Abstract
Diamond-Blackfan anemia is a congenital erythroid hypoplasia and is associated with physical malformations and a predisposition to cancer. Twenty-five percent of patients with Diamond-Blackfan anemia have mutations in a gene encoding ribosomal protein S19 (RPS19). Through overexpression of RPS19 using a lentiviral vector with the spleen focus-forming virus promoter, we demonstrated that the Diamond-Blackfan anemia phenotype can be successfully treated in Rps19-deficient mice. In our present study, we assessed the efficacy of a clinically relevant promoter, the human elongation factor 1α short promoter, with or without the locus control region of the β-globin gene for treatment of RPS19-deficient Diamond-Blackfan anemia. The findings demonstrate that these vectors rescue the proliferation defect and improve erythroid development of transduced RPS19-deficient bone marrow cells. Remarkably, bone marrow failure and severe anemia in Rps19-deficient mice was cured with enforced expression of RPS19 driven by the elongation factor 1α short promoter. We also demonstrate that RPS19-deficient bone marrow cells can be transduced and these cells have the capacity to repopulate bone marrow in long-term reconstituted mice. Our results collectively demonstrate the feasibility to cure RPS19-deficient Diamond-Blackfan anemia using lentiviral vectors with cellular promoters that possess a reduced risk of insertional mutagenesis.
Keywords: lentiviral vectors, gene therapy, Diamond-Blackfan anemia
Diamond-Blackfan anemia is a congenital erythroid hypoplasia. Twenty-five percent of patients have mutations in a gene encoding ribosomal protein S19. Using an RPS19-deficient mouse model, Debnath et al. demonstrate the feasibility to cure RPS19-deficient Diamond-Blackfan anemia by means of lentiviral vectors with cellular promoters that possess a reduced risk of insertional mutagenesis.
Introduction
Diamond-Blackfan anemia (DBA) is a rare inherited bone marrow failure disorder with pure red blood cell aplasia manifesting early in life. The hematological profile of patients with DBA shows macrocytic anemia with reticulocytopenia, normal or decreased levels of neutrophils, and variable platelet counts.1 Patients with DBA also exhibit various non-hematological manifestations such as physical abnormalities and cancer predisposition.2, 3
In at least 60%–70% of cases, DBA is caused by functional haploinsufficiency of genes encoding for ribosomal proteins.4, 5, 6, 7, 8, 9, 10, 11 Recent studies have discovered that two genes, erythroid transcriptional factor GATA1 and TSR2 (a direct binding partner of RPS26), can cause the DBA phenotype.12, 13, 14, 15 Twenty-five percent of patients have mutations in a gene coding ribosomal protein S19 (RPS19).4 For given mutations all reported patients are heterozygous. Furthermore, in most cases, the mutations are predicted to result in haploinsufficiency of the respective ribosomal protein.16, 17 Corticosteroids are the main therapeutic option in DBA.3 Around 80% of patients initially respond to corticosteroids, but only 40% of patients sustain the therapeutic response and the remaining 40% need chronic blood transfusion. Twenty percent of patients go into spontaneous remission and maintain acceptable hemoglobin levels without therapeutic intervention. Allogeneic bone marrow transplantation is currently the only curative treatment available for patients with DBA.18
Our previous studies demonstrated that enforced expression of RPS19 improves the proliferation, erythroid colony-forming potential, and differentiation of patient-derived RPS19-deficient hematopoietic progenitor cells in vitro.19, 20 Moreover, RPS19 overexpression enhances the engraftment and erythroid differentiation of patient-derived hematopoietic stem cells and progenitor cells when transplanted into immune-compromised mice.21 Collectively, these studies suggest that gene therapy may be a future therapeutic modality in the treatment of RPS19-deficient DBA. In our proof-of-principle study using lentiviral vectors harboring the spleen focus-forming virus (SFFV) promoter and a codon-optimized human RPS19 cDNA followed by the internal ribosomal entry site (IRES) and GFP (SFFV-RPS19), we showed that the DBA phenotype of Rps19-deficient mice can be successfully treated.22
In the current study, we assessed the efficacy of clinically relevant promoters to drive the therapeutic gene. To this effect, we designed lentiviral vectors harboring a codon-optimized human RPS19 cDNA driven by the shortened version of the human elongation factor 1α (EFS) promoter. Lentiviral vectors with the EFS promoter are shown to have a significantly decreased risk of insertional mutagenesis,23, 24 and no evidence of clonal dominance was reported during clinical trials of gene therapy for severe combined immunodeficiency X1 (SCID-X1) using the EFS promoter.25
The EFS promoter was followed by IRES and GFP (EFS-RPS19), while a vector without the RPS19 cDNA was used as a control (EFS-Spacer). To assess the therapeutic potential of the EFS-RPS19 vector in vivo, we transduced c-Kit-enriched bone marrow cells from control and uninduced small hairpin RNA (shRNA)-D mice and these were injected into lethally irradiated wild-type mice. The recipients transplanted with the EFS-Spacer transduced shRNA-D bone marrow showed a dramatic decrease in blood cellularity that led to death after a few weeks, while the recipients transduced with EFS-RPS19 shRNA-D bone marrow exhibited close to normal blood cellularity. These results demonstrate that EFS promoter-driven enforced expression of RPS19 can cure severe anemia and bone marrow failure in RPS19-deficient mice.
Results
Enforced Expression of RPS19 by the EFS Promoter in Rps19-Deficient Bone Marrow Cells Improves Proliferation and Erythroid Development In Vitro
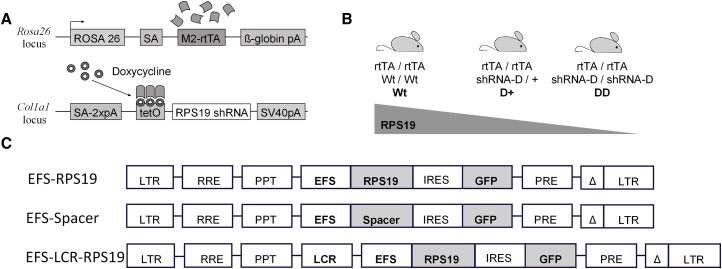
We have shown that enforced expression of RPS19 expands erythroid development in RPS19-deficient patients with DBA.19, 20, 21 In our previous study using lentiviral vectors driven by the SFFV promoter, we showed that the DBA phenotype of mice can be successfully treated.22 In this study, we assessed the efficacy of clinically relevant promoters like the EFS promoter in our mouse model of RPS19-deficient DBA. Briefly, this model contains an Rps19-targeting shRNA (shRNA-D) that is expressed under a doxycycline-responsive promoter located downstream of the collagen A1 gene (Figure 1A). Experimental animals were bred to be either heterozygous (D+) or homozygous (DD) for the shRNA in order to generate two models with intermediate or severe Rps19 deficiency, respectively (Figure 1B). To correct the Rps19 deficiency, we developed self-inactivating (SIN) lentiviral vectors harboring a codon-optimized human RPS19 cDNA driven by the internal EFS promoter, followed by IRES and GFP (EFS-RPS19) with or without a β-globin locus control region (LCR) cassette (Figure 1C). The codon-optimized RPS19 cDNA was further modified to prevent its recognition and downregulation by the Rps19-targeting shRNA used. A similar vector without the RPS19 cDNA was used as a control vector (EFS-Spacer).22, 26, 27, 28
Figure 1.
Mouse Model for RPS-Deficient DBA and SIN Lentiviral Vectors for DBA Gene Therapy
Transgenic mice containing a doxycycline-regulatable Rps19-targeting shRNA allow an inducible and graded downregulation of Rps19. (A) Overview of modified loci. Black arrowheads indicate TSSs. (B) Breeding strategy to adjust the level of Rps19 downregulation. (C) EFS-RPS19 vector, codon-optimized human RPS19 cDNA was constructed under the control of the human elongation factor 1α short (EFS) promoter and inserted into a lentiviral vector. Following the RPS19 cDNA, an internal ribosomal entry site (IRES), a GFP sequence, and improved post-transcriptional regulatory element (Pre*) were inserted. EFS-Spacer vector, in which the RPS19 cDNA was replaced with an equally long non-coding spacer sequence, was used as a control. The LCR-EFS-RPS19 vector, where in locus control region of β-globin gene was inserted before the EFS promoter. LTR, long terminal repeat; pA, polyadenylation signal; PPT, polypurine tract; RRE, Rev response element; SA, splice acceptor; Wt, wild-type.
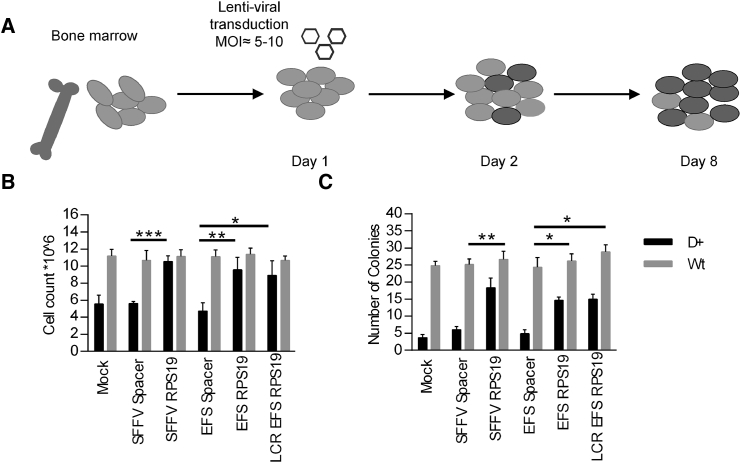
To assess the functionality of these vectors, we cultured transduced c-Kit-enriched bone marrow (BM) cells from control and heterozygous RPS19 shRNA (D+) mice in liquid cultures in the presence of doxycycline (Figure 2A). Based on the percentage of GFP+ cells, the initial transduction efficiency was between 20% and 40% on average (Figure 3B). D+ cells transduced with the EFS-Spacer control vector failed to expand during 7 days of culture after transduction (Figure 2B). In contrast, the EFS-RPS19 and LCR-EFS-RPS19 vectors mediated a 2-fold increase in total cell number compared to the EFS-Spacer vector.
Figure 2.
Enforced Expression of RPS19 Derived from the EFS Promoter Is Sufficient to Rescue the DBA Phenotype In Vitro
c-Kit-enriched hematopoietic progenitors (0.5 × 106) from the BM of uninduced mice were transduced and seeded in liquid culture or methyl cellulose in the presence of doxycycline. (A) Experimental design. (B) Total cell counts on day 8 after growth in liquid culture. (C) Total erythroid colony counts in methyl cellulose cultures (M3436) in the presence of doxycycline on day 14. Data shown in (B) and (C) represent the average of three independent experiments with three technical replicates. *p < 0.05; ***p < 0.001. Wt, wild-type.
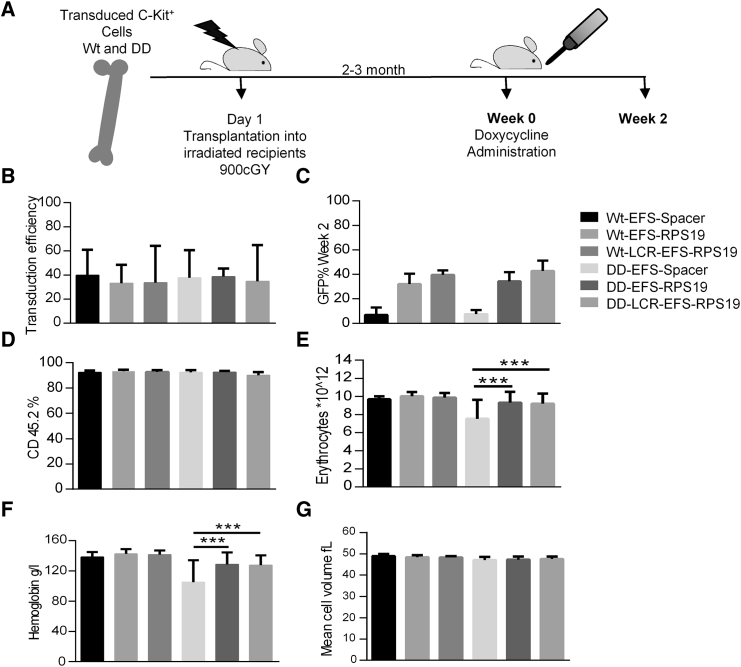
Figure 3.
Enforced Expression of RPS19 Derived from the EFS Promoter Is Sufficient to Rescue the Acute DBA Phenotype In Vivo
Enforced expression of RPS19 results in short-term rescue of the hematological defect in RPS19-deficient mice. (A) Experimental strategy to validate the short-term therapeutic potential of EFS-RPS19 and LCR-EFS-RPS19 vectors. (B) Transduction efficiency is shown. (C and D) GFP reconstitution and donor reconstitution are shown. (E–G) Erythrocyte number (E), hemoglobin concentration (F), and mean corpuscular value (G) (n = 20–21). Error bars represent the SD. ***p < 0.001. MCV, mean corpuscular value; Wt, wild-type.
Next we quantified the erythroid colony-forming potential of transduced c-Kit-enriched BM cells from control and D+ mice in methyl cellulose cultures in the presence of doxycycline for 14 days (Figure 2C). The findings demonstrate that the EFS-RPS19 and LCR-EFS-RPS19 vectors mediated a 3-fold increase in the total number of erythroid colonies compared to the EFS-Spacer vector.
Enforced Expression of RPS19 by the EFS Promoter Is Sufficient to Rescue the DBA Phenotype In Vivo
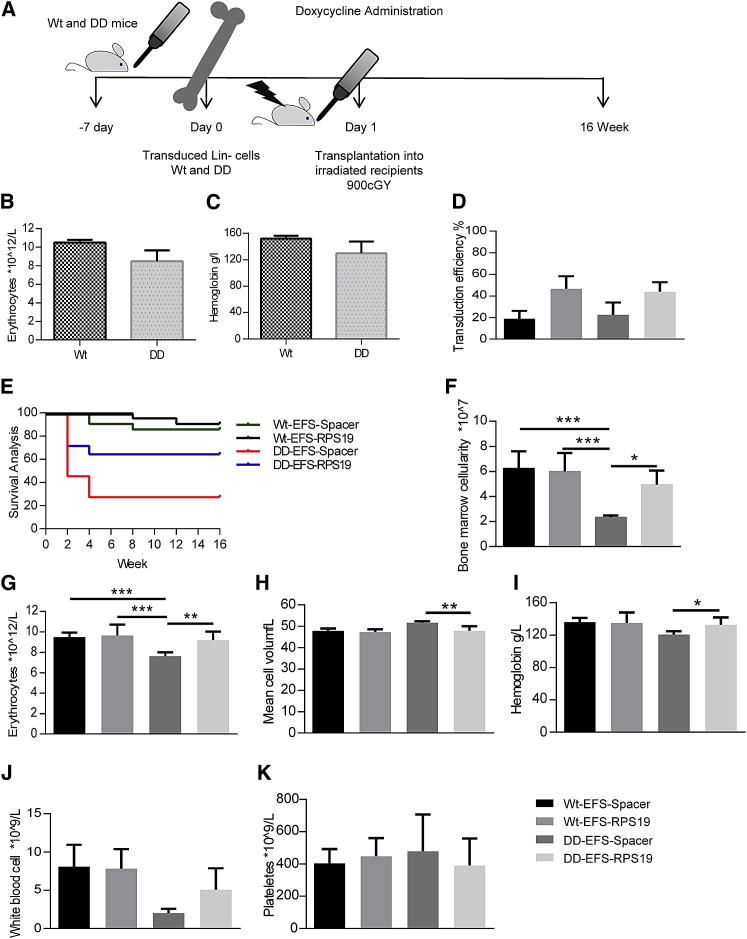
Subsequently, we probed whether EFS-RPS19 and LCR-EFS-RPS19 vectors generate a sufficient amount of RPS19 in vivo in order to assay the therapeutic efficacy. Doxycycline administration to transplanted recipients with the homozygous RPS19 shRNA (DD) genotype causes acute and lethal BM failure, while D+ recipients (one RPS19 shRNA allele) develop mild chronic anemia.28 Since DD mice develop lethal BM failure shortly after doxycycline administration, we chose this model to test the efficacy of gene correction to rigorously assess whether the lethal phenotype could be rescued and the mice cured. Uninduced BM cells from the control and DD mice were transduced with the vectors, and the transduced cells were transplanted into wild-type recipient mice. Following engraftment and stable donor-derived regeneration of the hematopoietic system, recipient mice were administered doxycycline to downregulate endogenous Rps19 in order to induce the disease (Figure 3A). Since we showed previously that the hematopoietic phenotype in Rps19-deficient mice is autonomous to the blood system, we decided to use lethally irradiated wild-type recipients.28
Before transplantation, initial transduction efficiencies with therapeutic and control vectors were measured based on the percentage of GFP+ cells and were between 20% and 40% on average (Figure 3B). After 2 weeks of doxycycline treatment, most of the mice receiving DD BM transduced with EFS-Spacer vector died due to dramatic BM failure (data not shown). At this time point, all groups showed high overall donor reconstitution, confirming the absence of recipient-derived hematopoiesis (Figures 3C and 3D). We demonstrated that the recipients transplanted with the EFS-RPS19 or LCR-EFS-RPS19 DD BM had normal blood cellularity (Figures 3E and 3F).
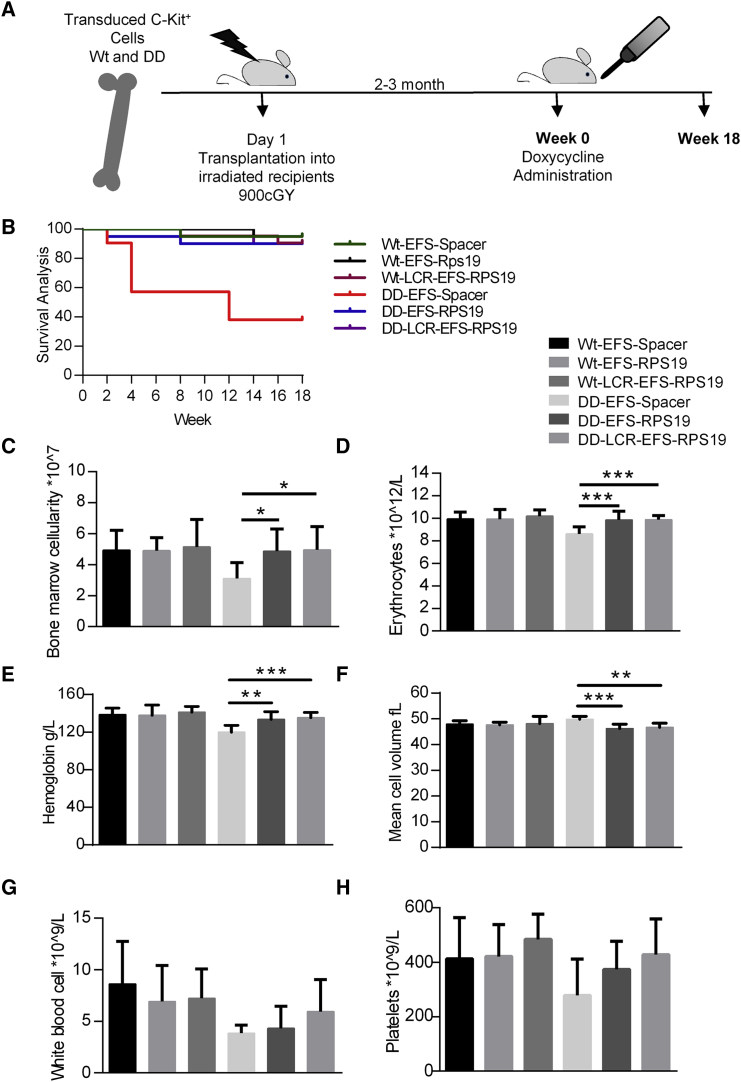
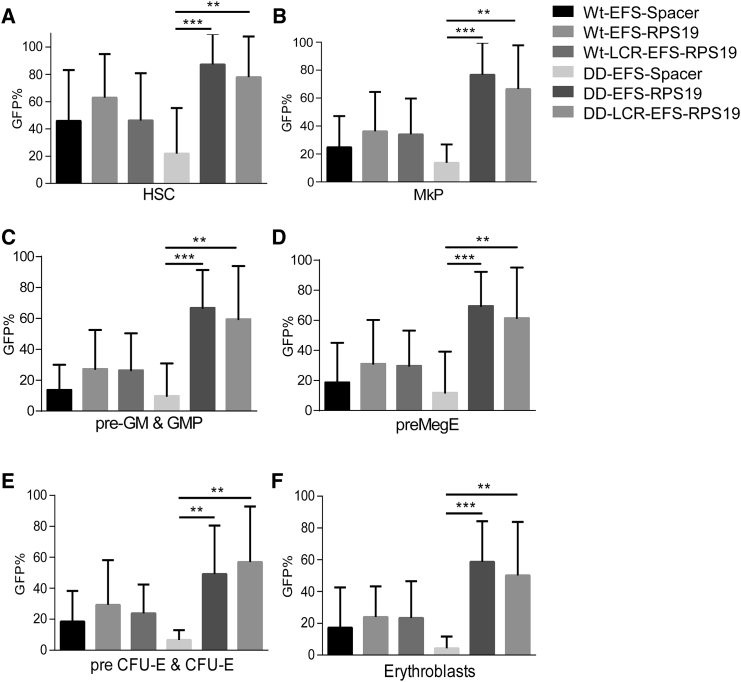
Doxycycline administration for 18 weeks was used as the time point to assess long-term efficacy (Figure 4A). Most recipients with DD BM transduced with EFS-Spacer vectors died, but the remaining surviving recipients exhibited macrocytic anemia and a decrease in erythrocyte numbers, hemoglobin value, and platelet counts (Figures 4B–4H). Remarkably, recipients transplanted with the EFS-RPS19 or LCR-EFS-RPS19 DD BM had normal blood cellularity and BM cellularity (Figures 4C–4H). Additionally, we analyzed the samples by fluorescence-activated cell sorting (FACS) to allow fractionation of the myeloid-erythroid compartment in the BM.28, 29 The mean percentage of GFP+ cells was substantially higher in recipients with EFS-RPS19 or LCR-EFS-RPS19 DD BM than in the other groups, indicating the competitive advantage of gene-corrected cells in the hematopoietic hierarchy (Figure 5).
Figure 4.
Enforced Expression of RPS19 Derived from the EFS Promoter Is Sufficient to Rescue the DBA Phenotype In Vivo
Enforced expression of RPS19 results in long-term rescue of the hematological defect in RPS19-deficient mice. (A) Experimental strategy to validate the long-term therapeutic potential of EFS-RPS19 and LCR-EFS-RPS19 vectors. (B) Survival curve. (C) BM cellularity after 18 weeks of doxycycline induction. (D–H) Erythrocyte number (D), hemoglobin concentration (E), mean corpuscular value (F), white blood cell count (G), and platelet number (H) after 18 weeks of doxycycline induction (n = 20–21). Error bars represent the SD. *p < 0.05; **p < 0.01; ***p < 0.001. MCV, mean corpuscular value; Wt, wild-type.
Figure 5.
Gene-Corrected Rps19-Deficient Cells Gain a Competitive Advantage Resulting in Increased Contribution to Hematopoiesis In Vivo
(A–F) The percentage of transduced cells in the HSC (A), MkP (B), pre-GM (granulocyte macrophage) and granulocyte macrophage progenitors (GMP) (C), preMegE (D), and preCFU-E (E) and colony forming unit (CFU)-E erythroblast compartment (F) (n = 16–24 per group). Error bars represent the SD. *p < 0.05; **p < 0.01; ***p < 0.001. Wt, wild-type.
RPS19-Deficient BM Cells Transduced with RPS19 Vectors Provide Long-Term Reconstitution
We asked whether doxycycline-induced, Rps19-defcient BM cells transduced with RPS19 lentiviral vectors can result in long-term engraftment in doxycycline-induced lethally irradiated wild-type recipient mice (Figure 6A). To this end, DD and control mice were induced with doxycycline for 1 week and erythrocyte numbers and hemoglobin levels were measured to confirm the DBA phenotype (Figures 6B and 6C). BM cells from induced mice were transduced and transplanted into doxycycline-induced lethally irradiated mice. Initial transduction efficiencies with therapeutic and control vectors were measured based on the percentage of GFP+ cells and were between 20% and 50% (Figure 6D). Most of the mice receiving DD BM transduced with EFS-Spacer failed to engraft and did not survive beyond 2–3 weeks after transplantation. Almost 60% of the mice receiving DD BM with corrected EFS-RPS19 vector survived and showed long-term engraftment (Figure 6E). We assessed long-term engraftment and the hematopoietic contribution of mice with gene-corrected DD BM at 16 weeks post-transplantation. At this point, these mice exhibited improved BM cellularity and recovery of erythrocyte numbers, hemoglobin levels, and platelet counts (Figures 6F–6K).
Figure 6.
Rps19-Deficient BM Cells Can Be Transduced and the Transduced Cells Provide Long-Term Reconstitution
Rps19-deficient BM can be transduced; after genetic correction, these cells show long-term engraftment in lethally irradiated wild-type mice. (A) Experimental strategy to validate the long-term reconstitution capacity of corrected Rps19-deficient cells. (B–F) Pre-transplant Wt and DD mice erythrocyte numbers (B), hemoglobin concentration (C), transduction efficiency (D), survival curve (E), and BM cellularity (F) after 16 weeks of doxycycline induction. (G–K) Erythrocyte number (G), mean corpuscular volume (H), hemoglobin concentration (I), white blood cell count (J), and platelet number (K) after 16 weeks of doxycycline induction (n = 20–28). Error bars represent the SD. *p < 0.05; **p < 0.01; ***p < 0.001. MCV, mean corpuscular volume; Wt, wild-type.
Gene-Corrected Rps19-Deficient Cells Show Polyclonal Hematopoiesis and Have a Typical Lentiviral Insertion Profile
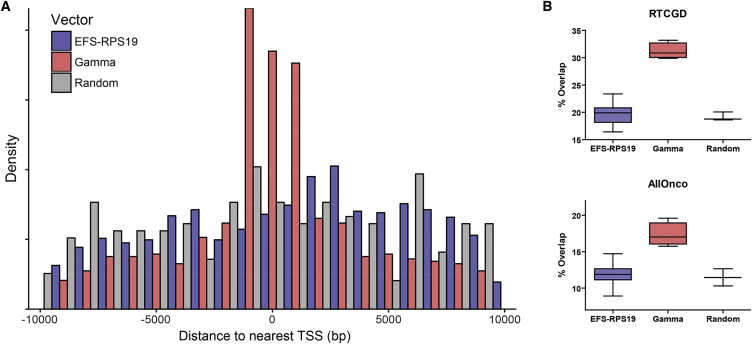
The risk of insertional mutagenesis is a major concern regarding the future clinical use of lentiviral vectors. To assess the safety of the EFS-RPS19 vector integration profile as well as the clonal dynamics of the transduced cells, we performed insertion site analysis of DNA from BM cells of four mice per vector group obtained from recipients after 16–18 weeks of doxycycline administration. Integration sites per vector group (Wt-EFS-Spacer [WES], Wt-.EFS-RPS19 [WER], Wt-LCR.-EFS-RPS19 [WLER], DD-EFS-RPS19 [DER], and DD-LCR-EFS.RPS19 [DLER]) were analyzed by linear amplification-mediated (LAM) PCR followed by Ion Torrent sequencing. A total of 2.88 × 106 sequences were processed, clustered for homology (increasing the read count of individual insertions), trimmed for the remaining vector sequences, and aligned to the murine genome. The 2.18 × 105 sequence reads were assigned to 5,420 individual insertions. Despite the known limitations in terms of absolute quantification of amplicon sequencing in integration site analysis,30 we use the read count as a surrogate marker for clonal abundance. We investigated the insertion profile in the different mice for the number of hits close to transcriptional start sites (TSSs) of genes, the clonal diversity,31 common insertion sites (CISs), and overlaps with cancer gene databases. Detailed information for each mouse is provided in Table S1 and Figures S1–S3. We did not observe a tendency to preferentially integrate within a 10-kb window around the TSSs of genes (Figure 7A). The overlap of EFS-RP19 insertions with the Retroviral Tagged Cancer Gene Database (RTCGD)32 or the All Onco cancer gene list33 was not different from a randomized control dataset (Figure 7B). We did not observe a significant difference in clonal diversity between the vector groups. However, six mice had a lower sequence diversity (Figure S2F) compared to all other treated animals. For two of these mice (DER1 and DLER3), we observed a dominant insertion within genes (Malt1 and Cdh26) listed in the RTCGD database. Both genes were found only once in an artificial B-cell lymphoma mouse model during insertional mutagenesis screens.34 From the overlap of gene symbols close to insertion sites and cancer gene databases alone, we cannot conclude a functional relationship exists between vector integration and increased clonal abundance. As we also cannot exclude a proliferation advantage due to insertional mutagenesis, we depict overlaps with four reference databases for those insertions with a read count above the 97.5 percentile of all reads (Table S2) and for all detected CISs (Table S3). A chi-square analysis revealed no statistical differences for the overlap with cancer gene databases between the vector groups. When we checked for common high-risk insertions in or near Prdm16, Mecom, Notch1, Lmo2, Setbp1, Ccnd2, Sox4, and Tal1, we found either no hits (Lmo2, Tal1) or only read contributions ≤ 0.58% (n = 19 of 5,420 sequences).
Figure 7.
EFS-RPS19 Integrations Do Not Cluster around the TSS and Show No Increased Overlap with Cancer Gene Databases
(A) Density plot showing the frequency of integrations in a 10-kb window around the transcriptional start site. As we found no statistical differences between the different EFS-RPS19 vectors, we combined them in one group (blue) and compared them to a gamma retroviral integration profile (red) or a randomized dataset (gray). (B) The overlap of gene symbols closest to the insertion sites with either the Retroviral Tagged Cancer Gene Database (RTCGD) or the AllOnco cancer gene list of the EFS-RPS19 vectors was not different from that of a randomized dataset. The increased overlap of the gamma retroviral integration dataset is shown for comparison.
Discussion
Here, we demonstrate the efficacy of RPS19 lentiviral vectors using clinically relevant promoters to correct lethal BM failure in Rps19-deficient mice. We show that the EFS promoter can express enough RPS19 to correct RPS19-deficient BM failure and the EFS-driven RPS19 single-gene vector can be used in a clinical gene therapy trial for RPS19-deficient DBA. Previously, we demonstrated that enforced expression of RPS19 improves the proliferation, erythroid colony-forming potential, and differentiation of patient-derived RPS19-deficient hematopoietic progenitor cells in vitro.19, 20 Using xenograft models, we have also shown that overexpression of RPS19 enhances the engraftment and erythroid differentiation of patient-derived hematopoietic stem and progenitor cells.21 In our proof-of-principle study using lentiviral vectors driven by the SFFV promoter and harboring a codon-optimized human RPS19 cDNA followed by IRES and GFP, we showed that the DBA phenotype of mice can be successfully treated.22
In the current study, we decided to utilize the ubiquitously expressed EFS promoter with or without the LCR of the β-globin gene for treatment of RPS19-deficient DBA. We have shown that these vectors rescue the proliferation defect and erythroid development of transduced c-Kit+ DD BM cells in vitro. The induction of Rps19 deficiency in recipient mice with DD BM generated lethal BM failure. Remarkably, the BM failure generated by DD BM was cured with EFS-RPS19. Since quite high levels of RPS19 are needed to correct the RPS19 deficiency by transgenesis, we were concerned that the EFS promoter might not generate sufficient levels of RPS19 in erythroid progenitors to correct the anemia. Therefore, we included vectors containing the EFS plus the β-globin LCR. However, the findings show that the EFS promoter without the β-globin LCR generates sufficient levels of RPS19 to cure anemia and BM failure in RPS19-deficient mice.
Additionally, we demonstrated that RPS19-deficient BM cells can be transduced and these cells survived the transduction procedure and had the capacity to repopulate the BM. However, most of the studies were performed with transduced shRNA-D/D BM transplanted into normal recipients. RPS19 deficiency was induced once the recipients had a stable graft. This is a justified, since we have previously shown that anemia and BM failure in the induced mice is due to the deficiency in the hematopoietic cells and not to a failure of the niche cells.28 If the recipients have Rps19 deficiency in all cells before transplantation of the transduced cells, some of the Rps19-deficient mice will not tolerate the combined toxicity of doxycycline Rps19 downregulation and radiation. However, the majority of the Rps19-deficient mice survived this procedure, as mentioned above.
In this study, we have shown that our RPS19-deficient mouse model is valuable and suitable for testing gene therapy using viral vectors with the RPS19 gene. However, it should be emphasized that this model is different from haploinsufficiency in patients with DBA, which is based on mutations in the RPS19 gene (most often point mutations or small deletions). In the mice used here, haploinsufficiency is generated by RNAi that is induced postnatally. Haploinsufficiency generates most of the hematological symptoms found in DBA in mice but not the physical abnormalities found in a large fraction of patients. Haploinsufficiency in mice causes reduced proliferation and erythroid development, which can be corrected by overexpression of RPS19. A similar effect was seen in cells from patients with RPS19-deficient DBA. Upon overexpression of RPS19 in the cells from patients, cellular proliferation and erythroid development were greatly improved.19, 20
Of course, it is clear that RPS19 vectors can only be used to treat patients with RPS19-deficient DBA. Therefore, patients with mutations in other ribosomal protein genes or the GATA1 gene cannot be treated with RPS19 vectors. Recently, mutations in GATA1 were found in a few patients with DBA. The GATA1 gene in humans produces two mRNAs, one long and one short. Patients with DBA could not produce the long form of GATA1.12, 13 Mice produce only the long form of GATA1; therefore, it will be difficult to evaluate the possibility of using mice as experimental animals in the development of GATA1 gene therapy for human GATA1-deficient patients with DBA.
The data presented in Figure 5 show that the RPS19 vectors increase the production of hematopoietic stem cells (HSCs) and early progenitor cells after overexpression in RPS19-deficient hematopoietic cells. In competitive transplantation experiments, we showed previously that RPS19-deficient HSCs have a competitive disadvantage compared to normal HSCs.28 Collectively, these data suggest that RPS19-deficient HSCs treated with RPS19 vectors may have a competitive advantage compared with untreated cells. It is therefore possible that gene therapy of RPS19-deficient DBA may be performed with little or no BM ablation before transplantation of the gene-corrected cells, due to the possible competitive advantage of these cells. However, the need for ablation in a clinical gene therapy setting must be investigated further in order to design clinical gene therapy trials with minimal risks for patients.
Significantly, by designing a codon-optimized RPS19 cDNA driven by the EFS promoter, we have succeeded in generating a clinically relevant vector system that allows high enough RPS19 expression for functional correction of anemia and BM failure in Rps19-deficient mice. Our studies assessing the efficacy of clinically relevant EFS promoters show a less likely risk of causing insertional oncogenesis.23 Further, our studies using EFS-RPS19 or LCR.EFS-RPS19 vectors that are safer than vectors with viral promoters, but can nevertheless generate sufficient RPS19 expression to correct the pathophysiology of DBA. In normal cells, ribosomal protein production is tightly regulated and physiological and excess protein is subjected to proteasomal degradation.35 Because of this mechanistic regulation of ribosomal protein, ectopic expression of RPS19 possesses a very low risk of promoting uncontrolled growth. In our study, we did not observe any hematologic abnormalities due to enforced expression of RPS19. Our results collectively demonstrate the feasibility of clinical gene therapy to cure RPS19-deficient patients with DBA in the future using EFS promoter-driven enforced expression of RPS19.
Materials and Methods
Lentiviral Vector Constructs
SIN lentiviral vectors used in this study were derived from pRRL.PPT.PGK.GFP pre-vector.36 A codon-optimized human RPS19 cDNA was designed and inserted downstream of the EFS promoter with or without the LCR of the β-globin gene (LCR).26, 27, 36 Following the RPS19 cDNA, the IRES, GFP, and improved post-transcriptional regulatory element (pre*) were inserted. Two vectors were obtained without the LCR pRRL.PPT.EFS.RPS19co.iresGFP.pre* vector (hereafter, EFS-RPS19) and with the LCR pRRL.PPT.LCR.EFS.RPS19co.iresGFP.pre* vector (hereafter, LCR-EFS-RPS19). Lentiviral vectors were produced by the Vector Unit at Lund University. Briefly, standard calcium phosphate transfection of 293T cells was used with the helper plasmid cytomegalovirus (pCMV)ΔR8.91 and pMD2.G—VSV-G envelop expressing plasmid (pMDG). The lentivirus-containing supernatant was harvested 24 hr after transfection, and the lentivirus was concentrated by ultracentrifugation at 25,000 rpm (SW32 rotor, Beckman L-70 Ultracentrifuge; Beckman Coulter) for 90 min at 4°C. Pellets were resuspended in serum-free medium (StemSpan SFEM; Stemcell Technologies) and stored at −80°C. Lentivirus titer was assessed by FACS for the transfer of GFP to HT1080 cells.
Mice
The mouse models are engineered to contain a doxycycline-regulatable Rps19-targeting shRNA (shRNA-D) located downstream of the collagen A1 locus, allowing dose-dependent downregulation of Rps19 expression.28 To generate two models with intermediate or severe Rps19 deficiency, transgenic animals were bred either heterozygous or homozygous for shRNA-D, respectively. RPS19 deficiency was induced by feeding the mice doxycycline in drinking water (1 mg/mL or 2 mg/mL doxycycline; Sigma-Aldrich) supplemented with 10 mg/mL sucrose (Sigma-Aldrich). Mice were maintained at the Lund University animal facility and all animal experiments were performed with consent from the Lund University animal ethics committee.
Blood and BM Analysis
Peripheral blood was collected from the tail vein into microvette tubes (Sarstedt) and was analyzed using sysmex XE-5000. Erythrocytes were lysed using ammonium chloride for 10 min at room temperature. To evaluate the contribution toward various blood lineages following BM transplantation, samples were stained with the following antibodies for 30 min on ice in the dark: CD45.1 (110730; Biolegend) CD45.2 (47-0454-82; eBioscience), B220 (103208; Biolegend), B220 (103212; Biolegend), CD3 (100312; Biolegend), CD11b (101208; Biolegend), and Gr1 (108408; Biolegend). Experiments were performed using a FACS Canto II cytometer (BD Biosciences) and were analyzed by FlowJo software (version 10.0.2; Tree Star). FACS analysis of the myeloerythroid compartment in BM was performed.28, 29 BM cells were isolated by crushing the femur and tibia in PBS containing 2% fetal bovine serum (FBS) (Gibco). Fresh cells were stained with the following antibodies: CD41 (12-0411-83; eBioscience), GR1 (115910; Biolegend), CD11b (101210; Biolegend), B220 (103210; Biolegend), CD3 (100310; Biolegend), c-Kit (47-1171-82; eBioscience), CD105 (120404; Biolegend), and Sca-1 (122520; Biolegend). Streptavidin was purchased from Life Technologies (Q10101MP). Propidium iodide (Life Technologies) was used to exclude dead cells. Experiments were performed using a FACS LSR II cytometer (Becton Dickinson) and were analyzed by FlowJo software (version 10.0.2).
Transduction and Transplantation of Hematopoietic Cells
c-Kit+-expressing cells were enriched from BM of transgenic mice (CD45.2) using CD117 microbeads and a magnetic-activated cell sorting (MACS) separation column (Miltenyi) and were pre-stimulated in serum-free StemSpan serum-free expansion medium (SFEM) (Stemcell Technologies), supplemented with penicillin/streptomycin (P/S; Gibco), murine stem cell factor (mSCF) (100 ng/mL; PeproTech),and human thrombopoietin (hTPO; 50 ng/mL; PeproTech) in six-well plates (non-tissue culture treated; BD) for 1 day at 0.5 × 106 cells/mL. Retronectin-coated (20 ng/mL; Takara) six-well plates were preloaded with the viral vectors (100 μL/well corresponding to a MOI of 10–20) and 1 million cells were seeded into each well in 3 mL pre-stimulation medium. After incubation for 1 day, 0.5 × 106 bulk transduced cells were transplanted in 500 μL PBS into the tail vein of irradiated (900 cGy) wild-type mice recipients (CD45.1 or CD45.1/45.2).
Transduction and Transplantation of RPS19-Deficient Hematopoietic Cells
Lineage-negative (Lin−) cells were enriched from the BM of doxycycline-induced transgenic mice (CD45.2) using Lineage microbeads and a MACS separation column (Miltenyi). Retronectin-coated (20 ng/mL; Takara) six-well plates were preloaded with the viral vectors (100 μL/well corresponding to a MOI of 10–20) and 1 million cells were seeded into each well in 3 mL serum-free StemSpan SFEM medium supplemented with P/S, mSCF (100 ng/mL), hTPO (50 ng/mL), and doxycycline (1 μg/mL; Sigma-Aldrich) in six-well plates (non-tissue culture treated; BD) for 1 day 0.5 × 106 cells/mL. After incubation for 1 day, 0.5 × 106 bulk transduced cells and 1 × 106 untransduced Lin+ cells were transplanted in 300 μL PBS into the tail vein of lethally irradiated (900 cGy) wild-type mice recipients (CD45.1 or CD45.1/45.2).
Cell Culture
c-Kit+-expressing cells were enriched using CD117 microbeads and a MACS separation column (Miltenyi) and retronectin-coated (20 ng/mL; Takara), pre-stimulated in serum-free StemSpan SFEM medium supplemented with P/S, mSCF (100 ng/mL), and hTPO (50 ng/mL) in six-well plates (non-tissue culture treated; BD) for 1 day 0.5 × 106 cells/mL. Twelve-well plates were preloaded with the viral vectors (50 μL/well corresponding to a MOI of 10–20) and 0.5 × 106 cells were seeded into each well in 1 mL serum-free StemSpan SFEM medium supplemented with P/S, mSCF (100 ng/mL), murine interleukin (IL)-3 (mIL-3; 10 ng/mL; PeproTech), and erythropoietin (EPO; 2 U/mL; Janssen-Cilag) with or without doxycycline (1 μg/mL). Light microscopy was used to evaluate the proliferation of culture after 6 days. For the burst forming unit-erythroid (BFU-E) assay, 40 × 103 c-Kit+ transduced cells were seeded in 1.5 mL M3436 methylcellulose (Stemcell Technologies) with doxycycline (1 μg/mL) and colonies were scored on day 14.
Insertion Site Analysis
We used 300 ng genomic DNA of whole BM cells isolated 18 weeks after transplantation. Samples were processed by LAM PCR as described by Schmidt et al.37 with modifications. For digestion, samples were split into three separate reactions with 5 U of CutSmart enzymes MlucI, MseI, and HindPI (the latter two with heat inactivation) from New England Biolabs (NEB). After digestion, samples were combined for nested PCR steps. The first nested PCR was performed with a forward primer binding to the SIN-LTR of the vectors (IT-IS-FW-PCR1: 5′-GTGGGTTTTCCAGTCACACTGCTCTTCCGATCTTCCCTCAGACCCTTTTAGTCA-3′) and a reverse primer recognizing the linker cassette (IT-IS-RV-PCR1: 5′-TTCGTTGGGAGTGAATTAGCC AGTGGCACAGCAGTTAGG-3′). The vector- and linker-specific sequences are underlined, and the italic sequence represents a tail homologous to the primers used for Ion Torrent sequencing, as described previously.38 Bioinformatics processing with custom Perl, R, and visual basic scripts involved barcode primer assignment, trimming, clustering, filtering, and Methods for Analyzing ViRal Integration Clusters analysis tool (MAVRIC) alignment.39 For CIS analysis, we followed the suggestions by Wu et al.,40 considering only five or more insertions in a 50-kb window. Distance to the TSSs was analyzed using the information from the MAVRIC alignments in combination with a customized R script (ggplot2; geom_histogram with the following parameters: aes y = density and binwidth = 1,000).
Control Datasets for Integration Site Analysis
The gamma retroviral integrations used for comparison originate from Lin− cell cultures (n = 4) transduced with RSF91.41, 42 DNA was harvested 4 days after transduction. The LAM-PCR procedure and next-generation sequencing were as described above. The randomized control datasets (n = 3) were produced by generating artificial chromosomal positions using the shuffle command (seed = 100, 101, and 102) of BEDtools.43 The shuffled BED files contained 2,000 genomic positions (500-bp window size) randomly distributed among the murine genome (NCBI47/mm9) as a function of the chromosome size. The BED files were converted to FASTA format and processed by MAVRIC with parameters identical to the biological insertion site data of EFS-RPS19 or the gamma retroviral vector.
Statistical Analysis
One-way ANOVA with the Tukey multiple-comparison test was used to determine statistical significance using GraphPad Prism (version 6; GraphPad Software).
Author Contributions
S.K. conceptualized the project and directed the research; S.D., K.S., M.R., J.C., and M.D. performed the experiments; S.D., P.J., M.R., H.G.B., J.F., A.S., and S.K. analyzed the data; and S.D., P.J., M.R., A.S., and S.K. wrote the manuscript.
Conflicts of Interest
The authors declare no competing financial interests.
Acknowledgments
The authors thank Beata Lindqvist for lentivirus production and Amol Ugale, Karolina Komorowska, and Abdul Ghani Alattar for technical assistance. This work was supported by a Hemato-Linne grant from the Swedish Research Council Linnaeus, grants from the Swedish Cancer Society and the Swedish Children’s Cancer Society (to S.K.), the Tobias Prize awarded by the Royal Swedish Academy of Sciences financed by the Tobias Foundation, a clinical research grant from Lund University Hospital (to S.K.), and European Union (EU) project grants STEMEXPAND and PERSIST. J.F. was supported by the Diamond Blackfan Anemia Foundation. H.B.G. is supported by the Great Ormond Street Hospital Children’s Charity and by the National Institute of Health Research Biomedical Research Centre at Great Ormond Street Hospital and University College London. A.S. and M.R. are supported by the Deutsche Forschungsgemeinschaft (DFG) (REBIRTH Cluster of Excellence and SFB738 projects).
Footnotes
Supplemental Information includes three figures and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.ymthe.2017.04.002.
Supplemental Information
References
- 1.Willig T.N., Niemeyer C.M., Leblanc T., Tiemann C., Robert A., Budde J., Lambiliotte A., Kohne E., Souillet G., Eber S. Identification of new prognosis factors from the clinical and epidemiologic analysis of a registry of 229 Diamond-Blackfan anemia patients. DBA group of Société d’Hématologie et d’Immunologie Pédiatrique (SHIP), Gesellshaft für Pädiatrische Onkologie und Hämatologie (GPOH), and the European Society for Pediatric Hematology and Immunology (ESPHI) Pediatr. Res. 1999;46:553–561. doi: 10.1203/00006450-199911000-00011. [DOI] [PubMed] [Google Scholar]
- 2.Orfali K.A., Ohene-Abuakwa Y., Ball S.E. Diamond Blackfan anaemia in the UK: clinical and genetic heterogeneity. Br. J. Haematol. 2004;125:243–252. doi: 10.1111/j.1365-2141.2004.04890.x. [DOI] [PubMed] [Google Scholar]
- 3.Lipton J.M., Atsidaftos E., Zyskind I., Vlachos A. Improving clinical care and elucidating the pathophysiology of Diamond Blackfan anemia: an update from the Diamond Blackfan Anemia Registry. Pediatr. Blood Cancer. 2006;46:558–564. doi: 10.1002/pbc.20642. [DOI] [PubMed] [Google Scholar]
- 4.Draptchinskaia N., Gustavsson P., Andersson B., Pettersson M., Willig T.N., Dianzani I., Ball S., Tchernia G., Klar J., Matsson H. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat. Genet. 1999;21:169–175. doi: 10.1038/5951. [DOI] [PubMed] [Google Scholar]
- 5.Gazda H.T., Grabowska A., Merida-Long L.B., Latawiec E., Schneider H.E., Lipton J.M., Vlachos A., Atsidaftos E., Ball S.E., Orfali K.A. Ribosomal protein S24 gene is mutated in Diamond-Blackfan anemia. Am. J. Hum. Genet. 2006;79:1110–1118. doi: 10.1086/510020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cmejla R., Cmejlova J., Handrkova H., Petrak J., Pospisilova D. Ribosomal protein S17 gene (RPS17) is mutated in Diamond-Blackfan anemia. Hum. Mutat. 2007;28:1178–1182. doi: 10.1002/humu.20608. [DOI] [PubMed] [Google Scholar]
- 7.Farrar J.E., Nater M., Caywood E., McDevitt M.A., Kowalski J., Takemoto C.M., Talbot C.C., Jr., Meltzer P., Esposito D., Beggs A.H. Abnormalities of the large ribosomal subunit protein, Rpl35a, in Diamond-Blackfan anemia. Blood. 2008;112:1582–1592. doi: 10.1182/blood-2008-02-140012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gazda H.T., Sheen M.R., Vlachos A., Choesmel V., O’Donohue M.F., Schneider H., Darras N., Hasman C., Sieff C.A., Newburger P.E. Ribosomal protein L5 and L11 mutations are associated with cleft palate and abnormal thumbs in Diamond-Blackfan anemia patients. Am. J. Hum. Genet. 2008;83:769–780. doi: 10.1016/j.ajhg.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doherty L., Sheen M.R., Vlachos A., Choesmel V., O’Donohue M.F., Clinton C., Schneider H.E., Sieff C.A., Newburger P.E., Ball S.E. Ribosomal protein genes RPS10 and RPS26 are commonly mutated in Diamond-Blackfan anemia. Am. J. Hum. Genet. 2010;86:222–228. doi: 10.1016/j.ajhg.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gazda H.T., Preti M., Sheen M.R., O’Donohue M.F., Vlachos A., Davies S.M., Kattamis A., Doherty L., Landowski M., Buros C. Frameshift mutation in p53 regulator RPL26 is associated with multiple physical abnormalities and a specific pre-ribosomal RNA processing defect in Diamond-Blackfan anemia. Hum. Mutat. 2012;33:1037–1044. doi: 10.1002/humu.22081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Landowski M., O’Donohue M.F., Buros C., Ghazvinian R., Montel-Lehry N., Vlachos A., Sieff C.A., Newburger P.E., Niewiadomska E., Matysiak M. Novel deletion of RPL15 identified by array-comparative genomic hybridization in Diamond-Blackfan anemia. Hum. Genet. 2013;132:1265–1274. doi: 10.1007/s00439-013-1326-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sankaran V.G., Ghazvinian R., Do R., Thiru P., Vergilio J.A., Beggs A.H., Sieff C.A., Orkin S.H., Nathan D.G., Lander E.S., Gazda H.T. Exome sequencing identifies GATA1 mutations resulting in Diamond-Blackfan anemia. J. Clin. Invest. 2012;122:2439–2443. doi: 10.1172/JCI63597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ludwig L.S., Gazda H.T., Eng J.C., Eichhorn S.W., Thiru P., Ghazvinian R., George T.I., Gotlib J.R., Beggs A.H., Sieff C.A. Altered translation of GATA1 in Diamond-Blackfan anemia. Nat. Med. 2014;20:748–753. doi: 10.1038/nm.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klar J., Khalfallah A., Arzoo P.S., Gazda H.T., Dahl N. Recurrent GATA1 mutations in Diamond-Blackfan anaemia. Br. J. Haematol. 2014;166:949–951. doi: 10.1111/bjh.12919. [DOI] [PubMed] [Google Scholar]
- 15.Gripp K.W., Curry C., Olney A.H., Sandoval C., Fisher J., Chong J.X., Pilchman L., Sahraoui R., Stabley D.L., Sol-Church K., UW Center for Mendelian Genomics Diamond-Blackfan anemia with mandibulofacial dystostosis is heterogeneous, including the novel DBA genes TSR2 and RPS28. Am. J. Med. Genet. A. 2014;164A:2240–2249. doi: 10.1002/ajmg.a.36633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Willig T.N., Draptchinskaia N., Dianzani I., Ball S., Niemeyer C., Ramenghi U., Orfali K., Gustavsson P., Garelli E., Brusco A. Mutations in ribosomal protein S19 gene and Diamond Blackfan anemia: wide variations in phenotypic expression. Blood. 1999;94:4294–4306. [PubMed] [Google Scholar]
- 17.Angelini M., Cannata S., Mercaldo V., Gibello L., Santoro C., Dianzani I., Loreni F. Missense mutations associated with Diamond-Blackfan anemia affect the assembly of ribosomal protein S19 into the ribosome. Hum. Mol. Genet. 2007;16:1720–1727. doi: 10.1093/hmg/ddm120. [DOI] [PubMed] [Google Scholar]
- 18.Vlachos A., Federman N., Reyes-Haley C., Abramson J., Lipton J.M. Hematopoietic stem cell transplantation for Diamond Blackfan anemia: a report from the Diamond Blackfan Anemia Registry. Bone Marrow Transplant. 2001;27:381–386. doi: 10.1038/sj.bmt.1702784. [DOI] [PubMed] [Google Scholar]
- 19.Hamaguchi I., Ooka A., Brun A., Richter J., Dahl N., Karlsson S. Gene transfer improves erythroid development in ribosomal protein S19-deficient Diamond-Blackfan anemia. Blood. 2002;100:2724–2731. doi: 10.1182/blood.V100.8.2724. [DOI] [PubMed] [Google Scholar]
- 20.Hamaguchi I., Flygare J., Nishiura H., Brun A.C., Ooka A., Kiefer T., Ma Z., Dahl N., Richter J., Karlsson S. Proliferation deficiency of multipotent hematopoietic progenitors in ribosomal protein S19 (RPS19)-deficient diamond-Blackfan anemia improves following RPS19 gene transfer. Mol. Ther. 2003;7:613–622. doi: 10.1016/s1525-0016(03)00091-1. [DOI] [PubMed] [Google Scholar]
- 21.Flygare J., Olsson K., Richter J., Karlsson S. Gene therapy of Diamond Blackfan anemia CD34(+) cells leads to improved erythroid development and engraftment following transplantation. Exp. Hematol. 2008;36:1428–1435. doi: 10.1016/j.exphem.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 22.Jaako P., Debnath S., Olsson K., Modlich U., Rothe M., Schambach A., Flygare J., Karlsson S. Gene therapy cures the anemia and lethal bone marrow failure in a mouse model of RPS19-deficient Diamond-Blackfan anemia. Haematologica. 2014;99:1792–1798. doi: 10.3324/haematol.2014.111195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zychlinski D., Schambach A., Modlich U., Maetzig T., Meyer J., Grassman E., Mishra A., Baum C. Physiological promoters reduce the genotoxic risk of integrating gene vectors. Mol. Ther. 2008;16:718–725. doi: 10.1038/mt.2008.5. [DOI] [PubMed] [Google Scholar]
- 24.Zhang F., Frost A.R., Blundell M.P., Bales O., Antoniou M.N., Thrasher A.J. A ubiquitous chromatin opening element (UCOE) confers resistance to DNA methylation-mediated silencing of lentiviral vectors. Mol. Ther. 2010;18:1640–1649. doi: 10.1038/mt.2010.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hacein-Bey-Abina S., Pai S.Y., Gaspar H.B., Armant M., Berry C.C., Blanche S., Bleesing J., Blondeau J., de Boer H., Buckland K.F. A modified γ-retrovirus vector for X-linked severe combined immunodeficiency. N. Engl. J. Med. 2014;371:1407–1417. doi: 10.1056/NEJMoa1404588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coci E.G., Maetzig T., Zychlinski D., Rothe M., Suerth J.D., Klein C., Schambach A. Novel self-inactivating vectors for reconstitution of Wiskott-Aldrich syndrome. Curr. Gene Ther. 2015;15:245–254. doi: 10.2174/1566523215666150126120327. [DOI] [PubMed] [Google Scholar]
- 27.Montiel-Equihua C.A., Zhang L., Knight S., Saadeh H., Scholz S., Carmo M., Alonso-Ferrero M.E., Blundell M.P., Monkeviciute A., Schulz R. The β-globin locus control region in combination with the EF1α short promoter allows enhanced lentiviral vector-mediated erythroid gene expression with conserved multilineage activity. Mol. Ther. 2012;20:1400–1409. doi: 10.1038/mt.2012.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jaako P., Flygare J., Olsson K., Quere R., Ehinger M., Henson A., Ellis S., Schambach A., Baum C., Richter J. Mice with ribosomal protein S19 deficiency develop bone marrow failure and symptoms like patients with Diamond-Blackfan anemia. Blood. 2011;118:6087–6096. doi: 10.1182/blood-2011-08-371963. [DOI] [PubMed] [Google Scholar]
- 29.Jaako P., Debnath S., Olsson K., Bryder D., Flygare J., Karlsson S. Dietary L-leucine improves the anemia in a mouse model for Diamond-Blackfan anemia. Blood. 2012;120:2225–2228. doi: 10.1182/blood-2012-05-431437. [DOI] [PubMed] [Google Scholar]
- 30.Brugman M.H., Suerth J.D., Rothe M., Suerbaum S., Schambach A., Modlich U., Kustikova O., Baum C. Evaluating a ligation-mediated PCR and pyrosequencing method for the detection of clonal contribution in polyclonal retrovirally transduced samples. Hum. Gene Ther. Methods. 2013;24:68–79. doi: 10.1089/hgtb.2012.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shannon C.E. The mathematical theory of communication. MD Comput. 1963;4:306–317. [PubMed] [Google Scholar]
- 32.Akagi K., Suzuki T., Stephens R.M., Jenkins N.A., Copeland N.G. RTCGD: Retroviral Tagged Cancer Gene Database. Nucleic Acids Res. 2004;32:D523–D527. doi: 10.1093/nar/gkh013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sadelain M., Papapetrou E.P., Bushman F.D. Safe harbours for the integration of new DNA in the human genome. Nat. Rev. Cancer. 2011;12:51–58. doi: 10.1038/nrc3179. [DOI] [PubMed] [Google Scholar]
- 34.Bijl J., Sauvageau M., Thompson A., Sauvageau G. High incidence of proviral integrations in the Hoxa locus in a new model of E2a-PBX1-induced B-cell leukemia. Genes Dev. 2005;19:224–233. doi: 10.1101/gad.1268505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lam Y.W., Lamond A.I., Mann M., Andersen J.S. Analysis of nucleolar protein dynamics reveals the nuclear degradation of ribosomal proteins. Curr. Biol. 2007;17:749–760. doi: 10.1016/j.cub.2007.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dull T., Zufferey R., Kelly M., Mandel R.J., Nguyen M., Trono D., Naldini L. A third-generation lentivirus vector with a conditional packaging system. J. Virol. 1998;72:8463–8471. doi: 10.1128/jvi.72.11.8463-8471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmidt M., Schwarzwaelder K., Bartholomae C., Zaoui K., Ball C., Pilz I., Braun S., Glimm H., von Kalle C. High-resolution insertion-site analysis by linear amplification-mediated PCR (LAM-PCR) Nat. Methods. 2007;4:1051–1057. doi: 10.1038/nmeth1103. [DOI] [PubMed] [Google Scholar]
- 38.Selich A., Daudert J., Hass R., Philipp F., von Kaisenberg C., Paul G., Cornils K., Fehse B., Rittinghausen S., Schambach A., Rothe M. Massive clonal selection and transiently contributing clones during expansion of mesenchymal stem cell cultures revealed by lentiviral RGB-barcode technology. Stem Cells Transl. Med. 2016;5:591–601. doi: 10.5966/sctm.2015-0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rittelmeyer I., Rothe M., Brugman M.H., Iken M., Schambach A., Manns M.P., Baum C., Modlich U., Ott M. Hepatic lentiviral gene transfer is associated with clonal selection, but not with tumor formation in serially transplanted rodents. Hepatology. 2013;58:397–408. doi: 10.1002/hep.26204. [DOI] [PubMed] [Google Scholar]
- 40.Wu X., Luke B.T., Burgess S.M. Redefining the common insertion site. Virology. 2006;344:292–295. doi: 10.1016/j.virol.2005.08.047. [DOI] [PubMed] [Google Scholar]
- 41.Hildinger M., Abel K.L., Ostertag W., Baum C. Design of 5′ untranslated sequences in retroviral vectors developed for medical use. J. Virol. 1999;73:4083–4089. doi: 10.1128/jvi.73.5.4083-4089.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schambach A., Wodrich H., Hildinger M., Bohne J., Kräusslich H.G., Baum C. Context dependence of different modules for posttranscriptional enhancement of gene expression from retroviral vectors. Mol. Ther. 2000;2:435–445. doi: 10.1006/mthe.2000.0191. [DOI] [PubMed] [Google Scholar]
- 43.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.