Abstract
Most orthopoxviruses, including vaccinia virus (VACV), contain genes in the E3L and K3L families. The protein products of these genes have been shown to combat PKR, a host defense pathway. Interestingly, ectromelia virus (ECTV) contains an E3L ortholog but does not possess an intact K3L gene. Here, we gained insight into how ECTV can still efficiently evade PKR despite lacking K3L. Relative to VACV, we found that ECTV-infected BS-C-1 cells accumulated considerably less double-stranded (ds) RNA, which was due to lower mRNA levels and less transcriptional read-through of some genes by ECTV. The abundance of dsRNA in VACV-infected cells, detected using a monoclonal antibody, was able to activate the RNase L pathway at late time points post-infection. Historically, the study of transcription by orthopoxviruses has largely focused on VACV as a model. Our data suggest that there could be more to learn by studying other members of this genus.
Keywords: ectromelia virus, mousepox virus, vaccinia virus, double-stranded RNA, dsRNA, E3L gene, K3L gene, innate immune evasion, IBT
Introduction
Ectromelia virus (ECTV; also referred to as “mousepox virus”) is a double-stranded DNA virus in the Poxviridae family. ECTV is a natural pathogen of mice and infects through abrasions in the skin (Fenner, 1947). Following initial replication at the site of infection, the virus disseminates to multiple organs over the course of several days (Esteban and Buller, 2005). Among the mice that survive the initial infection, characteristic pock lesions often manifest on the skin (Esteban and Buller, 2005; Fenner and Mortimer, 2006) in a similar fashion to the disease manifestations of humans infected with smallpox (Breman and Henderson, 2002). Vaccinia virus (VACV), which is the most studied poxvirus, was used to eradicate smallpox during vaccination campaigns. Despite a high degree of homology between these two viruses (Gubser et al., 2004), their host ranges are quite distinct (Buller and Palumbo, 1991). In mice, VACV infection is fatal only under certain experimental conditions and routes of infection (e.g., intranasal). In contrast, ECTV infection of susceptible mouse strains consistently results in death independent of the infection route and even with a very low initial inoculum (Parker et al., 2009; Xu et al., 2008).
The formation of double-stranded RNA (dsRNA) is a hallmark of numerous viral infections (Weber et al., 2006). For this reason, dsRNA is a major pathogen-associated molecule used by the immune system to detect invading viruses and initiate the appropriate anti-viral response. With respect to poxviruses, it has been known for several decades that VACV forms large amounts of dsRNA, especially during late times (i.e., post-DNA replication) of the infection cycle (Boone et al., 1979; Colby and Duesberg, 1969; Colby et al., 1971; Duesberg and Colby, 1969). VACV forms dsRNA because its genes are arranged such that there is minimal intergenic space. Also, adjacent genes are often transcribed in a convergent fashion, meaning that one gene might be transcribed from left to right and a neighboring gene from right to left. This phenomenon occurs when regions of the genome are transcribed from opposite strands of DNA (Broyles, 2003). Furthermore, the VACV RNA polymerase does not efficiently terminate at the ends of many viral genes – particularly for intermediate and late genes – which causes the transcription machinery to “read-through” into the next gene. This leads to heterogeneous 3’ termini on many viral transcripts (Xiang et al., 2000; 1998). Together, the above described events result in large amounts of mRNA with complementary ends, which allows for dsRNA formation in the cytoplasm. The anti-poxvirus drug isatin beta-thiosemicarbazone (IBT) has been shown to artificially increase the amount of dsRNA formed within VACV-infected cells by promoting read-through (Cresawn et al., 2007).
There are two important anti-viral pathways in cells that are induced by the presence of dsRNA: PKR and 2’5’ OAS/RNase L. Both can result in the termination of viral protein synthesis and also often lead to apoptosis of the infected cell – thus blunting virus dissemination within the host organism. Because dsRNA is a potent trigger of anti-viral immunity, viruses have evolved strategies to evade detection by the host (Seet et al., 2003). This study principally focused on the poxvirus immune evasion gene E3L and to a lesser extent K3L. These genes are involved in blunting the anti-viral response by PKR and RNase L.
Both ECTV and VACV possess a gene in the E3L family, which is conserved among most members of the Chordopoxvirinae (Bratke et al., 2013; Haller et al., 2014; Myskiw et al., 2011). The E3L gene encodes for a protein product (termed E3) with an amino-terminal Z-DNA-binding domain (Kim et al., 2003) and a carboxy-terminal dsRNA-binding domain (Chang and Jacobs, 1993). Interestingly, this protein was initially discovered because of its ability to bind to Poly(I:C), a synthetic dsRNA mimic, from extracts of VACV-infected cells (Chang et al., 1992; Watson et al., 1991). E3 has been shown to inhibit activation of host defenses such as PKR and RNase L, which is likely due in large part to its ability to sequester dsRNA (Langland et al., 2006; Rivas et al., 1998; Xiang et al., 2002; Zhang et al., 2008). Moreover, the dsRNA-binding domain is necessary for the virulence of VACV upon infection of mice (Brandt and Jacobs, 2001). E3L can also be an important host range gene since its deletion renders some poxviruses unable to replicate in cells derived from certain animal species. For example, VACV with a deletion of E3L can no longer replicate in HeLa or Vero cells (Beattie et al., 1996) and Myxoma virus relies on its E3 ortholog to replicate in cells from diverse species (Rahman et al., 2013).
The protein encoded by the K3L gene (termed K3) displays sequence homology to the eukaryotic translation initiation factor 2 alpha (eIF2α) and – similar to E3 – acts to dampen the downstream effects of PKR (Beattie et al., 1991; Carroll et al., 1993). Phosphorylation of eIF2α by activated PKR results in blunted cap-dependent translation in the cell. The most accepted model for the function of K3 is that it acts as a decoy of eIF2α to compete for binding to PKR (Davies et al., 1992). Therefore, K3 likely acts to prevent the cessation of host cell translation. Most orthopoxviruses, including VACV, possess a gene within the K3L family (Bratke et al., 2013). Interestingly, ECTV does not encode a full-length version of K3 because its open reading frame contains a premature stop codon and two deletions, thus yielding a highly truncated protein product that is predicted to be non-functional (Bratke et al., 2013; Chen et al., 2003; Smith and Alcami, 2002).
In this study, we demonstrate that VACV infection of BS-C-1 cells, which are derived from a non-human primate, leads to the accumulation of considerably more dsRNA than ECTV. The ability to detect dsRNA with a monoclonal antibody is likely impacted by the presence of E3. Our data point to several mechanisms that impact the prevalence of dsRNA. Relative to VACV, we find that ECTV displays a slower replication cycle, generally lower mRNA abundance, and less transcriptional read-through for a subset of its post-replicative genes. ECTV may generate less dsRNA to avoid innate immune detection as a result of containing a defunct K3L gene.
Materials and methods
Cells and culture methods
The following wild-type cell lines were used in this study: BS-C-1 (ATTC# CCL-26) and RK13 (ATTC# CCL-37). Two cell types were also utilized that constitutively expressed the E3 protein of VACV: BS-C-1+E3L and RK13+E3L (both generously supplied by Dr. Stefan Rothenburg). The RK13+E3L cells have been previously described in published reports (Hand et al., 2015; Rahman et al., 2013). The BS-C-1+E3L cells were constructed in a similar manner. However, unlike the E3 protein produced in the RK13+E3L cells, E3 in the BS-C-1 cells has mCherry fused at the C-terminus to aid in visualization using fluorescence microscopy. Both E3-expressing cell lines equivalently rescue the replication of VACVΔE3L (data not shown). Both of these stably transfected cells were maintained in the continuous presence of 500 µg/mL Geneticin/G418 (Gemini BioProducts) to preserve high E3 levels, which were consistently ≥90% positive as determined by flow cytometry. All cell lines were cultured in Dulbecco’s Modified Eagle Medium (DMEM; Invitrogen) supplemented with 10% fetal bovine serum (FBS; Gemini BioProducts), and penicillin-streptomycin (Gemini BioProducts). The DMEM contained 4mM L-glutamine, 1 mM sodium pyruvate, and 4.5 mg/mL D-glucose from the manufacturer. All cells were maintained at 37°C in a 6% CO2 incubator and split when they reached approximately 80–90% confluency. In some experiments, cells were kept at 30°C and 6% CO2.
Viruses
The following viruses were used during the course of this work: ECTV wild-type (Moscow strain; gift of Dr. Laurence Eisenlohr), ECTV expressing GFP [Moscow background; gift of Dr. Luis Sigal (Fang et al., 2008)], ECTV containing the VACV E3L gene, VACV wild8 type (Western Reserve strain; gift of Dr. Laurence Eisenlohr), VACV expressing GFP (Western Reserve background; BEI resources NR-624), VACVΔE3LΔK3L (Copenhagen background), and Cowpox virus (CPXV) expressing GFP (gift of Dr. Grant McFadden). VACVΔE3LΔK3L, kindly provided by Dr. Stefan Rothenburg, was constructed using standard homologous recombination techniques. This mutant virus expresses green fluorescent protein (GFP); more detailed information concerning this virus can be found in a previously published report (Brennan et al., 2014). We constructed a mutant ECTV (Moscow background) that contains the E3L gene of VACV (Western Reserve) in place of its native E3L. This mutant, labeled as ECTVΔE3L+VACV E3L, was constructed using ECTVΔE3L, which contained the coding sequence for GFP in place of the E3L gene (data not shown). A plasmid was constructed using the GeneArt Synthesis service (Invitrogen) that contained the sequence of VACV E3L surrounded by approximately 500 bp of sequence upstream and downstream of the ECTV E3L locus. After the initial infection/transfection stage, a GFP-negative plaque was isolated and subsequently underwent four rounds of passage at which point 100% of plaques did not express GFP. The E3L gene of this virus was sequenced (data not shown) and displayed the desired sequence for VACV E3L. In some experiments, cytosine β-D-arabinofuranoside (AraC; Sigma-Aldrich) was used at 50 #g/mL to block viral DNA replication and prevent the transcription of intermediate and late genes. All wild-type viruses (plus or minus GFP) were titered using normal BS-C-1 cells. VACVΔE3LΔK3L was titered using BS-C-1+E3L cells.
Fluorescence microscopy
Fluorescence microscopy was carried out using a Zeiss Axiostar plus epifluorescence microscope and images were captured with an Optronics camera system. All images were prepared for publication using ImageJ software. Cells were grown in four-well chamber slides (Thermo Scientific Nunc Lab-Tek II), infected with either ECTV or VACV (MOI=10), and fixed at the indicated time points with 5% formalin for 10 min. at room temperature. For virus factory analysis, glass coverslips were then immediately mounted onto the slide with ProLong Gold antifade reagent with 4’-6-diamidino-2-phenylindole (DAPI; Invitrogen) and allowed to cure overnight prior to visualization and analysis. For dsRNA staining, cells were fixed, permeabilized in acetone (Fisher Scientific) at −20° C for 10 min., incubated in blocking buffer [2% bovine serum albumin (BSA; Gemini BioProducts) and 2% FBS (Gemini BioProducts) in 1× PBS] for 45 min., and then incubated for an additional 45 min. with anti-dsRNA monoclonal antibody (J2 clone; Scicons). After washing with 1× PBS, Alexa Fluor 568 goat anti-Mouse IgG2a (Invitrogen) diluted in blocking buffer was applied for 35 min. Glass coverslips were then mounted as described above with ProLong Gold antifade reagent containing DAPI. For surface B5 staining, unpermeabilized cells were incubated - after fixation - in blocking buffer for 45 min. and then an anti-B5 monoclonal antibody (BEI resources NR-553; clone VMC-22) was added for 45 min. After washing with 1× PBS, Alexa Fluor 568 goat anti-Mouse IgG1 (Invitrogen) diluted in blocking buffer was applied to cells for 35 min. Finally, glass coverslips were mounted with ProLong Gold antifade reagent with DAPI. In some experiments, RK13 and RK13+E3L cells grown in chamber slides were transfected with 1 µg of Poly(I:C) (Sigma-Aldrich) prior to staining for dsRNA, as described above. As a control, the cells were also transfected separately with Poly(I:C) conjugated to fluorescein (InvivoGen). All transfections were carried out using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s instructions.
Flow cytometry
Flow cytometry data were collected using a FACSCalibur instrument (BD Biosciences) equipped with red and blue lasers. A minimum of 50,000 total cells were collected for each sample. The acquired data were analyzed using FCS Express 4 Flow Cytometry (De Novo Software; version 4.07). To measure the levels of dsRNA, infected cells (MOI=10) were harvested via trypsin digestion and washed once with 1× PBS containing 1% BSA (Gemini BioProducts). Cells were then fixed and permeabilized for 20 min. using the Cytofix/Cytoperm kit (BD Biosciences). After this fixation step, cells were washed twice with 1× Perm/Wash Buffer (BD Biosciences) before adding the J2 anti-dsRNA monoclonal antibody (Scicons). After 45 min. of incubation at room temperature, the cells were washed once with 1× Perm/Wash Buffer followed by the addition of anti-mouse IgG2a APC (eBioscience) for 45 min. A final wash with 1× Perm/Wash Buffer was performed prior to the addition of 2% paraformaldehyde (Electron Microscopy Sciences).
Plaque reduction assay
Plaque reduction assays were performed to determine the effect of isatin beta-thiosemicarbazone (IBT) on virus growth and subsequent plaque formation. Cells were seeded in six-well plates, allowed to incubate overnight, and were confluent the following day. The cells were then infected with approximately 75–100 plaque forming units of either ECTV or VACV in growth media containing a reduced FBS concentration (2% instead of 5%). After 1.5 hours to allow for virus adsorption and entry, the infection media was exchanged for standard growth media. IBT was then added to the cells at the following concentrations: 0, 1, 5, 10, 15, and 45 µM. Plaques were visualized using a 0.5% crystal violet solution after two days of incubation for VACV and five days for ECTV.
RNase L activity assay
BS-C-1 or BS-C-1+E3L cells were plated in 6-well culture plates one day prior and in sufficient numbers so as to be 80–90% confluent at the time of infection. Cells were either infected (MOI=5) with the indicated viruses or mock treated. Total RNA was isolated at the indicated times post-infection using the PureLink RNA Mini kit (Invitrogen) following the manufacturer’s instructions. RNA electrophoresis was performed using a “bleach gel” (Aranda et al., 2012), which contained 1% agarose and 1% bleach (Clorox; 6% sodium hypochlorite) in a TBE buffer-based gel. Each lane contained 4 µg of RNA. The gel was stained with ethidium bromide (2 µg/ml) to visualize the 28S and 18S rRNA bands.
Real-time PCR
BS-C-1 cells were plated in 6-well culture plates one day prior and in sufficient numbers so as to be 80–90% confluent at the time of infection. Cells were either infected (MOI=5) with ECTV or VACV or mock treated. Total RNA was isolated at the indicated times post-infection using the PureLink RNA Mini kit (Invitrogen) following the manufacturer’s instructions. A total of 2 µg of RNA was treated with 2U of RQ1 RNase-free DNase (Promega Corporation) according to manufacturer’s instructions. Then 1 µg of DNase-treated RNA was converted to cDNA using the High Capacity RNA-to-cDNA kit (Applied Biosystems) according to the manufacturer’s instructions. qPCR was carried out in 20 µl reactions using the Roche Light-Cycler 96 instrument. Each reaction contained 10 µl of 2× Essential Green Master Mix (Roche), 0.5 µM forward and reverse primers, and 0.2 µl of cDNA. qPCR was initiated with one cycle of 600 sec. at 95°C, followed by 45 cycles of 10 sec. at 95°C, 10 sec. at 58°C and 10 sec. at 72°C. This was followed by a melt curve to determine the Tm of each amplicon. F17R transcripts were detected using the following primer pairs: F17R-Forward (5’-TAG ATA AAC CCT CAT CGC CCG-3’) and F17R-Reverse (5’- TTT GTA GCA TGT CCG TCC TCA -3’). H4L transcripts were detected using the following primer pairs: H4L-Forward (5’- TGC GAG AAG GGA TCG TTG G-3’) and H4L-Reverse (5’- TGG GTT CTA ACG CCG ATC TC -3’). GAPDH was used as the internal control and for standardization among samples in a similar manner as Arndt, et al. (Arndt et al., 2016). Pre-designed primers to detect GAPDH transcripts from rhesus monkey were purchased from Bio-Rad (qMccCID0018506).
RNA Dot Blot
BS-C-1 cells were infected (MOI=10) with ECTV or VACV. At 24 hrs. post-infection, total RNA was isolated using the RNeasy Plus Mini Kit (Qiagen) and 5 µg RNA was spotted on a Zeta-Probe GT Membrane (Bio-Rad). RNA was fixed to the membrane by UV irradiation using the Stratalinker UV Crosslinker 1800 (Stratagene). For dsRNA detection, the membrane was incubated in RNase free Tris-buffered saline containing 0.1% Tween20 (TBS/T) and 1% acetylated bovine serum albumin (Sigma-Aldrich Chemie GmbH) for one hour and further incubated in 1% Ac-BSA/TBS/T containing the dsRNA-binding antibody J2 for 1 hour. After washing three times with TBS/T, the membrane was incubated in 1% Ac-BSA/TBS/T containing HRP goat anti-mouse IgG (Jackson ImmunoResearch Laboratories) for 1 hour and then washed three times with TBS/T. Finally, the membrane was incubated with Clarity Western ECL Blotting Substrate (Bio-Rad) for 5 min. Chemiluminescent signals were detected by using a ChemiDoc MP Imaging System (Bio-Rad) and analyzed by ImageLab software version 5.0 (Bio-Rad). For specific RNA detection, 3 µg total RNA was dissolved in 10 mM NaOH, 1 mM EDTA and spotted on a Zeta-Probe GT Membrane (Bio-Rad). Then the spots were washed with Low Stringency Wash Solution from the NorthernMax-Gly Kit (Thermo Fisher Scientific), which is equivalent to 2 × SSC, 0.1 % SDS. Fixation of RNA to the positively-charged nylon membrane and subsequent detection of specific RNA was performed as described for Northern Blot below.
Generation of Biotin-labeled RNA probes
Isolation of total RNA from MVA, conversion to cDNA, design of primer pairs, synthesis of oligonucleotides and RT-PCR were carried out as described previously (Price et al., 2014). PCR for H4L were performed with DNA from ECTV. Sequences of oligonucleotides and PCR product sizes are listed in Table 1. PCR products from E3L, 078R/G8R, 047R/F17R, and H4L were cloned into the TA-vector pCR2.1 (Invitrogen). Plasmids were linearized with the restriction enzyme SpeI and subsequently purified by phenol/chloroform extraction, precipitated with ethanol, and dissolved in RNase free water. These plasmids were then used to produce biotin-labeled RNA probes using the AmpliScribe T7-Flash Biotin-RNA Transcription Kit (Epicentre) according to the manufacturer’s instructions. To synthesize the β-actin RNA probe, cDNA from U937 cells (ECACC 85011440) was amplified using the sense primer 5’-CCT CGC CTT TGC CGA TCC-3’ and the antisense primer 5’-GGA TCT TCA TGA GGT AGT CAG TC- 3’ yielding a PCR product with a size of 626 bp (Raff et al., 1997). This PCR product was cloned in sense direction into pT-Adv (Clontech). The plasmid pT-Adv-β-actin was linearized with the restriction enzyme BstEII and used as template to produce a 103 bp long biotinylated β-actin antisense RNA by in vitro transcription, which was performed as previously described (Lehmann et al., 2001).
Table 1.
Summary of gene sizes and oligonucleotides used to produce RNA probes for Northern blot analysis.
| Target gene | Size of ORF (bp) |
Primers | oligonucleotide sequence (5’ to 3’) | product size (bp) |
|---|---|---|---|---|
| MVA/VACV/ECTV 050L/E3L/EVM043 | 573 | E3L-F | TTACTAGGCCCCACTGATTC | 406 |
| E3L-R | GTTCTGACGCAGAGATTGTG | |||
| MVA/VACV/ECTV 078R/G8R/EVM070 | 783 | G8R-F | AATGTAGACTCGACGGATGAG | 484 |
| G8R-R | ACGCATCAGTATTACCAGGAG | |||
| MVA/VACV/ECTV 047R/F17R/EVM040 | 306 | F17R-F | CATTTTGCATCTGCTCATACTC | 262 |
| F17R-R | TAGCCGCGAACATATTTTGTAG | |||
| MVA/VACV/ECTV 094L/H4L/EVM086 | 2,388 | H4L-F | TAGAATTTGCGAGAAGGGATC | 341 |
| H4L-R | GTCACACGTTAGCTCTTTGAAG |
Northern Blot
BS-C-1 cells were infected at the indicated MOI with ECTV or VACV or mock treated. Total RNA was isolated at the indicated times post-infection using the RNeasy Plus Mini Kit (Qiagen), mixed with an equal volume of Glyoxal loading dye, and heated at 50°C for 1 hour. Separation, blotting, and hybridization of RNA were performed with the NorthernMax-Gly Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Briefly, 5 µg total RNA was separated using a 1.2% agarose gel. Equal loading of RNA for all lanes in all experiments was confirmed via ethidium bromide visualization of the 28S and 18S rRNA bands (data not shown). The gel was blotted onto an Ambion BrightStar-Plus positively-charged nylon membrane by downward capillary transfer. RNA was fixed to the membrane by UV irradiation using the Stratalinker UV Crosslinker 1800 (Stratagene). Hybridization with specific Biotin-labeled RNA probes were performed in ULTRAhyb buffer at 68°C for 14 hours. After washing, the nylon membrane was further processed with the North2South Chemiluminescent Hybridization and Detection Kit (Thermo Fisher Scientific). Positive signals were detected by a ChemiDoc MP System and analyzed using Image Lab software (Bio-Rad).
Results
VACV produces higher levels of detectable dsRNA than ECTV
While closely related, ECTV and VACV differ greatly in host range and in vivo pathogenesis and result in distinct levels of inflammation upon infection of mice via the footpad route (Siciliano et al., 2014). Additionally, ECTV – unlike VACV – does not express a functional K3 protein (Smith and Alcamí, 2002). Since dsRNA is a potent pathogen-associated danger signal to the mammalian immune system, we hypothesized there might be detectable differences in the formation of this molecule in cells infected by these two viruses. To measure levels of dsRNA within infected cells, we made use of the well-characterized mouse monoclonal antibody named J2 that specifically recognizes dsRNA but not ssRNA or DNA (Burgess and Mohr, 2015; Schönborn et al., 1991; Weber et al., 2006).
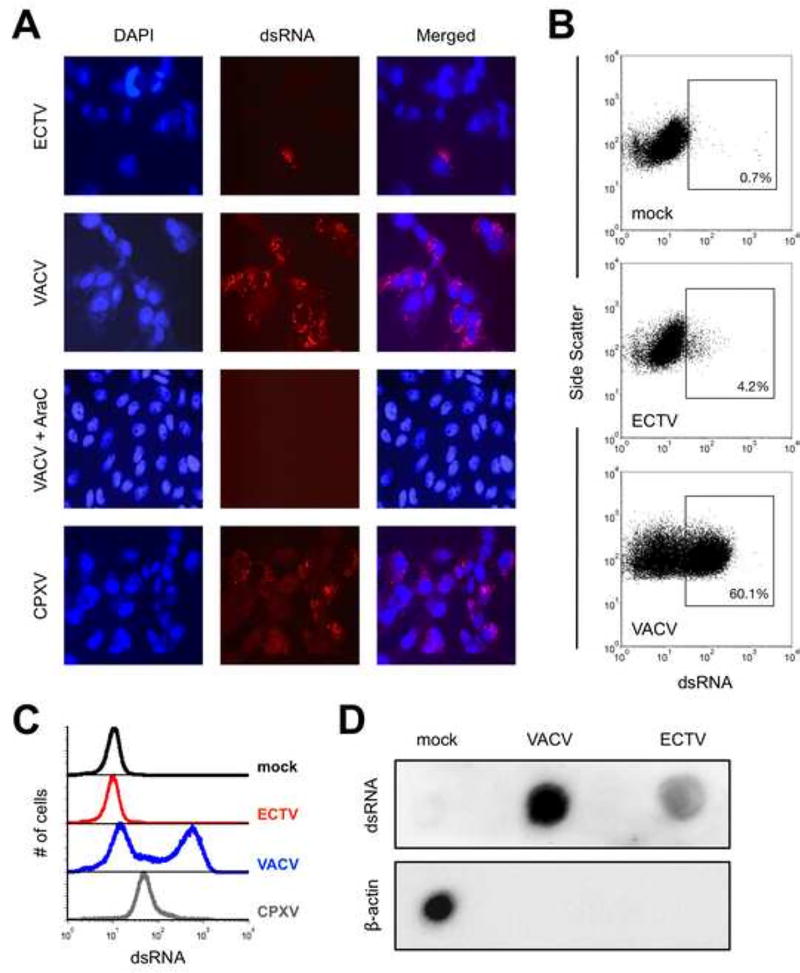
We began by examining dsRNA levels in BS-C-1 cells that had been infected for 24 hrs. with either ECTV or VACV. Using fluorescence microscopy, we found that only a small fraction of ECTV-infected cells displayed dsRNA staining whereas the majority of VACV-infected cells were dsRNA-positive (Fig. 1A). Similar results were also obtained using Vero cells (data not shown). Most of the dsRNA colocalized with viral factories as identified via cytoplasmic DAPI staining (Fig. 1A). Treatment of VACV-infected cells with AraC, which is known to inhibit DNA replication and subsequent intermediate and late gene transcription, abrogated our ability to detect dsRNA (Fig. 1A). This suggests that formation of dsRNA was mostly the result of post-replicative gene transcription, consistent with prior studies (Colby and Duesberg, 1969; Duesberg and Colby, 1969).
Figure 1. The accumulation of dsRNA greatly varies after infection with different orthopoxviruses.
(A) BS-C-1 cells were infected (MOI=10) with ECTV, VACV, or CPXV for 24 hrs. prior to staining for dsRNA (red) and cell nuclei/virus factories (DAPI; blue). Images were merged using ImageJ software, are at 400× total magnification, and representative of three independent trials. AraC was added at the time of infection where indicated. (B) BS-C-1 cells were infected (MOI=10) with either ECTV or VACV for 24 hrs. prior to staining intracellularly for dsRNA and analyzed using flow cytometry. Positive gates were drawn based upon the mock condition. The depicted data are representative of four independent trials. (C) Flow cytometry was used to measure dsRNA levels within infected cells as above. The data are shown in histogram format and representative of four independent trials. (D) BS-C-1 cells were infected (MOI=10) with ECTV or VACV. RNA was isolated at 24 hrs. post-infection using the RNeasy Plus Mini Kit (Qiagen) and 5 µg RNA was spotted onto a membrane. dsRNA was detected using the J2 antibody and beta-actin mRNA was detected using a biotin-labeled antisense RNA as described in the methods.
Additionally, we investigated the formation of dsRNA using flow cytometry similar to Arndt and colleagues (Arndt et al., 2016). In accordance with the fluorescence microscopy data, we consistently observed increased accumulation of dsRNA in VACV-infected BS-C-1 cells compared with ECTV (Figs. 1B and 1C). In order to verify the formation of dsRNA in a protein-free environment, we next carried out an RNA dot blot using purified total RNA from infected cells and from mock-infected cells as control. Using the J2 antibody, we found that RNA isolated from VACV-infected cells contained substantially more dsRNA compared to ECTV-infected cells (Fig. 1D). Importantly, this result conclusively demonstrated that ECTV formed less dsRNA in an independent assay. As previously shown for VACV (Rice and Roberts, 1983), host beta-actin mRNA was completely absent from the pool of RNA derived from cells infected with either virus (Fig. 1D).
Cowpox virus also produces higher levels of detectable dsRNA than ECTV
All identified orthopoxviruses and clade II poxviruses have been found to possess a gene in the K3L family except for ECTV and monkeypox virus (MPXV) (Bratke et al., 2013). A recent report by Arndt, et al. demonstrated that MPXV produced considerably less dsRNA compared to VACV (Arndt et al., 2016). We find it to be an noteworthy observation that both ECTV and MPXV have been shown [in this work and (Arndt et al., 2016), respectively] to accumulate significantly less dsRNA during their replication cycles relative to VACV. Perhaps poxviruses that contain a functional K3L ortholog – in addition to a dsRNA-binding protein, such as E3 – have less selective pressure to constrain the formation of dsRNA. Therefore, we hypothesized that cowpox virus (CPXV), a K3L-containing orthopoxvirus similar to VACV, also produces detectable amounts of dsRNA in our assay format. Indeed, we detected higher levels of dsRNA in CPXV-infected cells compared to ECTV-infected cells (Fig. 1A). However, while dsRNA was detected in most CPXV-infected cells, the quantity on a per-cell basis (i.e., median fluorescence intensity, MFI) was reduced compared to VACV (Fig. 1C). Thus, the presence of a K3 ortholog correlates with increased dsRNA detection relative to orthopoxviruses lacking this protein.
ECTV demonstrates protracted infection kinetics relative to VACV
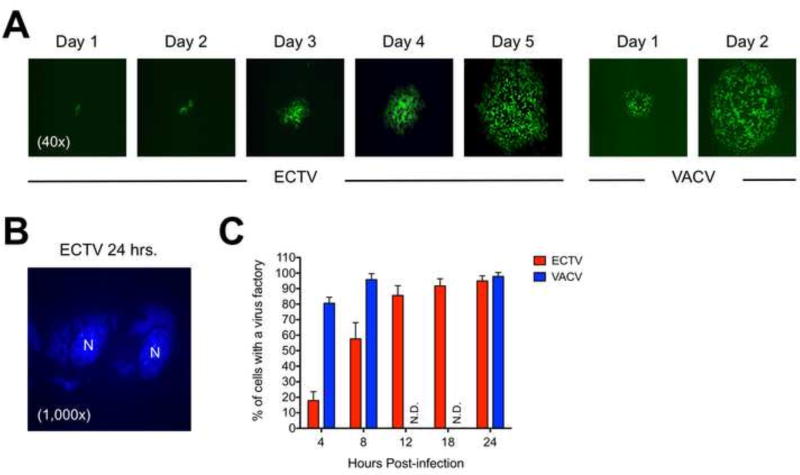
It has been known for some time that VACV plaques can be visualized by standard crystal violet staining of cell monolayers by 48 hrs. post-infection (Earl et al., 2001) but often at earlier time points as well, such as 36 hrs. (Roper, 2006). Conversely, ECTV plaques are commonly not visualized until after a minimum of 4 or 5 days of incubation (Chen et al., 1992; Hand et al., 2015; Lynn et al., 2012; Panchanathan et al., 2005). To demonstrate the differences in plaque formation kinetics between ECTV and VACV, representative images are shown in Figure 2A for GFP-expressing viruses. We also examined the rate of virus factory formation using DAPI staining to visualize the appearance of cytoplasmic viral DNA (representative stain shown in Fig. 2B). At only 4 hrs. post-infection, approximately 80% of VACV-infected cells contained discernible virus factories whereas less than 20% of ECTV-infected cells did so at this time point (Fig. 2C). It was not until about 12 hrs. post-infection that factory formation for ECTV was equivalent to the levels observed with VACV. Overall, these findings are similar to a prior report (Lynn et al., 2012). Therefore, ECTV exhibits delayed replication kinetics relative to VACV, which may have marked effects on viral dsRNA accumulation within cells.
Figure 2. ECTV exhibits delayed plaque formation and genome replication relative to VACV.
(A) Confluent monolayers of BS-C-1 cells were initially infected at a low MOI (~0.001) to allow for proper visualization of plaque development over time. The otherwise wild-type viruses were engineered to express GFP. The images were acquired using an epifluorescence microscope. All images are at 4× total magnification and representative of ECTV and VACV plaques at each time point. (B) A representative image showing the presence of cytoplasmic virus factories visualized by DAPI staining. Cell nuclei are labeled as “N” in the image, which is at 1,000× total magnification. (C) BS-C-1 cells were infected (MOI=10) with either ECTV or VACV for various lengths of time. For each time point, 150 randomly viewed cells within each infection condition were scored as either positive or negative for the presence of a virus factory of any size. The bars depict the average values and the error bars represent the standard deviations of two independent experiments. N.D., not determined.
VACV infection at 30°C displays a slower replication cycle and less dsRNA accumulation relative to growth at 37°C
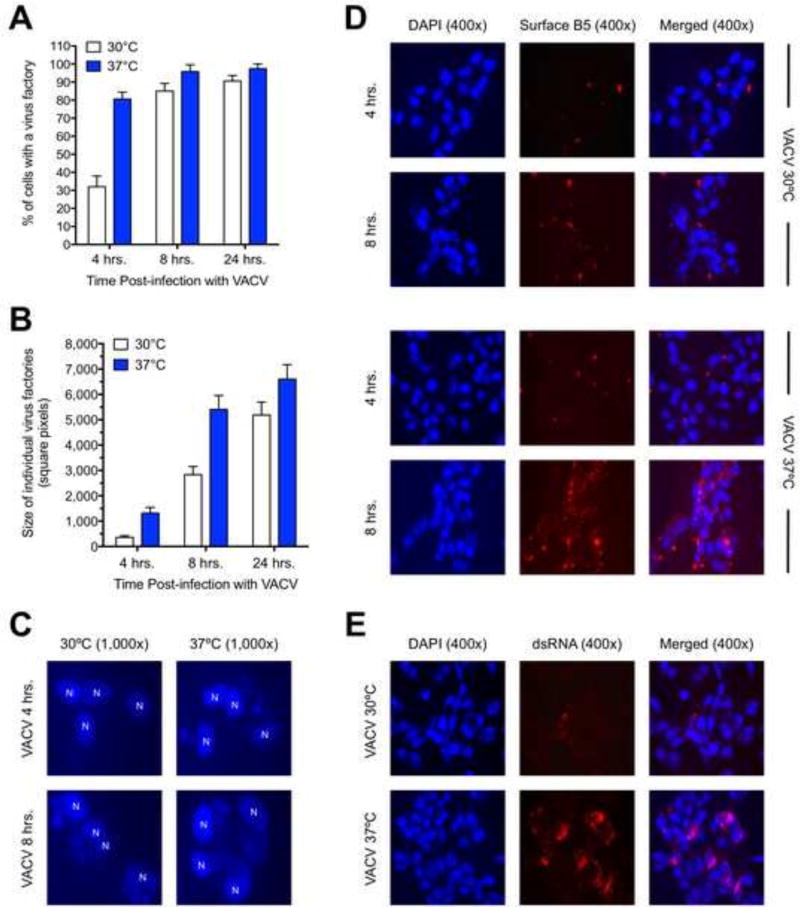
We hypothesized that one reason ECTV forms less dsRNA was due to its delayed replication kinetics in BS-C-1 cells compared with VACV. To address the effect of replication rate on dsRNA accumulation, we attempted to artificially slow down the replication cycle of VACV by performing infections at 30°C. By performing an infection time course, we found that both the manifestation (Fig. 3A) and size (Fig. 3B) of virus factories were significantly delayed by incubating VACV at 30°C compared to 37°C. Representative images of virus factories obtained via DAPI staining are shown in Figure 3C. We also examined cell surface expression of the viral B5 protein, which is a late (i.e., post-replicative) infection event since treatment of cells with AraC prevented positive staining (data not shown). Consistent with the virus factory data, we observed a marked delay in the appearance and relative abundance of cell surface B5 when VACV infections were performed at 30°C (Fig. 3D). The lower temperature also had an effect on the overall size of virus plaques, which were noticeably smaller at 30°C (data not shown). Thus, infection of cells at a lower temperature caused a delay in the molecular events of the replication cycle and this also had an impact at the phenotypic level with respect to plaque formation. These data are consistent with a previously published report (Condit and Motyczka, 1981). Next, we examined dsRNA accumulation in VACV-infected BS-C-1 cells incubated at the two different temperatures. We witnessed a significant decrease in dsRNA staining at 30°C compared to 37°C (Fig. 3E). Therefore, our data suggest that the rate of progression through the replication cycle can have an impact on the amount of dsRNA produced by poxviruses.
Figure 3. VACV exhibits delayed replication kinetics and less dsRNA abundance when grown at 30°C compared to 37°C.
(A) BS-C-1 cells were infected (MOI=10) with VACV and then incubated at either 30°C or 37°C for the indicated times. For each condition, 150 randomly viewed cells were scored as either positive or negative for the presence of a virus factory of any size. The bars depict the average values and the error bars represent the standard deviations of three independent trials. (B) Using the same data from the previous figure panel, images of virus factories were acquired and then analyzed using ImageJ software. The total size of 25 randomly selected virus factories was determined for each condition. The bars depict the average values and the error bars represent the standard deviations. (C) Representative images of virus factories are shown for the 4 hr. and 8 hr. time points at both temperatures. Cell nuclei are labeled as “N” in the images, which are at 1,000× total magnification. (D) BS-C-1 cells were infected (MOI=10) with VACV and then incubated at either 30°C or 37°C for 4 or 8 hrs. Cells were stained for B5 protein at the cell surface (red) and cell nuclei/virus factories (DAPI; blue). Images were merged using ImageJ software, are at 400× total magnification, and representative of two independent trials. (E) BS-C-1 cells were infected (MOI=10) with VACV and then incubated at either 30°C or 37°C for 24 hrs. Cells were stained for dsRNA (red) and cell nuclei/virus factories (DAPI; blue). Images were merged using ImageJ software, are at 400× total magnification, and representative of three independent trials.
ECTV gene expression is lower and some gene transcripts are shorter relative to VACV
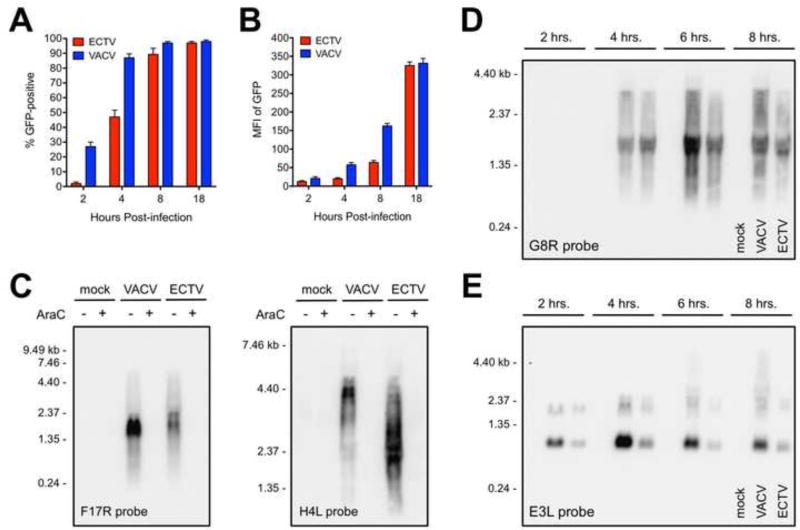
The virus factory data and plaque formation time course showed that ECTV has a delayed replication cycle relative to VACV. We next looked at more detail into the gene expression kinetics of these viruses. First, we made use of the recombinant versions of ECTV and VACV containing the GFP gene under the control of the same p7.5 early/late promoter. As shown in Figure 4A, the proportion of GFP-positive cells was delayed for ECTV. Equivalent levels of GFP positivity were not observed until 8 hrs. post-infection. The amount of fluorescence on a per-cell basis within ECTV-infected cells was also markedly lower compared to VACV (Fig. 4B). An equivalent MFI for GFP was not observed until after 12 hrs. post-infection.
Figure 4. ECTV exhibits reduced mRNA abundance and less read-through of some genes relative to VACV.
BS-C-1 cells were infected (MOI=10) with recombinant versions of ECTV and VACV containing the GFP gene under the control of the same promoter. At the indicated times post-infection, cells were isolated and analyzed for (A) GFP positivity and (B) median fluorescence intensity (MFI) using flow cytometry. The bars depict the average values and the error bars represent the standard deviations of three independent experiments. (C) Total RNA was isolated from infected (MOI=5) BS-C-1 cells at 18 hrs. post-infection. RNA probes specific for F17R (left panel) and H4L (right panel) were used in a Northern blot assay. AraC was included in some conditions at the time of infection. (D and E) Northern blot analysis of G8R and E3L transcripts was carried out in a time course format using a similar procedure as above. (C–E) Each lane contained 5 µg of total RNA. Data are representative of two independent trials. Sizes (in kilobases, kb) of the RNA standards are indicated.
Expression of two representative late genes, F17R and H4L (Yang et al., 2011), was assessed by Northern blot to determine the sizes and relative abundance of viral genes at 18 hrs. post-infection. The ECTV and VACV F17R transcripts were both about 2 kb in size, which is substantially longer than the predicted 306 bp open reading frame (Fig. 4C, left panel; Table 1). However, the relative amount of ECTV F17R transcripts was reduced compared to VACV. The lengths of the dominant ECTV H4L mRNAs corresponded well with the predicted 2,388 bp open reading frame (Fig. 4C, right panel; Table 1). Conversely, the main VACV H4L band was approximately 4.4 kb in size, thus indicating considerable read-through of this gene, as expected for a VACV post-replicative gene (Fig. 4C, right panel). In contrast to the F17R data, the abundance of H4L transcripts was similar between the two viruses at 18 hrs.
Previously it was shown that intermediate and even early RNA of VACV can form dsRNA (Ludwig et al., 2006; Willis et al., 2011). Therefore, expression of a representative intermediate gene, G8R, was assessed by Northern blot in a time course format. To synchronize the infections across multiple wells, cells were incubated at 4°C for 30 min. in the presence of virus (MOI=10). Warm medium (37°C) was then added to the plates before incubating them at 37°C. RNA isolations were performed at 2, 4, 6, and 8 hrs. post-infection. The analysis revealed that G8R was equivalently expressed in VACV- and ECTV-infected cells at 4 hrs. post-infection (Fig. 4D). The G8R transcripts were roughly 1.5 kb in size, which is twice the length of the predicted 783 bp open reading frame (Fig. 4D; Table 1). At 6 hrs., the levels of G8R mRNA strongly increased and additional longer transcripts were detected only in VACV-infected cells (Fig. 4D). These longer G8R transcripts remained present at 8 hrs. but were still largely absent from ECTV-infected cells (Fig. 4D). Finally, we examined the levels of E3L mRNA, which is expressed immediately after infection at early time points. In VACV-infected cells, E3L transcripts reached maximum levels at 4 hrs. post-infection and decreased thereafter (Fig. 4E). The relative amount of E3L mRNA in ECTV-infected cells at each time point was consistently lower compared to VACV (Fig. 4E). E3L mRNA was not detected for either virus at 18 hrs. (data not shown).
Our Northern blot data show that gene expression by ECTV was delayed and of lower abundance compared to VACV. The lower levels of gene expression were corroborated using real-time PCR analysis of F17R and H4L transcripts. We observed significantly lower levels of mRNA from both genes within ECTV-infected cells compared to VACV during the initial phase of infection. On average, ECTV F17R and H4L mRNA levels were approximately 3-fold and 8-fold lower than VACV at 4 hrs. post-infection, respectively (Supplemental Figure 1). Thus, the reduced mRNA abundance of many ECTV genes would be predicted to negatively impact the ability to form dsRNA.
The E3 protein may affect the ability of the J2 antibody to detect dsRNA
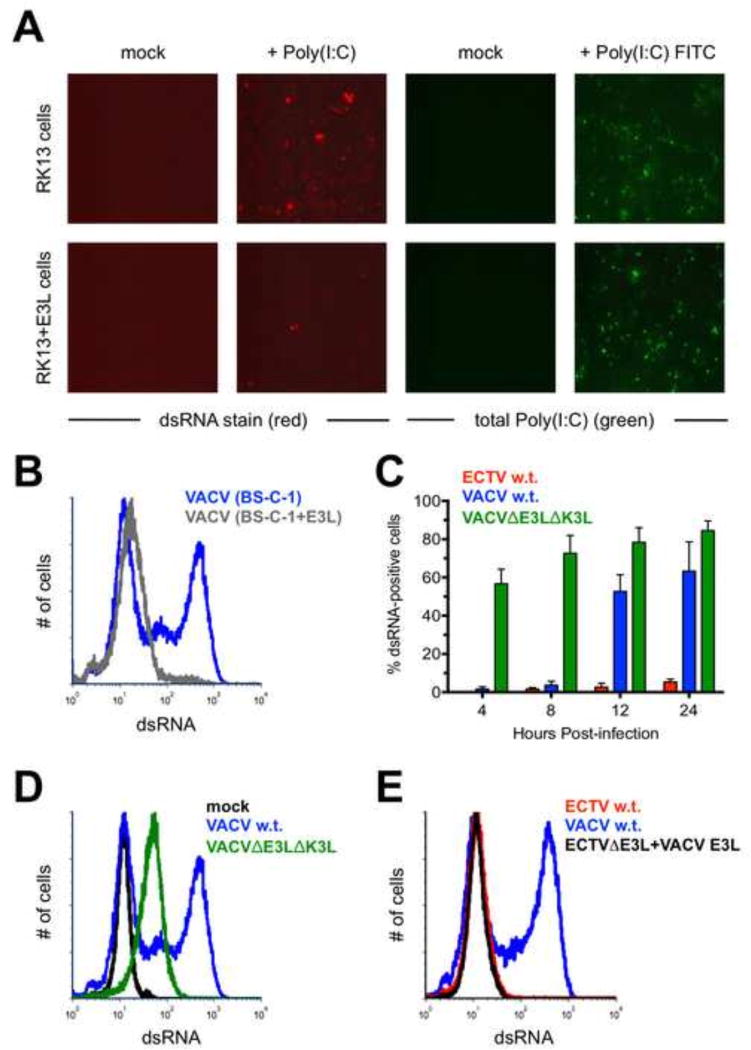
We next turned our attention to the E3 protein, which has been shown to bind to dsRNA in vitro. This protein was initially discovered because of the ability of extracts from VACV-infected cells to bind to Poly(I:C), a dsRNA mimic (Chang et al., 1992; Watson et al., 1991). It was subsequently shown to contain a carboxy-terminal dsRNA-binding domain (Chang and Jacobs, 1993). Due to the ability of E3 to bind to dsRNA, we hypothesized that it may be able to prevent binding of the J2 antibody to its epitope. We performed several experiments to address this point. First, we made use of wild-type RK13 cells and a version of this cell line that constitutively expresses VACV E3 (Rahman et al., 2013). We transfected 1 µg of Poly(I:C) into each cell type and stained for dsRNA using fluorescence microscopy. We found that the RK13+E3L cells displayed less dsRNA positivity than the parent cells (Fig. 5A). Equivalent transfection efficiency between the two cell lines was confirmed using Poly(I:C) conjugated to fluorescein (Fig. 5A). Using flow cytometry, we then examined dsRNA levels within wild-type BS-C-1 cells and BS-C-1+E3L cells infected by ECTV or VACV. We could not use the RK13 cells described above for this assay because rabbit cells are not permissive for ECTV (Hand et al., 2015). Consistent with the Poly(I:C) results above, the ability to detect dsRNA following infection with VACV was considerably blunted in the BS-C-1+E3L cells compared with the E3-negative parent cells (Fig. 5B). Thus, our data support the hypothesis that the J2 antibody is unable to detect dsRNA that is already bound by E3.
Figure 5. The E3 protein may prevent recognition of dsRNA by the J2 antibody.
(A) RK13 and RK13+E3L cells were transfected with 1 µg of Poly(I:C) or Poly(I:C) conjugated to fluorescein. The cells were stained for dsRNA (red) approximately 16 hrs. later. Images are at 400× total magnification and representative of two independent trials. (B) BS-C-1 or BS-C-1+E3L cells were infected (MOI=10) with either ECTV or VACV for 24 hrs. prior to staining for dsRNA. The flow cytometric data are depicted in histogram format and representative of three independent trials. (C) BS-C-1 cells were infected (MOI=10) with ECTV, wild-type VACV, or VACVΔE3LΔK3L for various lengths of time prior to staining for dsRNA using flow cytometry. The bars depict the average values and the error bars represent the standard deviations of three independent trials for each time point. (D) Representative flow cytometric data in histogram format are shown for BS-C-1 cells infected (MOI=10) with the indicated viruses. The cells were stained for dsRNA after 24 hrs. (E) BS-C-1 cells were infected (MOI=10) with VACV, wildtype ECTV, or a mutant ECTV in which the native E3L was replaced with the E3L gene from VACV. The cells were stained for dsRNA after 24 hrs. The flow cytometric data are depicted in histogram format and representative of two independent trials.
To further support this notion, we used a VACV mutant lacking the E3L gene. We speculated that this virus, which also had a deletion of the K3L gene (VACVΔE3LΔK3L), would produce higher levels of detectable dsRNA because the E3 protein was absent. By performing an infection time course, this is indeed what we observed (Fig. 5C). At 4 hrs. post-infection with VACVΔE3LΔK3L, approximately 50% of all BS-C-1 cells were dsRNA-positive and this amount increased at 8, 12, and 24 hrs. to reach about 85%. Conversely, we could not detect an appreciable dsRNA signal until 12 hrs. post-infection with wild-type VACV. A higher proportion of cells stained positive for dsRNA with the mutant VACV but the median fluorescence intensity was lower than the signal produced in cells infected with wild-type virus (Fig. 5D). This is likely because the replication cycle of VACVΔE3LΔK3L is blunted upon infection of non-E3 expressing cell lines (Hand et al., 2015), which resulted in lower overall levels of mRNA production. In summary, it appears that VACV produced dsRNA much earlier than 12 hrs. post-infection. However, the dsRNA was not detectable due to the presence of E3 and its apparent ability to mask recognition by J2. Additionally, it should be noted that we did not detect a considerable difference in the total amount of E3 protein within VACV- or ECTV-infected cells using flow cytometry or Western blotting (data not shown).
A BLAST comparison of E3 from ECTV and VACV revealed 91% sequence identity at the amino acid level. Since these orthologs are not identical, it is formally possible that ECTV E3 is somehow better able to prevent binding of J2 to dsRNA than VACV E3. In order to examine this possibility, we created an ECTV mutant containing the gene sequence for VACV E3L in place of the native E3L locus. This mutant was identical to wild-type ECTV with respect to the appearance of dsRNA following infection of BS-C-1 cells (Fig. 5E). Therefore, the disparity in dsRNA accumulation between infected cells was not due to the sequence differences between E3 from ECTV and VACV. Overall, our data demonstrate that E3 shields dsRNA from recognition by J2. The dsRNA detected in our assay setup is most likely in a state not bound by E3 (i.e., “free” dsRNA) or at least not completely saturated with this protein.
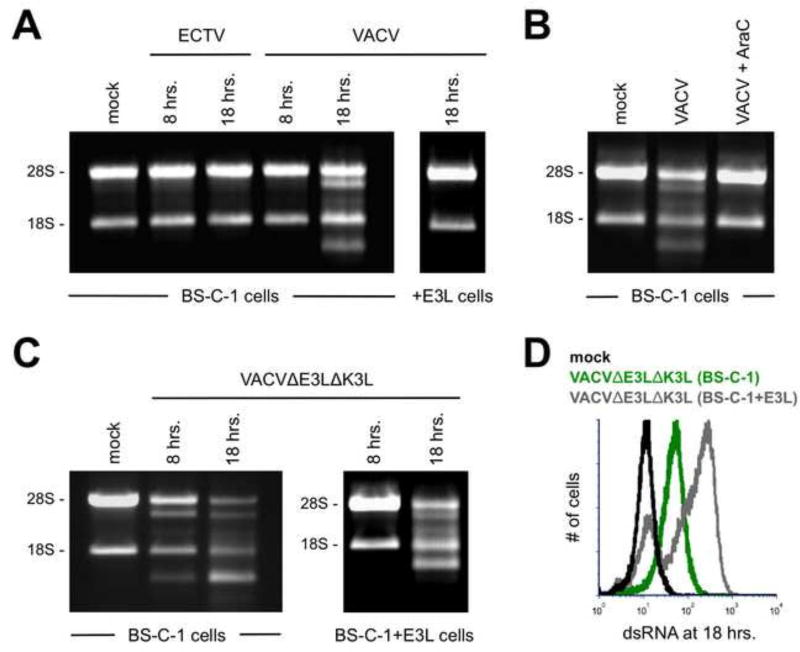
The 2’5’ OAS/RNase L pathway is activated at late time points only within VACV-infected cells
It is well established that dsRNA activates the 2’5’ OAS/RNase L pathway. This activation can be conveniently detected by monitoring the breakdown of rRNA via electrophoresis of total RNA isolated from cells (Silverman et al., 1983; Wreschner et al., 1981). Our data support a model by which free dsRNA is generated at late time points in the replication cycle of VACV – but not ECTV – in BS-C-1 cells. Therefore, we reasoned that this unbound dsRNA may be able to activate RNase L. As expected, we did not observe cleavage of rRNA in ECTV-infected BS-C-1 cells at any time point (Fig. 6A). Conversely, we did observe the breakdown of rRNA in VACV-infected cells at 18 hrs. post-infection (Fig. 6A). However, RNase L activity was not detected when BS-C-1+E3L cells were used (Fig. 6A). Importantly, RNase L activation correlated well with the time points at which dsRNA became detectable in VACV-infected cells (refer to Fig. 5C). Consistent with the loss of dsRNA staining upon treatment with AraC (Fig. 1A), the presence of this drug abrogated RNase L activation in VACV-infected cells (Fig. 6B). Infection of normal BS-C-1 cells with VACVΔE3LΔK3L predictably led to the activation of RNase L at both 8 hrs. and 18 hrs. post-infection (Fig. 6C). However, VACVΔE3LΔK3L infection of BS-C-1+E3L cells resulted in rRNA degradation only at the late time point (Fig. 6C). This was likely due to higher levels of dsRNA detected at 18 hrs. following VACVΔE3LΔK3L infection of BS-C-1+E3L cells relative to normal BS-C-1 cells (Fig. 6D). It appears that the replication capacity of VACVΔE3LΔK3L is rescued in BS-C-1+E3L cells such that the mutant replicates in a similar manner in the complementing cell line to how wild-type VACV replicates in normal BS-C-1 cells. In summary, we observed the activation of RNase L only at time points and for infection conditions that displayed detectable dsRNA using the J2 antibody. Notably, these data also provide additional confirmation (albeit indirect) that VACV produces higher amounts of dsRNA than ECTV at late time points in the replication cycle. This dsRNA is in a form (i.e., likely not bound by E3) that can trigger RNase L.
Figure 6. RNase L activation is observed only in VACV-infected cells at late time points.
(A) BS-C-1 or BS-C-1+E3L cells were infected (MOI=5) with either ECTV or VACV. Total RNA was isolated at the indicated time points and separated using a 1% bleach TBE gel. Each lane contained 4 µg of RNA. The data are representative of three independent trials. (B) BS-C-1 cells were infected (MOI=5) with VACV either in the presence of absence of AraC. RNA breakdown was detected as above at 18 hrs. post-infection. (C) BS-C-1 or BS-C-1+E3L cells were infected (MOI=5) with VACVΔE3LΔK3L. RNA breakdown was detected as above at the indicated time points. (D) BS-C-1 or BS-C-1+E3L cells were infected (MOI=10) with VACVΔE3LΔK3L for 18 hrs. prior to staining for dsRNA. The flow cytometric data are depicted in histogram format and representative of two independent trials.
ECTV and VACV are similarly sensitive to the anti-poxvirus drug IBT
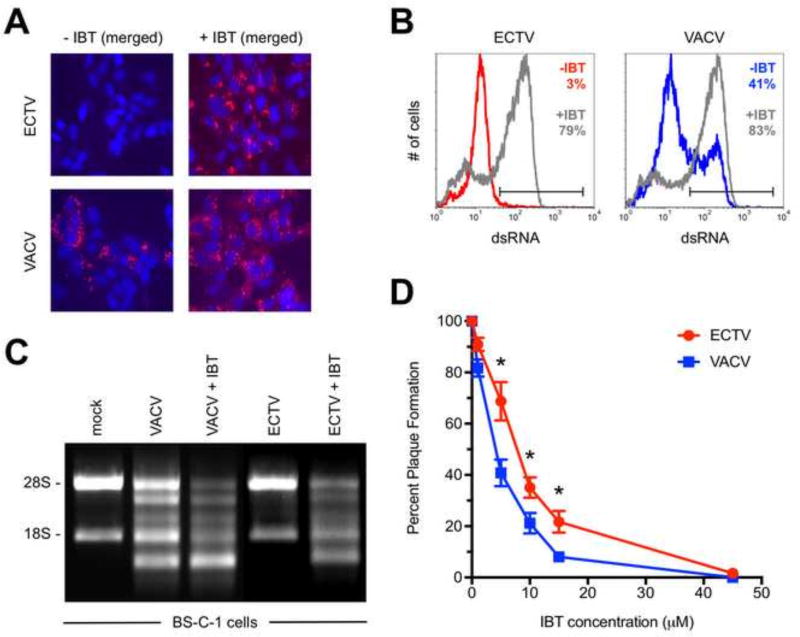
Prior work has shown that treatment of VACV-infected cells with IBT leads to elongated viral transcripts during intermediate and late time points, which leads to greater formation of dsRNA (Bayliss and Condit, 1993; Prins et al., 2004). Therefore, IBT treatment should lead to higher dsRNA levels in our assay. Indeed, we observed a significant increase in the frequency of dsRNA-positive cells following infection with VACV in the presence of 45 µM IBT (Figs. 7A and 7B), which led to an increase in RNase L activity in VACV-infected cells (Fig. 7C). Interestingly, we also found a dramatic increase in dsRNA staining for ECTV-infected cells that had been IBT treated (Figs. 7A and 7B). We were able to detect rRNA breakdown in ECTV-infected cells in the presence of IBT whereas no RNase L activity was observed in the absence of the drug (Figs. 6A and 7C).
Figure 7. Both ECTV and VACV are sensitive to the anti-poxvirus drug IBT.
(A) BS-C-1 cells were infected (MOI=10) with either ECTV or VACV for 24 hrs. prior to staining for dsRNA (red) and cell nuclei/virus factories (DAPI; blue). IBT (45 µM) was added at the time of infection. Due to space constraints, only the merged images are shown, which were prepared using ImageJ software. All images are at 400× total magnification and representative of three independent trials. (B) BS-C-1 cells were infected (MOI=10) with either ECTV or VACV for 24 hrs. prior to staining for dsRNA. IBT (45 µM) was added at the time of infection. The flow cytometric data are depicted in histogram format and representative of three independent trials. Percentages represent the fraction of cells within the positive gate. (C) RNase L activation assays are shown for BS-C-1 cells infected (MOI=5) with ECTV or VACV either in the presence of absence of 45 µM IBT. Total RNA was isolated at 18 hrs. post-infection and run on a 1% bleach TBE gel. The data are representative of two independent trials. (D) Plaque reduction assays were performed to determine the effect of IBT on virus replication. Confluent monolayers of BS-C-1 cells in six-well plates were infected with approximately 50 plaque forming units of either ECTV or VACV. IBT was added to the cells at the indicated concentrations. Plaques were visualized using a 0.5% crystal violet solution after two days of incubation for VACV and five days for ECTV. The indicated percentages are relative to the no drug control wells. Symbols depict the average values and each error bar represents the standard deviation of the mean. The data are an assembly of eight independent assays. Statistical analysis [performed using GraphPad Prism software (version 7.0a)] was carried out using the Mann-Whitney test. * denotes a p value <0.05.
Since decreased amounts of dsRNA during VACV infection has been associated with resistance to IBT (Cresawn et al., 2007; Prins et al., 2004), we determined if ECTV was more resistant to this drug than VACV by performing plaque reduction assays. ECTV was slightly more resistant to IBT at low concentrations of the drug but both viruses were fully inhibited at 45 µM (Fig. 7D). A recent study reported that MPXV formed less dsRNA than VACV and was naturally resistant to IBT (Arndt et al., 2016). This group found that a much higher amount of IBT (120 µM) was required to fully inhibit MPXV in a similar assay format. Therefore, ECTV and MPXV both form less detectable dsRNA than VACV but the mechanisms underlying this phenomenon may be distinct.
Discussion
In this study, we show that ECTV forms less dsRNA than VACV after infection of BS-C-1 cells. We plan to examine cells derived from other species in the future. We believe that the reduced dsRNA production is likely a compensatory mechanism because ECTV does not produce a full-length, functional K3 protein (Chen et al., 2003; Smith and Alcami, 2002). The deduced protein sequence for the now defunct ECTV K3 is quite similar to VACV K3 (Bratke et al., 2013) and the latter has been shown to be a potent inhibitor of mouse PKR (Rothenburg et al., 2009). Therefore, if the K3L gene of ECTV was not mutated, the encoded protein product would likely inhibit mouse PKR. While it is true that ECTV lacks other known host range genes (Bratke et al., 2013), the link between dsRNA and PKR is straightforward and well established (Langland et al., 2006). Hence, by lacking K3, ECTV seemingly relies on E3 as its main defense against PKR activation. Conversely, most other orthopoxviruses have a “two-pronged” approach against PKR by producing both E3 and K3. By accumulating lower levels of dsRNA, there is less of this potent innate immune trigger within ECTV-infected cells, which likely made the loss of K3 less detrimental during the evolutionary history of this virus. All identified orthopoxviruses and clade II poxviruses have been found to possess a K3L ortholog except for ECTV and MPXV (Bratke et al., 2013). Therefore, we find it to be an interesting correlation that both viruses have now been shown [in this work and (Arndt et al., 2016)] to accumulate considerably less dsRNA during their replication cycles compared to VACV.
The formation of dsRNA is thought to be common during the replication cycle of poxviruses due to their genome layout and mechanisms of transcription. As outlined above, dsRNA is largely the result of overlapping, complementary mRNA produced from tightly packed genes that are oriented in opposite directions with respect to the direction of transcription. The poxvirus protein E3 contains a conserved dsRNA-binding domain (Chang and Jacobs, 1993) that acts to shield recognition of this molecule from cellular defenses, such as PKR and RNase L. In this study, we made use of the monoclonal antibody J2, which binds to dsRNA of at least 40 bp in length (Bonin et al., 2000; Schönborn et al., 1991). We reasoned that if E3 is able to mask dsRNA from recognition by host cell proteins, then it seems likely it could also prevent binding of J2 to its epitope. Our data support this hypothesis. For example, VACV infection of BS-C-1 cells constitutively expressing E3 results in a significantly reduced ability to detect dsRNA relative to infection of wild-type, E3-negative cells. Therefore, we postulate that the J2 antibody is principally detecting “free” dsRNA (i.e., not bound by E3) within poxvirus-infected cells. Interestingly, a recent report showed that the J2 antibody could be used to pull-down a E3-GFP fusion protein in an immunoprecipitation assay (Liu and Moss, 2016). The authors used a human cell line at 13 hrs. post-infection for this experiment. These results indicate that under certain conditions J2 and E3 can bind simultaneously to the same segment of dsRNA. To reconcile our data with that of Liu and Moss, we posit that the degree of saturation of a region of dsRNA by E3 could influence the binding capacity of J2. Moreover, we detect less dsRNA in human-derived cells (e.g., HeLa or 293T) infected with VACV compared to cells from non-human primates (data not shown). This point further reinforces the need to perform follow-up studies using cells derived from other species.
An infection time course using wild-type VACV and a mutant lacking E3L revealed that VACV produces dsRNA well before we could detect it with the J2 antibody. Between 8 and 12 hrs. post-infection with wild-type VACV, we observed that levels of dsRNA transitioned from essentially undetectable to greater than 50% of the cells staining positive. Thus, it appears that E3 shields dsRNA from detection for a limited duration. If sufficient dsRNA is produced during replication, then a threshold is crossed by which additional dsRNA cannot be sequestered by the available pool of E3. Presumably, this is of little consequence for VACV because the replication cycle is relatively short and K3 is present to act as an additional layer of defense against the PKR pathway. It has been shown in numerous reports that wild-type VACV does not readily activate PKR during infection of various cell lines (Hand et al., 2015; Langland and Jacobs, 2004; Watson et al., 1991; Zhang et al., 2008). VACV also has other mechanisms to avoid innate immune detection, such as the inhibition of NF-κB signaling (Shisler and Jin, 2004).
The RNA polymerase of VACV does not efficiently terminate at the end of many viral genes – particularly for intermediate and late genes – which causes the transcription machinery to “read-through” into neighboring genes (Broyles, 2003). This leads to heterogeneous 3’ termini on many viral transcripts (Xiang et al., 2000; 1998) and greatly increases the likelihood of dsRNA formation through complementary base pairing. Arguably, the most direct mechanism for ECTV to produce less free dsRNA would be to increase the efficiency of transcription termination. This would produce mRNAs of less heterogeneous lengths, result in shorter transcripts, and reduce the opportunity for dsRNA formation. Our Northern blot data clearly show that some post-replicative transcripts in ECTV are indeed shorter compared to VACV. Interestingly, the length of F17R transcripts were actually slightly longer for ECTV relative to VACV. Therefore, it does not appear that ECTV terminates transcription more efficiently for all post-replicative genes in a universal manner. Future work will focus on understanding this apparent dichotomy in more detail.
It has been recently reported that monkeypox virus (MPXV) accumulates less free dsRNA and, unlike ECTV and VACV, is naturally resistant to IBT (Arndt et al., 2016). Arndt and colleagues found that a much higher amount of IBT (120 µM) was required to fully inhibit MPXV. These authors also observed an increase in dsRNA staining using the J2 antibody in VACV-infected – but not MPXV-infected – cells treated with IBT relative to the no drug control. While we found similar results for VACV-infected cells, we observed a striking increase in dsRNA positivity within ECTV-infected cells treated with IBT compared to untreated cells. Despite the fact that both ECTV and MPXV accumulate less dsRNA relative to VACV, the underlying mechanisms causing this may be distinct. It has been shown that IBT-resistant mutants of VACV form shorter transcripts during infection in the absence of the drug (Cresawn et al., 2007; Cresawn and Condit, 2007). Therefore, it is possible that MPXV displays universally shorter (i.e., less heterogeneous) post-replicative transcripts. While we did find that some ECTV transcripts are shorter than VACV, it does not appear this feature holds true for every gene.
In addition to the less heterogeneity of some post-replicative transcripts, we show that ECTV displays a protracted replication cycle and reduced mRNA abundance, which also impacts dsRNA accumulation. When we performed VACV infections at 30°C instead of 37°C, we found markedly lower levels of dsRNA in cells incubated at the lower temperature. In fact, the dsRNA staining was virtually indistinguishable between ECTV grown at 37°C compared to VACV at 30°C. Importantly, we demonstrated that the replication kinetics of VACV were noticeably delayed at the lower temperature. Growth at the lower temperature likely slowed the transcriptional kinetics of VACV and reduced overall mRNA levels. By replicating more slowly than VACV, it appears that ECTV is able to keep its dsRNA at levels low enough such that the available pool of E3 can sequester it.
Other factors in addition to replication rate and transcript length may also contribute to the reduction in the amount of dsRNA within ECTV-infected cells. For example, it was recently shown that a host RNA exonuclease, Xrn1, modulates dsRNA accumulation during VACV infection (Burgess and Mohr, 2015). Levels of dsRNA increased significantly within infected cells compared to the controls when Xrn1 was depleted. Therefore, the slower replication cycle of ECTV may allow host RNA decay enzymes, such as Xrn1, to “keep up” with viral mRNA turnover before dsRNA can accumulate. Alternatively, the mRNA decapping enzymes of ECTV may operate more efficiently than those of VACV, which would lead to an enhanced ability to degrade mRNAs from early genes before post-replicative gene mRNAs can base pair with them. Additional research into these possible mechanisms is warranted.
In conclusion, we find that ECTV forms less free dsRNA than VACV. The latter has historically been the main model system to study the details of transcription and RNA metabolism among orthopoxviruses. However, our data suggest that there could be more to learn by studying other members of this genus. The formation of dsRNA, which is a major signal to the host cell that a viral infection is underway, may require differential regulation if the full array of immune evasion proteins is altered or absent.
Supplementary Material
BS-C-1 cells were infected (MOI=5) with ECTV or VACV. Real-time reverse-transcriptase PCR was performed using gene-specific primers for F17R and H4L. The data points represent duplicate reactions from two independent infections and RNA isolations. Statistical analysis [performed using GraphPad Prism software (version 7.0a)] was carried out using the Mann-Whitney test. * denotes a p value <0.05.
Research highlights for VIRO-17-290R1.
Ectromelia virus forms less dsRNA compared to Vaccinia virus in BS-C-1 cells
Ectromelia virus produces generally less mRNA and shorter transcripts
Because Ectromelia virus lacks K3, it may have evolved to constrain dsRNA formation
Acknowledgments
As indicated in the methods section, some reagents were obtained through the NIH Biodefense and Emerging Infections Research Resources Repository. We are grateful to Dr. Richard Condit for providing a stock of IBT and thank Christine Brandmüller (Institute for Infectious Diseases and Zoonoses) for excellent technical assistance. This work was supported through the Albright Creative Research Experience (ACRE) program and the Kreider Award [both are internal grants at Albright College], NIH grant R01 AI110542, and the German Center for Infection Research (DZIF).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aranda PS, LaJoie DM, Jorcyk CL. Bleach gel: a simple agarose gel for analyzing RNA quality. Electrophoresis. 2012;33:366–369. doi: 10.1002/elps.201100335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt WD, White SD, Johnson BP, Huynh T, Liao J, Harrington H, Cotsmire S, Kibler KV, Langland J, Jacobs BL. Monkeypox virus induces the synthesis of less dsRNA than vaccinia virus, and is more resistant to the anti-poxvirus drug, IBT, than vaccinia virus. Virology. 2016;497:125–135. doi: 10.1016/j.virol.2016.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayliss CD, Condit RC. Temperature-sensitive mutants in the vaccinia virus A18R gene increase double-stranded RNA synthesis as a result of aberrant viral transcription. Virology. 1993;194:254–262. doi: 10.1006/viro.1993.1256. [DOI] [PubMed] [Google Scholar]
- Beattie E, Kauffman EB, Martinez H, Perkus ME, Jacobs BL, Paoletti E, Tartaglia J. Host-range restriction of vaccinia virus E3L-specific deletion mutants. Virus Genes. 1996;12:89–94. doi: 10.1007/BF00370005. [DOI] [PubMed] [Google Scholar]
- Beattie E, Tartaglia J, Paoletti E. Vaccinia virus-encoded eIF-2 alpha homolog abrogates the antiviral effect of interferon. Virology. 1991;183:419–422. doi: 10.1016/0042-6822(91)90158-8. [DOI] [PubMed] [Google Scholar]
- Bonin M, Oberstrass J, Lukacs N, Ewert K, Oesterschulze E, Kassing R, Nellen W. Determination of preferential binding sites for anti-dsRNA antibodies on double-stranded RNA by scanning force microscopy. RNA. 2000;6:563–570. doi: 10.1017/s1355838200992318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone RF, Parr RP, Moss B. Intermolecular duplexes formed from polyadenylylated vaccinia virus RNA. Journal of Virology. 1979;30:365–374. doi: 10.1128/jvi.30.1.365-374.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt TA, Jacobs BL. Both carboxy- and amino-terminal domains of the vaccinia virus interferon resistance gene, E3L, are required for pathogenesis in a mouse model. Journal of Virology. 2001;75:850–856. doi: 10.1128/JVI.75.2.850-856.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratke KA, McLysaght A, Rothenburg S. A survey of host range genes in poxvirus genomes. Infect. Genet. Evol. 2013;14:406–425. doi: 10.1016/j.meegid.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breman JG, Henderson DA. Diagnosis and management of smallpox. N. Engl. J. Med. 2002;346:1300–1308. doi: 10.1056/NEJMra020025. [DOI] [PubMed] [Google Scholar]
- Brennan G, Kitzman JO, Rothenburg S, Shendure J, Geballe AP. Adaptive gene amplification as an intermediate step in the expansion of virus host range. PLoS Pathog. 2014;10:e1004002. doi: 10.1371/journal.ppat.1004002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broyles SS. Vaccinia virus transcription. J. Gen. Virol. 2003;84:2293–2303. doi: 10.1099/vir.0.18942-0. [DOI] [PubMed] [Google Scholar]
- Buller RM, Palumbo GJ. Poxvirus pathogenesis. Microbiol. Rev. 1991;55:80–122. doi: 10.1128/mr.55.1.80-122.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess HM, Mohr I. Cellular 5“-3” mRNA exonuclease Xrn1 controls double-stranded RNA accumulation and anti-viral responses. Cell Host and Microbe. 2015;17:332–344. doi: 10.1016/j.chom.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll K, Elroy-Stein O, Moss B, Jagus R. Recombinant vaccinia virus K3L gene product prevents activation of double-stranded RNA-dependent, initiation factor 2 alpha-specific protein kinase. Journal of Biological Chemistry. 1993;268:12837–12842. [PubMed] [Google Scholar]
- Chang HW, Jacobs BL. Identification of a conserved motif that is necessary for binding of the vaccinia virus E3L gene products to double-stranded RNA. Virology. 1993;194:537–547. doi: 10.1006/viro.1993.1292. [DOI] [PubMed] [Google Scholar]
- Chang HW, Watson JC, Jacobs BL. The E3L gene of vaccinia virus encodes an inhibitor of the interferon-induced, double-stranded RNA-dependent protein kinase. Proc. Natl. Acad. Sci. U.S.A. 1992;89:4825–4829. doi: 10.1073/pnas.89.11.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N, Danila MI, Feng Z, Buller RML, Wang C, Han X, Lefkowitz EJ, Upton C. The genomic sequence of ectromelia virus, the causative agent of mousepox. Virology. 2003;317:165–186. doi: 10.1016/S0042-6822(03)00520-8. [DOI] [PubMed] [Google Scholar]
- Chen W, Drillien R, Spehner D, Buller RM. Restricted replication of ectromelia virus in cell culture correlates with mutations in virus-encoded host range gene. Virology. 1992;187:433–442. doi: 10.1016/0042-6822(92)90445-u. [DOI] [PubMed] [Google Scholar]
- Colby C, Duesberg PH. Double-stranded RNA in vaccinia virus infected cells. Nature. 1969;222:940–944. doi: 10.1038/222940a0. [DOI] [PubMed] [Google Scholar]
- Colby C, Jurale C, Kates JR. Mechanism of synthesis of vaccinia virus double-stranded ribonucleic acid in vivo and in vitro. Journal of Virology. 1971;7:71–76. doi: 10.1128/jvi.7.1.71-76.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condit RC, Motyczka A. Isolation and preliminary characterization of temperature-sensitive mutants of vaccinia virus. Virology. 1981;113:224–241. doi: 10.1016/0042-6822(81)90150-1. [DOI] [PubMed] [Google Scholar]
- Cresawn SG, Condit RC. A targeted approach to identification of vaccinia virus postreplicative transcription elongation factors: genetic evidence for a role of the H5R gene in vaccinia transcription. Virology. 2007;363:333–341. doi: 10.1016/j.virol.2007.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cresawn SG, Prins C, Latner DR, Condit RC. Mapping and phenotypic analysis of spontaneous isatin-beta-thiosemicarbazone resistant mutants of vaccinia virus. Virology. 2007;363:319–332. doi: 10.1016/j.virol.2007.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies MV, Elroy-Stein O, Jagus R, Moss B, Kaufman RJ. The vaccinia virus K3L gene product potentiates translation by inhibiting double-stranded-RNA-activated protein kinase and phosphorylation of the alpha subunit of eukaryotic initiation factor 2. Journal of Virology. 1992;66:1943–1950. doi: 10.1128/jvi.66.4.1943-1950.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duesberg PH, Colby C. On the biosynthesis and structure of double-stranded RNA in vaccinia virus-infected cells. Proc. Natl. Acad. Sci. U.S.A. 1969;64:396–403. doi: 10.1073/pnas.64.1.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl PL, Cooper N, Wyatt LS, Moss B, Carroll MW. Preparation of cell cultures and vaccinia virus stocks. Curr Protoc Mol Biol. 2001;16(16.16–16.16.13) doi: 10.1002/0471142727.mb1616s43. [DOI] [PubMed] [Google Scholar]
- Esteban DJ, Buller RML. Ectromelia virus: the causative agent of mousepox. J. Gen. Virol. 2005;86:2645–2659. doi: 10.1099/vir.0.81090-0. [DOI] [PubMed] [Google Scholar]
- Fang M, Lanier LL, Sigal LJ. A role for NKG2D in NK cell-mediated resistance to poxvirus disease. PLoS Pathog. 2008;4:e30. doi: 10.1371/journal.ppat.0040030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenner F. Studies in infectious ectromelia in mice; natural transmission; the portal of entry of the virus. Aust J Exp Biol Med Sci. 1947;25:275–282. doi: 10.1038/icb.1947.39. [DOI] [PubMed] [Google Scholar]
- Fenner F, Mortimer P. Classic paper: Fenner on the exanthemata. Rev. Med. Virol. 2006;16:353–363. doi: 10.1002/rmv.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubser C, Hué S, Kellam P, Smith GL. Poxvirus genomes: a phylogenetic analysis. J. Gen. Virol. 2004;85:105–117. doi: 10.1099/vir.0.19565-0. [DOI] [PubMed] [Google Scholar]
- Haller SL, Peng C, McFadden G, Rothenburg S. Poxviruses and the evolution of host range and virulence. Infect. Genet. Evol. 2014;21:15–40. doi: 10.1016/j.meegid.2013.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hand ES, Haller SL, Peng C, Rothenburg S, Hersperger AR. Ectopic expression of vaccinia virus E3 and K3 cannot rescue ectromelia virus replication in rabbit RK13 cells. PLoS ONE. 2015;10:e0119189. doi: 10.1371/journal.pone.0119189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y-G, Muralinath M, Brandt T, Pearcy M, Hauns K, Lowenhaupt K, Jacobs BL, Rich A. A role for Z-DNA binding in vaccinia virus pathogenesis. Proc. Natl. Acad. Sci. U.S.A. 2003;100:6974–6979. doi: 10.1073/pnas.0431131100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langland JO, Cameron JM, Heck MC, Jancovich JK, Jacobs BL. Inhibition of PKR by RNA and DNA viruses. Virus Res. 2006;119:100–110. doi: 10.3201/eid1204.051181. [DOI] [PubMed] [Google Scholar]
- Langland JO, Jacobs BL. Inhibition of PKR by vaccinia virus: role of the N- and C-terminal domains of E3L. Virology. 2004;324:419–429. doi: 10.1016/j.virol.2004.03.012. [DOI] [PubMed] [Google Scholar]
- Lehmann MH, Schreiber S, Vogelsang H, Sigusch HH. Constitutive expression of MCP-1 and RANTES in the human histiocytic lymphoma cell line U-937. Immunol. Lett. 2001;76:111–113. doi: 10.1016/s0165-2478(01)00178-x. [DOI] [PubMed] [Google Scholar]
- Liu R, Moss B. Opposing Roles of Double-Stranded RNA Effector Pathways and Viral Defense Proteins Revealed with CRISPR/Cas9 Knock-Out Cell Lines and Vaccinia Virus Mutants. Journal of Virology. 2016 doi: 10.1128/JVI.00869-16. JVI.00869–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig H, Suezer Y, Waibler Z, Kalinke U, Schnierle BS, Sutter G. Double-stranded RNA-binding protein E3 controls translation of viral intermediate RNA, marking an essential step in the life cycle of modified vaccinia virus Ankara. J. Gen. Virol. 2006;87:1145–1155. doi: 10.1099/vir.0.81623-0. [DOI] [PubMed] [Google Scholar]
- Lynn H, Horsington J, Ter LK, Han S, Chew YL, Diefenbach RJ, Way M, Chaudhri G, Karupiah G, Newsome TP. Loss of Cytoskeletal Transport during Egress Critically Attenuates Ectromelia Virus Infection In Vivo. Journal of Virology. 2012;86:7427–7443. doi: 10.1128/JVI.06636-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myskiw C, Arsenio J, Hammett C, van Bruggen R, Deschambault Y, Beausoleil N, Babiuk S, Cao J. Comparative analysis of poxvirus orthologues of the vaccinia virus E3 protein: modulation of protein kinase R activity, cytokine responses, and virus pathogenicity. Journal of Virology. 2011;85:12280–12291. doi: 10.1128/JVI.05505-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchanathan V, Chaudhri G, Karupiah G. Interferon function is not required for recovery from a secondary poxvirus infection. Proc. Natl. Acad. Sci. U.S.A. 2005;102:12921–12926. doi: 10.1073/pnas.0505180102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker S, Siddiqui AM, Oberle C, Hembrador E, Lanier R, Painter G, Robertson A, Buller RM. Mousepox in the C57BL/6 strain provides an improved model for evaluating anti-poxvirus therapies. Virology. 2009;385:11–21. doi: 10.1016/j.virol.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price PJR, Luckow B, Torres-Domínguez LE, Brandmüller C, Zorn J, Kirschning CJ, Sutter G, Lehmann MH. Chemokine (C-C Motif) receptor 1 is required for efficient recruitment of neutrophils during respiratory infection with modified vaccinia virus Ankara. Journal of Virology. 2014;88:10840–10850. doi: 10.1128/JVI.01524-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prins C, Cresawn SG, Condit RC. An isatin-beta-thiosemicarbazone-resistant vaccinia virus containing a mutation in the second largest subunit of the viral RNA polymerase is defective in transcription elongation. J. Biol. Chem. 2004;279:44858–44871. doi: 10.1074/jbc.M408167200. [DOI] [PubMed] [Google Scholar]
- Raff T, van der Giet M, Endemann D, Wiederholt T, Paul M. Design and testing of beta-actin primers for RT-PCR that do not co-amplify processed pseudogenes. BioTechniques. 1997 doi: 10.2144/97233st02. [DOI] [PubMed] [Google Scholar]
- Rahman MM, Liu J, Chan WM, Rothenburg S, McFadden G. Myxoma Virus Protein M029 Is a Dual Function Immunomodulator that Inhibits PKR and Also Conscripts RHA/DHX9 to Promote Expanded Host Tropism and Viral Replication. PLoS Pathog. 2013;9:e1003465. doi: 10.1371/journal.ppat.1003465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice AP, Roberts BE. Vaccinia virus induces cellular mRNA degradation. Journal of Virology. 1983;47:529–539. doi: 10.1128/jvi.47.3.529-539.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas C, Gil J, Mĕlková Z, Esteban M, Díaz-Guerra M. Vaccinia virus E3L protein is an inhibitor of the interferon (i.f.n.)-induced 2–5A synthetase enzyme. Virology. 1998;243:406–414. doi: 10.1006/viro.1998.9072. [DOI] [PubMed] [Google Scholar]
- Roper RL. Characterization of the vaccinia virus A35R protein and its role in virulence. Journal of Virology. 2006;80:306–313. doi: 10.1128/JVI.80.1.306-313.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenburg S, Seo EJ, Gibbs JS, Dever TE, Dittmar K. Rapid evolution of protein kinase PKR alters sensitivity to viral inhibitors. Nat. Struct. Mol. Biol. 2009;16:63–70. doi: 10.1038/nsmb.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönborn J, Oberstrass J, Breyel E, Tittgen J, Schumacher J, Lukacs N. Monoclonal antibodies to double-stranded RNA as probes of RNA structure in crude nucleic acid extracts. Nucleic Acids Res. 1991;19:2993–3000. doi: 10.1093/nar/19.11.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seet BT, Johnston JB, Brunetti CR, Barrett JW, Everett H, Cameron C, Sypula J, Nazarian SH, Lucas A, McFadden G. Poxviruses and immune evasion. Annu. Rev. Immunol. 2003;21:377–423. doi: 10.1146/annurev.immunol.21.120601.141049. [DOI] [PubMed] [Google Scholar]
- Shisler JL, Jin X-L. The vaccinia virus K1L gene product inhibits host NF-kappaB activation by preventing IkappaBalpha degradation. Journal of Virology. 2004;78:3553–3560. doi: 10.1128/JVI.78.7.3553-3560.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siciliano NA, Hersperger AR, Lacuanan AM, Xu R-H, Sidney J, Sette A, Sigal LJ, Eisenlohr LC. Impact of distinct poxvirus infections on the specificities and functionalities of CD4+ T cell responses. Journal of Virology. 2014;88:10078–10091. doi: 10.1128/JVI.01150-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman RH, Skehel JJ, James TC, Wreschner DH, Kerr IM. rRNA cleavage as an index of ppp(A2'p)nA activity in interferon-treated encephalomyocarditis virus-infected cells. Journal of Virology. 1983;46:1051–1055. doi: 10.1128/jvi.46.3.1051-1055.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith VP, Alcami A. Inhibition of Interferons by Ectromelia Virus. Journal of Virology. 2002;76:1124–1134. doi: 10.1128/JVI.76.3.1124-1134.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith VP, Alcamí A. Inhibition of interferons by ectromelia virus. Journal of Virology. 2002;76:1124–1134. doi: 10.1128/JVI.76.3.1124-1134.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JC, Chang HW, Jacobs BL. Characterization of a vaccinia virus-encoded double-stranded RNA-binding protein that may be involved in inhibition of the double-stranded RNA-dependent protein kinase. Virology. 1991;185:206–216. doi: 10.1016/0042-6822(91)90768-7. [DOI] [PubMed] [Google Scholar]
- Weber F, Wagner V, Rasmussen SB, Hartmann R, Paludan SR. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. Journal of Virology. 2006;80:5059–5064. doi: 10.1128/JVI.80.10.5059-5064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis KL, Langland JO, Shisler JL. Viral double-stranded RNAs from vaccinia virus early or intermediate gene transcripts possess PKR activating function, resulting in NF-kappaB activation, when the K1 protein is absent or mutated. Journal of Biological Chemistry. 2011;286:7765–7778. doi: 10.1074/jbc.M110.194704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wreschner DH, James TC, Silverman RH, Kerr IM. Ribosomal RNA cleavage, nuclease activation and 2–5A(ppp(A2'p)nA) in interferon-treated cells. Nucleic Acids Res. 1981;9:1571–1581. doi: 10.1093/nar/9.7.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Condit RC, Vijaysri S, Jacobs B, Williams BRG, Silverman RH. Blockade of interferon induction and action by the E3L double-stranded RNA binding proteins of vaccinia virus. Journal of Virology. 2002;76:5251–5259. doi: 10.1128/JVI.76.10.5251-5259.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Latner DR, Niles EG, Condit RC. Transcription elongation activity of the vaccinia virus J3 protein in vivo is independent of poly(A) polymerase stimulation. Virology. 2000;269:356–369. doi: 10.1006/viro.2000.0242. [DOI] [PubMed] [Google Scholar]
- Xiang Y, Simpson DA, Spiegel J, Zhou A, Silverman RH, Condit RC. The vaccinia virus A18R DNA helicase is a postreplicative negative transcription elongation factor. Journal of Virology. 1998;72:7012–7023. doi: 10.1128/jvi.72.9.7012-7023.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu RH, Cohen M, Tang Y, Lazear E, Whitbeck JC, Eisenberg RJ, Cohen GH, Sigal LJ. The orthopoxvirus type I IFN binding protein is essential for virulence and an effective target for vaccination. Journal of Experimental Medicine. 2008;205:981–992. doi: 10.1084/jem.20071854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Reynolds SE, Martens CA, Bruno DP, Porcella SF, Moss B. Expression profiling of the intermediate and late stages of poxvirus replication. Journal of Virology. 2011;85:9899–9908. doi: 10.1128/JVI.05446-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Jacobs BL, Samuel CE. Loss of protein kinase PKR expression in human HeLa cells complements the vaccinia virus E3L deletion mutant phenotype by restoration of viral protein synthesis. Journal of Virology. 2008;82:840–848. doi: 10.1128/JVI.01891-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
BS-C-1 cells were infected (MOI=5) with ECTV or VACV. Real-time reverse-transcriptase PCR was performed using gene-specific primers for F17R and H4L. The data points represent duplicate reactions from two independent infections and RNA isolations. Statistical analysis [performed using GraphPad Prism software (version 7.0a)] was carried out using the Mann-Whitney test. * denotes a p value <0.05.