Abstract
Holins comprise the most diverse functional group of proteins known. They are small bacteriophage-encoded proteins that accumulate during the period of late-protein synthesis after infection and cause lysis of the host cell at a precise genetically programmed time. It is unknown how holins achieve temporal precision, but a conserved feature of their function is that energy poisons subvert the normal scheduling mechanism and instantly trigger membrane disruption. On this basis, timing has been proposed to involve a progressive decrease in the energized state of the membrane until a critical triggering level is reached. Here, we report that membrane integrity is not compromised after the induction of holin synthesis until seconds before lysis. The proton motive force was monitored by the rotation of individual cells tethered by a single flagellum. The results suggest an alternative explanation for the lysis “clock,” in which holin concentrations build to a critical level that leads to formation of an oligomeric complex that disrupts the membrane.
For most bacteriophages, lysis of the host requires at least two proteins: an endolysin and a holin (1, 2). Endolysins are generally soluble proteins with one or more muralytic activities against the three different types of covalent bonds (glycosidic, amide, and peptide) of the peptidoglycan polymer of the cell wall. Holins are encoded by genes in at least 35 different families with no detectable orthologous relationships.
After accumulating and oligomerizing in the membrane throughout the period of late-gene expression, holins suddenly trigger to form a lesion that permeabilizes the membrane. This event ends macromolecular synthesis and thus effectively terminates the infection. Moreover, the disruption of the membrane directly leads to lysis by allowing endolysin to attack the peptidoglycan. Holin function is thus the sole direct determinant of the length of the infective cycle and the yield of progeny phage. Holins are subject to opposing evolutionary forces. On the one hand, there is pressure to extend the vegetative cycle to allow continued accumulation of virions at a linear rate. On the other hand, there is pressure to trigger lysis earlier to release progeny phage particles that can infect new hosts and potentially yield exponential increases in phage numbers (1–3).
Two holin genes have been analyzed extensively at the genetic level: λS and T4t (4–6). In both cases, many missense alleles causing altered lysis timing have been identified, suggesting that holins should be amenable to evolutionary optimization of the scheduling of lysis. Despite the unprecedented diversity, conserved function, and genetic tractability of holin genes, however, neither the physical nature of the membrane lesion nor the manner in which this simplest of biological timing mechanisms operates is known.
The phenomenon of premature lysis was discovered by Doermann (7) long before the existence of holins was known, and it has provided the only clue to the mechanism of triggering. Cells infected with phage T2 were observed to lyse prematurely after the addition of cyanide. Such premature lysis has been seen with every other phage known to use holin–endolysin-mediated lysis and can be triggered with a variety of other energy poisons that collapse the protein-motive force (pmf) (8–10). This uniform response has suggested a timing mechanism in which holin accumulation causes a proton (or other ion) leak and gradually titrates the pmf until a triggering threshold is reached (10–12).
Here, we present a study in which the rotational velocity of individual cells tethered by a single flagellum was monitored continuously after induction of holin synthesis. In these conditions, the rotational velocity, which is directly proportional to the pmf to at least −160 mV(13), is a measure of how well the integrity of the membrane is maintained as the holin accumulates in the bilayer and how much the pmf decreases before lysis ensues. The results are discussed in terms of a model for lysis timing in which holin accumulates to a critical concentration that initiates formation of a highly oligomeric membrane-disruption complex.
Materials and Methods
Media.
LB and tryptone broth (TB) media are from J. H. Miller (14). Kanamycin, chloramphenicol, and ampicillin were used at 40, 40, and 100 μg/ml, respectively, in solid medium to select for plasmid-borne antibiotic resistance markers. The same concentrations were used for liquid medium except for chloramphenicol, which was used at 30 μg/ml.
Strains and Plasmid Construction.
The motile strain RY8797 is an ara+ lac+ lacIq eda+ΔcheA derivative of the strain RP437 (15), which is wild type for motility. Construction of plasmid pS105, which has the λ late promoter, pR′, serving the lysis gene cassette, S105 RRzRz1 (Fig. 1), has been described previously (16). S105, a modified version of the S gene, encodes the active holin, S105. The start codon for the antiholin S107 (17) is eliminated in this construct. Several plasmids carrying different S105 alleles were used in this study (Table 1) (18, 19). The plasmid pS105A52G, containing the early lysis allele S105A52G , was used for most tethering assays and lysis inductions (20). To construct plasmid pS105A52GΔR, which is deleted for the RRzRz1 gene cluster, plasmid pS105A52G was cut with AatII and religated. To construct plasmid pQ, the Q gene was PCR-amplified from λ DNA. The PCR fragment was digested with KpnI and ClaI and cloned under control of the Plac/ara-1 promoter in the KanR plasmid pZS-24* (21). The primer pair used for PCR amplification specifies KpnI and ClaI restriction sites immediately upstream of the start codon and downstream of the stop codon of Q. Transcription of the Q gene from the lac/ara hybrid promoter is tightly repressed and can be induced by the addition of isopropyl β-d-thiogalactopyranoside (IPTG) and arabinose. The plasmids pZA32-R and pZA32-R/Rz/Rz1 were constructed for expression of the λ R gene and RRzRz1 gene cluster, respectively. λ R and RRzRz1 were amplified by PCR by using the plasmid pS105 as template and primers that generate unique KpnI and XbaI restriction sites upstream and downstream of the amplified genes. The PCR products were digested with the enzymes KpnI and XbaI and subcloned under PLlac-O1 promoter control in the ampicillin-resistance vector pZE12-luc (21). The resultant plasmid constructs, pZE12-R and pZE12-R/Rz/Rz1, were digested with XhoI and AvrII. The fragments containing the PLlac-O1 promoter and the lysis gene R or the lysis gene cluster RRzRz1 were inserted into the vector pZA32-luc (21). The resulting plasmids, pZA32-R and pZA32-R/Rz/Rz1, are CamR and contain λ R or RRzRz1 under control of the PLlac-O1 promoter, which is inducible by IPTG. Plasmids pQ, pS105 (and its isogenic derivatives), and pZA32-R or pZA32-R/Rz/Rz1 confer resistance to three different antibiotics and have different origins of replication, and they are therefore all compatible in the same cell.
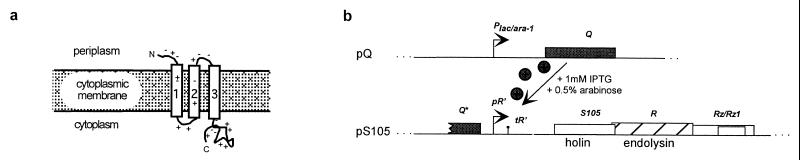
Figure 1.
(a) Topology of the λ S holin. It is not known how the holin oligomerizes to form the permeabilizing lethal membrane lesion. (b) Scheme for inducible expression of the cloned lysis genes. The λ antiterminator gene Q is cloned in the plasmid pQ under control of a lac/ara hybrid promoter, which is inducible with IPTG and arabinose. The λ lysis genes lie in an overlapping cluster in the λ lysis cassette located downstream of the late gene promoter, pR′, on the compatible plasmid pS105. Expression of the lysis genes follows transactivation of pR′ by Q.
Table 1.
Lysis times and interval between stop of rotation and actual cell lysis
| S allele† | Time from induction until lysis in broth culture, min‡ | Experiment 1
|
Experiment 2
|
||
|---|---|---|---|---|---|
| Time from induction until lysis for tethered cells, min§ | Time from rotation stop until lysis, sec¶ | Time from induction until lysis for tethered cells, min§ | Time from rotation stop until lysis, sec¶ | ||
| Sam7 | No lysis | No lysis | No lysis | No lysis | No lysis |
| S | 45 | 104 ± 8 (15) | 19 ± 6 (15) | ||
| S105 | 40 | 64 ± 7 (15) | 13 ± 4 (15) | ||
| SA52G | 10 | 19 ± 2 (5) | 26 ± 7 (5) | ||
| S105A52G | 15 | 25 ± 3 (20) | 32 ± 9 (20) | ||
| SC51S | 10 | 27 ± 4 (15) | 40 ± 6 (15) | ||
| S105C51S | 15 | 27 ± 1 (23) | 26 ± 7 (23) | 31 ± 4 (15) | 39 ± 8 (15) |
The motile strain RY8797pQ carrying pS105 or isogenic derivatives of this plasmid was used in all experiments.
Cells were cultured in LB medium at 37°C. At an A550 of 0.2, expression of the lysis genes was induced by the addition of IPTG and arabinose. The turbidity of the culture was followed until the A550 dropped below 0.05. The time when the turbidity of the culture started to decline was defined as the lysis time.
‡ Cells were grown and induced as described in Materials and Methods and tethered to glass coverslips. The time until lysis for individual cells was determined by microscopic observation. The number of cells analyzed is shown in parentheses.
Time between the halt of rotation and lysis of tethered cells.
Tethering Assay, Microscopic Analysis, and Video Recordings.
The cheA mutant strain RY8797, in which the flagellum rotates only counterclockwise, was used for all experiments. Overnight cultures of strain RY8797pQ carrying pS105 or an isogenic derivative were grown in TB medium at 30°C. For most of the tethering assays, strain RY8797pQpS105A52G was used to reduce the time between induction and lysis. Freshly grown overnight cultures were diluted 33-fold into TB medium and grown at room temperature (≈22°C) until the culture reached a density of A550 = 0.2. Cells were sheared and tethered with a flagellar filament-specific antibody to a glass coverslip, as described previously (22). The behavior of the tethered cells was observed in a flow chamber (23) by reverse phase-contrast microscopy. Before recording the baseline behavior of tethered cells on videotape, cells were equilibrated for several minutes by the flow of prewarmed (38°C) TB medium at a flow rate of ≈0.3 ml/min. At t = 0, expression of the lysis genes was induced by the flow of TB medium containing 1 mM IPTG and 0.5% arabinose. Every 5 min, the flow was stopped for 30 sec to record the rotation of individual cells. The flow was stopped permanently when lysis of the first cells was observed (20 min after expression of the S105A52G allele or 45 min after induction of the S105 allele).
The rotation speed of each cell at each time point was normalized to its speed during the previous interval of stopped flow (from 5 min earlier). This approach was adopted because many cells failed to spin for the entire experiment, because they became stuck or first began spinning during the experiment, or they became detached from the coverslip. This normalization underestimates the overall decrease in speed over the course of the experiment, but it does not affect the comparison between the control and experimental cells. The primary cause of the progressive slowing of rotation is the elongation of all tethered cells, induced and untreated, as an unavoidable consequence of doing the experiment under conditions that permit protein synthesis. At least seven cells were examined at each time point, and the mean normalized velocities and standard deviations were calculated.
To determine the effect of the protonophore dinitrophenol (DNP), the rotation speed of tethered cells was measured before and after the flow of prewarmed TB medium containing DNP at concentrations between 0 and 1,000 μM. DNP stock solutions were prepared in ethanol and diluted 50-fold in TB medium to the desired concentration. The ratio of the rotation speeds before and after DNP addition was calculated as described above for 20 cells, averaged and plotted with standard deviations. The Dazzle* Digital Video Creator and Photo Maker and the Ulead Video Studio, Version 4.0 (Dazzle Multimedia, Fremont, CA) were used to digitize the recordings and to extract single frames.
Lysis Curves and DNP Triggering.
Cultures of the strain RY8797pQpS105 were grown overnight in TB medium at 37°C. The cultures were diluted 250-fold into fresh TB medium and grown at 37°C to A550 = 0.2. Expression of the lysis genes was induced by the addition of 1 mM IPTG and 0.5% arabinose. Twenty minutes after induction, DNP was added to final concentrations between 0 and 1,500 μM. After DNP addition, the turbidity of the cultures was monitored as A550 until cell lysis was complete.
Results
The pmf Remains Constant Until Seconds Before Holin Triggering.
S105, the active holin of phage λ, contains three helical transmembrane domains in its 105 residues [Fig. 1a (19)] and accumulates in the membrane from the onset of late-gene expression, at about 8 min, until lysis, at about 45 min (Table 1) (24, 25). To test the leakage model experimentally, we monitored the pmf of individual Escherichia coli cells as a function of time after the induction of S synthesis by recording the rotational speed of cells tethered by a flagellum attached to a surface (26). The speed of rotation of tethered cells is directly proportional to the proton motive force up to at least −160 mV and probably through the entire physiological range (13). [A fully energized E. coli cell maintains its pmf at −180 to −200 mV (27)].
The lysis-gene cassette of phage λ cloned behind its cognate pR′ promoter on a medium-copy plasmid was regulated by a compatible plasmid in which the expression of Q, encoding the λ late-gene activator, is under the control of an inducible promoter (Fig. 1b). Motile cells that contained these plasmids were tethered to a glass coverslip by a single flagellum, by using antiserum against the flagellar filament. Because reversals in the direction of flagellar rotation complicate measurements of rotational velocity, a cheA mutant, in which the flagella rotate only counterclockwise, was used in these experiments.
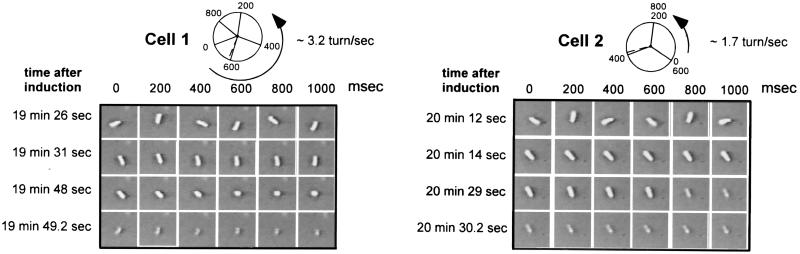
Tethered cells were equilibrated in the flow chamber, and synthesis of the S protein was induced with IPTG and arabinose. On induction of cells carrying the wild-type S105 gene, almost all of the cells lysed over the next hour (Table 1). Lysis occurred within 30 min in a strain containing the fast-lysis S105A52G allele (Fig. 2), which, in the context of the intact phage, causes lysis so early in the phage lytic cycle that there is no significant yield of progeny phage (5). However, after induction, individual cells maintained their rotational velocity until rotation stopped abruptly within seconds before lysis (Figs. 3 and 4a). Cells expressing a functional S allele and cells expressing a null allele of S behaved identically from the time of induction until the sudden paralysis and death of the former. The same abrupt halt in rotation was seen in cells synthesizing wild-type S105 or the S105A52G mutant protein (Table 1).
Figure 2.
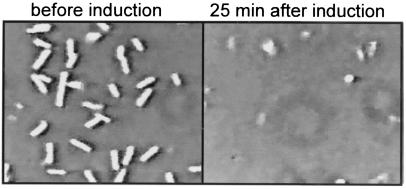
Typical microscope field as seen in a tethering assay before and after induction of the λ lysis genes. Bacterial cells of the strain RY8797pQpS105A52G were grown, tethered, and induced as described in Materials and Methods. A field of tethered cells is shown 1 min before induction (Left) and 25 min after induction (Right).
Figure 3.
Lysis of individual cells. Cells were tethered to coverslips as described in Fig. 2. At t = 0, expression of the lysis genes from the plasmid pS105A52G was induced by IPTG and arabinose. Single frames were chosen from the recordings of two representative cells and are depicted here to illustrate the process of cell lysis. Starting from the time point indicated (Left), single frames were captured every 200 ms. After induction of the lysis genes, the tethered cells rotate at high and constant speed (first row). About 20 min after induction, rotation of the cell abruptly slows and stops completely within 1–3 sec (second row). Cell lysis, because of digestion of the cell wall by the λ R endolysin, occurs within several seconds after the sudden stop in rotation (third and fourth rows). A digitized recording (quicktime movie) can be found as Movie 1, which is published as supplemental data on the PNAS web site, www.pnas.org.
Figure 4.
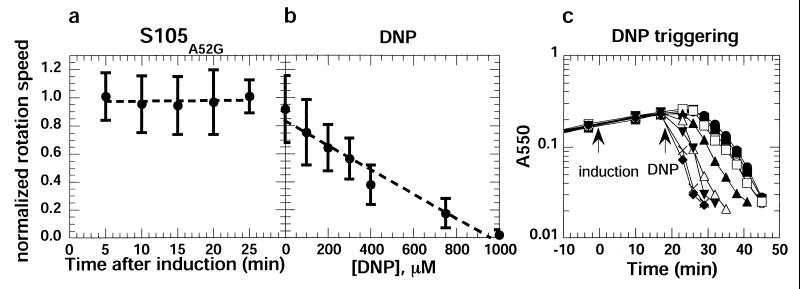
Rotation speeds after induction of the S gene or addition of DNP. (a) Cells of strain RY8797pQpS105A52G were tethered to a glass coverslip, induced at t = 0, recorded, and analyzed for rotation speed, as described in Materials and Methods. The mean of the normalized rotation speeds for 10 individual cells at each time is plotted, except for the 25-min time point, where the value is for seven surviving cells. The vertical bars indicate one standard deviation. (b) RY8797 cells were tethered to glass slides, equilibrated with prewarmed TB medium, and exposed to a 5-min flow of prewarmed TB medium containing DNP at the indicated concentrations. The rotation speed of at least 20 individual cells was determined 5 min after the addition of DNP and normalized to the rotation speed determined before addition. The vertical bars indicate one standard deviation. (c) DNP-induced triggering of S. Cultures of strain RY8797pQpS105 were grown in TB medium at 37°C to an A550 = 0.2 and induced at t = 0 with IPTG and arabinose. At t = 20 min, DNP was added to a final concentration of 0 μM (●), 5 μM (○), 50 μM (■), 100 μM (□), 200 μM (▴), 400 μM (Δ), 600 μM (▾), 1,000 μM (×), and 1,500 μM (⧫). After DNP addition, the turbidity of the cultures was monitored as A550 until lysis was complete.
The Interval Between Holin Triggering and Lysis Is Determined by Lysozyme Activity.
The halt in rotation was quickly followed by catastrophic lysis, visualized by a sudden loss of refractivity (Fig. 3). The interval between the cessation of motion and lysis was about 19 sec for the parental S allele and about 40 sec for S105A52G (Table 1). The longer interval between paralysis and lysis in cells expressing S105A52G was because of the lower level of endolysin present at the time of triggering, because an increased supply of endolysin provided in trans shortened the lysis delay without affecting the time at which the cells stopped spinning (Table 2).
Table 2.
Interval between the stop of rotation and lysis as a function of endolysin content
| Plasmids† | Time from induction until lysis for tethered cells, min‡ | Time from rotation stop until lysis, sec§ |
|---|---|---|
| pS105A52G | 25.1 ± 2.0 (15) | 37.9 ± 9.9 (15) |
| pZA32-luc | ||
| pS105A52GΔR | 27.1 ± 2.7 (15) | 11.5 ± 3.1 (15) |
| pZA32-R/Rz/Rz1 | ||
| pS105A52GΔR | 27.5 ± 2.0 (15) | 12.8 ± 4.4 (15) |
| pZA32R |
The indicated plasmids were introduced into strain RY8797pQ. Transformants carrying the three plasmids were grown, tethered to glass coverslips, and induced with IPTG and arabinose as described in Materials and Methods.
Lysis times for individual cells were determined; mean values are shown. The number of individual cells analyzed is given in parentheses.
Time after rotation stop until cell lysis.
The S Holin Is Triggered at 50% of the Normal pmf Across the E. coli Membrane.
The most straightforward interpretation of these results is that the accumulation of S does not decrease the pmf until the instant of triggering. Alternatively, the cell could compensate for leakage caused by the accumulating holin by increasing the rate of proton efflux by the respiratory chain. We tested whether the cells had a reserve capacity for generating pmf by measuring rotation speed as a function of the concentration of the protonophore uncoupler DNP. The angular velocity, and thus the pmf, decreased monotonically with increasing concentrations of DNP (Fig. 4b). However, DNP was increasingly less effective in triggering holin function at concentrations below 400 μM (Fig. 4c), suggesting that the trigger point for holins is set at about 50% of the normal value of the pmf, or −90 to −100 mV.
Discussion
We conclude that holins form their lethal membrane lesion without causing prior degradation of membrane integrity, at least in terms of the energy state of the cell; that is, holins kill without warning. Thus, the ability to trigger holin function prematurely by addition of an energy poison does not reflect the existence of an energy-draining component in the pathway to lysis. The simplest scheme for holin timing is a critical-concentration model. This scheme defines, in effect, a two-dimensional precipitation of the holin molecules that occurs when the amount of holins exceeds a critical level in the fluid bilayer. The model predicts that this precipitation event is opposed by the energized state of the membrane. Moreover, it requires that the newly formed foci of aggregation constitute membrane domains with reduced mechanical integrity that lead to local disruption of the membrane. Once this event occurs at one site, the transmembrane pmf collapses, triggering global precipitation of holins and massive membrane disruption.
The inhibitory effect of the membrane potential on lysis guarantees that the cell remains fully energized and healthy throughout the lytic cycle. The rapid triggering of lysis by depolarization may also confer a selective advantage. Many phages cause a transient depolarization of the membrane when they inject their DNA into the cell (28, 29). This transient depolarization would trigger the lethal function of holins already accumulated in the membrane, thus aborting the new infection and permitting the primary phage to achieve a modest yield of progeny uncontaminated with the secondary phage. The triggering of holins by membrane depolarization may serve as an emergency over-ride of the normal lysis timing that optimizes phage yield for the existing environmental and host conditions (1, 3, 30, 31).
The critical-concentration model is challenged by the result of a previous experiment. The model predicts that if two unrelated holins with different lysis times accumulate in the membrane, they will form separate mass action pools, provided they do not specifically interact. In the model proposed here, the holin with the lowest triggering concentration should determine the lysis time. However, we previously showed that two unrelated and very dissimilar holins, λ S and T4 T, each associated with a characteristic triggering time, lead to earlier lysis when they are produced together than when either holin is produced alone to the same extent as in the coexpression experiment (12). This observation led to the hypothesis that there is a common pathway for holin triggering. The possibility that each holin titrates out the pmf by causing proton leakage can be ruled out by the results presented here. Thus, we must either propose that two unrelated holins populate the same mass-action pool or postulate that there is a common target for holin action, distinct from the energy state of the membrane.
Several considerations make it unlikely that there is a common protein target. First, there is extreme diversity among holins. A cardinal example of convergent evolution, holins constitute the most heterogeneous group of proteins that share one function, with at least 35 distinct unrelated families with no discernible conserved sequence motif (1). Moreover, the best characterized holin gene, λ S, has been shown to be lethal in widely divergent bacteria and even in yeast (32). Finally, purified S105 protein can disrupt protein-free liposomes, whereas a lysis-defective mutant protein, S105A52V, cannot (16) (J. Deaton and R.Y., unpublished work). A common mass-action pool might be achieved if holins form nonspecific oligomeric structures that can act cooperatively, in effect to coprecipitate. This notion predicts that the unrelated T and S holins physically interact at some point in the pathway to lysis. In any case, despite their simple structure, construction of a complete blueprint for the “works” of these remarkable and ancient protein “clocks” remains elusive.
Supplementary Material
Acknowledgments
We thank I.-N. Wang for construction of the pQ plasmid, J. S. Parkinson for the cheA strain, G. Gadda for his help in counting the rotation speed for numerous cells, and members of the Young and Manson laboratories for technical assistance, encouragement, and critical discussion. A.G. was a doctoral student of the Institute of Microbiology, University of Vienna, Vienna, under the supervision of U. Bläsi, whose advice and critical contribution are gratefully acknowledged. This work was done in partial fulfillment of the requirements for the doctoral degree for A.G. Support for this work was derived from Public Health Service Grant GM27099, a grant from the Robert A. Welch Foundation to R.Y., and by Army Research Office Grant DAAG55–97-1–0380 to M.D.M.
Abbreviations
- pmf
proton-motive force
- TB
tryptone broth
- DNP
dinitrophenol
- IPTG
isopropyl β-d-thiogalactopyranoside
References
- 1.Wang I-N, Smith D L, Young R. Annu Rev Microbiol. 2000;54:799–825. doi: 10.1146/annurev.micro.54.1.799. [DOI] [PubMed] [Google Scholar]
- 2.Young R, Wang I-N, Roof W D. Trends Microbiol. 2000;8:120–128. doi: 10.1016/s0966-842x(00)01705-4. [DOI] [PubMed] [Google Scholar]
- 3.Wang I-N, Dykhuizen D E, Slobodkin L B. Evol Ecol. 1996;10:545–558. [Google Scholar]
- 4.Raab R, Neal G, Sohaskey C, Smith J, Young R. J Mol Biol. 1988;199:95–105. doi: 10.1016/0022-2836(88)90381-6. [DOI] [PubMed] [Google Scholar]
- 5.Johnson-Boaz R, Chang C-Y, Young R. Mol Microbiol. 1994;13:495–504. doi: 10.1111/j.1365-2958.1994.tb00444.x. [DOI] [PubMed] [Google Scholar]
- 6.Ramanculov E R, Young R. Gene. 2001;265:25–36. doi: 10.1016/s0378-1119(01)00365-1. [DOI] [PubMed] [Google Scholar]
- 7.Doermann A H. J Gen Physiol. 1952;35:645–656. doi: 10.1085/jgp.35.4.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Josslin R. Virology. 1971;44:101–107. doi: 10.1016/0042-6822(71)90157-7. [DOI] [PubMed] [Google Scholar]
- 9.Reader R W, Siminovitch L. Virology. 1971;43:607–622. doi: 10.1016/0042-6822(71)90286-8. [DOI] [PubMed] [Google Scholar]
- 10.Young R. Microbiol Rev. 1992;56:430–481. doi: 10.1128/mr.56.3.430-481.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garrett J M, Young R. J Virol. 1982;44:886–892. doi: 10.1128/jvi.44.3.886-892.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramanculov E R, Young R. Mol Gen Genet. 2001;265:345–353. doi: 10.1007/s004380000422. [DOI] [PubMed] [Google Scholar]
- 13.Fung D C, Berg H C. Nature (London) 1995;375:809–812. doi: 10.1038/375809a0. [DOI] [PubMed] [Google Scholar]
- 14.Miller J H. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1972. [Google Scholar]
- 15.Parkinson J S, Houts S E. J Bacteriol. 1982;151:106–113. doi: 10.1128/jb.151.1.106-113.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith D L, Struck D K, Scholtz J M, Young R. J Bacteriol. 1998;180:2531–2540. doi: 10.1128/jb.180.9.2531-2540.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bläsi U, Chang C-Y, Zagotta M T, Nam K, Young R. EMBO J. 1990;9:981–989. doi: 10.1002/j.1460-2075.1990.tb08200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith D L, Young R. J Bacteriol. 1998;180:4199–4211. doi: 10.1128/jb.180.16.4199-4211.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gründling A, Bläsi U, Young R. J Biol Chem. 2000;275:769–776. doi: 10.1074/jbc.275.2.769. [DOI] [PubMed] [Google Scholar]
- 20.Gründling A, Smith D L, Bläsi U, Young R. J Bacteriol. 2000;182:6075–6081. doi: 10.1128/jb.182.21.6075-6081.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lutz R, Bujard H. Nucleic Acids Res. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garza A G, Biran R, Wohlschlegel J A, Manson M D. J Mol Biol. 1996;258:270–285. doi: 10.1006/jmbi.1996.0249. [DOI] [PubMed] [Google Scholar]
- 23.Berg H C, Block S M. J Gen Microbiol. 1984;130:2915–2920. doi: 10.1099/00221287-130-11-2915. [DOI] [PubMed] [Google Scholar]
- 24.Smith D L, Chang C-Y, Young R. Gene Expression. 1998;7:39–52. [PMC free article] [PubMed] [Google Scholar]
- 25.Chang C-Y, Nam K, Young R. J Bacteriol. 1995;177:3283–3294. doi: 10.1128/jb.177.11.3283-3294.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Silverman M, Simon M. Nature (London) 1974;249:73–74. doi: 10.1038/249073a0. [DOI] [PubMed] [Google Scholar]
- 27.Harold F M, Maloney P C. In: Escherichia coli and Salmonella Cellular and Molecular Biology. Neidhardt F C, Curtiss R I, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Washington, DC: Am. Soc. Microbiol. Press; 1996. pp. 283–306. [Google Scholar]
- 28.Goldberg E B, Grinius L, Letellier L. In: Molecular Biology of Bacteriophage T4. Karam J D, Drake J W, Kreuzer K N, Mosig G, Hall D H, Eiserling F A, Black L W, Spicer E K, Kutter E, Carlson K, et al., editors. Washington, DC: Am. Soc. Microbiol. Press; 1994. pp. 347–357. [Google Scholar]
- 29.Labedan B, Letellier L. Proc Natl Acad Sci USA. 1981;78:215–219. doi: 10.1073/pnas.78.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abedon S T. In: Molecular Biology of Bacteriophage T4. Karam J D, Drake J W, Kreuzer K N, Mosig G, Hall D H, Eiserling F A, Black L W, Spicer E K, Kutter E, Carlson K, et al., editors. Washington, DC: Am. Soc. Microbiol. Press; 1994. pp. 397–405. [Google Scholar]
- 31.Abedon S T. Microbiol Ecol. 1989;18:79–88. doi: 10.1007/BF02030117. [DOI] [PubMed] [Google Scholar]
- 32.Garrett J, Bruno C, Young R. J Bacteriol. 1990;172:7275–7277. doi: 10.1128/jb.172.12.7275-7277.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.