Abstract
Although patients with “severe” asthma tend to be characterized by ongoing symptoms and airway inflammation despite treatment with high doses of inhaled and systemic corticosteroids, there is increasing recognition of marked phenotypic heterogeneity within affected patients. While “precision medicine” approaches for patients with severe asthma are needed, there are many hurdles that must be overcome in daily practice. The National Heart, Lung and Blood Institute's Severe Asthma Research Program (SARP) has been at the forefront of phenotype discovery in severe asthma for the past decade. SARP, along with other international groups, have described clinical severe asthma phenotypes in both adults and children that can be evaluated in the clinical setting. While these clinical phenotypes provide a good “starting point” for addressing disease heterogeneity in severe asthma in everyday practice, more efforts are needed to understand how these phenotypes relate to underlying disease mechanisms and pharmacological treatment responses. This review highlights the clinical asthma phenotypes identified to date, their associations with underlying endotypes and potential biomarkers, and remaining knowledge gaps that must be addressed before precision medicine can become a reality for patients with severe asthma.
Keywords: Severe asthma phenotypes, SARP, endotype, precision medicine, biomarker, cluster analysis
Introduction
Although the majority of patients with asthma in the United States experience symptom improvement with the initiation of inhaled corticosteroid (ICS) therapy, asthma control remains suboptimal and nearly 50% of these patients experience at least one asthma exacerbation each year.1 While the reasons underlying poor asthma control are multifactorial and include medication access and compliance,2 there is a relatively small subset of adults and children with “severe” or “refractory” asthma who have ongoing symptoms and airway inflammation despite daily receipt of high doses of ICS and even systemic corticosteroids.3 These patients with severe asthma are at increased risk for medication-related side effects4 and are also are more likely than patients with milder forms of asthma to experience recurrent and potentially life-threatening exacerbations that significantly impair quality of life.5 Consequently, severe asthma may account for up to 50% of all asthma-related costs due to frequent healthcare encounters as well as numerous prescription medications and missed days from school and work.6, 7
Although national asthma treatment guidelines have proven useful in standardizing care approaches and improving outcomes,8, 9 there is increasing recognition of phenotypic heterogeneity in patients with asthma that is particularly marked in those with severe disease. Given recent mandates for more personalized and more efficient medicine, “precision medicine” for patients with severe asthma is needed; particularly since the existing evidence base for severe asthma care is quite limited.3 However, there are many hurdles that must be overcome. The first hurdle is to accurately and easily characterize a given individual's severe asthma and assign a “phenotype” for the application of personalized therapeutic approaches (i.e., precision medicine). In this view, a “phenotype” is defined as observable characteristics that may or may not be associated with underlying disease mechanisms. An example of this is a patient with ongoing symptoms and severely obstructed patterns of lung function, which may be due to a variety of inflammatory factors including alterations in glucocorticoid receptor signaling and function, increased airway matrix deposition, or alternatively, progressive viral insults with impaired innate immune responses. The second hurdle is to ultimately refine these phenotyping efforts to measure or make inferences about basic pathophysiologic and biologic mechanisms (i.e., “endotypes”) that underlie the disease and ultimately guide therapy. The goal is to derive clinical phenotypes that clearly translate to biological endotypes, without excessive imaging and laboratory testing, for the purpose of time-efficient and resource-efficient precision pharmacological treatment. As more biological therapies become available, the final hurdle is to select the most appropriate first-line treatment.
The National Heart, Lung and Blood Institute's Severe Asthma Research Program (SARP) has been at the forefront of phenotype discovery in severe asthma for the past decade and has described clinical severe asthma phenotypes in both adults and children that can be evaluated in the clinical setting. These clinical phenotypes provide a good “starting point” for addressing disease heterogeneity in severe asthma in everyday practice. However, more efforts are needed to understand how these phenotypes relate to underlying disease treatment responses and underlying disease mechanisms. This review highlights the clinical asthma phenotypes identified in SARP, their associations with underlying endotypes, and remaining knowledge gaps that must be addressed before precision medicine can be a reality for patients with severe asthma.
Clinical asthma phenotypes in SARP
SARP is a multi-center network in the United States focused on the clinical, biological, and genetic attributes of severe versus non-severe asthma in adults and children. Since the initiation of SARP in 2001, each participating clinical site utilized uniform procedures that permitted rigorous yet consistent characterization of participants, including standardized medical history questionnaires, pulmonary function testing, methacholine challenge, and biomarker collection.10, 11 Because consensus definitions of “severe” asthma were lacking prior to 2000, the early SARP program adopted the definition of severe asthma proposed by an American Thoracic Society (ATS) Workshop,12 which required: 1) treatment with continuous high-dose ICS or continuous systemic corticosteroids and 2) at least two minor criteria which demonstrated poor asthma control or life-threatening disease. This definition advanced the concept of severe asthma as a biological disease entity associated with corticosteroid insensitivity and a number of resulting publications highlighted the unique clinical and intrinsic attributes of this group as compared to patients with milder disease.13, 14
Despite global differences between “severe” and “non-severe” asthma in SARP, significant heterogeneity was present in both groups, prompting further exploration of phenotypes irrespective of asthma severity definitions. Using unsupervised cluster analyses, subgroups of patients with asthma emerged.15, 16 In SARP adults, 5 phenotypic “clusters” emerged that were distinguished primarily by lung function and the age of asthma onset.16 The most “severe” asthma subjects were assigned to clusters 3, 4 and 5 that were associated with frequent use of oral corticosteroids for asthma exacerbations, hospitalizations for severe near-fatal asthma and increased medication requirements (Table 1).16 A similar cluster analysis in SARP children identified 4 phenotypic clusters that differed primarily according to asthma duration, the number of asthma controller medications and lung function.15 The most “severe” asthmatic children were assigned to clusters 3 and 4, which had the highest prevalence of comorbidity and symptom burden similar to the adults (Table 1).15 Pre-bronchodilator FEV1 % predicted and age of asthma onset were important in differentiating severe asthma clinical phenotypes in both the adult and pediatric analyses, emphasizing that onset of asthma prior to puberty is characteristic of the better understood “classic” allergic asthma phenotypes (Figure 1).
Table 1. Adult and Pediatric “Severe” Asthma Clusters Identified by the Severe Asthma Research Program (SARP).
| Early-onset allergic asthma | Later-onset adult asthma | ||||
|---|---|---|---|---|---|
| Pediatric Severe Asthma Cluster 3 | Pediatric Severe Asthma Cluster 4 | Adult Severe Asthma Cluster 4 | Adult Severe Asthma Cluster 5 | Adult Severe Asthma Cluster 3 | |
| Cluster Description | Co-morbid, difficult-to-treat asthma | Refractory asthma with airflow obstruction | “Classic” Early onset severe allergic asthma | Asthma with chronic airflow obstruction (COPD) | Obese asthma with high impairment, but normal lung function |
| Asthma onset | Infancy | Toddler to preschool years | Preschool or early school-age years, before puberty | Teenage years or adulthood, after puberty | Adulthood |
| Aeroallergen sensitization | Highly prevalent with multiple sensitization | Highly prevalent with multiple sensitization | Highly prevalent with multiple sensitization | Less prevalent | Less prevalent |
| Lung function | Reversible airflow obstruction | Partially reversible airflow obstruction | Partially reversible airflow obstruction | Severe, less reversible airflow obstruction | Borderline normal airflow obstruction |
| Asthma medications | Multiple controller medications, high-dose ICS, daily OCS | Multiple controller medications, high-dose ICS | Multiple controller medications, high-dose ICS, daily OCS | Multiple controller medications, high-dose ICS, daily OCS | Multiple controller medications, high-dose ICS |
| Healthcare utilization, Past Year | Multiple OCS bursts, acute visits, hospitalizations | Multiple OCS bursts, acute visits, hospitalizations | Multiple OCS bursts, acute visits | Multiple OCS bursts, acute visits, hospitalizations | Multiple OCS bursts, acute visits |
| Co-morbid features | Sinus disease, gastroesophageal reflux, obesity | Less frequent co-morbidity | Less frequent co-morbidity | Pneumonia, hypertension, obesity | Sinus disease, hypertension, obesity |
Adapted from Reference 15 (Pediatric Cluster Analysis) and, 16 (Adult Cluster Analysis).
Abbreviations: ICS= inhaled corticosteroid, OCS = chronic oral corticosteroid
Figure 1.
A schematic of the adult and pediatric severe asthma phenotypes identified in the SARP cluster analyses.15,16 Heterogeneity within each phenotype is indicated by the shape of the diamond; the “width” represents heterogeneity in the age of asthma onset (median line, IQR), the “height” symbolizes heterogeneity of lung function within each group (IQR). Note that the majority of severe asthma subjects had abnormal baseline lung function when bronchodilators were appropriately withhold prior to spirometry, reinforcing the importance of extended lung function testing in the clinic.
Pediatric Severe Asthma Clinical Phenotypes
Overall, children with severe asthma are characterized by ongoing symptoms and frequent exacerbations that impair daily functioning and quality of life.10, 17Although not all children with severe asthma are atopic, the majority of children with severe asthma (>80%) have sensitization to aeroallergens, in contrast to adults.18 Airflow limitation and air trapping with incomplete reversal after bronchodilator administration are other prominent features19 that may worsen during adolescence20 and increase the risk of chronic obstructive pulmonary disease in later adulthood.21, 22
Yet despite these commonalities, the phenotypes identified in the SARP pediatric cluster analysis argue for differing risk factors for asthma severity (Table 1). Children in Cluster 3 (20% of children studied) had an early age of asthma onset, atopic features, the highest prevalence of comorbidity, increased airway hyperresponsiveness to methacholine and airflow limitation. By contrast, children in Cluster 4 (18% of children studied) had the most advanced disease, with an early age of symptom onset accompanied by atopic features, less comorbidity, partially reversible and more advanced airflow obstruction and the greatest burden of symptoms and associated medication use.15 Although Clusters 3 and 4 did not completely conform to definitions of asthma severity proposed by current treatment guidelines, replication in a separate pediatric asthma population demonstrated that they were associated with differential and limited response to asthma therapies.23 Whereas children in Cluster 4 had best response with fluticasone/salmeterol, children in cluster 3 had the least efficacy to available guideline-based asthma treatment.23 However, that analysis was limited by relatively few children with highly symptomatic asthma and a focus on available asthma treatments, as opposed to novel biologics.
Adult Severe Asthma Clinical Phenotypes
The three severe asthma clusters in the adult SARP analysis represent the spectrum of severe early onset allergic asthma (Cluster 4), adolescent/early adult onset asthma with less reversible/chronic airflow obstruction (Cluster 5) and late onset, less allergic asthma, mostly obese individuals with high disease impairment but normal lung function (Cluster 3).16 Multiple other cluster analyses in different asthma cohorts have identified adult clinical phenotypes remarkably similar to those described in the SARP cluster analyses, suggesting that the phenotypes are fairly generalizable across different centers and internationally (Table 2).24-30 For example, Haldar et al. identified 4 clusters of severe, refractory asthma distinguished by age of onset, atopy, and obesity.24 Schatz et al. also noted similar clusters in an adolescent and adult severe or difficult-to-treat asthma population that were not only distinguished by age of onset and atopic features, but also by race and aspirin sensitivity.27
Table 2. “Severe” Asthma Clusters Identified by Other Investigators outside of the Severe Asthma Research Program (SARP).
| Reference | Population | Age group | N | Number of clusters identified | Number of “severe” clusters | Description of “severe” clusters |
|---|---|---|---|---|---|---|
| Haldar et al. (2008)24 | Secondary care, refractory asthma | Adults | 187 | 4 | 4 |
|
| Kim et al. (2013)25 | Secondary care, asthma | Adults | 2,567 | 4 | 1 |
|
| Amelink et al. (2013)26 | Secondary care, adult-onsetasthma | Adults | 200 | 3 | 2 |
|
| Schatz et al. (2014)27 | Severe or difficult-to-treat asthma | Children | 518 | 5 | 5 |
|
| Schatz et al. (2014)27 | Severe or difficult-to-treat asthma | Adolescents and adults | 3,612 | 5 | 5 |
|
| Newby et al. (2014)28 | Severe, refractory asthma | Adults | 349 | 5 | 5 |
|
| Zaihra et al. (2016)29 | Secondary care, difficult-to-treat asthma | Adults | 125 | 4 | 3 |
|
| Lefaudeux et al. (2017)30 | Mild-to-severe asthma | Adults | 418 | 4 | 4 |
|
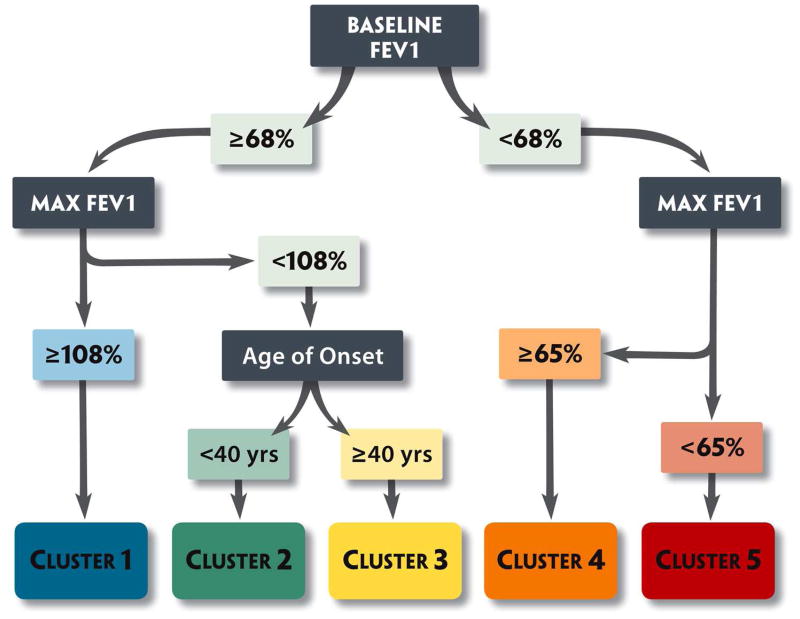
The four SARP phenotypes identified in the cluster analysis have also been replicated in “real life” clinics31, 32 and other cohorts33, 34 using a 3-variable algorithm that incorporates pre- and post-bronchodilator % predicted FEV1 and age of asthma onset to assign individual patients to these clusters (Figure 2). Reassuringly, from a practical sense, the described phenotypes resemble patients we actually see everyday in the clinic31, 32 and several studies have suggested that nearly half of patients demonstrate cluster stability over time, although the severe asthma cluster phenotypes (not surprisingly) appear to be slightly less stable temporally.28, 29, 34 As such, the challenge is how best to use these cross sectional clinical phenotypes to guide pharmacotherapy and effect clinical outcomes such as exacerbation rates and progressive lung function decline.
Figure 2.
SARP algorithm for cluster assignment in subjects > 12 years of age.16 Using three variables (1) Baseline FEV1 (with a bronchodilator withhold), (2) Maximal “Max” FEV1 (after 6-8 puffs of albuterol) and (3) age of onset of asthma, subjects can be assigned to the five clinical asthma phenotypes. Clusters 3, 4 and 5 are the severe asthma clusters; Color Key: Yellow, late onset nonatopic asthma; Orange, severe atopic asthma; Red, severe asthma with fixed airflow.
Pathobiology and Clinical Phenotypes
Ideally, distinct pathobiologic mechanisms (endotypes) would underpin clinical phenotypes and thus allow therapy to be targeted to the specific underlying biology, but this has not been so easily proven due to biologic heterogeneity. Sputum analysis in SARP adults had previously shown that subjects with concurrent elevations in sputum eosinophils and neutrophils had the most severe asthma,35 but this pattern does not differentiate severe asthma subjects from clinical clusters 4 and 5 (Figure 3).18 A subsequent SARP cluster analyses that aimed to reconcile the clinical phenotypes with biology by incorporating blood eosinophils and sputum granulocytes into a new cluster analysis also showed heterogeneity of inflammatory cell patterns within clusters with no clear distinction between the severe clusters.36 A third SARP cluster analysis that incorporated bronchoalveolar lavage inflammatory variables identified clusters that clinically overlap with the previously published results, but showed increased exhaled nitric oxide and airway lavage eosinophil counts in patients with severe early onset atopic asthma supporting a strong role for Type-2 inflammatory mechanisms in this group.37 This analysis confirmed an older age of onset and less atopic asthma phenotype, but again found that airway eosinophils and neutrophils did not clearly distinguish this group.37
Figure 3.
Sputum inflammatory profiles in Adult SARP severe asthma cluster phenotypes show heterogeneity within each cluster with no clear predominant pattern. Inflammatory profiles were determined for individual subjects based on % eosinophils (≥ 2%) and % neutrophils ≥ 40% in induced sputum as previously described.36 Color Key: Blue, paucigranulocytic; Green, neutrophil predominant; Red, eosinophil predominant; Purple, mixed granulocytic.
Similar analyses that have included biomarkers with clinical characteristics in other cohorts have likewise found heterogeneity within cluster groups.29 A recent study involving sputum proteomics and transcriptomics noted differences in expression profiles between cluster groups, but elevations in sputum eosinophils were not isolated to one cluster but rather 3 of the 4 clusters identified.30 Furthermore, the clusters in that report did not differ with regard to sputum neutrophil counts, exhaled nitric oxide or serum IgE levels.30 These studies suggest that while clinical phenotypes do inform asthma heterogeneity and can be easily applied using algorithms, they are not yet refined endotypes and thus, not precise enough to guide targeted immunomodulator therapy without a “biologic” marker to reveal underlying biologic heterogeneity.
Biomarkers associated with Clinical Phenotypes
All of the current biomarkers available in clinical practice are focused on Type-2 “allergic” inflammation [blood eosinophils, exhaled nitric oxide (FeNO), sputum eosinophils]. Sputum eosinophils have been the “gold standard” Type-2 inflammatory biomarker,38 but performing sputum analysis for inflammatory cellular profiles in clinic is challenging. Unfortunately, while blood eosinophils, serum total IgE and exhaled nitric oxide are easily obtained biomarkers, the correlation between these “noninvasive” measures and sputum eosinophils is suboptimal, making them poor surrogate measures of airway eosinophilia.39, 40 While several studies have used management approaches guided by these biomarkers and shown improved asthma outcomes, 41, 42 these older studies contain very few patients with severe asthma making applicability of these approaches to severe asthma unclear.
In fact, these Type-2 inflammatory biomarkers in blood, sputum or exhaled gases are associated with subtypes of all clinical asthma phenotypes in SARP, not specifically with the severe asthma phenotypes (Figures 3, 4).15, 16, 18, 36 In the adult SARP severe asthma clusters (Figure 3), 20% of each cluster showed a bland inflammatory cell profile with few eosinophils and neutrophils, while 20-30% had a mixed granulocytic pattern with elevations of both eosinophils and neutrophils.16, 18, 36 Similarly, in the pediatric severe asthma clusters (Figure 4), 20-30% of subjects did not have elevated blood eosinophils or exhaled nitric oxide (FeNO) while 30-40% showed increased blood eosinophils and FeNO in the severe asthma clusters.15, 18 Again, neither the biomarkers nor the pattern of the biomarkers was specific for the severe asthma phenotypes. This variability in biomarkers within the SARP severe asthma clinical phenotypes is due to pathobiologic heterogeneity that underlies the clinical phenotype. We instead propose that biomarkers be used to identify patients within a cluster who are most likely to be responders to a given immunomodulator as a disease modifier, not to define a specific clinical phenotype.43
Figure 4.
Association between exhaled nitric oxide values and blood eosinophils in the SARP pediatric severe asthma clinical clusters.15 Percentages reflect the percentage of children in the cluster. Although there was modest agreement between the biomarkers, heterogeneity in biomarker presentation was present in both clusters.
Using Clinical Phenotypes (+ biomarkers) in Clinical Trials
Most industry sponsored biologic clinical trials have effectively used the above approach by selecting study subjects that resemble the severe asthma clinical phenotypes of SARP (high intensity controller medication regimens, frequent exacerbations and low lung function), but then require an elevated biomarker (in blood, exhaled gas or sputum) that should identify patients most likely to be responders to a specific immunomodulator within those clinical phenotypes. The biologics in recently published clinical trials target Type-2 inflammatory pathways and restrict participation to subjects with elevated Type-2 biomarkers (blood or sputum eosinophils or FeNO). Post-hoc responder analyses to “group” biomarkers to better understand which patients have the greatest benefit from a biologic immunomodulator have led to an understanding that concurrent elevations in more than one Type-2 inflammatory biomarker may further identify “high responder” patients.44
Perhaps more relevant to this review, however, is a post-hoc cluster analysis of the DREAM study (Dose-Ranging Efficacy And safety with Mepolizumab study) that showed that a combination of a biomarker (blood eosinophils), airway physiology (magnitude of bronchodilator reversibility) and a clinical characteristic (Body Mass Index) could accurately identify four patient clusters with different magnitudes of rate reductions in asthma exacerbations when treated with mepolizumab.45 The cluster with a largest reduction in asthma exacerbations (67% reduction) was described as mostly obese women with reported age of onset in late adolescence or adulthood and less atopy by serum testing with poor baseline lung function. That clinical description is very similar indeed to SARP Cluster 5 (Table 1) in which 50% of patients had elevated sputum eosinophil counts (Figure 3). While it could be argued that the reduction in exacerbations in the multidimensional DREAM study group was not much greater than that observed when using sole biomarker directed anti-IL5 therapy without clinical input. these results provide some vision for how severe asthma management might be refined in the future.
A clinical cluster phenotype with limited biomarkers might identify the “best responder” for a pharmacologic therapy in the clinic while we await refinement of true endotypes needed for precision medicine. Figure 5 presents a possible approach for selection of a patient for a biologic therapy, using a combined approach of phenotyping by clinical characteristics together with a biomarker. Ultimately, this approach requires testing and prospective evaluation. The National Heart, Lung and Blood Institute's soon to be funded multicenter PrecISE network (Precision Interventions for Severe and/or Exacerbation Prone Asthma) will use patient phenotypes, endotypes and biomarkers to conduct adaptive clinical trials with sequential therapies to determine the most effective pathway to precision medicine in severe asthma.
Figure 5.
Potential approach for selection of biological therapy in severe asthma patients, utilizing phenotyping by clinical characteristics and biomarker assessment. IgE = immunoglobulin E, IL = interleukin, CRTH2 = chemokine receptor homologous molecule expressed on Th2 lymphocyte, PDE4 = phosphodiesterase 4
Conclusions
Ideally, severe asthma phenotypes should be easy to identify in the clinic and be indicative of pathobiologic mechanisms (endotypes) that can guide precision medicine. Although the clinical severe asthma phenotypes identified by SARP and others provide a good “starting point” for addressing disease heterogeneity in severe asthma in everyday practice, the association between clinical phenotype and endotype today is imprecise. Some biomarkers can be measured in clinic as potential surrogate measures of underlying pathobiologic mechanisms, but they are too simplistic and nonspecific for a given clinical phenotype to guide therapy in isolation of clinical characteristics. Current clinical trials of immunomodulators are essentially utilizing SARP-like clinical severe asthma cluster phenotypes with selected biomarkers to identify “responder” phenotypes, not just a biomarker alone. The continued replication of similar severe asthma phenotypes around the world and ongoing –omic approaches to pinpoint the pathobiologic mechanisms that underlie these phenotypes is “real-time” refining the endotypes that are needed before precision medicine and targeted pharmacological treatment of severe asthma can become a reality in the clinic. The goal of the upcoming PreCISE network is to guide precision medicine for severe asthma patients in the future. Clinical asthma phenotypes such as those described by the Severe Asthma Research Program, however can be a good “starting” tool for clinicians in practice today.
Abbreviations used
- ATS
American Thoracic Society
- FeNO
Fractional Excretion of nitric oxide
- ICS
Inhaled corticosteroid
- OCS
Oral corticosteroid
- SARP
Severe Asthma Research Program
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ward BW, Clarke TC, Freeman G, Schiller JS. Early release of selected estimates based on data from the January-June 2016 National Health Interview Survey. Centers for Disease Control and Prevention, National Center for Health Statistics. 2016 Nov; [Google Scholar]
- 2.Slejko JF, Ghushchyan VH, Sucher B, Globe DR, Lin SL, Globe G, et al. Asthma control in the United States, 2008-2010: indicators of poor asthma control. J Allergy Clin Immunol. 2014;133:1579–87. doi: 10.1016/j.jaci.2013.10.028. [DOI] [PubMed] [Google Scholar]
- 3.Chung KF, Wenzel SE, Brozek JL, Bush A, Castro M, Sterk PJ, et al. International ERS/ATS guidelines on definition, evaluation and treatment of severe asthma. Eur Respir J. 2014;43:343–73. doi: 10.1183/09031936.00202013. [DOI] [PubMed] [Google Scholar]
- 4.Wysocki K, Park SY, Bleecker E, Busse W, Castro M, Chung KF, et al. Characterization of factors associated with systemic corticosteroid use in severe asthma: data from the Severe Asthma Research Program. J Allergy Clin Immunol. 2014;133:915–8. doi: 10.1016/j.jaci.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moorman JE, Akinbami LJ, Bailey CM, Zahran HS, King ME, Johnson CA, et al. National surveillance of asthma: United States, 2001-2010. Vital Health Stat. 2012;3:1–58. [PubMed] [Google Scholar]
- 6.Ivanova JI, Bergman R, Birnbaum HG, Colice GL, Silverman RA, McLaurin K. Effect of asthma exacerbations on health care costs among asthmatic patients with moderate and severe persistent asthma. J Allergy Clin Immunol. 2012;129:1229–35. doi: 10.1016/j.jaci.2012.01.039. [DOI] [PubMed] [Google Scholar]
- 7.Szefler SJ, Zeiger RS, Haselkorn T, Mink DR, Kamath TV, Fish JE, et al. Economic burden of impairment in children with severe or difficult-to-treat asthma. Ann Allergy Asthma Immunol. 2011;107:110–9 e1. doi: 10.1016/j.anai.2011.04.008. [DOI] [PubMed] [Google Scholar]
- 8.Hasegawa K, Tsugawa Y, Clark S, Eastin CD, Gabriel S, Herrera V, et al. Improving Quality of Acute Asthma Care in US Hospitals: Changes Between 1999-2000 and 2012-2013. Chest. 2016;150:112–22. doi: 10.1016/j.chest.2016.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okelo SO, Butz AM, Sharma R, Diette GB, Pitts SI, King TM, et al. Interventions to modify health care provider adherence to asthma guidelines: a systematic review. Pediatrics. 2013;132:517–34. doi: 10.1542/peds.2013-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fitzpatrick AM, Gaston BM, Erzurum SC, Teague WG, National Institutes of Health/National Heart L, Blood Institute Severe Asthma Research P Features of severe asthma in school-age children: Atopy and increased exhaled nitric oxide. J Allergy Clin Immunol. 2006;118:1218–25. doi: 10.1016/j.jaci.2006.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moore WC, Bleecker ER, Curran-Everett D, Erzurum SC, Ameredes BT, Bacharier L, et al. Characterization of the severe asthma phenotype by the National Heart, Lung, and Blood Institute's Severe Asthma Research Program. J Allergy Clin Immunol. 2007;119:405–13. doi: 10.1016/j.jaci.2006.11.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Proceedings of the ATS workshop on refractory asthma: current understanding, recommendations, and unanswered questions. American Thoracic Society. Am J Respir Crit Care Med. 2000;162:2341–51. doi: 10.1164/ajrccm.162.6.ats9-00. [DOI] [PubMed] [Google Scholar]
- 13.Jarjour NN, Erzurum SC, Bleecker ER, Calhoun WJ, Castro M, Comhair SA, et al. Severe asthma: lessons learned from the National Heart, Lung, and Blood Institute Severe Asthma Research Program. Am J Respir Crit Care Med. 2012;185:356–62. doi: 10.1164/rccm.201107-1317PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fitzpatrick AM. Severe Asthma in Children: Lessons Learned and Future Directions. J Allergy Clin Immunol Pract. 2016;4:11–9. doi: 10.1016/j.jaip.2015.10.008. quiz 20-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fitzpatrick AM, Teague WG, Meyers DA, Peters SP, Li X, Li H, et al. Heterogeneity of severe asthma in childhood: confirmation by cluster analysis of children in the National Institutes of Health/National Heart, Lung, and Blood Institute Severe Asthma Research Program. J Allergy Clin Immunol. 2011;127:382–9. e1–13. doi: 10.1016/j.jaci.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moore WC, Meyers DA, Wenzel SE, Teague WG, Li H, Li X, et al. Identification of asthma phenotypes using cluster analysis in the Severe Asthma Research Program. Am J Respir Crit Care Med. 2010;181:315–23. doi: 10.1164/rccm.200906-0896OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Denlinger LC, Phillips BR, Ramratnam S, Ross K, Bhakta NR, Cardet JC, et al. Inflammatory and Comorbid Features of Patients with Severe Asthma and Frequent Exacerbations. Am J Respir Crit Care Med. 2017;195:302–13. doi: 10.1164/rccm.201602-0419OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moore WC, Fitzpatrick AM, Li X, Hastie AT, Li H, Meyers DA, et al. Clinical heterogeneity in the severe asthma research program. Ann Am Thorac Soc. 2013;10(Suppl):S118–24. doi: 10.1513/AnnalsATS.201309-307AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sorkness RL, Teague WG, Penugonda M, Fitzpatrick AM, National Institutes of Health NHL, Blood Institute's Severe Asthma Research P Sex dependence of airflow limitation and air trapping in children with severe asthma. J Allergy Clin Immunol. 2011;127:1073–4. doi: 10.1016/j.jaci.2010.12.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fitzpatrick AM, Teague WG, National Institutes of Health/National Heart L, Blood Institute's Severe Asthma Research P Progressive airflow limitation is a feature of children with severe asthma. J Allergy Clin Immunol. 2011;127:282–4. doi: 10.1016/j.jaci.2010.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tai A, Tran H, Roberts M, Clarke N, Wilson J, Robertson CF. The association between childhood asthma and adult chronic obstructive pulmonary disease. Thorax. 2014;69:805–10. doi: 10.1136/thoraxjnl-2013-204815. [DOI] [PubMed] [Google Scholar]
- 22.Tai A, Tran H, Roberts M, Clarke N, Wilson J, Robertson CF. Trends in eczema, rhinitis, and rye grass sensitization in a longitudinal asthma cohort. Ann Allergy Asthma Immunol. 2014;112:437–40. doi: 10.1016/j.anai.2014.03.005. [DOI] [PubMed] [Google Scholar]
- 23.Chang TS, Lemanske RF, Jr, Mauger DT, Fitzpatrick AM, Sorkness CA, Szefler SJ, et al. Childhood asthma clusters and response to therapy in clinical trials. J Allergy Clin Immunol. 2014;133:363–9. doi: 10.1016/j.jaci.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haldar P, Pavord ID, Shaw DE, Berry MA, Thomas M, Brightling CE, et al. Cluster analysis and clinical asthma phenotypes. Am J Respir Crit Care Med. 2008;178:218–24. doi: 10.1164/rccm.200711-1754OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim TB, Jang AS, Kwon HS, Park JS, Chang YS, Cho SH, et al. Identification of asthma clusters in two independent Korean adult asthma cohorts. Eur Respir J. 2013;41:1308–14. doi: 10.1183/09031936.00100811. [DOI] [PubMed] [Google Scholar]
- 26.Amelink M, de Nijs SB, de Groot JC, van Tilburg PM, van Spiegel PI, Krouwels FH, et al. Three phenotypes of adult-onset asthma. Allergy. 2013;68:674–80. doi: 10.1111/all.12136. [DOI] [PubMed] [Google Scholar]
- 27.Schatz M, Hsu JW, Zeiger RS, Chen W, Dorenbaum A, Chipps BE, et al. Phenotypes determined by cluster analysis in severe or difficult-to-treat asthma. J Allergy Clin Immunol. 2014;133:1549–56. doi: 10.1016/j.jaci.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 28.Newby C, Heaney LG, Menzies-Gow A, Niven RM, Mansur A, Bucknall C, et al. Statistical cluster analysis of the British Thoracic Society Severe refractory Asthma Registry: clinical outcomes and phenotype stability. PLoS One. 2014;9:e102987. doi: 10.1371/journal.pone.0102987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zaihra T, Walsh CJ, Ahmed S, Fugere C, Hamid QA, Olivenstein R, et al. Phenotyping of difficult asthma using longitudinal physiological and biomarker measurements reveals significant differences in stability between clusters. BMC Pulm Med. 2016;16:74. doi: 10.1186/s12890-016-0232-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lefaudeux D, De Meulder B, Loza MJ, Peffer N, Rowe A, Baribaud F, et al. U-BIOPRED clinical adult asthma clusters linked to a subset of sputum omics. J Allergy Clin Immunol. 2016 doi: 10.1016/j.jaci.2016.08.048. [DOI] [PubMed] [Google Scholar]
- 31.Patrawalla P, Kazeros A, Rogers L, Shao Y, Liu M, Fernandez-Beros ME, et al. Application of the asthma phenotype algorithm from the Severe Asthma Research Program to an urban population. PLoS One. 2012;7:e44540. doi: 10.1371/journal.pone.0044540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuhlen JL, Jr, Wahlquist AE, Nietert PJ, Bains SN. Identification of asthma phenotypes in a tertiary care medical center. Am J Med Sci. 2014;348:480–5. doi: 10.1097/MAJ.0000000000000346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bourdin A, Molinari N, Vachier I, Varrin M, Marin G, Gamez AS, et al. Prognostic value of cluster analysis of severe asthma phenotypes. J Allergy Clin Immunol. 2014;134:1043–50. doi: 10.1016/j.jaci.2014.04.038. [DOI] [PubMed] [Google Scholar]
- 34.Kupczyk M, Dahlen B, Sterk PJ, Nizankowska-Mogilnicka E, Papi A, Bel EH, et al. Stability of phenotypes defined by physiological variables and biomarkers in adults with asthma. Allergy. 2014;69:1198–204. doi: 10.1111/all.12445. [DOI] [PubMed] [Google Scholar]
- 35.Hastie AT, Moore WC, Meyers DA, Vestal PL, Li H, Peters SP, et al. Analyses of asthma severity phenotypes and inflammatory proteins in subjects stratified by sputum granulocytes. J Allergy Clin Immunol. 2010;125:1028–36 e13. doi: 10.1016/j.jaci.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moore WC, Hastie AT, Li X, Li H, Busse WW, Jarjour NN, et al. Sputum neutrophil counts are associated with more severe asthma phenotypes using cluster analysis. J Allergy Clin Immunol. 2014;133:1557–63 e5. doi: 10.1016/j.jaci.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu W, Bleecker E, Moore W, Busse WW, Castro M, Chung KF, et al. Unsupervised phenotyping of Severe Asthma Research Program participants using expanded lung data. J Allergy Clin Immunol. 2014;133:1280–8. doi: 10.1016/j.jaci.2013.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coumou H, Bel EH. Improving the diagnosis of eosinophilic asthma. Expert Rev Respir Med. 2016;10:1093–103. doi: 10.1080/17476348.2017.1236688. [DOI] [PubMed] [Google Scholar]
- 39.Fleming L, Tsartsali L, Wilson N, Regamey N, Bush A. Longitudinal relationship between sputum eosinophils and exhaled nitric oxide in children with asthma. Am J Respir Crit Care Med. 2013;188:400–2. doi: 10.1164/rccm.201212-2156LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hastie AT, Moore WC, Li H, Rector BM, Ortega VE, Pascual RM, et al. Biomarker surrogates do not accurately predict sputum eosinophil and neutrophil percentages in asthmatic subjects. J Allergy Clin Immunol. 2013;132:72–80. doi: 10.1016/j.jaci.2013.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith AD, Cowan JO, Brassett KP, Herbison GP, Taylor DR. Use of exhaled nitric oxide measurements to guide treatment in chronic asthma. N Engl J Med. 2005;352:2163–73. doi: 10.1056/NEJMoa043596. [DOI] [PubMed] [Google Scholar]
- 42.Green RH, Brightling CE, McKenna S, Hargadon B, Parker D, Bradding P, et al. Asthma exacerbations and sputum eosinophil counts: a randomised controlled trial. Lancet. 2002;360:1715–21. doi: 10.1016/S0140-6736(02)11679-5. [DOI] [PubMed] [Google Scholar]
- 43.Opina MT, Moore WC. Phenotype-Driven Therapeutics in Severe Asthma. Curr Allergy Asthma Rep. 2017;17:10. doi: 10.1007/s11882-017-0678-1. [DOI] [PubMed] [Google Scholar]
- 44.Hanania NA, Wenzel S, Rosen K, Hsieh HJ, Mosesova S, Choy DF, et al. Exploring the effects of omalizumab in allergic asthma: an analysis of biomarkers in the EXTRA study. Am J Respir Crit Care Med. 2013;187:804–11. doi: 10.1164/rccm.201208-1414OC. [DOI] [PubMed] [Google Scholar]
- 45.Ortega H, Li H, Suruki R, Albers F, Gordon D, Yancey S. Cluster analysis and characterization of response to mepolizumab. A step closer to personalized medicine for patients with severe asthma. Ann Am Thorac Soc. 2014;11:1011–7. doi: 10.1513/AnnalsATS.201312-454OC. [DOI] [PubMed] [Google Scholar]