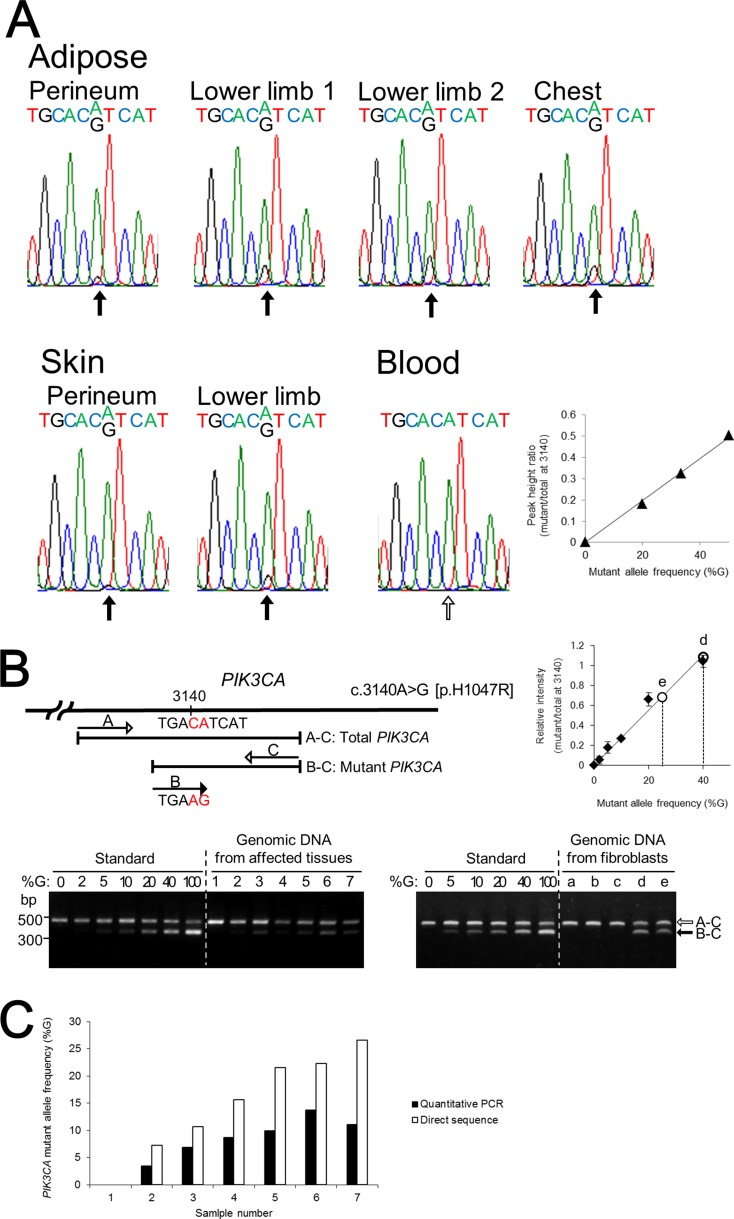
Figure 2. The identification and quantification of the mutation in PIK3CA.
(A) Direct nucleotide sequence analysis of PIK3CA was performed using isolated genomic DNA from blood and six different regions including subcutaneous adipose tissue and skin from the lower limbs. A mosaic mutation (c.3140A>G [p.H1047R]) in exon 20 was identified from affected adipose tissues and skin. The ratios of the mutant allele are different in the patient's affected tissues. Direct nucleotide sequence analysis was performed with plasmids containing normal or mutant PIK3CA fragment as standard reference materials. The right panel shows a standard curve for calculating PIK3CA mutant frequency by direct sequencing. (B) Multiplex PCR products of genomic DNA isolated from affected tissues, blood cells, and fibroblast cells were run through a 1.5% agarose gel and stained with ethidium bromide. Plasmids containing PCR products of wild-type and mutant PIK3CA exon 20 were used as standard reference materials. A 370-bp DNA fragment was generated from the mutant allele of PIK3CA (black arrow) and a 480-bp fragment was generated from both the wild-type and mutant PIK3CA alleles, as the internal control (white arrow). The sizes of the DNA markers are indicated on the left side. The upper right panel shows the standard curve for calculating PIK3CA mutant frequency by multiplex PCR. Lane: 1, blood; 2, skin from the perineum; 3, adipose tissue from the perineum; 4, skin from the lower limb; 5, adipose tissue from lower limb–1; 6, adipose tissue from the chest; 7, adipose tissue from lower limb–2; a, control fibroblast C2; b, control fibroblast C3; c, NHDF-c; d, PROS fibroblast from the skin of the lower limb; e, PROS fibroblast from the skin of the perineum. (C) PIK3CA mutant allele frequencies at nucleotide position 3140 in the patient's affected tissues and blood lymphocytes were calculated by quantitative multiplex PCR and direct sequencing. The x-axis labels are the same as in (B).