Abstract
The liver has a highly regenerative capacity. In the normal liver, hepatocytes proliferate to restore lost liver mass. However, when hepatocyte proliferation is impaired, biliary epithelial cells (BECs) activate and contribute to hepatocytes. We previously reported in zebrafish that upon severe hepatocyte ablation, BECs extensively contribute to regenerated hepatocytes. It was also speculated that BEC-driven liver regeneration might occur in another zebrafish liver injury model in which temporary knockdown of the mitochondrial import gene tomm22 by morpholino antisense oligonucleotides (MO) induces hepatocyte death. Given the importance of multiple BEC-driven liver regeneration models for better elucidating the mechanisms underlying innate liver regeneration in the diseased liver, we hypothesized that BECs would contribute to hepatocytes in tomm22 MO-injected larvae. In this MO-based liver injury model, by tracing the lineage of BECs, we found that BECs significantly contributed to hepatocytes. Moreover, we found that surviving, preexisting hepatocytes become BEC–hepatocyte hybrid cells in tomm22 MO-injected larvae. Intriguingly, both the inhibition of Wnt/β-catenin signaling and macrophage ablation suppressed the formation of the hybrid hepatocytes. This new liver injury model in which both hepatocytes and BECs contribute to regenerated hepatocytes will aid in better understanding the mechanisms of innate liver regeneration in the diseased liver.
Key words: Liver regeneration, Macrophage, Oval cells, Liver progenitor cells
INTRODUCTION
The liver is a crucial organ with a remarkable regenerative capacity. Even after removing two thirds of the liver mass, the liver can recover the lost mass and restore its function1. In this regeneration setting, hepatocytes proliferate to make more hepatocytes, thereby recovering the lost liver mass. However, when hepatocyte proliferation is impaired, biliary epithelial cells (BECs) contribute to hepatocytes2,3. Based on the source of regenerated hepatocytes, liver regeneration can be classified into either hepatocyte- or BEC-driven liver regeneration. In BEC-driven liver regeneration, BECs are first activated to form oval cells, also called ductular reactions, and then differentiate into hepatocytes. Multiple lineage-tracing studies in mice have shown that in diverse oval cell activation models, including 3,5-diethoxycarbonyl-1,4-dihydrocollidine (DDC), a choline-deficient ethionine-supplemented (CDE) diet, CCl4 administration, and α-naphthyl-isothiocyanate (ANIT) diet, regenerated hepatocytes are derived only from preexisting hepatocytes4–8, indicating the negligible contribution of BECs to hepatocytes. In contrast to these oval cell activation models, two severe liver injury models have recently been described in which BECs extensively give rise to hepatocytes: (1) a mouse model in which the E3 ubiquitin ligase gene Mdm2 is inducibly deleted in hepatocytes9 and (2) a zebrafish model in which hepatocytes are pharmacogenetically ablated10–12. In the mouse model, the Mdm2 deletion completely blocks hepatocyte proliferation and induces p53-mediated hepatocyte senescence and death, which triggers oval cell activation and their differentiation into hepatocytes9. In the zebrafish model, the transgenic zebrafish lines that express bacterial nitroreductase (NTR) enzyme under the hepatocyte-specific fabp10a promoter allow for hepatocyte-specific ablation. The NTR enzyme converts a nontoxic drug, metronidazole (Mtz), into a cytotoxic drug, resulting in the ablation of only NTR-expressing cells13–16. Upon severe hepatocyte ablation in zebrafish larvae, BECs extensively contribute to hepatocytes, thereby leading to a full liver recovery10–12.
Given the correlation between oval cell numbers and disease severity in diseased human livers17 and a recent report that BECs appear to contribute to hepatocytes in regressed human cirrhotic livers18, it is important to understand the mechanisms underlying BEC-driven liver regeneration, which will provide insights into promoting innate liver regeneration in patients with advanced liver diseases. Although the zebrafish hepatocyte ablation and the mouse Mdm2 deletion models will contribute to the elucidation of the mechanisms underlying BEC-driven liver regeneration, additional liver injury models for this type of liver regeneration should further contribute to such elucidation. In fact, diverse oval cell activation models collectively contribute to the current understanding of the oval cell activation process19. Here we report a novel liver injury model resulting from the temporary knockdown of tomm22, in which BECs contribute to hepatocytes.
The mitochondrial import gene tomm22 zebrafish mutants exhibit hepatocyte-specific cell death, leading to the death of the animal20. Similarly, tomm22 knockdown using the morpholino antisense oligonucleotide (MO) approach initially results in hepatocyte death, but later new hepatocytes form and the liver recovers, following the gradual depletion of the MOs. Based on biliary marker expression, it was speculated that in tomm22 MO-injected larvae, BECs gave rise to the new hepatocytes; however, no lineage-tracing data were formulated to confirm this speculation20. Since hepatocyte death induces BEC-driven liver regeneration in the zebrafish hepatocyte ablation model10–12 and the mouse hepatocyte-specific Mdm2 deletion model9, we hypothesized that BECs contributed to the hepatocytes in tomm22 MO-injected larvae. To test this hypothesis, we performed lineage tracing experiments to unequivocally determine the origin of new hepatocytes in tomm22 MO-injected larvae. Surprisingly, we found that both preexisting hepatocytes and BECs contributed to the new hepatocytes in tomm22 MO-injected larvae. Moreover, using the tomm22 MO-based liver regeneration model, we investigated the role of Wnt/β-catenin signaling and macrophages in liver regeneration.
MATERIALS AND METHODS
Zebrafish Strains
Experiments were performed with the approval of the Institutional Animal Care and Use Committee (IACUC) at the University of Pittsburgh. Embryos and adult fish were raised and maintained under standard lab conditions21. We used the following transgenic lines: Tg(Tp1:H2B-mCherry) s939( 22 ), Tg(ubb:loxP-GFP-loxP-mCherry) cz1701( 23 ), Tg(Tp1:CreERT2) s951( 24 ), Tg(fabp10a:mAGFP-gmnn) pt608( 25 ), Tg(WRE:d2GFP) kyu1( 26 ), Tg(mpeg1:Gal4-VP16) gl24( 27 ), Tg(UAS:NTR-mCherry) c264( 14 ), Tg(fabp10a:CFP-NTR) s931( 12 ), Tg(fabp10a:GFP) as3( 28 ), and Tg(mpeg1:Dendra2) uwm12( 29 ).
Morpholino Injection
tomm22 MO (5′-GAGAAAGCTCCTGGATCGTAGCCAT-3′)20 was purchased from GeneTools (Philomath, OR, USA); 6 ng of tomm22 MO was injected into embryos at the one-cell stage.
Cre/loxP-Mediated Lineage Tracing, Macrophage Ablation, and Wnt/β-Catenin Suppression
For lineage tracing experiments, Tg(fabp10a:CFP-NTR);Tg(ubb:loxP-GFP-loxP-mCherry);Tg(Tp1:CreERT2) larvae were treated with 10 μM 4-hydroxytamoxifen (4-OHT) from 48 to 84 h postfertilization (hpf) for 36 h to induce Cre-mediated recombination. For macrophage ablation experiments, Tg(mpeg1:Gal4-VP16);Tg(UAS:NTR-mCherry) larvae were treated with 10 mM Mtz from 4 to 6 or 7 days postfertilization (dpf). For Wnt/β-catenin suppression experiments, larvae were treated with 10 μM XAV939 from 4 to 6 or 7 dpf.
Whole-Mount In Situ Hybridization and Immunostaining
Whole-mount in situ hybridization was performed as previously described30. We used the following probes: prox1a, foxa3, sepp1b, cp, and fabp10a. Whole-mount immunostaining was performed as previously described31, using the following antibodies: chicken polyclonal anti-GFP (1:1,000; Aves Labs, Tigard, OR, USA), rabbit polyclonal anti-Prox1 (1:150; GeneTex, Irvine, CA, USA), mouse monoclonal anti-Anxa4 (also named as 2F11; 1:100; Abcam, Cambridge, MA USA), rabbit polyclonal anti-DsRed (1:200; Clontech, Mountain View, CA, USA), rabbit monoclonal anti-DsRed (1:400; Allele Biotechnology, San Diego, CA, USA), goat polyclonal anti-Hnf4a (1:50; Santa Cruz Biotechnology, Dallas, TX, USA), mouse monoclonal anti-Bhmt (1:800; gift from Jinrong Peng at Zhejiang University, P.R. China), mouse monoclonal anti-Alcam (Zn5; 1:10; ZIRC, USA), and conjugated secondary antibodies, including Alexa Fluor 405, 488, 568, and 647 (1:500; Life Technologies, Grand Island, NY, USA).
TUNEL and EdU Assays
Apoptotic cell death was analyzed according to the protocol of the In Situ Cell Death Detection Kit, Fluorescein (Roche, Switzerland). Following whole-mount immunostaining, TUNEL was applied. The 5-ethynyl-2′-deoxyuridine (EdU) assay was performed using the protocol outlined in the Click-iT EdU Alexa Fluor 647 Imaging Kit (Life Technologies). Embryos were treated with egg water containing 10 mM EdU and 10% DMSO from 43 hpf for 1 h and harvested at 45 hpf for EdU staining.
Image Acquisition, Processing, and Statistical Analysis
Zeiss LSM700 confocal and Leica M205 FA epifluorescence microscopes were used to obtain image data. Confocal stacks were analyzed using the Zen 2009 software. All figures, labels, arrows, scale bars, and outlines were assembled or drawn using the Adobe Illustrator software. Unpaired two-tailed Student’s t-tests in the GraphPad Prism 5 software were used for statistical analysis. A value of p < 0.05 was considered statistically significant.
RESULTS
tomm22 Knockdown Initially Reduces Liver Size But Later the Liver Recovers
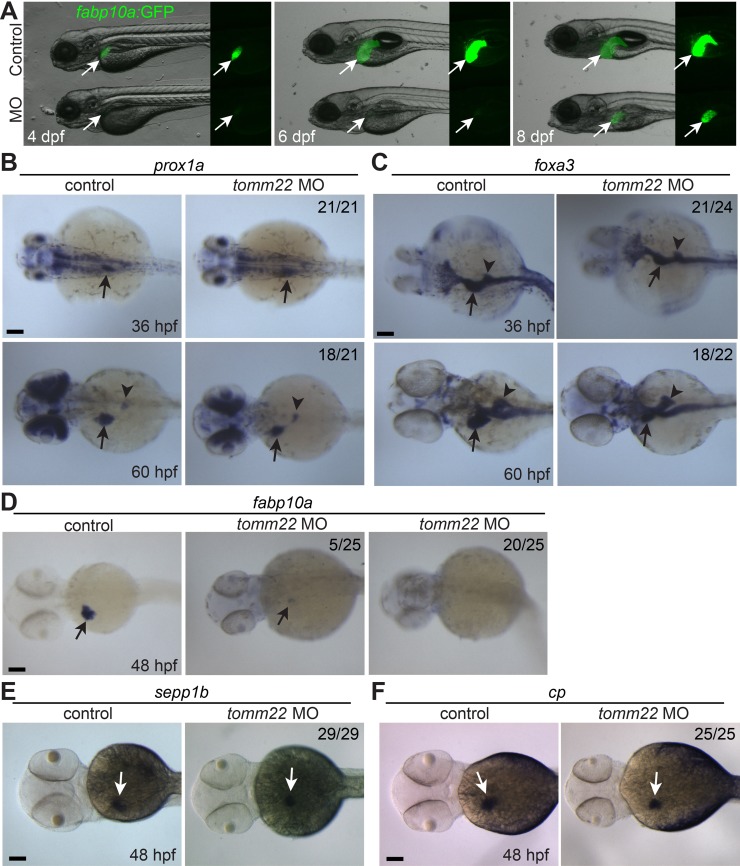
The recovery of the initial liver defect in tomm22 MO-injected larvae, but not in tomm22 mutants, makes this MO-based assay a novel liver regeneration model20. Before we determined the cellular source for this recovery, we carefully examined liver formation in tomm22 MO-injected embryos and larvae with multiple liver markers. Using the Tg(fabp10a:GFP) line that expresses GFP under the hepatocyte-specific fabp10a promoter28, we found that the liver size in the MO-injected larvae was smaller than that in the control larvae at least from 4 dpf (Fig. 1A, arrows). The intensity of fabp10a:GFP intrinsic fluorescence was also much weaker in the MO-injected larvae than in the controls until 6 dpf (Fig. 1A). However, at 8 dpf, not only did the fabp10a:GFP intensity in the MO-injected larvae recover and appear similar to controls, but the liver size was also greatly increased, compared to the 6-dpf MO-injected liver (Fig. 1A), as previously reported20. The MO-injected larvae grew into adults (data not shown), suggesting that the recovered liver is fully functional.
Figure 1.
tomm22 knockdown reduces liver size. (A) Fluorescence images showing hepatic fabp10a:GFP expression (arrows) in tomm22 MO-injected larvae. Note very weak fabp10a:GFP expression until 6 days postfertilization (dpf) but its strong expression at 8 dpf. (B–E) Whole-mount in situ hybridization images showing the expression of prox1a, foxa3, fabp10a, sepp1b, and cp in the MO-injected embryos. Numbers indicate the proportion of larvae exhibiting the representative expression shown. Arrows point to the liver bud or the liver; arrowheads point to the dorsal pancreas. Scale bars: 100 μm.
Given the smaller liver of the MO-injected larvae at 4 dpf, we determined whether the liver size was still smaller from the early liver developmental stages. To reveal the entire liver bud during the early stages, we examined the expression of the hepatoblast marker, prox1a, and the pan-endoderm marker, foxa3 32. Although, liver bud size was comparable between the control and the MO-injected embryos at 36 hpf, it was noticeably smaller in the MO-injected embryos at 60 hpf (Fig. 1B and C, arrows). In contrast, the pancreas appeared unaffected (Fig. 1C, arrowheads). These data indicate that the initial liver bud formation is grossly normal, but its subsequent growth is defective in tomm22 MO-injected embryos.
Given the very faint fabp10a:GFP expression in tomm22 MO-injected larvae at 4 and 6 dpf (Fig. 1A), we tested if hepatocyte differentiation was impaired in the MO-injected embryos by examining the expression of the hepatocyte markers fabp10a, sepp1b, and cp. fabp10a expression in the MO-injected embryos at 48 hpf was faint compared to the control embryos (Fig. 1D, arrows), confirming the fabp10a:GFP expression pattern. However, sepp1b and cp expression in the MO-injected embryos was much stronger than fabp10a expression in the MO-injected embryos and was comparable with their expression in the controls (Fig. 1D–F, arrows). These data suggest that hepatoblast differentiation into hepatocytes is normal in tomm22 MO-injected embryos, as observed in tomm22 mutants20.
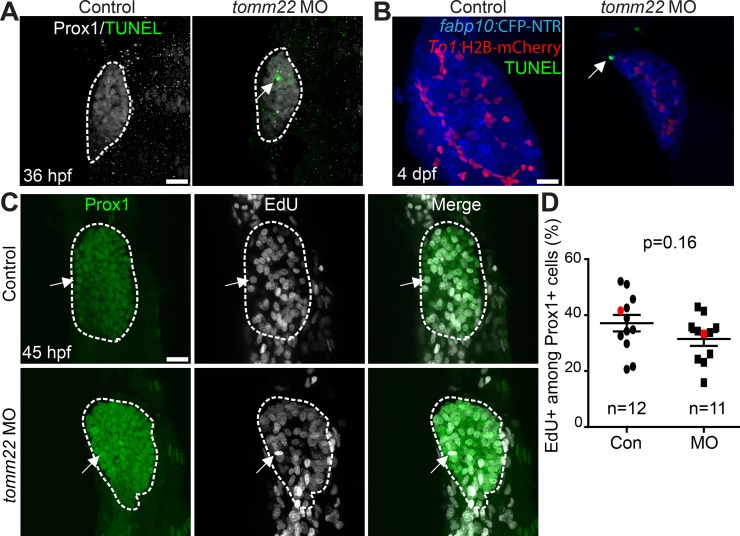
To determine why liver size was reduced upon tomm22 knockdown, we first examined cell death using terminal deoxynucleotidyl transferase (TdT) dUTP nick-end labeling (TUNEL). TUNEL+ dying cells were not observed in the control liver at 36 hpf or 4 dpf, whereas they were observed in the liver of the MO-injected embryos/larvae at 36 hpf (Fig. 2A) and 4 dpf (Fig. 2B). At 4 dpf, the TUNEL+ cells were hepatocytes, as assessed by hepatocyte fabp10a:CFP and BEC Tp1:H2B-mCherry expression (Fig. 2B, arrow). We next examined proliferation using EdU labeling. There was no significant difference in the percentage of EdU+ hepatic cells at 45 hpf between the control and the MO-injected embryos (Fig. 2C and D). These data suggest that the small liver in tomm22 MO-injected embryos is mainly due to increased cell death, consistent with the previous report20.
Figure 2.
Cell death and proliferation in tomm22 MO-injected larvae. (A) Confocal projection images showing Prox1 expression (gray) and terminal deoxynucleotidyl transferase (TdT) dUTP nick-end labeling (TUNEL) (green) in the liver bud (dashed lines) at 36 h postfertilization (hpf). Arrow points to TUNEL+ cells. (B) Confocal projection images showing TUNEL (green) and the expression of fabp10a:CFP-NTR (blue) and Tp1:H2B-mCherry (red) in the liver at 4 dpf. Arrow points to TUNEL+ hepatocytes. (C) Confocal projection images showing Prox1 expression (green) and 5-ethynyl-2′-deoxyuridine (EdU) labeling (gray) in the liver (dashed lines) at 45 hpf. Arrows point to EdU/Prox1 double-positive cells. (D) Graph showing the percentage of EdU+ cells among Prox1+ cells in the liver at 45 hpf. There was no significant difference in the proliferation rate between the control and tomm22 MO-injected larvae. Red marks indicate the embryos shown in (C); n indicates the number of larvae examined. Scale bars: 20 μm; error bars: ±SEM.
BECs Contribute to Hepatocytes in tomm22 MO-Injected Larvae
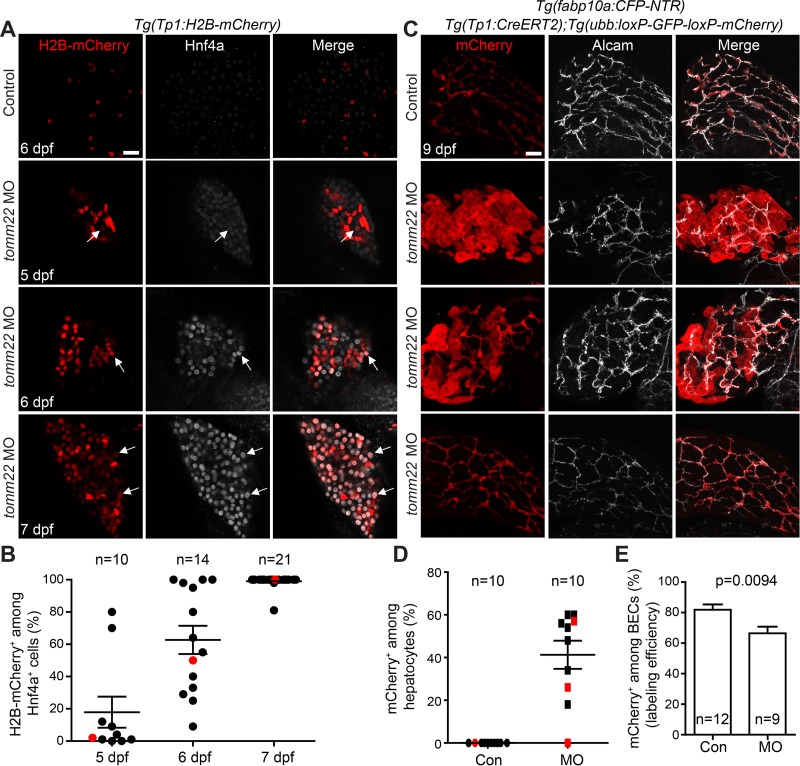
We next determined which cells contributed to the liver recovery in tomm22 MO-injected larvae. We first examined the hepatic expression of the hepatocyte marker, Hnf4a, and the BEC marker, Tp1:H2B-mCherry. The Tg(Tp1:H2B-mCherry) line expresses histone H2B (H2B) and mCherry fusion proteins under the promoter containing the notch-responsive element22. Since Notch signaling is active in BECs, but not in hepatocytes, Tp1:H2B-mCherry expression specifically reveals BECs in the liver33. Moreover, H2B makes the H2B-mCherry fusion protein very stable; thus, this line allows for the easy detection of BEC-derived cells even after Notch activity turns off. In fact, in the zebrafish hepatocyte ablation model, BECs exhibit strong H2B-mCherry expression, and BEC-derived hepatocytes exhibit weak H2B-mCherry expression in the regenerating liver12,25. In the control larvae, there were no Hnf4a/Tp1:H2B-mCherry double-positive cells at 5, 6, and 7 dpf (Fig. 3A and data not shown). In contrast, in the MO-injected larvae at 7 dpf, nearly all Hnf4a+ cells were positive for Tp1:H2B-mCherry (Fig. 3A and B), suggesting their biliary origin. To determine when these double-positive cells first appeared, we examined earlier stages and found that such double-positive cells appeared in a subset of the MO-injected larvae at 5 dpf (Fig. 3A, arrows). The formation of the double-positive cells can be explained in two ways: (1) Tp1:H2B-mCherry+ BECs turn on Hnf4a expression, suggesting BEC conversion to hepatocytes, or (2) Hnf4a+ hepatocytes turn on Tp1:H2B-mCherry expression, suggesting the formation of hybrid hepatocytes. To unambiguously determine the origin of hepatocytes in the recovering liver of the MO-injected larvae, we performed permanent lineage tracing experiments using the Cre/loxP system. To trace BEC lineage, we used the Tg(Tp1:CreERT2) line together with a Cre reporter line, Tg(ubb:loxP-GFP-STOP-loxP-mCherry), which expresses mCherry upon Cre-mediated excision of the STOP cassette23. 4-OHT was treated from 48 to 84 hpf, the stage before the appearance of Tp1:H2B-mCherry/Hnf4a double-positive cells. In the control larvae, only BECs were labeled with mCherry, as previously reported12. In sharp contrast, in the MO-injected larvae, numerous hepatocytes were labeled with mCherry at 9 dpf (Fig. 3C), indicating their biliary origin. However, about 40% of hepatocytes were labeled with mCherry with the range of 0–60% (Fig. 3D). Although 66% of the Cre-mediated labeling efficiency in the MO-injected larvae at 9 dpf (Fig. 3E) can explain, in part, such a low percentage, the percentage is still low compared with the percentage of Tp1:H2B-mCherry+ cells among Hnf4a+ cells (∼100%) at 7 dpf (Fig. 3D vs. Fig. 3B), suggesting the contribution of non-BECs to recovering hepatocytes in tomm22 MO-injected larvae. These lineage tracing data reveal that both BECs and non-BECs, most likely surviving hepatocytes, give rise to recovered hepatocytes in tomm22 MO-injected larvae.
Figure 3.
Biliary epithelial cells (BECs) give rise to hepatocytes in tomm22 MO-injected larvae. (A) Confocal single-optical section images showing Tp1:H2B-mCherry (red) and Hnf4a (gray) expression in the liver. Arrows point to H2B-mCherry/Hnf4a double-positive cells. (B) A graph showing the percentage of H2B-mCherry+ cells among Hnf4a+ cells in the livers of the tomm22 MO-injected larvae. Red dots indicate the larvae shown in (A). (C) Confocal projection images showing the hepatic expression of ubb:mCherry (red, Cre-labeled cells) and Alcam (gray, BECs) at 9 dpf. 4-OHT was treated from 48 to 84 hpf. (D) Graph showing the percentage of ubb:mCherry+ hepatocytes, which were derived from BECs. fabp10a:CFP-NTR expression was used to define hepatocytes. Red marks indicate the larvae shown in (C). (E) Graph showing the percentage of mCherry+ cells among Alcam+ BECs at 9 dpf, indicating Cre-mediated labeling efficiency. n indicates the number of larvae examined. Scale bars: 20 μm; error bars: ±SEM.
Surviving Hepatocytes Become Hybrid Hepatocytes in tomm22 MO-Injected Larvae
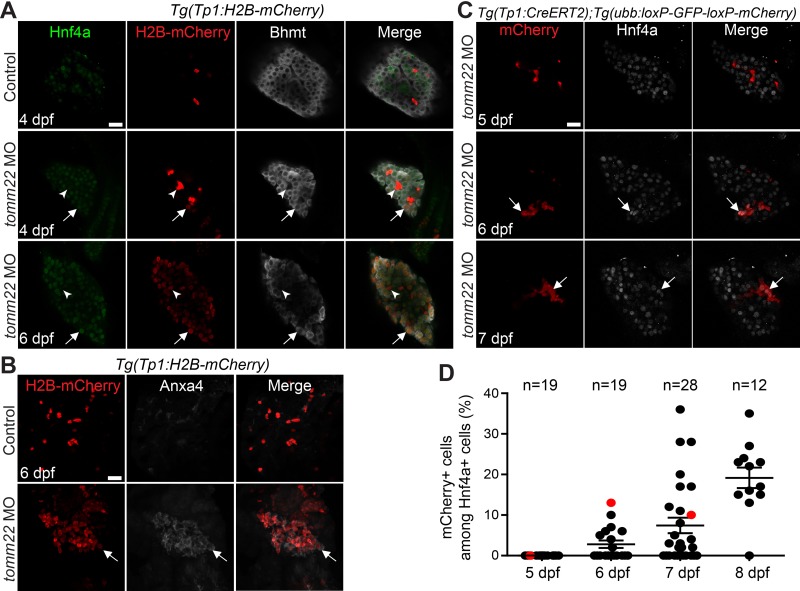
Our lineage tracing data suggest that surviving hepatocytes turned on Tp1:H2B-mCherry expression in tomm22 MO-injected larvae. Since Notch signaling is required and sufficient for biliary specification34,35 and is required for the conversion of hepatocytes to BECs7, the hepatocyte induction of Tp1:H2B-mCherry can be considered the formation of hybrid hepatocytes expressing biliary markers. We investigated if such hybrid cells indeed formed in tomm22 MO-injected larvae. We assumed that if hepatocytes start to express Tp1:H2B-mCherry, its expression should be very faint at the beginning. By focusing on such faint expression with the increase in gain in confocal microscopy, we could detect a few hepatocytes exhibiting the extremely faint expression of Tp1:H2B-mCherry in the MO-injected larvae at 4 dpf (Fig. 4A, arrows). The expression of the hepatocyte differentiation marker, Bhmt36, was used to distinguish hepatocytes from BECs. At 4 dpf, Tp1:H2B-mCherry expression in Bhmt+ hepatocytes was much weaker than its expression in Bhmt− BECs, whereas, at 6 dpf, its expression level was comparable with that in Bhmt− BECs (Fig. 4A, arrows vs. arrowheads). Since BEC markers are expressed in hybrid hepatocytes in mice4,5,7, we next examined the expression of another BEC marker, Anxa433, and found that Anxa4 was also expressed in most Tp1:H2B-mCherry+ cells in the MO-injected larvae at 6 dpf (Fig. 4B, arrows).
Figure 4.
Surviving hepatocytes become hybrid hepatocytes. (A) Confocal single-optical section images showing the expression of Hnf4a (green), Tp1:H2B-mCherry (red), and Bhmt (gray) in the liver. Arrows point to hepatocytes that express Tp1:H2B-mCherry; arrowheads point to BECs negative for Hnf4a and Bhmt. (B) Confocal projection images showing Tp1:H2B-mCherry (red) and Anxa4 (gray) expression in the liver. Arrows point to H2B-mCherry/Anxa4 double-positive cells. (C) Confocal single-optical section images showing the hepatic expression of ubb:mCherry (red, Cre-labeled cells) and Hnf4a (gray) in tomm22 MO-injected larvae. Arrows point to mCherry/Hnf4a double-positive cells. (D) Graph showing the percentage of ubb:mCherry+ cells among Hnf4a+ cells, which were derived from BECs. Red dots indicate the larvae shown in (C); n indicates the number of larvae examined. Scale bars: 20 μm; error bars: ±SEM.
If BECs or BEC-derived cells do not express Hnf4a at 4–5 dpf, it will indirectly indicate that the H2B-mCherry/Hnf4a double-positive cells at these stages are derived from hepatocytes. Thus, we performed BEC lineage tracing experiments and examined Hnf4a expression in lineage-traced, MO-injected larvae at 4–8 dpf. The Cre-labeled, mCherry+ cells did not express Hnf4a until 5 dpf, but from 6 dpf, a subset of these cells expressed Hnf4a (Fig. 4C and D). These data together with the Tp1:H2B-mCherry expression data indicate that surviving hepatocytes become hybrid hepatocytes and contribute to recovering hepatocytes in tomm22 MO-injected larvae.
Suppression of Wnt/β-Catenin Signaling Blocks Hepatocyte Proliferation and Represses the Formation of Hybrid Hepatocytes in tomm22 MO-Injected Larvae
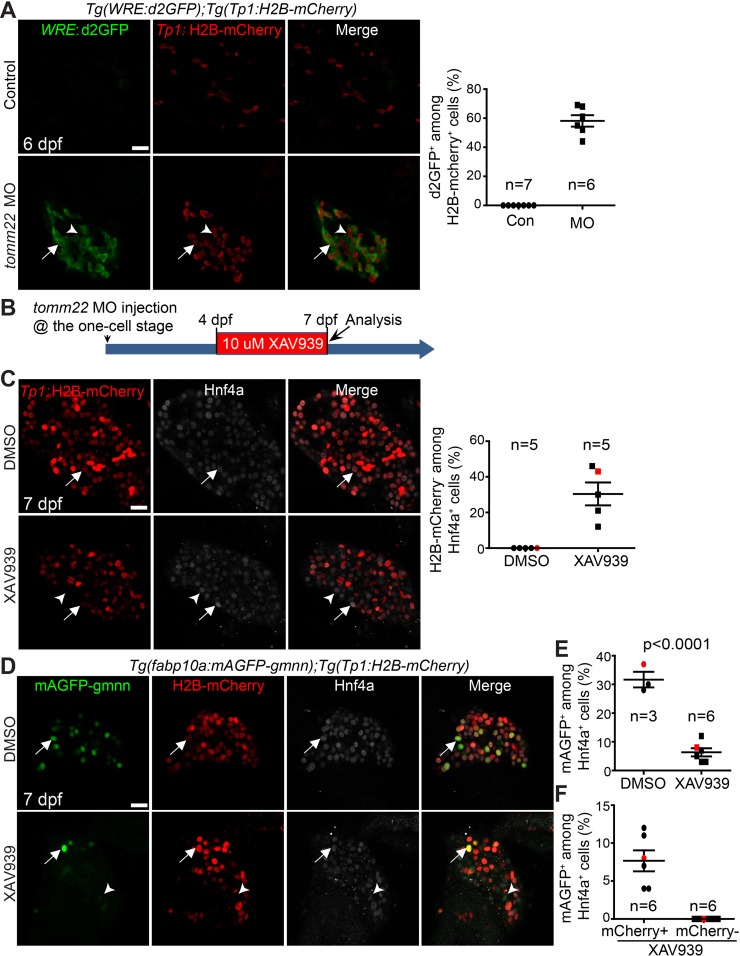
It was previously reported that Wnt/β-catenin signaling positively regulates liver recovery in tomm22 MO-injected larvae20. A Wnt ligand gene, wnt2bb, was upregulated in the livers of the MO-injected larvae, and the suppression of Wnt/β-catenin signaling reduced the size of the recovering livers of the MO-injected larvae20. Since Wnt/β-catenin signaling regulates hepatocyte proliferation during liver regeneration37, we sought to determine if the reduced liver recovery upon Wnt suppression was due to a proliferation defect. We first examined if Wnt activity was enhanced in tomm22 MO-injected larvae. Using a Wnt reporter line, Tg(WRE:d2GFP) 26, we observed a strong WRE:d2GFP expression in the livers of tomm22 MO-injected larvae at 6 dpf, but no expression in the control livers (Fig. 5A). About 60% of Tp1:H2B-mCherry+ cells exhibited Wnt activity in the livers of the MO-injected larvae. We next suppressed Wnt/β-catenin signaling by treating tomm22 MO-injected larvae from 4 to 7 dpf with a Wnt inhibitor, XAV939, which stimulates β-catenin degradation by stabilizing Axin38 (Fig. 5B). As reported previously20, the suppression of Wnt/β-catenin signaling reduced the size of the recovering liver in the MO-injected larvae. Consistent with the positive role of Wnt/β-catenin signaling in hepatocyte proliferation during liver regeneration37, XAV939 treatment greatly reduced hepatocyte proliferation in tomm22 MO-injected larvae at 7 dpf compared with the DMSO treatment control (Fig. 5D and E). For the proliferation assay, we used the Tg(fabp10a:mAGFP-gmnn) line25, which expresses green fluorescent proteins in hepatocytes that are in the S/G2/M, but not in the G0 or G1, phases of the cell cycle39. In addition to this proliferation phenotype, we intriguingly observed a novel phenotype, the presence of Hnf4a+ cells negative for Tp1:H2B-mCherry (Fig. 5C and D, arrowheads). About 30% of Hnf4a+ cells were negative for Tp1:H2B-mCherry in XAV939-treated, tomm22 MO-injected larvae at 7 dpf, whereas none of such cells were present in the DMSO-treated, MO-injected larvae (Fig. 5C). Since BEC-derived hepatocytes retain Tp1:H2B-mCherry expression, it is likely that a subset of surviving, preexisting hepatocytes failed to turn on Tp1:H2B-mCherry expression in the XAV939-treated, MO-injected larvae. Interestingly, the proliferation assay with the Tg(fabp10a:mAGFP-gmnn) line revealed that Tp1:H2B-mCherry−/Hnf4a+ hepatocytes were more sensitive to Wnt/β-catenin inhibition than Tp1:H2B-mCherry+/Hnf4a+ hepatocytes because the proliferation rate of the former cells was much lower than that of the latter (0% vs. 6%) (Fig. 5F). Altogether, these data suggest that in tomm22 MO-injected larvae, Wnt/β-catenin signaling promotes the formation of hybrid hepatocytes as well as hepatocyte proliferation.
Figure 5.
Suppression of Wnt/β-catenin signaling represses hepatocyte proliferation and the formation of hybrid hepatocytes in tomm22 MO-injected larvae. (A) Confocal single-optical section images showing WRE:d2GFP (green) and Tp1:H2B-mCherry (red) expression in the liver at 6 dpf. Arrows point to d2GFP/H2B-mCherry double-positive cells; arrowheads point to H2B-mCherry single-positive cells. Quantification of the percentage of d2GFP+ cells among H2B-mCherry+ cells is shown. (B) Scheme illustrating the period of XAV939 treatment for (C)–(F). (C) Confocal single-optical section images showing the hepatic expression of Tp1:H2B-mCherry (red) and Hnf4a (gray) at 7 dpf. Arrows point to H2B-mCherry/Hnf4a double-positive cells; arrowheads point to H2B-mCherry−/Hnf4a+ cells. Quantification of the percentage of H2B-mCherry− cells among Hnf4a+ cells is shown. (D) Confocal single-optical images showing the hepatic expression of fabp10a:mAGFP-gmnn (green), Tp1:H2B-mCherry (red), and Hnf4a (gray) at 7 dpf. Arrows point to mAGFP-gmnn/H2B-mCherry/Hnf4a triple-positive cells; arrowheads point to mAGFP-gmnn/Hnf4a double-positive cells. (E) Graph showing the percentage of mAGFP-gmnn+ cells among Hnf4a+ cells. Red marks indicate the larvae shown in (D). (F) Graph showing the percentage of mCherry+ or mCherry− proliferating hepatocytes. Red marks indicate the larvae shown in (D). n indicates the number of larvae examined. Scale bars: 20 μm; error bars: ±SEM.
Macrophage Ablation Impairs the Formation of Hybrid Hepatocytes in tomm22 MO-Injected Larvae
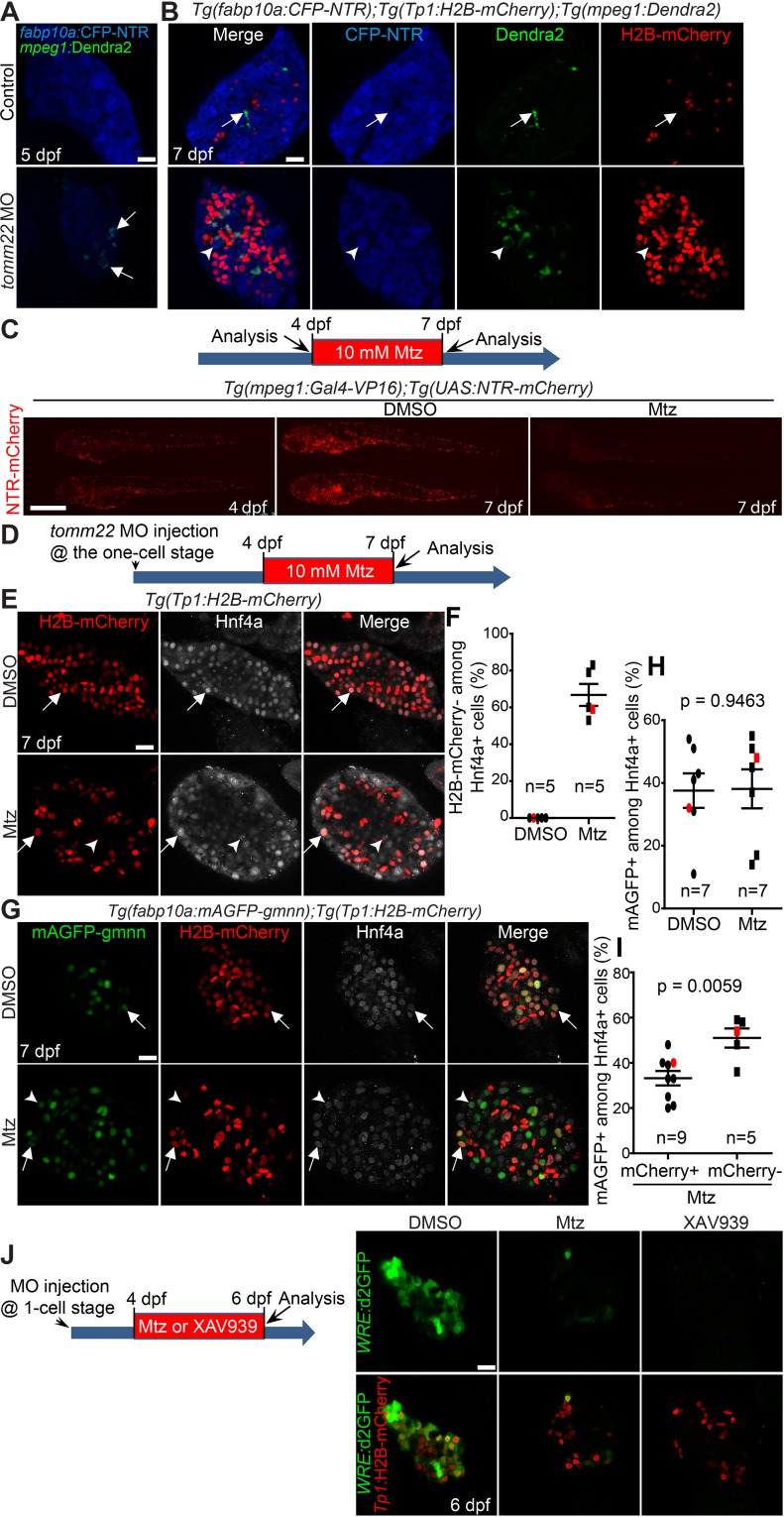
Given the positive role of macrophages in oval cell activation40 and oval cell differentiation into hepatocytes41 in mice, we sought to determine the role of macrophages in tomm22 MO-mediated liver regeneration. We first examined if macrophages were present in the livers of tomm22 MO-injected larvae. Using the Tg(mpeg1:Dendra2) line that expresses a fluorescent protein Dendra2 under the macrophage-specific mpeg1 promoter29, we first detected macrophages in the livers of tomm22 MO-injected larvae at 5 dpf (Fig. 6A, arrows) and observed more macrophages at 7 dpf (Fig. 6B). We often observed macrophages surrounding Tp1:H2B-mCherry− hepatocytes (Fig. 6B, arrowheads) but barely detected macrophages surrounding Tp1:H2B-mCherry+ hepatocytes. In the control liver, a few macrophages were also detected at 7 dpf, but their shape was different from that of the MO-injected larvae: macrophages in the control liver were elongated, whereas those in the MO-injected larvae were spherical (Fig. 6A and B). Given the increased number of macrophages in the livers of tomm22 MO-injected larvae and their shape change, we next investigated the effect of macrophage ablation on tomm22 MO-mediated liver regeneration. Using two transgenic lines that make NTR specifically expressed in macrophages42, Tg(mpeg1:Gal4-VP16) and Tg(UAS:NTR-mCherry), we ablated macrophages by treating the double transgenic larvae with Mtz. Since macrophages were detected in the livers of the MO-injected larvae from 5 dpf (Fig. 6A), we applied Mtz from 4 to 7 dpf and examined the liver at 7 dpf (Fig. 6D). This Mtz treatment greatly reduced the number of macrophages in the whole body of the transgenic larvae at 7 dpf, as assessed by NTR-mCherry intrinsic fluorescence (Fig. 6C), validating efficient macrophage ablation with the NTR/Mtz system. Intriguingly, we observed the presence of Tp1:H2B-mCherry−/Hnf4a+ hepatocytes following macrophage ablation in tomm22 MO-injected larvae (Fig. 6E, arrowheads), similar to our observations in XAV939-treated, MO-injected larvae (Fig. 5C). All Hnf4a+ cells were positive for Tp1:H2B-mCherry in the DMSO-treated, MO-injected larvae, whereas 67% of them were negative for Tp1:H2B-mCherry (Fig. 6F), suggesting a defect in the formation of hybrid hepatocytes. However, unlike XAV939 treatment, macrophage ablation did not reduce the proliferation of Hnf4a+ hepatocytes (Fig. 6G and H). Moreover, Tp1:H2B-mCherry−/Hnf4a+ hepatocytes proliferated more than Tp1:H2B-mCherry+/Hnf4a+ hepatocytes (Fig. 6I). Given the possibility that macrophages regulate Wnt/β-catenin signaling41, using the Tg(WRE:d2GFP) line, we examined Wnt activity in macrophage-ablated, tomm22 MO-injected larvae. Hepatic WRE:d2GFP expression was greatly reduced in the macrophage-ablated, MO-injected larvae compared with unablated, MO-injected larvae (Fig. 6J), supporting the possibility that macrophages regulate Wnt/β-catenin signaling during tomm22 MO-mediated liver regeneration. Altogether, these data suggest that during tomm22 MO-mediated liver regeneration, macrophages do not control hepatocyte proliferation but regulate the formation of hybrid hepatocytes, in part, via Wnt/β-catenin signaling.
Figure 6.
Macrophage ablation impairs the formation of hybrid hepatocytes in tomm22 MO-injected larvae. (A) Confocal single-optical section images showing the expression of mpeg1:Dendra2 (green, macrophages) and fabp10a:CFP-NTR (blue, hepatocytes) in the liver at 5 dpf. Arrows point to macrophages. (B) Confocal single-optical section images showing the expression of mpeg1:Dendra2 (green), Tp1:H2B-mCherry (red), and fabp10a:CFP-NTR (blue) in the liver at 7 dpf. Arrows point to macrophages in the control liver; arrowheads point to a macrophage engulfing a hepatocyte in tomm22 MO-injected larvae. (C) Fluorescence images showing macrophage NTR-mCherry expression before and after metronidazole (Mtz) treatment. Mtz (10 mM) was treated from 4 to 7 dpf. Note the great reduction in the number of NTR-mCherry+ macrophages in the Mtz-treated larvae at 7 dpf compared with the DMSO-treated controls. Lateral views, dorsal up, anterior to the left. Scale bar: 250 μm. (D) Scheme illustrating the period of Mtz treatment and harvest stage for macrophage ablation for (E–I). (E) Confocal single-optical section images showing the expression of Tp1:H2B-mCherry (red) and Hnf4a (gray) in the liver at 7 dpf. Arrows point to H2B-mCherry/Hnf4a double-positive cells; arrowheads point to Hnf4a single-positive cells. (F) Graph showing the percentage of H2B-mCherry− cells among Hnf4a+ cells. Red marks indicate the larvae shown in (E). (G) Confocal single-optical section images showing the expression of fabp10a:mAGFP-gmnn (green), Tp1:H2B-mCherry (red), and Hnf4a (gray) in the liver at 7 dpf. Arrows point to mAGFP-gmnn/H2B-mCherry/Hnf4a triple-positive cells; arrowheads point to mAGFP-gmnn+/H2B-mCherry−/Hnf4a+ cells. (H) Graph showing the percentage of fabp10a:mAGFP-gmnn+ cells among Hnf4a+ cells. Red marks indicate the larvae shown in (G). (I) Graph showing the percentage of H2B-mCherry+ or H2B-mCherry− proliferating Hnf4a+ cells. Red marks indicate the larvae shown in (G). n indicates the number of larvae examined. (J) Confocal single-optical section images showing the expression of WRE:d2GFP (green, Wnt activity) and Tp1:H2B-mCherry (red) in the liver at 6 dpf. Anti-GFP antibody was used to reveal WRE:d2GFP expression. Note the complete absence of hepatic d2GFP expression in the XAV939-treated, tomm22 MO-injected larvae, whereas its faint expression in macrophage-ablated, tomm22 MO-injected larvae. Scale bars: 20 μm, except (C); error bars: ±SEM.
DISCUSSION
In this study, we determined the origin of hepatic cells that contribute to recovering hepatocytes in tomm22 MO-injected larvae. By temporarily knocking down tomm22, we observed the initial reduction of liver size as a result of hepatocyte cell death. This liver damage not only activated BECs to give rise to hepatocytes but also induced the phenotypic change of surviving hepatocytes, generating hybrid hepatocytes. Upon MO dilution as larvae grow, hepatocytes arising from either surviving hepatocytes or BECs began to form properly, allowing for the survival of the MO-injected larvae. Using this MO-based liver regeneration model, we discovered that both Wnt/β-catenin signaling and macrophages control the formation of hybrid hepatocytes.
In tomm22 MO-injected larvae at 7 dpf, nearly all Hnf4a+ cells expressed Tp1:H2B-mCherry (Fig. 3B). However, permanent BEC lineage tracing data show that about 7% of Hnf4a+ cells, with the range of 0–36%, were derived from BECs at 7 dpf (Fig. 4D). Even considering ∼66% Cre labeling efficiency in the MO-injected larvae (Fig. 3E), these data suggest that a large subset of Hnf4a/Tp1:H2B-mCherry double-positive cells were derived from non-BECs. Our finding that most of the double-positive cells expressed the hepatocyte differentiation marker, Bhmt (Fig. 4A), and the BEC marker, Anxa4 (Fig. 4B), in the livers of tomm22 MO-injected larvae at 6 dpf supports that hepatocytes become hybrid hepatocytes in the MO-injected larvae. These hybrid hepatocytes were frequently observed in diverse murine liver injury models. In particular, hepatocyte lineage tracing studies in mice revealed that upon chronic liver injuries, a subset of hepatocytes convert to oval cells, which express both hepatocyte and biliary markers and later redifferentiate into hepatocytes4,6–8. This hepatocyte conversion and redifferentiation appear to occur in tomm22 MO-injected larvae as well.
We previously reported that injury levels determine the mode of liver regeneration between hepatocyte- and BEC-driven liver regeneration12. Upon mild hepatocyte ablation, only preexisting hepatocytes contribute to regenerating hepatocytes, whereas upon near-total hepatocyte ablation, BECs contribute to hepatocytes without the contribution of preexisting hepatocytes. Intriguingly, upon intermediate hepatocyte ablation, both hepatocytes and BECs appear to contribute to regenerating hepatocytes12. This relationship between liver injury levels and the mode of liver regeneration observed in zebrafish is supported by mouse studies. In mouse oval cell activation models, including CCl4 injection and DDC and CDE diet, preexisting hepatocytes, but not BECs, contribute to regenerating hepatocytes4–8. Since remaining hepatocytes actively proliferate in these liver injury models, liver injury levels can be considered “mild” in these models. However, when liver injury is very severe, that is, remaining hepatocytes cannot proliferate, BECs extensively contribute to regenerating hepatocytes. This phenomenon was recently observed in hepatocyte-specific Mdm2 knockout mice9. Given the dual contribution of surviving, preexisting hepatocytes and BECs to recovering hepatocytes in tomm22 MO-injected larvae, liver injury levels in this MO-based liver injury model can be considered “intermediate.” In fact, 2 ng of tomm22 MO injection appeared to elicit a mild liver injury because the liver size in the MO-injected larvae was smaller than in the controls but larger than in larvae injected with 6 ng of the MO. Importantly, in the larvae injected with 2 ng of the MO, we did not observe any BEC contribution to hepatocytes (data not shown), further supporting the relationship between liver injury levels and the mode of liver regeneration.
Wnt/β-catenin signaling promotes hepatocyte proliferation during liver regeneration37, and in chronic liver injury settings, such as the CDE diet, Wnt/β-catenin signaling also appears to promote the differentiation of liver progenitor cells into hepatocytes41. Here we confirmed the positive effect of Wnt/β-catenin signaling on hepatocyte proliferation in tomm22 MO-injected larvae by (1) showing the enhanced Wnt activity in the MO-injected livers and (2) reporting that hepatocyte proliferation was greatly reduced in XAV939-treated, MO-injected larvae compared with DMSO-treated, MO-injected larvae. Moreover, we found a novel phenotype in XAV939-treated, recovering livers: the presence of Hnf4a+ cells negative for Tp1:H2B-mCherry. Since BEC-derived Hnf4a+ cells retain Tp1:H2B-mCherry expression due to the long half-life of H2B-mCherry proteins, the Hnf4a single-positive cells should be derived from surviving hepatocytes. Given that nearly all Hnf4a+ cells were positive for Tp1:H2B-mCherry in tomm22 MO-injected larvae at 7 dpf (Fig. 3B), the presence of the single-positive cells suggests that the suppression of Wnt/β-catenin signaling represses Tp1:H2B-mCherry induction in hepatocytes (i.e., the formation of hybrid hepatocytes). This suggestion is supported by a recent mouse study showing that Wnt/β-catenin signaling induces the expression of biliary markers, such as Sox9, EpCAM, and CK19, in hepatocytes43. Intriguingly, we found a similar phenotype in the MO-injected larvae upon macrophage ablation. Our finding that hepatic Wnt activity was greatly reduced in tomm22 MO-injected larvae with macrophage ablated compared with the unablated MO-injected larvae (Fig. 6J) suggests that macrophages may promote the formation of hybrid hepatocytes, in part, via Wnt/β-catenin signaling. This hypothesis is consistent with a mouse study showing that in the CDE model, macrophage engulfment of dying hepatocytes and their debris induces Wnt3a expression41. However, the number of Hnf4a+ cells negative for Tp1:H2B-mCherry was doubled upon macrophage ablation compared with XAV939 treatment (67% vs. 30%), suggesting the involvement of other factors from macrophages in the formation of hybrid hepatocytes.
Despite the reduced Wnt activity in the recovering livers of the macrophage-ablated, MO-injected larvae, hepatocyte proliferation was not affected at all, making a sharp contrast with the great reduction of hepatocyte proliferation in the XAV939-treated, MO-injected larvae (Fig. 5E vs. Fig. 6H). The direct comparison of hepatic Wnt activity between these two cases revealed no Wnt activity in the XAV939-treated, MO-injected larvae but faint Wnt activity in the macrophage-ablated, MO-injected larvae (Fig. 6J). These data suggest that the remaining Wnt activity in the macrophage-ablated larvae may be sufficient for hepatocyte proliferation.
In summary, here we report an additional zebrafish liver injury model for BEC-driven liver regeneration. Together with the NTR/Mtz-mediated hepatocyte ablation model, this tomm22 MO-based liver injury model will help elucidate the mechanisms underlying BEC-driven liver regeneration, providing insights into augmenting innate liver regeneration in patients with advanced liver diseases.
ACKNOWLEDGMENTS
The authors thank Jinrong Peng for anti-Bhmt antibody, Neil Hukriede and Michael Tsang for discussion, and Mehwish Khaliq for the critical reading of the manuscript. The authors also thank Juhoon So for his confirmative experiments with tomm22 MO. The work was supported by a grant from the NIH to D.S. (R01DK101426). J.W. is a visiting research scholar at the University of Pittsburgh and was supported from the China Scholarship Council. The authors declare no conflicts of interest.
REFERENCES
- 1. Michalopoulos GK. Liver regeneration after partial hepatectomy: Critical analysis of mechanistic dilemmas. Am J Pathol. 2010;176(1):2–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Duncan AW, Dorrell C, Grompe M. Stem cells and liver regeneration. Gastroenterology 2009;137(2):466–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fausto N, Campbell JS. The role of hepatocytes and oval cells in liver regeneration and repopulation. Mech Dev. 2003;120(1):117–30. [DOI] [PubMed] [Google Scholar]
- 4. Yanger K, Knigin D, Zong YW, Maggs L, Gu GQ, Akiyama H, Pikarsky E, Stanger BZ. Adult hepatocytes are generated by self-duplication rather than stem cell differentiation. Cell Stem Cell 2014;15(3):340–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tarlow BD, Pelz C, Naugler WE, Wakefield L, Wilson EM, Finegold MJ, Grompe M. Bipotential adult liver progenitors are derived from chronically injured mature hepatocytes. Cell Stem Cell 2014;15(5):605–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schaub JR, Malato Y, Gormond C, Willenbring H. Evidence against a stem cell origin of new hepatocytes in a common mouse model of chronic liver injury. Cell Rep. 2014;8(4):933–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yanger K, Zong Y, Maggs LR, Shapira SN, Maddipati R, Aiello NM, Thung SN, Wells RG, Greenbaum LE, Stanger BZ. Robust cellular reprogramming occurs spontaneously during liver regeneration. Genes Dev. 2013;27(7):719–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Malato Y, Naqvi S, Schurmann N, Ng R, Wang B, Zape J, Kay MA, Grimm D, Willenbring H. Fate tracing of mature hepatocytes in mouse liver homeostasis and regeneration. J Clin Invest. 2011;121(12):4850–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lu W-Y, Bird TG, Boulter L, Tsuchiya A, Cole AM, Hay T, Guest RV, Wojtacha D, Man TY, Mackinnon A, Ridgway RA, Kendall T, Williams MJ, Jamieson T, Raven A, Hay DC, Iredale JP, Clarke AR, Sansom OJ, Forbes SJ. Hepatic progenitor cells of biliary origin with liver repopulation capacity. Nature Cell Biol. 2015;17(8):971–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Huang MB, Chang A, Choi M, Zhou D, Anania FA, Shin CH. Antagonistic interaction between Wnt and Notch activity modulates the regenerative capacity of a zebrafish fibrotic liver model. Hepatology 2014;60(5):1753–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. He J, Lu H, Zou Q, Luo L. Regeneration of liver after extreme hepatocyte loss occurs mainly via biliary transdifferentiation in zebrafish. Gastroenterology 2014;146(3):789–800.e8. [DOI] [PubMed] [Google Scholar]
- 12. Choi TY, Ninov N, Stainier DY, Shin D. Extensive conversion of hepatic biliary epithelial cells to hepatocytes after near total loss of hepatocytes in zebrafish. Gastroenterology 2014;146(3):776–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pisharath H, Rhee JM, Swanson MA, Leach SD, Parsons MJ. Targeted ablation of beta cells in the embryonic zebrafish pancreas using E. coli nitroreductase. Mech Dev. 2007;124(3):218–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Davison JM, Akitake CM, Goll MG, Rhee JM, Gosse N, Baier H, Halpern ME, Leach SD, Parsons MJ. Transactivation from Gal4-VP16 transgenic insertions for tissue-specific cell labeling and ablation in zebrafish. Dev Biol. 2007;304(2):811–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Curado S, Anderson RM, Jungblut B, Mumm J, Schroeter E, Stainier DYR. Conditional targeted cell ablation in zebrafish: A new tool for regeneration studies. Dev Dyn. 2007;236(4):1025–35. [DOI] [PubMed] [Google Scholar]
- 16. Curado S, Stainier DYR, Anderson RM. Nitroreductase-mediated cell/tissue ablation in zebrafish: A spatially and temporally controlled ablation method with applications in developmental and regeneration studies. Nat Protoc. 2008;3(6):948–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lowes KN, Brennan BA, Yeoh GC, Olynyk JK. Oval cell numbers in human chronic liver diseases are directly related to disease severity. Am J Pathol. 1999;154(2):537–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stueck AE, Wanless IR. Hepatocyte buds derived from progenitor cells repopulate regions of parenchymal extinction in human cirrhosis. Hepatology 2015;61(5):1696–707. [DOI] [PubMed] [Google Scholar]
- 19. Miyajima A, Tanaka M, Itoh T. Stem/progenitor cells in liver development, homeostasis, regeneration, and reprogramming. Cell Stem Cell 2014;14(5):561–74. [DOI] [PubMed] [Google Scholar]
- 20. Curado S, Ober EA, Walsh S, Cortes-Hernandez P, Verkade H, Koehler CM, Stainier DYR. The mitochondrial import gene tomm22 is specifically required for hepatocyte survival and provides a liver regeneration model. Dis Model Mech. 2010;3(7–8):486–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Westerfield M. The zebrafish book. A guide for the laboratory use of zebrafish (Danio rerio), 5th ed Eugene (OR): University of Oregon Press; 2007. [Google Scholar]
- 22. Ninov N, Borius M, Stainier DYR. Different levels of Notch signaling regulate quiescence, renewal and differentiation in pancreatic endocrine progenitors. Development 2012;139(9):1557–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mosimann C, Kaufman CK, Li P, Pugach EK, Tamplin OJ, Zon LI. Ubiquitous transgene expression and Cre-based recombination driven by the ubiquitin promoter in zebrafish. Development 2011;138(1):169–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ninov N, Hesselson D, Gut P, Zhou A, Fidelin K, Stainier DYR. Metabolic regulation of cellular plasticity in the pancreas. Curr Biol. 2013;23(13):1242–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ko S, Choi TY, Russell JO, So J, Monga SPS, Shin D. Bromodomain and extraterminal (BET) proteins regulate biliary-driven liver regeneration. J Hepatol. 2016;64(2):316–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shimizu N, Kawakami K, Ishitani T. Visualization and exploration of Tcf/Lef function using a highly responsive Wnt/beta-catenin signaling-reporter transgenic zebrafish. Dev Biol. 2012;370(1):71–85. [DOI] [PubMed] [Google Scholar]
- 27. Ellett F, Pase L, Hayman JW, Andrianopoulos A, Lieschke GJ. mpeg1 promoter transgenes direct macrophage-lineage expression in zebrafish. Blood 2011;117(4):E49–E56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Her GM, Chiang CC, Chen WY, Wu JL. In vivo studies of liver-type fatty acid binding protein (L-FABP) gene expression in liver of transgenic zebrafish (Danio rerio). FEBS Lett. 2003;538(1–3):125–33. [DOI] [PubMed] [Google Scholar]
- 29. Harvie EA, Green JM, Neely MN, Huttenlocher A. Innate immune response to Streptococcus iniae infection in zebrafish larvae. Infect Immun. 2013;81(1):110–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Alexander J, Stainier DY, Yelon D. Screening mosaic F1 females for mutations affecting zebrafish heart induction and patterning. Dev Genet. 1998;22(3):288–99. [DOI] [PubMed] [Google Scholar]
- 31. Dong PDS, Munson Ca, Norton W, Crosnier C, Pan X, Gong Z, Neumann CJ, Stainier DYR. Fgf10 regulates hepatopancreatic ductal system patterning and differentiation. Nat Genet. 2007;39(3):397–402. [DOI] [PubMed] [Google Scholar]
- 32. Field HA, Ober EA, Roeser T, Stainier DY. Formation of the digestive system in zebrafish. I. Liver morphogenesis. Dev Biol. 2003;253(2):279–90. [DOI] [PubMed] [Google Scholar]
- 33. Lorent K, Moore JC, Siekmann AF, Lawson N, Pack M. Reiterative use of the notch signal during zebrafish intrahepatic biliary development. Dev Dyn. 2010;239(3):855–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kodama Y, Hijikata M, Kageyama R, Shimotohno K, Chiba T. The role of notch signaling in the development of intrahepatic bile ducts. Gastroenterology 2004;127(6):1775–86. [DOI] [PubMed] [Google Scholar]
- 35. Zong Y, Panikkar A, Xu J, Antoniou A, Raynaud P, Lemaigre F, Stanger BZ. Notch signaling controls liver development by regulating biliary differentiation. Development 2009;136(10):1727–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yang SL, Aw SS, Chang CQ, Korzh S, Korzh V, Peng JR. Depletion of Bhmt elevates sonic hedgehog transcript level and increases beta-cell number in zebrafish. Endocrinology 2011;152(12):4706–17. [DOI] [PubMed] [Google Scholar]
- 37. Nejak-Bowen KN, Monga SPS. Beta-catenin signaling, liver regeneration and hepatocellular cancer: Sorting the good from the bad. Semin Cancer Biol. 2011;21(1):44–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Huang S-Ma, Mishina YM, Liu S, Cheung A, Stegmeier F, Michaud Ga, Charlat O, Wiellette E, Zhang Y, Wiessner S, Hild M, Shi X, Wilson CJ, Mickanin C, Myer V, Fazal A, Tomlinson R, Serluca F, Shao W, Cheng H, Shultz M, Rau C, Schirle M, Schlegl J, Ghidelli S, Fawell S, Lu C, Curtis D, Kirschner MW, Lengauer C, Finan PM, Tallarico JA, Bouwmeester T, Porter JA, Bauer A, Cong F. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature 2009;461(7264):614–20. [DOI] [PubMed] [Google Scholar]
- 39. Sugiyama M, Sakaue-Sawano A, Iimura T, Fukami K, Kitaguchi T, Kawakami K, Okamoto H, Higashijima SI, Miyawaki A. Illuminating cell-cycle progression in the developing zebrafish embryo. Proc Natl Acad Sci USA 2009;106(49):20812–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bird TGT, Lu WW-Y, Boulter L, Gordon-Keylock S, Ridgway Ra, Williams MJ, Taube J, Thomas Ja, Wojtacha D, Gambardella A and others. Bone marrow injection stimulates hepatic ductular reactions in the absence of injury via macrophage-mediated TWEAK signaling. Proc Natl Acad Sci USA 2013;110(16):6542–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Boulter L, Govaere O, Bird TG, Radulescu S, Ramachandran P, Pellicoro A, Ridgway RA, Seo SS, Spee B, Van Rooijen N and others. Macrophage-derived Wnt opposes Notch signaling to specify hepatic progenitor cell fate in chronic liver disease. Nat Med. 2012;18(4):572–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lorent K, Gong W, Koo KA, Waisbourd-Zinman O, Karjoo S, Zhao X, Sealy I, Kettleborough RN, Stemple DL, Windsor PA, Whittaker SJ, Porter JR, Wells RG, Pack M. Identification of a plant isoflavonoid that causes biliary atresia. Sci Transl Med. 2015;7(286):286ra67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Okabe H, Yang J, Sylakowski K, Yovchev M, Miyagawa Y, Nagarajan S, Chikina M, Thompson M, Oertel M, Baba H, Monga SP, Nejak-Bowen KN. Wnt signaling regulates hepatobiliary repair following cholestatic liver injury in mice. Hepatology 2016;64(5):1652–66. [DOI] [PMC free article] [PubMed] [Google Scholar]