Abstract
BRAF-V600E (1799T > A) is one of the most frequently reported driver mutations in multiple types of cancers, and patients with such mutations could benefit from selectively inactivating the mutant allele. Near this mutation site, there are two TTTN and one NGG protospacer-adjacent motifs (PAMs) for Cpf1 and Cas9 CRISPR nucleases, respectively. The 1799T > A substitution also leads to the occurrence of a novel NGNG PAM for the EQR variant of Cas9. We examined the editing efficacy and selectivity of Cpf1, Cas9, and EQR variant to this mutation site. Only Cpf1 demonstrated robust activity to induce specific disruption of only mutant BRAF, not wild-type sequence. Cas9 recognized and cut both normal and mutant alleles, and no obvious gene editing events were observed using EQR variant. Our results support the potential applicability of Cpf1 in precision medicine through highly specific inactivation of many other gain-of-function mutations.
Keywords: Cpf1, targeted therapy, BRAF V600E
Introduction
Genetic mutations are a hallmark of cancer development.1 Over the past decade, selective disruption or inhibition of the oncoproteins using small molecules and antibodies has been a mainstay of antitumor drug development effort, resulting in a handful of approved cancer therapies.2, 3 However, a major problem in clinical practice is that cancer cells can also quickly develop resistance to these targeted agents, leading to cancer recurrence and treatment failure.4, 5, 6 Moreover, for most of the mutated oncoproteins, such as Ras and β-catenin, it is difficult to design or screen a specific inhibitor.
In the past 3 years, one of the most powerful biotechnologies has been CRISPR/Cas9. It has become one of the most popular ways to perform targeted genetic manipulation. S. pyogenes Cas9 has been shown to be able to tolerate a variable number of mismatches between guide RNA (gRNA) and target DNA, especially the protospacer-adjacent motif (PAM)-distal regions.7, 8, 9 Hence, several strategies, including Fok1-dCas9, truncated gRNAs, and engineered Cas9, have been used to diminish the off-target effects of Cas9.10, 11, 12, 13 On the other hand, mismatches in the PAM sequence are less tolerated.7, 8, 9 This feature has also been used to specifically recognize and disrupt genome mutants, which coincidently lead to the occurrence of a novel PAM sequence.14, 15, 16 Several engineered Cas9 variants use different PAM sequences. For example, SpCas9 with VRER mutation recognizes NGCG PAM sequence, SpCas9 with EQR mutation uses NGAG as PAM sequence, and the PAM sequences of SpCas9 VQR variant are NGAN or NGNG.17 However, few studies have used these variants for genetic manipulation, and their activities and specificities need further validation.
Cpf1 nucleases have recently been identified as RNA-guided type V class II CRISPR-Cas systems.18, 19 Different from the guanosine-rich sequences recognized by Cas9, the PAM sequences of Cpf1 are thymidine-rich DNA sequences and are expected to broaden the scope of CRISPR genome editing target sites. More important, it has been recently shown that Cpf1 is highly specific, with substantial fewer off-target cleavage sites compared with Cas9. It could tolerate single or double mismatches in the 3′ PAM-distal region, but the bases in the 5′ PAM-proximal region are highly sensitive to single or double substitutions. The off-target effects are almost undetectable using targeted deep sequencing analyses.20, 21 Thus Cpf1 can be exploited to achieve precise gene modifications and genome editing.
BRAF-V600E is one of the most frequently reported driver mutations in multiple types of cancers.22 It has been reported to be present in all patients with hairy cell leukemia,23 95% of those with papillary craniopharyngioma,24 about 45% of those with papillary thyroid cancer,25 and ≥50% of those with melanoma.26 Several selective ATP-competitive small-molecule BRAF inhibitors, such as vemurafenib and dabrafenib, have been approval for the treatment of metastatic and unresectable BRAF-mutated melanomas.27, 28 However, the efficacy of BRAF inhibitors is limited to a subset of patients with BRAF-mutated metastatic melanoma, despite the abundance of BRAF mutations identified in thyroid, colorectal, glioblastoma, and non-small-cell lung cancers.22 Moreover, drug resistance and cancer relapse remain major problems in clinical practice.29, 30, 31
The purpose of the present study was therefore to ascertain the specificity and effectiveness of Cpf1 in targeted disruption of BRAF V600E, a well-known gain-of-function mutation.
Results
gRNA Design for Cpf1, Cas9, and Cas9-EQR
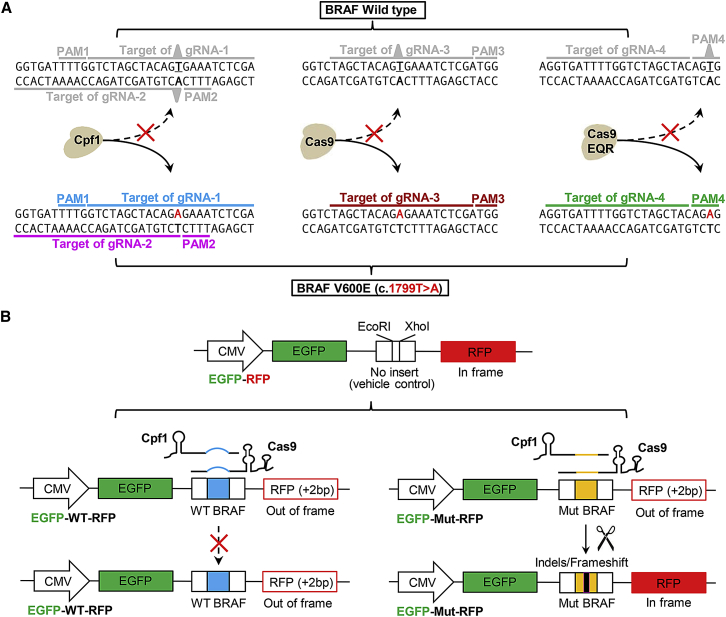
As shown in Figure 1A, near the BRAF mutation site (1799T > A), there are two PAM sequences (PAM1 and PAM2) of Cpf1, one PAM sequence of SpCas9, and a novel PAM (PAM4) of SpCas9-EQR variant. Thus, we designed four gRNAs with the aim of comparing the editing efficacy and selectivity of these nucleases.
Figure 1.
gRNA Design for Cpf1, Cas9, and Cas9-EQR Systems and Dual-Fluorescence Reporter Plasmids for Evaluation of Editing Efficacy and Selectivity
(A) Design of single-guide RNAs (sgRNAs) for Cpf1, Cas9, and Cas9-EQR systems targeting mutant BRAF allele, with a hypothesis that they cannot bind and cleave wild-type BRAF allele. Two gRNAs (gRNA-1 and gRNA-2) were designed for both AsCpf1 and LbCpf1. gRNA-3 was used for spCas9. Mutant BRAF forms a PAM of spCas9 EQR variant, which is absent from the wild-type BRAF, and thus gRNA-4 was designed for Cas9-EQR. (B) Schematic illustration of the GFP-RFP reporter plasmids for evaluation of the editing efficacy and selectivity. The reporter vector contains a CMV promoter and sequences encoding EGFP and RFP, which are separated by multiple cloning sites. gRNA binding regions and PAM motifs were inserted between EcoRI and XhoI sites, and this insertion caused RFP coding region to be shifted out of frame. Target cleavage and subsequent non-homologous end-joining repair can lead to sequence indels/frameshifts and bring the downstream RFP back in frame.
Evaluation of the Editing Efficacy and Selectivity of Cpf1, Cas9, and Cas9-EQR Using a Dual-Fluorescence Reporter System and Sanger Sequencing
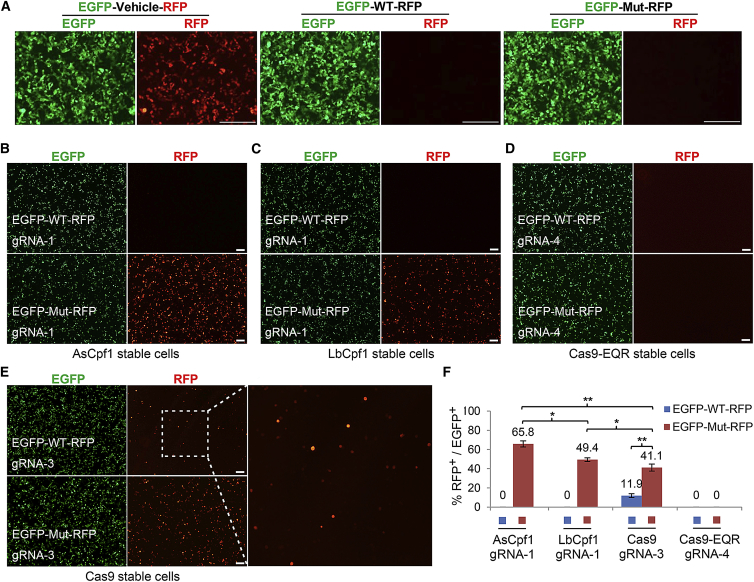
To facilitate the quantification and comparison of these nucleases, we first constructed a dual-fluorescence reporter system containing a cytomegalovirus (CMV) promoter and sequences encoding EGFP and RFP, which are separated by multiple cloning sites (Figure 1B). Sequences bearing BRAF 1799T > A mutation or the corresponding wild-type (WT) sequences were cloned into the reporter construct. After transfection in HEK293T cells, dual fluorescence can be observed in GFP-vehicle-RFP control group, whereas both GFP-WT-RFP and GFP-Mut-RFP groups show no fluorescence of RFP. After co-transfection of these reporter constructs with gRNAs in HEK293T cells, both AsCpf1 and LbCpf1 show robust cleavage activities only in mutant sequence, while Cas9 system cut not only mutant but also the WT allele, and no cleavage events were observed in Cas9-EQR group (Figures 2B–2E). Fluorescence-activated cell sorting (FACS) analysis indicated that AsCpf1-gRNA-1 possessed the highest activities in cleavage of mutant allele (Figure 2F). Of the two gRNAs (gRNA-1 and gRNA-2) for Cpf1, only gRNA1 showed robust activities, whereas the efficiency of AsCpf1-gRNA-2 was very weak, and no activity was detected using LbCpf1-gRNA-2 (data not shown). On the other hand, to exclude other unexpected reasons with respect to the failure of EQR variant, we also performed co-transfection of three plasmids (Cpf1/Cas9/Cas9-EQR + gRNAs + reporters) in normal HEK293T cells, and the results were almost the same as with the above co-transfection of two plasmids in nuclease stable cells (data not shown).
Figure 2.
Evaluation of Editing Efficacy and Selectivity in HEK293T Cells
(A) Transient transfection of GFP-vehicle-RFP control, GFP-WT-RFP reporter, and GFP-Mut-RFP reporter in HEK293T cells for 48 hr. (B–E) Evaluation of the editing efficacy and selectivity of the indicated CRISPR systems. All gRNAs were designed for targeting mutant BRAF allele. HEK293T cells constitutively expressing AsCpf1, LbCpf1, Cas9, or Cas9-EQR were established respectively by lentiviral transduction and puromycin selection. Images were acquired 60 hr after co-transfection with indicated GFP-RFP reporter vectors and gRNAs. (F) Statistical analysis of editing efficiency and selectivity by FACS. Error bars represent SD from experiments performed in triplicates. One-way ANOVA: *p < 0.05, **p < 0.01. WT, wild-type; Mut, mutant. The scale bar represents 200 μm.
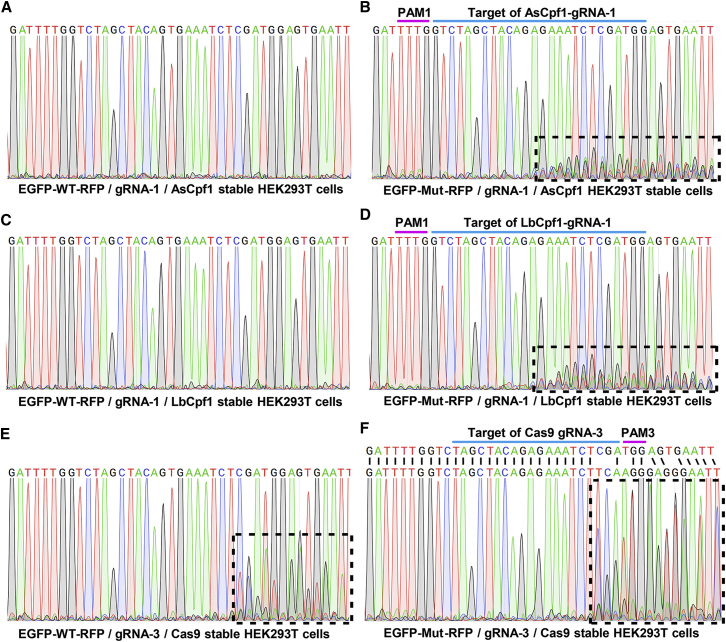
Sequence-specific disruption of Cpf1 was further validated by Sanger sequencing (Figure 3). As shown, obvious overlapping peaks were observed downstream of the cleavage sites only in EGFP-Mut-RFP/Cpf1 groups (Figures 3B and 3D), not in EGFP-WT-RFP/Cpf1 groups (Figures 3A and 3C). Consistent with the observation in dual-fluorescence transfection, mixed peaks were observed not only in the EGFP-Mut-RFP/Cas9 group (Figure 3F) but also in the EGFP-WT-RFP/Cas9 group (Figure 3E), indicating that Cas9 is tolerant to this single-point mismatch, and as predicted, no change was observed in Cas9-EQR groups. It is notable that although dual-fluorescence transfection and FACS analysis indicated that Cpf1 systems were more powerful, from the sequencing results, Cas9 system caused more dramatic sequence changes compared with Cpf1 systems.
Figure 3.
Representative Sequencing Results of Corresponding DNA Samples in HEK293T Cells
Sixty hours after co-transfection as described above, genome DNA was extracted and subjected to PCR amplification. PCR products were sequenced directly. All gRNAs were designed for targeting mutant BRAF allele as indicated. (A, C, and E) Sequencing results of gene editing of wild-type allele. (B, D, and F) Results of mutant allele. Regions with continuous overlapping peaks, caused by cleavage and subsequent non-homologous end-joining repair, are indicated with dashed boxes. Alignment to the mutant BRAF regions inserted in EGFP-Mut-RFP reporter is shown in (F).
Cpf1 Systems Can Efficiently Target and Disrupt Endogenous Mutant BRAF in A375 Cells
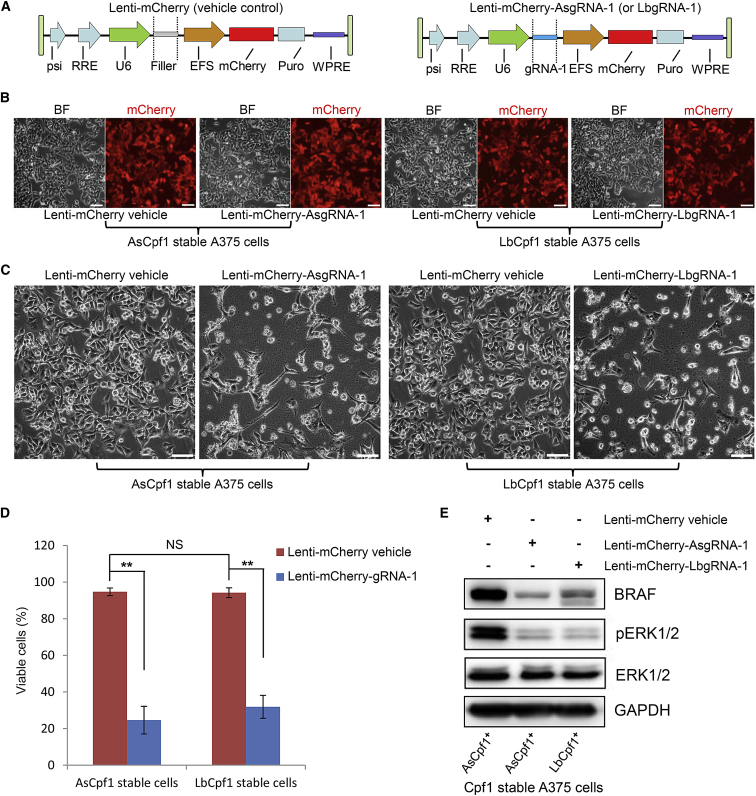
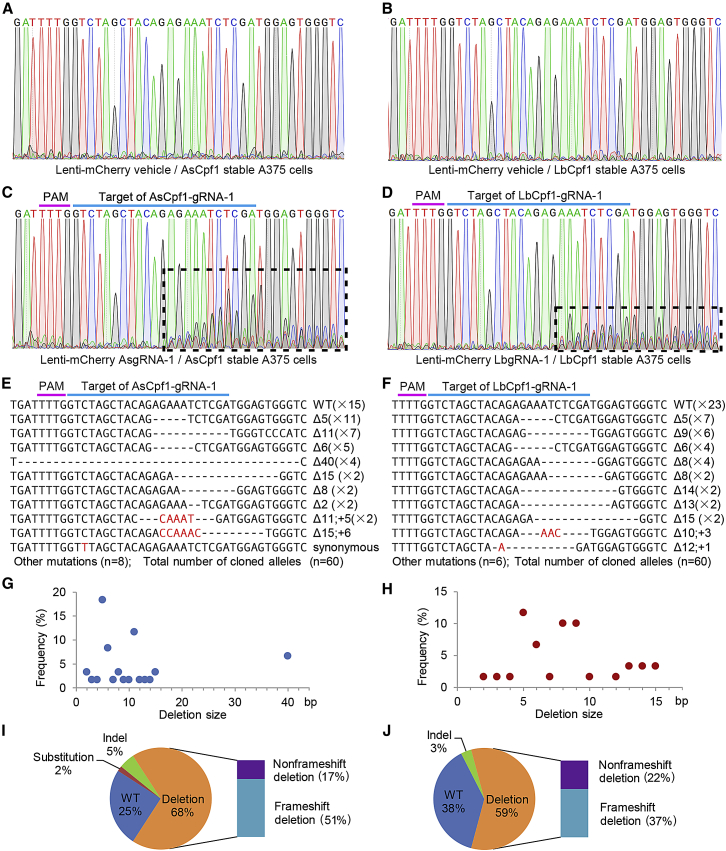
On the basis of the aforementioned results, we further tested the genome editing efficacy and selectivity of Cpf1 systems in A375 cells, a melanoma cell line with a homozygous BRAF V600E mutation. First, A375 cells constitutively expressing AsCpf1 or LbCpf1 were established respectively by lentiviral transduction and puromycin selection. Then we constructed lentiviral vectors for gRNAs expressing and fluorescently labeling (Figure 4A). Figure 4B shows highly efficient transduction of A375 Cpf1 stable cells with mCherry-gRNA-1 lentiviral vectors. Morphological changes, characterized by shrinkage and formation of cytoplasmic blebs, could be observed only in lenti-mCherry-gRNA-1 groups 3 days following transduction (data not shown). Significant cell death was observed in lenti-mCherry-gRNA-1 groups 7 days after transductions compared with vehicle control groups, and no obvious difference were observed in cell viability between the AsCpf1 and LbCpf1 groups (Figures 4C and 4D). We also used scramble gRNA and a gRNA targeting EGFP as negative controls, and no significant effects on cell viability were observed (data not shown). The expression of BRAF and pERK1/2 decreased dramatically following Cpf1-gRNAs treatment for 72 hr (Figure 4E). Targeted genome editing was further validated by Sanger sequencing (Figure 5). As shown, obvious overlapping peaks were observed downstream of the cleavage sites only in lenti-mCherry-gRNA-1/Cpf1 groups, not in vehicle control groups 72 hr following treatment (Figures 5A–5D). When the PCR products were subjected to TA cloning and sequencing 7 days after transduction, the majority of the mutations observed were small deletions (Figures 5E–5J). Of the five predicted potential off-target sites, no gene editing effects were detected by targeted sequencing in both AsCpf1 and LbCpf1 groups (Table S1).
Figure 4.
Evaluation of Editing Efficacy and Selectivity in Cpf1-gRNA-Treated A375, a Melanoma Cell Line with a Homozygous BRAF V600E Mutation
(A) Schematic illustration of the lentiviral vectors used for gRNA expressing and fluorescently labeling. (B) Highly efficient transduction of A375 stable cells with indicated lentiviral vectors. Images were acquired on day 2 following transduction. (C) Cpf1 stable cells were transducted with vehicle or gRNA-expression lentiviral vectors. Images were acquired on day 7 following transduction. A375 cells constitutively expressing AsCpf1 or LbCpf1 were established respectively by lentiviral transduction and puromycin selection. (D) Statistical analysis of cell viability by trypan blue exclusion test. Error bars represent SD from experiments performed in triplicate. One-way ANOVA analysis: NS, not significant; **p < 0.01. (E) Expression status of BRAF, ERK1/2, and pERK1/2 in A375 stable cells following Cpf1-gRNA treatment for 72 hr by western blotting. AsCpf1 positive stable A375 cells transducted with lenti-mCherry-vehicle were set as control group. GAPDH serves as a loading control. AsgRNA, gRNA for AsCpf1; LbgRNA, gRNA for LbCpf1. The scale bar represents 100 μm.
Figure 5.
Mutant Sequences Induced at the Cpf1-gRNA-1 Target Site in A375 Cells
Genome DNA was extracted after lentiviral transduction of A375 stable cells as described above. All gRNAs were designed for targeting mutant BRAF allele as indicated. Data are representative of three independent experiments. Mutant sequence induced by AsCpf1-gRNA-1 and LbCpf1-gRNA-1 are shown on the left and right, respectively. (A–D) PCR products harboring target region were sequenced directly 72 hr after transduction. Regions with continuous overlapping peaks, caused by cleavage and subsequent non-homologous end-joining repair, are indicated with dashed boxes. (E–J) PCR products harboring target region were subjected to TA cloning 7 days after transduction. In total, 60 clones from three independent samples (20 clones per independent sample) were analyzed by sequencing. (E and F) Representative mutant sequences are shown. The deleted bases are marked with dashes, and the inserted or substituted bases are shown in red. Mutation types and the occurrence of each sequence are indicated to the left of the alignment. Unmodified reads are marked as “WT.” (G and H) Size distribution of deletions identified at the target site. (I and J) Mutation profiling of the sequenced clones.
Discussion
Over the past decade, high-throughput next-generation sequencing has successfully identified a large number of somatic tumor-driver mutations and numerous SNPs associated with human diseases.32, 33 The CRISPR/Cas9 system has been investigated in an allele-specific manner, demonstrating its potential for treating dominant hereditary conditions and even cancers.14, 15, 16, 34 Several initial studies have shown that the 10–12 bp closest to the PAM, called the “seed sequence,” determine Cas9 specificity and are generally more important than the rest of the gRNA sequences.35, 36 However, more recent studies have revealed an unexpected complexity of Cas9-gRNA interaction and cleavage beyond the simple paradigm of site determination on the basis of the “seed” and PAM sequence.7, 8, 9, 37, 38, 39 It has been shown that most bases within the 20 bp target site provide varying degrees of specificity, and large variations were observed across target sites and cell types regarding the importance of base pairing at each position.37, 40 In the present study, as shown in Figure 1A, the 1799T > A substitution is located at a site 11 bp upstream of the PAM of Cas9 gRNA, 13 bp away from the PAM of Cpf1 gRNA-1, and 1 bp near the PAM of Cpf1 gRNA-2. As a result, while Cas9-gRNA-3 recognized and cut both normal and mutant allele, limited cleavage activity was observed with Cpf1-gRNA-2, and only Cpf1-gRNA-1 demonstrated robust activity to induce specific disruption of only mutant BRAF. Moreover, no gene editing effects were detected in the five predicted potential off-target sites by targeted sequencing both in AsCpf1 and in LbCpf1 groups. To further understand the general specificity of Cpf1, additional studies are needed to examine its specific recognition of more SNP loci.
Although the Cas9 system has been shown to tolerate a variable number of mismatches between gRNA and target DNA, it is sensitive to changes in PAM.7, 8, 35, 36 The BRAF 1799T > A point mutation generates a new PAM sequence of EQR variants of Cas9. Surprisingly, we failed to detect editing events in mutant allele using EQR variant of Cas9. To target a gene using the CRISPR system, we can design a handful of gRNAs and screen out the best candidate. To target a single point mutation, we often have limited choice in designing gRNA. In fact, of the two gRNAs of Cpf1 we designed for targeting V600E mutation, only gRNA-1 could efficiently perform genome editing. Thus, it is possible that EQR variant has very low or no activity in this locus using the present gRNA.
The repair mechanism after Cpf1-mediated DNA cleavage is limited. In the present study, although dual-fluorescence transfection and FACS analysis indicated that Cpf1 systems were more powerful, sequencing results showed that the Cas9 system caused more dramatic sequence changes compared with Cpf1 systems. It has been reported that LbCpf1, AsCpf1, and Cas9 induced different mutation signatures. Cpf1 rarely induced insertions, and consequently, in-frame mutations caused by deletions were more likely to be induced by Cpf1 than by Cas9.20, 21 Consistent with these findings, we also found that the majority of the mutations induced by Cpf1 were small deletions. On the other hand, although significant growth inhibition and cell death were observed, some clones survived and kept on proliferating after treatment. It is therefore likely that some WT clones or clones with 3, 6, or 9 nt or other triple-nucleotide combination deletions may survive and maintain the oncogenic phenotype. Thus, for future application of Cpf1 in gene disruption, in addition to off-target cleavage, strategies should emphasize improving out-of-frame editing efficiency. A comprehensive understanding of the cleavage and repair mechanisms of Cpf1 will help us use this nuclease more effectively.
Although CRISPR-Cas9 systems are widely used for genome editing, the range of sequences Cas9 can recognize is constrained by the need for a specific PAM. It can therefore be challenging to perform allele-specific alterations without a NGG PAM. LbCpf1 and AsCpf1 recognize and cleave target DNA sequences composed of the TTTN PAM sequence. Thus Cpf1-family proteins expand the targeting range of RNA-guided allele-specific genome editing.
Together, our results support the potential applicability of Cpf1 in the specific inactivation of gain-of-function mutations. Further studies are needed to understand the repairing mechanism after cleavage and to address its limitation of the requirement of a TTTN PAM.
Materials and Methods
Plasmids and Vector Construction
pcDNA3.1-hAsCpf1, pcDNA3.1-hLbCpf1, Crispr V2, VQR, and EQR Cas9 variants were acquired from Addgene (69982, 69988, 52961, 65771, and 65772, respectively). The open-reading frames of hAsCpf1, hLbCpf1, and mCherry were cloned into XbaI/BamHI sites of lentiCRISPR V2 vector, forming pLenti-hAsCpf1, pLenti-hLbCpf1, and pLenti-mCherry, respectively. The open-reading frames of VQR and EQR Cas9 variants were added to an SV40 nuclear localization sequence in their C termini by PCR and subsequently cloned into XbaI/BamHI sites of lentiCRISPR V2 vector, forming pLenti-VQR and pLenti-EQR, respectively. Oligonucleotide duplexes corresponding to spacer sequences were annealed and ligated into BbsI-digested phU6-gRNA (Addgene 53188) or BsmBI-digested pLenti-mCherry for gRNA expression of hAsCpf1, hLbCpf1, SpCas9, and Cas9 variants. The GFP-RFP reporter plasmids were constructed on the basis of the parental pEGFP-C1 vector. RFP was amplified from PDsRed-N1 and ligated into the KpnI/BamHI site of pEGFP-C1. Double-stranded oligonucleotides, containing BRAF c.1799T > A-derived WT and mutant target sites, were ligated between EGFP and RFP sequences, exploiting vector-derived XhoI and EcoRI sites. All the oligonucleotides for vector construction, PCR amplification, and sequencing are provided in the Supplemental Information.
Cell Culture, Lentivirus Production, and Transfection Conditions
HEK293T and A375 cell lines were purchased from ATCC and maintained at 37°C, saturated humidity, and 5% CO2 in high-glucose DMEM supplemented with 10% heat-inactivated fetal bovine serum and antibiotics (100 U/mL penicillin, 100 μg/mL streptomycin). Mycoplasma testing was performed every 3 months.
Lentiviral particles were produced in HEK293 cells in a 10 cm cell culture dish at 80%–90% confluence. Cells were transfected with 1 μg of pMD2.G, 3 μg of psPAX2 (Addgene 12259 and 12260), and 4 μg of lentiviral vector using 30 μg PEI (Sigma-Aldrich) per dish. Fresh medium was added 16 hr post-transfection, and viral-containing supernatant was collected 48 hr post-transfection, filtered through 0.45 μm filters, and centrifuged for 2 hr at 20,000 × g and 4°C. The virus pellets were resuspended in 100 μL serum-free DMEM and stored in aliquots at −80°C. Lentiviral transduction was performed in six-well plates at about 40%–60% confluence. Cells were transduced with 20 μL concentrated viral supernatant in 2 mL total volume supplemented with 8 μg/ml polybrene (Sigma). Cpf1, Cas9, or Cas9 variant expression stable cell lines (HEK293T and A375) were established by puromycin selection, and these cells were used for subsequent gene editing experiments. For lentiviral vector-mediated gene editing, Cpf1 or Cas9 stable cell line A375 was infected with concentrated viral supernatant containing pLenti-mCherry-gRNAs as described above. Transient plasmid transfection and gene editing were performed in six-well plates at 70%–90% confluence with Lipofectamine 3000 (Life Technologies) using 1.5 μg gRNA vectors and 1 μg GFP-RFP reporter per well according to the manufacturer’s instructions. For Cpf1, Cas9, and Cas9 variants, three-plasmid co-transfection in normal HEK293T with Lipofectamine 3000 was also performed by using 1 μg plasmids of Cas9 or Cas9 variants, 0.8 μg gRNA vectors, and 0.7 μg GFP-RFP reporter per well.
Western Blotting
Total cellular proteins were extracted in RIPA buffer supplemented with protease inhibitor cocktail and sodium orthovanadate (Sigma-Aldrich). Protein concentrations were determined by a BCA (bicinchoninic acid) protein assay kit. Equal amounts of protein were resolved on 10% SDS-PAGE gel. After blocking with 5% skimmed milk, membrane was incubated with the following primary antibodies at 4°C overnight: mouse monoclonal antibody to BRAF (Zen BioScience, 1:2000, 200532-4E1), mouse monoclonal antibody to MEK1 (Zen BioScience, 1:1,000, 200424), rabbit monoclonal antibody to ERK1/2 (Cell Signaling Technology, 1:1,000, 4695S), rabbit monoclonal antibody to pERK1/2 (Cell Signaling Technology, 1:2,000, 4370S), and rabbit polyclonal antibody to GADPH (BOSTER, 1:400, BA2913). The second antibody was purchased from Beyotime Biotech and used at a dilution of 1:2,000. Targeted protein bands were detected by enhanced chemiluminescence (ECL) and digital imaging (Clinx Science Instruments, Chemiscope 5300).
Cell Viability and FACS Analysis
Cell viability was assessed using the trypan blue cell viability assay. 7 days after transduction with lengti-mCherry or lenti-mCherry-gRNA, Cpf1 stable A375 cells were collected by trypsin digestion, centrifugation, and trypan blue staining according to the manufacturer’s instruction (Sigma-Aldrich). Target-specific cleavage activity was quantified by FACS assay of GFP+ cells within the populations of RFP+ cells using a BD FACSAria III system (Becton Dickinson). Assays were performed in triplicate. One-way ANOVA was used to determine statistical significance. A p value of less than 0.05 was considered to indicate statistical significance.
DNA Isolation and Sequencing
DNA extraction from A375 or HEK293T cells was performed by using QuickExtract DNA Extraction Solution (Epicenter). Potential off-target loci were predicted using CasOFFinder (http://www.rgenome.net/cas-offinder/). Primers flanking the BRAF target or potential off-target loci are listed in the Supplemental Information. PCR products were sequenced directly or were subcloned using ZTOPO-TA Zero Background Fast Cloning Kit (ZOMANBIO). DNA sequencing was performed in TSINGKE biotech.
Author Contributions
G.G. and A.T. designed the experiments. M.Y., H.W., Y.W., J.D., Y.T., and L.Z. performed the experiments. M.Y., H.W., and A.T. analyzed the data and wrote the manuscript.
Conflicts of Interest
We declare no competing financial interests.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grants 31471286 and 31171370) and the National Scientific and Technological Special Project for New Drugs Development (2015ZX09102010).
Footnotes
Supplemental Information includes Supplemental Materials and Methods and one table and can be found with this article online at http://dx.doi.org/10.1016/j.omtn.2017.05.009.
Contributor Information
Gang Guo, Email: guogang@scu.edu.cn.
Aiping Tong, Email: aipingtong@scu.edu.cn.
Supplemental Information
References
- 1.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Scott A.M., Wolchok J.D., Old L.J. Antibody therapy of cancer. Nat. Rev. Cancer. 2012;12:278–287. doi: 10.1038/nrc3236. [DOI] [PubMed] [Google Scholar]
- 3.Zhang J., Yang P.L., Gray N.S. Targeting cancer with small molecule kinase inhibitors. Nat. Rev. Cancer. 2009;9:28–39. doi: 10.1038/nrc2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shah N.P., Nicoll J.M., Nagar B., Gorre M.E., Paquette R.L., Kuriyan J., Sawyers C.L. Multiple BCR-ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer Cell. 2002;2:117–125. doi: 10.1016/s1535-6108(02)00096-x. [DOI] [PubMed] [Google Scholar]
- 5.Paez J.G., Jänne P.A., Lee J.C., Tracy S., Greulich H., Gabriel S., Herman P., Kaye F.J., Lindeman N., Boggon T.J. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 6.Choi Y.L., Soda M., Yamashita Y., Ueno T., Takashima J., Nakajima T., Yatabe Y., Takeuchi K., Hamada T., Haruta H., ALK Lung Cancer Study Group EML4-ALK mutations in lung cancer that confer resistance to ALK inhibitors. N. Engl. J. Med. 2010;363:1734–1739. doi: 10.1056/NEJMoa1007478. [DOI] [PubMed] [Google Scholar]
- 7.Fu Y., Foden J.A., Khayter C., Maeder M.L., Reyon D., Joung J.K., Sander J.D. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pattanayak V., Lin S., Guilinger J.P., Ma E., Doudna J.A., Liu D.R. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat. Biotechnol. 2013;31:839–843. doi: 10.1038/nbt.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin Y., Cradick T.J., Brown M.T., Deshmukh H., Ranjan P., Sarode N., Wile B.M., Vertino P.M., Stewart F.J., Bao G. CRISPR/Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNA sequences. Nucleic Acids Res. 2014;42:7473–7485. doi: 10.1093/nar/gku402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guilinger J.P., Thompson D.B., Liu D.R. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat. Biotechnol. 2014;32:577–582. doi: 10.1038/nbt.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu Y., Sander J.D., Reyon D., Cascio V.M., Joung J.K. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Slaymaker I.M., Gao L., Zetsche B., Scott D.A., Yan W.X., Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kleinstiver B.P., Pattanayak V., Prew M.S., Tsai S.Q., Nguyen N.T., Zheng Z., Joung J.K. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Courtney D.G., Moore J.E., Atkinson S.D., Maurizi E., Allen E.H., Pedrioli D.M.L., McLean W.H., Nesbit M.A., Moore C.B. CRISPR/Cas9 DNA cleavage at SNP-derived PAM enables both in vitro and in vivo KRT12 mutation-specific targeting. Gene Ther. 2016;23:108–112. doi: 10.1038/gt.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y., Mendiratta S., Ehrhardt K., Kashyap N., White M.A., Bleris L. Exploiting the CRISPR/Cas9 PAM constraint for single-nucleotide resolution interventions. PLoS ONE. 2016;11:e0144970. doi: 10.1371/journal.pone.0144970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shin J.W., Kim K.H., Chao M.J., Atwal R.S., Gillis T., MacDonald M.E., Gusella J.F., Lee J.M. Permanent inactivation of Huntington’s disease mutation by personalized allele-specific CRISPR/Cas9. Hum. Mol. Genet. 2016 doi: 10.1093/hmg/ddw286. Published online September 15, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kleinstiver B.P., Prew M.S., Tsai S.Q., Topkar V.V., Nguyen N.T., Zheng Z., Gonzales A.P., Li Z., Peterson R.T., Yeh J.R. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zetsche B., Gootenberg J.S., Abudayyeh O.O., Slaymaker I.M., Makarova K.S., Essletzbichler P., Volz S.E., Joung J., van der Oost J., Regev A. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fonfara I., Richter H., Bratovič M., Le Rhun A., Charpentier E. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature. 2016;532:517–521. doi: 10.1038/nature17945. [DOI] [PubMed] [Google Scholar]
- 20.Kim D., Kim J., Hur J.K., Been K.W., Yoon S.H., Kim J.S. Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat. Biotechnol. 2016;34:863–868. doi: 10.1038/nbt.3609. [DOI] [PubMed] [Google Scholar]
- 21.Kleinstiver B.P., Tsai S.Q., Prew M.S., Nguyen N.T., Welch M.M., Lopez J.M., McCaw Z.R., Aryee M.J., Joung J.K. Genome-wide specificities of CRISPR-Cas Cpf1 nucleases in human cells. Nat. Biotechnol. 2016;34:869–874. doi: 10.1038/nbt.3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holderfield M., Deuker M.M., McCormick F., McMahon M. Targeting RAF kinases for cancer therapy: BRAF-mutated melanoma and beyond. Nat. Rev. Cancer. 2014;14:455–467. doi: 10.1038/nrc3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arcaini L., Zibellini S., Boveri E., Riboni R., Rattotti S., Varettoni M., Guerrera M.L., Lucioni M., Tenore A., Merli M. The BRAF V600E mutation in hairy cell leukemia and other mature B-cell neoplasms. Blood. 2012;119:188–191. doi: 10.1182/blood-2011-08-368209. [DOI] [PubMed] [Google Scholar]
- 24.Brastianos P.K., Taylor-Weiner A., Manley P.E., Jones R.T., Dias-Santagata D., Thorner A.R., Lawrence M.S., Rodriguez F.J., Bernardo L.A., Schubert L. Exome sequencing identifies BRAF mutations in papillary craniopharyngiomas. Nat. Genet. 2014;46:161–165. doi: 10.1038/ng.2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xing M. BRAF mutation in thyroid cancer. Endocr. Relat. Cancer. 2005;12:245–262. doi: 10.1677/erc.1.0978. [DOI] [PubMed] [Google Scholar]
- 26.Davies H., Bignell G.R., Cox C., Stephens P., Edkins S., Clegg S., Teague J., Woffendin H., Garnett M.J., Bottomley W. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 27.Hauschild A., Grob J.J., Demidov L.V., Jouary T., Gutzmer R., Millward M., Rutkowski P., Blank C.U., Miller W.H., Jr., Kaempgen E. Dabrafenib in BRAF-mutated metastatic melanoma: a multicentre, open-label, phase 3 randomised controlled trial. Lancet. 2012;380:358–365. doi: 10.1016/S0140-6736(12)60868-X. [DOI] [PubMed] [Google Scholar]
- 28.Bollag G., Tsai J., Zhang J., Zhang C., Ibrahim P., Nolop K., Hirth P. Vemurafenib: the first drug approved for BRAF-mutant cancer. Nat. Rev. Drug Discov. 2012;11:873–886. doi: 10.1038/nrd3847. [DOI] [PubMed] [Google Scholar]
- 29.Das Thakur M., Salangsang F., Landman A.S., Sellers W.R., Pryer N.K., Levesque M.P., Dummer R., McMahon M., Stuart D.D. Modelling vemurafenib resistance in melanoma reveals a strategy to forestall drug resistance. Nature. 2013;494:251–255. doi: 10.1038/nature11814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miao B., Ji Z., Tan L., Taylor M., Zhang J., Choi H.G., Frederick D.T., Kumar R., Wargo J.A., Flaherty K.T. EPHA2 is a mediator of vemurafenib resistance and a novel therapeutic target in melanoma. Cancer Discov. 2015;5:274–287. doi: 10.1158/2159-8290.CD-14-0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paraiso K.H., Haarberg H.E., Wood E., Rebecca V.W., Chen Y.A., Xiang Y., Ribas A., Lo R.S., Weber J.S., Sondak V.K. The HSP90 inhibitor XL888 overcomes BRAF inhibitor resistance mediated through diverse mechanisms. Clin. Cancer Res. 2012;18:2502–2514. doi: 10.1158/1078-0432.CCR-11-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gonzaga-Jauregui C., Lupski J.R., Gibbs R.A. Human genome sequencing in health and disease. Annu. Rev. Med. 2012;63:35–61. doi: 10.1146/annurev-med-051010-162644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vogelstein B., Papadopoulos N., Velculescu V.E., Zhou S., Diaz L.A., Jr., Kinzler K.W. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gebler C., Lohoff T., Paszkowski-Rogacz M., Mircetic J., Chakraborty D., Camgoz A., Hamann M.V., Theis M., Thiede C., Buchholz F. Inactivation of cancer mutations utilizing CRISPR/Cas9. J. Natl. Cancer Inst. 2016;109:djw183. doi: 10.1093/jnci/djw183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu X., Kriz A.J., Sharp P.A. Target specificity of the CRISPR-Cas9 system. Quant. Biol. 2014;2:59–70. doi: 10.1007/s40484-014-0030-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fu B.X., Hansen L.L., Artiles K.L., Nonet M.L., Fire A.Z. Landscape of target:guide homology effects on Cas9-mediated cleavage. Nucleic Acids Res. 2014;42:13778–13787. doi: 10.1093/nar/gku1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carroll D. Staying on target with CRISPR-Cas. Nat. Biotechnol. 2013;31:807–809. doi: 10.1038/nbt.2684. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.