC6 construct containing both catalytic domains expressed in Escherichia coli with the p53K120Ac-based peptide substrate Ac-LHSGTAK(ac)SVT-COOH was measured using a discontinuous liquid chromatography-mass spectrometry (LC-MS) assay reported previously (Hai and Christianson, 2016). Briefly, 0.05 μM HDAC6 was incubated with 100 μM substrate in 20 mM HEPES (pH 7.5), 100 mM NaCl, 5 mM KCl, 1 mM MgCl2 for 20 min at room temperature, and the reaction was quenched by the addition of acetonitrile (equal volume to the reaction solution). The deacetylation reaction mixtures were analysed by LC-MS using a Waters SQD equipped with an Acquity UPLC (Waters, Milford, MA, USA) and quantified using the standard curves generated from the mass signals of the corresponding deacetylated synthetic peptide (Ac-LHSGTAKSVT-COOH). As a negative control, the assay was run in the presence of 10 μM of the HDAC6 specific inhibitor ACY-1215. All assays were performed in triplicate. The peptide was custom-synthesized by Genscript. The human HDAC6 construct was expressed and purified as previously reported 28.
Chromatin Immunoprecipitation (ChIP)
ChIP was performed as we have previously described 19. The following antibodies were used to perform ChIP: ARID1A (Santa Cruz), RNA polymerase II (Santa Cruz), H3Ac (Active Motif) and BRG1 (Santa Cruz). An isotype matched IgG was used as a negative control. ChIP DNA was analysed by quantitative PCR against the promoter of the human HDAC6 gene. All primer sequences in Table S1. For single site PCR, the primers for position −880 upstream of transcription starting site were used.
Immunofluorescence and immunohistochemical staining
Immunofluorescence was performed after 48hrs as indicated by fixing samples in 4% paraformaldehyde and permeabilizing with 0.5% Triton-X. Samples were incubated with primary antibodies for 2 hours at room temperature, highly cross absorbed secondary antibodies (Invitrogen) for 1 hour at room temperature and mounted with prolong anti-fade reagent (Invitrogen). Immuno-stained cells were imaged using a Leica Confocal microscope. Immunohistochemical staining was performed as we have described previously on consecutive sections from xenografted tumours dissected from control or ACY1215 treated immunocompromised nude female mice 19, 39. Expression of the stained markers was scored using a histologic score (H score) as previously described 40.
Mitochondrial isolation
Mitochondria were isolated using the Mitochondria Isolation Kit (Thermo Fisher Scientific). Isolation was performed according to manufacturers’ instructions using a “B” dounce homogenizer. Protein was isolated from purified mitochondria as described in immunoblotting section. Cytosolic fraction was collect for immunoblot analysis.
Intrabursal orthotopic xenograft models in vivo
The protocols were approved by the Institutional Animal Care and Use Committee (IACUC). All mouse experiments were conducted according to ethical regulations. For in vivo experiments, the sample size of 6 mice per group was determined based on the data shown from in vitro experiments. Intrabursal orthotopic xenograft was performed as described previously 19, 39. Briefly, 1 × 106 luciferase-expressing TOV21G or RMG1 cells were unilaterally injected into the ovarian bursa sac of 6–8-week- old female immunocompromised mice (n=6 per group). Two weeks after injection, tumours were visualized by injecting luciferin (i.p.: 4 mg/mice) resuspended in PBS and imaged with an In Vivo Imaging System (IVIS). The mice were then randomized into two groups based on luciferase activity and treated with vehicle control (2% DMSO/30% PEG 300/ddH2O) or ACY1215 (50 mg/kg daily) for three weeks 35 and imaged for luciferase activity. Images were analysed using Live Imaging 4.0 software. Imaging analysis was performed blindly but not randomly. At end of the experiments, tumours were surgically dissected and tumour burden was calculated based on tumour weight. Intraperitoneally disseminated tumour nodules were quantified.
Arid1a−/−/Pik3caH1047R genetic clear cell ovarian tumour mouse model
All experiments were approved by IACUC. Transgenic mice with latent mutations in Arid1a and Pik3ca were generated by crossing Arid1aflox/flox mice (kindly provided by Dr. Wang, U. Michigan 41 and crossed onto a C57BL/6J background for 9 generations) with R26-Pik3caH1047R mice carrying inducible Pik3ca mutations (Jackson Laboratory, Jax#016977). Administration of intrabursal adeno-Cre, performed as previously reported 42, induce ovarian clear cell carcinoma in ~45 days, which is similar to a previous report 6. All mice were maintained in specific pathogen-free barrier facilities. To induce tumorigenesis, 6–10 weeks old Pik3caH1047R/Arid1aflox/flox female mice were intrabursally injected adenovirus-Cre as previously described. Mice were randomized and treated with ACY1215 (50mg/kg) or vehicle control for 21 days as previously published 35. Following treatment mice were sacrificed and the reproductive tracts were removed. The changes in volumes of tumours formed on the injected ovary were calculated against the contrary side non-injected ovary from the same mice.
Statistical analysis and reproducibility
Experiments were repeated 3 times unless otherwise stated. The representative images were shown unless otherwise stated. Statistical analysis was performed using GraphPad Prism 5 (GraphPad) for Mac OS. Quantitative data are expressed as mean ± S.E.M. unless otherwise stated. Analysis of variance (ANOVA) with Fisher’s Least Significant Difference (LSD) was used to identify significant differences in multiple comparisons. For all statistical analyses, the level of significance was set at 0.05. For correlation studies, Pearson correlation was used for calculating P and r value GraphPad Prism 5 (GraphPad) for Mac OS. Imaging analysis was performed blindly but not randomly. Animal experiments were randomized. There was no exclusion from the experiments.
Data availability
Gene expression profiling data based on RNA-seq have been deposited in the Gene Expression Ominibus (GEO) under accession codes GSE84405. Previously published RNA-seq data for ARID1A wildtype or mutated human ovarian clear cell carcinoma specimens that were re-analysed here are available at the European Genome-Phenome Archive (EGAS) under accession code EGAS 00000000075 5. For correlation between ARID1A and HDAC6 expression, gene expression data obtained from GEO (under accession code: GSE36139) for clear cell and endometrioid ovarian cancer cell lines in cancer cell line encyclopedia 25 and a microarray database obtained from GEO (under accession code: GSE29450) for profiling gene expression in laser capture microdissected human clear cell ovarian tumour specimens and ovarian surface epithelial cells were used 26. ARID1A chromatin immunoprecipitation followed by next generation sequencing (ChIP-seq) and input tracks at the human HDAC6 gene promoter were based on ChIP-seq data (GEO Accession Number: GSE69568) 27.
Source data used for statistical analyses of Figures 1b, 1f, 1h, 2b–c, 2e, 2g, 2j, 3b–c, 3g–i, 4a, 4c–d, 4f, 4h–i, 4k–p, 5a, 5d–e, 6b, 6d, 7b, 7d, 7f–g, 8a, 8c, 8e, 8g, and Supplementary Figs 1a–c, 2b–c, 2e–f, 2h–i, 3, 4b–f, 4h–j, 5a, 5c–d, 6c, 8b–c, 8e, 8f–g and 8i are provided as Supplementary Table 6 (Statistics source data). All other data supporting the findings of this study are available upon request.
Supplementary Material
Acknowledgments
We thank C. Kadoch for the ARID1A CRISPR plasmid and K. Payne and T. Fukumoto for technical assistance. This work was supported by US National Institutes of Health grants (R01CA160331, R01CA163377 and R01CA202919 to R. Z., K99CA194318 to B.G.B., K99CA194309 to K.M.A. and R01GM49758 to D.W.C.), US Department of Defense (OC140632P1 and OC150446 to R. Z.), an Ovarian Cancer Research Fund (OCRF) program project (to R. Z.) and The Jayne Koskinas & Ted Giovanis Breast Cancer Research Consortium at Wistar (to R. Z.). Support of Core Facilities was provided by Cancer Centre Support Grant (CCSG) CA010815 to The Wistar Institute.
Footnotes
Author Contributions
B.G.B., S.W., P.H.P., Y.H., K.M.A., Y.W., A.V.A. performed the experiments, analysed data. B.G.B. and R.Z. designed the experiments. A.V.K. performed the bioinformatics analysis. F.J.R., J.R.C.-G., W.Z., D.W.S. participated in the experimental design, K.R.C. and Y.Z. contributed key reagents. D.G.H., D.W.C. and R.Z. supervised studies. B.G.B., K.M.A., Y.W., K.R.C., D.W.C. and R.Z. wrote the manuscript. R.Z. conceived the study.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Wilson BG, Roberts CW. SWI/SNF nucleosome remodellers and cancer. Nature reviews. Cancer. 2011;11:481–492. doi: 10.1038/nrc3068. [DOI] [PubMed] [Google Scholar]
- 2.Kadoch C, et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet. 2013;45:592–601. doi: 10.1038/ng.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lawrence MS, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones S, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010;330:228–231. doi: 10.1126/science.1196333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wiegand KC, et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med. 2010;363:1532–1543. doi: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chandler RL, et al. Coexistent ARID1A-PIK3CA mutations promote ovarian clear-cell tumorigenesis through pro-tumorigenic inflammatory cytokine signalling. Nature communications. 2015;6:6118. doi: 10.1038/ncomms7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guan B, et al. Roles of deletion of Arid1a, a tumor suppressor, in mouse ovarian tumorigenesis. J Natl Cancer Inst. 2014;106 doi: 10.1093/jnci/dju146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhai Y, et al. Arid1a inactivation in an Apc- and Pten-defective mouse ovarian cancer model enhances epithelial differentiation and prolongs survival. J Pathol. 2016;238:21–30. doi: 10.1002/path.4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guan B, Wang TL, Shih Ie M. ARID1A, a factor that promotes formation of SWI/SNF-mediated chromatin remodeling, is a tumor suppressor in gynecologic cancers. Cancer research. 2011;71:6718–6727. doi: 10.1158/0008-5472.CAN-11-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guan B, Gao M, Wu CH, Wang TL, Shih Ie M. Functional analysis of in-frame indel ARID1A mutations reveals new regulatory mechanisms of its tumor suppressor functions. Neoplasia. 2012;14:986–993. doi: 10.1593/neo.121218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ye S, et al. Clinicopathologic Significance of HNF-1beta, AIRD1A, and PIK3CA Expression in Ovarian Clear Cell Carcinoma: A Tissue Microarray Study of 130 Cases. Medicine (Baltimore) 2016;95:e3003. doi: 10.1097/MD.0000000000003003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kobel M, et al. Differences in tumor type in low-stage versus high-stage ovarian carcinomas. International journal of gynecological pathology : official journal of the International Society of Gynecological Pathologists. 2010;29:203–211. doi: 10.1097/PGP.0b013e3181c042b6. [DOI] [PubMed] [Google Scholar]
- 13.Chan JK, et al. Do clear cell ovarian carcinomas have poorer prognosis compared to other epithelial cell types? A study of 1411 clear cell ovarian cancers. Gynecologic oncology. 2008;109:370–376. doi: 10.1016/j.ygyno.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 14.Mackay HJ, et al. Prognostic relevance of uncommon ovarian histology in women with stage III/IV epithelial ovarian cancer. International journal of gynecological cancer : official journal of the International Gynecological Cancer Society. 2010;20:945–952. doi: 10.1111/IGC.0b013e3181dd0110. [DOI] [PubMed] [Google Scholar]
- 15.Saito T, Katabuchi H. Annual Report of the Committee on Gynecologic Oncology, Japan Society of Obstetrics and Gynecology: Patient Annual Report for 2013 and Treatment Annual Report for 2008. J Obstet Gynaecol Res. 2016;42:1069–1079. doi: 10.1111/jog.13043. [DOI] [PubMed] [Google Scholar]
- 16.Li Y, Shin D, Kwon SH. Histone deacetylase 6 plays a role as a distinct regulator of diverse cellular processes. The FEBS journal. 2013;280:775–793. doi: 10.1111/febs.12079. [DOI] [PubMed] [Google Scholar]
- 17.Bazzaro M, et al. Ubiquitin proteasome system stress underlies synergistic killing of ovarian cancer cells by bortezomib and a novel HDAC6 inhibitor. Clin Cancer Res. 2008;14:7340–7347. doi: 10.1158/1078-0432.CCR-08-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Santo L, et al. Preclinical activity, pharmacodynamic, and pharmacokinetic properties of a selective HDAC6 inhibitor, ACY-1215, in combination with bortezomib in multiple myeloma. Blood. 2012;119:2579–2589. doi: 10.1182/blood-2011-10-387365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bitler BG, et al. Synthetic lethality by targeting EZH2 methyltransferase activity in ARID1A-mutated cancers. Nat Med. 2015;21:231–238. doi: 10.1038/nm.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheung HW, et al. Systematic investigation of genetic vulnerabilities across cancer cell lines reveals lineage-specific dependencies in ovarian cancer. Proc Natl Acad Sci U S A. 2011;108:12372–12377. doi: 10.1073/pnas.1109363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawaguchi Y, et al. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell. 2003;115:727–738. doi: 10.1016/s0092-8674(03)00939-5. [DOI] [PubMed] [Google Scholar]
- 22.Marks PA. Discovery and development of SAHA as an anticancer agent. Oncogene. 2007;26:1351–1356. doi: 10.1038/sj.onc.1210204. [DOI] [PubMed] [Google Scholar]
- 23.Haggarty SJ, Koeller KM, Wong JC, Grozinger CM, Schreiber SL. Domain-selective small-molecule inhibitor of histone deacetylase 6 (HDAC6)-mediated tubulin deacetylation. Proc Natl Acad Sci U S A. 2003;100:4389–4394. doi: 10.1073/pnas.0430973100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strasser A, O’Connor L, Dixit VM. Apoptosis signaling. Annu Rev Biochem. 2000;69:217–245. doi: 10.1146/annurev.biochem.69.1.217. [DOI] [PubMed] [Google Scholar]
- 25.Barretina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stany MP, et al. Identification of novel therapeutic targets in microdissected clear cell ovarian cancers. PLoS One. 2011;6:e21121. doi: 10.1371/journal.pone.0021121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raab JR, Resnick S, Magnuson T. Genome-Wide Transcriptional Regulation Mediated by Biochemically Distinct SWI/SNF Complexes. PLoS Genet. 2015;11:e1005748. doi: 10.1371/journal.pgen.1005748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hai Y, Christianson DW. Histone deacetylase 6 structure and molecular basis of catalysis and inhibition. Nat Chem Biol. 2016;12:741–747. doi: 10.1038/nchembio.2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sykes SM, et al. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol Cell. 2006;24:841–851. doi: 10.1016/j.molcel.2006.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sykes SM, Stanek TJ, Frank A, Murphy ME, McMahon SB. Acetylation of the DNA binding domain regulates transcription-independent apoptosis by p53. J Biol Chem. 2009;284:20197–20205. doi: 10.1074/jbc.M109.026096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mellert HS, McMahon SB. Biochemical pathways that regulate acetyltransferase and deacetylase activity in mammalian cells. Trends Biochem Sci. 2009;34:571–578. doi: 10.1016/j.tibs.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang Y, Luo J, Zhang W, Gu W. Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol Cell. 2006;24:827–839. doi: 10.1016/j.molcel.2006.11.021. [DOI] [PubMed] [Google Scholar]
- 33.Seidel C, Schnekenburger M, Dicato M, Diederich M. Histone deacetylase 6 in health and disease. Epigenomics. 2015;7:103–118. doi: 10.2217/epi.14.69. [DOI] [PubMed] [Google Scholar]
- 34.Chen X, Wong JY, Wong P, Radany EH. Low-dose valproic acid enhances radiosensitivity of prostate cancer through acetylated p53-dependent modulation of mitochondrial membrane potential and apoptosis. Mol Cancer Res. 2011;9:448–461. doi: 10.1158/1541-7786.MCR-10-0471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Putcha P, et al. HDAC6 activity is a non-oncogene addiction hub for inflammatory breast cancers. Breast Cancer Res. 2015;17:149. doi: 10.1186/s13058-015-0658-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cho KR, Shih Ie M. Ovarian cancer. Annu Rev Pathol. 2009;4:287–313. doi: 10.1146/annurev.pathol.4.110807.092246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- 38.Aird KM, et al. Suppression of nucleotide metabolism underlies the establishment and maintenance of oncogene-induced senescence. Cell reports. 2013;3:1252–1265. doi: 10.1016/j.celrep.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bitler BG, et al. Wnt5a suppresses epithelial ovarian cancer by promoting cellular senescence. Cancer research. 2011;71:6184–6194. doi: 10.1158/0008-5472.CAN-11-1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCarty KS, Jr, et al. Cancer research. 1986;46:4244s–4248s. [PubMed] [Google Scholar]
- 41.Gao X, et al. ES cell pluripotency and germ-layer formation require the SWI/SNF chromatin remodeling component BAF250a. Proc Natl Acad Sci U S A. 2008;105:6656–6661. doi: 10.1073/pnas.0801802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scarlett UK, et al. Ovarian cancer progression is controlled by phenotypic changes in dendritic cells. J Exp Med. 2012;209:495–506. doi: 10.1084/jem.20111413. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Gene expression profiling data based on RNA-seq have been deposited in the Gene Expression Ominibus (GEO) under accession codes GSE84405. Previously published RNA-seq data for ARID1A wildtype or mutated human ovarian clear cell carcinoma specimens that were re-analysed here are available at the European Genome-Phenome Archive (EGAS) under accession code EGAS 00000000075 5. For correlation between ARID1A and HDAC6 expression, gene expression data obtained from GEO (under accession code: GSE36139) for clear cell and endometrioid ovarian cancer cell lines in cancer cell line encyclopedia 25 and a microarray database obtained from GEO (under accession code: GSE29450) for profiling gene expression in laser capture microdissected human clear cell ovarian tumour specimens and ovarian surface epithelial cells were used 26. ARID1A chromatin immunoprecipitation followed by next generation sequencing (ChIP-seq) and input tracks at the human HDAC6 gene promoter were based on ChIP-seq data (GEO Accession Number: GSE69568) 27.
Source data used for statistical analyses of Figures 1b, 1f, 1h, 2b–c, 2e, 2g, 2j, 3b–c, 3g–i, 4a, 4c–d, 4f, 4h–i, 4k–p, 5a, 5d–e, 6b, 6d, 7b, 7d, 7f–g, 8a, 8c, 8e, 8g, and Supplementary Figs 1a–c, 2b–c, 2e–f, 2h–i, 3, 4b–f, 4h–j, 5a, 5c–d, 6c, 8b–c, 8e, 8f–g and 8i are provided as Supplementary Table 6 (Statistics source data). All other data supporting the findings of this study are available upon request.