Abstract
Recent mass spectrometry maps of the human interactome independently support the existence of a large multiprotein complex, dubbed ‘Commander’. Broadly conserved across animals and ubiquitously expressed in nearly every human cell type examined thus far, Commander likely plays a fundamental cellular function, akin to other ubiquitous machines involved in expression, proteostasis, and trafficking. Experiments on individual subunits support roles in endosomal protein sorting, including the trafficking of Notch proteins, copper transporters, and lipoprotein receptors. Commander is critical for vertebrate embryogenesis, and defects in the complex and its interaction partners disrupt craniofacial, brain, and heart development. Here, we review the synergy between large-scale proteomic efforts and focused studies in the discovery of Commander, describe its composition, structure, and function, and discuss how it illustrates the power of systems biology. Based on 3D modeling and biochemical data, we draw strong parallels between Commander and the retromer cargo-recognition complex, laying a foundation for future research into Commander’s role in human developmental disorders.
Graphical Abstract
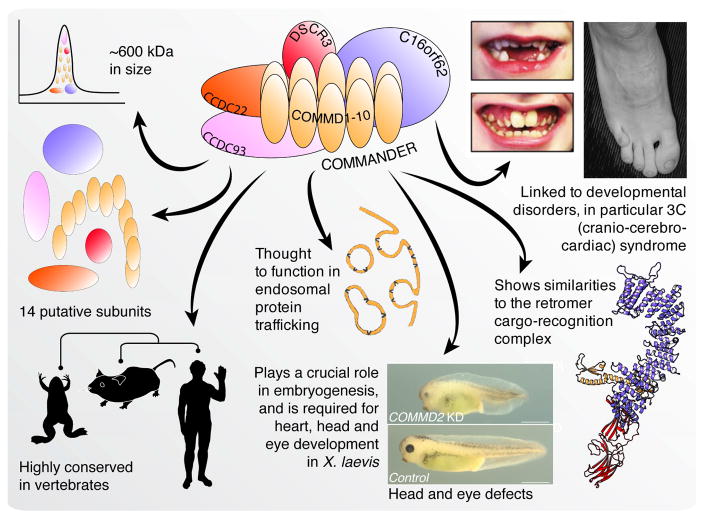
Large-scale exploratory studies of the mammalian interactome are providing unparalleled information about the architecture of biological pathways and protein complexes (Wan et al., 2015; Dey et al., 2015; Hein et al., 2015; Huttlin et al., 2015; Li et al., 2014). When considered together, these networks identify overlapping parts of an entirely new and highly conserved metazoan multiprotein complex that we have dubbed ‘Commander’ (Fig. 1 and Box 1), named because it contains up to ten copper metabolism Murr1 domains (COMMDs). Current data leads us to estimate that the Commander complex comprises up to 14 different proteins that equal approximately 600 kDa in mass. Commander genes are highly conserved in metazoa and near-ubiquitously expressed across tissue types (Burstein et al., 2005; Consortium et al., 2014; Wang et al., 2015). Although the molecular function of Commander genes is largely unknown, they are linked to endosomal protein sorting (Bartuzi et al., 2016; Li et al., 2015; Phillips-Krawczak et al., 2015), as well as proinflammatory signaling (Maine et al., 2007; Starokadomskyy et al., 2013), and hypoxia adaptation (van de Sluis et al., 2010; van de Sluis et al., 2007). Defects in Commander and its interaction partners are additionally implicated in Ritscher-Schinzel (RS)/3C (cranio-cerebro-cardiac) syndrome (Elliott et al., 2013; Kolanczyk et al., 2015; Voineagu et al., 2012), a rare developmental disorder characterized by craniofacial abnormalities, cerebellar brain anomalies, congenital heart defects and intellectual disability (Ritscher et al., 1987).
Figure 1.
Overview of the Commander complex. Commander is thought to contain up to 14 subunits equaling approximately 600 kDa in mass (Wan et al., 2015, Drew et al., 2017), is highly conserved in vertebrates, and functions in endosomal protein trafficking (Bartuzi et al., 2016; Li et al., 2015; Phillips-Krawczak et al., 2015). It also plays a crucial role in embryogenesis (Wan et al., 2015), and is linked to developmental disorders (Kolanczyk et al., 2015; Ritscher et al., 1987; Starokadomskyy et al., 2013). We suggest that Commander may function as a retromer-like sorter of cargo proteins within the endosomal system.
Box 1. Lessons from Commander.
The Commander complex serves as a general example of the power of systems biology to uncover new biological phenomena by first visualizing the ‘big picture’, then focusing on novel elements revealed from this vantage. Such holistic, top-down approaches highlight entire systems, and connect nicely with mechanistic and clinical studies on genes and processes of interest. System-wide studies additionally provide alternative contextualization for previously undertaken gene-specific studies. The latter provide context, mechanistic, and disease-related links, leading to rapid generation and refinement of hypotheses about the system as a whole.
There are often clear trade-offs between large-scale and focused approaches. For example, the risk of losing information by simply not probing for the correct interaction is vastly reduced when many experiments are integrated in a quantitative discovery framework, with global control over false-positives, but the finer details of cellular machinery may be missed.
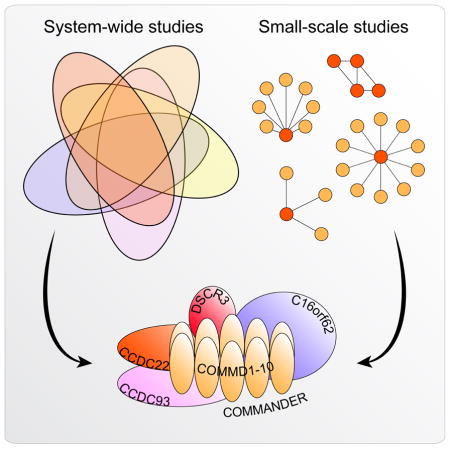
In the case of Commander, there was a mutually beneficial convergence of these discovery-based proteome-wide and specific smaller-scale approaches. Here, proteome-wide interactome studies, combining results from nearly 10,000 individual mass spectrometry experiments, served to create an overview map of protein complexes in human and animal cells. The ‘strength in numbers’ of the large-scale proteomics uncovered a large, poorly characterized cellular machine; comparison of the full complex with prior small-scale studies mutually supported and reinforced both datasets, and the resulting syntheses across these studies should now help guide targeted biochemical and structural studies to determine complex stoichiometry, architecture, and function.
This Review presents the combined evidence from large-scale systems-wide and small-scale proteomics experiments that Commander functions as a physically associated and fully integrated complex. More generally, the discovery and characterization of Commander serve as an example of the power of systems biology to uncover new biological machines and phenomena (Box 1). We describe the conservation and abundance of Commander, and preliminary functional analyses of complex subunits at both the molecular and organism level. Finally, we speculate on the similarities between the 3D-structure, architecture, function, and molecular mechanism of Commander to retromer and Ndc80, and the potential role of the complex in developmental disease.
Discovery of the 14-subunit Commander complex by large-scale proteomics and phylogenetic profiling efforts
Several recently published human interaction networks describe interrelated pieces of the Commander complex (Wan et al., 2015; Dey et al., 2015; Hein et al., 2015; Huttlin et al., 2015; Li et al., 2014). The independent approaches used by these studies, which apply a variety of experimental and bioinformatic strategies to build global molecular interaction maps, make this evidence especially compelling. Taken together, these provide an initial set of candidate proteins that comprise Commander (Fig. 2).
Figure 2.
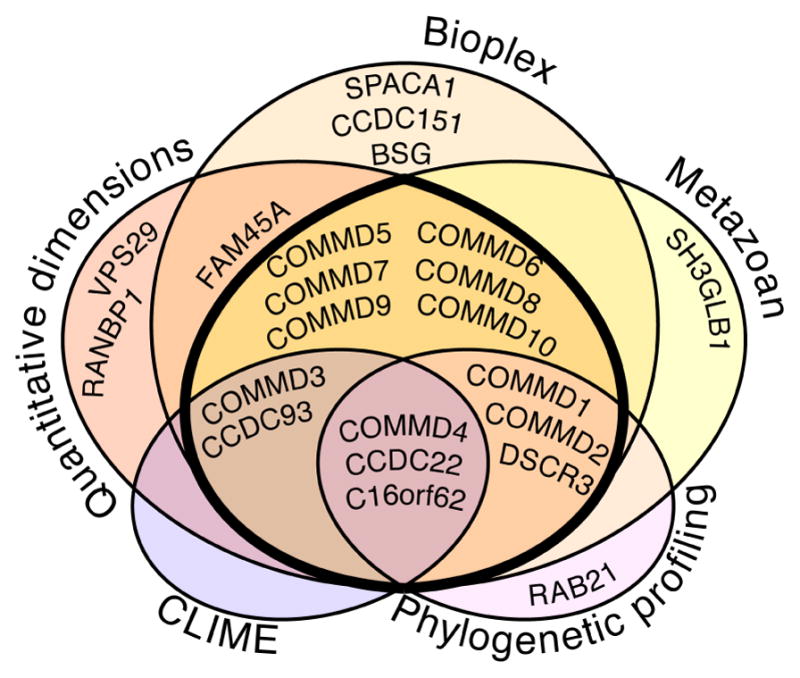
Putative Commander subunits. Recent human interaction networks describe overlapping pieces of the Commander complex. Three are experimentally derived and include a metazoan complex map (Wan et al., 2015, Drew et al., 2017), the ‘Bioplex’ network (Huttlin et al., 2015) and an interactome in ‘quantitative dimensions’ (Hein et al., 2015). Two are computational networks calculated using phylogenetic profiling (Dey et al., 2015) or ‘clustering by inferred models of evolution’ (CLIME) (Li et al., 2014). The 14 protein subunits identified by at least three studies are considered in this proposal as the strongest candidates to be core components of Commander (thick black line).
A putative form of Commander was identified by our lab in a set of extensive experiments to examine the composition of soluble multiprotein complexes among diverse metazoan models (Wan et al., 2015, Drew et al., 2017). These involved biochemical fractionation of native macromolecular assemblies from different species followed by quantitative mass spectrometry to determine protein complex membership and produce a map of protein complexes common to animals. The results suggested that Commander, one of the largest novel complexes to be uncovered in this study, comprises up to 15 different structurally uncharacterized subunits (Fig. 2). Ten of these, COMMD1-10, are structural and functional homologs from the COMMD family. They are defined by a unique and conserved C-terminal COMM domain that varies from 68-77 residues and mediates the formation of multimeric COMMD complexes (Burstein et al., 2005). The COMMDs also contain a more variable N-terminal α-helical domain of 18-151 residues (Sommerhalter et al., 2007). Other Commander components identified in this study include two coiled-coil domain proteins CCDC22 and CCDC93, and the protein C16orf62, all of unknown function, the endophillin-B1 SH3GLB1, and the Downs Syndrome-related protein DSCR3.
Using perhaps the largest set of affinity purification-mass spectrometry (AP-MS) experiments to date, Huttlin et al. profiled the BioPlex (biophysical interactions of ORFEOME-derived complexes) network (Huttlin et al., 2015). This involved 2,594 AP-MS experiments performed in human cell culture to characterize 23,744 interactions and connect 7,668 proteins. Although the BioPlex network of the human interactome is calculated by a different method to the metazoan complex map from co-fractionation-MS experiments, a complex almost equivalent to Commander was identified as a community of 18 proteins (Fig. 2).
In a third independent large-scale proteomics study, Hein et al. applied a technique called quantitative bacterial artificial chromosome GFP interactomics (QUBIC) to generate a library of 1,125 HeLa cell lines expressing GFP-tagged bait proteins that express under near-endogenous levels (Hein et al., 2015). By using these for GFP immunoprecipitation (IP)-MS experiments, the authors were able to detect specific interactions, estimate interaction stoichiometries, and measure cellular abundances within the human interactome, recovering a total of 28,504 interactions between 5,462 proteins. Several GFP-labeled bait proteins (CCDC22, CCDC93 and the Ran-binding protein RANBP1) pulled down prey proteins consistent with the Commander subunits predicted by the other two experimental interactome studies (Fig. 2). At the time, this group was part of the largest assembly found in the study with no current annotation in the CORUM database (Ruepp et al., 2010).
Commander components have also been independently predicted by several large-scale bioinformatics studies (Dey et al., 2015; Li et al., 2014). Dey et al. applied phylogenetic profiling to determine functional links between human genes (Dey et al., 2015). This approach assumes that genes that function together are gained and lost together in evolution (Gabaldon and Koonin, 2013; Pellegrini et al., 1999). In this study, an extended phylogenetic analysis was used to compare the evolutionary history of orthologous groups of genes, including the many gene duplications present in the human genome. The authors report a set of co-evolving human orthogroup phylogenetic (hOP) modules that represent predictions of functional units, one of which contains putative Commander subunits COMMD1, COMMD2, COMMD4, CCDC22, C16orf62 and DSCR3 (Fig. 2). These predictions match those from an alternative phylogenetic profiling approach to identify new components of biological pathways called ‘clustering by inferred models of evolution’ (CLIME) (Li et al., 2014). CLIME uses an input gene set of a known pathway to predict new genes that share the same evolutionary distribution, and places Commander subunits COMMD3, COMMD4, CCCDC22, CCDC93, and C16orf62 in the same evolutionary module (Fig. 2). It is interesting to note that although these bioinformatic analyses identify fewer potential Commander subunits compared to the experimental approaches, the particularly strong co-evolution of certain components suggests they represent a stable sub-complex.
We suggest that the strongest candidates as core components of Commander are the 14 protein subunits identified by at least three or more of the independent studies profiling the human interactome (Fig. 2). These are COMMD1-10, CCDC22, CCDC93, C16orf62 and DSCR3 (Table 1). Structural modeling suggests that the COMM domain adopts a fold most similar to a pleckstrin homology domain, with an anti-parallel β-sheet followed by a C-terminal α-helix (Table 1). CCDC22 and CCDC93 are predicted to be coiled-coiled helical proteins that share common ancestry with the microtubule associated proteins (MAPs) NDC80 and NUF2, and several proteins that form part of the intraflagellar transport (IFT) system key to the assembly and maintenance of microtubule-based cilia and flagella in eukaryotes (Rosenbaum and Witman, 2002; Schou et al., 2014). This suggests CCDC22 and CCDC93 might also form microtubule-bound complexes. C16orf62 is predicted to contain α-helical repeats, and DSCR3 is predicted to adopt an arrestin fold (Table 1). Both C16orf62 and DSCR3 are predicted to share strong structural homology with subunits in the retromer cargo recognition complex, as we address in more detail in a later section.
Table 1.
Candidate Commander protein subunits. The subunit name, residue number, molecular weight, predicted pI, and predicted structure are shown.
Candidate Commander complex subunits
| Subunit | Number of residues | Molecular weight (kDa) | Theoretical pI# | Predicted structure* |
|---|---|---|---|---|
| COMMD1 | 190 | 21.2 | 5.9 |
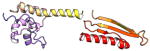
|
| COMMD2 | 199 | 22.7 | 6.2 |
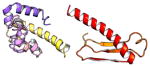
|
| COMMD3 | 195 | 22.2 | 5.6 |
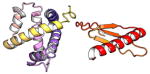
|
| COMMD4 | 199 | 21.8 | 6.9 |
|
| COMMD5 | 224 | 24.7 | 6.5 |
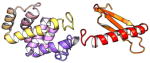
|
| COMMD6 | 85 | 9.6 | 5.7 |
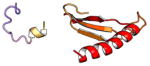
|
| COMMD7 | 200 | 22.5 | 5.7 |

|
| COMMD8 | 183 | 21.1 | 5.3 |
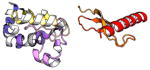
|
| COMMD9 | 198 | 21.8 | 5.6 |
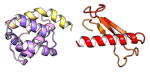
|
| COMMD10 | 202 | 23.0 | 6.1 |
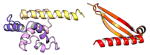
|
| CCDC22 | 627 | 70.8 | 6.3 |
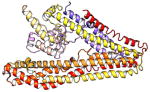
|
| CCDC93 | 631 | 73.2 | 8.2 |

|
| DSCR3 | 297 | 33.0 | 7.6 |

|
| C16orf62 | 963 | 109.6 | 6.8 |
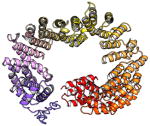
|
Theoretical pIs were calculated from the amino acid sequence using the ProtParam tool within ExPaSy (Wilkins et al., 1999).
Protein structures shown are models generated by I-TASSER (Iterative Threading ASSEmbly Refinement) (Yang et al., 2015) or Robetta (Rosetta Comparative Modeling and Ab Initio Modeling) (Raman et al., 2009), which combine template-based and ab-initio modeling. Where both models were calculated we found they were in good agreement. Structure prediction for the COMMDs was performed on the individual N-and C-terminal domains to maximize accuracy. We did not model the relative orientation of the domains and they are depicted for maximum clarity only. PDB structures are shown for the N-terminal domains of COMMD1 (PDB = 2H2M) and COMMD9 (4OE9), which represent the only experimentally available structural information Commander subunits. The COMMD domains are predicted to adopt a fold similar to a pleckstrin homology domain, in agreement with a previous ab initio model calculated for COMMD1 (Burkhead et al., 2009).
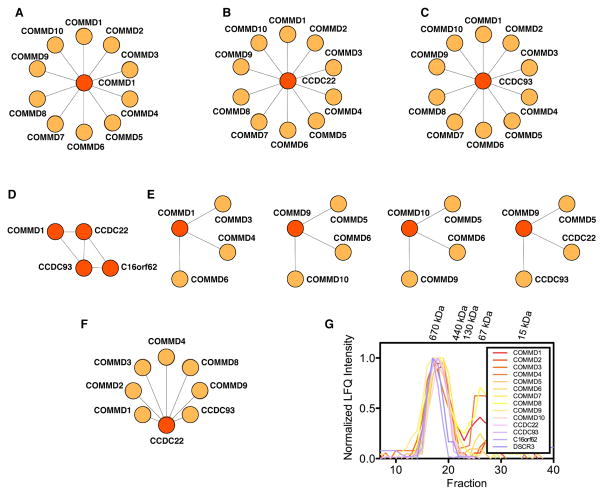
The results of these high-throughput screens are supported by more focused biochemical and cell biology experiments, many of which represent detailed work undertaken independently by the Burstein group (Bartuzi et al., 2016; Li et al., 2015; Mao et al., 2011; Phillips-Krawczak et al., 2015; Starokadomskyy et al., 2013; van de Sluis et al., 2010). A number of smaller scale co-immunoprecipitation (co-IP) assays have been reported that are consistent with the presence of stable Commander sub-complexes (Fig. 3A–F). All 10 COMMDs can apparently interact with each other through COMMD1 (Burstein et al., 2005) (Fig 3A), individually bind to CCDC22 (Starokadomskyy et al., 2013) (Fig. 3B) and uniformly precipitate CCDC93 (Phillips-Krawczak et al., 2015) (Fig. 3C). Most strikingly, a three-subunit complex dubbed ‘CCC’ of COMMD-CCDC22-CCDC93 has been well documented (Phillips-Krawczak et al., 2015, Li et al., 2015, Bartuzi et al., 2016). The CCC complex further interacts with C16orf62 and functions in endosomal trafficking (Phillips-Krawczak et al., 2015) (Fig. 3D). CCC was first characterized containing COMMD1 (Phillips-Krawczak et al., 2015), but other COMMD proteins have been observed in combination with CCDC22 and CCDC93 in later studies (Fig. 3E) (Bartuzi et al., 2016; Li et al., 2015). This makes Commander/CCC potentially very similar in composition; in this review, for simplicity’s sake (and acknowledging that such definitions may evolve over time), we use the nomenclature Commander to refer to the full, putatively 14-member, complex (Fig. 2), which contains the CCC subcomplex and its COMMD variants, as well as DSCR3 and C16orf62. We have independently verified many of the novel protein-protein interactions in the putative Commander complex by AP-MS experiments in human cell lines (Fig. 3F) (Wan et al., 2015).
Figure 3.
Evidence for Commander from small-scale proteomic studies. (A)-(F) Examples of published bait (red) and prey (orange) experiments with Commander subunits. (A) COMMD1 interacts with itself and all other COMMD proteins (Burstein et al., 2005). (B) CCDC22 binds COMMD1-10 (Starokadomskyy et al., 2013). (C) CCDC93 interacts with COMMD1-10 (Phillips-Krawczak et al., 2015). (D) A CCC complex of COMMD1-CCDC22-CCDC93 bound to C16orf62 has been extensively biochemically characterized, and is involved in endosomal protein trafficking (Bartuzi et al., 2016; Li et al., 2015; Phillips-Krawczak et al., 2015). (E) Other combinations of COMMDs have been seen to preferentially interact, for example COMMD3, -4 and -6 were identified in an early COMMD1 tandem affinity purification (TAP) screen (Burstein et al., 2005; Starokadomskyy et al., 2013). (F) We have verified several of the novel protein-protein interactions in the Commander complex by AP-MS (Wan et al., 2015). (G) Size exclusion chromatography of Commander measured by MS (Kirkwood et al., 2013). Elution profiles of all 14 putative Commander subunits indicate that they co-elute as a single entity of approximately 600 kDa in mass.
Proteomic data that characterize native human protein complexes based on size independently corroborate the hypothesis that the putative Commander components form a stable complex (Fig. 3G) (Wan et al., 2015; Kirkwood et al., 2013). In these experiments, native size-exclusion chromatography (SEC) is combined with mass-spectrometry analysis to characterize soluble protein complexes isolated from human osteosarcoma cells (Kirkwood et al., 2013). These data show the 14 most likely Commander components elute together with a mass of ~600 kDa (Fig. 3G). Interestingly, a stable oligomeric complex of approximately 600 kDa containing endogenous COMMD1 was reported in a membrane fraction separated from human HepG2 (liver cancer) cells and analyzed by blue native polyacrylamide gel electrophoresis (BN-PAGE) (Burkhead et al., 2009), further suggesting that this corresponds to the molecular weight of endogenous human Commander. This mass of 600 kDa is slightly higher than the combined molecular weight of 497 kDa for the 14 putative Commander components (Table 1), and additional experiments are needed to determine if these subunits have a 1:1 stoichiometry within the complex, or if additional proteins identified in the human interaction networks are involved (Fig. 2). Indeed, several of the COMMDs have been reported to oligomerize (Burstein et al., 2005; Burkhead et al., 2009), making it possible that Commander functions as a set of similarly-sized heterogeneous complexes of several core subunits with stoichiometries of more than one. In agreement with this, studies by the Burstein lab indicate that the CCC complex performs different functions with other combinations of COMMDs aside from COMMD1 (Bartuzi et al., 2016; Li et al., 2015; Phillips-Krawczak et al., 2015).
Commander’s broad evolutionary conservation and ubiquitous gene expression suggest a fundamental cellular role in animals
Current sequence annotations indicate that Commander genes are of old eukaryotic origin and derive from unicellular eukaryotes (Fig. 4A). However, almost all have been lost in major eukaryotic clades, notably protists, fungi, and flowering plants. The subunits are highly conserved across metazoa, but with exceptions, including the loss of all COMMDs except COMMD4 in worm (Caenorhabditis elegans), and COMMD1, 6, 7 and 9 in fly (Drosophila melanogaster; Fig. 4A). These smaller Commander subunit groupings may represent less intricate versions of the human complex, and may give clues to its architecture and assembly. Notably, Dictyostelium discoideum (slime mold) contains orthologues of the entire putative complex (Fig. 4A), which along with a near-complete complex in the protozoan parasite Trichomonas vaginalis, suggests Commander is an ancient (> 1 billion years old) complex that has been lost or significantly diverged in fungi and many protists and plants.
Figure 4.
Commander is evolutionarily conserved and broadly expressed across human tissues. (A) Phylogenetic profiles for the presence and absence of each of the Commander genes were calculated for each of 66 species’ proteomes (Altenhoff et al., 2016). Dark grey = present; light grey = absent. (B) RNA and protein expression profiles for Commander genes in human cells. Tissue types are shown for which transcripts (source: FANTOM5 project) and protein (source: PaxDB) data are readily available. For RNA expression profiles, abundant = 2–150 and low = < 2 FPKM. For protein expression, abundant = 2–250 ppm and low = < 2 ppm. FPKM = Fragments Per Kilobase of transcript per Million mapped reads. (C) Subcellular localization of predicted Commander proteins as reported in Refs (Lindskog, 2015) (a), (Mao et al., 2011) (b), (Burkhead et al., 2009) (c), (Phillips-Krawczak et al., 2015) (d), (Drevillon et al., 2011) (e), (Chang et al., 2011) (f), (Li et al., 2015) (g), (de Bie et al., 2006) (h), and (Hu et al., 2006) (i).
Old complexes tend to be broadly expressed and abundant, contain proteins with fewer domains and are enriched for human disease interactions (Wan et al., 2015; Greco and Cristea, 2016; Marsh and Teichmann, 2015). In contrast, metazoan-only (i.e. new, and approximately 500 million years old) complexes are more specifically expressed, contain multidomain proteins, are enriched for cancer related-proteins, and tend to be involved in cell adhesion, organization and differentiation (Wan et al., 2015). Commander genes are indeed strongly and ubiquitously expressed throughout human tissues at both the mRNA (Consortium et al., 2014) and protein (Wang et al., 2015) level (Fig. 4B). This trend and the deep evolutionary conservation argue that Commander is likely a complex of central importance involved in fundamental cellular function.
The reported subcellular localization of Commander varies. Subunits have been observed using immunofluorescence in human cells in more than one compartment, including the nucleus, cytoplasm and vesicles (Fig. 4C). This could mean that Commander proteins exist as full complexes, subcomplexes, and monomeric species in different cellular compartments. In addition, the COMMDs have two predicted nuclear export signals in the COMMD domain, suggesting that their transport from the nucleus to the cytosol is highly regulated (Muller et al., 2009). The observation of most Commander proteins in vesicles, endosomes and/or membranes is consistent with a reported role of many Commander subunits in cellular trafficking, as we next discuss.
The apparent pleiotropic functions of Commander proteins
We can reconsider previously reported functional data on individual Commander proteins in the context of them operating as a large, multi-protein complex. COMMD1 is the most studied of the Commander proteins, and is linked to a diverse range of cellular processes including proinflammatory signaling (Burstein et al., 2005; Maine and Burstein, 2007; Starokadomskyy et al., 2013), hypoxia adaptation (van de Sluis et al., 2010; van de Sluis et al., 2007), copper metabolism (Phillips-Krawczak et al., 2015; van De Sluis et al., 2002), ubiquitination (Drevillon et al., 2011; Ganesh et al., 2003; Maine et al., 2007), and electrolyte transport (Biasio et al., 2004). COMMD1 has been identified as a therapeutic target in anti-inflammatory pathways (Bartuzi et al., 2013; de Becdelievre et al., 2013; de Bie et al., 2006; Ganesh et al., 2003; Maine et al., 2007), in hypoxia-related tumor invasion (van de Sluis et al., 2010), and in cystic fibrosis (de Becdelievre et al., 2013; Drevillon et al., 2011). Interestingly, with the exception of inflammation and hypoxic response, many reported functions of Commander proteins can be linked to the trafficking of transmembrane proteins. In particular, almost all of the predicted Commander subunits are directly implicated in protein sorting processes within the endosomal system (Bartuzi et al., 2016; Burkhead et al., 2009; Drevillon et al., 2011; Harbour et al., 2012; Li et al., 2015; Phillips-Krawczak et al., 2015), suggesting a key role for the whole complex in intracellular trafficking. We discuss a potential role for Commander in ubiquitination-related inflammatory pathways, hypoxia, and endosomal trafficking in more detail below.
The COMMDs regulate the activity of NF-κB, a proinflammatory multiprotein transcriptional complex that controls almost 400 genes involved in immune responses, immune system development, and cell survival and proliferation (Bartuzi et al., 2013; Maine and Burstein, 2007). Specifically, NF-κB is down-regulated by the COMMDs, a necessary response after an inflammatory episode to prevent chronic inflammation. While all COMMD proteins are reported to interact distinctly with NF-κB (Burstein et al., 2005), a detailed mechanism of NF-κB regulation has only been proposed for COMMD1. This involves NF-κB ubiquitination and proteasomal degradation in the nucleus through an interaction of COMMD1 with a multimeric E3 ubiquitin ligase complex (Maine et al., 2007). It has been suggested that the mechanisms by which each COMMD regulates NF-κB are distinct (Bartuzi et al., 2013). We suggest a role for Commander in this ubiquitination-controlled anti-inflammatory response should also be considered. Consistent with this, Commander subunit CCDC22 is required for the ubiquitination of IκB, an inhibitor of the NF-κB pathway (Starokadomskyy et al., 2013).
Gene inactivation studies in mice have shown that COMMD1 mediates hypoxia-inducible factor 1 (HIF-1) (van de Sluis et al., 2007), a heterodimeric transcription factor that regulates oxygen homeostasis (Gordan and Simon, 2007). HIF-1 controls energy metabolism, angiogenesis, erythropoiesis, and critical events during embryogenesis (Gordan and Simon, 2007). It plays a physiological role in the cellular adaptation to hypoxia, which occurs when available oxygen falls below 5 %, and a patho-physiological role in cancer, where local hypoxia in rapidly growing tumors is thought to lead to HIF activation. COMMD1 inhibits HIF-mediated gene expression in cancer cells by physically associating with the amino terminus of HIF-1α, a subunit of the HIF heterodimer, preventing its dimerization and subsequent DNA binding and transcriptional activation (van de Sluis et al., 2010; van de Sluis et al., 2007). This suggests suppression of COMMD1 as a therapeutic avenue in cancer. Interestingly, studies have shown that there is extensive cross talk between HIF and NF-κB pathways (Rius et al., 2008; van Uden et al., 2011), and although the involvement of other COMMDs in these processes is unstudied, taken as a whole, these data support a common role for Commander linking hypoxic response to innate immunity and inflammation.
Most strikingly, elegant studies on a substantial number of Commander components provide strong evidence that the complex plays a key role in endosomal protein sorting (Bartuzi et al., 2016; Drevillon et al., 2011; Harbour et al., 2012; Li et al., 2015; Phillips-Krawczak et al., 2015). This is consistent with the subcellular localization of many Commander proteins in vesicles and/or endosomes (Fig. 4C). The endosomal system comprises a number of distinct and dynamic vesicular compartments (endosomes) that sort and deliver membrane protein cargo between the plasma membrane and the Golgi apparatus (Seaman, 2008; Soldati and Schliwa, 2006). The movement of endosomes within the cell is regulated by their interactions with cytoskeleton polymers such as microtubules and actin filaments (Soldati and Schliwa, 2006). Protein complexes that function to recognize and deliver cargo are coupled to cytoskeleton systems to drive membrane fission and vesicle movement. Endosomal protein sorting also requires localized actin polymerization, possibly to generate a local force to facilitate the production and/or scission of endosomal tubules (Seaman et al., 2013). Commander subunits have been linked to key components in the endosomal protein-sorting machinery by co-IP, co-localization, phenotypic, and phylogenetic profiling studies (Bartuzi et al., 2016; Dey et al., 2015; Harbour et al., 2012; Kolanczyk et al., 2015; Li et al., 2015; Phillips-Krawczak et al., 2015). These include the Wiskott-Aldrich syndrome proteins and SCAR homologue (WASH) complex, which is the major endosomal actin-polymerization promoting complex (Derivery et al., 2009; Gomez and Billadeau, 2009; Jia et al., 2010; Seaman et al., 2013), and the retromer complex, whose primary role is to select cargo proteins for retrograde endosomal transport to the Golgi (Hierro et al., 2007; Seaman et al., 2013).
WASH and retromer work together to facilitate the correct trafficking of many membrane cargo proteins from either the endosome to Golgi or endosome to cell surface (Seaman et al., 2013). Specifically, WASH is recruited to endosomes by the retromer to promote the formation of filamentous (F-) actin patches localized on endosomes, generating filaments that segregate membrane proteins (Harbour et al., 2012; Helfer et al., 2013; Jia et al., 2012) (Fig. 5A). These branched actin patches define discrete domains into which specific proteins are sorted for transport to their destinations (Seaman et al., 2013). Thus, WASH function is linked to the cargo protein-sorting and membrane-tubulation activity of retromer. In the next sections we outline evidence to suggest that Commander is a retromer-like sorter of cargo proteins within the endosomal system. Analogous to retromer, we hypothesize that Commander likely recruits WASH to facilitate cargo trafficking (Fig. 5A). The predicted fundamental role of the Commander complex as a key piece in the endosomal trafficking machinery helps to explain the apparent pleiotropic function of its subunits, its deep conservation (Fig. 4A), and its broad expression (Fig. 4B).
Figure 5.
Structural modeling and functional studies suggest roles for Commander. (A) Models for WASH-dependent endosomal protein trafficking by retromer (top) (based on Seaman et al., 2013) and Commander (bottom). (B) & (C) Structural data for the retromer cargo recognition complex. (B) Negative stain electron microscopy images of the human retromer cargo recognition complex (VPS26-VPS29-VPS35) of five representative structural classes. Images are reproduced from Hierro et al., 2007. (C) Small angle X-ray scattering (SAXS) model of the retromer VPS26 (purple)-VPS29 (green)-VPS35 (pink) complex, reproduced from Lucas at al., 2016. (D) Structural comparison of the retromer cargo recognition complex and core Commander subunits. Top: A structural model of a partial retromer cargo recognition complex (VPS26, red; SNX3, orange; and VPS35, purple) based on crystal structures (2R17 and 5F0J), electron microscopy, and SAXS (Hierro et al., 2007; Lucas at al., 2016). Bottom: A proposed structural model for core components of the Commander complex (DSCR3, red; COMMD1, orange; and C16orf62, purple) based on their similarities to retromer subunits. For homologous domains, the probability score calculated by HHPred (Soding, 2005) is shown. This represents the likelihood that they are true homologs when using Pfam as a database of template hidden Markov models (HMMs) (Finn et al., 2014). A score of greater than 95 % indicates that the homology is nearly certain. Crystal structures are available for the N- and C-terminus of human VPS35 (VPS35-N, residues 9-462, PDB = 5F0J; VPS35-C, residues 483-780, PDB = 2R17) and human VPS26 (PDB = 2FAU). These were used as templates to generate models of C16orf62 (C16orf62-N, residues 182-620); C16orf62-C, residues 629-926) and DSCR3, respectively, using MODELER (Sali and Blundell, 1993). Based on the predicted presence of a phosphoinositide-binding domain in the COMMDs, we suggest they occupy a homologous binding pocket to SNX3. (D) A structural comparison of the N-terminal domains of IFT81 (residues 1-109; PDB = 4LVP) and human NDC80 (residues 11-114; PDB = 2IGP) to the N-terminal domains of Commander CCDC22 (residues 1-109) and CCDC93 (residues 12-119), respectively. Homology scores were calculated by HHPred, and template structures were used to calculate models of CCDC22 and CCDC93 as described in (C). It is possible that the CH-domains in CCDC22 and CCDC93 function in microtubule binding. Interestingly, the C-terminal domains of both CCDC22 and CCDC93 show strong homology to tropomyosin (99 % and 95 %, respectively; PDB = 1C1G). (E) Phenotypic traits of defective Commander. Morpholino knockdown of COMMD2 or COMMD3 in X. laevis embryos causes defective head and eye development (left), and COMMD2/3 knockdown animals show altered neural patterning (right), including a posterior shift or loss of expression of mid-brain marker EN2 and of KROX20 (EGR1), the latter specifically in rhombomeres R3/R5. Images reproduced from Wan et al., 2015.
Commander’s role in trafficking and transport
Commander subunits COMMD1, CCDC22, CCDC93 and C16orf62 have recently been shown to function together to recognize and regulate endosomal cargo proteins such as copper transporters (Phillips-Krawczak et al., 2015), Notch receptors (Li et al., 2015) and lipoprotein receptors (Bartuzi et al., 2016). Three of these subunits form the CCC complex identified by the Burstein lab (Phillips-Krawczak et al., 2015) (Fig. 3D), a likely sub-complex of Commander. An earlier study on the role of COMMD1-mediated ubiquitination in the regulation of CFTR trafficking in cystic fibrosis also proposed that COMMD1 has a specific function within the endocytic machinery (Drevillon et al., 2011).
Trafficking of copper transporters by the CCC complex involves early endosomal colocalization with WASH, retromer, and the copper transport protein ATP7A (Phillips-Krawczak et al., 2015). WASH is a pentameric complex of WASH-1, FAM21, strumpellin, SWIP and CCDC53 (Jia et al., 2010), while mammalian retromer comprises a cargo-recognition complex and a sorting nexin (SNX) complex, each with distinct roles in endosome-to-Golgi protein retrieval (Seaman, 2008). The retromer cargo-recognition complex of three vacuolar protein sorting (VPS) proteins, VPS26, VPS29 and VPS35, is defined as such because VPS35 directly binds to the sorting motifs in cargo proteins (Nothwehr et al., 2000). SNXs are a family of proteins defined by the presence of a phospholipid-binding Phox (PX-) domain (Teasdale and Collins, 2012). The SNX complex plays a structural role in deforming the endosomal membrane to generate cargo-loaded tubules for transport (Cullen and Korswagen, 2012). Several types of SNX proteins interact with retromer: SNX3, a sorting nexin consisting only of the PX domain (Harrison et al., 2014); the SNX-BAR SNX1/2-SNX5/6 heterodimers, which have an additional bar domain (Rojas et al., 2007; Wassmer et al., 2007); and the SNX-FERM protein SNX27, which has an additional PDZ and FERM domain (Steinberg et al., 2013).
Retromer recruits WASH to endosomes by an interaction between WASH subunit FAM21 and retromer subunit VPS35 (Derivery et al., 2009; Gomez and Billadeau, 2009; Harbour et al., 2012; Helfer et al., 2013; Seaman et al., 2013) (Fig. 5A). It is thought that this interaction enables WASH to generate F-actin-driven force to increase the efficiency of tubule scission by the retromer SNX complex (Gomez and Billadeau, 2009). Key to the predicted function of the Commander complex, the Burstein lab have shown by silencing, localization and co-IP experiments that FAM21 is also required for the recruitment of the CCC complex to the endosome (Phillips-Krawczak et al., 2015). It follows that Commander, like retromer, may function in concert with the WASH complex (Fig. 5A), as we discuss further below. Consistent with this, CLIME predicts that Commander subunits COMMD3, COMMD4, CCDC22, CCDC93 and C16orf62, co-evolved with the WASH complex, indicating a combined functional role (Li et al., 2014).
Proteomic, biochemical, and 3D structural evidence suggesting Commander functions as a retromer-like sorter of endosomal proteins
Analysis of the CCC complex by the Burstein group using deletion constructs and IPs suggest an architecture where the C-termini regions of CCDC22 and CCDC93 interact with the middle of the unstructured tail region of FAM21 (Phillips-Krawczak et al., 2015) (Fig. 5A). In addition, The COMMDs appear to interact using their C-terminal COMM domain with CCDC22 (Starokadomskyy et al., 2013), an interaction that is independent of CCDC93 and C16orf62 (Phillips-Krawczak et al., 2015). IPs have also shown that the interaction of CCDC22 and CCDC93 with FAM21 does not happen at the same time as the interaction of FAM21 with the retromer subunit VPS35 (Harbour et al., 2012). We have consolidated structural, proteomic and functional evidence to suggest that Commander in its entirety forms a retromer-like complex that binds to FAM21 of WASH to traffic endosomal proteins (Fig. 5A–D).
A structural model for the architecture of the retromer cargo-recognition complex has been proposed based on data from crystal structures, electron microscopy, and SAXS (Hierro et al., 2007; Lucas et al, 2016) (Fig. 5B–D). Our preliminary structural analysis shows that COMMD1, C16orfC2 and DSCR3 have strikingly similar properties to a partial retromer cargo recognition complex of VPS26, VPS35, and SNX3 (Hierro et al., 2007; Lucas et al, 2016) (Fig. 5D). Commander protein DSCR3 and retromer VPS26 are direct homologs (Aubry et al., 2009; Shi et al., 2006), and likely act at the plasma membrane to sort specific cargos into endocytic vesicles (Zhang et al., 1997). The crystal structure of human VPS26 has been solved and adopts an arrestin fold (PDB = 2FAU) (Shi et al., 2006). We used the HHpred server for template-based structure prediction, which uses Hidden Markov Models (HMMs) to determine remote homologs (Soding, 2005), to show that VPS26 (327 residues) is the top match for DSCR3 (297 residues) with a probability score of 100 % that the two are homologs. Likewise, retromer VPS35 (796 residues), whose amino (PDB = 5F0J) and carboxy (PDB = 2R17) termini adopt an α-helical solenoid (Hierro et al., 2007; Lucas et al, 2016), is the most likely structural homolog (probability = 100 %) for the similarly-sized Commander C16orf62 (963 residues).
Although not predicted to be direct structural homologs, the SNXs and the COMMDs share some striking similarities. Based on these, we hypothesize they have parallel functional roles, and occupy equivalent binding sites, in retromer and Commander, respectively (Fig.s 5A, D). The predicted pleckstrin homology (PH-) fold of the COMM domains (Burkhead et al., 2009) (Table 1) is functionally analogous to the Phox homology (PX-) domain found in the SNXs (Cullen and Korswagen, 2012). Both PH- and PX-domains are phosphoinositide-binding domains that target proteins to phospholipid-enriched membranes (Lemmon, 2008). The COMM domains in Commander may therefore have a membrane recruitment role similar to the retromer SNX complex. In good agreement with this hypothesis, COMMD1 binds with high specificity to the lipid phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P2; PIP2), an interaction that is mediated by the COMM domain (Burkhead et al., 2009). It has been suggested that different combinations of retromer-SNX interactions give rise to different cargo selection specificities via higher order assemblies with different morphologies (Lucas et al, 2016). A similar scenario can be imagined for different combinations of COMMDs in Commander.
Finally, functional studies on the role of the CCC complex in Notch signaling report similar phenotypes in cells deficient in Commander components CCDC93 and C16orf62 and the retromer subunit VPS35, linking both Commander and retromer to Notch trafficking. Taken together, these structural, proteomic and functional similarities between Commander and retromer lead us to hypothesize that core Commander subunits COMMD-DSCR3-C16orf62 function in a complex of similar architecture to the retromer cargo-recognition complex of SNX3-VPS26-VPS35 (Fig. 5D). This model should be helpful in determining the specific events in the endosomal sorting process that are regulated by Commander, and the functional contributions of specific subunits.
It is interesting to note that in a recent bioinformatics study, Commander subunits CCDC22 and CCDC93 were classified into a previously unidentified protein family of 11 proteins defined by a novel N-terminal calponin homology (CH) domain (Schou et al., 2014). Proteins within this family, which also include several IFT-B intraflagellar transport proteins, are predicted to share yeast- Ndc80 and NuF2 as common ancestors, and have been implicated in actin and microtubule binding (Korenbaum and Rivero, 2002; Schou et al., 2014). We built homology models of the N-terminal domains of Commander CCDC22 and CCDC93 using the structures of IFT81 (PDB = 4LVP) and Ndc80 (PDB = 2IGP) as templates, respectively, which have close to 100% probability of homology (Fig. 5D). We suggest that CCDC22 and CCDC93 might function as direct points of contact between Commander and actin, perhaps by forming an actin-binding module similar to the Ndc80-NuF2 (Ciferri et al., 2008) and IFT81-IFT74 (Bhogaraju et al., 2013) complexes (Fig. 5D). This model can be used to direct further experiments into the architecture and function of Commander.
Strong links between Commander and its interactome in developmental diseases
Reverse genetic experiments (Wan et al., 2015) (Fig. 5E), combined with studies on individual Commander subunits (Kolanczyk et al., 2015; van de Sluis et al., 2007; Voineagu et al., 2012), provide useful insights into role of the complex in human development and disease. A clear importance of Commander proteins in vertebrate embryonic development has been demonstrated. In mouse, individual homozygous knockouts (KOs) of COMMD1, COMMD6, COMMD9 and COMMD10 are reported to cause embryonic-lethal phenotypes, and in some cases complex cardiovascular abnormalities (Bartuzi et al., 2013; Brown and Moore, 2012; Eppig et al., 2015; Li et al., 2015; van de Sluis et al., 2007). Interestingly, notable differences in the phenotype for embryonic lethality in KO COMMD1 and COMMD9 mice suggests non-redundant developmental functions for the two proteins, consistent with the idea of heterogeneous Commander assemblies (Li et al., 2015; van de Sluis et al., 2007). We have characterized several Commander genes in frog, which indicate the complex has an explicit role in tadpole eye and head development (Fig. 5F) (Wan et al., 2015). In particular, morpholino-oligonucleotide (MO) mediated gene knockdowns (KDs) of either COMMD2 or COMMD3 in frog embryos give phenotypes with defective head and eye development along with altered neural patterning. This includes defects in the patterning of the hindbrain, the precursor to the cerebellum (Fig. 5E). Taken together, these findings in mouse and frog point towards a crucial role for Commander in vertebrate embryogenesis and provide the basis for more detailed genetic studies.
Consistent with these studies, defects in one Commander gene are implicated in human Ritscher-Schinzel (RS)/3C (cranio-cerebro-cardiac) syndrome, a rare disorder characterized by intellectual disability, cerebellar brain malformations, congenital heart defects, and craniofacial abnormalities (Ritscher et al., 1987). Specifically, missense alleles T17A and Y557C in CCDC22 were identified as causes of RS/3C syndrome in human patients (Kolanczyk et al., 2015; Voineagu et al., 2012), who display profound developmental phenotypes similar to our Xenopus Commander KD animals. Combined with the deep conservation of the entire Commander complex, this implies all Commander genes as strong candidates in the etiology of RS/3C syndrome. KIAA0196 of the WASH complex, also known as strumpellin, has also been directly linked to RS/3C syndrome by the disease allele that leads to a substitution of the last 99 amino-acids of the protein (Elliott et al., 2013). This suggests that the endosomal sorting processes governed by the interplay of the Commander and WASH (Fig. 5A) are key to developmental processes, and disruptions in this machinery may cause disorders such as RS/3C-syndrome. This is consistent with the documented crucial role of the CCC complex in the vesicular sorting and delivery of Notch proteins to the cell surface (Li et al., 2015), which are transmembrane receptors involved in many stages of development by forming a highly conserved signaling pathway between cells (High and Epstein, 2008). The links between COMMD1 and anti-inflammatory pathways (Bartuzi et al., 2013; de Becdelievre et al., 2013; de Bie et al., 2006; Ganesh et al., 2003; Maine et al., 2007), hypoxia-related tumor invasion (van de Sluis et al., 2010), and endosomal trafficking of the CFTR-gene (de Becdelievre et al., 2013; Drevillon et al., 2011) warrant further research into the role of Commander and its protein-sorting function in diseases such as cancer and cystic fibrosis.
Future outlook
We have presented an overview of recent proteome-wide human interactome experiments that together with gene-focused studies have uncovered a large new multiprotein complex called Commander. Commander is a deeply conserved, macromolecular machine of unknown architecture, and contains genes that are vital for vertebrate embryogenesis and are implicated in developmental disease. The preliminary evidence we have summarized here suggests that Commander functions as an ancient complex of central importance in intracellular protein trafficking. By combining insights from both large- and small-scale studies, we have proposed a model of the Commander complex to help guide future studies into its structure and function. We expect future research to provide crucial mechanistic information about the role of the complex in developmental defects and rare disorders, and also chronic diseases such as cancer and cystic fibrosis.
Acknowledgments
The authors acknowledge funding from the National Institutes of Health (DP1 GM106408, R01 DK110520, R01 HD085901, R35 GM122480), the National Science Foundation, and the Welch Foundation (F1515).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altenhoff AM, Boeckmann B, Capella-Gutierrez S, Dalquen DA, DeLuca T, Forslund K, Huerta-Cepas J, Linard B, Pereira C, Pryszcz LP, et al. Standardized benchmarking in the quest for orthologs. Nature Methods. 2016;13:425–430. doi: 10.1038/nmeth.3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubry L, Guetta D, Klein G. The arrestin fold: variations on a theme. Current Genomics. 2009;10:133–142. doi: 10.2174/138920209787847014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartuzi P, Billadeau DD, Favier R, Rong S, Dekker D, Fedoseienko A, Fieten H, Wijers M, Levels JH, Huijkman N, et al. CCC- and WASH-mediated endosomal sorting of LDLR is required for normal clearance of circulating LDL. Nature Communications. 2016;7:10961. doi: 10.1038/ncomms10961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartuzi P, Hofker MH, van de Sluis B. Tuning NF-kappaB activity: a touch of COMMD proteins. Biochimica et Biophysica Acta. 2013;1832:2315–2321. doi: 10.1016/j.bbadis.2013.09.014. [DOI] [PubMed] [Google Scholar]
- Bhogaraju S, Cajanek L, Fort C, Blisnick T, Weber K, Taschner M, Mizuno N, Lamla S, Bastin P, Nigg EA, et al. Molecular Basis of Tubulin Transport Within the Cilium by IFT74 and IFT81. Science. 2013;341:1009–1012. doi: 10.1126/science.1240985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biasio W, Chang T, McIntosh CJ, McDonald FJ. Identification of Murr1 as a regulator of the human delta epithelial sodium channel. The Journal of Biological Chemistry. 2004;279:5429–5434. doi: 10.1074/jbc.M311155200. [DOI] [PubMed] [Google Scholar]
- Brown SDM, Moore MW. The International Mouse Phenotyping Consortium: past and future perspectives on mouse phenotyping. Mamm Genome. 2012;23:632–640. doi: 10.1007/s00335-012-9427-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhead JL, Morgan CT, Shinde U, Haddock G, Lutsenko S. COMMD1 forms oligomeric complexes targeted to the endocytic membranes via specific interactions with phosphatidylinositol 4,5-bisphosphate. The Journal of Biological Chemistry. 2009;284:696–707. doi: 10.1074/jbc.M804766200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burstein E, Hoberg JE, Wilkinson AS, Rumble JM, Csomos RA, Komarck CM, Maine GN, Wilkinson JC, Mayo MW, Duckett CS. COMMD proteins, a novel family of structural and functional homologs of MURR1. The Journal of Biological Chemistry. 2005;280:22222–22232. doi: 10.1074/jbc.M501928200. [DOI] [PubMed] [Google Scholar]
- Chang T, Ke Y, Ly K, McDonald FJ. COMMD1 regulates the delta epithelial sodium channel (deltaENaC) through trafficking and ubiquitination. Biochemical and Biophysical Research Communications. 2011;411:506–511. doi: 10.1016/j.bbrc.2011.06.149. [DOI] [PubMed] [Google Scholar]
- Ciferri C, Pasqualato S, Screpanti E, Varetti G, Santaguida S, Dos Reis G, Maiolica A, Polka J, De Luca JG, De Wulf P, et al. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 2008;133:427–439. doi: 10.1016/j.cell.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium, F., the, RP., Clst. Forrest AR, Kawaji H, Rehli M, Baillie JK, de Hoon MJ, Haberle V, Lassmann T, et al. A promoter-level mammalian expression atlas. Nature. 2014;507:462–470. doi: 10.1038/nature13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen PJ, Korswagen HC. Sorting nexins provide diversity for retromer-dependent trafficking events. Nature Cell Biology. 2012;14:29–37. doi: 10.1038/ncb2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Becdelievre A, Rocca J, Aissat A, Drevillon L, Moutereau S, Le Gouvello S, Hinzpeter A, Tarze A, Fanen P. COMMD1 modulates noxious inflammation in cystic fibrosis. The International Journal of Biochemistry & Cell Biology. 2013;45:2402–2409. doi: 10.1016/j.biocel.2013.07.012. [DOI] [PubMed] [Google Scholar]
- de Bie P, de Sluis B, Burstein E, Duran KJ, Berger R, Duckett CS, Wijmenga C, Klomp LWJ. Characterization of COMMD protein-protein interactions in NF-kappa B signalling. Biochemical Journal. 2006;398:63–71. doi: 10.1042/BJ20051664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derivery E, Sousa C, Gautier JJ, Lombard B, Loew D, Gautreau A. The Arp2/3 Activator WASH Controls the Fission of Endosomes through a Large Multiprotein Complex. Developmental Cell. 2009;17:712–723. doi: 10.1016/j.devcel.2009.09.010. [DOI] [PubMed] [Google Scholar]
- Dey G, Jaimovich A, Collins SR, Seki A, Meyer T. Systematic Discovery of Human Gene Function and Principles of Modular Organization through Phylogenetic Profiling. Cell Reports. 2015;10:993–1006. doi: 10.1016/j.celrep.2015.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drevillon L, Tanguy G, Hinzpeter A, Arous N, de Becdelievre A, Aissat A, Tarze A, Goossens M, Fanen P. COMMD1-mediated ubiquitination regulates CFTR trafficking. PloS ONE. 2011;6:e18334. doi: 10.1371/journal.pone.0018334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew K, Lee C, Huizar RL, Tu F, Borgeson B, McWhite CD, Ma Y, Wallingford JB, Marcotte EM. A synthesis of over 9, 000 mass spectrometry experiments reveals the core set of human protein complexes. bioRxiv. 2017 doi: 10.15252/msb.20167490. https://doi.org/10.1101/092361. [DOI] [PMC free article] [PubMed]
- Elliott AM, Simard LR, Coghlan G, Chudley AE, Chodirker BN, Greenberg CR, Burch T, Ly V, Hatch GM, Zelinski T. A novel mutation in KIAA0196: identification of a gene involved in Ritscher-Schinzel/3C syndrome in a First Nations cohort. J Med Genet. 2013;50:819–822. doi: 10.1136/jmedgenet-2013-101715. [DOI] [PubMed] [Google Scholar]
- Eppig JT, Blake JA, Bult CJ, Kadin JA, Richardson JE, Grp MGD. The Mouse Genome Database (MGD): facilitating mouse as a model for human biology and disease. Nucleic Acids Research. 2015;43:D726–D736. doi: 10.1093/nar/gku967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, et al. Pfam: the protein families database. Nucleic Acids Research. 2014;42:D222–230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabaldon T, Koonin EV. Functional and evolutionary implications of gene orthology. Nature Reviews Genetics. 2013;14:360–366. doi: 10.1038/nrg3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesh L, Burstein E, Guha-Niyogi A, Louder MK, Mascola JR, Klomp LW, Wijmenga C, Duckett CS, Nabel GJ. The gene product Murr1 restricts HIV-1 replication in resting CD4+ lymphocytes. Nature. 2003;426:853–857. doi: 10.1038/nature02171. [DOI] [PubMed] [Google Scholar]
- Gomez TS, Billadeau DD. A FAM21-containing WASH complex regulates retromer-dependent sorting. Developmental Cell. 2009;17:699–711. doi: 10.1016/j.devcel.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordan JD, Simon MC. Hypoxia-inducible factors: central regulators of the tumor phenotype. Current Opinion in Genetics & Development. 2007;17:71–77. doi: 10.1016/j.gde.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco TM, Cristea IM. The Biochemical Evolution of Protein Complexes. Trends in Biochemical Sciences. 2016;41:4–6. doi: 10.1016/j.tibs.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbour ME, Breusegem SY, Seaman MN. Recruitment of the endosomal WASH complex is mediated by the extended ‘tail’ of Fam21 binding to the retromer protein Vps35. The Biochemical Journal. 2012;442:209–220. doi: 10.1042/BJ20111761. [DOI] [PubMed] [Google Scholar]
- Harrison MS, Hung CS, Liu TT, Christiano R, Walther TC, Burd CG. A mechanism for retromer endosomal coat complex assembly with cargo. Proc Natl Acad Sci USA. 2014;111:267–272. doi: 10.1073/pnas.1316482111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein MY, Hubner NC, Poser I, Cox J, Nagaraj N, Toyoda Y, Gak IA, Weisswange I, Mansfeld J, Buchholz F, et al. A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell. 2015;163:712–723. doi: 10.1016/j.cell.2015.09.053. [DOI] [PubMed] [Google Scholar]
- Helfer E, Harbour ME, Henriot V, Lakisic G, Sousa-Blin C, Volceanov L, Seaman MN, Gautreau A. Endosomal recruitment of the WASH complex: active sequences and mutations impairing interaction with the retromer. Biology of the Cell. 2013;105:191–207. doi: 10.1111/boc.201200038. [DOI] [PubMed] [Google Scholar]
- Hierro A, Rojas AL, Rojas R, Murthy N, Effantin G, Kajava AV, Steven AC, Bonifacino JS, Hurley JH. Functional architecture of the retromer cargo-recognition complex. Nature. 2007;449:1063–1067. doi: 10.1038/nature06216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- High FA, Epstein JA. The multifaceted role of Notch in cardiac development and disease. Nature Reviews Genetics. 2008;9:49–61. doi: 10.1038/nrg2279. [DOI] [PubMed] [Google Scholar]
- Hu YH, Warnatz HJ, Vanhecke D, Wagner F, Fiebitz A, Thamm S, Kahlem P, Lehrach H, Yaspo ML, Janitz M. Cell array-based intracellular localization screening reveals novel functional features of human chromosome 21 proteins. BMC Genomics. 2006;7:155. doi: 10.1186/1471-2164-7-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttlin EL, Ting L, Bruckner RJ, Gebreab F, Gygi MP, Szpyt J, Tam S, Zarraga G, Colby G, Baltier K, et al. The BioPlex Network: A Systematic Exploration of the Human Interactome. Cell. 2015;162:425–440. doi: 10.1016/j.cell.2015.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia D, Gomez TS, Billadeau DD, Rosen MK. Multiple repeat elements within the FAM21 tail link the WASH actin regulatory complex to the retromer. Molecular Biology of the Cell. 2012;23:2352–2361. doi: 10.1091/mbc.E11-12-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia D, Gomez TS, Metlagel Z, Umetani J, Otwinowski Z, Rosen MK, Billadeau DD. WASH and WAVE actin regulators of the Wiskott-Aldrich syndrome protein (WASP) family are controlled by analogous structurally related complexes. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:10442–10447. doi: 10.1073/pnas.0913293107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkwood KJ, Ahmad Y, Larance M, Lamond AI. Characterization of native protein complexes and protein isoform variation using size-fractionation-based quantitative proteomics. Molecular & Cellular Proteomics: MCP. 2013;12:3851–3873. doi: 10.1074/mcp.M113.032367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolanczyk M, Krawitz P, Hecht J, Hupalowska A, Miaczynska M, Marschner K, Schlack C, Emmerich D, Kobus K, Kornak U, et al. Missense variant in CCDC22 causes X-linked recessive intellectual disability with features of Ritscher-Schinzel/3C syndrome. European Journal of Human Genetics. 2015;23:633–638. doi: 10.1038/ejhg.2014.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korenbaum E, Rivero F. Calponin homology domains at a glance. Journal of Cell Science. 2002;115:3543–3545. doi: 10.1242/jcs.00003. [DOI] [PubMed] [Google Scholar]
- Lemmon MA. Membrane recognition by phospholipid-binding domains. Nature Reviews Molecular Cell Biology. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- Li H, Koo Y, Mao X, Sifuentes-Dominguez L, Morris LL, Jia D, Miyata N, Faulkner RA, van Deursen JM, Vooijs M, et al. Endosomal sorting of Notch receptors through COMMD9-dependent pathways modulates Notch signaling. The Journal of Cell Biology. 2015;211:605–617. doi: 10.1083/jcb.201505108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Calvo SE, Gutman R, Liu JS, Mootha VK. Expansion of biological pathways based on evolutionary inference. Cell. 2014;158:213–225. doi: 10.1016/j.cell.2014.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindskog C. The potential clinical impact of the tissue-based map of the human proteome. Expert Rev Proteomic. 2015;12:213–215. doi: 10.1586/14789450.2015.1040771. [DOI] [PubMed] [Google Scholar]
- Lucas M, Gershlick DC, Vidaurrazaga A, Rojas AL, Bonifacino JS, Hierro A. Structural mechanism for cargo recognition by the retromer complex. Cell. 2016;167:1623–1635. doi: 10.1016/j.cell.2016.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maine GN, Burstein E. COMMD proteins and the control of the NF kappa B pathway. Cell Cycle. 2007;6:672–676. doi: 10.4161/cc.6.6.3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maine GN, Mao X, Komarck CM, Burstein E. COMMD1 promotes the ubiquitination of NF-kappaB subunits through a cullin-containing ubiquitin ligase. The EMBO Journal. 2007;26:436–447. doi: 10.1038/sj.emboj.7601489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X, Gluck N, Chen B, Starokadomskyy P, Li H, Maine GN, Burstein E. COMMD1 (copper metabolism MURR1 domain-containing protein 1) regulates Cullin RING ligases by preventing CAND1 (Cullin-associated Nedd8-dissociated protein 1) binding. J Biol Chem. 2011;286:32355–32365. doi: 10.1074/jbc.M111.278408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh JA, Teichmann SA. Structure, dynamics, assembly, and evolution of protein complexes. Annual Review of Biochemistry. 2015;84:551–575. doi: 10.1146/annurev-biochem-060614-034142. [DOI] [PubMed] [Google Scholar]
- Muller PA, van de Sluis B, Groot AJ, Verbeek D, Vonk WI, Maine GN, Burstein E, Wijmenga C, Vooijs M, Reits E, et al. Nuclear-cytosolic transport of COMMD1 regulates NF-kappaB and HIF-1 activity. Traffic. 2009;10:514–527. doi: 10.1111/j.1600-0854.2009.00892.x. [DOI] [PubMed] [Google Scholar]
- Nothwehr SF, Ha SA, Bruinsma P. Sorting of yeast membrane proteins into an endosome-to-Golgi pathway involves direct interaction of their cytosolic domains with Vps35p. The Journal of Cell Biology. 2000;151:297–310. doi: 10.1083/jcb.151.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini M, Marcotte EM, Thompson MJ, Eisenberg D, Yeates TO. Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:4285–4288. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips-Krawczak CA, Singla A, Starokadomskyy P, Deng Z, Osborne DG, Li H, Dick CJ, Gomez TS, Koenecke M, Zhang JS, et al. COMMD1 is linked to the WASH complex and regulates endosomal trafficking of the copper transporter ATP7A. Molecular Biology of the Cell. 2015;26:91–103. doi: 10.1091/mbc.E14-06-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman S, Vernon R, Thompson J, Tyka M, Sadreyev R, Pei J, Kim D, Kellogg E, DiMaio F, Lange O, et al. Structure prediction for CASP8 with all-atom refinement using Rosetta. Proteins. 2009;77(Suppl 9):89–99. doi: 10.1002/prot.22540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritscher D, Schinzel A, Boltshauser E, Briner J, Arbenz U, Sigg P. Dandy-Walker(like) malformation, atrio-ventricular septal defect and a similar pattern of minor anomalies in 2 sisters: a new syndrome? American Journal of Medical Genetics. 1987;26:481–491. doi: 10.1002/ajmg.1320260227. [DOI] [PubMed] [Google Scholar]
- Rius J, Guma M, Schachtrup C, Akassoglou K, Zinkernagel AS, Nizet V, Johnson RS, Haddad GG, Karin M. NF-kappaB links innate immunity to the hypoxic response through transcriptional regulation of HIF-1alpha. Nature. 2008;453:807–811. doi: 10.1038/nature06905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas R, Kametaka S, Haft CR, Bonifacino JS. Interchangeable but essential functions of SNX1 and SNX2 in the association of retromer with endosomes and the trafficking of mannose 6-phosphate receptors. Mol Cell Biol. 2007;27:1112–1124. doi: 10.1128/MCB.00156-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum JL, Witman GB. Intraflagellar transport. Nature reviews Molecular cell biology. 2002;3:813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- Ruepp A, Waegele B, Lechner M, Brauner B, Dunger-Kaltenbach I, Fobo G, Frishman G, Montrone C, Mewes HW. CORUM: the comprehensive resource of mammalian protein complexes--2009. Nucleic Acids Research. 2010;38:D497–501. doi: 10.1093/nar/gkp914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. Journal of Molecular Biology. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Schou KB, Andersen JS, Pedersen LB. A divergent calponin homology (NN-CH) domain defines a novel family: implications for evolution of ciliary IFT complex B proteins. Bioinformatics. 2014;30:899–902. doi: 10.1093/bioinformatics/btt661. [DOI] [PubMed] [Google Scholar]
- Seaman MN. Endosome protein sorting: motifs and machinery. Cellular and Molecular Life Sciences. 2008;65:2842–2858. doi: 10.1007/s00018-008-8354-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman MN, Gautreau A, Billadeau DD. Retromer-mediated endosomal protein sorting: all WASHed up! Trends in Cell Biology. 2013;23:522–528. doi: 10.1016/j.tcb.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Rojas R, Bonifacino JS, Hurley JH. The retromer subunit Vps26 has an arrestin fold and binds Vps35 through its C-terminal domain. Nature Structural & Molecular Biology. 2006;13:540–548. doi: 10.1038/nsmb1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- Soldati T, Schliwa M. Powering membrane traffic in endocytosis and recycling. Nature Reviews Molecular Cell Biology. 2006;7:897–908. doi: 10.1038/nrm2060. [DOI] [PubMed] [Google Scholar]
- Sommerhalter M, Zhang Y, Rosenzweig AC. Solution structure of the COMMD1 N-terminal domain. Journal of Molecular Biology. 2007;365:715–721. doi: 10.1016/j.jmb.2006.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starokadomskyy P, Gluck N, Li H, Chen B, Wallis M, Maine GN, Mao X, Zaidi IW, Hein MY, McDonald FJ, et al. CCDC22 deficiency in humans blunts activation of proinflammatory NF-kappaB signaling. The Journal of Clinical Investigation. 2013;123:2244–2256. doi: 10.1172/JCI66466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg F, Gallon M, Winfield M, Thomas EC, Bell AJ, Heesom KJ, Tavaré JM, Cullen PJ. A global analysis of SNX27-retromer assembly and cargo specificity reveals a function in glucose and metal ion transport. Nat Cell Biol. 2013;15:461–471. doi: 10.1038/ncb2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teasdale RD, Collins BM. Insights into the PX (phox-homology) domain and SNX (sorting nexin) protein families: Structures, functions and roles in disease. Biochem J. 2012;441:39–59. doi: 10.1042/BJ20111226. [DOI] [PubMed] [Google Scholar]
- van de Sluis B, Mao X, Zhai Y, Groot AJ, Vermeulen JF, van der Wall E, van Diest PJ, Hofker MH, Wijmenga C, Klomp LW, et al. COMMD1 disrupts HIF-1alpha/beta dimerization and inhibits human tumor cell invasion. The Journal of Clinical Investigation. 2010;120:2119–2130. doi: 10.1172/JCI40583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Sluis B, Muller P, Duran K, Chen A, Groot AJ, Klomp LW, Liu PP, Wijmenga C. Increased activity of hypoxia-inducible factor 1 is associated with early embryonic lethality in Commd1 null mice. Molecular and Cellular Biology. 2007;27:4142–4156. doi: 10.1128/MCB.01932-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van De Sluis B, Rothuizen J, Pearson PL, van Oost BA, Wijmenga C. Identification of a new copper metabolism gene by positional cloning in a purebred dog population. Human Molecular Genetics. 2002;11:165–173. doi: 10.1093/hmg/11.2.165. [DOI] [PubMed] [Google Scholar]
- van Uden P, Kenneth NS, Webster R, Muller HA, Mudie S, Rocha S. Evolutionary conserved regulation of HIF-1beta by NF-kappaB. PLoS Genetics. 2011;7:e1001285. doi: 10.1371/journal.pgen.1001285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voineagu I, Huang L, Winden K, Lazaro M, Haan E, Nelson J, McGaughran J, Nguyen LS, Friend K, Hackett A, et al. CCDC22: a novel candidate gene for syndromic X-linked intellectual disability. Molecular Psychiatry. 2012;17:4–7. doi: 10.1038/mp.2011.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan C, Borgeson B, Phanse S, Tu F, Drew K, Clark G, Xiong X, Kagan O, Kwan J, Berzginov A, et al. Panorama of ancient metazoan macromolecular complexes. Nature. 2015;525:339–344. doi: 10.1038/nature14877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Herrmann CJ, Simonovic M, Szklarczyk D, von Mering C. Version 4.0 of PaxDb: Protein abundance data, integrated across model organisms, tissues, and cell-lines. Proteomics. 2015;15:3163–3168. doi: 10.1002/pmic.201400441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassmer T, Attar N, Bujny MV, Oakley J, Traer CJ, Cullen PJ. A loss-of-function screen reveals SNX5 and SNX6 as potential components of the mammalian retromer. J Cell Sci. 2007;120:45–54. doi: 10.1242/jcs.03302. [DOI] [PubMed] [Google Scholar]
- Wilkins MR, Gasteiger E, Bairoch A, Sanchez JC, Williams KL, Appel RD, Hochstrasser DF. Protein identification and analysis tools in the ExPASy server. Methods Mol Biol. 1999;112:531–552. doi: 10.1385/1-59259-584-7:531. [DOI] [PubMed] [Google Scholar]
- Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. The I-TASSER Suite: protein structure and function prediction. Nature Methods. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Barak LS, Winkler KE, Caron MG, Ferguson SS. A central role for beta-arrestins and clathrin-coated vesicle-mediated endocytosis in beta2-adrenergic receptor resensitization. Differential regulation of receptor resensitization in two distinct cell types. The Journal of Biological Chemistry. 1997;272:27005–27014. doi: 10.1074/jbc.272.43.27005. [DOI] [PubMed] [Google Scholar]