Abstract
Castration-resistant prostate cancer (CRPC) that occurs after the failure of androgen deprivation therapy is the leading cause of deaths in prostate cancer patients. Thus, there is an obvious and urgent need to fully understand the mechanism of CRPC and discover novel therapeutic targets. Long noncoding RNAs (lncRNAs) are crucial regulators in many human cancers, yet their potential roles and molecular mechanisms in CRPC are poorly understood. In this study, we discovered that an lncRNA HOXD-AS1 is highly expressed in CRPC cells and correlated closely with Gleason score, T stage, lymph nodes metastasis, and progression-free survival. Knockdown of HOXD-AS1 inhibited the proliferation and chemo-resistance of CRPC cells in vitro and in vivo. Furthermore, we identified several cell cycle, chemo-resistance, and castration-resistance-related genes, including PLK1, AURKA, CDC25C, FOXM1, and UBE2C, that were activated transcriptionally by HOXD-AS1. Further investigation revealed that HOXD-AS1 recruited WDR5 to directly regulate the expression of target genes by mediating histone H3 lysine 4 tri-methylation (H3K4me3). In conclusion, our findings indicate that HOXD-AS1 promotes proliferation, castration resistance, and chemo-resistance in prostate cancer by recruiting WDR5. This sheds a new insight into the regulation of CRPC by lncRNA and provides a potential approach for the treatment of CRPC.
Keywords: castration-resistant prostate cancer, CRPC, long noncoding RNA HOXD-AS1, cell cycle, chemo-resistance, WDR5, PLK1, AURKA
Graphical Abstract
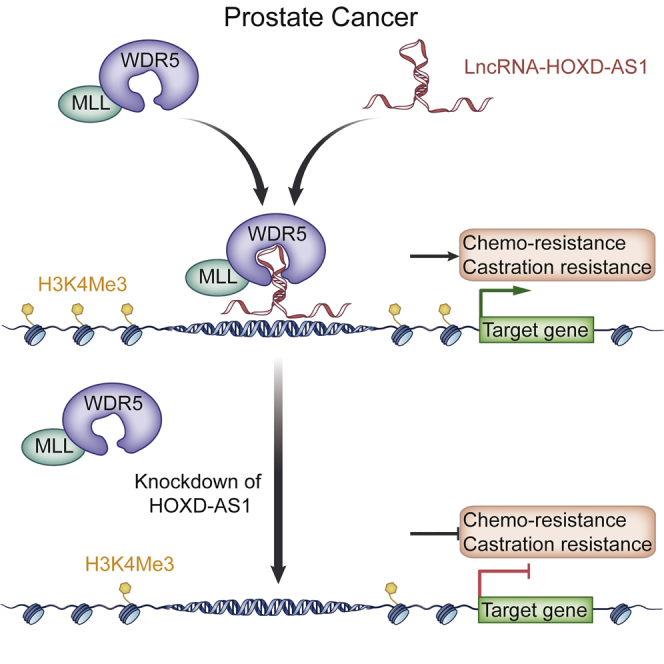
Huang, Lin, and colleagues show that long noncoding RNA HOXD-AS1 is upregulated in castration-resistant prostate cancer (CRPC) and correlated with disease progression. HOXD-AS1 promotes proliferation, castration resistance, and chemo-resistance of prostate cancer cells via interacting with WDR5, which in turn activates the transcription of downstream genes.
Introduction
Prostate cancer (PCa) is the most frequently diagnosed malignancy and the second leading cause of male cancer-related death in the United States.1 Androgen deprivation therapy (ADT) is the first-line treatment for patients with advanced PCa. Despite initial remission, nearly all patients inevitably progress to castration-resistant PCa (CRPC).2, 3 Paclitaxel-based chemotherapy or other systemic therapy is proven to decrease prostate-specific antigen (PSA) levels and palliate symptoms; however, the survival benefit is limited.4 Accumulating evidence suggests that persistent androgen receptor (AR) pathway signaling activation, alternative growth pathway activation (such as phosphatidylinositol 3-kinase [PI3K]/Akt/mTOR pathway), aberrant expression of anti-apoptotic proteins, and deregulation of cell-cycle-related genes are involved in the development of CRPC.5, 6 However, many of the key elements in the transition from androgen-dependent (AD) to androgen-independent (AI) cancer remain poorly understood. Identification of genes involved in this transition might allow the identification of novel therapeutic strategies for CRPC.
Long noncoding RNAs (lncRNAs) are RNA molecules that are longer than 200 nt but lack protein coding potential.7 Emerging evidence has revealed that lncRNAs play key roles in physiological and pathological processes, including embryonic development, organ formation, and tumorigenesis.7, 8, 9 Aberrant expression of lncRNAs has been observed in many types of cancers and may play important roles in regulating proliferation, chemo-resistance, and metastasis of cancer cells.10, 11, 12 Transcriptome sequencing and microarray analysis of clinical samples have discovered that numerous lncRNAs are deregulated in PCa,13 some of which are involved in the initiation and progression of PCa by acting as oncogenes or tumor suppressors. For example, lncRNA SChLAP1 is overexpressed in PCa and promotes PCa aggressiveness by antagonizing the SWI/SNF chromatin modification complex.14 A recent report has found that the lncRNA HOTAIR promotes AR signaling in an androgen-independent manner.15 To date, however, the roles of lncRNAs in CRPC remain largely unexplored. Thus, an improved understanding of lncRNAs in CRPC could help to develop better therapeutic strategies.
LncRNA HOXD-AS1, also known as HAGLR, is an evolutionary conserved non-protein coding transcript encoded by the HOXD gene cluster. HOXD-AS1 is upregulated upon retinoid acid stimulation and might control the expression of angiogenesis and inflammation-associated genes in neuroblastoma.16 HOXD-AS1 overexpression is also noted in bladder cancer tissue and cell lines, correlated with histological grade and TNM stage. Knockdown of HOXD-AS1 inhibits bladder cancer cell proliferation and migration in vitro.17 However, the function and mechanism of HOXD-AS1 in PCa remain unknown.
Current studies have shown that many lncRNAs bind to chromatin remodeling complexes, such as polycomb repressive complexes (PRCs) and mixed-lineage leukemia 1 (MLL1) to modulate downstream gene expression, serving as scaffolds of the histone modification complex.18 The WD repeat domain 5 (WDR5), a key subunit of MLL1, has been proven to associate with numerous lncRNAs to preserve the activation of chromatin.19 Furthermore, WDR5 is involved in multiple stages of tumorgenesis.20 WDR5 promotes proliferation, chemo-resistance, and self-renewal in bladder cancer cells by mediating H3K4 tri-methylation.20 More importantly, WDR5 is overexpressed in PCa tissue and enhances PCa cells androgen signaling by affecting phosphorylation and methylation on H3, thus maintaining an active chromatin state of androgen-responsive genes; however, the mechanism of how WDR5 is recruited to the target genes remains unclear.21 Thus, it would be interesting to investigate whether lncRNAs recruit WDR5 to exert its effects in PCa.
To address this, we used a microarray and database analysis to identify a castration-resistant-related lncRNA termed HOXD-AS1. We also investigated the function and mechanism of HOXD-AS1 in PCa cells. Our findings strongly suggest that HOXD-AS1 participates in PCa progression and is a multi-functional and promising therapeutic target.
Results
HOXD-AS1 Is Identified as a CRPC-Related lncRNA
To study the aberrantly expressed lncRNAs in CRPC cells, we first constructed two LNCaP castration-resistant sublines that are considered to best simulate the clinical progression of CRPC.22, 23 First, we generated LNCaP-Bic by continuously exposing cells to bicalutamide for more than 12 months. Second, LNCaP-AI was established by sustained passaging in cultural medium supplied with androgen-deprived serum (Figure S1A). The LNCaP-Bic and LNCaP-AI cells were resistant to bicalutamide and could proliferate persistently under androgen ablation conditions (Figure S1B and S1C). Consistent with previous studies,22, 23, 24 the mRNA and protein expression of AR, c-Myc, and bcl-2, and the mRNA expression of AR-V7 were significantly upregulated, whereas PSA was downregulated in LNCaP-Bic and LNCaP-AI cells (Figure S1D and S1E).
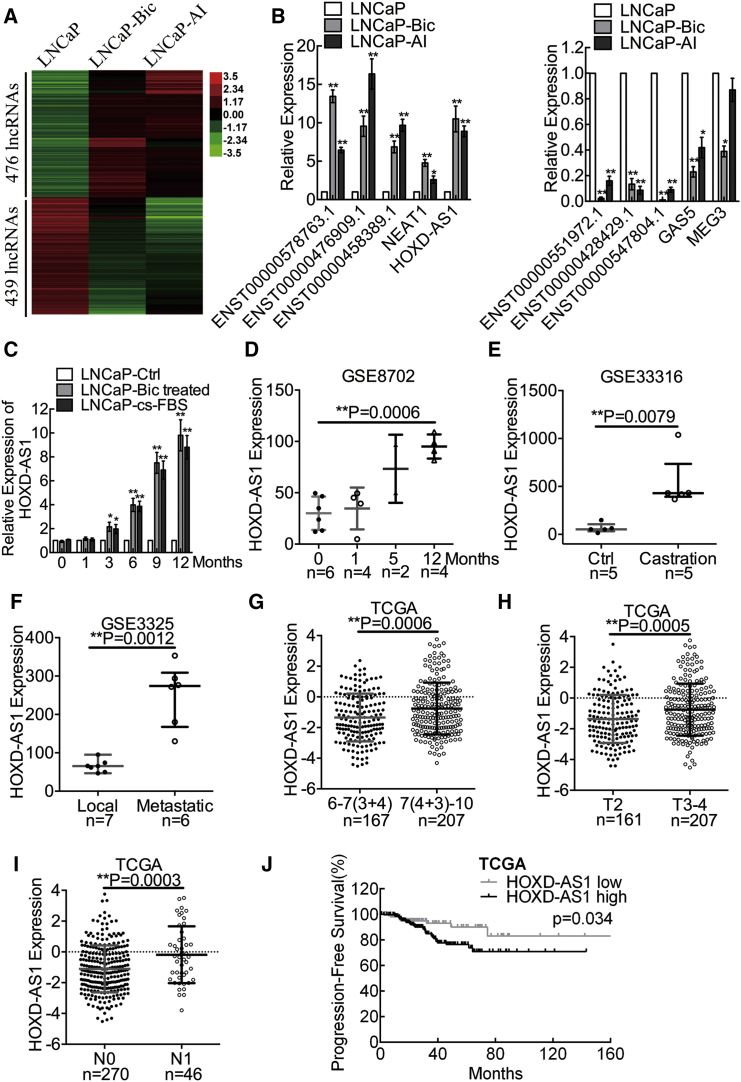
Next, we performed microarray analysis to identify differentially expressed lncRNAs in the transition from androgen-dependent to androgen-independent PCa cells. We identified 476 upregulated lncRNAs and 439 downregulated lncRNAs in CRPC cell lines LNCaP-Bic and LNCaP-AI compared with the parental LNCaP cells (Figure 1A). Furthermore, we used real-time qPCR to validate the findings in our microarray data. Initially, we focused on upregulated and downregulated lncRNAs with >5-fold changes (Figure 1B). Interestingly, we found that certain lncRNAs that were reported to play a role in PCa (NEAT1, GAS5, and MEG3) were upregulated or downregulated.25, 26, 27 Additionally, we observed that the expression of HOXD-AS1 increased gradually with prolonged androgen ablation (Figure 1C), and HOXD-AS1 was also overexpressed in androgen-independent PC-3 cells compared with LNCaP cells (Figure S1F). To identify the key lncRNA that modulates CRPC, we analyzed D’Antonio et al.’s datasets28 and found that HOXD-AS1 expression increased in LNCaP cells in a time course of androgen ablation (Figure 1D). Similarly, the HOXD-AS1 level also increased significantly in surgical castrated mice PCa xenografts compared with normal mice29 (Figure 1E). Moreover, overexpression of HOXD-AS1 was identified in metastatic PCa specimens compared with localized tumors in PCa patients30 (Figure 1F). Collectively, these data suggest that HOXD-AS1 might be a pivotal regulator in CRPC.
Figure 1.
HOXD-AS1 Is Identified as a Castration-Resistant Prostate-Cancer-Related lncRNA, Associates with Prostate Cancer Clinical Characteristics, and Predicts Disease Prognosis
(A) The differentially expressed lncRNAs in LNCaP versus LNCaP-Bic and LNCaP-AI were detected using a microarray. (B) The results from microarray analysis were validated by real-time qPCR. The results are presented as the means ± SD of values obtained in three independent experiments. (C) The expression of HOXD-AS1 in LNCaP cells treated with either bicalutamide or androgen ablation at different points in time was detected by real-time qPCR. The results are presented as the means ± SD of values obtained in three independent experiments. (D) GEO analysis of HOXD-AS1 expression in LNCaP cells under androgen ablation. The whiskers indicate means ± SD in the plots. (E) GEO analysis of HOXD-AS1 expression in castrated mice xenografts. The whiskers indicate median ± quartile in the plots. (F) GEO analysis of HOXD-AS1 expression in metastatic PCa versus localized PCa. The whiskers indicate medians ± quartile in the plots. (G) The expression of HOXD-AS1 in Gleason score 6–7(3+4) versus Gleason score 7(4+3)–10 PCa from TCGA database. The whiskers indicate means ± SD in the plots. (H) The expression of HOXD-AS1 in T2 versus T3-4 PCa. The whiskers indicate means ± SD in the plots. (I) The expression of HOXD-AS1 in N0 versus N1 PCa. The whiskers indicate means ± SD in the plots. (J) The progression-free survival rates of the 309 PCa patients were compared in the HOXD-AS1-low and HOXD-AS1-high groups. −1.8 was used as cutoff value in the survival analysis. The total number of patients was 374, 368, and 316 in each TCGA analysis, respectively. Patients with unavailable profiles were excluded before each analysis. See also Figure S2. *p < 0.05; **p < 0.01.
HOXD-AS1 Associates with PCa Clinical Characteristics and Predicts Disease Prognosis
To investigate whether HOXD-AS1 was involved in clinical PCa progression, we analyzed a large-scale RNA-sequencing (RNA-seq) dataset and the corresponding clinical information from The Cancer Genome Atlas (TCGA) and The Atlas of Noncoding RNAs in Cancer (TANRIC).31, 32 A total of 374 PCa profiles were included. We found that the expression level of HOXD-AS1 did not change significantly between benign tissues and PCa tissues (Figure S2). However, the HOXD-AS1 level was significantly higher in patients with a Gleason score of 7(4+3)–10 compared with 6–7(3+4), in T3-4 tumors compared with T2 tumors, and tumors with positive lymph node metastasis (Figures 1G–1I). We also found that the expression of HOXD-AS1 was correlated with Gleason score, T stage, and lymph node status (Table 1).
Table 1.
Association between HOXD-AS1 Expression and the Clinicopathological Features of TCGA Prostate Cancer Patients, n = 374
| Characteristics | Cases, n (%) | χ2 | p Value | |
|---|---|---|---|---|
| HOXD-AS1 expression | Low | High | ||
| Age, year (n = 368) | 0.596 | 0.44 | ||
| ≤60 | 58 (16) | 105 (29) | ||
| >60 | 81 (22) | 124 (33) | ||
| Gleason score (n = 374) | 7.285 | 0.007a | ||
| 6–7 (3+4) | 76 (20) | 91 (24) | ||
| 7 (4+3)–10 | 66 (18) | 141 (38) | ||
| Tumor stage (n = 368) | 6.367 | 0.013a | ||
| T2 | 72 (20) | 89 (24) | ||
| T3-4 | 66 (18) | 141 (38) | ||
| Lymph nodes status (n = 316) | 4.419 | 0.036a | ||
| Negative | 102 (32) | 168 (54) | ||
| Positive | 10 (3) | 36 (11) | ||
p < 0.05 was considered significant. Patients with no available clinical data were excluded from analysis with respect to the comparison of HOXD-AS1 expression.
Next, Kaplan-Meier survival analysis of 309 cases of TCGA PCa patients showed significantly reduced progression-free survival in PCa patients with increased HOXD-AS1 expression, as compared with patients with low HOXD-AS1 expression (p = 0.034; Figure 1J). To further evaluate the prognostic factors associated with progression-free survival in the 309 cases of TCGA PCa patients, we first carried out univariate analysis using age, tumor stage, Gleason score, lymph node status, and HOXD-AS1 expression as parameters. HOXD-AS1 expression and tumor stage were associated significantly with progression-free survival (p = 0.014 and 0.023, respectively; Table 2). Furthermore, the variables associated with survival by univariate analyses were adopted as covariates in the multivariate analyses, which revealed that high HOXD-AS1 expression in addition to tumor stage was an independent predictor of shorter progression-free survival (p = 0.005 and 0.009, respectively; Table 2). These findings clearly demonstrate the potential of HOXD-AS1 as a marker of poor prognosis in PCa.
Table 2.
Univariate and Multivariate Analyses of Factors Associated with Progression-Free Survival in 309 PCa Patients in TCGA
| Variable | Univariate |
Multivariate |
||||
|---|---|---|---|---|---|---|
| HRa | 95% CI | p Value | HRa | 95% CI | p Value | |
| Age, years (>60/≤60) | 0.912 | 0.496–1.677 | 0.768 | NA | ||
| Gleason score [7(4+3)–10 / 6–7(3+4)] | 0.853 | 0.624–1.167 | 0.321 | NA | ||
| Tumor stage (T3-4/T2) | 0.453 | 0.241–0.853 | 0.014b | 0.399 | 0.211–0.754 | 0.005b |
| Nodal metastasis (N1/N0) | 0.809 | 0.528–1.241 | 0.331 | NA | ||
| HOXD-AS1 (high/low) | 2.453 | 1.131–5.318 | 0.023b | 2.827 | 1.297–6.161 | 0.009b |
Univariate and multivariate analyses were performed. Cox proportional hazards regression model was used. Variables associated with survival by univariate analyses were adopted as covariates in multivariate analyses.
HR > 1, risk for death increased; HR < 1, risk for death reduced.
Significant p values.
Knockdown of HOXD-AS1 Inhibits PCa Cell Proliferation by Inducing G2/M Cell Cycle Arrest
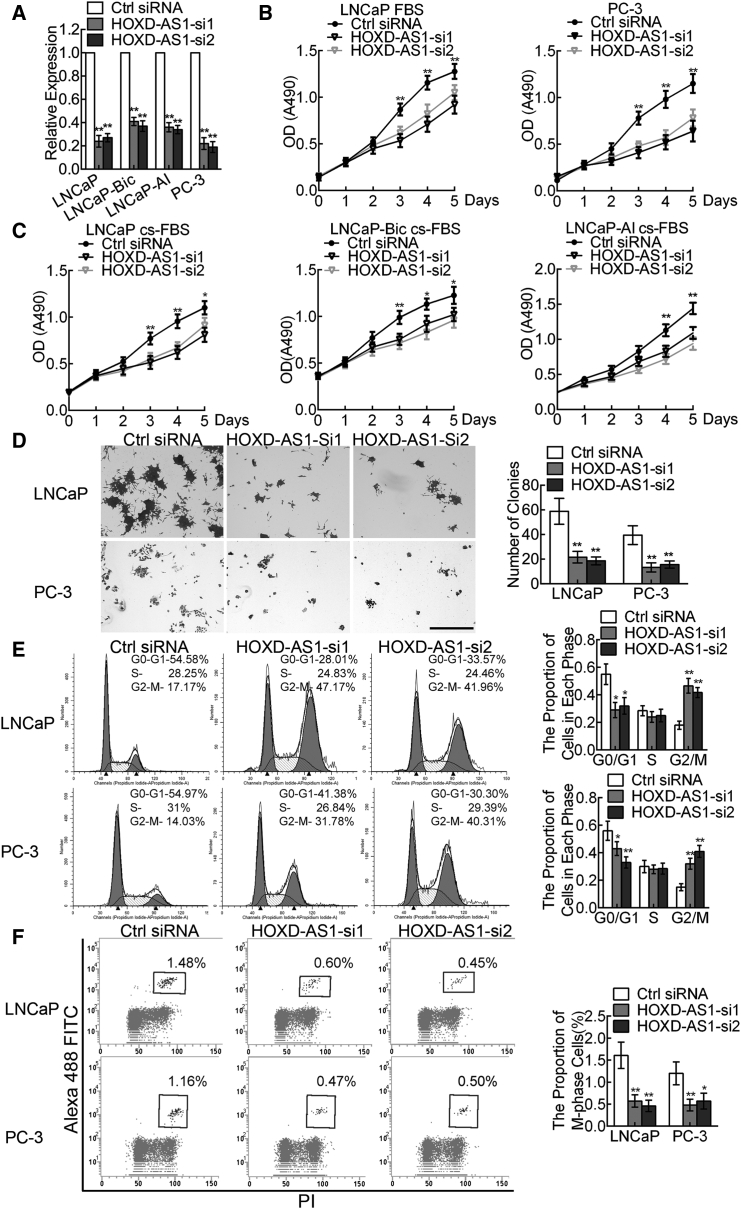
To further examine the functions of HOXD-AS1 in PCa progression, we knocked down HOXD-AS1 expression in PCa cells using two small interfering RNAs (siRNAs). Real-time qPCR showed that HOXD-AS1 was remarkably downregulated in LNCaP, LNCaP-Bic, LNCaP-AI, and PC-3 cells transfected with the HOXD-AS1 siRNAs compared with those transfected with the control siRNA (Figure 2A). HOXD-AS1 depletion not only decreased proliferation of LNCaP and PC-3 cells in normal medium (Figure 2B), but also inhibited the growth of androgen-sensitive LNCaP and castration-resistant LNCaP-Bic and LNCaP-AI cells in androgen ablated cultural medium (Figure 2C). Consistent with the cell growth data, HOXD-AS1 knockdown cells formed significantly fewer and smaller colonies compared with the control cells (Figure 2D).
Figure 2.
Knockdown of HOXD-AS1 Inhibits Prostate Cancer Cell Proliferation by Inducing G2/M Cell Cycle Arrest
(A) Efficiency of HOXD-AS1 knockdown in LNCaP, LNCaP-Bic, LNCaP-AI, and PC-3 cells by two siRNAs was verified by real-time qPCR. (B) Influence of HOXD-AS1 knockdown on viability of LNCaP and PC-3 cells in normal cultural medium measured by the MTT assay. The results are presented as the means ± SD of values obtained in three independent experiments. (C) The effect of HOXD-AS1 knockdown on LNCaP, LNCaP-Bic, and LNCaP-AI cells in androgen-deprived cultural medium. The results are presented as the means ± SD of values obtained in three independent experiments. (D) Effect of HOXD-AS1 knockdown on colony formation was measured in LNCaP and PC-3 cells. Scale bar, 600 μm. The histogram showed the mean ± SD of colonies from three independent experiments. (E) LNCaP and PC-3 cells were transfected with HOXD-AS1 siRNAs or control siRNA for 48 hr, and the cell cycles were analyzed by flow cytometry. (F) HOXD-AS1 silencing LNCaP and PC-3 cells were incubated with phospho-histone H3 antibody and analyzed by flow cytometry. Percentages (%) of cell populations at different stages of cell cycles are listed within the panels. All histograms show the percentage (%) of cell populations from three independent experiments. See also Figures S3 and S4. *p < 0.05; **p < 0.01.
Next, we performed flow cytometry assays to determine whether HOXD-AS1 was involved in the cell cycle regulation. Interestingly, HOXD-AS1 silencing increased the cell population in the G2/M phase dramatically and reduced the cell population in the G0/G1 phase in LNCaP, LNCaP-Bic, LNCaP-AI, and PC-3 cells (Figure 2E; Figure S3). To further determine the effect of HOXD-AS1 on the G2/M phase of PCa cells, we used phospho-histone H3 as a marker of mitosis. Interestingly, HOXD-AS1 knockdown significantly decreased the proportion of mitotic cells compared with the control group (Figure 2F). Moreover, treatment with nocodazole, which synchronized PCa cells at M-phase, further confirmed a decreased proportion of mitotic cells after HOXD-AS1 silencing (Figure S4). Collectively, our data indicate that knockdown of HOXD-AS1 inhibits PCa cell proliferation by inducing G2/M cell cycle arrest, predominantly by blocking entry into mitosis.
Downregulation of HOXD-AS1 Represses Castration Resistance and Chemo-Resistance of PCa Cells
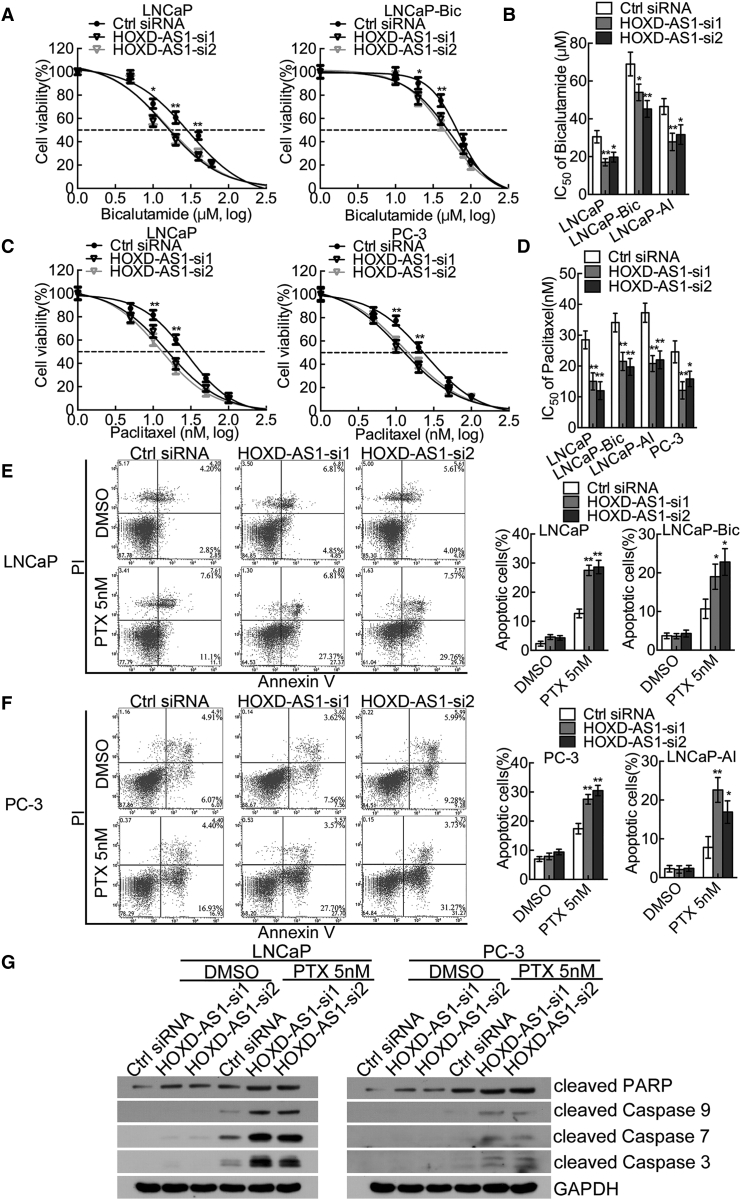
Bicalutamide is one of the most widely used ADT drugs in PCa. However, androgen-sensitive PCa eventually progresses into CRPC and no longer responds to bicalutamide. We explored whether HOXD-AS1 regulated the resistance to bicalutamide in PCa cells using the methyl thiazolyl tetrazolium (MTT) assay. Interestingly, HOXD-AS1 depletion sensitized LNCaP, LNCaP-Bic, and LNCaP-AI to bicalutamide treatment and produced a lower bicalutamide half inhibition concentration (IC50) compared with that of the control cells (Figures 3A and 3B; Figure S5A), indicating that HOXD-AS1 contributes to the castration resistance of PCa.
Figure 3.
Downregulation of HOXD-AS1 Represses Castration Resistance and Chemo-Resistance of Prostate Cancer Cells
(A) The viability of LNCaP and LNCaP-Bic cells transfected with HOXD-AS1 siRNAs or control siRNA, treated with bicalutamide for 120 hr and analyzed by MTT assay. (B) Calculation of IC50 of bicalutamide in LNCaP, LNCaP-Bic, and LNCaP-AI, four-parameter logistic curve (best-fit solution, nonlinear regression dynamic fitting), and normality tests are used (GraphPad Prism 5). (C) The viability of LNCaP and PC-3 cells transfected with HOXD-AS1 siRNAs or control siRNA, treated with paclitaxel for 48 hr and analyzed by MTT assay. (D) Calculation of IC50 of paclitaxel in LNCaP, LNCaP-Bic, LNCaP-AI, and PC-3 cells. (E and F) The LNCaP (E) and PC-3 (F) cells transfected with control or HOXD-AS1 siRNAs were treated with 5 nM paclitaxel for 48 hr. The percentage of apoptotic cells was analyzed by flow cytometer. The histogram showed the percentage (%) of late and early apoptotic cells from three independent experiments. (G) HOXD-AS1-depleted LNCaP and PC-3 cells were treated with either DMSO or 5 nM paclitaxel for 48 hr, and the expression of cleaved PARP and cleaved caspase-3, -7, and -9 were detected by western blotting. See also Figure S5. *p < 0.05; **p < 0.01.
Chemo-resistance to paclitaxel in PCa is correlated closely with disease progression and prognosis.33 Therefore, we investigated the role of HOXD-AS1 in chemo-resistance via MTT assays, flow cytometry, and western blotting. As shown in Figures 3C and 3D and Figure S5B, HOXD-AS1 knockdown diminished the resistance to paclitaxel and decreased the paclitaxel IC50 in LNCaP, LNCaP-Bic, LNCaP-AI, and PC-3 cells. We then quantified apoptosis by staining cells with Annexin V and propidium iodide (PI). HOXD-AS1 knockdown alone induced moderate levels of apoptosis, whereas the percentage of apoptotic cells increased significantly under paclitaxel treatment in LNCaP, LNCaP-Bic, LNCaP-AI, and PC-3 cells (Figure 3E and 3F; Figure S5C). Furthermore, we detected the apoptosis markers, such as cleaved PARP and caspase levels in LNCaP and PC-3 cells. Cells transfected with HOXD-AS1 siRNAs displayed an increased protein expression of cleaved PARP and cleaved caspase-3, -7, and -9 under paclitaxel treatment (Figure 3G). These data described above suggest that HOXD-AS1 plays a critical role in castration resistance and chemo-resistance of PCa.
Repression of HOXD-AS1 Suppresses Tumorigenicity and Chemo-Resistance of PCa In Vivo
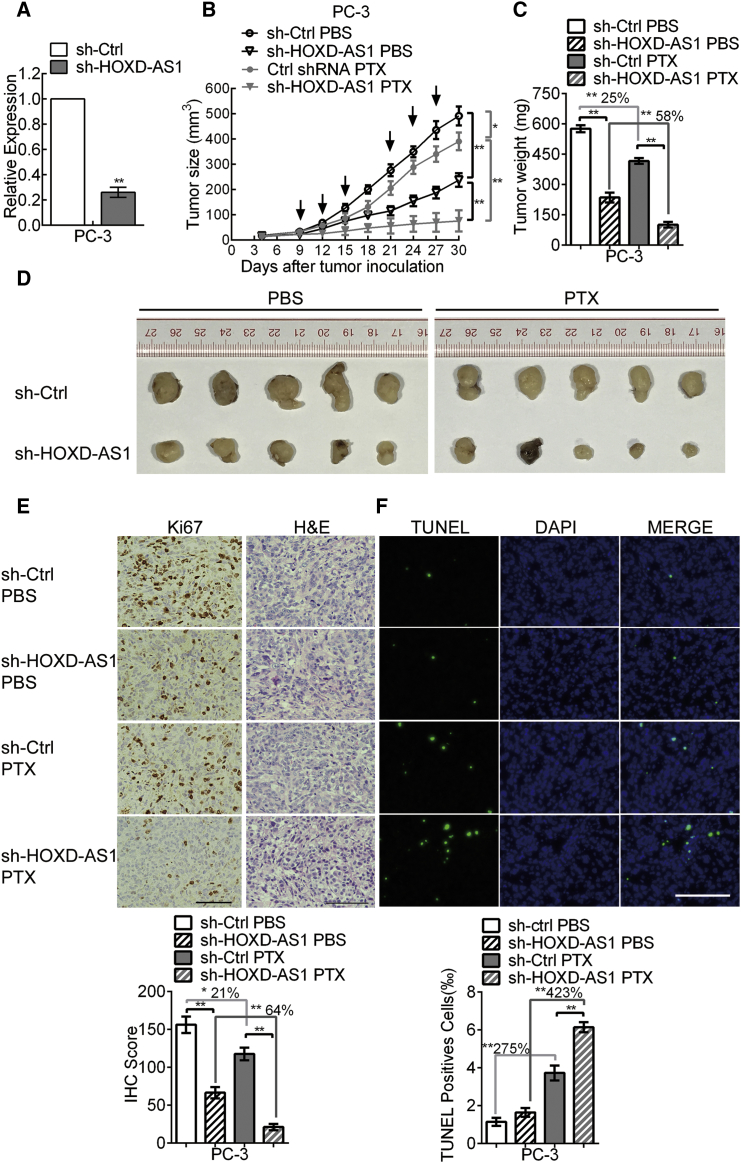
To further evaluate the effects of HOXD-AS1 on PCa cell tumorigenesis and chemo-resistance in vivo, we generated HOXD-AS1 stable knockdown PC-3 cells by lentivirus (Figure 4A). HOXD-AS1 knockdown PC-3 cells or control cells were then injected subcutaneously into BALB/c male nude mice. One week after tumor cells inoculation, the nude mice bearing PC-3-sh-Control or PC-3-sh-HOXD-AS1 xenografts were selected randomly for treatment with paclitaxel or PBS as reported previously.34, 35 Tumor growth in the HOXD-AS1 knockdown group was suppressed prominently compared with the control group (Figures 4B–4D). The weight of tumors from the HOXD-AS1 knockdown group was significantly lower than those of the control group (Figure 4C). Interestingly, the gap between the HOXD-AS1 knockdown and control groups was much more significant in the group treated with paclitaxel, as compared with the group given PBS (Figures 4B–4D). Moreover, the tumors derived from the HOXD-AS1 knockdown group exhibited lower expression of proliferation marker Ki67 and higher proportion of terminal deoxynucleotidyl transferase (TdT) dUTP nick-end labeling (TUNEL)-positive cells than the control group, and was especially significant under paclitaxel treatment (Figures 4E and 4F). These results indicate that HOXD-AS1 depletion inhibits PCa cell growth and chemo-resistance in vivo.
Figure 4.
Downregulation of HOXD-AS1 Suppresses Tumorigenicity and Chemo-Resistance of Prostate Cancer Cells In Vivo
(A) Construction of HOXD-AS1 stable knockdown PC-3 cells. The results are presented as the means ± SD of values obtained in three independent experiments. (B) The volume of tumors was measured every 3 days. The arrows indicate the time of PBS or paclitaxel administration. The results are presented as the means ± SD of values (n = 5). (C) The weight of tumors was measured after the tumors were surgically dissected. The results are presented as the means ± SD of values (n = 5). (D) Images of surgically dissected tumors of each group. (E) The expression of Ki67 in the tumor was examined by IHC. (F) The apoptosis in the tumor was detected by TUNEL assay. The scale bars in IHC and TUNEL images represent 50 μm. The histograms showed the score of IHC and proportion of TUNEL-positive cells in each group. The percentage in the histograms indicates the decreased or increased percentage in the PTX group compared with the PBS group. *p < 0.05; **p < 0.01.
The Target Genes of HOXD-AS1 Are Identified in PCa Cells
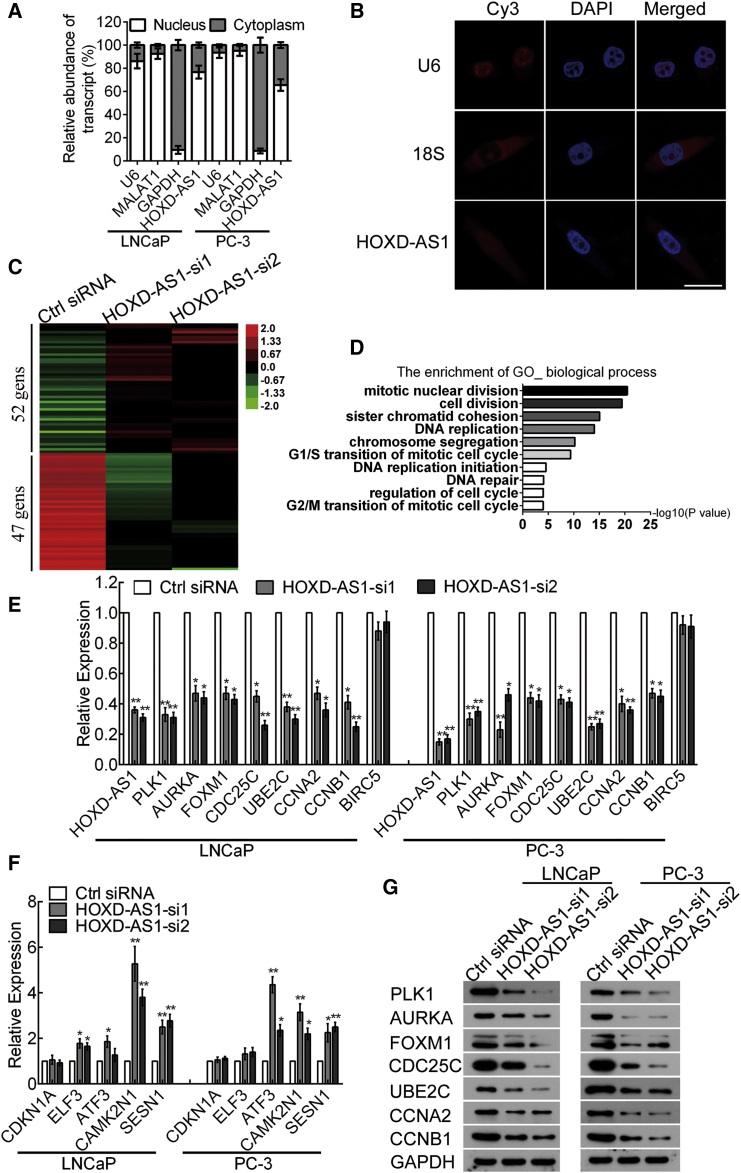
The subcellular localization of lncRNA is associated closely with its biological function. We conducted cellular fractionation assays and found that HOXD-AS1 was enriched in the nuclear fraction in LNCaP and PC3 cells (Figure 5A). Similarly, RNA fluorescence in situ hybridization (RNA-FISH) in PC-3 cells further confirmed that HOXD-AS1 was distributed mainly in the nucleus (Figure 5B). LncRNAs located in the nucleus are usually associated with transcriptional regulation.8, 10 To investigate the mechanism of HOXD-AS1 in PCa, we performed a microarray analysis to identify the target genes of HOXD-AS1 in LNCaP cells. Interestingly, in agreement with our functional assays, we found that several genes involved in the cell cycle, chemo-resistance, and castration resistance changed significantly. Fifty-two upregulated genes and 47 downregulated genes were found in the HOXD-AS1 knockdown groups compared with the control group (Figure 5C). Gene ontology (GO) analysis revealed that the genes regulated by HOXD-AS1 were enriched in mitotic nuclear division, cell division, DNA replication transcription, and DNA repair (Figure 5D). We then validated the expression of these genes in LNCaP, LNCaP-Bic, LNCaP-AI, and PC-3 cells transfected with control or HOXD-AS1 siRNA by real-time qPCR. The mRNA expression of PKL1, AURKA, FOXM1, CDC25C, UBE2C, CCNA2, and CCNB1, which facilitate mitosis,36, 37 G2/M cell cycle transition,38 microtube stabilization,39 and AR signaling,40 were downregulated significantly. In contrast, the mRNA expression of ATF3, CAMK2N1, and SESN1, which inhibit the growth of cancer cells,41, 42, 43 were increased in HOXD-AS1 silenced cells (Figures 5E and 5F; Figures S6A and S6B). Furthermore, consistent with their mRNA level, we found that the protein expression of PKL1, AURKA, FOXM1, CDC25C, UBE2C, CCNA2, and CCNB1 were repressed significantly upon HOXD-AS1 knockdown (Figure 5G). Collectively, these data suggest that HOXD-AS1 regulates PCa by modulating the expression of a series of cell cycle, chemo-resistance, and castration-resistance-related genes.
Figure 5.
The Target Genes of HOXD-AS1 Are Identified in Prostate Cancer Cells
(A) Nuclear fraction experiment and real-time qPCR detected the abundance of HOXD-AS1 in the nucleus and cytoplasm. GAPDH is the positive control for cytoplasm, and MALAT1 and U6 are the positive control for nucleus. The results are presented as the means ± SD of values obtained in three independent experiments. (B) The subcellular distribution of HOXD-AS1 was visualized by RNA fluorescence in situ hybridization (FISH) in PC3 cells. 18S was the positive control for cytoplasm, and U6 was the positive control for the nucleus. The scale bar represents 10 μm. (C) A heatmap represents mRNA expression levels in the LNCaP cells transfected with control or HOXD-AS1 siRNA for 48 hr. Each column represents the indicated sample, and each row indicates one mRNA. Red and green colors indicate high and low expression, respectively. (D) Gene ontology (GO) analysis was performed to identify the enrichment of the biological process. (E and F) The downregulated (E) and upregulated (F) genes in the microarray were verified in LNCaP and PC-3 cells by real-time qPCR. The results are presented as the means ± SD of values obtained in three independent experiments. (E) The expression of HOXD-AS1 target genes was detected by western blotting. GAPDH were used as internal controls. See also Figures S6 and S7. *p < 0.05; **p < 0.01.
HOXD-AS1 Binds to WDR5 and Activates Gene Expression by Mediating H3 Lysine 4 Tri-methylation at the Promoter Region of Target Genes
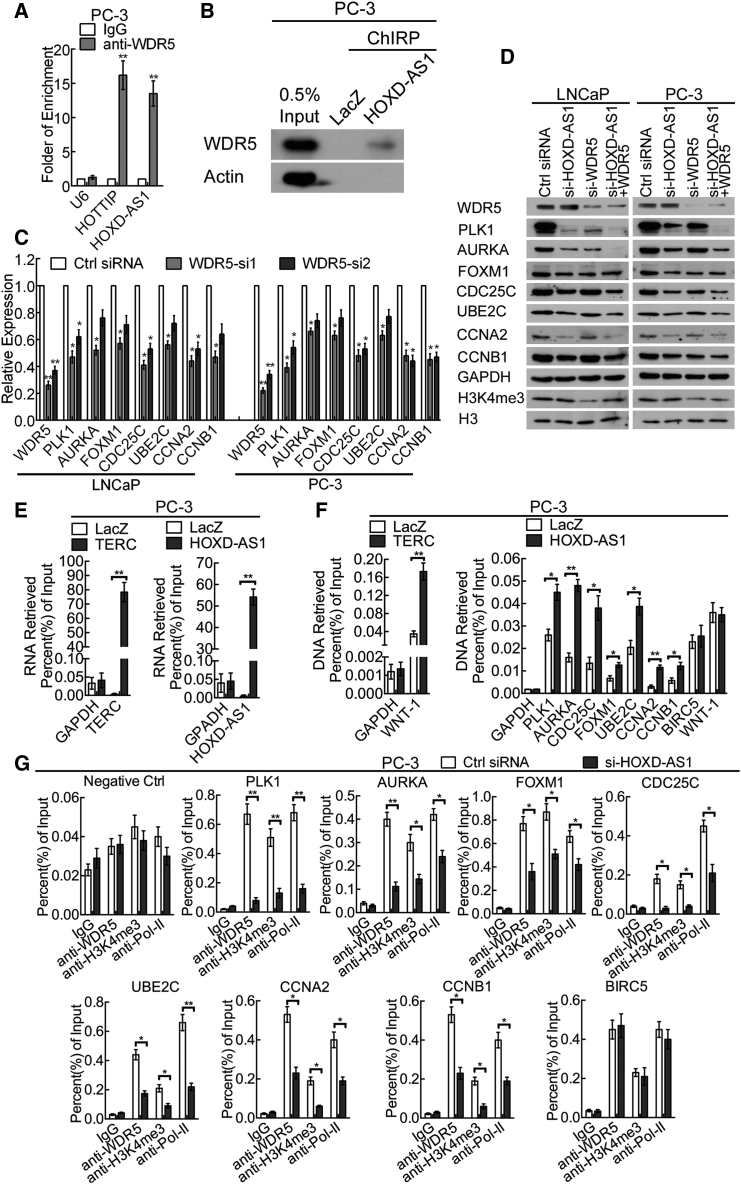
A recent study of RNA immunoprecipitation-sequencing (RIP-seq) analysis has suggested that many lncRNAs bind to WDR5, which is essential for WDR5 to “turn on” genetic transcription.19 Considering that HOXD-AS1 is located mainly in the nucleus, and more genes were downregulated than upregulated after HOXD-AS1 knockdown, we conducted an RIP assay in PC-3 cells to investigate whether HOXD-AS1 bound to WDR5. We found 13.5 times enrichment of HOXD-AS1 and 16.2 times enrichment of lncRNA HOTTIP using an anti-WDR5 antibody compared with using IgG (Figure 6A). LncRNA HOTTIP is a proven WDR5-binding lncRNA.18 No obvious enrichment was observed for the non-specific control U6. Furthermore, we also conducted a chromatin isolation by RNA purification (ChIRP) experiment in PC-3 cells to provide direct evidence that HOXD-AS1 associated with WDR5. Consistent with our RIP results, we found that WDR5 protein was enriched by HOXD-AS1 probes, but not the negative control LacZ probes, as detected by western blotting (Figure 6B). These results indicate that HOXD-AS1 interacts with the WDR5 protein.
Figure 6.
HOXD-AS1 Binds to WDR5 and Activates Gene Expression by Mediating H3 Lysine 4 Tri-methylation at the Promoter Region of Target Genes
(A) Real-time qPCR analysis of HOXD-AS1 and lncRNA HOTTIP in RNA immunoprecipitation assay of PC-3 cells using anti-WDR5, RNA enrichment was determined relative to the non-immune IgG control. U6 was used as a non-specific control. The results are presented as the means ± SD of values obtained in three independent experiments. (B) WDR5 was detected from the retrieved ChIRP protein of PC-3 cells by western blotting. Actin was detected as a non-specific control; LacZ was used as a negative control probe set. (C) HOXD-AS1 target genes were detected by real-time qPCR in WDR5 knockdown LNCaP and PC-3 cells. The results are presented as the means ± SD of values obtained in three independent experiments. (D) The effect of combined knockdown of HOXD-AS1 and WDR5 on the HOXD-AS1 target genes, as compared with silencing each of HOXD-AS1 or WDR5, or control siRNA, as assessed by western blotting. GAPDH were used as internal controls, and H3 was used as a control for H3K4me3. (E) ChIRP experiment was conducted using PC-3 cells. TERC and HOXD-AS1 were detected by ChIRP-real time qPCR. The TERC probe set was used as a ChIRP positive control; GAPDH was detected as a non-specific control. The values are normalized to the input and presented as the means ± SD. (F) The promoters of target genes of TERC and HOXD-AS1 were detected by ChIRP real-time qPCR. WNT-1 was used as a positive control of the TERC probe set; GAPDH promoter was detected as a non-specific control. WNT-1 was detected as a non-specific control of the HOXD-AS1 probe set. The values are normalized to the input and presented as the means ± SD. (G) ChIP analysis of IgG, WDR5, H3K4me3, and RNA polymerase-II status of candidate HOXD-AS1 target genes in PC-3 cells after knockdown assay. The values are normalized to input and presented as the means ± SD. See also Figure S8. *p < 0.05; **p < 0.01.
Next, we explored whether WDR5 regulated the target genes of HOXD-AS1. WDR5 siRNAs were transfected into LNCaP and PC-3 cells, and real-time qPCR was conducted to examine the transcriptional changes of PKL1, AURKA, FOXM1, CDC25C, UBE2C, CCNA2, and CCNB1. The expression of these genes was inhibited (Figure 6C), similarly to the HOXD-AS1-silenced PCa cells. Moreover, we inhibited the expression of HOXD-AS1 and WDR5 separately or together in LNCaP and PC-3 cells. Western blotting showed that the protein levels of PLK1, AURKA, CDC25C, and CCNB1 decreased significantly in the HOXD-AS1 and WDR5 combined knockdown group compared with knockdown of either of them alone. However, only a moderate decrease was observed for FOXM1, UBE2C, and CCNA2. In addition, WDR5 silencing significantly reduced the histone 3 lysine4 tri-methylation (Figure 6D). To identify whether the biological function of HOXD-AS1 was dependent on WDR5 in PCa cells, we overexpressed HOXD-AS1 and then downregulated WDR5 in androgen-dependent LNCaP cells (Figures S8A and S8B). Surprisingly, we found that enforced expression of HOXD-AS1 enhanced the proliferation, castration resistance, and chemo-resistance in LNCaP cells, whereas knockdown of WDR5 abolished the effect of HOXD-AS1 (Figures S8C–S8G). Taken together, these results suggest that HOXD-AS1 exerts its regulatory function in a WDR5-dependent manner.
To provide direct evidence that HOXD-AS1 associated with the promoter regions of its target genes, we performed a ChIRP experiment and detected the enrichment of specific regulatory regions by real-time qPCR. We observed obvious enrichment of the regulatory region of PLK1, AURKA, CDC25C, FOXM1, UBE2C, CCNA2, and CCNB1 using HOXD-AS1 probes, but no enrichment of GAPDH, WNT1, and BIRC5, as compared with LacZ probes. TERC and its downstream gene WNT-1 served as positive control for RNA and DNA enrichment (Figures 6E and 6F). To further confirm that HOXD-AS1 activated the transcription of target genes directly by binding WDR5 and mediating histone H3 lysine 4 tri-methylation (H3K4me3), we performed a chromatin immunoprecipitation (ChIP) assay in control or HOXD-AS1 knockdown cells. HOXD-AS1 knockdown resulted in decreased location of WDR5, H3K4me3, and RNA polymerase-II levels in the promoter regions of PKL1, AURKA, FOXM1, CDC25C, UBE2C, CCNA2, and CCNB1, but not in the negative control and BIRC5, suggesting that downregulation of these genes was regulated directly by HOXD-AS1 (Figure 6G). Taken together, these data indicate that HOXD-AS1 activates the transcription of target genes directly in PCa by recruiting WDR5 to mediate H3K4me3 at their promoter region.
Discussion
In the current study, we demonstrated the function and mechanism of HOXD-AS1 in CRPC. We used a microarray to identify dysfunctional lncRNAs in the transition from androgen-dependent to androgen-independent PCa. The lncRNAs NEAT1, GAS5, and MEG3 were upregulated or downregulated in our microarray. Overexpression of lncRNA NEAT1 is associated with PCa progression and confers castration resistance in PCa cells.25 Meanwhile, lncRNA GAS5 and lncRNA MEG3 are found downregulated in PCa tissue, suppressing proliferation and inducing apoptosis in PCa cells.26, 27 These findings confirmed the reliability of our cell model and microarray analysis. Furthermore, re-analysis of GEO datasets and 374 TCGA PCa profiles provided solid evidence that HOXD-AS1 is overexpressed in CRPC cell lines and in vivo castration tumor models. Clinical correlation analysis revealed overexpression of HOXD-AS1 correlated closely with higher Gleason score, T stage, lymph node metastasis, and poor prognosis in PCa patients. Similarly, HOXD-AS1 correlates with tumor size, grade, and TNM stage in bladder cancer.17 Taken together, HOXD-AS1 is involved in the PCa progression, especially the transition to a castration-defiant state.
LncRNAs participate in multiple biological processes crucial for PCa development, including proliferation,44 metastasis,45 and AR signaling.46 Here, we found that knockdown of HOXD-AS1 inhibited PCa cell proliferation in vitro and tumor growth in vivo by inducing the G2 to M phase cell cycle arrest. Consistent with our results, a recent study has found that knockdown of HOXD-AS1 inhibits bladder cancer proliferation in vitro17; however, the in vivo function of HOXD-AS1 and the underlying mechanism is not defined. Through a microarray analysis and real-time qPCR we identified several cell cycle modulators that were regulated by HOXD-AS1, including PLK1, AURKA, FOXM1, CDC25C, UBE2C, CCNA2, and CCNB1. These proteins are key regulators involved in various steps in G2 and M phase, such as mitosis entry,38 spindle assembly,36, 47 proper sister-chromatin segregation,36 and mitosis progression.36, 37, 38, 47, 48 These results suggest that HOXD-AS1 regulates PCa cell cycle by modulating a series of G2 and M phase-related genes.
Resistance to androgen deprivation is a conundrum in the clinical treatment of PCa. Persistent androgen signaling accounts for the most important mechanism in CRPC.5, 6 In our study, we found that the resistance to bicalutamide was impaired significantly in PCa cells after HOXD-AS1 depletion. Importantly, the target genes of HOXD-AS1, PLK1, AURKA, and UBE2C are also overexpressed in CRPC and promote CRPC development.40, 49, 50, 51, 52 A recent study has revealed that PLK1 provides growth signaling for PCa cells under androgen deprivation by activating the PI3K/AKT/mTOR pathway.49 It has also been reported that PLK1 promotes AR signaling by increasing intra-tumoral androgen biosynthesis.40, 49 Moreover, AURKA promotes neuroendocrine differentiation of PCa (NEPC) cells,50, 51 which leads to therapeutic resistance and a more aggressive phenotype of PCa.53 Mechanistically, AURKA facilitates neuroendocrine differentiation via stabilizing N-MYC and regulating the expression of the AR splicing variant AR-V7.51, 54 The transcription of UBE2C is selectively activated by AR and AR-V7 in CRPC,55, 56 and UBE2C depletion inhibits the proliferation of CRPC cells.56 Overexpression of CDC25C is found in PCa and correlated with disease progression.57 Knockdown of CHK2 sensitizes PCa cells to androgen stimulation in a CDC25C-dependent manner.58 It has been discovered that FOXM1 and CENPF drive PCa malignant progression by synergistically regulating the PI3K signaling.59 Taken together, our results indicate that HOXD-AS1 promotes the castration resistance of PCa cells through regulating the expression of PLK1, AURKA, CDC25C, and UBE2C.
Paclitaxel is the first-line drug in CRPC chemo-therapy, and resistance to paclitaxel is closely associated with the aberrant expression of anti-apoptotic proteins in PCa cells.60, 61 In the current study, we discovered that knockdown of HOXD-AS1 sensitized PCa cells to paclitaxel in vitro and in vivo. Similarly, downregulation of HOXD-AS1 increases the apoptosis of bladder cancer cells, but the detail mechanism remains uncovered.17 In our study, we identified several chemo-resistance-related genes including PLK1, AURKA, CDC25C, and FOXM1 that were activated by HOXD-AS1. Recent studies have revealed that downregulation of PLK1 and CDC25C enhances the chemo-sensitivity of paclitaxel in PCa and breast cancer.39, 62, 63 Moreover, AURKA overexpression confers resistance of paclitaxel by impairing the cell cycle checkpoint,64 whereas FOXM1 contributes to the chemo-resistance of paclitaxel in nasopharyngeal carcinoma and breast cancer cells.65, 66 Collectively, these findings support that HOXD-AS1 promotes the chemo-therapy resistance in PCa cells by regulating PLK1, AURKA, CDC25C, and FOXM1, and might represent a multi-functional target for drug development to PCa.
LncRNAs bind to chromatin modification complexes and serve as molecular scaffolds to mediate gene activation or repression.18, 67 WDR5, a component of the MLL1 complex, interacts frequently with lncRNAs and promotes gene expression by mediating H3K4me3.18, 19 Here, we found that HOXD-AS1 was enriched in the nucleus and binds to WDR5. Coincidentally, WDR5 knockdown also repressed the expression of HOXD-AS1 target genes. Additionally, a combined knockdown of HOXD-AS1 and WDR5 augmented the inhibition of HOXD-AS1-regulated genes, whereas the in vitro effect of enforced HOXD-AS1 expression could be abrogated by WDR5 silencing. This observation further supported that HOXD-AS1 binds to WDR5 to regulate its target genes. Chromatin-interacting lncRNAs recognize target genes by forming the RNA-DNA triplex.68 To provide that HOXD-AS1 interacted directly with target genes and bound to WDR5, we conducted ChIRP and found that HOXD-AS1 was associated with the regulatory region of target genes and WDR5. More importantly, ChIP assay confirmed that HOXD-AS1 regulated downstream gene activation by interacting with WDR5, which in turn mediated H3K4me3 at the promoters of target genes. Thus, our results demonstrate that HOXD-AS1 interacts directly with the promoter region of target genes and mediates H3K4me3 to activate transcription via binding to WDR5.
Overcoming castration resistance has always been the Gordian knot in PCa treatment. Fortunately, research during recent decades has brought us to the dawn of curing CRPC, because numerous biological targets have been discovered. Compared with protein coding genes and miRNAs, lncRNAs are emerging as novel therapeutic targets in cancer treatment.10 Delivering antisense oligonucleotides targeting MALAT1 has prevented lung cancer metastasis successfully in vivo.69 Additionally, enforced expression of GAS5 inhibits glioma growth and prolongs survival in vivo by targeting miR-222.70 In this paper, we observed that knockdown of HOXD-AS1 impeded proliferation and chemo-resistance of PCa significantly both in vitro and in vivo. Further exploration identified genes that are directly activated by HOXD-AS1, including PLK1, AURKA, FOXM1, CDC25C, and UBE2C. These genes have been identified as crucial regulators in CRPC and might represent cancer target. PLK1 and AURKA inhibitors are available and show inspiring in vitro effect in PCa.49, 71 Collectively, these results indicate that HOXD-AS1 has great potential as a multi-potent therapeutic target.
In summary, we showed that HOXD-AS1 promotes proliferation and chemo-resistance of CRPC by recruiting WDR5. The in vivo and in vitro results demonstrated that HOXD-AS1-based gene therapy improved the CRPC therapeutic efficacy. Therefore, HOXD-AS1 is a novel and promising therapeutic target for the management of CRPC.
Materials and Methods
Cell Culture
The cell lines used in this study included the human PCa cells LNCaP and PC-3 (ATCC), and the CRPC-like cell LNCaP-Bic and LNCaP-AI. LNCaP cells were cultured in RPMI-1640 (GIBCO), PC-3 cells were cultured in F-12K media (GIBCO), supplemented with 10% FBS (fetal bovine serum; Shanghai ExCell Biology), and LNCaP-AI cells were cultured in phenol red free RPMI-1640 containing 10% charcoal stripped FBS (GIBCO), whereas LNCaP-Bic were cultured with 20 μM bicalutamide (Sigma). All media were supplemented with 1% penicillin/streptomycin. LNCaP cells were cultured in phenol red free medium with charcoal stripped FBS for at least 2 days before any experiment. Cells were grown in a humidified atmosphere of 5% CO2 at 37°C.
Construction of Castration-Resistant LNCaP Sublines
LNCaP cells were purchased from ATCC. To construct LNCaP-Bic cells, we cultured LNCaP cells in phenol red free RPMI-1640 containing 10% FBS. Bicalutamide was supplemented at a starting concentration of 5 μM, with a weekly increment of 50% concentration, ultimately being maintained at 20 μM. To construct LNCaP-AI cells, we cultured LNCaP cells in phenol red free RPMI-1640 containing 10% charcoal stripped FBS (cs-FBS). Both sublines were passaged every 5 days at an ∼1:2–1:4 ratio for 12 months, and the cultural medium was refreshed thrice per week. The LNCaP cells began to undergo obvious growth inhibition 1 week after androgen ablation and the inhibition continued for ∼6–7 months; then the proliferation accelerated gradually until reaching a comparable rate of normal LNCaP cells. Because LNCaP and its sublines were very loosely attached to the cultural flasks, the flasks should be handled carefully and avoid unnecessary turbulence; the cells should be kept without any disturbance for at least 48 hr after each passaging.
Microarray Analysis
Total RNA was extracted using TRIzol reagent and was further purified using QIAGEN RNeasy Mini Kit according to the manufacturer’s instructions. The LncRNA+ mRNA Human Gene Expression Microarray V4.0 (CapitalBio) was used to investigate the differential expressed lncRNAs in LNCaP and its sublines. For detection of the expression of genes after HOXD-AS1 knockdown, the PrimeView Human Gene Expression Array (Affymetrix) was used in this study, and the analysis was performed by CapitalBio Corporation according to the manufacturer’s instructions. The arrays were scanned on a GeneChip Scanner 3000, and the data were analyzed using GeneChip Operating software (GCOS 1.4). All primary data in microarray analysis have been uploaded to the Gene Expression Omnibus (GEO): GSE93929 and GSE93928.
The TCGA Data Mining
Patients’ clinical profiles in the TCGA prostate adenocarcinoma cohort31 are available at TCGA https://cancergenome.nih.gov/. The expression of HOXD-AS1 in PCa was obtained from TANRIC32 (http://ibl.mdanderson.org/tanric/_design/basic/query.html). The characteristics and clinicopathological features of the patients are listed in in Table S1. The TCGA prostate adenocarcinoma cohort comprising 374 patients was used for the analysis. Patients with no available clinical data were excluded from analysis in respective comparison of HOXD-AS1 expression. For the survival, univariate, and multivariate analyses, 309 cases with complete follow-up information were included; the rest were excluded.
RNAi
siRNA oligos targeting HOXD-AS1 (5′-GAAAGAAGGACCAAAGUAATT-3′, 5′-GCCCUUUCUGACCUGCUUATT-3′), WDR5 (5′-GCUCAGAGGAUAACCUUGUTT-3′, 5′-CCCAGUCCAACCUUAUUGUTT-3′), and negative control siRNAs were purchased from GenePharma. siRNA transfections were performed with 75 nM siRNA and Lipofectamine RNAiMAX (Life Technologies) as described previously.72
Stable HOXD-AS1 Knockdown Cell Lines
The pLKO.1 TRC cloning vector (Addgene plasmid 10878) was used to generate a short hairpin RNA (shRNA) against HOXD-AS1 (5′-GAAAGAAGGACCAAAGTAATG-3′) or negative control (5′-CCTAAGGTTAAGTCGCCCTCG-3′). Lentivirus production and infection were conducted as described previously.73
Cell Proliferation Assay
The MTT (MTS, Promega) colorimetric assay was used to detect cell viability. Cells transfected with control or HOXD-AS1 siRNA were seeded in 96-well plates at a density of 2 × 103 cells/well. Then the absorbance was measured at a wavelength of 490 nm for 5 days using a SpectraMax M5 (Molecular Devices).
For the colony formation assay, the cells were seeded in a 96-well plate at a density of 500 cells/well after siRNA transfection. Approximately 7 days later, the clones were washed with 1× PBS and stained with crystal violet for approximately 20 min. The clones were then imaged and quantified.
For the cell cycle analysis, 48 hr after transfection, the cells were harvested and fixed in 70% ice-cold ethanol, followed by RNase A treatment, and stained with 50 μg/mL PI. For the analysis of M-phase cells, fixed cells were collected and treated with 0.5% Triton PBS, then incubated with anti-phospho-H3 antibody (Cell Signaling Technology) at 1:50 dilution. All analyses were conducted on a FACSCalibur BD flow cytometer. The data were collected and processed using the BD FACSuite analysis software.
Chemosensitivity Assay
The cells transfected with control or HOXD-AS1 siRNAs were treated with 0, 10, 20, 40, 60, 100 μM of bicalutamide (Sigma) for 120 hr or 0, 5, 10, 20, 50, 100 nM of paclitaxel (Selleck) for 48 hrs. Cell viability was measured using the MTT assay. The cell viability was measured using the same method as MTT assay. For calculation of IC50, data were fitted in GraphPad Prism 5 (GraphPad Software), and dose-response curve was plotted using the equation log(inhibitor) versus response-variable slope. This is also called a four-parameter dose-response curve: Y = bottom + (top-bottom)/ (1+10ˆ [(log IC50-X) *HillSlope]).20
Apoptosis Analysis
The cells transfected with control or HOXD-AS1 siRNAs were treated with 0.1% DMSO or 5 nM paclitaxel (in DMSO) for 48 hr. The cells were then collected, washed with PBS, and the cell apoptosis was analyzed with Annexin V-fluorescein isothiocyanate (FITC) and PI (Biotool) staining in a FACSCalibur BD flow cytometer.
The TUNEL assay was conducted by using the In Situ Cell Death Detection Kit (Roche), following the manufacturer’s instructions.
RNA Isolation and Real-Time qPCR
Total RNA was extracted from cells using TRIzol reagent (TaKaRa Biotechnology) according to the manufacturer’s protocol. Total RNA was reverse transcribed with a PrimerScript RT-PCR kit (Takara Biotechnology). Real-time qPCR was conducted using a standard SYBR Green PCR kit (Roche) protocol with a CFX real-time instrument (Bio-Rad). The relative expression was calculated using the 2−ΔΔCt method.74 The transcription level of GAPDH was used as an internal control. All specific primers are listed in Table S2.
Western Blotting
Western blotting was performed as previously described.20 Primary antibodies specific to cleaved PARP, cleaved caspase-9, cleaved caspase-7, cleaved caspase-3, AURKA, CDC25C, FOXM1, CCNA2, CCNB1, H3, PSA, c-Myc, BCL-2, GAPDH (1:1,000; Cell Signaling Technology), PLK1, UBE2C, WDR5, H3K4me3, SESN1 (1:1,000; Abcam), AR (1:200; Santa Cruz), and CAMK2N1 (1:1,000; Proteintech) were used. The blots were then incubated with goat anti-rabbit or anti-mouse secondary antibody (Cell Signaling Technology) and visualized using enhanced chemiluminescence. The full images of all western blots were provided at the end of the Supplemental Information.
Mouse Xenograft Experiments
All of the animal care and experimental procedures were approved by the Institutional Animal Care and Use Committee of Sun Yat-sen University. The animal experiments were carried out in accordance with institutional guidelines. Male BALB/c nude mice (4–5 weeks old) were purchased from the Experimental Animal Center of Sun Yat-sen University and housed in specific pathogen-free (SPF) barrier facilities. A total of 3 × 106 cells mixed with an equal volume of Matrigel (Corning) were injected subcutaneously on the right side of the dorsum and five mice were used. After 1 week of tumor cells inoculation, the nude mice bearing PC-3-sh-control or PC-3-sh-HOXD-AS1 xenografts were selected randomly for treatment with paclitaxel (10 mg/kg) or PBS control administered thrice per week by intraperitoneal injection during weeks 2 and 4 as previously reported.34, 35 The size of the tumor was measured every 3 days. Thirty days post-implantation, the mice were euthanized and tumors were dissected surgically. The tumor specimens were fixed in 4% paraformaldehyde.
IHC Staining and Scoring Analyses
This experiment and immunohistochemistry (IHC) score calculations were conducted as described previously.73 Anti-Ki67 antibodies (1:500; Zhongshan Bio-Tech) were used to detect the expression of Ki67 in mouse tumors. Images were visualized using a Nikon ECLIPSE Ti microscope system and processed with Nikon software.
RNA FISH
The FISH kit was purchased from Ribo Bio (Guangzhou), and the experiment is performed according to the manufacturer’s instructions and visualized by a confocal microscope (Zeiss). In brief, the cells were seeded and fixed with 4% paraformaldehyde, and treated with 0.5% Triton in PBS followed by pre-hybridization. They were then hybridized at 5 μM probe concentration overnight. The CY3-labeled U6, 18S, probes were provided by the Ribo Bio (Guangzhou), and the HOXD-AS1 probe was synthesized by Sangon HOXD-AS1 probe 5′-CGCATCTCTATTTGGTTTGA-3′.
Nuclear Fraction
The cellular fraction was isolated as described previously.67 In brief, 107 cells were harvested, resuspended in 1 mL of ice-cold RNase-free PBS, 1 mL of buffer C1 (1.28M sucrose, 40 mM Tris-HCl [pH 7.5], 20mM MgCl2, 4% Triton X-100), and 3 mL of RNase-free water, and incubated for 15 min on ice. Then cells were centrifuged for 15 min at 2,500 rpm; the supernatant containing cytoplasmic constituent and the nuclear pellet were kept for RNA extraction.
RNA Immunoprecipitation
RNA immunoprecipitation (RIP) was performed using the EZ-Magna RIP kit (Millipore) according to the manufacturer’s instructions and described previously.67 In brief, 107 cells were lysed with RIP lysis buffer with one freeze-thaw cycle. Cell extracts were coimmunoprecipitated with anti-WDR5 (Abcam), and the retrieved RNA was subjected to real-time qPCR analysis. Normal rabbit IgG was used as a negative control. For real-time qPCR analysis, HOTTIP was used as a positive control, and U6 was used as a non-specific control.
ChIP
Cells were transfected with an equal portion of mixed HOXD-AS1 siRNAs or control siRNA for 48 hr. ChIP was conducted using an EZ-Magna ChIP A/G kit (Millipore) according to the manufacturer’s instructions and the previously reported detailed protocol.20, 73 Anti-WDR5 and anti-H3K4me3 were purchased from Abcam. Normal mouse IgG was used as a negative control, and anti-RNA polymerase-II (pol-II; Millipore) was used as a positive control antibody. Primers for ChIP-qPCR were listed in Table S3.
Chromatin Isolation by RNA Purification
A Magna ChIRP RNA Interactome Kit was purchased from Millipore (Millipore), and the experiment was conducted according to the manufacturer’s instructions. In brief, 3′ end Biotin-TEG modified-DNA probes against HOXD-AS1 and TERC were synthesized by Sangon. The sequences of the probes are available in Table S4. A total of 2 × 107 PC-3 cells was cross-linked for each hybrid reaction. Then the cell lysate was sonicated to shear the chromatin to ∼100–500 bp fragments. The sonicated cell lysates were hybridized with a mixture of biotinylated DNA probes for 4 hr at 37°C. Then the binding complexes were recovered using streptavidin-conjugated magnet beads. Finally, DNA, RNA, and protein were eluted and purified from the beads for real-time qPCR or western blotting analyses. The LacZ probe set was provided along with the kit to serve as a negative control probe. The primers used for real-time qPCR are listed in Table S5.
Statistical Analyses
Data are presented as the mean ± SD of three independent experiments. Two-tailed Student’s t tests and one-way analysis of variance were used to evaluate the data. Cumulative survival time was calculated using the Kaplan-Meier method and analyzed by the log rank test. A multivariate Cox proportional hazards model was used to estimate the adjusted hazard ratios (HRs) and 95% confidence intervals (CIs) and to identify independent prognostic factors. All statistical analyses were performed with SPSS 19.0. Differences were considered statistically significant at p < 0.05 and p < 0.01.
Author Contributions
J. Huang and T.L. designed the study, analyzed data, and wrote the manuscript. P.G. and X.C. performed the initial experimental design, participated in the experiment, performed data analysis, and wrote the initial manuscript. R.X., W.X., and M.Y. performed the animal experiments. J. Han and C.C. analyzed the clinical data. B.W. and W.D. performed the IHC and TUNEL assays. Z.C. and J.J. performed the experiment for cell cycle analysis. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare that they have no conflict of interests.
Acknowledgments
This study was funded by the National Natural Science Foundation of China (grants 81572514, U1301221, 81472384, 81402106, 81372729, 81272808, 81172431, and 81402106); National Natural Science Foundation of Guangdong (grants 2016A030313321, 2016A030313244, 2015A030311011, 2015A030310122, S2013020012671, and 2015A030310122); Science and Technology Program of Guangzhou (grants 201604020156 and 201604020177); “Three Big Constructions” funds of Sun Yat-sen University (J. Huang and T.L.); Specialized Research Fund for the Doctoral Program of Higher Education (grant 20130171110073 to T.L.); Fundamental Research Funds for the Central Universities (J. Huang); Project Supported by Guangdong Province Higher Vocational Colleges & Schools Pearl River Scholar Funded Scheme (T.L.); Elite Young Scholars Program of Sun Yat-Sen Memorial Hospital (grant J201401 to T.L.); Sun Yat-sen Initiative Program for Scientific Research (grant YXQH201708 to X.C.); Cultivation of Major Projects and Emerging, Interdisciplinary Fund, Sun Yat-Sen University (grant 16ykjc18); Yat-Sen Scholarship for Young Scientist (to B.W.); National Clinical Key Specialty Construction Project for Department of Urology and Department of Oncology; Key Laboratory of Malignant Tumor Gene Regulation and Target Therapy of Guangdong Higher Education Institutes, Sun-Yat-Sen University (grant KLB09001); and Key Laboratory of Malignant Tumor Molecular Mechanism and Translational Medicine of Guangzhou Bureau of Science and Information Technology (grant [2013]163).
Footnotes
Supplemental Information includes Supplemental Materials and Methods, eight figures, and five tables and can be found with this article online at http://dx.doi.org/10.1016/j.ymthe.2017.04.016.
Contributor Information
Jian Huang, Email: urolhj@sina.com.
Tianxin Lin, Email: tianxinl@sina.com.
Supplemental Information
References
- 1.Siegel R., Ma J., Zou Z., Jemal A. Cancer statistics, 2014. CA Cancer J. Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Wong Y.N., Ferraldeschi R., Attard G., de Bono J. Evolution of androgen receptor targeted therapy for advanced prostate cancer. Nat. Rev. Clin. Oncol. 2014;11:365–376. doi: 10.1038/nrclinonc.2014.72. [DOI] [PubMed] [Google Scholar]
- 3.Katzenwadel A., Wolf P. Androgen deprivation of prostate cancer: leading to a therapeutic dead end. Cancer Lett. 2015;367:12–17. doi: 10.1016/j.canlet.2015.06.021. [DOI] [PubMed] [Google Scholar]
- 4.Dayyani F., Gallick G.E., Logothetis C.J., Corn P.G. Novel therapies for metastatic castrate-resistant prostate cancer. J. Natl. Cancer Inst. 2011;103:1665–1675. doi: 10.1093/jnci/djr362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karantanos T., Corn P.G., Thompson T.C. Prostate cancer progression after androgen deprivation therapy: mechanisms of castrate resistance and novel therapeutic approaches. Oncogene. 2013;32:5501–5511. doi: 10.1038/onc.2013.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zong Y., Goldstein A.S. Adaptation or selection—mechanisms of castration-resistant prostate cancer. Nat. Rev. Urol. 2013;10:90–98. doi: 10.1038/nrurol.2012.237. [DOI] [PubMed] [Google Scholar]
- 7.Schmitz S.U., Grote P., Herrmann B.G. Mechanisms of long noncoding RNA function in development and disease. Cell. Mol. Life Sci. 2016;73:2491–2509. doi: 10.1007/s00018-016-2174-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perry R.B., Ulitsky I. The functions of long noncoding RNAs in development and stem cells. Development. 2016;143:3882–3894. doi: 10.1242/dev.140962. [DOI] [PubMed] [Google Scholar]
- 9.Huarte M. The emerging role of lncRNAs in cancer. Nat. Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 10.Gutschner T., Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9:703–719. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yue B., Cai D., Liu C., Fang C., Yan D. Linc00152 functions as a competing endogenous RNA to confer oxaliplatin resistance and holds prognostic values in colon cancer. Mol. Ther. 2016;24:2064–2077. doi: 10.1038/mt.2016.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng J., Liu X., Wang P., Xue Y., Ma J., Qu C., Liu Y. CRNDE promotes malignant progression of glioma by attenuating miR-384/PIWIL4/STAT3 axis. Mol. Ther. 2016;24:1199–1215. doi: 10.1038/mt.2016.71. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Prensner J.R., Iyer M.K., Balbin O.A., Dhanasekaran S.M., Cao Q., Brenner J.C., Laxman B., Asangani I.A., Grasso C.S., Kominsky H.D. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat. Biotechnol. 2011;29:742–749. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prensner J.R., Iyer M.K., Sahu A., Asangani I.A., Cao Q., Patel L., Vergara I.A., Davicioni E., Erho N., Ghadessi M. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat. Genet. 2013;45:1392–1398. doi: 10.1038/ng.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang A., Zhao J.C., Kim J., Fong K.W., Yang Y.A., Chakravarti D., Mo Y.Y., Yu J. LncRNA HOTAIR enhances the androgen-receptor-mediated transcriptional program and drives castration-resistant prostate cancer. Cell Rep. 2015;13:209–221. doi: 10.1016/j.celrep.2015.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yarmishyn A.A., Batagov A.O., Tan J.Z., Sundaram G.M., Sampath P., Kuznetsov V.A., Kurochkin I.V. HOXD-AS1 is a novel lncRNA encoded in HOXD cluster and a marker of neuroblastoma progression revealed via integrative analysis of noncoding transcriptome. BMC Genomics. 2014;15(Suppl 9):S7. doi: 10.1186/1471-2164-15-S9-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li J., Zhuang C., Liu Y., Chen M., Chen Y., Chen Z., He A., Lin J., Zhan Y., Liu L. Synthetic tetracycline-controllable shRNA targeting long non-coding RNA HOXD-AS1 inhibits the progression of bladder cancer. J. Exp. Clin. Cancer Res. 2016;35:99. doi: 10.1186/s13046-016-0372-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang K.C., Yang Y.W., Liu B., Sanyal A., Corces-Zimmerman R., Chen Y., Lajoie B.R., Protacio A., Flynn R.A., Gupta R.A. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang Y.W., Flynn R.A., Chen Y., Qu K., Wan B., Wang K.C., Lei M., Chang H.Y. Essential role of lncRNA binding for WDR5 maintenance of active chromatin and embryonic stem cell pluripotency. eLife. 2014;3:e02046. doi: 10.7554/eLife.02046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen X., Xie W., Gu P., Cai Q., Wang B., Xie Y., Dong W., He W., Zhong G., Lin T., Huang J. Upregulated WDR5 promotes proliferation, self-renewal and chemoresistance in bladder cancer via mediating H3K4 trimethylation. Sci. Rep. 2015;5:8293. doi: 10.1038/srep08293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim J.Y., Banerjee T., Vinckevicius A., Luo Q., Parker J.B., Baker M.R., Radhakrishnan I., Wei J.J., Barish G.D., Chakravarti D. A role for WDR5 in integrating threonine 11 phosphorylation to lysine 4 methylation on histone H3 during androgen signaling and in prostate cancer. Mol. Cell. 2014;54:613–625. doi: 10.1016/j.molcel.2014.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kokontis J., Takakura K., Hay N., Liao S. Increased androgen receptor activity and altered c-myc expression in prostate cancer cells after long-term androgen deprivation. Cancer Res. 1994;54:1566–1573. [PubMed] [Google Scholar]
- 23.Pignatta S., Arienti C., Zoli W., Di Donato M., Castoria G., Gabucci E., Casadio V., Falconi M., De Giorgi U., Silvestrini R., Tesei A. Prolonged exposure to (R)-bicalutamide generates a LNCaP subclone with alteration of mitochondrial genome. Mol. Cell. Endocrinol. 2014;382:314–324. doi: 10.1016/j.mce.2013.10.022. [DOI] [PubMed] [Google Scholar]
- 24.Raffo A.J., Perlman H., Chen M.W., Day M.L., Streitman J.S., Buttyan R. Overexpression of bcl-2 protects prostate cancer cells from apoptosis in vitro and confers resistance to androgen depletion in vivo. Cancer Res. 1995;55:4438–4445. [PubMed] [Google Scholar]
- 25.Chakravarty D., Sboner A., Nair S.S., Giannopoulou E., Li R., Hennig S., Mosquera J.M., Pauwels J., Park K., Kossai M. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat. Commun. 2014;5:5383. doi: 10.1038/ncomms6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xue D., Zhou C., Lu H., Xu R., Xu X., He X. LncRNA GAS5 inhibits proliferation and progression of prostate cancer by targeting miR-103 through AKT/mTOR signaling pathway. Tumour Biol. 2016 doi: 10.1007/s13277-016-5429-8. Published online October 14, 2016. [DOI] [PubMed] [Google Scholar]
- 27.Luo G., Wang M., Wu X., Tao D., Xiao X., Wang L., Min F., Zeng F., Jiang G. Long non-coding RNA MEG3 inhibits cell proliferation and induces apoptosis in prostate cancer. Cell. Physiol. Biochem. 2015;37:2209–2220. doi: 10.1159/000438577. [DOI] [PubMed] [Google Scholar]
- 28.D’Antonio J.M., Ma C., Monzon F.A., Pflug B.R. Longitudinal analysis of androgen deprivation of prostate cancer cells identifies pathways to androgen independence. Prostate. 2008;68:698–714. doi: 10.1002/pros.20677. [DOI] [PubMed] [Google Scholar]
- 29.Sun Y., Wang B.E., Leong K.G., Yue P., Li L., Jhunjhunwala S., Chen D., Seo K., Modrusan Z., Gao W.Q. Androgen deprivation causes epithelial-mesenchymal transition in the prostate: implications for androgen-deprivation therapy. Cancer Res. 2012;72:527–536. doi: 10.1158/0008-5472.CAN-11-3004. [DOI] [PubMed] [Google Scholar]
- 30.Varambally S., Yu J., Laxman B., Rhodes D.R., Mehra R., Tomlins S.A., Shah R.B., Chandran U., Monzon F.A., Becich M.J. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell. 2005;8:393–406. doi: 10.1016/j.ccr.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 31.Cancer Genome Atlas Research Network The molecular taxonomy of primary prostate cancer. Cell. 2015;163:1011–1025. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li J., Han L., Roebuck P., Diao L., Liu L., Yuan Y., Weinstein J.N., Liang H. TANRIC: an interactive open platform to explore the function of lncRNAs in cancer. Cancer Res. 2015;75:3728–3737. doi: 10.1158/0008-5472.CAN-15-0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim J.J., Yin B., Christudass C.S., Terada N., Rajagopalan K., Fabry B., Lee D.Y., Shiraishi T., Getzenberg R.H., Veltri R.W. Acquisition of paclitaxel resistance is associated with a more aggressive and invasive phenotype in prostate cancer. J. Cell. Biochem. 2013;114:1286–1293. doi: 10.1002/jcb.24464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hershberger P.A., Yu W.D., Modzelewski R.A., Rueger R.M., Johnson C.S., Trump D.L. Calcitriol (1,25-dihydroxycholecalciferol) enhances paclitaxel antitumor activity in vitro and in vivo and accelerates paclitaxel-induced apoptosis. Clin. Cancer Res. 2001;7:1043–1051. [PubMed] [Google Scholar]
- 35.Sowery R.D., Hadaschik B.A., So A.I., Zoubeidi A., Fazli L., Hurtado-Coll A., Gleave M.E. Clusterin knockdown using the antisense oligonucleotide OGX-011 re-sensitizes docetaxel-refractory prostate cancer PC-3 cells to chemotherapy. BJU Int. 2008;102:389–397. doi: 10.1111/j.1464-410X.2008.07618.x. [DOI] [PubMed] [Google Scholar]
- 36.Asteriti I.A., De Mattia F., Guarguaglini G. Cross-talk between AURKA and Plk1 in mitotic entry and spindle assembly. Front. Oncol. 2015;5:283. doi: 10.3389/fonc.2015.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wierstra I. The transcription factor FOXM1 (Forkhead box M1): proliferation-specific expression, transcription factor function, target genes, mouse models, and normal biological roles. Adv. Cancer Res. 2013;118:97–398. doi: 10.1016/B978-0-12-407173-5.00004-2. [DOI] [PubMed] [Google Scholar]
- 38.Bouldin C.M., Kimelman D. Cdc25 and the importance of G2 control: insights from developmental biology. Cell Cycle. 2014;13:2165–2171. doi: 10.4161/cc.29537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hou X., Li Z., Huang W., Li J., Staiger C., Kuang S., Ratliff T., Liu X. Plk1-dependent microtubule dynamics promotes androgen receptor signaling in prostate cancer. Prostate. 2013;73:1352–1363. doi: 10.1002/pros.22683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Z., Chen L., Wang H., Ahmad N., Liu X. Inhibition of Plk1 represses androgen signaling pathway in castration-resistant prostate cancer. Cell Cycle. 2015;14:2142–2148. doi: 10.1080/15384101.2015.1041689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Z., Xu D., Ding H.F., Kim J., Zhang J., Hai T., Yan C. Loss of ATF3 promotes Akt activation and prostate cancer development in a Pten knockout mouse model. Oncogene. 2015;34:4975–4984. doi: 10.1038/onc.2014.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang T., Guo S., Liu Z., Wu L., Li M., Yang J., Chen R., Liu X., Xu H., Cai S. CAMK2N1 inhibits prostate cancer progression through androgen receptor-dependent signaling. Oncotarget. 2014;5:10293–10306. doi: 10.18632/oncotarget.2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang G., Jones S.J., Marra M.A., Sadar M.D. Identification of genes targeted by the androgen and PKA signaling pathways in prostate cancer cells. Oncogene. 2006;25:7311–7323. doi: 10.1038/sj.onc.1209715. [DOI] [PubMed] [Google Scholar]
- 44.Kino T., Hurt D.E., Ichijo T., Nader N., Chrousos G.P. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal. 2010;3:ra8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu M., Chen Q., Liu X., Sun Q., Zhao X., Deng R., Wang Y., Huang J., Xu M., Yan J., Yu J. lncRNA H19/miR-675 axis represses prostate cancer metastasis by targeting TGFBI. FEBS J. 2014;281:3766–3775. doi: 10.1111/febs.12902. [DOI] [PubMed] [Google Scholar]
- 46.Yang L., Lin C., Jin C., Yang J.C., Tanasa B., Li W., Merkurjev D., Ohgi K.A., Meng D., Zhang J. lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature. 2013;500:598–602. doi: 10.1038/nature12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kumar S., Sharma A.R., Sharma G., Chakraborty C., Kim J. PLK-1: angel or devil for cell cycle progression. Biochim. Biophys. Acta. 2016;1865:190–203. doi: 10.1016/j.bbcan.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 48.Hao Z., Zhang H., Cowell J. Ubiquitin-conjugating enzyme UBE2C: molecular biology, role in tumorigenesis, and potential as a biomarker. Tumour Biol. 2012;33:723–730. doi: 10.1007/s13277-011-0291-1. [DOI] [PubMed] [Google Scholar]
- 49.Zhang Z., Hou X., Shao C., Li J., Cheng J.X., Kuang S., Ahmad N., Ratliff T., Liu X. Plk1 inhibition enhances the efficacy of androgen signaling blockade in castration-resistant prostate cancer. Cancer Res. 2014;74:6635–6647. doi: 10.1158/0008-5472.CAN-14-1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Beltran H., Rickman D.S., Park K., Chae S.S., Sboner A., MacDonald T.Y., Wang Y., Sheikh K.L., Terry S., Tagawa S.T. Molecular characterization of neuroendocrine prostate cancer and identification of new drug targets. Cancer Discov. 2011;1:487–495. doi: 10.1158/2159-8290.CD-11-0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dardenne E., Beltran H., Benelli M., Gayvert K., Berger A., Puca L., Cyrta J., Sboner A., Noorzad Z., MacDonald T. N-Myc induces an EZH2-mediated transcriptional program driving neuroendocrine prostate cancer. Cancer Cell. 2016;30:563–577. doi: 10.1016/j.ccell.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen Z., Zhang C., Wu D., Chen H., Rorick A., Zhang X., Wang Q. Phospho-MED1-enhanced UBE2C locus looping drives castration-resistant prostate cancer growth. EMBO J. 2011;30:2405–2419. doi: 10.1038/emboj.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Santoni M., Conti A., Burattini L., Berardi R., Scarpelli M., Cheng L., Lopez-Beltran A., Cascinu S., Montironi R. Neuroendocrine differentiation in prostate cancer: novel morphological insights and future therapeutic perspectives. Biochim. Biophys. Acta. 2014;1846:630–637. doi: 10.1016/j.bbcan.2014.10.008. [DOI] [PubMed] [Google Scholar]
- 54.Jones D., Noble M., Wedge S.R., Robson C.N., Gaughan L. Aurora A regulates expression of AR-V7 in models of castrate resistant prostate cancer. Sci. Rep. 2017;7:40957. doi: 10.1038/srep40957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hu R., Lu C., Mostaghel E.A., Yegnasubramanian S., Gurel M., Tannahill C., Edwards J., Isaacs W.B., Nelson P.S., Bluemn E. Distinct transcriptional programs mediated by the ligand-dependent full-length androgen receptor and its splice variants in castration-resistant prostate cancer. Cancer Res. 2012;72:3457–3462. doi: 10.1158/0008-5472.CAN-11-3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Q., Li W., Zhang Y., Yuan X., Xu K., Yu J., Chen Z., Beroukhim R., Wang H., Lupien M. Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell. 2009;138:245–256. doi: 10.1016/j.cell.2009.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ozen M., Ittmann M. Increased expression and activity of CDC25C phosphatase and an alternatively spliced variant in prostate cancer. Clin. Cancer Res. 2005;11:4701–4706. doi: 10.1158/1078-0432.CCR-04-2551. [DOI] [PubMed] [Google Scholar]
- 58.Ta H.Q., Ivey M.L., Frierson H.F., Jr., Conaway M.R., Dziegielewski J., Larner J.M., Gioeli D. Checkpoint kinase 2 negatively regulates androgen sensitivity and prostate cancer cell growth. Cancer Res. 2015;75:5093–5105. doi: 10.1158/0008-5472.CAN-15-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Aytes A., Mitrofanova A., Lefebvre C., Alvarez M.J., Castillo-Martin M., Zheng T., Eastham J.A., Gopalan A., Pienta K.J., Shen M.M. Cross-species regulatory network analysis identifies a synergistic interaction between FOXM1 and CENPF that drives prostate cancer malignancy. Cancer Cell. 2014;25:638–651. doi: 10.1016/j.ccr.2014.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang M., Mukherjee N., Bermudez R.S., Latham D.E., Delaney M.A., Zietman A.L., Shipley W.U., Chakravarti A. Adenovirus-mediated inhibition of survivin expression sensitizes human prostate cancer cells to paclitaxel in vitro and in vivo. Prostate. 2005;64:293–302. doi: 10.1002/pros.20263. [DOI] [PubMed] [Google Scholar]
- 61.Yamanaka K., Rocchi P., Miyake H., Fazli L., So A., Zangemeister-Wittke U., Gleave M.E. Induction of apoptosis and enhancement of chemosensitivity in human prostate cancer LNCaP cells using bispecific antisense oligonucleotide targeting Bcl-2 and Bcl-xL genes. BJU Int. 2006;97:1300–1308. doi: 10.1111/j.1464-410X.2006.06147.x. [DOI] [PubMed] [Google Scholar]
- 62.Spänkuch B., Heim S., Kurunci-Csacsko E., Lindenau C., Yuan J., Kaufmann M., Strebhardt K. Down-regulation of Polo-like kinase 1 elevates drug sensitivity of breast cancer cells in vitro and in vivo. Cancer Res. 2006;66:5836–5846. doi: 10.1158/0008-5472.CAN-06-0343. [DOI] [PubMed] [Google Scholar]
- 63.Al Nakouzi N., Cotteret S., Commo F., Gaudin C., Rajpar S., Dessen P., Vielh P., Fizazi K., Chauchereau A. Targeting CDC25C, PLK1 and CHEK1 to overcome Docetaxel resistance induced by loss of LZTS1 in prostate cancer. Oncotarget. 2014;5:667–678. doi: 10.18632/oncotarget.1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Anand S., Penrhyn-Lowe S., Venkitaraman A.R. AURORA-A amplification overrides the mitotic spindle assembly checkpoint, inducing resistance to Taxol. Cancer Cell. 2003;3:51–62. doi: 10.1016/s1535-6108(02)00235-0. [DOI] [PubMed] [Google Scholar]
- 65.Hou Y., Zhu Q., Li Z., Peng Y., Yu X., Yuan B., Liu Y., Liu Y., Yin L., Peng Y. The FOXM1-ABCC5 axis contributes to paclitaxel resistance in nasopharyngeal carcinoma cells. Cell Death Dis. 2017;8:e2659. doi: 10.1038/cddis.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Carr J.R., Park H.J., Wang Z., Kiefer M.M., Raychaudhuri P. FoxM1 mediates resistance to herceptin and paclitaxel. Cancer Res. 2010;70:5054–5063. doi: 10.1158/0008-5472.CAN-10-0545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.He W., Cai Q., Sun F., Zhong G., Wang P., Liu H., Luo J., Yu H., Huang J., Lin T. linc-UBC1 physically associates with polycomb repressive complex 2 (PRC2) and acts as a negative prognostic factor for lymph node metastasis and survival in bladder cancer. Biochim. Biophys. Acta. 2013;1832:1528–1537. doi: 10.1016/j.bbadis.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 68.Mondal T., Subhash S., Vaid R., Enroth S., Uday S., Reinius B., Mitra S., Mohammed A., James A.R., Hoberg E. MEG3 long noncoding RNA regulates the TGF-β pathway genes through formation of RNA-DNA triplex structures. Nat. Commun. 2015;6:7743. doi: 10.1038/ncomms8743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gutschner T., Hämmerle M., Eissmann M., Hsu J., Kim Y., Hung G., Revenko A., Arun G., Stentrup M., Gross M. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73:1180–1189. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhao X., Wang P., Liu J., Zheng J., Liu Y., Chen J., Xue Y. Gas5 exerts tumor-suppressive functions in human glioma cells by targeting miR-222. Mol. Ther. 2015;23:1899–1911. doi: 10.1038/mt.2015.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee E.C., Frolov A., Li R., Ayala G., Greenberg N.M. Targeting Aurora kinases for the treatment of prostate cancer. Cancer Res. 2006;66:4996–5002. doi: 10.1158/0008-5472.CAN-05-2796. [DOI] [PubMed] [Google Scholar]
- 72.Fan X., Chen X., Deng W., Zhong G., Cai Q., Lin T. Up-regulated microRNA-143 in cancer stem cells differentiation promotes prostate cancer cells metastasis by modulating FNDC3B expression. BMC Cancer. 2013;13:61. doi: 10.1186/1471-2407-13-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen X., Gu P., Xie R., Han J., Liu H., Wang B., Xie W., Xie W., Zhong G., Chen C. Heterogeneous nuclear ribonucleoprotein K is associated with poor prognosis and regulates proliferation and apoptosis in bladder cancer. J. Cell. Mol. Med. 2016 doi: 10.1111/jcmm.12999. Published online November 10, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jiang J., Chen X., Liu H., Shao J., Xie R., Gu P., Duan C. Polypyrimidine Tract-Binding Protein 1 promotes proliferation, migration and invasion in clear-cell renal cell carcinoma by regulating alternative splicing of PKM. Am. J. Cancer Res. 2017;7:245–259. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.