Abstract
Nano technology is a cutting edge science which is now effectively used in the field of cancer biology. Smart Flare gold nanoparticles are now used often for differential gene expression analysis. In this manuscript we are reporting the use of micro RNA miR 146a and onco gene EZH2 Smart Flare probes to study their expression in different prostate cancer cell lines and the effect of novel Rhenium compounds on these genes using a flow cytometer and a Fluorescence microscope. Our results showed this novel nanotechnology can be effectively used in cancer biology to successfully detect the effect of novel drugs on oncogenes and could be a very useful tool for next generation of cancer researchers.
Keywords: Nanoparticles, micro RNA, gene expression
INTRODUCTION
MicroRNAs (often abbreviated as miRNA) are a class of short (20–24bp) noncoding RNAs which are involved in the recognition and post-translational repression or degradation of specific mRNAs. MiRNAs have been associated with biological processes in mammals such as cancer, insulin secretion, cell differentiation and viral infections. miRNAs are transcribed by RNA polymerase II, the primary transcript is cleaved by the ribonuclease III enzyme Drosha which is further cleaved by Dicer ribonuclease to generate the mature miRNA and antisense miRNA products. The mature miRNA is then incorporated into the RISC (RNA-induced silencing complex) which recognizes the target mRNA and results in translational inhibition or destabilization [1].
miR-146a is thought to be a mediator of inflammation along with another microRNA, mir-155. The expression of miR-146a is upregulated by inflammatory factors such as interleukin 1 and tumor necrosis factor-alpha, miR-146a dysregulates a number of targets which are mostly involved in toll-like receptor pathways that bring about a cytokine response as part of the innate immune system [2, 3], miR-146a operates in a feedback system or "negative regulatory loop" [4] to finely tune inflammatory responses [5]. miR-146a was found to significantly overexpressed in gastric cancer and breast cancer tissues [6, 7].
RNA molecules play crucial roles in cells such as coding, decoding, regulation, and expression of genes, yet they are much more difficult to study. Smart Flares are nanoparticle-based probes for the detection and imaging of RNA in live cells. Smart Flare ranked second in The Scientist top ten 2013 innovations, with one of the judges, Kevin Lustig, commenting “These new RNA detection probes can be used to visualize RNA expression in live cells at the single-cell level.” The following year, Smart Flare won an R&D100 award. The technology comes from Chad Mirkin’s lab at Northwestern University. Chad Mirkin is the winner of numerous prestigious prizes and a science adviser to the President of the United States. The scientific articles introducing the Smart Flare concept (under the name of Nano-Flare) were published in the Journal of the American Chemical Society in 2007. In 2013, the Smart Flare technology was licensed to EMD Millipore. A molecular sensor needs a detection mechanism to work. The principle of the Smart Flare is explained where a capture oligonucleotide (i.e. DNA) is bound to the gold nanoparticles. A reporter strand is bound to the capture strand. The reporter strand carries a fluorophore but that fluorophore does not emit light because it is too close to the gold (the fluorescence is “quenched”). In the presence of the target RNA, the reporter strand is replaced by the target RNA and therefore released, quenching stops, and fluorescence is detected by a flow cytometer.
Novel Rhenium based compounds are now being tested as anti cancer drugs in many cancer research laboratories as better alternatives to platinum compounds. These compounds could be cheaper, more effective with less drug resistance than other chemotherapeutic drugs.
In this study, we tested the expression of miR 146 alpha in several prostate cancer (PCa) cell lines derived from both African American (AA) and Caucasian (CA) patients in comparison to normal prostate cells by using Smart flare technology. We also studied the effects of two novel rhenium compounds called PC1 and RPR2 (Figure 1) on the miR 146a and oncogene EZH2 by using these gold nanoparticles as described in this manuscript.
Figure 1.
The structural formulas of PC-1 and RPR-2 and miR 146a secondary structure.
MATERIALS AND METHODS
Cell Culture and Maintenance
LNCAP (CA PCa), PC3 (CA PCa), RWPE1 (Healthy prostate cell line), MDA (AA PCa) cell lines were obtained from American Type Culture Collection. The cell lines were grown and maintained in Roswell Park Memorial Institute Medium (RPMI 1640). Cells were stored at 5% CO2 in 25cm2 filter cap flasks in a laboratory CO2 incubator and were additionally maintained with 10% Fetal Bovine Serum (FBS) and 100µl/ml antibiotic/ anti mycotic (penicillin/ streptomycin/ amphotericin B).
Rhenium compounds PC1 and RPR2 were synthesized as described previously [8, 9].
Smart Flare Experiments
Cells were grown and incubated at 37°C @ 5% CO2 in 35mm cell culture dishes until fully confluent. 10µm of rhenium compound PC-1 was added to the dishes and allowed to incubate for 48 hours, but after 24 hours 1µl of miRNA146a gold nano probe was diluted into 20µl of 1× PBS and added to the cells and allowed to incubate overnight. The next day, cells were then removed and scraped using a sterile cell scraper and pipetted in microcentrifuge tubes. Pipetted cells were then centrifuged at 12,000 × g for 10 minutes until cell pellet was visible. Pelleted cells were then suspended using 1× PBS and then diluted 1:1000. Using a Guava® EasyCyte Single Sample Flow Cytometer, gene expression was analyzed. A BIO RAD (USA) fluorescence microscope was used to take pictures of the gold nano particle transfected cells. The experiment with the EZH2 oncogene was similarly done for analyzing its expression in Rhenium drug RPR1 treated and untreated PC3 prostate cancer cells.
RESULTS
The Smart Flare analysis of different prostate cancer cell lines derived from both African American and Caucasian patients showed increased expression of miR 146a in the cancer cell lines than the normal cells (Figure 2). Rhenium compound PC1 lowered miR 146a expression, however, the Rhenium compound RPR2 failed to lower the expression of oncogene EZH2 in PC3 prostate cancer cell lines (Figures 3, 4a, b).
Figure 2.
Guava Smart Flare technology done on prostate cancer cell lines and RWPE1/CRL 2854 normal cell line. Experiment proving that miRNA 146a is upregulated more in cancer cells than normal.
Figure 3.
Guava Smart Flare experiment showing that the rhenium compound PC1 effectively decreases the level of miRNA 146a in PC3 prostate cancer cell lines.
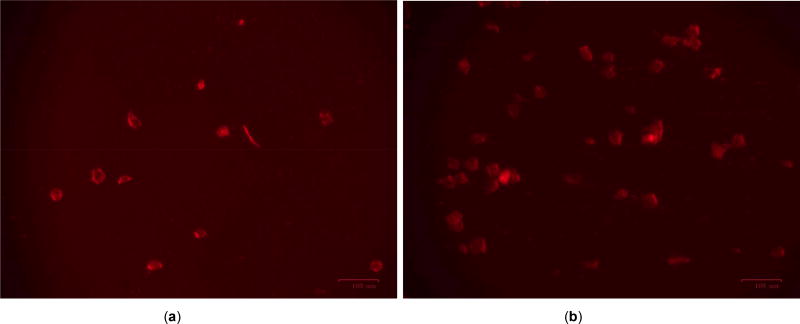
Figure 4.
a shows DMSO treated EZH2 gold nano particle transfected and b the same with Rhenium treated. No significant change in fluorescence concludes that Rhenium compound RPR1 might not be effective in lowering onco gene EZH2 expression.
DISCUSSION
In previous literature, together with our findings, suggests that miR-146a may be an important oncogene in prostate cancer. A recent study in breast cancer cells showed that miR-146a was significantly upregulated in invasive cell lines when compared with normal and less invasive cell lines [9]. Previous studies have also shown that abnormal expression of miR-146a is associated with paradoxical roles as an oncogene or a tumor suppressor in different human cancer tissues. This variation seems to be context dependent in terms of tissue type as well as signaling networks, miR-146a has been shown to function as an oncogene in cervical cancer [10] and anaplastic thyroid carcinoma [11] but as a tumor suppressor in pancreatic cancer [12] and gastric cancer [13]. Within breast cancer, overexpression of miR-146a has been shown to be oncogenic by increasing the proliferation of breast cancer cells and to function as a tumor suppressor [14] by impairing the invasion and migration capacity when overexpressed in some breast cancer cell lines. These observations reconfirm that miRs are not only expressed in tissue specific manner but are also a part of the intricate network involving direct interactions and cross talks with other genes, microRNAs and signaling molecules. In addition, each microRNA can have multiple targets and each target can be regulated by multiple microRNAs (i.e. signal integration). In this study we showed that using nanotechnology of Smart flare gold nanoparticles, we can very easily and quickly analyze gene expression by using Flowcytometry and Fluorescence microscopy. We found miR 146a to be consistently upregulated in all prostate cancer cell lines we tested and also could be suppressed by Rhenium drug PC1. Even though our experiments with the Rhenium compound RPR2 in downregulating oncogene EZH2 was not successful, however, we established the fact that the Smartflare technology could be applied in effectively testing anti- cancer drugs much quicker with less efforts than using techniques like RNA isolation, cDNA synthesis, REAL-TIME PCR etc which we currently use for gene expression analysis. Thus Smart Flare novel gold nanoparticles could revolutionize the field of differential gene expression studies and drug discovery [15–17].
Acknowledgments
Supported by a NIH Grant# T34 GM100831 and a NSF-LSAMP award to Elizabeth City State University and a NCI disability Supplement Research award to Dr. H. Banerjee.
References
- 1.Liu Z, Xiao B, Tang B, Li B, Li N, Zhu E, et al. Up-regulated microRNA-146a negatively modulate Helicobacter pylori-induced inflammatory response in human gastric epithelial cells. Microbes Infect. 2010;12(11):854–63. doi: 10.1016/j.micinf.2010.06.002. https://doi.org/10.1016/j.micinf.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 2.Sonkoly E, Stahle M, Pivarcsi A. MicroRNAs and immunity: novel players in the regulation of normal immune function and inflammation. Semin Cancer Biol. 2008;18(2):131–40. doi: 10.1016/j.semcancer.2008.01.005. https://doi.org/10.1016/j.semcancer.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 3.Williams AE, Perry MM, Moschos SA, Larner-Svensson HM, Lindsay MA. Role of miRNA-146a in the regulation of the innate immune response and cancer. Biochem Soc Trans. 2008;36(6):1211–15. doi: 10.1042/BST0361211. https://doi.org/10.1042/BST0361211. [DOI] [PubMed] [Google Scholar]
- 4.Philippidou D, Schmitt M, Moser D, Margue C, Nazarov PV, Muller A, et al. Signatures of microRNAs and selected microRNA target genes in human melanoma. Cancer Res. 2010;70(10):4163–73. doi: 10.1158/0008-5472.CAN-09-4512. https://doi.org/10.1158/0008-5472.CAN-09-4512. [DOI] [PubMed] [Google Scholar]
- 5.Zhong H, Wang HR, Yang S, Zhong JH, Wang T, Wang C, et al. Targeting Smad4 linksmicroRNA-146a to the TGF-beta pathway during retinoid acid induction in acute promyelocytic leukemia cell line. Int J Hematol. 2010;92(1):129–35. doi: 10.1007/s12185-010-0626-5. https://doi.org/10.1007/s12185-010-0626-5. [DOI] [PubMed] [Google Scholar]
- 6.Wang LH, Kim SH, Lee JH, Choi YL, Kim YC, Park TS, et al. Inactivation of SMAD4 tumor suppressor gene during gastric carcinoma progression. Clin Cancer Res. 2007;13(1):102–10. doi: 10.1158/1078-0432.CCR-06-1467. https://doi.org/10.1158/1078-0432.CCR-06-1467. [DOI] [PubMed] [Google Scholar]
- 7.He H, Jazdzewski K, Li W, Liyanarachchi S, Nagy R, Volinia S, et al. The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci USA. 2005;102(52):19075–80. doi: 10.1073/pnas.0509603102. https://doi.org/10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mbagu MK, Kebulu DN, Winstead A, Pramanik SK, Banerjee HN, Iwunze MO, et al. Fac-tricarbonyl(pentylcarbonato)(α-diimine)rhenium complexes: One-pot synthesis, characterization, fluorescence studies, and cytotoxic activity against human MDA-MB-231 breast, CCl-227 colon and BxPC-3 pancreatic carcinoma cell lines. Inorg Chem Commun. 2012;21:35–38. https://doi.org/10.1016/j.inoche.2012.04.004. [Google Scholar]
- 9.Brown DA, Kimari DM, Duzs-Moore AM, Budzichowski TA, Ho DM, Mandal SK. Reactions of dirhenium heptoxide with manganese(I) and rhenium(I) hydrido, alkoxo, methylcarbonato, carbonato-bridged, and methoxymethyl complexes. The X-ray structures of fac-(CO)3(dppp)MnOReO3 and fac-(CO)3(dppp)ReOReO3. J Organomet Chem. 2002;658(1–2):88–93. https://doi.org/10.1016/S0022-328X(02)01632-7. [Google Scholar]
- 10.Wang X, Tang S, Le S-Y, Lu R, Rader JS, Meyers C, et al. Aberrant expression of oncogenic and tumor-suppressive microRNAs in cervical cancer is required for cancer cell growth. PLoS One [internet] 2008;3(7):e2557. doi: 10.1371/journal.pone.0002557. https://doi.org/10.1371/journal.pone.0002557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Volinia S, Calin GA, Liu C-G, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103(7):2257–61. doi: 10.1073/pnas.0510565103. https://doi.org/10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin SL, Chiang A, Chang D, Ying SY. Loss of mir-146a function in hormone-refractory prostate cancer. RNA. 2008;14(3):417–24. doi: 10.1261/rna.874808. https://doi.org/10.1261/rna.874808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jazdzewski K, Murray EL, Franssila K, Jarzab B, Schoenberg DR, de la CA. Common SNP in pre-miR-146a decreasesmmature miR expression and predisposes to papillary thyroid carcinoma. Proc Natl Acad Sci USA. 2008;105(20):7269–74. doi: 10.1073/pnas.0802682105. https://doi.org/10.1073/pnas.0802682105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hurst DR, Edmonds MD, Scott GK, Benz CC, Vaidya KS, Welch DR. Breast cancer metastasis suppressor 1 upregulates miR-146, which suppresses breast cancer metastasis. Cancer Res. 2009;69(4):1279–83. doi: 10.1158/0008-5472.CAN-08-3559. https://doi.org/10.1158/0008-5472.CAN-08-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhaumik D, Scott GK, Schokrpur S, Patil CK, Campisi J, Benz CC. Expression of microRNA-146 suppresses NF-kappaB activity with reduction of metastatic potential in breast cancer cells. Oncogene. 2008;27(42):5643–7. doi: 10.1038/onc.2008.171. https://doi.org/10.1038/onc.2008.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McClellan S, Slamecka J, Howze P, Thompson L, Finan M, Rocconi R, et al. mRNA detection in living cells: A next generation cancer stem cell identification technique. Methods. 2015;82:47–54. doi: 10.1016/j.ymeth.2015.04.022. https://doi.org/10.1016/j.ymeth.2015.04.022. [DOI] [PubMed] [Google Scholar]
- 17.Amatya VJ, Mawas AS, Kushitani K, Mohi El-Din MM, Takeshima Y. Differential microRNA expression profiling of mesothelioma and expression analysis of miR-1 and miR-214 in mesothelioma. Int J Oncol. 2016;48(4):1599–607. doi: 10.3892/ijo.2016.3358. https://doi.org/10.3892/ijo.2016.3358. [DOI] [PubMed] [Google Scholar]