Abstract
We aimed to identify a panel of circulating plasma microRNAs that can predict EGFR mutation status and monitor epidermal growth factor receptor-tyrosine kinase inhibitor treatment in patients with non-small cell lung cancer. Microarrays were performed for the preliminary screening of dysregulated microRNAs in 9 EGFR mutation-positive patients versus healthy controls. MiR-107 was upregulated and miR-195 was downregulated in the exon 19 deletion versus wild-type group. The areas under the receiver operating characteristic curves for miR-107, miR-195, and a panel of these 2 microRNAs were 0.72, 0.75, and 0.74, with sensitivities and specificities of 64.7% and 76.6%, 71.8% and 69.1%, and 71.7% and 78.9%, respectively. MiR-122 was significantly upregulated in the p.L858R versus wild-type group. An area under the receiver operative characteristic curve of 0.75 suggests that miR-122 might be a specific biomarker for patients with the p.L858R mutation. In addition, dynamic changes in these 3 microRNAs were also found to correlate with responses to epidermal growth factor receptor-tyrosine kinase inhibitor treatment, indicating that circulating plasma microRNAs may represent potential biomarkers for monitoring epidermal growth factor receptor-tyrosine kinase inhibitor treatment. This study demonstrates the prospective application of circulating plasma microRNAs as potential non-invasive, convenient biomarkers for patients with EGFR-sensitive mutations.
Keywords: circulating microRNA, EGFR mutation status, epidermal growth factor receptor-tyrosine kinase inhibitor, non-small cell lung cancer, tumor marker
INTRODUCTION
As one of the most frequently diagnosed cancers, lung cancer continues to represent the first leading cause of cancer-related mortality worldwide [1, 2]. Non-small cell lung cancer (NSCLC) accounts for approximately 80.0–85.0% of all lung cancers and remains a significant public health problem in China [1]. The majority of NSCLC patients are diagnosed at advanced stages with poor prognoses, especially in those treated with traditional chemotherapy regimens [3].
To date, the EGFR gene remains the most important oncogenic driver of NSCLC and treatment-naïve patients with advanced NSCLC harboring specific EGFR-sensitive mutations (principally EGFR exon 19 deletions and EGFR exon 21 p.L858R point mutations that account for approximately 45.0% and 40.0% of patients, respectively) are recommended to receive first-line epidermal growth factor receptor-tyrosine kinase inhibitors (EGFR-TKIs) as per the National Comprehensive Cancer Network guidelines [4–6]. The efficacy of EGFR-TKIs has been proven in several large-scale randomized clinical trials, especially for Asian, female, non-smoking individuals and adenocarcinoma patients [7]. Specifically, the EGFR exon 19 deletion was reported to be associated with a longer progression-free survival (PFS) compared to the EGFR p.L858R point mutation [8]. Therefore, it is crucial to identify the genotype of the tumor after the histopathological classification is determined to predict the sensitivity or resistance to an increasing number of EGFR-TKIs.
In a clinical setting, obtaining adequate tumor specimens for pathological evaluation and specific molecular analyses are decisive prerequisites for establishing an optimal, individualized treatment regimen for the patient. Tumor biopsy specimens can sometimes be difficult to obtain from certain patients. Tumor pathology and genotyping are often determined prior to commencing first-line EGFR-TKI treatment. However, re-biopsy during or after the time of progressive disease (PD) on EGFR-TKI treatment to continuously monitor EGFR-sensitive mutations and to analyze the mechanism(s) of resistance to EGFR-TKI treatment remains a significant challenge and appears to be unrealistic. Hence, as a non-invasive test, liquid biopsy has warranted wide attention in recent years with its unique advantage of simultaneously capturing multiple sites of tumor growth and testing EGFR mutation status, which could assist oncological clinicians in the timely adjustment of therapeutic strategies for NSCLC patients [9, 10]. Therefore, it is imperative to explore non-invasive, convenient, and economical tumor markers to predict EGFR mutation status and to monitor EGFR-TKI treatment in NSCLC patients.
MicroRNAs comprise a large family of small (approximately 21–25 nucleotides in length) endogenous, non-coding RNAs that negatively regulate gene expression at post-transcriptional level via inhibition of target messenger RNAs by pairing with the complementary sequences in the 3′ untranslated region [11, 12]. MicroRNAs exert a wide range of biological functions, including early tumorigenesis and tumor progression. It has been reported that circulating microRNAs are packaged into microparticles or are associated with RNA binding proteins and lipoprotein complexes, making them ideal candidates for tumor biomarkers owing to their high stability in body fluids [13, 14]. Accumulating evidence has proven that circulating microRNA signatures in human plasma or serum may serve as disease fingerprints and novel molecular markers for NSCLC [15–17]. However, associations between circulating microRNAs in plasma and EGFR mutation status and their application for monitoring EGFR-TKI treatment and disease progression have not been systematically studied. Therefore, we aimed to identify a panel of circulating plasma microRNAs that can distinguish between NSCLC patients with EGFR-sensitive mutations and EGFR wild-type patients and to explore the potential of this microRNA panel to monitor tumor responses to EGFR-TKI treatment.
RESULTS
Patient characteristics
Between December 2014 and April 2016, we recruited 153 patients with pathologically confirmed NSCLC and 41 healthy controls, with a median age of 56.1 (range, 45–78) years and a male to female ratio of 0.71. The clinical characteristics of the patients are summarized in Table 1.
Table 1. Clinical characteristics of NSCLC patients (n = 153).
| Characteristic | NSCLC patients | p-valueb | |||
|---|---|---|---|---|---|
| EGFR19DEL (n = 64) | EGFRp.L858R (n = 36) | EGFRMUT(+)a (n = 100) | EGFRWT (n = 53) | ||
| Age (years), mean (range) | 56.3 (37–76) | 61.1 (34–80) | 57.9 (34–80) | 59.0 (34–84) | 0.537 |
| Sex, n (%) | |||||
| M | 29 (45.3) | 17 (47.2) | 46 (46.0) | 26 (49.1) | |
| F | 35 (54.7) | 19 (52.8) | 54 (54.0) | 27 (50.9) | 0.424 |
| ECOG PS, n (%) | |||||
| 0 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| 1 | 64 (100.0) | 36 (100.0) | 100 (100.0) | 53 (100.0) | |
| 2 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | N/A |
| Complications, n (%) | |||||
| Y | 17 (26.6) | 9 (25.0) | 26 (26.0) | 18 (34.0) | |
| N | 47 (73.4) | 27 (75.0) | 74 (74.0) | 35 (66.0) | 0.198 |
| Smoker, n (%) | |||||
| Y | 22 (34.4) | 13 (36.1) | 35 (35.0) | 28 (52.8) | |
| N | 42 (65.6) | 23 (63.9) | 65 (65.0) | 25 (47.2) | 0.025* |
| FH of cancer, n (%) | |||||
| Y | 8 (12.5) | 5 (13.9) | 13 (13.0) | 10 (18.9) | |
| N | 56 (87.5) | 31 (86.1) | 87 (87.0) | 43 (81.1) | 0.231 |
| cStage, n (%) | |||||
| IIIA–B | 7 (10.9) | 3 (8.3) | 10 (10.0) | 9 (17.0) | |
| IV | 57 (89.1) | 33 (91.7) | 90 (90.0) | 44 (83.0) | 0.161 |
| Histology, n (%) | |||||
| ADC | 59 (92.2) | 34 (94.4) | 93 (93.0) | 51 (96.2) | |
| ADC+SCC | 5 (7.8) | 2 (5.6) | 7 (7.0) | 2 (3.8) | 0.340 |
* p < 0.05
a EGFR19DEL and EGFRp.L858R patients combined
bEGFRMUT(+) versus EGFRWT
Abbreviations: 19DEL, exon 19 deletion; ADC, adenocarcinoma; cStage, clinical stage; ECOG, Eastern Cooperative Oncology Group; F, female; FH, family history; M, male; MUT(+), mutation-positive; N, no; N/A, not applicable; PS, performance status; SCC, squamous cell carcinoma; WT, wild-type; Y, yes
Briefly, plasma samples were collected from 64 EGFR exon 19 deletion, 36 EGFR p.L858R mutation, and 53 EGFR wild-type patients in the Department of Lung Cancer and 41 healthy controls from the Physical Examination Centre at our institution. With the exception of smoking status, no significant differences in clinical characteristics were observed between the NSCLC patients and healthy controls or between subgroups of the NSCLC patients stratified according to EGFR mutation status. Groups containing patients with EGFR-sensitive mutations (EGFR exon 19 deletions or EGFR p.L858R point mutations) had fewer smokers than the group containing EGFR wild-type patients (p < 0.05). Blood samples of 36 of the 64 EGFR exon 19 deletion patients were dynamically collected at hospital visits at 1, 3, and 5 months during EGFR-TKI treatment. In addition, blood samples of 12 patients were also collected at the time of PD on EGFR-TKI treatment (Table 2).
Table 2. Clinical characteristics of EGFR exon 19 deletion patients with NSCLC who had dynamic plasma collected (n = 36).
| Characteristic | Patients (n = 36) |
|---|---|
| Age (years), mean (range) | 51.3 (41–72) |
| Sex, n (%) | |
| M | 17 (47.2) |
| F | 19 (52.8) |
| ECOG PS, n (%) | |
| 0 | 0 (0.0) |
| 1 | 36 (100.0) |
| 2 | 0 (0.0) |
| Complications, n (%) | |
| Y | 9 (25.0) |
| N | 27 (75.0) |
| Smoker, n (%) | |
| Y | 7 (19.4) |
| N | 29 (80.6) |
| FH of cancer, n (%) | |
| Y | 3 (8.3) |
| N | 33 (91.7) |
| cStage, n (%) | |
| IIIA–B | 2 (5.5) |
| IV | 34 (94.5) |
| Histology, n (%) | |
| ADC | 35 (97.2) |
| ADC+SCC | 1 (2.8) |
| EGFR-TKI treatment, n (%) | |
| Gefitinib | 19 (52.8) |
| Erlotinib | 1 (2.8) |
| Icotinib | 16 (44.4) |
| Line of EGFR-TKI treatment, n (%) | |
| First | 28 (77.8) |
| Second | 8 (22.2) |
| Best response to EGFR-TKI treatment, n (%) | |
| CR | 1 (2.8) |
| PR | 23 (63.9) |
| SD | 11 (30.5) |
| PD | 1 (2.8) |
| Sample collected during EGFR-TKI treatment, n (%) | |
| Y | 36 (100.0) |
| N | 0 (0.0) |
| Sample collected at the time of PD on EGFR-TKI treatment, n (%) | |
| Y | 12 (33.3) |
| N | 24 (66.7) |
Abbreviations: ADC, adenocarcinoma; CR, complete response; cStage, clinical stage; ECOG, Eastern Cooperative Oncology Group; F, female; FH, family history; M, male; N, no; PS, performance status; SCC, squamous cell carcinoma; Y, yes
Microarray analysis of candidate plasma microRNAs for EGFR-sensitive mutations
Plasma samples of 3 EGFR exon 19 deletion, 3 EGFR p.L858R mutation, and 3 EGFR wild-type patients and 4 healthy controls were selected for Agilent microRNA microarray analysis to detect differences in the expression levels of circulating plasma microRNAs (n = 2,568) between the above cohorts (Gene Expression Omnibus accession number: GSE93300).
Compared to healthy controls, 19 microRNAs were upregulated and 57 microRNAs were downregulated in the plasma of all 9 NSCLC patients (fold-change >2.0, p < 0.05) (Figure 1). Further analysis of the 76 dysregulated microRNAs was performed to select candidate microRNAs between NSCLC patients with different EGFR-sensitive mutations. Compared to the EGFR wild-type group, 14 microRNAs were up-regulated and 7 microRNAs were downregulated in EGFR exon 19 deletion group, and 2 microRNAs were upregulated and 3 microRNAs were downregulated in the EGFR p.L858R mutation group (fold-change >2.0, p < 0.05; Table 3 and Table 4). It is noteworthy that relative to the EGFR wild-type group, the EGFR exon 19 deletion group contained more differently expressed microRNAs than the EGFR p.L858R mutation group. Indeed, it has been reported that EGFR exon 19 deletion patients are associated with a longer PFS compared to EGFR p.L858R mutation patients [8]. Thus, we primarily focused on EGFR exon 19 deletion patients in our subsequent analyses.
Figure 1. Hierarchical clustering analysis of circulating microRNA expression in plasma from patients with NSCLC versus healthy controls.
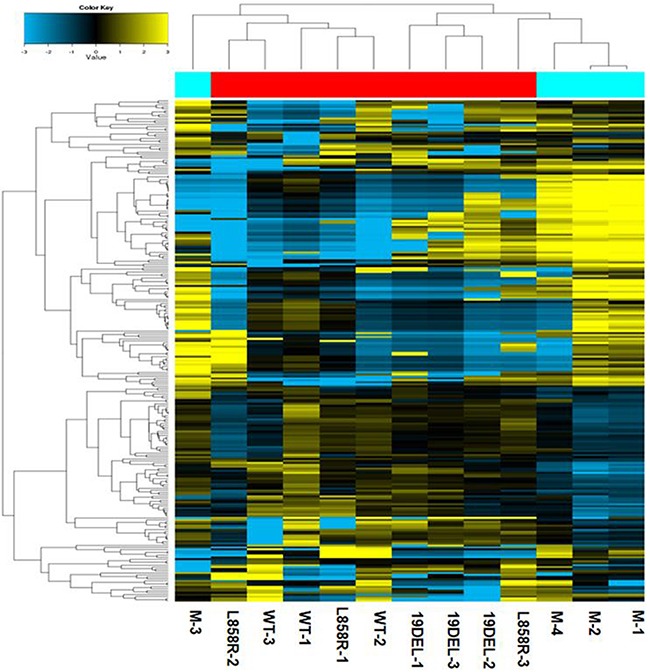
M-1, M-2, M-3, and M-4 represent the healthy controls.
Table 3. Microarray analysis of the differential expression of circulating plasma microRNAs in EGFR exon 19 deletion versus EGFR wild-type NSCLC patients.
| MicroRNA | Fold-change | Regulation | p-value |
|---|---|---|---|
| miR-25-3p | 36.0 | Up | 0.002* |
| miR-15b-5p | 14.9 | Up | 0.005* |
| let-7b-5p | 10.6 | Up | 0.020* |
| miR-21-5p | 16.1 | Up | 0.046* |
| miR-4306 | 42.6 | Up | 0.001* |
| miR30d-5p | 34.5 | Up | 0.006* |
| let-7i-5p | 27.5 | Up | 0.003* |
| miR-19a-3p | 27.7 | Up | 0.002* |
| miR-17-5p | 12.3 | Up | 0.010* |
| miR-20a-5p | 31.1 | Up | 0.003* |
| miR-3195 | 8.5 | Up | 0.038* |
| miR-24-3p | 32.9 | Up | 0.008* |
| miR-15a-5p | 24.6 | Up | 0.004* |
| miR-107 | 30.2 | Up | 0.002* |
| miR-6751-3p | 32.9 | Down | 0.046* |
| miR-6779-3p | 14.3 | Down | 0.022* |
| miR-6858-5p | 30.0 | Down | 0.036* |
| miR-7106-5p | 21.3 | Down | 0.033* |
| miR-6797-3p | 17.4 | Down | 0.030* |
| miR-483-3p | 30.2 | Down | 0.039* |
| miR-619-5p | 10.9 | Down | 0.021* |
* p < 0.05
Table 4. Microarray analysis of the differential expression of circulating plasma microRNAs in EGFR p.L858R mutation versus EGFR wild-type NSCLC patients.
| MicroRNA | Fold-change | Regulation | p-value |
|---|---|---|---|
| miR-1229 | 21.9 | Up | 0.017* |
| miR-3141 | 9.3 | Up | 0.027* |
| miR-4281 | 2.8 | Down | 0.010* |
| miR-4516 | 3.9 | Down | 0.008* |
| miR-3663-3p | 3.0 | Down | 0.035* |
* p < 0.05
Quantitative real-time reverse-transcription polymerase chain reaction (qRT-PCR) validation of candidate stable circulating plasma microRNAs
In total, 20 NSCLC patients (EGFR exon 19 deletion [n = 12] and EGFR wild-type [n = 8]) and 8 healthy controls were selected for further screening of candidate microRNAs correlating with EGFR exon 19 deletion mutation status. Ten microRNAs were screened out from the above microarray data set (miR-107, miR-19a-3p, miR-20a-5p, miR-21-5p, miR-24-3p, miR-25-3p, and miR-30d-5p) and published literature (miR122, miR-125a-5p, and miR-195) [15–17] using qRT-PCR in a pre-experiment. The 5S ribosomal RNA (5SrRNA) was used as an endogenous control to normalize the raw quantification cycle (Ct) values. Subtraction of the Ct value of the target microRNA from 5SrRNA (ΔCt) was performed and converted to 2−ΔCt to assess alterations in microRNA expression levels between the cohorts. Fold-changes were calculated using the 2−Δ(ΔCt) method, where Δ(ΔCt) = (CtmiR – Ct5SrRNA)EGFR-MUT(+) – (CtmiR – Ct5SrRNA)EGFR-WT. Table 5 displays the relative expression levels of the 10 plasma microRNAs in the EGFR exon 19 deletion versus EGFR wild-type group. We found that miR-19a-3p, miR-20a-5p, miR-21-5p, miR-24-3p, miR-25-3p, and miR-30d-5p were not significantly differentially expressed in the EGFR exon 19 deletion versus EGFR wild-type group. Therefore, 4 microRNAs (miR-107, miR-122, miR-125a-5p, and miR-195) were selected for further investigation.
Table 5. Pre-experimental analysis of the differential expression of 10 selected circulating plasma microRNAs in EGFR exon 19 deletion versus EGFR wild-type NSCLC patients.
| MicroRNA | Fold-changea | Regulation | p-value |
|---|---|---|---|
| miR-19a-3p | 4.1 | N/A | 0.070 |
| miR-20a-5p | 5.7 | N/A | 0.052 |
| miR-21-5p | 4.8 | N/A | 0.107 |
| miR-24-3p | 4.0 | N/A | 0.119 |
| miR-25-3p | 5.7 | N/A | 0.052 |
| miR-30d-5p | 4.1 | N/A | 0.098 |
| miR-107 | 14.5 | Up | 0.002* |
| miR-122 | 6.3 | Up | 0.005* |
| miR-125a-5p | 6.6 | Up | 0.017* |
| miR-195 | 9.0 | Down | 0.002* |
* p < 0.05
aCalculated using the equation 2−Δ (ΔCt), where Δ(ΔCt) = (CtmiR – Ct5SrRNA)EGFR-MUT(+) – (CtmiR – Ct5SrRNA)EGFR-WT
Abbreviations: N/A, not applicable.
Dysregulated circulating plasma microRNAs for EGFR-sensitive mutations
To further investigate the dysregulated circulating plasma microRNAs in NSCLC EGFR mutation-positive (EGFR exon 19 deletion or EGFR p.L858R mutation) patients, we performed qRT-PCR with 5SrRNA as the endogenous control for the EGFR exon 19 deletion, EGFR p.L858R mutation, and EGFR wild-type groups. MiR-125a-5p was screened out for exhibiting no statistically significant differences between the cohorts. MiR-107 was significantly upregulated in the EGFR exon 19 deletion, EGFR p.L858R mutation, and EGFR mutation-positive groups compared to the EGFR wild-type group (p < 0.05; Figure 2A and Table 6). Conversely, no statistically significant differences were observed between the EGFR exon 19 deletion and EGFR p.L858R mutation groups. MiR-122 was significantly upregulated in the EGFR p.L858R mutation group compared to the EGFR wild-type or EGFR mutation-positive groups (p < 0.05). Conversely, no statistically significant differences were observed between the EGFR exon 19 deletion and EGFR p.L858R mutation or EGFR wild-type groups (p > 0.05; Figure 2B and Table 6). MiR-195 was significantly downregulated in the EGFR exon 19 deletion group compared to the EGFR wild-type group (p < 0.05), although no statistically significant differences were observed between the other groups (p > 0.05; Figure 2C and Table 6).
Figure 2.
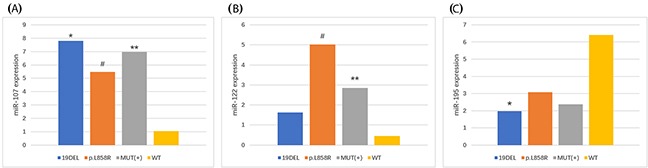
(A) MiR-107, (B) miR-122, and (C) miR-195 expression in NSCLC patients with different EGFR-sensitive mutations. *EGFR exon 19 deletion (19DEL) vs. EGFR wild-type (WT) patients (fold-change: miR-107, 7.44 and miR-195, 3.25; p < 0.05). #EGFR p.L858R mutation vs. EGFRWT patients (fold-change: miR-107, 5.22 and miR-122, 11.29; p < 0.05). *EGFR mutation-positive (MUT[+]) vs. EGFRWT patients (fold-change: miR-107, 6.64 and miR-122, 6.41; p < 0.05).
Table 6. Relative expression of miR-107, miR-122, and miR-195 in EGFR exon 19 deletion (EGFR19DEL), EGFR p.L858R mutation (EGFRp.L858R), and EGFR wild-type (EGFRWT) patients with NSCLC (n = 153).
| microRNA | Normalized expression (mean ± SEM)a | p-value | ||||||
|---|---|---|---|---|---|---|---|---|
| EGFR19DEL(n = 64) | EGFRp.L858R (n = 36) | EGFRMUT(+b (n = 100) | EGFRWT(n = 53) | 19DEL vs. WT | p.L858R vs. WT | MUT(+)b vs. WT | 19DEL vs. p.L858R | |
| miR-107 | 7.80 ± 2.88 | 5.47 ± 2.05 | 6.96 ± 1.98 | 1.05 ± 0.41 | 0.024* | 0.041* | 0.033* | 0.513 |
| miR-122 | 1.62 ± 1.12 | 5.02 ± 2.28 | 2.85 ± 1.10 | 0.45 ± 0.28 | 0.310 | 0.018* | 0.036* | 0.136 |
| miR-195 | 1.98 ± 0.84 | 3.08 ± 0.97 | 2.37 ± 0.64 | 6.41 ± 2.01 | 0.033* | 0.199 | 0.061 | 0.349 |
* p < 0.05
aData expressed as the mean (2−ΔCt) ± SEM
bEGFR19DEL and EGFRp.L858R patients combined
Abbreviations: SEM, standard error of the mean
Diagnostic analysis of circulating plasma microRNAs for EGFR mutation-positive NSCLC patients
Receiver operating characteristic (ROC) curve analysis was performed to evaluate the potential applications of circulating plasma miR-107, miR-122, and miR-195 as diagnostic markers for NSCLC patients with EGFR-sensitive mutations. MiR-107 exhibited a significant difference between EGFR mutation-positive and EGFR wild-type patients. In a comparison between the EGFR exon 19 deletion and EGFR wild-type groups, miR-107 was associated with an area under the ROC curve (AUC) value of 0.72 (95.0% confidence interval [CI]: 0.62–0.81), with a sensitivity of 64.7% and a specificity of 76.6% at a cutoff of 0.097 (Figure 3A). Meanwhile, in a comparison between the EGFR p.L858R mutation and EGFR wild-type groups, miR-107 was associated with an AUC value of 0.77 (95.0% CI: 0.68–0.87), with a sensitivity of 64.2% and a specificity of 80.6% at a cutoff of 0.153 (Figure 3B). With respect to miR-122, the AUC value was 0.75 (95.0% CI: 0.64–0.85), with a sensitivity of 73.6% and a specificity of 63.9% at a cutoff of 0.124, for the comparison between the EGFR p.L858R mutation and EGFR wild-type groups (Figure 4A). Nevertheless, despite the significant difference in miR-122 expression between the EGFR mutation-positive and EGFR wild-type groups, ROC curve analysis demonstrated that miR-122 was unable to distinguish between the two groups (AUC value: 0.61, 95.0% CI: 0.52–0.69; Figure 4B). However, miR-195 could distinguish between the EGFR exon 19 deletion and EGFR wild-type groups. The AUC value was 0.75 (95.0% CI: 0.60–0.79), with a sensitivity of 71.8% and a specificity of 69.1% at a cutoff of 0.876 (Figure 5).
Figure 3.
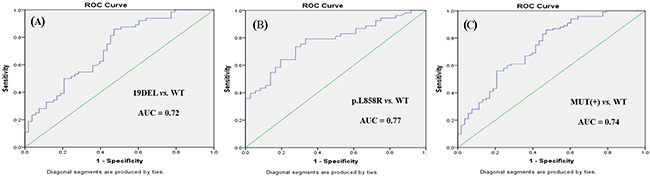
ROC curves for miR-107 in (A) EGFR exon 19 deletion (19DEL), (B) EGFR p.L858R mutation, and (C) EGFR mutation-positive (MUT[+]) patients with NSCLC versus EGFR wild-type (WT) patients.
Figure 4.
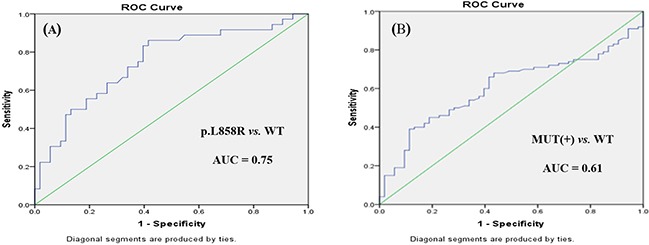
ROC curves for miR-122 in (A) EGFR p.L858R mutation and (B) EGFR mutation-positive (MUT[+]) patients with NSCLC versus EGFR wild-type (WT) patients.
Figure 5. ROC curve for miR-195 in EGFR exon 19 deletion (19DEL) versus EGFR wild-type (WT) patients.
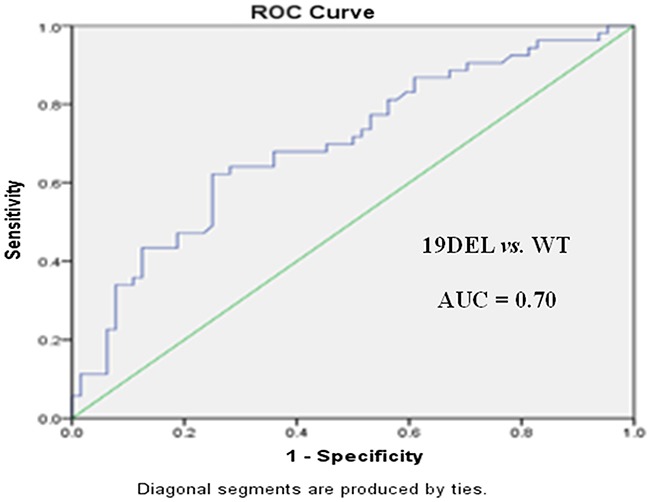
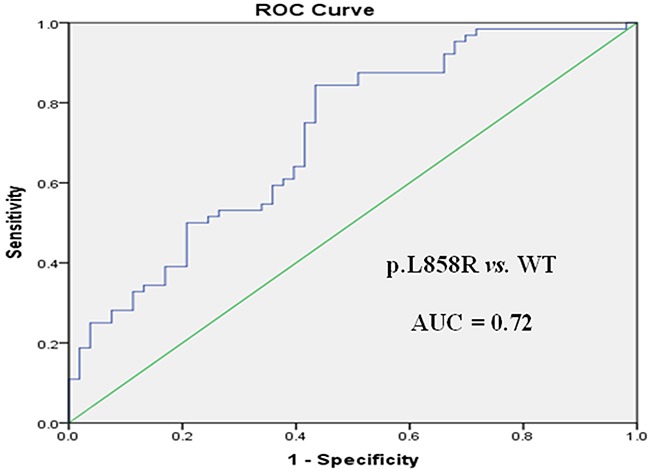
To further evaluate the diagnostic potential of combinations of miR-107, miR-122, and miR-195, as panels, we performed logistic regression analysis combined with ROC curve analysis for these 3 microRNAs. A panel of miR-107 and miR-195 produced an AUC value of 0.74 (95.0% CI: 0.65–0.83), with a sensitivity of 71.7% and a specificity of 78.9% at a cutoff of -0.057, for the comparison between the EGFR exon 19 deletion and EGFR wild-type groups (Figure 6 and Table 7). This was higher than the AUC and sensitivity and specificity values of miR-107 or miR-195 alone as potential diagnostic markers. As for the comparison between the EGFR p.L858R mutation and EGFR wild-type groups, a panel of miR-107 and miR-122 produced an AUC value of 0.78 (95.0% CI: 0.69–0.88), with a sensitivity of 67.1% and a specificity of 82.5% at a cutoff of 0.321 (Figure 7 and Table 7). This was higher than the AUC value of miR-107 or miR-122 alone as markers to distinguish between these two groups.
Figure 6. ROC curve for miR-107 and miR-195 in EGFR exon 19 deletion (19DEL) versus EGFR wild-type (WT) patients.
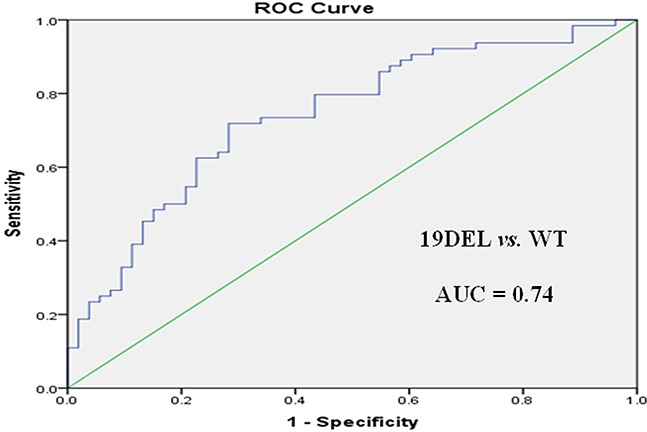
Table 7. Measures of the diagnostic potential of the differentiation of NSCLC patients with EGFR exon 19 deletion (EGFR19DEL) or EGFR p.L858R mutations (EGFRp.L858R) versus EGFR wild-type (EGFRWT) patients.
| Variable | EGFR19DEL vs. EGFRWT | EGFRp.L858R vs. EGFRWT | ||||
|---|---|---|---|---|---|---|
| miR-107 | miR-195 | miR-107 + miR-195 | miR-107 | miR-122 | miR-107 + miR-122 | |
| AUC | 0.72 | 0.75 | 0.74 | 0.77 | 0.75 | 0.78 |
| Sensitivity | 64.7% | 71.8% | 71.7% | 64.2% | 73.6% | 67.1% |
| Specificity | 76.6% | 69.1% | 78.9% | 80.6% | 63.9% | 82.5% |
| Cutoff | 0.097 | 0.876 | −0.057 | 0.153 | 0.124 | 0.321 |
| PPV | 82.1% | 79.0% | 91.4% | 80.0% | 83.7% | 81.8% |
| NPV | 76.8% | 80.3% | 80.3% | 89.8% | 86.9% | 88.3% |
Abbreviations: PPV, positive predictive value; NPV, negative predictive value.
Figure 7. ROC curve for miR-107 and miR-122 in EGFR exon 19 deletion (19DEL) versus EGFR wild-type (WT) patients.
Meanwhile, we also estimated the positive and negative predictive values for distinguishing between NSCLC patients harboring differentEGFR-sensitive mutations. In a comparison between the EGFR exon 19 deletion and EGFR wild-type groups, the positive and negative predictive values of a panel of miR-107 and miR-195 were 91.4% and 80.3%, respectively (Table 7). With respect to the EGFR p.L858R mutation versus EGFR wild-type group, the positive and negative predictive values of a panel of miR-107 and miR-122 were 81.8% and 88.3%, respectively. These results also indicate the potential diagnostic capability of the selected microRNAs.
Univariate and multivariate analyses of clinicopathological variables associated with EGFR-sensitive mutations
In the univariate analysis, smoking status, miR-107, and miR-195 significantly correlated with EGFR mutation status (EGFR exon 19 deletion or p.L858R mutation versus EGFR wild-type) (Table 8). A multivariate analysis was subsequently performed to identify associations between miR-107, miR-195, and clinicopathological variables (sex, age, complications, and smoking status) and EGFR-sensitive mutations. Since we are in the Department of Medical Oncology, all of the enrolled patients were diagnosed with locally advanced or advanced stage disease. Meanwhile, the NSCLC patients we screened in this study were all newly diagnosed with an Eastern Cooperative Oncology Group performance status of 1. Consequently, clinical stage and Eastern Cooperative Oncology Group performance status were excluded from the multivariate analysis. The final results revealed that miR-107 and smoking status are important diagnostic predictors of EGFR-sensitive mutations. NSCLC patients with high levels of miR-107 expression and no prior history of smoking were more likely to harbor EGFR exon 19 deletions or EGFR p.L858R mutations.
Table 8. Univariate and multivariate analyses of clinicopathological variables associated with EGFR-sensitive mutations.
| Variable | Hazard ratioa | 95.0% CI | p-value |
|---|---|---|---|
| Univariate analysis | |||
| Age | −0.25 | −4.05-3.13 | 0.811 |
| Sex (male vs. female) | 1.60 | N/A | 0.236 |
| Complications (no vs. yes) | 0.67 | N/A | 0.478 |
| Smoker (no vs. yes) | 10.58 | N/A | 0.002* |
| miR-107 | 2.15 | 0.49–11.34 | 0.033* |
| miR-122 | 1.58 | −0.60–5.40 | 0.116 |
| miR-195 | −2.36 | −7.41–-0.66 | 0.019* |
| Multivariate analysis | |||
| Age | 2.03 | 0.93–1.01 | 0.154 |
| Sex (male vs. female) | 1.08 | 0.63–4.62 | 0.299 |
| Complications (no vs. yes) | 2.55 | 0.86–4.53 | 0.110 |
| Smoker (no vs. yes) | 9.44 | 1.85–16.25 | 0.002* |
| miR-107 | 4.08 | 0.78–1.00 | 0.043* |
| miR-122 | 0.69 | 0.79–1.10 | 0.405 |
| miR-195 | 2.81 | 0.99–1.07 | 0.094 |
| Further multivariate analysis | |||
| Smoker (no vs. yes) | 9.90 | 1.55–6.67 | 0.002* |
| miR-107 | 5.56 | 0.78–0.98 | 0.018* |
* p < 0.05
aChi-square values for categorical variables and t-test values for continuous variables in the univariate analysis and Wald values for variables in the multivariate analysis
Abbreviations: N/A, not applicable
Circulating plasma microRNAs as potential markers for monitoring EGFR-TKI treatment
As mentioned above, we collected plasma samples for a subset of patients harboring EGFR-sensitive mutations during EGFR-TKI treatment. The clinical characteristics and responses to EGFR-TKI treatment are summarized in Table 2. The median PFS was 8.0 (range, 1.0–31.0) months. To elucidate the fluctuations in circulating microRNAs with the course of EGFR-TKI treatment, subtraction of the ΔCt before EGFR-TKI treatment with that during or after the time of PD on EGFR-TKI treatment was analyzed. According to the Response Evaluation Criteria in Solid Tumors (version 1.1) [18], only one patient exhibited the poor response, PD. Consequently, the PD group was not analyzed in this comparison. As illustrated in Figure 8A, the ΔCt of miR-107 in patients with a complete or partial response (CR/PR) to EGFR-TKI treatment appeared to increase more sharply in comparison to that of patients with stable disease (SD) and this proved to be statistically significant (p < 0.05). Nevertheless, no statistically significant difference was detected between patients with a PFS of >8.0 months compared to patients with a PFS of ≤8.0 months for EGFR-TKI treatment (p > 0.05). Similarly, subtraction of the ΔCt for miR-195 revealed a significant difference between the CR/PR and SD groups (p < 0.05). This suggested that miR-195 expression in patients exhibiting a CR or PR to EGFR-TKI treatment could be significantly downregulated compared to patients exhibiting SD in response to EGFR-TKI treatment (Figure 8B). Meanwhile, for 12 patients, plasma was collected at the time of PD on EGFR-TKI treatment. Figure 9A and Figure 9B indicate that no matter the response of the tumor (CR, PR, SD, or PD), miR-107 and miR-195 expression levels at the time of PD exhibit a tendency to return to the levels before EGFR-TKI treatment in the majority of patients.
Figure 8.
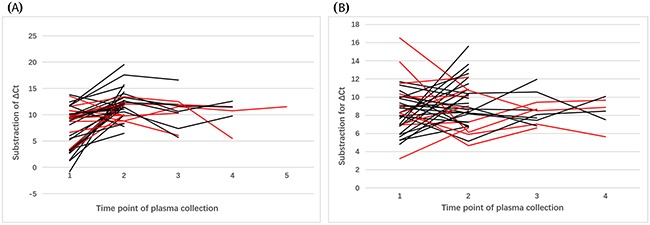
Dynamic changes in the expression of (A) miR-107 and (B) miR-195 in EGFR exon 19 deletion patients with NSCLC during EGFR-TKI treatment (n = 36). Patients with a complete or partial response are represented by the black lines and patients with stable disease are represented by the red lines.
Figure 9.
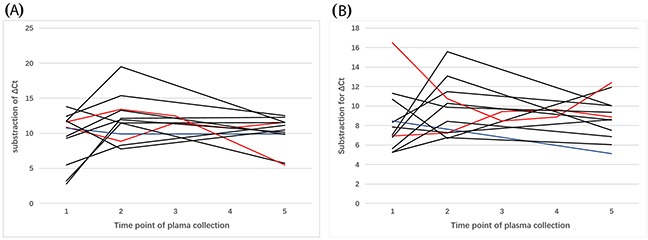
Dynamic changes in the expression of (A) miR-107 and (B) miR-195 in EGFR exon 19 deletion patients with NSCLC during and at the time of progressive disease on EGFR-TKI treatment (n = 12). Patients with a complete or partial response are represented by the black lines, patients with stable disease are represented by the red lines, and patients with progressive disease are represented by the blue lines.
DISCUSSION
“Precision medicine” warrants further attention in the individualized treatment of NSCLC, especially small molecular targeted therapies that require rapid and accurate molecular genotyping. EGFR-TKIs remain the most important targeted drugs for NSCLC patients with EGFR-sensitive mutations and have been recommended, for a number of years, as the standard first-line treatment for these patients by the National Comprehensive Cancer Network [4]. Therefore, to ascertain EGFR mutation status after pathological diagnosis is crucial. While, in a real clinical setting, it may not be possible to obtain a tumor specimen from certain patients for pathological examination or it may not be possible to obtain adequate tissue for further molecular analyses, it may also be difficult and unrealistic for patients with NSCLC to undergo re-biopsy after multi-line therapy. Therefore, as a non-invasive test, liquid biopsy has the potential to complement tissue biopsies and is fairly convenient to monitor responses to EGFR-TKI treatment and drug resistance [19, 20]. Circulating tumor DNA (ctDNA) has been approved by the Committee for Medicinal Products for Human Use of the European Medicines Agency [21] to assess EGFR mutation status in patients with NSCLC for whom it is not possible to obtain a tumor sample. Indeed, ctDNA exhibits high accuracy for EGFR mutation status analysis and is commercially available. Meanwhile, ctDNA analysis has been applied in clinical practice. The sensitivity of ctDNA has been reported to range from 70.0–75.0%, which may be promoted by highly sensitive technologies that are relatively costly for the patients (e.g., digital droplet PCR and next generation sequencing) [21]. Therefore, it is imperative to explore non-invasive, convenient, and economical tumor markers as supplements to predict EGFR mutation status and to monitor EGFR-TKI treatment in NSCLC patients. Given their stability in plasma and serum, microRNAs are considered a novel class of non-invasive biomarkers for various types of malignancies [22]. In the present study, we comprehensively evaluated the potential of 3 circulating plasma microRNAs (miR-107, miR-122, and miR-195) as potential novel markers of EGFR mutation status in NSCLC patients.
No related information was available as to the two major specific EGFR mutation genotypes (EGFR exon 19 deletions or EGFR p.L858R mutations) that are independently associated with circulating microRNAs. As observed in the clinic and in a study comparing icotinib with gefitinib in previously treated patients with advanced NSCLC [23], the EGFR exon 19 deletion was associated with a better response to treatment compared to the EGFR p.L858R mutation. To identify potential markers that could distinguish between EGFR mutation-positive and EGFR wild-type patients and between patients with different EGFR-sensitive mutations (EGFR exon 19 deletions and EGFR p.L858R mutations) would be worthwhile in basic and clinical research. Indeed, in the present study, 3 circulating plasma microRNAs (miR-107, miR-122, and miR-195) were significantly dysregulated in EGFR mutation-positive patients compared to EGFR wild-type patients.
Initially, microRNA expression profiles were analyzed in the EGFR mutation-positive, EGFR wild-type, and healthy control cohorts. Seventy-six dysregulated microRNAs were detected in NSCLC patients compared to healthy controls. Further analysis of the 76 dysregulated microRNAs was performed according to the different EGFR mutation genotypes. Twenty-one and 5 dysregulated microRNAs were detected in EGFR exon 19 deletion and EGFR p.L858R mutation patients, respectively, compared to EGFR wild-type patients. All 5 dysregulated microRNAs in EGFR p.L858R mutation patients were newly discordant microRNAs with large serial numbers (Table 4). Consequently, this cohort of patients was not investigated further. Our study primarily focused on EGFR exon 19 deletion patients. This group of patients benefited most from EGFR-TKI treatment. According to the microarray data and related published literature [15–17], 10 microRNAs were screened out for further investigation. Finally, 3 microRNAs (miR-107, miR-122, and miR-195) were identified as being significantly dysregulated.
MiR-107 was significantly upregulated (fold-change: 6.64, p < 0.05) in EGFR mutation-positive patients compared to EGFR wild-type patients. With respect to specific EGFR mutation genotypes, miR-107 was upregulated in EGFR exon 19 deletion and EGFR p.L858R mutation patients compared to EGFR wild-type patients (fold-change: 7.44 and 5.22, respectively). However, no significant difference was observed between EGFR exon 19 deletion and EGFR p.L858R mutation patients (p > 0.05). Meanwhile, ROC curve analysis revealed that miR-107 had promising diagnostic potential for distinguishing between EGFR mutation-positive and EGFR wild-type patients. There are a limited number of studies indicating that miR-107 is dysregulated in NSCLC tissues and cell lines. However, our study is the first to identify miR-107 as a potential marker for EGFR mutation-positive patients. As demonstrated in previous studies [24, 25], miR-107 expression is reduced in NSCLC tissues compared to paired normal tissues. MiR-107 functions as a tumor suppressor in NSCLC by targeting brain-derived neurotrophic factor and indirectly regulating the PI3K/AKT signaling pathway. EGFR activation elicits its effects via several downstream signaling pathways, including the PI3K/AKT/mTOR pathway [26]. Thus, we speculate that miR-107 is closely associated with the EGFR pathway and could be a new specific marker for EGFR-sensitive mutations. In addition, this could be indirectly validated by its dynamic changes during EGFR-TKI treatment.
MiR-122 and miR-195 have been reported [27, 28] to be dysregulated in EGFR mutation-positive patients compared to EGFR wild-type patients. However, no study has investigated the differential expression of these 2 circulating microRNAs between specific EGFR mutation genotypes. In the present study, miR-122 was significantly upregulated in EGFR p.L858R mutation patients compared to EGFR wild-type patients. However, no significant difference was detected between EGFR exon 19 deletion and EGFR wild-type patients. ROC curve analysis demonstrated that miR-122 may serve as a potential specific marker for EGFR p.L858R mutation patients (AUC: 0.75). In addition, miR-195, which was significantly downregulated in EGFR exon 19 deletion patients compared to EGFR wild-type patients, may serve as a potential marker of NSCLC in EGFR exon 19 deletion patients (AUC: 0.75). These findings are in agreement with a number of studies focusing on the association of circulating microRNAs with EGFR-sensitive mutations. Qiang et al. [27] revealed that miR-122 and miR-195 could distinguish between EGFR mutation-positive and EGFR wild-type patients, and miR-195 was associated with poor differentiation. Another study [28] suggested that circulating miR-122 and miR-195 may have prognostic significance in predicting EGFR mutation status and overall survival in female, non-smoking lung adenocarcinoma patients. However, these two studies [27, 28] did not analyze the association between circulating microRNAs and specific EGFR mutation genotypes.
Furthermore, we analyzed the diagnostic potential of a combination of microRNAs as a biomarker panel. A combination of miR-107 and miR-195 for discriminating EGFR exon 19 deletion patients from EGFR wild-type patients produced similar AUC values and higher sensitivity and specificity values than miR-107 or miR-195 alone in the ROC curve analysis. Meanwhile, a panel of miR-107 and miR-122 also produced higher AUC and specificity values for discriminating EGFR p.L858R mutation patients from EGFR wild-type patients than miR-107 or miR-122 alone; although sensitivity values were lower than miR-122 alone. Taken together, these findings suggest that it may be better to have a panel of 2 microRNAs as markers to distinguish EGFR mutation-positive patients from EGFR wild-type patients. In addition, multivariate analysis revealed that miR-107 and smoking status are important diagnostic predictors for EGFR-sensitive mutations and NSCLC patients with high miR-107 expression levels and no prior history of smoking are more likely to have EGFR exon 19 deletion or EGFR p.L858R mutations. It has been proven that the distinct profiles of oncogenic mutations, especially EGFR, are different between smoking and non-smoking NSCLC patients and this is consistent with the findings of the present study. We are the first to report on miR-107 as a potential marker for EGFR-sensitive mutations (EGFR exon 19 deletions and EGFR p.L858R mutations). To our knowledge, no study has focused on the dynamic changes of circulating plasma microRNAs during the course of EGFR-TKI treatment. Our present findings revealed that the extent of ΔCt variation for miR-107 and miR-195 was significantly correlated with the response to EGFR-TKI treatment, while no significant differences were detected between patients with different PFS times on EGFR-TKI treatment. From the limited number of patients with plasma collected at the time of PD during EGFR-TKI treatment, we found that irrespective of the response of the tumor (CR, PR, SD, or PD), miR-107 and miR-195 expression levels at the time of PD exhibited a tendency to return to pre-treatment levels in the majority of patients. This suggests that circulating plasma microRNAs may have the potential to serve as markers for monitoring EGFR-TKI treatment. In this study, we have only preliminarily observed the objective phenomenon of dynamic changes of microRNAs during EGFR-TKI treatment in a small number of patients from a single institution. Therefore, it is warranted to have well designed large scale studies of patients and to design experiments on tumor cells to determine the precise mechanism(s) of the relationship between microRNA change and EGFR expression in plasma after EGFR-TKI treatment.
This study is limited by the fact that it was performed at a single institution with specific NSCLC patients (e.g., with relatively advanced clinical stages and without a sufficiently large scale of enrolled samples). Further studies are warranted with larger sample sizes and different characteristics to validate the diagnostic potential of circulating plasma microRNAs (miR-107, miR-122, and miR-195) for EGFR-sensitive mutations. In addition, cytological experiments on EGFR signaling pathways are required to elucidate the specific functions of these circulating plasma microRNAs. Beyond that, similar studies that have been reported to date that have presented conflicting findings. This may be correlated with ethnical diversity, relatively small sample sizes, and different study technologies. Therefore, it is essential to conduct multicentral studies with standardized methodological protocols to achieve more reliable results.
In conclusion, the present study revealed that upregulation of circulating miR-107 may serve as a potential marker for EGFR-sensitive mutations, including EGFR exon 19 deletions and EGFR p.L858R mutations. In addition, miR-122 may serve as an EGFR p.L858R mutation specific marker and miR-195 as an EGFR exon 19 deletion specific marker for distinguishing between these patients and EGFR wild-type patients. ROC curve analysis suggested that a combined panel of miR-107 and miR-195 or miR-107 and miR-122 had the potential to be a diagnostic marker for EGFR exon 19 deletion or EGFR p.L858R mutation patients, respectively. Dynamic changes in these 3 microRNAs were also detected during EGFR-TKI treatment, revealing that circulating plasma microRNAs may serve as potential markers for monitoring EGFR-TKI treatment. Our findings highlight the prospective application of circulating plasma microRNAs as non-invasive, convenient, and economical markers for EGFR-sensitive mutations.
MATERIALS AND METHODS
Patient enrollment
This study was approved by the Ethics Committee of the Affiliated Hospital of the Academy of Military Medical Science, Beijing, China (No. 2012-11-171). NSCLC patients (n = 153) were recruited from the Department of Lung Cancer and age and sex-matched healthy controls (n = 41) from the Physical Examination Center between December 2014 and April 2016. All study participants have provided informed written consent. Research was conducted in accordance with the 1964 Declaration of Helsinki and its later amendments. The enrolled NSCLC patients were all newly diagnosed with no previous tumor-related therapy and the pathological diagnosis was confirmed by two independent pathologists. Clinical staging was also assessed by two independent professional oncologists according to the American Joint Committee on Cancer/Union for International Cancer Control tumor-node-metastasis staging system (seventh edition) [29]. The performance status of the NSCLC patients was 0–2 according to the criteria of the Eastern Cooperative Oncology Group [30]. All NSCLC patients harboring EGFR-sensitive mutations (EGFR exon 19 deletions or p.L858R mutations) in tumor tissues received first or second-line EGFR-TKI treatment (gefitinib [250.0 mg], erlotinib [150.0 mg], or icotinib [125.0 mg] daily). Tumor responses were assessed by two independent professional oncologists in accordance with the latest Response Evaluation Criteria in Solid Tumors (version 1.1) [18].
Plasma collection
For all participants enrolled in the study, 7.0 mL fasted peripheral venous blood samples were collected between 07:00 and 08:00 AM. The plasma was separated within 30 minutes of collection by centrifugation at 4,500 g for 10 minutes at 4.0°C and stored immediately at -80.0°C for further analysis.
Total RNA extraction from plasma samples
Total RNA containing microRNAs was isolated from 200.0 μL of plasma using QIAzol Lysis Reagent (catalogue no. 217184; QIAGEN Inc., Valencia, CA, USA) and purified using the miRNeasy Serum/Plasma Kit (catalogue no. 217184; QIAGEN Inc., Valencia, CA, USA) in accordance with the manufacturer's guidelines.
Plasma microRNA profiling
Human microRNA Microarrays (Release 21.0; Agilent Technologies Ltd., Santa Clara, CA, USA) containing 2,568 mature microRNA sequences were performed for preliminary screening of dysregulated circulating plasma microRNAs in NSCLC patients with different EGFR mutation genotypes. Raw data were extracted using Agilent's Feature Extraction software (version 10.7; Agilent Technologies Ltd., Santa Clara, CA, USA) and normalized using GeneSpring software (Agilent Technologies Ltd., Santa Clara, USA) with the normalization of labeling spike-in RNA and hybridization spike-in RNA according to the manufacturer's protocol.
qRT-PCR of selected microRNAs
The expression levels of selected microRNAs were analyzed by qRT-PCR using the miScript SYBR Green PCR Kit (catalogue no. 218073; QIAGEN Inc., Valencia, CA, USA). Poly(A)-tailed RNA was used in the qRT-PCR reactions and 5SrRNA was selected as the internal control for data normalization. As previously described [31, 32], a 15.0 μL reaction volume containing total RNA, 0.2 μL of ribonucleotide adenosine triphosphate, 1.5 μL of 10X poly(A) polymerase chain reaction buffer, and 0.2 μL of poly(A) polymerase (catalogue no. M0276L; New England BioLabs Inc., Ipswich, MA, USA) was incubated at 42.0°C for 60 minutes, 70.0°C for 5 minutes, and immediately placed on ice to terminate the reaction. The poly(A)-tailed total RNA was reverse transcribed using 1.0 μg of miR reverse transcription primer (5′-GCG AGC ACA GAA TTA ATA CGA CTC ACT ATA GG(t)18VN-3′), 1.0 μL of ImProm reverse transcriptase (Promega Biotech Co., Ltd., Beijing, China), and reverse transcriptase buffer, according to the manufacturer's protocol. Lastly, qRT-PCR was performed using the miScript SYBR Green PCR Kit (QIAGEN Inc., Valencia, CA, USA). The primer sequences were as follows: 5SrRNA, forward primer: 5′-TCT GAT CTC GGA AGC TAA GCA-3′, reverse primer: 5′-CCT ACA GCA CCC GGT ATT CC-3′; miR-107, forward primer: 5′-AGC AGC ATT GTA CAG GGC TAT CA-3′; miR-122, forward primer: 5′-TGG AGT GTG ACA ATG GTG TTT G-3′; miR-125a-5p, forward primer: 5′-TCC CTG AGA CCC TTT AAC CTG TGA-3′; and miR-195, forward primer: 5′-TAG CAG CAC AGA AAT ATT GGC-3′. All 4 microRNAs shared a common reverse primer (5′-GCG AGC ACA GAA TTA ATA CGA C-3′). QRT-PCR was performed in triplicate in 15.0 μL reaction volumes of PCR master mix using the PCR program on the Stratagene Mx3005P Real-Time PCR Detection System (Agilent Technologies Ltd., Santa Clara, CA, USA). Denaturation was performed at 95.0°C for 10 minutes, followed by 45 cycles of 95.0°C for 15 seconds and 60.0°C for 60 seconds. The raw Ct data were normalized to the expression levels of 5SrRNA using the 2−Δ(ΔCt) method.
Statistical analyses
The expression of each target microRNA was measured in triplicate and the results were expressed as the mean ΔCt (normalized against 5SrRNA that exhibited relatively stable expression levels across all plasma samples). Subtraction of the Ct value of the target microRNA from 5SrRNA (ΔCt) was calculated. A high ΔCt corresponded to a low microRNA expression level. The ΔCt was then converted to 2−ΔCt to assess alterations in microRNA expression levels between different groups and the fold-change was calculated using the 2−Δ(ΔCt) method. The relative expression of each microRNA was presented as the mean and standard error of the mean. Differences in microRNA expression levels between groups were compared using unpaired Student's t-tests or a one-way analysis of variance. Chi-square tests were used for qualitative data. ROC curve analysis was performed to provide an estimate of the target microRNAs’ abilities to discriminate between NSCLC patients with different EGFR mutation genotypes. All statistical analyses were conducted using Statistical Package for the Social Sciences for Windows, software version 23.0 (IBM Corp., Armonk, NY, USA). A p < 0.05 was considered statistically significant.
Abbreviations
- 5SrRNA
5S ribosomal RNA
- AUC
area under the ROC curve
- CI
confidence interval
- CR
complete response
- Ct
quantification cycle
- ctDNA
circulating tumor DNA
- EGFR-TKI
epidermal growth factor receptor-tyrosine kinase inhibitor
- NSCLC
non-small cell lung cancer
- PD
progressive disease
- PFS
progression-free survival
- PR
partial response
- qRT-PCR
quantitative real-time reverse-transcription polymerase chain reaction
- ROC
receiver operating characteristic
- SD
stable disease.
Footnotes
Author contributions
Study concept, design, and supervision: X. Liu; experimental work: L.Q. and L.L.; data collection: C.T., H.Q., X. Li., H.W., J.L., W.W., S.Y., L.W., G.Z., P.L., Y.L., M.Z., and H.G.; technical assistance: X.Z. and H.F.; and supervision of the drafting of the manuscript: S.S.
CONFLICTS OF INTEREST
The authors declare no actual or potential conflicts of interest.
FUNDING
This study was supported by a grant from the Capital Medical Development Research Fund (No. 2007-3042) and the National Key Foundation for Exploring Scientific Instrument of China (No. 2011YQ170067).
REFERENCES
- 1.Chen W, Zheng R, Zeng H, Zhang S. The incidence and mortality of major cancers in China, 2012. Chin J Cancer. 2016;35:73. doi: 10.1186/s40880-016-0137-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rafiemanesh H, Mehtarpour M, Khani F, Hesami SM, Shamlou R, Towhidi F, Salehiniya H, Makhsosi BR, Moini A. Epidemiology, incidence and mortality of lung cancer and their relationship with the development index in the world. J Thorac Dis. 2016;8:1094–1102. doi: 10.21037/jtd.2016.03.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Howlander N, Noone AM, Krapcho M, Garshell J, Miller D, Altekruse SF. SEER Cancer Statistics Review, 1975–2012. 2015 Available from: http://seer.cancer.gov/csr/1975_2012/
- 4.NCCN Clinical Practice Guidelines in Oncology Version 1. 2017 Available from: http://nccn.org/professionals/physician_gls/pdf/nscl.pdf.
- 5.Farago AF, Snyder EL, Jacks T. SnapShot: Lung cancer models. Cell. 2012;149:246–246.e1. doi: 10.1016/j.cell.2012.03.015. [DOI] [PubMed] [Google Scholar]
- 6.Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7:169–181. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 7.Wang J, Nong J, Jia H, Qin N, Li X, Zhang H, Zhang Q, Zhang Z, Zhang S. Efficacy and predictors of EGFR tyrosine kinase inhibitors in Chinese advanced lung adenocarcinoma: analyses of 253 cases from a single institute. Oncol Res. 2014;21:237–246. doi: 10.3727/096504014X13907540404833. [DOI] [PubMed] [Google Scholar]
- 8.Liang W, Sheng J, Wu X, Zhang Y, Kang S, Zhang L. Exon 19 deletion association with progression-free survival compared to L858R mutation at exon 21 in treatment with first-line EGFR-TKIs: A meta-analysis of subgroup data from eight phase III randomized controlled trials. J Clin Oncol. 2014;32(15_suppl):8107–8107. [Google Scholar]
- 9.Lin CC, Huang WL, Wei F, Su WC, Wong DT. Emerging platforms using liquid biopsy to detect EGFR mutations in lung cancer. Expert Rev Mol Diagn. 2015;15:1427–1440. doi: 10.1586/14737159.2015.1094379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Douillard JY, Ostoros G, Cobo M, Ciuleanu T, Cole R, McWalter G, Walker J, Dearden S, Webster A, Milenkova T, McCormack R. Gefitinib treatment in EGFR mutated caucasian NSCLC: circulating-free tumor DNA as a surrogate for determination of EGFR status. J Thorac Oncol. 2014;9:1345–1353. doi: 10.1097/JTO.0000000000000263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Price C, Chen J. MicroRNAs in Cancer Biology and Therapy: Current Status and Perspectives. Genes Dis. 2014;1:53–63. doi: 10.1016/j.gendis.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 13.Berger F, Reiser MF. Micro-RNAs as potential new molecular biomarkers in oncology: have they reached relevance for the clinical imaging sciences? Theranostics. 2013;3:943–952. doi: 10.7150/thno.7445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 15.Markou A, Sourvinou I, Vorkas PA, Yousef GM, Lianidou E. Clinical evaluation of microRNA expression profiling in non small cell lung cancer. Lung cancer. 2013;81:388–396. doi: 10.1016/j.lungcan.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 16.Aushev VN, Zborovskaya IB, Laktionov KK, Girard N, Cros MP, Herceg Z, Krutovskikh V. Comparisons of microRNA patterns in plasma before and after tumor removal reveal new biomarkers of lung squamous cell carcinoma. PLoS one. 2013;8:e78649. doi: 10.1371/journal.pone.0078649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franchina T, Amodeo V, Bronte G, Savio G, Ricciardi GR, Picciotto M, Russo A, Giordano A, Adamo V. Circulating miR-22, miR-24 and miR-34a as novel predictive biomarkers to pemetrexed-based chemotherapy in advanced non-small cell lung cancer. J Cell Physiol. 2014;229:97–99. doi: 10.1002/jcp.24422. [DOI] [PubMed] [Google Scholar]
- 18.Nishino M, Jackman DM, Hatabu H, Yeap BY, Cioffredi LA, Yap JT, Jänne PA, Johnson BE, Ven den Abbeele AD. New Response Evaluation Criteria in Solid Tumors (RECIST) guidelines for advanced non-small cell lung cancer: comparison with original RECIST and impact on assessment of tumor response to targeted therapy. AJR Am J Roentgenol. 2010;195:W221–W228. doi: 10.2214/AJR.09.3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Masroor M, Mir R, Javid J, Prasant Y, Imtiyaz A, Mariyam Z, Mohan A, Ray PC, Saxena A. Cell Free EGFR mRNA Expression and Implications for Survival and Metastasis in Non-Small Cell Lung Cancer Cases. Asian Pac J Cancer Prev. 2015;16:6445–6449. doi: 10.7314/apjcp.2015.16.15.6445. [DOI] [PubMed] [Google Scholar]
- 20.Hofman P. Liquid biopsy for early detection of lung cancer. Curr Opin Oncol. 2017;29:73–78. doi: 10.1097/CCO.0000000000000343. [DOI] [PubMed] [Google Scholar]
- 21.Yu XM, Wu YC, Liu X, Huang XC, Hou XX, Wang JL, Cheng XL, Mao WM, Ling ZQ. Cell-Free RNA Content in Peripheral Blood as Potential Biomarkers for Detecting Circulating Tumor Cells in Non-Small Cell Lung Carcinoma. Int J Mol Sci. 2016:17. doi: 10.3390/ijms17111845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bianchi F, Nicassio F, Veronesi G, di Fiore PP. Circulating microRNAs: next-generation biomarkers for early lung cancer detection. Ecancer medical science. 2012;6:246. doi: 10.3332/ecancer.2012.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi Y, Zhang L, Liu X, Zhou C, Zhang L, Zhang S, Wang D, Li Q, Qin S, Hu C, Zhang Y, Chen J, Cheng Y, et al. Icotinib versus gefitinib in previously treated advanced non-small-cell lung cancer (ICOGEN): a randomized, double-blind phase 3 non-inferiority trial. Lancet Oncol. 2013;14:953–961. doi: 10.1016/S1470-2045(13)70355-3. [DOI] [PubMed] [Google Scholar]
- 24.Xia H, Li Y, Lv X. MicroRNA-107 inhibits tumor growth and metastasis by targeting the BDNF-mediated PI3K/AKT pathway in human non-small lung cancer. Int J Oncol. 2016;49:1325–1333. doi: 10.3892/ijo.2016.3628. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Zhong KZ, Chen WW, Hu XY, Jiang AL, Zhao J. Clinicopathological and prognostic significance of microRNA-107 in human non small cell lung cancer. Int J Clin Exp Pathol. 2014;7:4545–4551. [PMC free article] [PubMed] [Google Scholar]
- 26.Wu DW, Wu TC, Chen CY, Lee H. PAK1 Is a Novel Therapeutic Target in Tyrosine Kinase Inhibitor-Resistant Lung Adenocarcinoma Activated by the PI3K/AKT Signaling Regardless of EGFR Mutation. Clin Cancer Res. 2016;22:5370–5382. doi: 10.1158/1078-0432.CCR-15-2724. [DOI] [PubMed] [Google Scholar]
- 27.Zhao Q, Cao J, Wu YC, Liu X, Han J, Huang XC, Jiang LH, Hou XX, Mao WM, Ling ZQ. Circulating miRNAs is a potential marker for gefitinib sensitivity and correlation with EGFR mutational status in human lung cancers. Am J Cancer Res. 2015;5:1692–1705. [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang H, Su Y, Xu F, Kong J, Yu H, Qian B. Circulating microRNAs in relation to EGFR status and survival of lung adenocarcinoma in female non-smokers. PLoS One. 2013;8:e81408. doi: 10.1371/journal.pone.0081408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boffa DJ, Greene FL. Reacting to changes in staging designations in the 7th edition of the AJCC staging manual. Ann Surg Oncol. 2011;18:1–3. doi: 10.1245/s10434-010-1427-z. [DOI] [PubMed] [Google Scholar]
- 30.Buccheri G, Ferrigno D. Karnofsky and ECOG performance status in lung cancer: Equivalence, construct validity, and predictive validity. Lung Cancer. 1994;11:87–96. [Google Scholar]
- 31.Lv G, Hu Z, Tie Y, Du J, Fu H, Gao X, Zheng X. MicroRNA-451 regulates activating transcription factor 2 expression and inhibits liver cancer cell migration. Oncol Rep. 2014;32:1021–1028. doi: 10.3892/or.2014.3296. [DOI] [PubMed] [Google Scholar]
- 32.Fu H, Tie Y, Xu C, Zhang Z, Zhu J, Shi Y, Jiang H, Sun Z, Zheng X. Identification of human fetal liver miRNAs by a novel method. FEBS Lett. 2005;579:3849–3854. doi: 10.1016/j.febslet.2005.05.064. [DOI] [PubMed] [Google Scholar]


