Abstract
An emerging body of evidence indicates that post-transcriptional gene regulation not only relies on the linear sequence of messenger RNAs but also on their folding into intricate secondary structures and on chemical modification of the RNA bases. These features, which are highly dynamic and interdependent, exert direct control over the transcriptome thereby influencing many aspects of cell function. Here, we consider that coupling of RNA modifications and structure actively shapes RNA-protein interactions through individual steps of gene expression.
Introduction
The life of mRNAs involves intricate coordination of precursor mRNA (pre-mRNA) processing events, including capping, splicing and polyadenylation, as well as the export of mature transcripts to the cytoplasm for translation. How do cells exert tight spatiotemporal control over these multi-step processes? Coupling of nuclear and cytosolic events can be accomplished through two distinct mechanisms. First, nucleocytoplasmic shuttling RNA binding proteins (RBPs) can remain bound to their target transcripts to influence downstream processes. An excellent example of this is serine/arginine-rich (SR) proteins, RBPs initially identified for their roles in splicing1 but later found to serve important roles in transcription elongation, poly(A) site selection, mRNA export, and translation1–5. Second, nuclear processing including splicing and polyadenylation, as well as dynamic changes in RNA chemical modifications and secondary structures (discussed below), can facilitate downstream RBP associations in the cytoplasm.
As RBPs link multiple steps of gene expression, factors governing their binding to target RNAs must be considered to understand how coupling is achieved. The mRNA sequence is often insufficient to drive protein binding, as RBP-consensus motifs are highly overrepresented compared to the incidence of binding6–8. What additional features are responsible for determining the specificity of protein-RNA interactions in cells? Several recent studies have revealed that post-transcriptional modification of RNA through the addition or removal of chemical groups (termed the epitranscriptome) can expand the information encoded by RNA9. As we discuss below, RNA modifications, which are dynamic and reversible, can modulate protein-RNA interactions and mediate rapid responses to environmental changes. In addition to the panoply of chemical modifications, mRNAs also fold into intricate secondary structures. Until recently, the structural aspect of the transcriptome was largely unexplored owing to technical limitations. However, the development of new transcriptome-wide approaches to study RNA structure in vivo has unraveled their plasticity, complexity, and functionality10–12.
In this Opinion article, we discuss recent findings that demonstrate the functional coupling and interdependence of mRNA modifications and structures, and provide an updated view of gene expression that incorporates the dynamics and physiological relevance of these RNA features.
RNA Modification: The Epitranscriptome
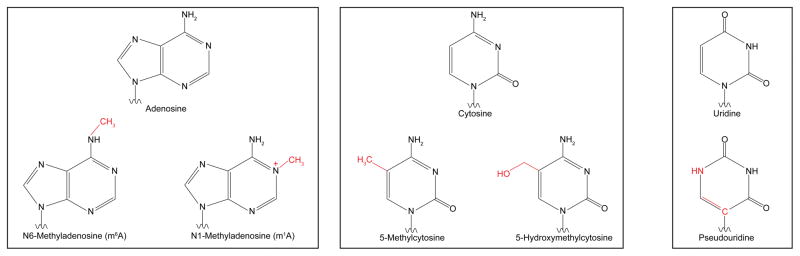
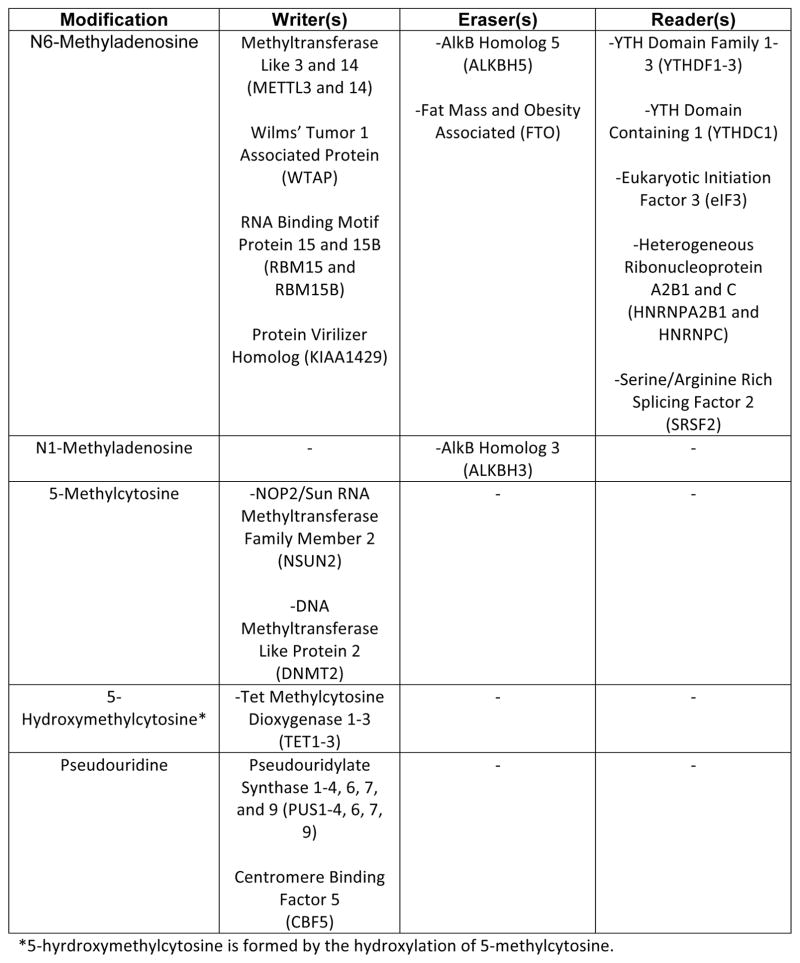
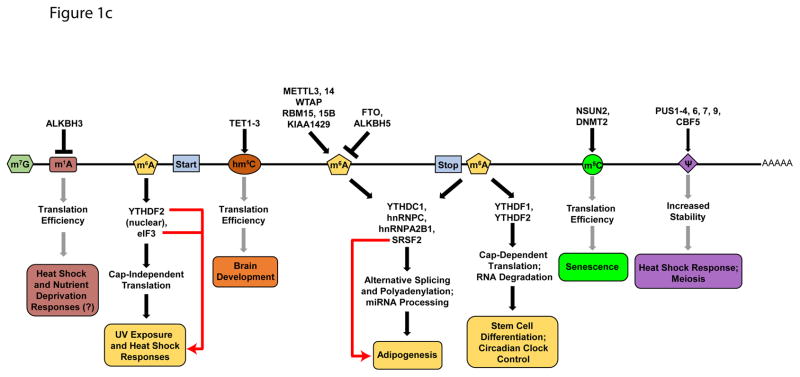
The epitranscriptome is shaped through the activity of evolutionarily conserved factors that encode (writers), decode (readers), and remove (erasers) various chemical modifications on RNA bases (Figure 1a & b). More than 100 distinct modifications have been found in cellular RNAs13, with the list continuing to grow. While most modifications—including N6-methyladenosine (m6A), 5-methylcytosine (m5C), N1-methyladenosine (m1A), pseudouridine (ψ), as well as adenosine-to-inosine and cytosine-to-uridine editing (not discussed here, see reviews for details14, 15)—were originally identified in highly abundant rRNAs, tRNAs, and small nuclear RNA (snRNAs)16, 17, recent advances in sequencing and mass spectrometry approaches have allowed for their detection and characterization in relatively lowly expressed mRNAs. Genome-wide analyses coupled with gain- and loss-of-function studies of readers, writers, and erasers have (i) expanded the repertoire of mRNA modifications in eukaryotic transcriptomes, (ii) annotated their abundance and distribution along transcripts, (iii) mapped the modification sites conserved between species, (iv) unraveled changes in modification abundance and position in response to environmental changes, (v) demonstrated their impact on mRNA processing, stability and translation, and (vi) revealed physiological consequences of modification dynamics (Figure 1c).
FIGURE 1. RNA Modifications and Their Biological Functions.
a | Chemical structures of unmodified RNA bases (top) and the modified structures (below). b | Table of writers, readers, and erasers of RNA modifications. c | Biological effects of RNA modifications. In response to stress, N1-methyladenosine (m1A) and N6-methyladenosine (m6A) levels in 5′UTRs are dynamically altered. m1A promotes cap-dependent translation through an unknown reader, while 5′UTR m6A facilitates cap-independent translation through interactions with eIF3. 5′UTR m6A is protected from demethylation by YTHDF2. In coding sequences, 5-hydroxymethylcytosine (hm5C) increases the translation efficiency of modified transcripts via an unknown mechanism. hm5C levels are highest in the brain, and ablation of TET genes affects brain development in Drosophila melanogaster. m6A in coding regions affects RNA processing by modulating the binding of YTHDC1, hnRNPC, hnRNPA2B1, and SRSF2. Removal of m6A by FTO antagonizes SRSF2 binding and is necessary for adipogenesis. 3′UTR m6A also affects RNA processing via the aforementioned readers, as well as cap-dependent translation via YTHDF1 and RNA degradation via YTHDF2. Regulation of RNA stability by m6A is crucial for stem cell differentiation and circadian clock control. 5-methylcytosine (m5C) is linked to translational control of senescence-related genes. Pseudouridine (ψ) in 3′ UTRs has been shown to increase the stability of target transcripts during heat shock. Gray arrows are used for modifications with no known readers. Red arrows connect readers to biological processes they are directly associated with.
RNA Modifications Expand the Splicing Code
Amongst internal mRNA modifications, m6A is the most prevalent—accounting for ~80% of all RNA base methylations in eukaryotes18, 19. Emerging evidence demonstrates that m6A deposition—catalyzed by a writer complex consisting of methyltransferase-like 3 and 14 (METTL3 and METTL14), Wilm’s tumor 1-associated protein (WTAP), protein virilizer homolog (KIAA1429), and RNA-binding motif protein 15 and 15B (RBM15 and RBM15B)18–26—has important roles in regulating pre-mRNA processing. In mammals, m6A is enriched in 3′ untranslated regions (UTRs), introns, and alternatively spliced exons18, 19, 27, and depletion of METTL3, WTAP, or the m6A eraser fat mass and obesity-associated (FTO)28, results in splicing defects for hundreds of genes18, 25, 29. A portion of these events occurs in 3′UTRs and last exons, suggesting m6A also impacts polyadenylation. Indeed, higher m6A density near proximal poly(A) sites correlates with decreased usage of those sites for polyadenylation, whereas m6A at sites upstream of and near distal poly(A) sites correlates with increased usage in a subset of mRNAs27.
The sizeable proportion (~30%) of m6A found in introns indicates it is likely deposited co-transcriptionally and prior to splicing23, 30. Likewise, co-localization of m6A methyltransferases and demethylases with pre-mRNA splicing factors in the splicing-associated nuclear speckles suggests an intrinsic connection between m6A and splicing23, 31. Furthermore, deficiency of the m6A demethylase AlkB homolog 5 (ALKBH5) disrupts the recruitment of the splicing factors SRSF1, SRSF2, and the SRSF Protein Kinase 1 (SRPK1) to nuclear speckles31. As phosphorylation of SR proteins by SRPKs and their localization are intimately linked with splicing32, 33, these results partially explain how changes in m6A could affect global splice site choices. In another study, FTO-regulated m6A sites were found to display a marked overlap (60–80%) with exonic splicing enhancer sequences bound by SRSF229. Strikingly, SRSF2 binding and exon inclusion are inversely correlated to FTO expression, indicating that m6A potentiates SRSF2 binding and splicing regulation. However, it is unclear whether this is a direct effect or if m6A-binding proteins modulate SRSF2 binding. The m6A reader YTH domain containing 1 (YTHDC1) presents an example supporting the second scenario. Through binding to m6A, YTHDC1 modulates thousands of splicing events through the promotion or inhibition of transcript binding by SRSF3 or SRSF10, respectively34. Splicing factors can also directly bind to m6A-containing sequences. HNRNPA2B1, a member of the heterogeneous nuclear ribonucleoprotein (hnRNP) family of splicing factors, directly binds to m6A-bearing regions near exons regulated by METTL3 and promotes splicing in the same direction35. Thus, m6A serves as a docking site allowing protein-RNA interactions that can directly or indirectly influence pre-mRNA processing.
Modifications and miRNAs Cooperate to Control mRNA Decay
In addition to affecting pre-mRNA processing, RNA modifications control mRNA decay. The development of chemical labeling techniques has led to the transcriptome-wide mapping of ψ in yeast and human mRNAs36–39. Pseudouridylation of mRNAs is catalyzed by the pseudouridylate synthase (PUS) family of enzymes. Upon heat shock in yeast, PUS7 catalyzes ψ formation at over 200 distinct sites39. PUS7 targets became more highly expressed compared to those in PUS7-deficient cells, suggesting ψ actively regulates mRNA stability. Conversely, depletion of m6A writer METTL3 results in a global increase in target mRNA half-life40, 41. In mouse and human embryonic stem cells, this stabilization occurs in pluripotency genes resulting in increased abundance and failure to exit naïve pluripotency41, 42. These effects are regulated by the m6A reader YTH domain family 2 (YTHDF2) and by the RBP human antigen R (HuR), with opposing consequences. YTHDF2 binding to m6A recruits the CCR4-NOT deadenylase complex43 and facilitates mRNA transport to processing bodies44, sites of mRNA decay45. However, conflicting functions for YTHDF2 in stabilizing HIV mRNA and YTHDF2-tethered reporter constructs were published recently46, raising questions about the role of YTHDF2 in destabilizing mRNAs. By contrast, HuR stabilizes these mRNAs by binding in their 3′UTRs, thereby blocking miRNA binding30. Thus, decreased methylation following METTL3 depletion promotes HuR binding and stabilizes target transcripts by blocking miRNA-dependent mRNA decay.
The spatial proximity of m6A and miRNA-binding sites in 3′UTRs raises an interesting question—do miRNAs guide m6A writers to methylate specific adenosines? Recently, miRNAs were shown to modulate METTL3 binding to transcripts through a sequence-dependent pairing mechanism and confer methylation specificity independently of the miRNA-interacting Argonaute (AGO) proteins47. Although intriguing, this result has yet to be validated by other groups. METTL3 also methylates many primary miRNAs (pri-miRNAs), and is obligatory for their processing by the Microprocessor Complex Protein DGCR848. This is mediated by HNRNPA2B1, which recruits DGCR8 to pri-miRNAs48. Additionally—although the abundance of m5C in mRNA is low—transcriptome-wide profiling shows substantial overlap between m5C sites and Ago1–4 binding sites49, suggesting a role in miRNA-mediated silencing. Collectively, these studies highlight the dynamic cooperation between the epitranscriptome and miRNAs to modulate mRNA decay.
Epitranscriptome Directs Translation During Stress
Translational regulation is a highly dynamic process that facilitates rapid responses to environmental stimuli owing to the ample availability of translationally-competent mRNA pools. Translation initiation can be cap-dependent or cap-independent50, and the epitranscriptome has profound effects on both mechanisms.
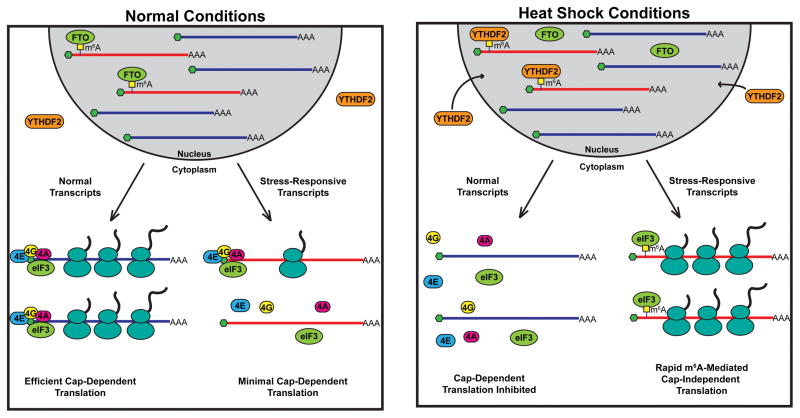
In human cells, the m6A reader YTHDF1 promotes cap-dependent translation initiation through associations with eukaryotic initiation factor 3 (eIF3) and other RBPs51. However, YTHDF1 has also been shown to be interchangeable with YTHDF2 and YTHDF3 in regulating mRNA decay43, 46, 52, suggestive of a role outside of translation. Although m6A levels are low in 5′UTRs, heat shock significantly increases m6A in 5′UTRs of heat-shock responsive genes in mammalian cells53, 54. Heat shock inhibits cap-dependent translation55 but, remarkably, a single m6A residue in 5′UTRs is sufficient to facilitate cap-independent translation of heat-shock responsive genes, including heat shock protein 70 (HSP70)53 (Figure 2). Under normal conditions, FTO removes m6A from the 5′UTRs of HSP70 and other heat shock-responsive genes, preventing their cap-independent translation. During heat shock, YTHDF2 translocates from the cytoplasm to the nucleus and binds m6A residues in the 5′UTR of heat shock-responsive genes, preventing demethylation by FTO and promoting rapid cap-independent translation54. In this fashion, 5′UTR m6A deposition ensures the production of proteins that are essential for survival during heat shock when general translation is halted.
FIGURE 2. Stress Induces N6-methyladenosine Driven Cap-Independent Translation.
Under normal conditions (left), N6-methyladenosine (m6A) within the 5′UTRs of stress-responsive transcripts is removed by the eraser fat mass and obesity-associated (FTO). Upon export to the cytoplasm, the unmodified transcripts undergo efficient cap-dependent translation, whereas the demethylated transcripts encoding stress-responsive genes are minimally translated. Upon heat shock (right), the m6A reader YTH domain family 2 (YTHDF2) translocates from the cytoplasm to the nucleus. Nuclear YTHDF2 then binds to 5′UTR m6A in the stress-responsive mRNAs and prevents them from FTO-mediated demethylation. These conditions inhibit cap-dependent translation, thus impairing protein production from unmodified transcripts. However, the m6A residues within the 5′ UTRs of stress responsive genes are directly bound by eukaryotic initiation factor 3 (eIF3), facilitating rapid cap-independent translation.
m5C and its derivative hm5C are also implicated in translation regulation. Expression of the m5C writer NOP2/Sun RNA methyltransferase family member 2 (NSUN2) is tightly regulated during the cell cycle and its overexpression delays replicative senescence through methylation of Cyclin-dependent kinase 1 (CDK1) and the CDK inhibitor 1B (p27KIP1) transcripts. Deposition of m5C in the CDK1 3′UTR enhances its translation, while m5C in the 5′UTR of p27KIP1 represses its translation, resulting in increased cellular proliferation56, 57. Deposition of hm5C by methylcytosine dioxygenases58 in mRNA correlates with higher association with polyribosomes in Drosophila melanogaster59. Recently, the existence of another modification, m1A, was discovered60, 61. Interestingly, m1A displays tissue specificity and plasticity in response to nitrogen deprivation in yeast60 as well as multiple stimuli in mammalian cells60, 61. Most modified transcripts harbor a single m1A in their 5′UTRs, and m1A-bearing transcripts have higher translation efficiency and protein levels compared to unmodified transcripts. Finally, for some transcripts pseudouridylation can affect their translation through alternative decoding of pseudouridylated codons, which may improve stress tolerance62.
While much work remains, these studies clearly demonstrate the impact of RNA modifications on different steps of gene expression. A natural question that arises is how do modifications link various nuclear and cytosolic processes. As for m6A, the discrete localization of readers along with their distinct protein-protein interaction domains seems to allow their interaction with different downstream effector proteins and regulate specific events in the nucleus and cytoplasm29, 34, 35, 44, 51, 54.
RNA Structure: Higher-Order Organization
Unlike DNA, which universally adopts a double-helix conformation, RNA forms extensive intramolecular interactions to fold into a vast array of complex structures63. RNA structure is highly dynamic, governed by factors such as temperature, cellular energy state, ATP-dependent RNA helicases, chaperone proteins and other RBPs64. RNA structures enable a myriad of functions, including encoding genetic information and catalyzing chemical reactions65, 66. The dynamic nature of RNA structure is illustrated by splicing, during which structural rearrangements of snRNAs permit the recognition of splice sites and branch point sequences in pre-mRNAs and facilitate the stepwise assembly of spliceosomes67. Numerous studies have confirmed that mRNAs also contain structured regions, although the degree of folding is significantly lower than that of noncoding RNAs. The advent of chemical modification-based procedures and RBP–RNA cross-linking methods11, 12, 68, 69 has allowed for transcriptome-wide mapping of RNA structures in different species. These studies have unveiled a previously unappreciated and complex layer of gene regulation.
Structure Shapes Protein Binding to Affect Splicing
RNA folding begins as the transcript is being synthesized by RNA polymerase II (Pol II)70–72. Transcript elongation by Pol II is not continuous, but rather entails periods of active elongation interrupted by pauses and backtracking. The formation of RNA structures, such as hairpins, on the nascent transcript can create physical barriers that prevent backtracking and promote forward elongation by Pol II70, 71. As splicing and polyadenylation are dependent upon Pol II elongation kinetics73, 74, RNA structure–elongation coupling likely affects downstream RNA processing.
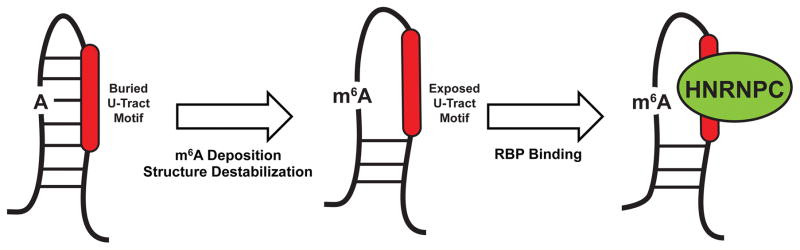
RNA structures may also regulate RNA processing in a direct manner through three distinct mechanisms. First, local RNA structure variations can promote or inhibit the binding of RBPs8. Secondary structure surrounding the consensus motifs for the splicing factors RNA binding protein, Fox-1 homolog 2 (RBFOX2) and Muscleblind Like 1 (MBNL1) is a key determinant of whether these sites will be bound in vivo. Moreover, motifs near evolutionarily conserved alternative exons are more single-stranded and exhibit stronger RBP binding than species-specific alternative exons or constitutive exons8. Distinct structural features also occur at 5′ and 3′ splice sites, strong and weak splice sites, and polyadenylation signals, indicating that local structures regulate RNA processing globally68, 75, 76. Second, longer-range RNA structures could facilitate splicing regulation by bringing distal regulatory regions into close proximity of target exons. An example of this mechanism is presented by RBFOX2. Over half of RBFOX2 binding sites are found over 500 nucleotides (nt) away from any annotated exons77. Regulation from these deep intronic sites is enabled through the formation of long-range intronic structures that deliver RBFOX2 close to target exons (Figure 3A). Moreover, recent studies utilizing methods that detect long-range RNA duplexes have revealed that up to 40% of transcript structures span more than 300nt10, 78, 79. Thus, the RNA structure-based proximity system observed for RBFOX2 may represent a mechanism common to other RBPs. Third, RNA modifications may act to stabilize or disrupt structural elements and influence RBP accessibility. Such a mechanism has been identified to regulate the binding of HNRNPC80, an RBP involved in mRNA stability and processing7, 81, 82. U-tract sequences recognized by HNRNPC are often buried within stem structures that prevent binding. Adenosines in these stems are often N6-methylated, which destabilizes the stem structure, exposes the U-tract, and allows HNRNPC binding (Figure 3B). Thousands of m6A structural switches were identified, with the expression of nearly 2,000 genes and over 100 splicing events being regulated by HNRNPC and METTL3–METTL1480. Taken together, these studies indicate that the structural landscape of the transcriptome is an essential mediator of RNA processing, and yet another factor that must be fully understood to complete the assembly of the splicing code.
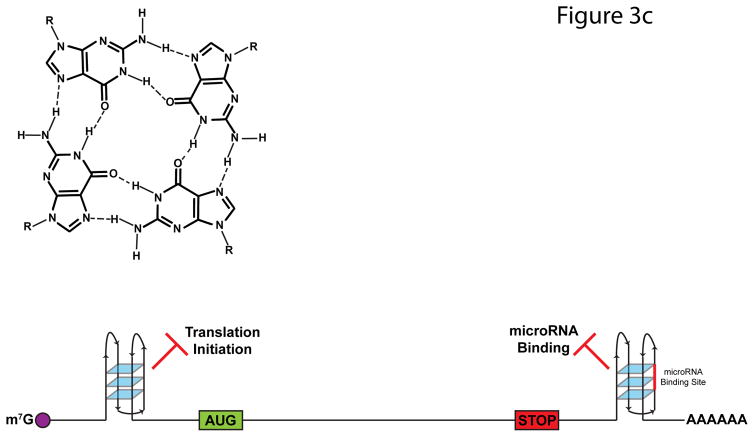
FIGURE 3. Impact of RNA Structures on Alternative Splicing.
a | A majority of RNA binding protein, Fox 2 homolog (RBFOX2) binding sites are over 500 nucleotides away from the nearest exon. To allow splicing regulation from these distal binding sites, base-pairing interactions occur within the intron, which bring RBFOX2 into close proximity to its target exons. b | N6-methyladenosine (m6A) deposition destabilizes base-pairing interactions and disrupts local secondary structure. In doing so, m6A can expose previously buried RNA-binding motifs to RNA-binding proteins, such as the U-tracts bound by heterogeneous ribonucleoprotein C (HNRNPC). The figure is reproduced, with permission, from REF. 80. c | G-qaudruplexes (RGQs) form from the stacking of three or more G-quartets (top). In 5′ untranslated regions (UTRs), RGQs impede the initiation and scanning steps of translation. In 3′UTRs, RGQs can block access to microRNA (miRNA) binding sites (shown in red), thus preventing miRNA-mediated decay and translational repression. These structures are dynamic and can be unwound by RNA helicases.
Structure Tunes Translation and Stability
RNA structure has also been linked to translational regulation. Across plants, animals, and fungi, the ~5nt region surrounding the start codon displays a significant lack of structure11, 68, 76, 83, 84. This feature is enriched in genes with high translation efficiencies and absent in those with low efficiencies11, 68, 76, 83, 84, indicating that start codon accessibility promotes translation. RNA secondary structure is also highly dynamic in response to external stimuli. Upon the transition from 30 to 37ºC in yeast, over 25,000 bases clustered at ~2,000 sites in mRNAs specifically unpair84. These heat-sensitive bases are enriched in 5′UTRs, regions in which structure is known to influence translation85. Future studies may determine if temperature-sensitive RNA structural elements indeed regulate eukaryotic gene expression.
Another structural element that has gained attention in the past decade are RNA G-quadruplexes (RGQs), four-stranded structures that result from the stacking of multiple planar G-quartets (Figure 3C) that are implicated in splicing, stability, and translation regulation. Although prevalent in vitro—especially in 5′ and 3′ UTRs—recent in vivo genome-wide analysis has revealed that RGQs are globally unfolded in mammalian cells86. Nonetheless, many individual examples of functionally important RGQs have been published85, 87. Moreover, inhibition of the RGQ-resolving RNA helicase eIF4A in T-cell acute lymphoblastic leukemia (T-ALL) cells induces apoptosis and delayed tumor growth due to decreased translation of oncogenes such as MYC, NOTCH, and BCL288. Transcripts affected by eIF4A inhibition contain longer 5′UTRs on average, and display a strong enrichment for the 12-nucleotide RGQ-forming motif (CGG)4. Thus, eIF4A’s tumorigenic properties may be due to its ability to unwind translation-inhibiting RGQs in the 5′UTRs of oncogenes. In addition to 5′ structures, RGQs in the 3′UTRs of mRNAs can also affect mRNA translation and stability through the occlusion of miRNA binding sites89.
The degradation of mRNAs is primarily controlled by the exosome, a multiprotein exonuclease90. Exosome-mediated degradation requires a single-stranded region at the 3′ end of mRNAs, providing the potential for RNA structure to affect transcript stability. Indeed, 3′ UTR structures involving the poly(A) tail are the primary determinants of transcript stability in yeast, with alternative polyadenylation isoforms from the same gene displaying drastically different half-lives91, 92, although stability differences between 3′UTR isoforms are subtler in mammalian cells93. Also, the formation of 3′ triple helical structures in nonpolyadenylated mammalian and viral long noncoding RNAs (lncRNAs) drastically increases their half-lives by preventing exonuclease access to the 3′ end (BOX 1). During heat shock in yeast, RNA structures with low melting temperatures are rapidly degraded by the exosome as the 3′ end becomes accessible84. As discussed above, ψ deposition increases upon heat shock and increases target mRNA stability39. As ψ-A base pairs are more stable than U-A base pairs62, it is possible that ψ increases mRNA half-lives by preventing the melting of RNA structures that protect the 3′ end at increased temperatures. In contrast to ψ, m6A acts as a “spring-loaded” modification, disrupting RNA duplexes due to the adoption of an unfavorable high-energy conformation94, 95. Indeed, transcriptome-wide structural analysis revealed a distinct structural profile at m6A sites consistent with unpaired RNA11. In addition to its role in miRNA-mediated decay, the strong enrichment of m6A in 3′UTRs suggests it may also promote transcript degradation by destabilizing 3′-protecting structural elements. Interestingly, 3′UTR structures are not always protective. Transcriptome-wide analysis of Staufen 1 (STAU1), an RBP that binds double-stranded RNA and regulates mRNA stability96, revealed it binds to thousands of duplexes in 3′UTRs12. Depletion of STAU1 significantly increased the stability of mRNAs containing these 3′ duplexes, demonstrating that these structures promote transcript degradation.
BOX 1. Triple Helices.
Canonical transcript maturation involves 3′ end cleavage and the addition of a poly(A) tail of up to 250 nucleotides. A poly(A) tail is present in the majority of eukaryotic mRNAs, and is essential for proper nucleocytoplasmic export, translation, and confers stability by protecting the coding information from degradation by exonucleases100. Interestingly, a sizeable portion (15–25%) of annotated mRNAs and long noncoding RNAs (lncRNAs) are not polyadenylated or are bimorphic, with some transcripts containing and others lacking a poly(A) tail101, raising the question of how these transcripts are stabilized and—in the case of mRNAs—translated. One such group of non-polyadenylated mRNAs belong to the intronless replication-dependent histone genes. These mRNAs are instead stabilized by highly conserved stem-loop structures that form in their 3′UTRs, which are also required for their translation102. This provides direct evidence that structures at the 3′ end of transcripts can functionally replace the poly(A) tail in both RNA turnover and translation.
The triple helix structure that forms at the expression and nuclear retention element (ENE) in the 3′ end of the Kaposi’s sarcoma-associated herpesvirus lncRNA PAN (polyadenylated nuclear) protects the transcript from rapid nuclear deadenylation-dependent decay103–105. Recently, a pair of studies discovered similar ENE elements in two mammalian nonpolyadenylated lncRNAs, metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) and multiple endocrine neoplasia-β (MENβ)106, 107. Both MALAT1 and MENβ are not processed by the canonical cleavage and polyadenylation machinery, and instead contain 3′ terminal tRNA-like structures that are recognized and cleaved by RNase P108, 109. Following cleavage, genomically encoded A-rich tract at the 3′ ends of both lncRNAs form triple-helical interactions nearly identical to those formed in the PAN RNA106, 107. These structures are essential for MALAT1 and MENβ stability, as deletions or destabilizing mutations made to these structures drastically reduce the transcripts’ half-lives. Strikingly, placement of the MALAT1 and MENβ 3′ structures onto polyadenylation-deficient reporter genes not only significantly stabilizes the mRNAs but also facilitates efficient translation107, suggesting such RNA structures may have a wider role in the translation and/or stabilization of other poly(A)-lacking transcripts. Recently, bioinformatics analysis has identified similar structures that stabilize hundreds of transposon transcripts in fungi, providing the first evidence of triple helices in mRNAs110.
Future Perspectives
An increasing number of studies have recognized that dynamic changes in RNA structures and modifications are indispensable features of eukaryotic gene expression. Recent findings support a new paradigm in which these features promote the coordination of individual gene expression processes by modulating access to specific RBPs and facilitating active transcriptome reprogramming in response to varying cellular conditions. Current RNA structural data represent only brief snapshots in time. However, it is likely that mRNAs are dynamically refolded as they progress throughout their life cycle, entering distinct subcellular compartments and associating with different RBPs. These structural rearrangements may affect downstream processes, facilitating remodeling of the mRNA-protein complex through the occlusion, exposure, or formation of RBP binding sites. Thus, the tracking of individual transcript structures throughout their life cycle will be necessary to fully understand the impact of mRNA structure. Future studies should discern how external cues fine-tune the structural and chemical variations within transcripts to produce a coherent biological response. For instance, do different signaling pathways recruit discrete epitranscriptome readers, writers or erasers as effectors to influence fates of individual mRNAs? Efforts to map the epitranscriptome at single nucleotide resolution also must continue to identify the exact nucleotides modified in each transcript, define how these sites are selected, and determine if the dynamics are achieved through active control of writers versus erasers.
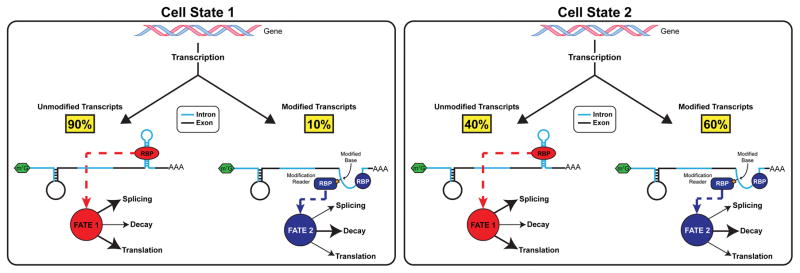
Another important aspect yet to be addressed is the dynamics of RNA modification stoichiometry. Epitranscriptome studies today deal mostly with modification sites, not the fraction of each site that is modified. Low-throughput analysis of m6A modification sites in mRNA and viral RNA shows that no m6A site is 100% modified97–99. Changes in the modification fraction at individual sites may also represent a dynamic parameter of RNA modification biology. Since modifications can affect mRNA structure and/or recruitment of RBPs, fractional modification at one specific site would generate two distinct mRNA species that differ only in their structures or bound reader proteins (Figure 4). Therefore, fractional modification could represent another mechanism to generate functional diversity from the same RNA transcript. High-throughput methods that can determine the modification fraction are urgently needed to address this aspect of the epitranscriptome. This is especially important for identifying the functional residues in heavily modified transcripts. For instance, the X-inactive specific transcript (XIST) lncRNA is m6A methylated at nearly 80 distinct sites—more than any other RNA—and these modifications are necessary for recruiting YTHDC1 and transcriptional silencing of genes on the X chromosome24. However, the percentage of each adenosine that is modified and in turn, which modified bases or combinations of modified bases are required for this function is unknown.
FIGURE 4. Dynamics of Modification Fractions and Stoichiometry.
Within the pool of transcripts from a given gene, only a fraction of each site is modified. For instance, under certain cellular conditions, a small percentage of the transcripts may contain a base modification (Cell State 1, left panel). By causing structural rearrangements and directly recruiting modification reader proteins, the modified pool of pre-mRNAs has a different cellular fate, which could result in increased turnover as shown (though alternate fates are possible). As cellular conditions change, modification stoichiometry may change as well, resulting in a higher (Cell State 2, right panel) or lower percentage of modified transcripts due to alterations in reader, writer, and eraser activities or expression.
Finally, we also need to explore further how structural and chemical alterations cooperate to allow functional coupling of different steps of the mRNA life cycle. In addition to the functions provided by direct reader proteins, RNA modifications may be one of the primary drivers of structural plasticity within the transcriptome by stabilizing or disrupting base-pairing interactions. As such, the phenotypes observed upon perturbations to modification writers or erasers may be—at least in part—due to global changes in the RNA structural topography.
Acknowledgments
A.K. is supported by grants from the US National Institute of Health (R01HL126845), March of Dimes (5-FY14-112), and the Center for Advanced Study at the University of Illinois. T.P. is supported by the US National Institutes of Health (R01GM113194). The authors apologize to those whose work could not be cited because of lack of space.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
References
- 1.Howard JM, Sanford JR. The RNAissance family: SR proteins as multifaceted regulators of gene expression. Wiley Interdiscip Rev RNA. 2015;6:93–110. doi: 10.1002/wrna.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang Y, Gattoni R, Stevenin J, Steitz JA. SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol Cell. 2003;11:837–43. doi: 10.1016/s1097-2765(03)00089-3. [DOI] [PubMed] [Google Scholar]
- 3.Muller-McNicoll M, et al. SR proteins are NXF1 adaptors that link alternative RNA processing to mRNA export. Genes Dev. 2016;30:553–66. doi: 10.1101/gad.276477.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ni JZ, et al. Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev. 2007;21:708–18. doi: 10.1101/gad.1525507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh G, et al. The cellular EJC interactome reveals higher-order mRNP structure and an EJC-SR protein nexus. Cell. 2012;151:750–64. doi: 10.1016/j.cell.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hafner M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–41. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Konig J, et al. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol. 2010;17:909–15. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taliaferro JM, et al. RNA Sequence Context Effects Measured In Vitro Predict In Vivo Protein Binding and Regulation. Mol Cell. 2016;64:294–306. doi: 10.1016/j.molcel.2016.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilbert WV, Bell TA, Schaening C. Messenger RNA modifications: Form, distribution, and function. Science. 2016;352:1408–12. doi: 10.1126/science.aad8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu Z, et al. RNA Duplex Map in Living Cells Reveals Higher-Order Transcriptome Structure. Cell. 2016;165:1267–79. doi: 10.1016/j.cell.2016.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spitale RC, et al. Structural imprints in vivo decode RNA regulatory mechanisms. Nature. 2015;519:486–90. doi: 10.1038/nature14263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sugimoto Y, et al. hiCLIP reveals the in vivo atlas of mRNA secondary structures recognized by Staufen 1. Nature. 2015;519:491–4. doi: 10.1038/nature14280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cantara WA, et al. The RNA Modification Database, RNAMDB: 2011 update. Nucleic Acids Res. 2011;39:D195–201. doi: 10.1093/nar/gkq1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nishikura K. A-to-I editing of coding and non-coding RNAs by ADARs. Nat Rev Mol Cell Biol. 2016;17:83–96. doi: 10.1038/nrm.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blanc V, Davidson NO. APOBEC-1-mediated RNA editing. Wiley Interdiscip Rev Syst Biol Med. 2010;2:594–602. doi: 10.1002/wsbm.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.El Yacoubi B, Bailly M, de Crecy-Lagard V. Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Annu Rev Genet. 2012;46:69–95. doi: 10.1146/annurev-genet-110711-155641. [DOI] [PubMed] [Google Scholar]
- 17.Karijolich J, Kantartzis A, Yu YT. RNA modifications: a mechanism that modulates gene expression. Methods Mol Biol. 2010;629:1–19. doi: 10.1007/978-1-60761-657-3_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dominissini D, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–6. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 19.Meyer KD, et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–46. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwartz S, et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Rep. 2014;8:284–96. doi: 10.1016/j.celrep.2014.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Agarwala SD, Blitzblau HG, Hochwagen A, Fink GR. RNA methylation by the MIS complex regulates a cell fate decision in yeast. PLoS Genet. 2012;8:e1002732. doi: 10.1371/journal.pgen.1002732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horiuchi K, et al. Identification of Wilms’ tumor 1-associating protein complex and its role in alternative splicing and the cell cycle. J Biol Chem. 2013;288:33292–302. doi: 10.1074/jbc.M113.500397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu J, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10:93–5. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patil DP, et al. m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016 doi: 10.1038/nature19342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ping XL, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24:177–89. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhong S, et al. MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell. 2008;20:1278–88. doi: 10.1105/tpc.108.058883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ke S, et al. A majority of m6A residues are in the last exons, allowing the potential for 3′ UTR regulation. Genes Dev. 2015 doi: 10.1101/gad.269415.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jia G, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885–7. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao X, et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014;24:1403–19. doi: 10.1038/cr.2014.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y, et al. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol. 2014;16:191–8. doi: 10.1038/ncb2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng G, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhong XY, Ding JH, Adams JA, Ghosh G, Fu XD. Regulation of SR protein phosphorylation and alternative splicing by modulating kinetic interactions of SRPK1 with molecular chaperones. Genes Dev. 2009;23:482–95. doi: 10.1101/gad.1752109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhong XY, Wang P, Han J, Rosenfeld MG, Fu XD. SR proteins in vertical integration of gene expression from transcription to RNA processing to translation. Mol Cell. 2009;35:1–10. doi: 10.1016/j.molcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiao W, et al. Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Mol Cell. 2016;61:507–19. doi: 10.1016/j.molcel.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 35.Alarcon CR, et al. HNRNPA2B1 Is a Mediator of m(6)A-Dependent Nuclear RNA Processing Events. Cell. 2015;162:1299–308. doi: 10.1016/j.cell.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carlile TM, et al. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature. 2014;515:143–6. doi: 10.1038/nature13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li X, et al. Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat Chem Biol. 2015;11:592–7. doi: 10.1038/nchembio.1836. [DOI] [PubMed] [Google Scholar]
- 38.Lovejoy AF, Riordan DP, Brown PO. Transcriptome-wide mapping of pseudouridines: pseudouridine synthases modify specific mRNAs in S. cerevisiae. PLoS One. 2014;9:e110799. doi: 10.1371/journal.pone.0110799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwartz S, et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell. 2014;159:148–62. doi: 10.1016/j.cell.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sommer S, Lavi U, Darnell JE., Jr The absolute frequency of labeled N-6-methyladenosine in HeLa cell messenger RNA decreases with label time. J Mol Biol. 1978;124:487–99. doi: 10.1016/0022-2836(78)90183-3. [DOI] [PubMed] [Google Scholar]
- 41.Batista PJ, et al. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. 2014;15:707–19. doi: 10.1016/j.stem.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Geula S, et al. Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science. 2015;347:1002–6. doi: 10.1126/science.1261417. [DOI] [PubMed] [Google Scholar]
- 43.Du H, et al. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun. 2016;7:12626. doi: 10.1038/ncomms12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang X, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–20. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–8. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kennedy EM, et al. Posttranscriptional m(6)A Editing of HIV-1 mRNAs Enhances Viral Gene Expression. Cell Host Microbe. 2016;19:675–85. doi: 10.1016/j.chom.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen T, et al. m(6)A RNA methylation is regulated by microRNAs and promotes reprogramming to pluripotency. Cell Stem Cell. 2015;16:289–301. doi: 10.1016/j.stem.2015.01.016. [DOI] [PubMed] [Google Scholar]
- 48.Alarcon CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF. N6-methyladenosine marks primary microRNAs for processing. Nature. 2015;519:482–5. doi: 10.1038/nature14281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Squires JE, et al. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012;40:5023–33. doi: 10.1093/nar/gks144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Merrick WC. Cap-dependent and cap-independent translation in eukaryotic systems. Gene. 2004;332:1–11. doi: 10.1016/j.gene.2004.02.051. [DOI] [PubMed] [Google Scholar]
- 51.Wang X, et al. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell. 2015;161:1388–99. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tirumuru N, et al. N(6)-methyladenosine of HIV-1 RNA regulates viral infection and HIV-1 Gag protein expression. Elife. 2016;5 doi: 10.7554/eLife.15528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meyer KD, et al. 5′ UTR m(6)A Promotes Cap-Independent Translation. Cell. 2015;163:999–1010. doi: 10.1016/j.cell.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou J, et al. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature. 2015;526:591–4. doi: 10.1038/nature15377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Feigenblum D, Schneider RJ. Cap-binding protein (eukaryotic initiation factor 4E) and 4E-inactivating protein BP-1 independently regulate cap-dependent translation. Mol Cell Biol. 1996;16:5450–7. doi: 10.1128/mcb.16.10.5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tang H, et al. NSun2 delays replicative senescence by repressing p27 (KIP1) translation and elevating CDK1 translation. Aging (Albany NY) 2015;7:1143–58. doi: 10.18632/aging.100860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xing J, et al. NSun2 Promotes Cell Growth via Elevating Cyclin-Dependent Kinase 1 Translation. Mol Cell Biol. 2015;35:4043–52. doi: 10.1128/MCB.00742-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fu L, et al. Tet-mediated formation of 5-hydroxymethylcytosine in RNA. J Am Chem Soc. 2014;136:11582–5. doi: 10.1021/ja505305z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Delatte B, et al. RNA biochemistry. Transcriptome-wide distribution and function of RNA hydroxymethylcytosine. Science. 2016;351:282–5. doi: 10.1126/science.aac5253. [DOI] [PubMed] [Google Scholar]
- 60.Dominissini D, et al. The dynamic N-methyladenosine methylome in eukaryotic messenger RNA. Nature. 2016 doi: 10.1038/nature16998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li X, et al. Transcriptome-wide mapping reveals reversible and dynamic N(1)-methyladenosine methylome. Nat Chem Biol. 2016;12:311–6. doi: 10.1038/nchembio.2040. [DOI] [PubMed] [Google Scholar]
- 62.Karijolich J, Yi C, Yu YT. Transcriptome-wide dynamics of RNA pseudouridylation. Nat Rev Mol Cell Biol. 2015;16:581–5. doi: 10.1038/nrm4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wan Y, Kertesz M, Spitale RC, Segal E, Chang HY. Understanding the transcriptome through RNA structure. Nat Rev Genet. 2011;12:641–55. doi: 10.1038/nrg3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dethoff EA, Chugh J, Mustoe AM, Al-Hashimi HM. Functional complexity and regulation through RNA dynamics. Nature. 2012;482:322–30. doi: 10.1038/nature10885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 66.Sharp PA. The centrality of RNA. Cell. 2009;136:577–80. doi: 10.1016/j.cell.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 67.Smith DJ, Query CC, Konarska MM. “Nought may endure but mutability”: spliceosome dynamics and the regulation of splicing. Mol Cell. 2008;30:657–66. doi: 10.1016/j.molcel.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ding Y, et al. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature. 2014;505:696–700. doi: 10.1038/nature12756. [DOI] [PubMed] [Google Scholar]
- 69.Rouskin S, Zubradt M, Washietl S, Kellis M, Weissman JS. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature. 2014;505:701–5. doi: 10.1038/nature12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu SR, Hu CG, Zhang JZ. Regulatory effects of cotranscriptional RNA structure formation and transitions. Wiley Interdiscip Rev RNA. 2016 doi: 10.1002/wrna.1350. [DOI] [PubMed] [Google Scholar]
- 71.Zhang J, Landick R. A Two-Way Street: Regulatory Interplay between RNA Polymerase and Nascent RNA Structure. Trends Biochem Sci. 2016 doi: 10.1016/j.tibs.2015.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pan T, Sosnick T. RNA folding during transcription. Annu Rev Biophys Biomol Struct. 2006;35:161–75. doi: 10.1146/annurev.biophys.35.040405.102053. [DOI] [PubMed] [Google Scholar]
- 73.de la Mata M, et al. A slow RNA polymerase II affects alternative splicing in vivo. Mol Cell. 2003;12:525–32. doi: 10.1016/j.molcel.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 74.Pinto PA, et al. RNA polymerase II kinetics in polo polyadenylation signal selection. EMBO J. 2011;30:2431–44. doi: 10.1038/emboj.2011.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jin Y, Yang Y, Zhang P. New insights into RNA secondary structure in the alternative splicing of pre-mRNAs. RNA Biol. 2011;8:450–7. doi: 10.4161/rna.8.3.15388. [DOI] [PubMed] [Google Scholar]
- 76.Wan Y, et al. Landscape and variation of RNA secondary structure across the human transcriptome. Nature. 2014;505:706–9. doi: 10.1038/nature12946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lovci MT, et al. Rbfox proteins regulate alternative mRNA splicing through evolutionarily conserved RNA bridges. Nat Struct Mol Biol. 2013;20:1434–42. doi: 10.1038/nsmb.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Aw JG, et al. In Vivo Mapping of Eukaryotic RNA Interactomes Reveals Principles of Higher-Order Organization and Regulation. Mol Cell. 2016;62:603–17. doi: 10.1016/j.molcel.2016.04.028. [DOI] [PubMed] [Google Scholar]
- 79.Sharma E, Sterne-Weiler T, O’Hanlon D, Blencowe BJ. Global Mapping of Human RNA-RNA Interactions. Mol Cell. 2016;62:618–26. doi: 10.1016/j.molcel.2016.04.030. [DOI] [PubMed] [Google Scholar]
- 80.Liu N, et al. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560–4. doi: 10.1038/nature14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McCloskey A, Taniguchi I, Shinmyozu K, Ohno M. hnRNP C tetramer measures RNA length to classify RNA polymerase II transcripts for export. Science. 2012;335:1643–6. doi: 10.1126/science.1218469. [DOI] [PubMed] [Google Scholar]
- 82.Zarnack K, et al. Direct competition between hnRNP C and U2AF65 protects the transcriptome from the exonization of Alu elements. Cell. 2013;152:453–66. doi: 10.1016/j.cell.2012.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kertesz M, et al. Genome-wide measurement of RNA secondary structure in yeast. Nature. 2010;467:103–7. doi: 10.1038/nature09322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wan Y, et al. Genome-wide measurement of RNA folding energies. Mol Cell. 2012;48:169–81. doi: 10.1016/j.molcel.2012.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bugaut A, Balasubramanian S. 5′-UTR RNA G-quadruplexes: translation regulation and targeting. Nucleic Acids Res. 2012;40:4727–41. doi: 10.1093/nar/gks068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Guo JU, Bartel DP. RNA G-quadruplexes are globally unfolded in eukaryotic cells and depleted in bacteria. Science. 2016;353 doi: 10.1126/science.aaf5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Millevoi S, Moine H, Vagner S. G-quadruplexes in RNA biology. Wiley Interdiscip Rev RNA. 2012;3:495–507. doi: 10.1002/wrna.1113. [DOI] [PubMed] [Google Scholar]
- 88.Wolfe AL, et al. RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nature. 2014;513:65–70. doi: 10.1038/nature13485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kenny PJ, et al. MOV10 and FMRP regulate AGO2 association with microRNA recognition elements. Cell Rep. 2014;9:1729–41. doi: 10.1016/j.celrep.2014.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Houseley J, LaCava J, Tollervey D. RNA-quality control by the exosome. Nat Rev Mol Cell Biol. 2006;7:529–39. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- 91.Geisberg JV, Moqtaderi Z, Fan X, Ozsolak F, Struhl K. Global analysis of mRNA isoform half-lives reveals stabilizing and destabilizing elements in yeast. Cell. 2014;156:812–24. doi: 10.1016/j.cell.2013.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Moqtaderi Z, Geisberg JV, Struhl K. Secondary structures involving the poly(A) tail and other 3′ sequences are major determinants of mRNA isoform stability in yeast. Microb Cell. 2014;1:137–139. doi: 10.15698/mic2014.04.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Spies N, Burge CB, Bartel DP. 3′ UTR-isoform choice has limited influence on the stability and translational efficiency of most mRNAs in mouse fibroblasts. Genome Res. 2013;23:2078–90. doi: 10.1101/gr.156919.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kierzek E, Kierzek R. The thermodynamic stability of RNA duplexes and hairpins containing N6-alkyladenosines and 2-methylthio-N6-alkyladenosines. Nucleic Acids Res. 2003;31:4472–80. doi: 10.1093/nar/gkg633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Roost C, et al. Structure and thermodynamics of N6-methyladenosine in RNA: a spring-loaded base modification. J Am Chem Soc. 2015;137:2107–15. doi: 10.1021/ja513080v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Heraud-Farlow JE, Kiebler MA. The multifunctional Staufen proteins: conserved roles from neurogenesis to synaptic plasticity. Trends Neurosci. 2014;37:470–9. doi: 10.1016/j.tins.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Csepany T, Lin A, Baldick CJ, Jr, Beemon K. Sequence specificity of mRNA N6-adenosine methyltransferase. J Biol Chem. 1990;265:20117–22. [PubMed] [Google Scholar]
- 98.Horowitz S, Horowitz A, Nilsen TW, Munns TW, Rottman FM. Mapping of N6-methyladenosine residues in bovine prolactin mRNA. Proc Natl Acad Sci U S A. 1984;81:5667–71. doi: 10.1073/pnas.81.18.5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liu N, et al. Probing N6-methyladenosine RNA modification status at single nucleotide resolution in mRNA and long noncoding RNA. RNA. 2013;19:1848–56. doi: 10.1261/rna.041178.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 101.Yang L, Duff MO, Graveley BR, Carmichael GG, Chen LL. Genomewide characterization of non-polyadenylated RNAs. Genome Biol. 2011;12:R16. doi: 10.1186/gb-2011-12-2-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Marzluff WF, Wagner EJ, Duronio RJ. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat Rev Genet. 2008;9:843–54. doi: 10.1038/nrg2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Conrad NK, Mili S, Marshall EL, Shu MD, Steitz JA. Identification of a rapid mammalian deadenylation-dependent decay pathway and its inhibition by a viral RNA element. Mol Cell. 2006;24:943–53. doi: 10.1016/j.molcel.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 104.Conrad NK, Shu MD, Uyhazi KE, Steitz JA. Mutational analysis of a viral RNA element that counteracts rapid RNA decay by interaction with the polyadenylate tail. Proc Natl Acad Sci U S A. 2007;104:10412–7. doi: 10.1073/pnas.0704187104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Conrad NK, Steitz JA. A Kaposi’s sarcoma virus RNA element that increases the nuclear abundance of intronless transcripts. EMBO J. 2005;24:1831–41. doi: 10.1038/sj.emboj.7600662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Brown JA, Valenstein ML, Yario TA, Tycowski KT, Steitz JA. Formation of triple-helical structures by the 3′-end sequences of MALAT1 and MENbeta noncoding RNAs. Proc Natl Acad Sci U S A. 2012;109:19202–7. doi: 10.1073/pnas.1217338109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wilusz JE, et al. A triple helix stabilizes the 3′ ends of long noncoding RNAs that lack poly(A) tails. Genes Dev. 2012;26:2392–407. doi: 10.1101/gad.204438.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sunwoo H, et al. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009;19:347–59. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wilusz JE, Freier SM, Spector DL. 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008;135:919–32. doi: 10.1016/j.cell.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tycowski KT, Shu MD, Steitz JA. Myriad Triple-Helix-Forming Structures in the Transposable Element RNAs of Plants and Fungi. Cell Rep. 2016;15:1266–76. doi: 10.1016/j.celrep.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]