Abstract
Müller glia, the most abundant glia of vertebrate retina, have an elaborate morphology characterized by a vertical stalk that spans the retina and branches in each retinal layer. Müller glia play diverse, critical roles in retinal homeostasis, which are presumably enabled by their complex anatomy. However, much remains unknown, particularly in mouse, about the anatomical arrangement of Müller cells and their arbors, and how these features arise in development. Here we use membrane-targeted fluorescent proteins to reveal the fine structure of mouse Müller arbors. We find sublayer-specific arbor specializations within the inner plexiform layer (IPL) that occur consistently at defined laminar locations. We then characterize Müller glia spatial patterning, revealing how individual cells collaborate to form a pan-retinal network. Müller cells, unlike neurons, are spread across the retina with homogenous density, and their arbor sizes change little with eccentricity. Using “Brainbow” methods to label neighboring cells in different colors, we find that Müller glia tile retinal space with minimal overlap. The shape of their arbors is irregular but non-random, suggesting local interactions between neighboring cells determine their territories. Finally, we identify a developmental window at postnatal days 6–9 when Müller arbors first colonize the synaptic layers beginning in stereotyped IPL sublaminae. Together, our study defines the anatomical arrangement of mouse Müller glia and their network in the radial and tangential planes of the retina, in development and adulthood. The local precision of Müller glia organization suggests that their morphology is sculpted by specific cell-cell interactions with neurons and each other.
Keywords: development, glia, mosaic, patterning, Glast-CreER, Brainbow, tiling, sox9, RRID: AB_2239761, RRID: AB_2534023, RRID: AB_2079751, RRID: AB_10000340, RRID: AB_90755, RRID: AB_303507, RRID: AB_393571, RRID: AB_2068506
INTRODUCTION
The mammalian retina is a stratified sheet of neural tissue comprising three layers of cell bodies separated by two layers of synapses. The neural circuits of the retina are organized in both the radial (vertical through the thickness of the retina) and tangential (horizontal, parallel to the retinal surface) planes. Radially, photoreceptors located in the outer nuclear layer (ONL) send information across layers to multiple downstream target neurons that act in parallel to process distinct aspects of the visual scene. These parallel circuits are then duplicated tangentially, across the extent of the retina, to build the visual field (Wassle and Boycott, 1991; Kay and Sanes, 2014). The organization of neurons in these two planes is accompanied by similar patterning of Müller glia, the most numerous glial cell type in the retina. These cells play a vital role in maintaining the health and integrity of the retinal tissue. Like protoplasmic astrocytes in the brain, Müller glia have diverse physiological functions including regulation of synapse development, neurovascular coupling, electrolyte balance, and cellular metabolism (Vecino et al., 2016). Müller glia also make important contributions to the structural integrity of the retina. Because of their crucial roles, Müller glia must be present in sufficient number at each retinotopic location, and in each layer, to support the function of the retinal circuitry at that position. However, much remains unknown about how individual Müller glia and the population as a whole are apportioned in the retinal volume. Understanding the anatomical arrangement of Müller glia in the adult retina, and how it arises during development, will provide important insight into the specific interactions between Müller glia and neurons that support visual function.
The morphology of Müller glia has been studied using Golgi impregnation and immunohistochemistry in a variety of vertebrate species, including rabbit, tree shrew, turtle, horse, rat, and mouse (Reichenbach and Wohlrab, 1983; Drager et al., 1984; Reichenbach and Wohlrab, 1986; Robinson and Dreher, 1990; Dreher et al., 1992). From these studies we know that Müller cells have a conserved bipolar morphology in the radial plane. Their somas reside at the center of the middle cell body layer, the inner nuclear layer (INL), from which radially oriented processes emerge to span the thickness of the neuroretina. As they traverse the retina, Müller glia adopt a distinct morphology at each retinal layer: 1) conically branching endfeet densely ensheathe neurons and blood vessels in the ganglion cell layer (GCL) and form the inner limiting membrane (ILM); 2) fine processes ramify in both synaptic layers, the inner and outer plexiform layers (IPL and OPL); 3) the vertical stalk divides to weave through the ONL, surrounding photoreceptor cell bodies and forming the outer limiting membrane (OLM); and 4) microvilli extend past the OLM to associate tightly with photoreceptor inner segments. The remarkable subcellular specialization of Müller glia across retinal layers presumably reflects the distinct functions they perform at each layer. For example, they subserve crucial structural roles at the limiting membranes, and in the plexiform layers they support synaptic function in a manner similar to brain protoplasmic astrocytes (Rasmussen, 1972; Reichenbach et al., 1989; Clarke and Barres, 2013; Reichenbach and Bringmann, 2013; Allen, 2014; Vecino et al., 2016). Given that the IPL can be further divided into sublaminae containing various types of synapses with different functions (Sanes and Zipursky, 2010), it is possible that Müller glia may also display sublaminar specializations that have previously not been appreciated.
In the tangential plane, Müller glia spatial patterning manifests in the retinotopic distribution of their cell bodies, the arborization of single cells, and the relationship between territories of neighboring cells. Cell body distribution has been studied in several species. In contrast to their conserved radial morphology, the density of Müller glia can vary by over six-fold depending on species and retinal location (Chao et al., 1997). In rabbit, for example, Müller glia are most dense along the visual streak in central retina where neurons are most numerous, and their density declines by threefold along the vertical meridian into the periphery (Robinson and Dreher, 1990). This finding supports the intuitively appealing idea that the number of glia is determined by the number of neurons they support. However, whether such a rule is universal or is limited to rabbit remains unexplored.
Compared to the arrangement of Müller glia cell bodies, we know much less about the fine-scale pattern of Müller arbors in the tangential plane. One reason for this is that Golgi studies are better suited to revealing single-cell anatomy than the cell-cell interactions that build Müller glia networks. Further, immunohistochemistry against common glial markers, such as glutamine synthetase and glial fibrillary acidic protein (GFAP), is a poor tool for revealing fine lateral processes. Because GFAP staining only labels ~15% of the volume of protoplasmic astrocytes (Bushong et al., 2002), there is reason to suspect that Müller glia morphology, particularly in the plexiform layers, has not been fully appreciated by staining for typical markers. Even with these limitations, the anatomical data to date suggest that Müller glia arbors form a continuous network filling each retinal layer (Reichenbach et al., 1989), as might be expected given the crucial functions of Müller glia and the potential deleterious consequences of leaving parts of the retina bereft of glia. How this network forms, and how individual cells contribute to it, is unclear. Brain astrocytes exhibit a phenomenon called “tiling” – they fill the neuropil with exclusive, minimally-overlapping arbors that create synaptic domains governed by single astrocytes (Bushong et al., 2002; Ogata and Kosaka, 2002; Halassa et al., 2007; Livet et al., 2007). Some types of retinal neurons also tile (Wassle et al., 1981), but whether Müller glia coordinate their territories in mammalian retina by a similar mechanism is not known.
Notably lacking from past Müller glia comparative anatomy studies is the mouse, an increasingly important model system for visual neurobiology (Huberman and Niell, 2011). Two previous studies have used vimentin immunohistochemistry to label mouse Müller glia (Drager et al., 1984; Chao et al., 1997), but the radial morphology of Müller glia has not been examined by Golgi or other cell-filling techniques, and the tangential patterning of cell bodies and arbors have not been investigated. Here we employ the molecular-genetic tools available in mouse to reveal the morphology of single Müller glia and the full Müller population, in adults and across development. We find that mouse Müller glia are distributed homogeneously across the retina, despite center-to-periphery differences in neuron numbers (Jeon et al., 1998). The arbors of neighboring Müller cells tile the synaptic layers, the cell body layers, and the OLM, creating a gap-free glial network that efficiently covers visual space. In the radial plane, mouse Müller glial morphology is similar to that of other species. However, by using membrane-targeted fluorophores to reveal fine details of Müller glia anatomy, we found that arbor morphology varies across IPL sublayers in a systematic manner. Finally, we document how these radial and tangential anatomical features arise in development. We show that Müller branching begins in the synaptic layers, and indeed in specific IPL sublayers that correspond to the layers with highest branch density in adults. Together, our results reveal new anatomical features of the Müller glia network, and highlight important ways in which mouse Müller glia are similar or different to those of other species. Furthermore, our observations suggest that local and highly specific cell-cell interactions, both amongst Müller glia themselves and with particular types of neurons, coordinate to guide morphological maturation of Müller glia in three dimensions.
MATERIALS AND METHODS
Mice
Juvenile and adult (postnatal days 35–50) mice of both sexes were used in this study. Wildtype C57BL/6 were purchased from Jackson Laboratory. GLAST-CreER and Rosa26mTmG on mixed C57BL/6 backgrounds were obtained from Jackson Laboratory (strains 012586, 007576). This study was performed with the approval of the Duke University IACUC.
GLAST-CreER mice express a Cre recombinase-estrogen receptor fusion protein (CreER) under control of a glia-specific promoter. The mTmG mouse strain expresses membrane-associated green fluorescent protein (GFP) in a Cre-dependent manner. To induce CreER-mediated recombination, GLAST-CreER; mTmG mice were injected with the estrogen receptor ligand tamoxifen (TMX; Sigma-Aldrich). TMX was dissolved in corn oil through sonicating at room temperature for 30 min to make a 20 mg/mL solution. Postnatal day (P) 5 mice were injected intraperitoneally with 100 µg of TMX for early Müller glia labeling, and P22 mice were injected with 100 mg/kg TMX either once or on three consecutive days to label mature Müller glia sparsely or densely, respectively.
Antibodies
| Antibody Name | Antigen | Antibody info | Dilution |
|---|---|---|---|
| Sox9 | KLH-conjugated peptide VPSIPQTHSPQHWEQPVYTQLTRP from human Sox9 | Millipore (AB5535) Rabbit polyclonal RRID: AB_2239761 | 1:4,000 |
| RNA-binding Protein with Multiple Splicing (RBPMS) | RBPMS 4–24: GGKAEKENTPSEANLQEEEVRCKLH | Rodriguez et al. (2014) Guinea pig polyclonal | 1:1,000 |
| GFP | GFP isolated directly from the jellyfish Aequorea victoria | Life Technologies (A10262) Chicken polyclonal RRID: AB_2534023 | 1:1,000 |
| mCherry | Recombinant His-tagged mCherry | Cai et al. (2013) Rabbit polyclonal | 1:3,000 |
| mKate | Recombinant His-tagged mKate2 | Cai et al. (2013) Guinea pig polyclonal | 1:1000 |
| Choline Acetyltransferase (ChAT) | Human placental enzyme | Millipore (AB144P) Goat polyclonal RRID:AB_2079751 | 1:400 |
| Calbindin | recombinant rat calbindin D-28k | Swant (CB38) Rabbit polyclonal RRID: AB_10000340 | 1:10,000 |
| Tyrosine Hydroxylase (TH) | Native tyrosine hydroxylase from rat pheochromocytoma | Millipore (AB1542) Sheep polyclonal RRID: AB_90755 | 1:3,000 |
| Protein Kinase C (PKC) | Purified bovine brain protein kinase C | Abcam (Ab31) Mouse monoclonal, clone MC5 RRID: AB_303507 | 1:500 |
| CD31 | 129/Sv mouse-derived endothelioma cell line tEnd.1 | BD Biosciences (550274) Rat monoclonal, clone MEC13.3 RRID: AB_393571 | 1:100 |
| Calretinin | Recombinant rat calretinin | Millipore (AB5054) Rabbit polyclonal RRID: AB_2068506 | 1:2,000 |
Antibody Characterization
Antibodies against endogenous proteins were used as markers of cell types and retinal lamina, and revealed cellular morphology and lamina specificity consistent with the literature. Sox9 antibody (Millipore AB5535, RRID: AB_2239761) labeled Müller glial nuclei, as previously reported (Poche et al., 2008). RBPMS immunolabeling was observed in RGCs, as previously reported for this antiserum (Rodriguez et al., 2014). ChAT antibody (Millipore AB144P, RRID: AB_2079751) recognized cholinergic amacrine neurons and their projections to the IPL as previously reported (Haverkamp and Wassle, 2000; Voinescu et al., 2009). Calbindin antibody (Swant CB38, RRID: AB_10000340) immunolabeled three IPL sublayers, including the two cholinergic sublayers, as previously reported (Haverkamp and Wassle, 2000). (TH antibody (Millipore AB1542, RRID: AB_90755) labeled amacrine cells with large somata and projections to a single stratum of the IPL, a pattern consistent with this and other TH antibodies (Voinescu et al., 2009). PKC antibody (Clone MC5, Abcam Ab31, RRID: AB_303507) labeled rod bipolar cells and their terminals in the IPL, as previously reported for this monoclonal antibody (Haverkamp et al., 2003). CD31 (clone MEC13.3, BD Biosciences 550274, RRID: AB_393571) labeled endothelial cells, as previously reported (Gnanaguru et al., 2013). Calretinin antibodies (Millipore AB5054, RRID: AB_2068506) labeled three IPL sublaminae, including the cholinergic sublayers and one layer between them, as previously reported (Haverkamp and Wassle, 2000).
For antibodies against exogenous fluorescent proteins (i.e. anti-GFP, anti-mCherry, and anti-mKate), specificity was confirmed by lack of signal in retinas not expressing those transgenes (data not shown).
Immunohistochemistry
Mice were deeply anesthetized with isoflurane, decapitated, eyes rapidly removed, and immersion fixed in 4% paraformaldehyde for 1.5 hours at 4°C. For flat-mount experiments, retinas were then dissected free of the eye, blocked for 2 hours at room temperature in PBS with 0.03% Triton X-100 (Sigma-Aldrich) and 3% Normal Donkey Serum (Jackson Immunoresearch). In a subset of experiments, retinal orientation was identified by marking the right retina with a hole made by a hypodermic needle in the superior retina. Retinas were then incubated with primary antibodies in the same blocking solution at 4°C for 5–7 days, washed 3–4 times with PBS, and then incubated with 1:1000 Donkey secondary antibodies (Jackson Immunoresearch) for 1 day. Retinas were then flat mounted by making 4 relieving cuts from the retinal periphery approximately two thirds of the way to the optic nerve head, mounted on cellulose membrane filters (Millipore), and coverslipped with Fluoromount-G (Southern Biotech).
For cross-section experiments, eyes were fixed on ice for 90 min in 4% paraformaldehyde in PBS. After fixation, the cornea was cut along the ora serrata to remove the anterior portion of the eye. The optic cup was then incubated in 30% sucrose in PBS for 1 hour and flash frozen in Tissue Freezing Medium (Triangle Biomedical Sciences). The frozen blocks were sectioned at 20 µm with a cryostat at 25°C. Sections were dried at room temperature and washed in PBS to dissolve the freezing medium. Washed sections were incubated with blocking solution at room temperature for 30 minutes. Sections were incubated with primary antibodies diluted in the blocking solution overnight at 4°C. Sections were then washed with PBS 3–4 times, before incubation with secondary antibodies for 2 hours at room temperature. After washing for 3–4 times, the sections were coverslipped with Fluoromount-G.
Intravitreal Virus Injections and Brainbow labeling
Adeno-associated virus (AAV) injection techniques were adapted from Cai et al. (2013). The two Brainbow AAV9 viruses, encoding farnesylated fluorescent proteins that are targeted to the plasma membrane (UPenn Vector Core), were mixed to 7.5 × 1012 genome copies per mL. Adult mice (P40-P50) were anesthetized with ketamine-xylazine by intraperitoneal injection. Propraracaine hydrochloride (0.5%) ophthalmic solution (Akorn) was applied to the eye to provide local anesthesia. A 30½ G needle was used to make an opening and 1µl of virus was injected with a 33 G blunt-ended needle intravitreally. Beginning a week after virus administration, 100 mg/kg TMX (prepared as above) was injected for 5 consecutive days. Tissue was collected 28 days after the last TMX injection.
The two Brainbow viruses produce stochastic Cre-dependent expression of the following fluorescent proteins: TagBFP, YFP, TFP, and mCherry (Cai et al., 2013). To amplify the endogenous fluorescent signals, we immunostained for the four fluorescent proteins using three antibodies: 1) anti-GFP, which recognizes YFP and TFP; 2) anti-mKate, which recognizes TagBFP; 3) anti-mCherry. The tissue was then imaged in three channels on the confocal microscope.
Confocal Imaging
Immunostained retinas were images on Olympus FV300 or Nikon A1 confocal laser scanning microscopes. For experiments with reported retinal eccentricities, a motorized translational stage (Prior ProScan III) integrated with the Nikon A1 microscope was used to record coordinates of the ONH and each field of view in order to calculate distance from the ONH.
Image Analysis
Images were analyzed in FIJI/ImageJ (NIH). The Plot Profile function was used to quantify laminar distributions of Müller glia and cell-type markers in maximum intensity projections of Z-stacks, with rectangular regions of interest (ROIs) placed around single or small clusters of GFP-positive Müller glia. Fluorescence intensity was then normalized to maximum and IPL depth normalized by percentage from GCL to INL. For RGC densities, RBPMS-positive cells were counted manually in maximum intensity Z-projections through the GCL. For Müller glia densities, Sox9-positive cells were counted in a semi-automated manner in Z-projections through the INL. Images were thresholded, and closely packed nuclei separated with the Watershed function. The Analyze Particles function was then used with a minimum size of 10 µm to count cells. Parameters for cell-counting were calibrated by comparison to a subset of manually counted images. Müller glia arbor sizes were measured in Z-projections through the OPL and IPL with the Polygonal Selection tool. ChAT immunostaining was used as a landmark to guide acquisition of IPL Z-stacks. Z-projections for quantification of IPL arbors were made through and including both ChAT-positive layers. For quantification of developmental IPL branching, cells were manually graded in a binary fashion as having or not having branches in each IPL sublamina, using calbindin immunoreactive strata as landmarks to divide the IPL into S1-S5 zones.
Density recovery profiles were calculated as described (Kay et al., 2012) from 60× images of Sox9-labeled retinal whole-mounts. Simulated cell arrays were generated using a custom MATLAB script that places cells into a two-dimensional region of user-defined size. Cell diameter and density can also be specified by the user. Cells are constrained from occupying the same location. The script is available at www.sites.duke.edu/kaylab.
Semi-Automated Chromatic Segmentation of Brainbow-labeled Cells
Cells infected with Brainbow viruses express GFP, mKate, and mCherry immunoreactivity in stochastically determined ratios. This produces variation in color between cells that can be used to delineate cell boundaries. Because we immunostained for the four virally encoded fluorescent proteins with three primary antibodies and imaged cells in three channels, cells should exhibit six fundamental colors: red, green, blue, magenta (red + blue), cyan (green + blue) and yellow (red + green). However, due to variations in infection count, transgene expression, and noise, the pixel values belonging to imaged cells occupy a range of colors around each fundamental color. We developed an algorithm to exploit this cell-to-cell variation by dividing the RGB color spectrum into user-specified ranges in order to segment pixels into cells by color. This algorithm exhibits flexibility for manual intervention and can benefit from parallel processing for memory and speed optimized implementation.
In order to reliably divide the color spectrum into the fundamental colors and their variations, our algorithm transforms the color model of Brainbow images from RGB to the Hue-Saturation-Intensity (HSI) model (Gonzalez and Woods, 2008). HSI represents color space in cylindrical coordinates, with hue the angular coordinate, saturation the radial coordinate, and intensity the axial coordinate. Using all three coordinates could conceivably best detect cell boundaries, but creates an excessively large search space since all of these parameters vary independently over all pixels. Since hue is the parameter that fundamentally represents different colors conferred by the different fluorophores, we used only the hue spectrum of the HSI color model to segment arbor territories.
The segmentation process begins by computing a normalized histogram of the hue channel of the image. The histogram reveals how many pixels have each hue value. Depending on which fluorophores are expressed by a given cell and at what levels, all pixels in one cell should spread around a specific fundamental hue value. The next step consists of providing a lower and upper bound to each fundamental color in the hue histogram to divide the spectrum into sections. Initially, the bounds are automatically generated based on the valley locations around each peak in the spectrum. If needed, to optimize segmentation accuracy, the user then has the option to adjust the position and width of the range of hues. After all relevant cells are segmented, cell area and overlapping area are quantified and segmentations are exported in any common image format (example: tif, png, jpeg) for further analysis.
The above algorithm was incorporated in an open-source software named “SegThresh” with an intuitive graphical user interface using MATLAB 2015a. To encourage further research in this area, the SegThresh software developed for this project is made publically available at http://www.duke.edu/~sf59/Wang_2016.htm.
Analysis of Müller Glia Territories with SegThresh
To analyze the overlap of Müller glia territories in the synaptic layers, three consecutive optical slices from 60× Z-stacks spaced 0.4µm apart were exported from Fiji as PNG files. All three files were then loaded into SegThesh and the middle one was designated the “base image” for segmentation. The boundaries created on the “base image” were then superimposed onto the upper and lower images to verify that segmentation accurately delineates cells at all three depths. In rare cases, when the program could not create the precise outlines for cells with long processes, boundaries were manually edited. Typically, ~40 cells were segmented from a single field of view of mid-peripheral retina. Müller cell territory size and overlap were quantified in a pairwise manner among cells that were clearly adjacent or touching. To be certain that the results were not skewed by selection of cells that did not actually touch, cell pairs were excluded from the analysis if their overlap was calculated to be zero. Percentage of overlap was measured by dividing the area of overlap by the overall area of the cell pair.
We tested the performance of the SegThresh algorithm through two control experiments. First, to make sure SegThresh accurately defines the perimeters of individual cells, we compared the territory areas of individual cells and overlap between pairs of cells generated by SegThresh to values measured from manually segmented cells and found no significant difference between the two (two-tailed t-test; arbor size n = 10 cells per group, p=0.21; overlap n = 10 cell pairs per group, p=0.63). Second, to ensure that SegThresh is capable of detecting a range of overlap values, we artificially created images with varying degrees of overlap. Cells were manually segmented in Adobe Illustrator and artificially superimposed onto one another. In test images with large degrees of overlap (n = 3), SegThresh could still segment the cells.
Generation and Analysis of Spatially Randomized Cell Territories
To test whether the local shape of cell territories affects coverage and overlap, we compared pairs of cells in real images to cell pairs obtained from images in which the cells were reflected along their horizontal axis. A subset of overlapping cell pairs was arbitrarily selected, and segmented outlines exported to Adobe Illustrator in .TIF format. The outlines were then flipped about the horizontal axis, preserving their relative horizontal positions. Only cell pairs that had measurable overlap both before and after flipping were included in the analysis. Overlapping area in both the real and the flipped images was then outlined with the freehand selection tool in ImageJ and the area measured.
Statistical Analysis
Descriptive statistics are reported mean ± standard error. All statistical analyses were performed in JMP 12 (SAS Institute).
RESULTS
Radial Morphology of Individual Müller Glia across Retinal Layers
We first sought to describe the cellular morphology of Müller glia in mouse retina. We reasoned that a membrane-targeted fluorescent protein might provide improved labeling of fine glial processes relative to immunohistochemical or cytosolic fluorescent markers used previously (Yang et al., 2011). We therefore expressed membrane-targeted GFP (mGFP) selectively in Müller glia by crossing GLAST-CreER mice to mTmG Cre-conditional reporter mice (Muzumdar et al., 2007), adjusting the dose of tamoxifen to drive CreER-mediated recombination in varying numbers of Müller glia. This labeling strategy succeeded in revealing fine details of Müller glia morphology. In retinas with extensive recombination, we observed a confluent network of Müller arbors that fills the volume of the retina in each layer (Fig. 1D,E). In retinas with sparse recombination, the morphology of individual Müller glia could be clearly visualized, from the thick vertical stalk to small branches and microvilli (Fig. 1B).
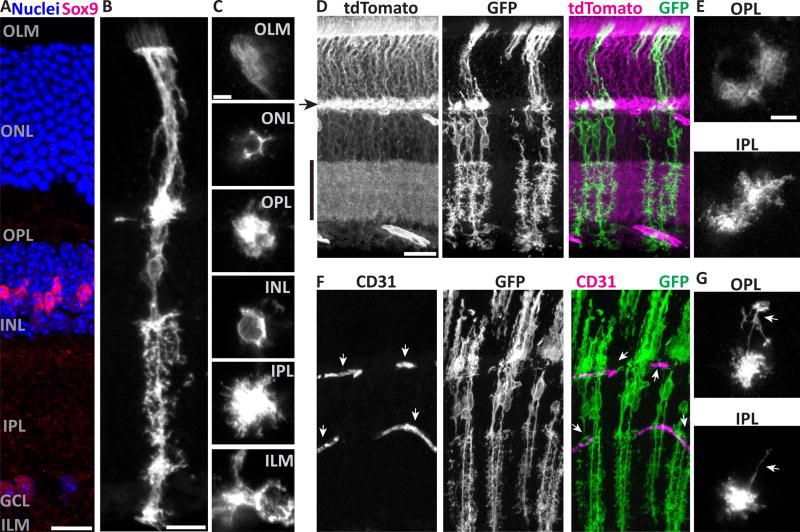
Figure 1. Müller cells have radially-oriented bipolar stalks with layer-specific branching morphology.
A) Cross-section of adult mouse retina stained with Sox9 to show Müller glia (MG) nuclei in INL, and counterstained with nuclear marker (Hoechst) to reveal major retinal layers. Text labels indicate cell body layers (GCL, INL, ONL), synaptic layers (IPL, OPL), and approximate location of limiting membranes (ILM, OLM). B,C) Morphology of individual mGFP-expressing MG from GLAST-CreER; mTmG mice, viewed in cross-section (B) or en face (C). C depicts the same cell imaged at different planes of a flat mount. Image in B is scaled to approximately match layers in A. Note morphological specializations at each layer: OLM, microvilli; ONL, processes intercalated between photoreceptor cell bodies; OPL and IPL, extensive fine branches; INL, MG cell soma; ILM, broad branches and endfeet. D,E) Retinas with dense MG labeling, showing confluence of MG arbors in synaptic layers and limiting membranes. D: Cross-section view; tdTomato fluorescence from unrecombined mTmG cells (left) counterstains synaptic layers (arrowhead, OPL; vertical bar, IPL). E: Flat mount en face view, showing confluent arbors of neighboring MG. F,G) MG branches are closely associated with CD31+ blood vessels (F, arrows). MG often have long horizontal branches (G, arrows) in the plexiform layers which can terminate on blood vessels. Scale bars (in µm): 15 (A,B), 5 (C), 25 (D,F), 10 (E,G).
To document how Müller glia morphology varies across retinal layers, we examined single cells in cross-sections, and by tracking them through Z-stacks spanning the entire thickness of retinal whole-mounts (Fig. 1B–C). We observed distinct morphologies at synaptic layers, cell body layers, and limiting membranes. First, in the synaptic layers (OPL and IPL), the central Müller stalks give rise to extensive, fine, bushy processes that encompass a volume of neuropil, reminiscent of brain protoplasmic astrocytes. While Müller glia territories in these layers surround the vertical stalk, they are not uniformly cylindrical (Fig. 1C). Indeed, individual processes were observed to protrude laterally, sometimes long distances, from the space-filling core of the Müller glia arbor (Fig. 1B,G). The longest side branches, which have been termed “horizontal fibers” in other species (Kreutzberg and Hussain, 1982), were frequently observed to terminate on blood vessels with endfoot-like structures (Fig. 1G). Second, in the cell body layers, Müller glia ensheath neuronal cell bodies. This is most obvious in the ONL, where Müller glia fill the space between photoreceptors, highlighting the neuronal cell bodies as lacunae surrounded by mGFP-labeled processes (Fig. 1B,C). Individual Müller glia sometimes completely surround one or more photoreceptors, but more often their arbors form an incomplete polygon around photoreceptor cell bodies (Fig. 1C), implying that adjacent Müller glia can cooperate to provide a photoreceptor with its glial sheath. Finally, at the limiting membranes, Müller glia arbors extend laterally from the central stalk, as in the synaptic layers, but with a distinct morphology. ILM branches can be broader than plexiform branches, and do not fill space in the way that plexiform branches do. Further, ILM branches are often accompanied by endfeet (Fig. 1C) that contact blood vessels (not shown). OLM branches remain confined to the outlines of photoreceptors. They are accompanied by microvilli that extend past the OLM; in other species these contact photoreceptor inner segments, and the morphology we observed in mouse is consistent with this as well (Fig. 1B,C). Together, our single-cell observations of mGFP-labeled cells reveal the trans-laminar fine structure of mouse Müller glia, highlighting morphological specializations that likely mediate their interactions with blood vessels, neurons, and each other.
Precise Sublaminar Organization of Müller Glia Arbors within the IPL
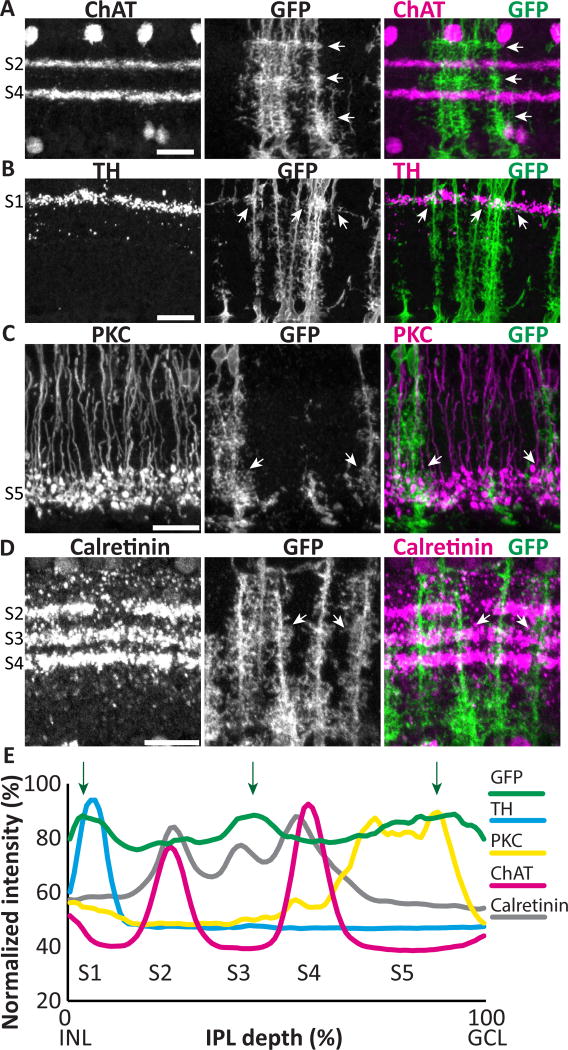
A prominent feature of retinal organization in the radial plane is the stratification of the IPL into sublaminae that contain the projections of distinct classes of neurons (Masland, 2001; Sanes and Zipursky, 2010). Given that individual Müller glia generate specialized morphologies across retinal layers, we wondered whether they would also show specialization at the sublaminar level. Indeed, it has long been known that Müller arbors are not uniform across the IPL, and in some species arbors appear to differ in a sublaminar fashion (Ramon y Cajal, 1972), but whether arbor variability occurs consistently in particular IPL sublaminae has not been examined. To address this question we assessed Müller arbor morphology in retinal cross-sections. While Müller glia formed a confluent network at all sublaminar positions, the density of their arbors appeared greater at the center and the edges of the IPL (Fig. 2A). Quantification of fluorescence intensity across the IPL confirmed this observation, as three small peaks were found at consistent IPL levels across animals (Fig. 2E). To ask whether these dense regions correspond to identified IPL strata, we co-stained for well-characterized sublaminar markers. First, we compared the density of mGFP-labeled Müller processes to the ChAT-positive starburst amacrine cell dendrites in sublaminae (S) 2 and S4. We found that the Müller glia-dense regions are located above, between, and below the ChAT layers, indicating that they lie in IPL regions commonly denoted as S1, S3, and S5 (Haverkamp and Wassle, 2000) (Fig. 2A). Therefore, we next co-labeled with markers of particular neuron subtypes that project to these regions. The dopaminergic amacrine cells, labeled with anti-TH antibodies, occupy a single stratum in S1. Double staining for TH and mGFP revealed that the Müller glia-dense S1 region is located in this sublayer (Fig. 2B). Similarly, the glia-dense region in S3 matches the sublayer defined by the arbors of calretinin-positive neurons (Fig. 2D). Finally, by labeling with antibodies against PKC that mark rod bipolar cell terminals, we found that the S5 Müller glia-dense region overlaps with rod bipolar projections (Fig 2C). These results indicate that mouse Müller processes do show IPL sublaminar specializations – while they branch throughout the IPL, they exhibit a small but consistent preference for branching more extensively in certain IPL sublamina (Fig. 2E).
Figure 2. Müller glia branching patterns differ across IPL sublaminae.
Adult GLAST-CreER; mTmG retinal cross-sections co-stained for IPL sublaminar markers and anti- GFP to reveal MG. A) ChAT staining divides the IPL into 2 ChAT-positive sublayers (S2, S4, labeled at left) and 3 ChAT-negative layers (S1, S3, S5). mGFP-labeled MG processes have heterogeneous density across the IPL, with highest density in S1, S3, and S5 (arrows). B–D) Dense MG arbors in S1, S3, and S5 co-localize with distinct classes of neurons: dendrites of TH-positive dopaminergic amacrine cells in S1 (B); axon terminals of PKC-positive rod bipolar cells in S5 (C); and dendrites of calretinin expressing neurons in S3 (D). Arrows show sites of colocalization. E) Quantification of fluorescence intensity by IPL depth for MG (GFP) (n = 31 measurements) and sublaminar neuronal markers. Müller branches are abundant across the entire IPL, but consistently show peaks in density in S1, S3, and S5 (arrows), which align with sublaminar marker peaks. Scale bars: 20 µm.
Spatial Organization of Müller Glia Cell Bodies in Mouse Retina
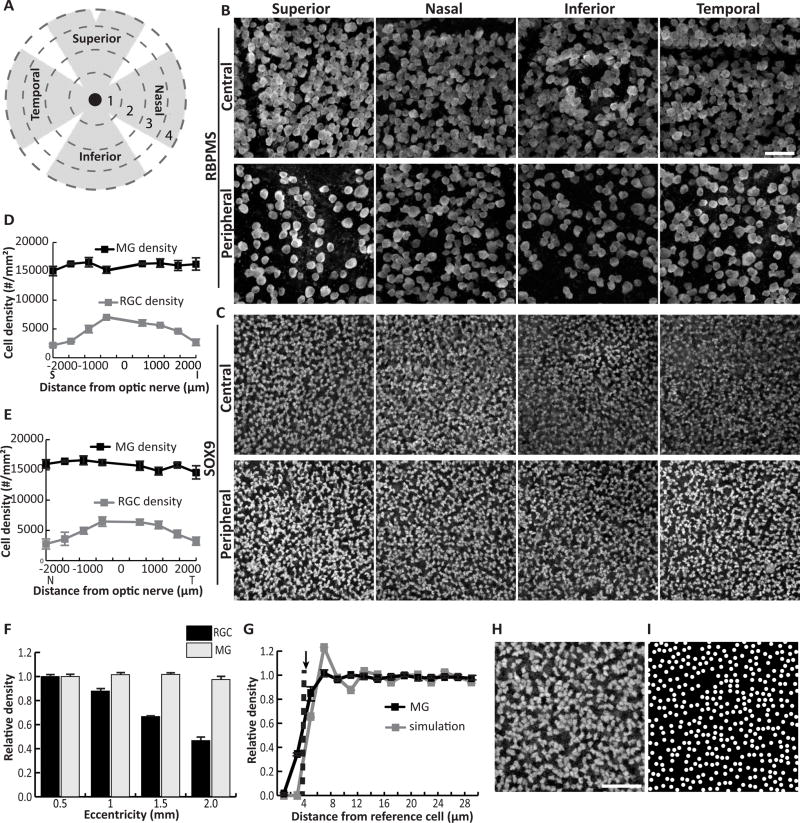
After detailing the anatomy of Müller cells along the radial axis of the retina, we next examined how the Müller population is distributed in the tangential plane. Since many functions of Müller glia are posited to involve support of neurons, we began by asking if retinal regions with more neurons might also have more Müller glia. In previous studies, the density of mouse retinal ganglion cells (RGCs) was found to be highest in central retina and to decline with eccentricity, a pattern that holds true for most other retinal neurons including photoreceptors (Jeon et al., 1998; Ortin-Martinez et al., 2014; Rodriguez et al., 2014). To ask if Müller glia also follow this pattern, we examined the spatial distribution of RGCs and Müller glia in young adult C57Bl/6 mouse retinas. Tissue was double immunostained for Sox9, a transcription factor that specifically marks Müller cell nuclei in the INL, and the RGC marker RBPMS (Rodriguez et al., 2014). Cells expressing each marker were counted in confocal images acquired from flat-mount retinas at four eccentricities (500, 1000, 1500, and 2000 µm from the optic nerve head (ONH)) in each quadrant of the retina (Fig. 3A–C). As expected, RGC density declined with eccentricity in all retinal quadrants, with the most dramatic decline in the superior quadrant (Fig. 3D–F). Significant main effects of eccentricity (F(3,32) = 119.02, p < 0.0001) and quadrant (F(3,32) = 4.60, p = 0.0087), and also a significant interaction (F(9, 32) = 6.04, p < 0.0001), were found for RGCs. By contrast, the density of Sox9-positive Müller glia was not affected by retinal location (eccentricity, F(3,32) = 1.89, p = 0.15; quadrant, F(3,32) = 1.20, p = 0.32; eccentricity × quadrant, F(9,32) = 1.09, p = 0.40) (Fig. 3D–F). The grand average Müller cell density was 15,890 cells/mm2, a value that did not differ significantly from any of the measured local densities (analysis of means, all pairwise comparisons p > 0.05) (Fig. 1D). We conclude that Müller glia are not sensitive to the local density of neurons, but rather are spread homogenously across the retina.
Figure 3. Müller cell bodies are arrayed across the mouse retina at even density.
A) Schematic indicating division of flat mounted retina into 4 quadrants and 4 eccentricities marked by dashed circles of expanding radii. B,C) Confocal images of RBPMS-stained RGCs in the GCL (B) and Sox9-stained MG in the INL (C) were taken at each eccentricity in each quadrant for 3 adult mouse retinas. Images from central (region 1) and peripheral (region 4) retina are shown. D,E) Density of RGCs and MG plotted against retinal eccentricity along vertical (D) and horizontal (E) meridians. F) Normalized density of MG and RGCs with eccentricity. MG density does not vary with distance from the optic nerve head, whereas RGC density in the far periphery is approximately half that of central retina. See Results for statistical analyses. G) Density recovery profile (DRP) for MG cell bodies (black solid line).. The radius of the MG exclusion zone (dashed line) closely matches the average minimum MG cell diameter (arrow). Gray line, DRP of simulated random arrays (n = 9) matched in cell size to minimum MG diameter and in density to MG array. The real and simulated DRPs are well matched. H) High-power view of MG array showing regions of tightly-packed interlocking cells separated by small gaps. I) A simulated random array, like those used to calculate DRP in (G), shows a cell distribution highly similar to the real MG array.. Scale bar: 50 µm (B,C), 40 µm (H,I).
To gain further insight into the rules governing Müller soma spatial organization, we examined their distribution on a finer spatial scale. Many retinal cell types are organized in so-called “mosaics”; that is, cell bodies are non-randomly placed in an orderly array to maximize their distance from all neighbors of the same class. Mosaics likely serve to distribute cell types across the retina for full visual field coverage (Kay and Sanes, 2014; Reese and Keeley, 2015). If a Müller glia mosaic exists, it should be possible to measure a so-called “exclusion zone,” an area around each cell where another of the same type is rarely found, presumably due to local cell-cell repulsion. By contrast, if the Müller glia array does not comprise a mosaic – that is, if cells are positioned randomly – no exclusion zone will be evident. To test whether Müller glia exhibit exclusion zones, we used the density recovery profile (DRP), a plot of cell densities within annuli of expanding radius centered on each cell in an array (Fig. 3G). The exclusion zone (also known as the effective radius of the DRP) is measured as the radius at which cell density remains below the average density for the entire population (Rodieck, 1991; Kay et al., 2012). This was calculated to be 3.86 ± 0.18 µm for Müller glia (n = 9 measurements from 3 mice). Because two cells cannot occupy the exact same physical location, the exclusion zone for a random array of cells approximates the cell diameter (Rodieck, 1991). For a nonrandom mosaic, by contrast, the exclusion zone will be larger. To ask how well the size of a Müller cell matches the DRP exclusion zone, we took into account that Müller somata have complex polygonal shapes (Fig. 3H), unlike neurons which tend to be spherical. Accordingly, we measured for each cell the maximum and minimum diameters – that is, the longest and shortest line segment traversing the cell (n = 60 cells from 3 mice). We found that the Müller glia exclusion zone was similar in size to the minimum cell diameter (4.46 ± 0.34 µm; max diameter = 6.59 ± 0.48 µm; Fig. 3G). This finding suggests that Müller cells do not form a mosaic, and that physical contact with a neighboring cell is the only limitation on Müller glia positioning.
To further test the idea that Müller cell arrangement is random, we asked whether a simulated random cell array could successfully model the real Müller glial array. We used a simple model in which circular cells matching the minimum diameter of Müller glia were placed at random positions, at a density matching the observed Müller glial density (Fig. 3D,E). Qualitatively, these random simulations resembled the observed cell distribution, with regions of closely-packed cells separated by larger gaps (Fig. 3H–I). Their DRP was also well matched to the observed DRP (Fig. 3G), suggesting that the random model captures key aspects of the actual cell distribution. By contrast, when cell diameter was set to the Müller glial maximum diameter, the simulated DRP curve was less tightly aligned to the Müller cell DRP curve (not shown). We conclude that the position of Müller cells is likely to be random, with the minimum cell diameter providing the major constraint on cell positioning. At low cell density, such a patterning rule could lead to a patchy array of cells that inefficiently covers retinal territory. But we observed that Müller glia density was high enough that most cells are in contact with their neighbors (Fig. 3C,H). While some gaps in the soma array are evident, we noticed that in many retinal regions the polygonal cell bodies often fit together in complementary patterns, leaving little space between cells (Fig. 3H). This observation suggests that while Müller glia are positioned randomly, some degree of order may be imposed on the soma array by the physical constraints of passive cell crowding.
Tiling of Müller Glia Arbors in Retinal Synaptic Layers
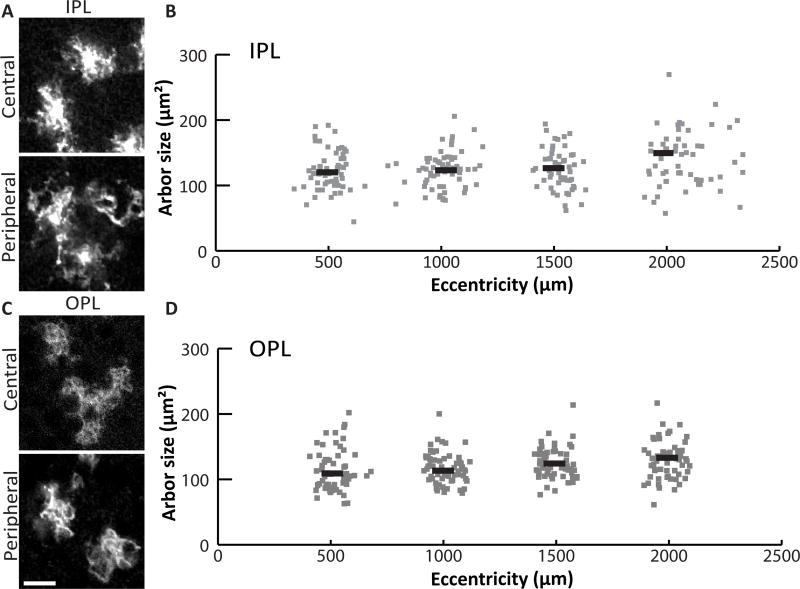
Given that Müller arbors form a confluent network in all retinal layers (Fig 1D,E), we next set out to understand how individual glial cells collaborate to cover retinal space in the tangential plane. One possibility is that, like brain astrocytes, each Müller cell covers a unique territory that minimally overlaps its neighbors’ – the pattern known as tiling. Alternatively, Müller glia might form an overlapping network, which could take several forms. For instance, their arbors might overlap at a constant rate, or vary in density to match regional differences in neuron density (Fig. 3) (Jeon et al., 1998). It is even possible that multiple Müller glia subtypes tile amongst themselves, but overlap densely with each other. To begin distinguishing among these possibilities, we measured the territory occupied by individual Müller arbors in retinal whole-mounts from sparsely-recombined GLAST-CreER; mTmG mice. We focused for this analysis on the plexiform layers, where the absence of cell bodies simplifies the task of measuring a single glial cell’s territory. We imaged mGFP+ Müller arbors in the plexiform layers and measured each cell’s distance from the ONH (Fig. 4). Even from qualitative observations (Fig. 4A,C), it was readily apparent that Müller glia do not show dramatic cell-to-cell differences in arbor size or morphology, arguing against the notion of multiple Müller glia cell types. Nor did we find substantial differences in arbor size across the retina. While we did detect an effect of eccentricity on Müller glia territories in IPL and OPL, the effect size was small (one-way ANOVA: IPL, F(3,226) = 3.61, p = 0.014; OPL, F(3, 234) = 4.01, p = 0.008. 95% confidence intervals: IPL, center 113.75 – 131.12 µm2, peripheral 131.45 – 148.81 µm2; OPL, center 106.79 – 120.62 µm2, peripheral 122.61 – 136.56 µm2) (Fig. 4B,D). Furthermore, none of the local area measurements differed significantly from the overall mean arbor area, indicating that arbor size differences, if any, are quite minor (ANOM, alpha 0.05). Given that Müller cell density is also consistent across the retina (Fig. 3), these results militate against models in which overlap is influenced by neuron density or other factors that vary between retinal regions.
Figure 4. The size of Müller glia territories is similar across the retina.
A) Arbors of sparsely labeled MG in the IPL of GLAST-CreER; mTmG flat mounted retina. B) MG arbor size was measured from Z-projections of confocal slices spanning IPL sublaminae S2-S4. Sizes are plotted against retinal eccentricity. Grey points are single cells; black lines represent the mean for 500 µm bins. C,D) MG arbors in the OPL were measured in the same way, using Z-projections spanning the entire OPL. Scale bar: 10 µm.
Since Müller cell density and arbor shape/size are all fairly uniform, our results suggest that arbor overlap is also likely to be relatively uniform – at least at a global level. But the single-cell analysis cannot rule out local variability in overlap rate. We therefore sought a method to directly measure overlap between adjacent cells. With single color labeling, overlap cannot be measured because of the difficulty in distinguishing where one cell ends and the next begins. Therefore, we decided to label Müller glia with multiple fluorescent proteins. To accomplish this, we used a “Brainbow” strategy for stochastic Cre-mediated combinatorial expression of fluorescent proteins (Livet et al., 2007; Cai et al., 2013). A mixture of adeno-associated viruses (AAV), bearing Brainbow cassettes coding for membrane-targeted GFP, mKate2, and mCherry derivatives (Cai et al., 2013), were injected into the eyes of adult GLAST-CreER mice. After Tamoxifen injection, a large subset of Müller glia expressed one or more fluorescent proteins, and individual cells could be readily discerned from their neighbors by their distinct hues (Fig. 5A–C). To analyze these spectrally complex images in an efficient and unbiased way, we developed an algorithm for segmenting individual cells on the basis of color (Fig. 5D,E). In this way, we could delineate the territories of individual Müller glia in Brainbow-labeled flat-mount retinas in a semi-automated manner (Fig. 5F). To test the algorithm’s accuracy, we compared Müller glia segmented by hand to ones segmented using our software and found a high degree of concordance between the two methods (data not shown). Additionally, we found that arbor areas measured from segmented Brainbow Müller glia were similar to those obtained from mGFP-expressing Müller glia (data not shown), further confirming the accuracy of our algorithm.
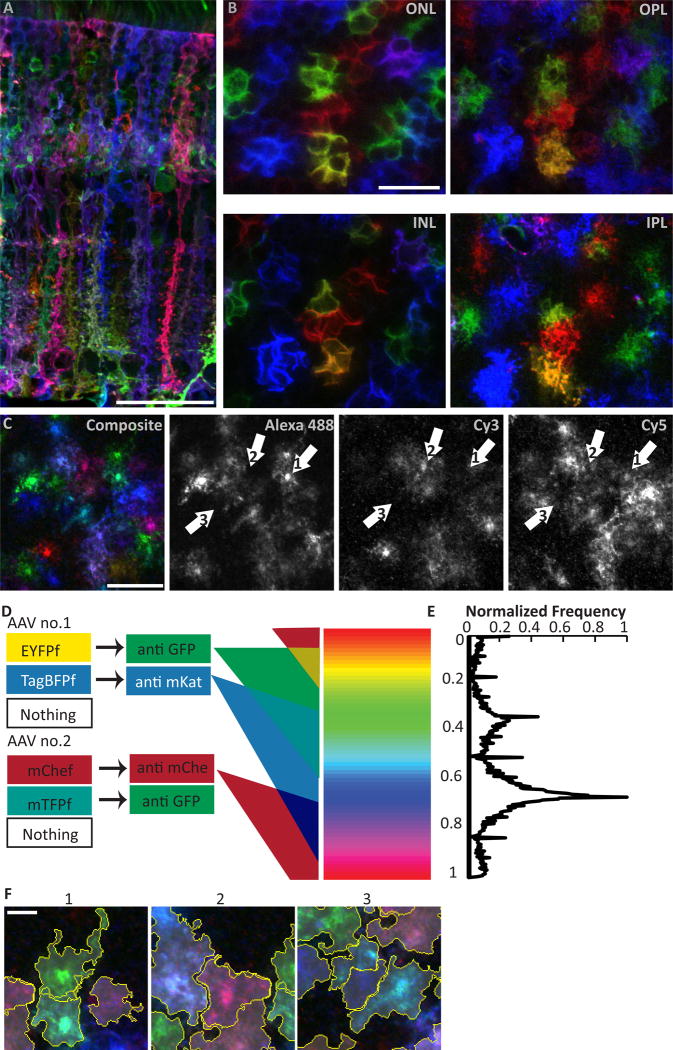
Figure 5. Combinatorial expression of multiple fluorescent proteins via the Brainbow system uniquely labels neighboring Müller cells.
A,B) Adult GLAST-CreER eyes infected with Brainbow AAVs viewed in cross-section (A) or flat mounts (B) where the same cells were imaged at different retinal layers (INL, cell somata; ONL, photoreceptor intercalation; OPL and IPL, synaptic layer arbors). Adjacent Müller glia (MG) are labeled in different hues and appear to occupy non-overlapping columnar domains. C) Splitting of Brainbow images into their 3 constituent color channels reveals that most MG express each fluorescent protein. They appear as different colors due to cell-specific levels of each fluorophore. D) Schematic of semi-automated chromatic segmentation approach. Left, possible protein expression outcomes for each brainbow virus. Center, antibodies that recognize each virally-encoded fluorescent protein. Triple-staining with these antibodies yields tissues labeled in three component colors. Due to copy number variation from variable infection count, parametric variation in fluorescence intensity in each channel generates a continuum of possible hues for each cell (right). E) Frequency distribution for hues of individual pixels of a Brainbow image. Y-axis is aligned to color spectrum in (D). Because the pixels comprising one cell tend to share a narrow range of hues, this histogram can be used for segmentation. F) Contiguous spatial segmentation of pixels by hue, as implemented in our algorithm, successfully delineates individual cells, even when adjacent to or overlapping with cells of similar color. Scale bars (in µm): 40 (A), 20 (B,C), 5 (F).
Using this Brainbow strategy, we examined the spatial relationship between the territories of neighboring Müller cells. Qualitatively, Müller arbors appeared to tessellate in the synaptic layers to cover the retina completely and efficiently, consistent with the tiling hypothesis (Fig. 5B,F). Unique and non-overlapping domains also appear to exist in other layers, as is evident in cross-sectional and en face views of the ONL (Fig. 5A,B). We quantified Müller arbor coverage in the IPL and OPL using our software and found only minimal (~4%) overlap between neighboring cells (Fig. 6A,E,F), further supporting the notion that Müller glia tile. The software was capable of detecting higher degrees of overlap, because when we used image editing software to artificially vary the amount of overlap between two cells of different colors, the algorithm still accurately segmented both cells. We did not detect differences in overlap between central and peripheral retina, suggesting that the small difference in measured arbor size between these regions (Fig. 4B,D) is not sufficient to influence tiling (OPL overlap: central 1.8 ± 1.1 %, peripheral 2.4 ± 2.2 %, t-test p = 0.53; IPL overlap: central 1.2 ± 0.9 %, peripheral 1.8 ± 1.6 %, p = 0.45).
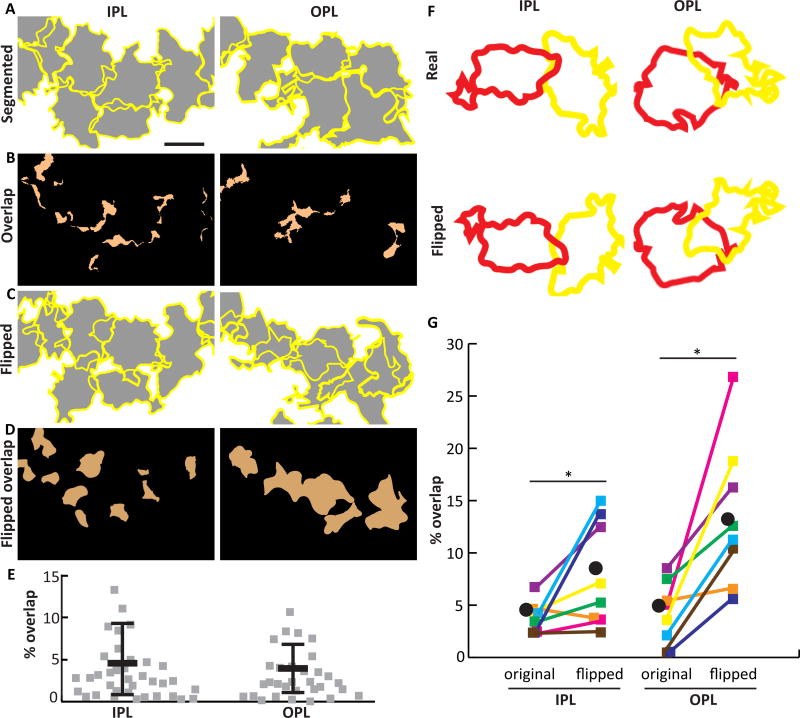
Figure 6. Müller cells establish minimally overlapping territories.
A) Z-projections of SegThresh-segmented Brainbow-labeled MG in IPL and OPL. Yellow, cell borders; gray, cell territories. B) The regions of A where two or more cells overlap are indicated in orange. C) Same fields of view as A, except that each cell has been flipped about its horizontal axis. D) Regions of overlap in flipped image. When cells are flipped, retinal coverage is decreased (more of the field of view is white in C than A), and overlap is increased (more orange in D than B), suggesting that the specific shapes of arbors relative to their neighbors is not random. E) Quantification of arbor overlap for interacting pairs of cells in the IPL and OPL. F,G) Pairwise comparison of arbor overlap for real and flipped neighboring cells (example pairs shown in F). Flipped territories demonstrate significantly more overlap than real territories (G). Colored squares, individual cell pairs; black circle, group mean. *IPL p=0.023; *OPL p=0.008 (see Results for statistical details). Scale bar: 10 µm (A–D).
To ask whether the observed overlap percentage might arise by chance, given the size of Müller glia arbors and the labeling density in our Brainbow samples, we compared real images to the same images in which each cell was flipped along one axis. This procedure randomizes relative cell position while preserving most other features of the real data, including arbor size, shape, and cell location (Wassle et al., 1981). Significantly less overlap was found in the real images than the flipped controls (Fig. 6C–F; OPL overlap: 4.2 ± 3.0% real vs. 13.5 ± 7.0% flipped, Wilcoxon signed-rank matched pair test, two-tailed p = 0.008; IPL: 3.8 ± 1.5% real vs. 7.9 ± 5.0% flipped, p = 0.023). This finding strongly suggests that the shapes and positions of Müller glia arbors are not arbitrary. Rather, they likely reflect local cell-cell interactions that act to minimize overlap of neighboring cells while still filling available space, as is seen in other systems where cells tile (Lefebvre et al., 2015). Together, our observations on Müller cell density, arbor size, and overlap support a model whereby a single type of Müller cell is distributed at constant density across the retina, with individual cells contributing to the complete coverage of retinal space by occupying unique territories.
Development of Müller Glia Arbors
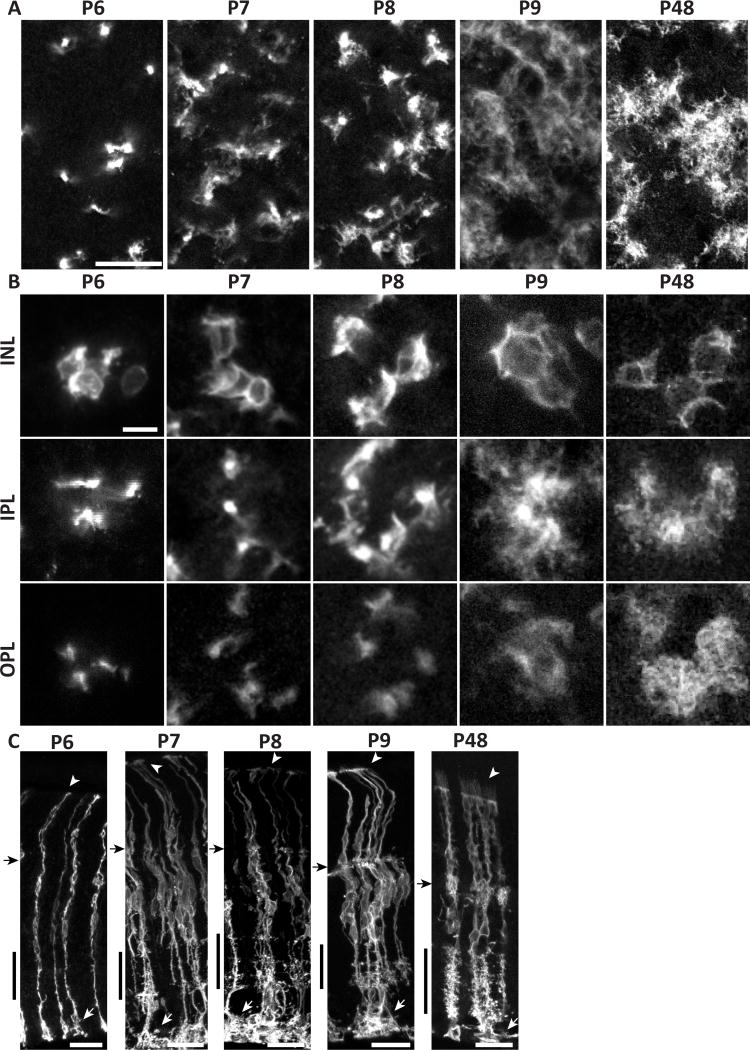
Our analysis of adult mouse retina indicates that Müller glia fill each retinal layer, and each synaptic sublayer, with morphologically specialized arbors. We next asked how these arbors arise during development. Müller glia are formed, starting around P6, by the differentiation of retinal neural progenitor cells (Nelson et al., 2011). Like Müller cells, these progenitors span the retina with endfeet at the limiting membranes, but they lack the extensive lateral arborizations of differentiated Müller glia. How the intricate radial morphology of Müller glia emerges from simple bipolar progenitors is not known. To investigate this process, we imaged mGFP-expressing Müller glia in retinas harvested at different postnatal ages. Through P6, the age at which differentiating progenitors first begin to express many of the genes that are characteristic of mature Müller glia (Nelson et al., 2011), most cells still resembled radial progenitors, with stalks devoid of lateral processes and no contact with neighboring cells (Fig. 7A,B). However, a few cells showed one or two lateral branches in the IPL (Fig. 7C). At P7, nearly all cells had lateral branches in the IPL, and the first cell-cell contacts were observed at this age. However, OPL branches did not emerge in the majority of cells until P8 (Fig.7B). With time, lateral processes continued to become more abundant, and ramification of fine processes gave Müller glia their space-filling morphology by P9 (Fig. 7A–C).
Figure 7. Morphological development of mouse Müller cells occurs rapidly between P7 and P9.
mGFP-expressing MG from densely-labeled GLAST-CreER; mTmG mice, viewed in flat mount (A,B) or cross-section (C). P6-P9 time course documents branch formation. A, lower-power view at the level of IPL S5. B, high-power views of cell clusters, imaged at 3 different planes: INL (cell bodies), IPL, and OPL. Note that all clusters contain cell somata that are adjacent and/or touching (INL panels). At P6, MG are largely bipolar with few lateral branches. Pioneering branches appear at P7, and approach adult (P48) levels of complexity by P9. Arbors assume space-filling morphology by P9, and maintain it to adulthood. ILM endfeet elaborate greatly starting at P7 (C, white arrows). Microvilli first appear beyond OLM by P9 (C, arrowheads). Black arrow in C, OPL; black vertical bar, IPL. Scale bars (in µm): 20 (A), 10 (B), 25 (C).
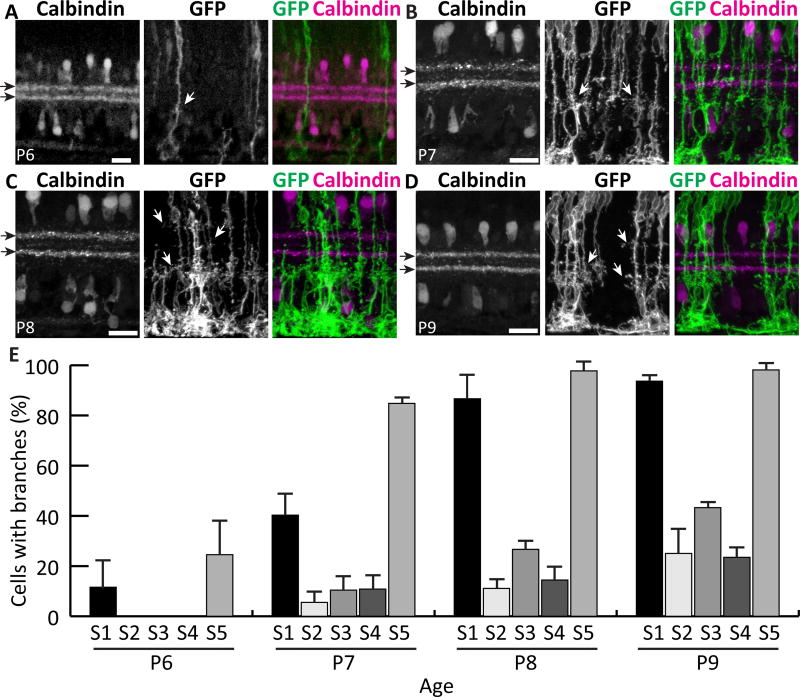
Over the course of this analysis we noticed that the pioneering lateral IPL branches appeared to emerge at stereotyped sublaminar locations. We therefore stained for IPL sublaminar markers in order to delineate the synaptic regions where Müller glia first arborize. Using calbindin immunofluorescence to define S2 and S4, we found that, at P6, the earliest branches emerge preferentially in S1 and S5 (Fig 8A,E). At P7, the proportion of cells with S1 and S5 branches increases; only a few cells have branches in S2-S4 (Fig. 8B,E). S5 branches are the most precocious; the vast majority of cells have branches in S5 by P7, but a similar proportion is not reached for S1 branches until a day later (Fig. 8E). During this developmental period the IPL is changing rapidly, with new neuronal projections arriving to create new sublayers, and new synapses forming at a high rate (Fisher, 1979). The finding of sublaminar specificity in the early arborization of Müller glia suggests that these glial arbors may influence, or be influenced by, the development of neuronal sublaminar projections. Furthermore, our results suggest an intriguing correlation between adult Müller glia morphology and their developmental history: the IPL sublayers where adult Müller glia branch most densely (Fig. 2) appear to be the same layers where they arborize earliest (Fig. 8E).
Figure 8. Müller cells first branch at stereotyped depths within the IPL.
Retinal cross sections of different developmental ages double-stained for mGFP-labeled MG and Calbindin to mark S2 and S4 (black arrows). A) At P6, most cells are unbranched, but occasional MG branches (white arrows) are seen in S1 and S5. B) At P7, branches in S1 and S5 become more abundant and the first branches in central IPL emerge. C,D) Between P8 (C) and P9 (D), branching in all layers continues to increase. Most cells branch in S1 and S5 by P9, but central layer branches continue to emerge after this time. E) Percentage of cells per retina that have branches in each IPL sublamina, measured at P6–9. Scale bars: 20 µm.
DISCUSSION
In the nervous system, structure reflects function as neurons and glia adopt elaborate morphologies to support their physiology. Müller glia are cells with intricate structures and diverse roles in health and disease. In this study, we characterize the spatial organization of Müller glia in the mouse retina for the first time. We show that individual Müller cells display precise radial specializations that correspond not only to retinal layers but also specific synaptic sublaminae. The population of Müller glia forms a confluent network that fills the retina, with each cell occupying an exclusive domain in the retina’s tangential plane. Finally, we identify a brief time period at the end of the first postnatal week during which Müller glia rapidly develop, with lateral processes emerging at stereotyped depths within the IPL. From our data in the mouse retina, we derive two organizational principles of Müller glia. First, in the radial plane, individual Müller cells bear specialized branches matched to their laminar and sublaminar position. Second, in the tangential plane, Müller glia are distributed at uniform density and coordinate with their neighbors to tile the neuropil with minimally overlapping arbor territories. These principles highlight anatomical features of Müller glia and their network which are likely to play important roles in their function.
Radial Morphology of Müller Glia: Laminar and Sublaminar Specialization
Using membrane-targeted fluorescent proteins, we examined the morphology of Müller arbors at each retinal layer. Overall, the morphology of mouse Müller glia was similar to other species. However, using tools available in mouse retina we were able to observe important new details concerning the fine laminar structure of Müller glia arbors. In the IPL, neurons devoted to particular visual processing tasks project to specific sublaminae, where they join into circuits. There are at least 10 different sublaminae (Masland, 2001; Sanes and Zipursky, 2010) several of which can be identified immunohistochemically based on the cell types that project there. Previous studies in other species have noted that Müller branch density can vary across the IPL, sometimes even in a manner that appears layer-specific (Ramon y Cajal, 1972; Robinson and Dreher, 1990), but whether these anatomical variations indeed occur in consistent IPL locations had not been addressed. Using lamina-specific markers we identified three IPL regions – perhaps single sublayers or perhaps multiple adjacent sublayers – that contain a higher density of Müller glia branches than the surrounding neuropil. Because Müller branching patterns were consistent relative to these independent markers, we conclude that Müller cells have sublaminar-specific specializations in the IPL. The layers containing the starburst amacrine cells, for instance, were relatively low in Müller arbor density, while those containing the dopaminergic amacrine cells and rod bipolar cells were relatively high. The reasons for these laminar disparities in branch density are not yet clear. It may be that distinct types of neurons interact differently with Müller cells, or that there are features of IPL sublayers such as synapse number or type that can alter Müller arbor density. It will be interesting to learn whether brain astrocytes might also have morphological specializations associated with particular types of neurons or synapses.
Factors Influencing Müller Glia Cell Density
In many mammalian species, neuron density varies across the retina due to specialization of different retinal zones for distinct functions (e.g. acuity vs. low light sensitivity) (Hughes, 1981; Stone and Johnston, 1981; Robinson and Dreher, 1990; Dreher et al., 1992; Rodriguez et al., 2014). Given the important role of Müller glia in metabolism, we decided to investigate whether neuron density correlates with Müller cell density. The topography of RGCs in mouse reflects that of other retinal neuron classes (Jeon et al., 1998), so we used RGC measurements as a proxy for overall neuron density. The RGC density curves we obtained are consistent with prior reports in mouse (Drager and Olsen, 1981; Galindo-Romero et al., 2011; Rodriguez et al., 2014) and rat (Nadal-Nicolas et al., 2009; Salinas-Navarro et al., 2009). We found that, whereas RGC density decreases with eccentricity, Müller glia are spread evenly across the mouse retina with a mean density of 15,890 cells mm−2 (Fig. 3). This figure is in close agreement with the estimate of 16,000 cells mm−2 reported by and Chao et al. (1997), and within an order of magnitude to the 12,000 cells mm−2 first reported (Drager et al., 1984). This produces local variation in the ratio of Müller glia to RGCs within the mouse retina, from 2.6:1 centrally to 5.3:1 peripherally. These findings strongly suggest that Müller density is not in fact sensitive to neuron density, but is instead determined by an independent mechanism.
Our results in mouse are in line with Müller density data from rabbit, rat, and guinea pig – in these species, glial density does vary with retinal eccentricity but the range of densities is ~7-fold less for Müller glia than for RGCs (Reichenbach and Wohlrab, 1986; Robinson and Dreher, 1990; Dreher et al., 1992). In mouse we find an even larger difference between Müller glia and RGCs, with no decrease in Müller density even in the far periphery and no relationship between the distributions of the two cell populations. Interestingly, Chao et al. (1997) found a wide range of neuron:Müller ratios across phylogeny, further undermining the notion that neuron density might be a simple determinant of glial density. While the precise mechanisms that regulate Müller cell density are yet to be determined, it is likely that factors intrinsic to progenitor cells, such as transcription factors and epigenetic regulators, play a crucial role in controlling how many neurons and glia are generated (Bassett and Wallace, 2012).
Spatial Arrangement of Müller Glia in the Tangential Plane
Müller glia arbors form a confluent network in all retinal layers, completely covering the tangential plane. Here we have investigated how individual Müller cell bodies and arbors are arranged to give rise to this network. We initially considered several possibilities, each of which would have suggested a different logic of network organization, implying different functions. First, the position of individual Müller glia somata, and/or the extent of their arbors, might have been arbitrary. This pattern would give rise to an inhomogeneous network in which the number of Müller glia covering any given point in retinal space is highly variable. However, our data were not consistent with this model. While Müller glia somata were not arranged in a mosaic, instead showing a random arrangement, our data favor the idea that their position is not entirely arbitrary. Even with a random arrangement, high cell density causes cell-cell packing. While Müller cells do not completely fill their layer of the INL – some gaps in their soma array are evident – many cells are squeezed tightly next to each other. In this arrangement, their relative position is not arbitrary because it is determined by the size of neighboring cells – specifically, in this case, the average minimum diameter of neighboring cells. Thus, it is possible that at least some degree of spatial order is imposed on the Müller soma array by passive cell-to-cell crowding. We observed this same cell density not only in adulthood (Fig. 3), but also at P9 when Müller cells are newly differentiated (data not shown) suggesting that this arrangement is a hallmark of the array from its inception. A few other retinal cell types, notably rods in the mouse and human retina (Curcio et al., 1990), appear to achieve orderly distribution according to this “crowding” rule. If packing is indeed important for imposing order, this would suggest that the laminar restriction of Müller glia somata to a single monolayer within the INL is an important contributor to their tangential patterning, because the number of Müller glia cell bodies is only sufficient to achieve tight packing if they are restricted in this way.
Second, we considered the possibility that the glial network varies in a retinotopic fashion to match the number of neurons present at a given retinal location. As we note in the previous section above, the local density of Müller glia somata did not vary in this way; however, if the size and/or overlap of their arbors were found to be higher in central retina, where neurons are most numerous, this would indicate that a denser glial network is required to support greater neuronal demand. Our data do not support this model, as neither arbor size nor overlap was found to vary much across the retina. Instead, our results provide strong evidence in favor of a third model, whereby Müller glial arbors are present at essentially constant density throughout the tangential plane. Using multi-color Brainbow labeling, we were able to assess the contribution of individual cells to the confluent Müller glia networks that exist in each retinal layer. We conclude from this analysis that mouse Müller glia tile the retina, coordinating their arbors to cover the plexiform layers (and likely the ONL and OLM as well) with minimal overlap between territories of adjacent cells (Fig. 6). Locally, adjacent Müller glia arbors closely conform to their neighbors’ borders even though their perimeters are irregularly shaped. These observations raise the possibility that local cell-cell interactions between Müller glia sculpt their territories, perhaps through homotypic repulsion.
One of our observations was potentially inconsistent with this conclusion: we found that peripheral Müller glia arbors are marginally larger than central ones. Given that Müller glia cell density does not vary across the retina (Fig. 3), this finding could indicate increased arbor overlap in peripheral retina, inconsistent with a tiling model. However, in our Brainbow experiments we did not find increased overlap in the periphery. We suspect that that the small arbor size difference arose from a sample handling artifact: when flat-mounting a cup-shaped retina, there are more stretching forces on the peripheral retinal tissue than the center. These stretching forces might cause peripheral arbors to appear larger. Based on the direct measurements of overlap we performed in our Brainbow tissue, we conclude that the center-to-periphery difference in arbor size is either artifactual or of insignificant magnitude to meaningfully affect overlap.
Our conclusion that Müller glia tile is similar to that of Williams et al. (2010), who examined Müller glia arbors in zebrafish. They photobleached single cells in animals that expressed GFP in all Müller glia, and found that the bleached area was comparable to the size of GFP-expressing Müller glia in sparsely labeled retinas. Their finding suggests that, at the population level, there is no consistent overlap of Müller glia processes within the plexiform layers, but this study did not directly measure overlap of adjacent Müller glia territories. Taken together with our mouse study, however, it appears likely that tiling may be typical of Müller glia across many vertebrate species from fish to mammals.
The tiled organization of the Müller glia network has several possible implications for its function. First, individual Müller glia must be capable of handling all of the many support and structural functions required at any given retinotopic location. This is the case even though the number and precise cell-type composition of retinal neurons might vary substantially across the retina. Second, the tiling pattern suggests that uniform arbor density is a salient feature of the Müller glia network. Other than the need to avoid gaps that entirely lack glial coverage, it is not clear what aspects of neural function might require uniform arbor coverage. Finally, the observation that mouse brain astrocytes tile (Bushong et al., 2002) has led to the hypothesis that the set of synapses defined by an astrocyte’s domain, or “synaptic island”, might be a functional processing unit, with astrocytes uniquely able to integrate and modulate activity therein (Halassa et al., 2007). The degree of redundancy in glial coverage may be regulated in a species-dependent manner, however; in visual cortex, the territories of mouse astrocytes are non-overlapping while those in ferret overlap substantially (Lopez-Hidalgo et al., 2016). We find that mouse Müller glia territories, like mouse brain astrocyte domains, coordinate to tile space with minimal overlap. Even if the specifics of the synaptic island hypothesis are not born out, it is clear that Müller glia territories delineate distinct neuronal domains where metabolism, synaptic development, and circuit function might be locally regulated by Müller glia.
Development of Müller Glial Arbors
Müller glia have a remarkably complex anatomy, but there have been few systematic efforts to understand when and how their key anatomical features arise during development. It is clear that Müller glia retain their basic radial glial morphology from the neural progenitors that give rise to them, but the cellular dynamics of their maturation remain largely unknown. We find here that mouse Müller glia make their first lateral branches in the IPL and OPL at P6–7. This is quite early in their maturation process, as Müller glia undergo their terminal mitosis between postnatal days 1 (P1) and 5 (Young, 1985), and only begin to display mature gene expression and immunohistochemical profiles around P6 (Hojo et al., 2000; Nelson et al., 2011). Over the next several days, the number and length of branches increases steadily until space-filling is achieved by P9. The period of Müller glia arbor differentiation coincides with a brief epoch of bone morphogenetic protein signaling, which might be part of the molecular mechanism that controls arbor formation (Ueki et al., 2015). P6–9 is also a time of intense synaptogenesis (Blanks et al., 1974; Fisher, 1979). The timing of Müller glia differentiation in the synaptic layers may therefore be related to synapse maturation, as is true for brain astrocytes (Clarke and Barres, 2013). If there is such a relationship, it will be interesting to learn whether Müller glia arbors sprout in response to the formation of synapses, whether synapses form in response to signals from newly-differentiated Müller glia, or some combination of both mechanisms.
We observed that Müller glia IPL branches first appear in stereotyped sublaminar locations. Pioneering branches initially colonize S1 and S5, and these substrata maintain an increased density of Müller glia processes into adulthood (Figs 2, 8). Why branches emerge in these locations remains unclear. It may be that these sublayers have more synapses, or different types of synapses. Alternatively, developing sublamina are marked by unique sets of cell surface and extracellular matrix molecules (Sanes and Zipursky, 2010; Baier, 2013; Lefebvre et al., 2015), which may play a molecular role in controlling Müller glia branch initiation, stability, or pruning. Unraveling the molecular basis of Müller glia arbor development, and their interactions with neurons during this process, will be an important future direction.
As P9 is the time when Müller glia begin extensively interacting with their neighbors, this is presumably the beginning of the developmental process that gives rise to tiling. In many cases, the process that produces tiling is homotypic repulsion, a mechanism whereby arbors of neighboring cells arrest each other’s growth upon contact (Lefebvre et al., 2015). There is precedent for homotypic repulsion sculpting the morphology of glia: photoablation of zebrafish Müller glia or mouse Schwann cells leads to expansion of neighboring cells into the vacated territory (Williams et al., 2010; Brill et al., 2011). However, these experiments cannot exclude release of an attractive cue by dying cells. Laser damage could also induce reactive gliosis, which in some circumstances can cause astrocytes to invade each other’s territories (Oberheim et al., 2008). Whether homotypic repulsion is indeed involved in mammalian Müller glia development and what molecules guide establishment of Müller glia territories will be an area for further study.
Conclusions
Our study provides the first thorough description of the spatial organization of Müller glia in mouse retina. We propose a model in which total and efficient glial coverage is achieved in development by: 1) generating a uniform high density of Müller cells at all retinal positions; 2) the tiling of Müller glia arbors through homotypic repulsion; and 3) bidirectional neuroglial interactions that influence Müller arbor branching across IPL sublamina. While mechanistic aspects of this model remain to be tested, the anatomy we report here is consistent with it. We expect that our results will provide an important baseline morphological foundation for future explorations of neuron-glia interactions in the retina.
Acknowledgments
Thanks to Nicholas Brecha (UCLA) for sharing RBPMS antibodies, Josh Sanes (Harvard) and Dawen Cai (University of Michigan) for sharing anti-mCherry and anti-mKate antibodies, Megan Stogsdill and Ari Pereira for mouse colony assistance.
Financial Support: Ruth K Broad Foundation Postdoctoral Fellowship (MO’S); Pew Charitable Trusts, E Matilda Ziegler Foundation for the Blind, Karl Kirchgessner Foundation, Alfred P. Sloan Foundation, McKnight Endowment Fund for Neuroscience, NEI Grant R01EY024694 (JNK); NSF GRFP DGE-1644868 (VMP), NEI Core Grant for Vision Research EY5722, Research to Prevent Blindness Unrestricted Grant (Duke University)
Role of Authors
All authors had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Study concept and design: JW, MLO’S, JNK. Acquisition of data: JW, MLO’S, VMP. Analysis and interpretation of data: JW, MLO’S, DM, VMP, SF, JNK. Drafting of the manuscript: MLO’S, JW, JNK. Critical revision of the manuscript for important intellectual content: JW, MLO’S, DM, SF, JNK. Statistical analysis: JW, MLO’S. Obtained funding: JNK. Study supervision: JNK.
Footnotes
Conflict of Interest Statement
The authors have no conflicting interests.
Literature Cited
- Allen NJ. Astrocyte regulation of synaptic behavior. Annu Rev Cell Dev Biol. 2014;30:439–463. doi: 10.1146/annurev-cellbio-100913-013053. [DOI] [PubMed] [Google Scholar]
- Baier H. Synaptic laminae in the visual system: molecular mechanisms forming layers of perception. Annu Rev Cell Dev Biol. 2013;29:385–416. doi: 10.1146/annurev-cellbio-101011-155748. [DOI] [PubMed] [Google Scholar]
- Bassett EA, Wallace VA. Cell fate determination in the vertebrate retina. Trends in neurosciences. 2012;35(9):565–573. doi: 10.1016/j.tins.2012.05.004. [DOI] [PubMed] [Google Scholar]
- Blanks JC, Adinolfi AM, Lolley RN. Synaptogenesis in the photoreceptor terminal of the mouse retina. J Comp Neurol. 1974;156(1):81–93. doi: 10.1002/cne.901560107. [DOI] [PubMed] [Google Scholar]
- Brill MS, Lichtman JW, Thompson W, Zuo Y, Misgeld T. Spatial constraints dictate glial territories at murine neuromuscular junctions. The Journal of cell biology. 2011;195(2):293–305. doi: 10.1083/jcb.201108005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushong EA, Martone ME, Jones YZ, Ellisman MH. Protoplasmic astrocytes in CA1 stratum radiatum occupy separate anatomical domains. J Neurosci. 2002;22(1):183–192. doi: 10.1523/JNEUROSCI.22-01-00183.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai D, Cohen KB, Luo T, Lichtman JW, Sanes JR. Improved tools for the Brainbow toolbox. Nat Methods. 2013;10(6):540–547. [PubMed] [Google Scholar]
- Chao TI, Grosche J, Friedrich KJ, Biedermann B, Francke M, Pannicke T, Reichelt W, Wulst M, Muhle C, Pritz-Hohmeier S, Kuhrt H, Faude F, Drommer W, Kasper M, Buse E, Reichenbach A. Comparative studies on mammalian Muller (retinal glial) cells. Journal of neurocytology. 1997;26(7):439–454. doi: 10.1023/a:1018525222826. [DOI] [PubMed] [Google Scholar]
- Clarke LE, Barres BA. Emerging roles of astrocytes in neural circuit development. Nat Rev Neurosci. 2013;14(5):311–321. doi: 10.1038/nrn3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curcio CA, Sloan KR, Kalina RE, Hendrickson AE. Human photoreceptor topography. J Comp Neurol. 1990;292(4):497–523. doi: 10.1002/cne.902920402. [DOI] [PubMed] [Google Scholar]
- Drager UC, Edwards DL, Barnstable CJ. Antibodies against filamentous components in discrete cell types of the mouse retina. J Neurosci. 1984;4(8):2025–2042. doi: 10.1523/JNEUROSCI.04-08-02025.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drager UC, Olsen JF. Ganglion cell distribution in the retina of the mouse. Invest Ophthalmol Vis Sci. 1981;20(3):285–293. [PubMed] [Google Scholar]
- Dreher Z, Robinson SR, Distler C. Muller cells in vascular and avascular retinae: a survey of seven mammals. J Comp Neurol. 1992;323(1):59–80. doi: 10.1002/cne.903230106. [DOI] [PubMed] [Google Scholar]
- Fisher LJ. Development of synaptic arrays in the inner plexiform layer of neonatal mouse retina. J Comp Neurol. 1979;187(2):359–372. doi: 10.1002/cne.901870207. [DOI] [PubMed] [Google Scholar]
- Galindo-Romero C, Aviles-Trigueros M, Jimenez-Lopez M, Valiente-Soriano FJ, Salinas-Navarro M, Nadal-Nicolas F, Villegas-Perez MP, Vidal-Sanz M, Agudo-Barriuso M. Axotomy-induced retinal ganglion cell death in adult mice: quantitative and topographic time course analyses. Exp Eye Res. 2011;92(5):377–387. doi: 10.1016/j.exer.2011.02.008. [DOI] [PubMed] [Google Scholar]
- Gnanaguru G, Bachay G, Biswas S, Pinzon-Duarte G, Hunter DD, Brunken WJ. Laminins containing the beta2 and gamma3 chains regulate astrocyte migration and angiogenesis in the retina. Development. 2013;140(9):2050–2060. doi: 10.1242/dev.087817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez RC, Woods RE. Digital Image Processing. Upper Saddle River, NJ: Prentice Hall; 2008. [Google Scholar]
- Halassa MM, Fellin T, Takano H, Dong JH, Haydon PG. Synaptic islands defined by the territory of a single astrocyte. J Neurosci. 2007;27(24):6473–6477. doi: 10.1523/JNEUROSCI.1419-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haverkamp S, Ghosh KK, Hirano AA, Wassle H. Immunocytochemical description of five bipolar cell types of the mouse retina. J Comp Neurol. 2003;455(4):463–476. doi: 10.1002/cne.10491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haverkamp S, Wassle H. Immunocytochemical analysis of the mouse retina. J Comp Neurol. 2000;424(1):1–23. [PubMed] [Google Scholar]
- Hojo M, Ohtsuka T, Hashimoto N, Gradwohl G, Guillemot F, Kageyama R. Glial cell fate specification modulated by the bHLH gene Hes5 in mouse retina. Development. 2000;127(12):2515–2522. doi: 10.1242/dev.127.12.2515. [DOI] [PubMed] [Google Scholar]
- Huberman AD, Niell CM. What can mice tell us about how vision works? Trends in neurosciences. 2011;34(9):464–473. doi: 10.1016/j.tins.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes A. Population magnitudes and distribution of the major modal classes of cat retinal ganglion cell as estimated from HRP filling and a systematic survey of the soma diameter spectra for classical neurones. J Comp Neurol. 1981;197(2):303–339. doi: 10.1002/cne.901970209. [DOI] [PubMed] [Google Scholar]
- Jeon CJ, Strettoi E, Masland RH. The major cell populations of the mouse retina. J Neurosci. 1998;18(21):8936–8946. doi: 10.1523/JNEUROSCI.18-21-08936.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay JN, Chu MW, Sanes JR. MEGF10 and MEGF11 mediate homotypic interactions required for mosaic spacing of retinal neurons. Nature. 2012;483(7390):465–469. doi: 10.1038/nature10877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay JN, Sanes JR. Development of Retinal Arbors and Synapses. In: Werner JS, Chalupa LM, editors. The New Visual Neurosciences. Cambridge, MA: The MIT Press; 2014. pp. 1291–1304. [Google Scholar]
- Kreutzberg GW, Hussain ST. Cytochemical heterogeneity of the glial plasma membrane: 5'-nucleotidase in retinal Muller cells. Journal of neurocytology. 1982;11(1):53–64. doi: 10.1007/BF01258004. [DOI] [PubMed] [Google Scholar]
- Lefebvre JL, Sanes JR, Kay JN. Development of dendritic form and function. Annu Rev Cell Dev Biol. 2015;31:741–777. doi: 10.1146/annurev-cellbio-100913-013020. [DOI] [PubMed] [Google Scholar]
- Livet J, Weissman TA, Kang H, Draft RW, Lu J, Bennis RA, Sanes JR, Lichtman JW. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450(7166):56–62. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- Lopez-Hidalgo M, Hoover WB, Schummers J. Spatial organization of astrocytes in ferret visual cortex. J Comp Neurol. 2016;524(17):3561–3576. doi: 10.1002/cne.24015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masland RH. The fundamental plan of the retina. Nature neuroscience. 2001;4(9):877–886. doi: 10.1038/nn0901-877. [DOI] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45(9):593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- Nadal-Nicolas FM, Jimenez-Lopez M, Sobrado-Calvo P, Nieto-Lopez L, Canovas-Martinez I, Salinas-Navarro M, Vidal-Sanz M, Agudo M. Brn3a as a marker of retinal ganglion cells: qualitative and quantitative time course studies in naive and optic nerve-injured retinas. Invest Ophthalmol Vis Sci. 2009;50(8):3860–3868. doi: 10.1167/iovs.08-3267. [DOI] [PubMed] [Google Scholar]
- Nelson BR, Ueki Y, Reardon S, Karl MO, Georgi S, Hartman BH, Lamba DA, Reh TA. Genome-wide analysis of Muller glial differentiation reveals a requirement for Notch signaling in postmitotic cells to maintain the glial fate. PLoS One. 2011;6(8):e22817. doi: 10.1371/journal.pone.0022817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberheim NA, Tian GF, Han X, Peng W, Takano T, Ransom B, Nedergaard M. Loss of astrocytic domain organization in the epileptic brain. J Neurosci. 2008;28(13):3264–3276. doi: 10.1523/JNEUROSCI.4980-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata K, Kosaka T. Structural and quantitative analysis of astrocytes in the mouse hippocampus. Neuroscience. 2002;113(1):221–233. doi: 10.1016/s0306-4522(02)00041-6. [DOI] [PubMed] [Google Scholar]
- Ortin-Martinez A, Nadal-Nicolas FM, Jimenez-Lopez M, Alburquerque-Bejar JJ, Nieto-Lopez L, Garcia-Ayuso D, Villegas-Perez MP, Vidal-Sanz M, Agudo-Barriuso M. Number and distribution of mouse retinal cone photoreceptors: differences between an albino (Swiss) and a pigmented (C57/BL6) strain. PLoS One. 2014;9(7):e102392. doi: 10.1371/journal.pone.0102392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poche RA, Furuta Y, Chaboissier MC, Schedl A, Behringer RR. Sox9 is expressed in mouse multipotent retinal progenitor cells and functions in Muller glial cell development. J Comp Neurol. 2008;510(3):237–250. doi: 10.1002/cne.21746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramon y Cajal S. In: The Structure of the Reina. Thorpe S, Glickstein M, translators. Springfield, IL: Charles Thomas; 1972. p. 196. [Google Scholar]
- Rasmussen KE. A morphometric study of the Muller cell cytoplasm in the rat retina. J Ultrastruct Res. 1972;39(5):413–429. doi: 10.1016/s0022-5320(72)90095-0. [DOI] [PubMed] [Google Scholar]
- Reese BE, Keeley PW. Design principles and developmental mechanisms underlying retinal mosaics. Biol Rev Camb Philos Soc. 2015;90(3):854–876. doi: 10.1111/brv.12139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichenbach A, Bringmann A. New functions of Muller cells. Glia. 2013;61(5):651–678. doi: 10.1002/glia.22477. [DOI] [PubMed] [Google Scholar]
- Reichenbach A, Schneider H, Leibnitz L, Reichelt W, Schaaf P, Schumann R. The structure of rabbit retinal Muller (glial) cells is adapted to the surrounding retinal layers. Anat Embryol (Berl) 1989;180(1):71–79. doi: 10.1007/BF00321902. [DOI] [PubMed] [Google Scholar]
- Reichenbach A, Wohlrab F. Quantitative properties of Muller cells in rabbit retina as revealed by histochemical demonstration of NADH-diaphorase activity. Graefes Arch Clin Exp Ophthalmol. 1983;220(2):81–83. doi: 10.1007/BF02133876. [DOI] [PubMed] [Google Scholar]
- Reichenbach A, Wohlrab F. Morphometric parameters of Muller (glial) cells dependent on their topographic localization in the nonmyelinated part of the rabbit retina. A consideration of functional aspects of radial glia. Journal of neurocytology. 1986;15(4):451–459. doi: 10.1007/BF01611728. [DOI] [PubMed] [Google Scholar]
- Robinson SR, Dreher Z. Muller cells in adult rabbit retinae: morphology, distribution and implications for function and development. J Comp Neurol. 1990;292(2):178–192. doi: 10.1002/cne.902920203. [DOI] [PubMed] [Google Scholar]
- Rodieck RW. The density recovery profile: a method for the analysis of points in the plane applicable to retinal studies. Vis Neurosci. 1991;6(2):95–111. doi: 10.1017/s095252380001049x. [DOI] [PubMed] [Google Scholar]
- Rodriguez AR, de Sevilla Muller LP, Brecha NC. The RNA binding protein RBPMS is a selective marker of ganglion cells in the mammalian retina. J Comp Neurol. 2014;522(6):1411–1443. doi: 10.1002/cne.23521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinas-Navarro M, Mayor-Torroglosa S, Jimenez-Lopez M, Aviles-Trigueros M, Holmes TM, Lund RD, Villegas-Perez MP, Vidal-Sanz M. A computerized analysis of the entire retinal ganglion cell population and its spatial distribution in adult rats. Vision Res. 2009;49(1):115–126. doi: 10.1016/j.visres.2008.09.029. [DOI] [PubMed] [Google Scholar]
- Sanes JR, Zipursky SL. Design principles of insect and vertebrate visual systems. Neuron. 2010;66(1):15–36. doi: 10.1016/j.neuron.2010.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone J, Johnston E. The topography of primate retina: a study of the human, bushbaby, and new- and old-world monkeys. J Comp Neurol. 1981;196(2):205–223. doi: 10.1002/cne.901960204. [DOI] [PubMed] [Google Scholar]
- Ueki Y, Wilken MS, Cox KE, Chipman LB, Bermingham-McDonogh O, Reh TA. transient wave of BMP signaling in the retina is necessary for Muller glial differentiation. Development. 2015;142(3):533–543. doi: 10.1242/dev.118745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecino E, Rodriguez FD, Ruzafa N, Pereiro X, Sharma SC. Glia-neuron interactions in the mammalian retina. Prog Retin Eye Res. 2016;51:1–40. doi: 10.1016/j.preteyeres.2015.06.003. [DOI] [PubMed] [Google Scholar]
- Voinescu PE, Kay JN, Sanes JR. Birthdays of retinal amacrine cell subtypes are systematically related to their molecular identity and soma position. J Comp Neurol. 2009;517(5):737–750. doi: 10.1002/cne.22200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassle H, Boycott BB. Functional architecture of the mammalian retina. Physiol Rev. 1991;71(2):447–480. doi: 10.1152/physrev.1991.71.2.447. [DOI] [PubMed] [Google Scholar]
- Wassle H, Peichl L, Boycott BB. Dendritic territories of cat retinal ganglion cells. Nature. 1981;292(5821):344–345. doi: 10.1038/292344a0. [DOI] [PubMed] [Google Scholar]
- Williams PR, Suzuki SC, Yoshimatsu T, Lawrence OT, Waldron SJ, Parsons MJ, Nonet ML, Wong RO. In vivo development of outer retinal synapses in the absence of glial contact. J Neurosci. 2010;30(36):11951–11961. doi: 10.1523/JNEUROSCI.3391-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Vidensky S, Jin L, Jie C, Lorenzini I, Frankl M, Rothstein JD. Molecular comparison of GLT1+ and ALDH1L1+ astrocytes in vivo in astroglial reporter mice. Glia. 2011;59(2):200–207. doi: 10.1002/glia.21089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RW. Cell differentiation in the retina of the mouse. Anat Rec. 1985;212(2):199–205. doi: 10.1002/ar.1092120215. [DOI] [PubMed] [Google Scholar]