Abstract
The ubiquitination pathway regulates growth, development, and stress responses in plants, and the U-box protein family of ubiquitin ligases has important roles in this pathway. Here, 64 putative U-box proteins were identified in the Medicago truncatula genome. In addition to the conserved U-box motif, other functional domains, such as the ARM, kinase, KAP, and WD40 domains, were also detected. Phylogenetic analysis of the M. truncatula U-box proteins grouped them into six subfamilies, and chromosomal mapping and synteny analyses indicated that tandem and segmental duplications may have contributed to the expansion and evolution of the U-box gene family in this species. Using RNA-seq data from M. truncatula seedlings subjected to three different abiotic stresses, we identified 33 stress-inducible plant U-box genes (MtPUBs). Specifically, 25 salinity-, 15 drought-, and 16 cold-regulated MtPUBs were detected. Among them, MtPUB10, MtPUB17, MtPUB18, MtPUB35, MtPUB42, and MtPUB44 responded to all three stress conditions. Expression profiling by qRT-PCR was consistent with the RNA-seq data, and stress-related elements were identified in the promoter regions. The present findings strongly indicate that U-box proteins play critical roles in abiotic stress response in M. truncatula.
Introduction
Ubiquitin-mediated proteolysis is required for most cellular processes, and the pathway is mediated by three sequential ubiquitination enzymes, E1, E2, and E3. E3 ubiquitin ligases are of particular importance as they confer substrate specificity that catalyzes the attachment of ubiquitin to protein targets [1,2]. The E3 ligases can be categorized into distinct families based on their protein domains (RING, HECT, or U-box domains) or mode of action [3,4]. The U-box E3 ligases, of which there are 64 members in Arabidopsis, were identified most recently and comprise the smallest E3 ligase family [5]. They have an approximately 70-amino-acid conserved U-box motif, which is present in U-box E3 ligases from yeast to humans [6]. A large expansion of the U-box gene family occurred in plants, which may be attributable to biological processes that are unique to the plant life cycle. It has been reported that plant U-box (PUB) proteins are largely involved in abiotic and biotic stress responses [7].
The Arabidopsis PUB protein AtCHIP plays an important role in temperature stress tolerance [8]. U-BOX17, another Arabidopsis PUB protein, and its tobacco homolog ACRE276 have been identified as positive regulators of cell death and defense [9], and subsequent studies yielded similar findings for the functions of these PUB proteins. AtPUB22 and AtPUB23 were found to have critical combinatory roles in response to drought stress [10], and they directly ubiquitinate RPN6, a 26S proteasome lid subunit, for subsequent degradation in Arabidopsis [7]. Similarly, AtPUB18 has a function linked to that of AtPUB19 in the negative regulation of ABA-mediated drought stress responses [11]. AtPUB13 acts as a node that connects flowering time regulation and salicylic acid (SA)-dependent defense signaling in Arabidopsis [12]. AtPUB30 acts in salt stress tolerance as a negative factor whose activity during germination is ABA independent [13]. The roles of PUBs in response to abiotic stresses have also been shown in other plants. For example, rice (Oryza sativa) Spotted leaf11 (Spl11) encodes a U-box-containing E3 ligase and negatively regulates plant cell death and defense [14]. OsPUB15 helps reduce cellular oxidative stress during seedling establishment [15], and its ARM repeat domain is essential for its physical interaction with the kinase domain of PID2 (PID2K), an interaction observed both in vitro and in vivo [16]. OsUPS, another gene encoding a U-box-containing E3 ligase, responds to phosphate starvation in rice [17]. In hot pepper (Capsicum annuum L. cv. Pukang), CaPUB1 has been implicated in counteracting dehydration and high-salinity stress [18].
Efforts have been made to characterize these U-box genes in plant species as well as algae. Thus far, 30 full-length U-box genes have been identified in the Chlamydomonas reinhardtii genome sequence [19]. In Arabidopsis and rice, 64 and 77 U-box genes have been identified, respectively [20,21]. However, U-box genes have not been studied in the model legume M. truncatula. Here, we present a comprehensive analysis of the genes encoding U-box family proteins in M. truncatula.
Materials and methods
Identification of PUB proteins
Putative PUB proteins were identified in the M. truncatula genome database (http://www.medicagohapmap.org/tools/Blastform) using the BLAST program and the amino acid sequences of published U-box proteins as queries. The proteins identified by the BLAST program were used for domain searches from the Pfam (http://www.sanger.ac.uk/Software/Pfam/) and SMART (http://smart.embl-heidelberg.de/) databases with an E-value cut-off level of 1.0 or 10. These cut-off values were recommended for more reliable search results. Using the Pfam/SMART databases, the C-terminal domain of each PUB protein was analyzed with an E-value cut-off level of 1.0.
Alignments, phylogenetic analysis, intron/exon organization, and localization of PUB genes on chromosomes
The U-box domain (PF00646) was obtained from the Pfam database, and HMMER 3.0 (http://hmmer.janelia.org/) was used for U-box motif identification in each PUB protein. Clustal X (version 2.0; http://www.clustal.org/) was used for the multiple sequence alignment of all predicted U-box protein motifs. A neighbor-joining (NJ) tree was constructed by MEGA (version 5.1; Tamura et al. 2011), using the p-distance method with gaps treated by pairwise deletion and a 1,000 bootstrap replicate. Intron/exon organization was determined using the M. truncatula genome database (http://www.medicagohapmap.org/home/view), and chromosomal maps were generated using the Genome Pixelizer Tcl/Tk script [www.atgc.org/GenomePixelizer (released 02/15/2002)]. Gene duplication was defined according to the following criteria: (1) The length of the sequence alignment covered ≥80% of the longest gene, and (2) the similarity of the aligned gene regions was ≥70% [22,23].
Promoter element analysis
To investigate cis elements in the promoter sequences of the U-box protein-encoding genes, the 1,500 bp DNA sequences upstream of the transcriptional start site were obtained from NCBI (http://www.ncbi.nlm.nih.gov/). The PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) was used to identify cis elements in the promoters and to collect data for the following: ABRE, ARE, AuxRR core, CGTCA motif, ERE, GARE motif, HSE, LTR, MBS, P Box, TC-rich repeat, TCA element, TGA element, and TGACG motif.
Plant materials and stress treatments
M. truncatula seeds were soaked with distilled water and placed on a plastic net floating on 1/4-strength Hoagland nutrient solution (1.0 mM Ca2+, 1.5 mM K+, 0.5 mM Mg2+, 0.25 mM NH4+, 3.5 mM NO3-, 0.25 mM H2PO4-, 0.523 mM SO42-, 22 μM Fe2+, 0.30 μM Cu2+, 0.8 μM Zn2+, 9 μM Mn2+, 46 μM BO33-, 0.1 μM MoO42-). After germination, seedlings were grown under the following conditions for 4 weeks: 22–24°C, 200 μmol m−2s−1 photosynthetic active radiation, and a photoperiod of 14/10 h (day/night).
Four weeks after germination, seedlings were subjected to various treatments. For drought treatment, the seedlings were transferred to dry Whatman 3MM paper in a sterile petri dish for 0, 2, 6, and 12 h. For cold treatment, the seedlings were transferred to 4°C for 0, 2, 6, and 12 h. For salt treatment, the seedlings were transferred to solutions containing 300 mM NaCl for 0, 2, 6, and 12 h. After treatment, the seedlings were harvested, immediately frozen in liquid nitrogen, and stored at -80°C for further analysis.
Statistical analysis
Experiments in the study were independently performed in triplicate. Each result in this study is the mean of at least three replicated treatments and each treatment contained at least 10 plants. The significant differences between treatments were statistically assessed by standard deviation and one-way analysis of variance (ANOVA). The data between differently treated groups were compared statistically by ANOVA, followed by the least significant difference (LSD) test if the ANOVA result was significant at P<0.05.
Library construction and sequencing
For RNA-seq analyses, RNA was extracted using Trizol. The 3’-tag digital gene expression libraries were prepared using the Illumina Gene Expression Sample Prep Kit based on the method described by Zhou et al. [24]. Deep sequencing were carried out using the Illumina HiSeq 3000 platform (Illumina, San Diego, CA, USA) following the manufacturer’s instructions by Genergy Biotechnology Co. Ltd. (Shanghai, China). The raw data comprised 100-bp paired-end sequences, and the cleaned reads were then mapped to Arabidopsis thaliana genome (TAIR10) using default settings of TOPHATv2.0.8. The duplicated reads were removed and alignments with MAPQ score > 20 were used for further analysis. RNA-seq alignments were processed using HTSeq-count, and differentially expressed genes were identified using DESeq with |log2 fold change| > 3.5.
Results
Identification and homology analysis of U-box proteins in M. truncatula
U-box domains (PF04564) were downloaded from the Pfam database and used as queries to identify U-box proteins in the M. truncatula genome database (http://www.medicagohapmap.org/tools/Blastform) using the BLAST program (HMMER 3.0, http://hmmer.janelia.org/). The identified proteins were used for a domain search of the Pfam (http://www.sanger.ac.uk/Software/Pfam/) and SMART (http://smart.embl-heidelberg.de/) databases with an E-value cut-off level of 1.0 or 10, which was recommended for more reliable search results. Using the Pfam/SMART databases, the C-terminal domains of each U-box protein with an E-value cut-off level of 1.0 were analyzed. We found 64 proteins containing at least one U-box motif in M. truncatula as annotated by the SMART/Pfam databases, and these proteins were designated as U-box proteins (MtPUB) (Table 1 and S1 Table). The isoelectric point (pI) bias of most of these U-box proteins was neutral. Only MtPUB10 and MtPUB11 had a pI greater than 10, and only MtPUB62 had a pI less than 5 (Table 1). Some of the genes encoding these U-box proteins had numerous introns; for example, MtPUB9, MtPUB39, MtPUB47, and MtPUB64 all had more than 10 introns (Table 1).
Table 1. Distribution of MtPUB genes in the Medicago truncatula genome.
| S.No | Gene_ID | Accession number | Other domain | Predicted protein (aa) |
Mol wt (kDa) | pI | Chromosome | No. of introns |
|---|---|---|---|---|---|---|---|---|
| 1 | MtPUB1 | Medtr1g017770.1 | Unknown | 434 | 48.39 | 6.88 | 1 | 0 |
| 2 | MtPUB2 | Medtr1g056840.1 | Unknown | 411 | 46.09 | 8.47 | 1 | 1 |
| 3 | MtPUB3 | Medtr1g056870.1 | Unknown | 437 | 48.80 | 6.95 | 1 | 0 |
| 4 | MtPUB4 | Medtr1g056880.1 | Unknown | 437 | 48.90 | 7.91 | 1 | 0 |
| 5 | MtPUB5 | Medtr1g056910.1 | Unknown | 406 | 46.25 | 8.54 | 1 | 0 |
| 6 | MtPUB6 | Medtr1g069845.1 | ARM | 608 | 66.67 | 6.52 | 1 | 4 |
| 7 | MtPUB7 | Medtr1g076400.1 | Unknown | 1013 | 112.48 | 5.09 | 1 | 3 |
| 8 | MtPUB8 | Medtr1g079450.1 | Unknown | 446 | 49.65 | 8.05 | 1 | 1 |
| 9 | MtPUB9 | Medtr1g090320.1 | WD40 | 1488 | 166.85 | 5.96 | 1 | 16 |
| 10 | MtPUB10 | Medtr1g093965.1 | Unknown | 200 | 21.84 | 10.05 | 1 | 3 |
| 11 | MtPUB11 | Medtr1g093995.1 | Unknown | 200 | 21.86 | 10.05 | 1 | 3 |
| 12 | MtPUB12 | Medtr1g094025.1 | Unknown | 296 | 33.24 | 8.14 | 1 | 3 |
| 13 | MtPUB13 | Medtr1g094215.1 | ARM | 447 | 48.10 | 6.13 | 1 | 3 |
| 14 | MtPUB14 | Medtr1g100820.1 | Kinase | 715 | 80.80 | 5.44 | 1 | 7 |
| 15 | MtPUB15 | Medtr2g007630.1 | Unknown | 259 | 28.73 | 9.58 | 2 | 3 |
| 16 | MtPUB16 | Medtr2g011140.1 | Unknown | 383 | 42.16 | 6.77 | 2 | 0 |
| 17 | MtPUB17 | Medtr2g018010.1 | ARM | 720 | 78.67 | 6.58 | 2 | 0 |
| 18 | MtPUB18 | Medtr2g087350.1 | Unknown | 403 | 45.33 | 8.61 | 2 | 0 |
| 19 | MtPUB19 | Medtr2g096850.1 | Kinase | 810 | 91.60 | 7.01 | 2 | 6 |
| 20 | MtPUB20 | Medtr3g008270.1 | Kinase | 797 | 88.60 | 6.48 | 3 | 9 |
| 21 | MtPUB21 | Medtr3g008280.1 | Kinase | 809 | 90.02 | 7.21 | 3 | 9 |
| 22 | MtPUB22 | Medtr3g065080.1 | Unknown | 439 | 49.13 | 8.08 | 3 | 0 |
| 23 | MtPUB23 | Medtr3g078160.1 | Unknown | 681 | 75.94 | 8.29 | 3 | 0 |
| 24 | MtPUB24 | Medtr3g078340.1 | ARM | 529 | 57.91 | 7.03 | 3 | 0 |
| 25 | MtPUB25 | Medtr3g085610.1 | KAP | 766 | 84.92 | 6.10 | 3 | 5 |
| 26 | MtPUB26 | Medtr3g095730.1 | Unknown | 419 | 46.77 | 8.92 | 3 | 0 |
| 27 | MtPUB27 | Medtr3g096370.1 | Unknown | 404 | 45.02 | 6.35 | 3 | 0 |
| 28 | MtPUB28 | Medtr3g115670.1 | HEAT | 814 | 89.47 | 5.06 | 3 | 3 |
| 29 | MtPUB29 | Medtr3g466220.1 | ARM | 836 | 90.68 | 5.45 | 3 | 3 |
| 30 | MtPUB30 | Medtr4g028960.1 | ARM | 701 | 76.42 | 6.83 | 4 | 0 |
| 31 | MtPUB31 | Medtr4g051515.1 | Unknown | 413 | 47.12 | 9.26 | 4 | 0 |
| 32 | MtPUB32 | Medtr4g063800.1 | ARM | 662 | 72.09 | 5.14 | 4 | 3 |
| 33 | MtPUB33 | Medtr4g085720.1 | Unknown | 410 | 45.35 | 7.81 | 4 | 0 |
| 34 | MtPUB34 | Medtr4g091880.1 | Unknown | 375 | 40.59 | 8.36 | 4 | 0 |
| 35 | MtPUB35 | Medtr4g107010.1 | ARM | 747 | 83.52 | 8.04 | 4 | 1 |
| 36 | MtPUB36 | Medtr4g125930.1 | Kinase | 764 | 85.49 | 6.00 | 4 | 8 |
| 37 | MtPUB37 | Medtr4g485520.1 | ARM | 652 | 70.78 | 7.01 | 4 | 3 |
| 38 | MtPUB38 | Medtr5g015210.1 | Unknown | 451 | 49.54 | 6.50 | 5 | 0 |
| 39 | MtPUB39 | Medtr5g015500.1 | Pro isomerase | 552 | 59.75 | 7.65 | 5 | 10 |
| 40 | MtPUB40 | Medtr5g020570.1 | KAP | 782 | 88.26 | 6.44 | 5 | 5 |
| 41 | MtPUB41 | Medtr5g032010.1 | Kinase | 808 | 92.93 | 7.98 | 5 | 8 |
| 42 | MtPUB42 | Medtr5g034440.1 | ARM | 689 | 76.82 | 7.19 | 5 | 0 |
| 43 | MtPUB43 | Medtr5g048050.1 | Unknown | 438 | 50.05 | 6.88 | 5 | 0 |
| 44 | MtPUB44 | Medtr5g077510.1 | Unknown | 442 | 49.45 | 8.53 | 5 | 0 |
| 45 | MtPUB45 | Medtr5g083030.1 | ARM | 694 | 76.93 | 6.78 | 5 | 0 |
| 46 | MtPUB46 | Medtr6g008170.1 | KAP | 554 | 61.48 | 8.22 | 6 | 0 |
| 47 | MtPUB47 | Medtr6g013690.1 | Ufd2p | 1047 | 11.80 | 5.47 | 6 | 15 |
| 48 | MtPUB48 | Medtr6g071340.1 | Unknown | 418 | 47.61 | 5.56 | 6 | 0 |
| 49 | MtPUB49 | Medtr7g005940.1 | Unknown | 1073 | 12.18 | 7.21 | 7 | 8 |
| 50 | MtPUB50 | Medtr7g053260.1 | Unknown | 459 | 51.53 | 8.39 | 7 | 1 |
| 51 | MtPUB51 | Medtr7g059405.1 | ARM | 634 | 69.82 | 6.27 | 7 | 4 |
| 52 | MtPUB52 | Medtr7g077780.1 | Kinase | 896 | 100.38 | 6.02 | 7 | 8 |
| 53 | MtPUB53 | Medtr7g078330.1 | ARM | 646 | 72.78 | 5.05 | 7 | 3 |
| 54 | MtPUB54 | Medtr7g097020.1 | ARM | 767 | 84.39 | 7.44 | 7 | 5 |
| 55 | MtPUB55 | Medtr7g106340.1 | Unknown | 421 | 46.96 | 8.71 | 7 | 0 |
| 56 | MtPUB56 | Medtr7g116600.1 | Unknown | 460 | 51.32 | 8.35 | 7 | 1 |
| 57 | MtPUB57 | Medtr7g117890.1 | ARM | 1001 | 111.25 | 5.30 | 7 | 4 |
| 58 | MtPUB58 | Medtr8g011720.1 | TPR | 277 | 31.95 | 6.38 | 8 | 7 |
| 59 | MtPUB59 | Medtr8g027140.1 | Unknown | 1006 | 112.03 | 5.62 | 8 | 4 |
| 60 | MtPUB60 | Medtr8g068530.1 | Kinase | 769 | 88.97 | 5.81 | 8 | 7 |
| 61 | MtPUB61 | Medtr8g077205.1 | KAP | 760 | 85.19 | 6.66 | 8 | 4 |
| 62 | MtPUB62 | Medtr8g080280.1 | Unknown | 767 | 85.42 | 4.87 | 8 | 5 |
| 63 | MtPUB63 | Medtr8g092870.1 | Unknown | 418 | 46.35 | 7.48 | 8 | 0 |
| 64 | MtPUB64 | Medtr8g103227.1 | WD40 | 1335 | 148.78 | 5.64 | 8 | 14 |
Analysis of the functional domains of the M. truncatula U-box proteins
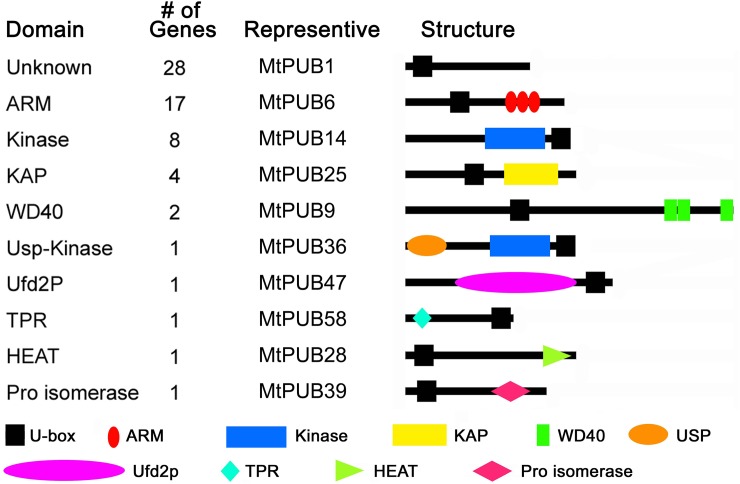
U-box proteins often contain several other functional domains at their N- or C-terminal regions. The SMART and Pfam database searches revealed that the U-box proteins contained several known or unknown conserved domains, which presumably participate in substrate recognition, and we designated these domains as functional domains (Fig 1). The types of functional domains in the U-box proteins are listed in Table 1. The 36 U-box proteins with one or more known functional domains were as follows, with the number in parentheses indicating the number of proteins: ARM(17), Armadillo/beta-catenin-like repeat; Kinase(8), protein tyrosine kinase; KAP(4), kinesin-associated protein; WD40(2), WD40 domain, G-beta repeat; USP-Kinase(1); Ufd2p(1), ubiquitin elongating factor core; TPR (1), TPR repeats; HEAT(1), HEAT repeats; and Pro isomerase(1), cyclophilin-type peptidyl-prolyl cis-trans isomerase/CLD. Some U-box proteins had no other obvious interaction domains or had a few rare or functionally uncertain domains; all of these were classified together as ‘Unknown’ (Fig 1).
Fig 1. Number and domain structure of U-box proteins in Medicago truncatula.
Shown on the left are the types of functional domains and the number of U-box proteins predicted to have those domains. The domain names are taken from the Pfam or SMART database. Domain abbreviations: Unknown, U-box proteins that have no obvious N- or C-terminal interaction domain or have rare or functionally uncertain domains; ARM, Armadillo/beta-catenin-like repeat; Kinase, protein tyrosine kinase; KAP, kinesin-associated protein; WD40, WD40 domain, G-beta repeat; USP-Kinase; Ufd2p, ubiquitin elongating factor core; TPR2, TPR repeats; HEAT, HEAT repeats; Pro isomerase, cyclophilin-type peptidyl-prolyl cis-trans isomerase/CLD.
Aside from the U-box motif, the ARM (Armadillo/beta-catenin-like repeat) domain, an approximately 40-amino-acid tandemly repeated sequence motif, was the most highly represented functional domain among the identified MtPUB proteins. In beta-catenin, these tandem repeats form a super-helix of helices that presumably mediates ligand interaction (Fig 1). U-box-ARM proteins have been reported in Arabidopsis. For example, AtPUB18 and AtPUB19 have related functions in negatively regulating ABA-mediated drought stress response [11]. The homologs of AtPUB18 and AtPUB19 in M. truncatula are MtPUB35 and MtPUB42 (S1 Fig). In Medicago truncatula, MtPUB35 and MtPUB42 have high sequence similarities with AtPUB18 and AtPUB19 (S1 Fig). MtPUB32 also has high sequence similarity with AtPUB13, which encodes a U-box-ARM protein that links the flowering time and SA-dependent defense signaling pathways in Arabidopsis [12] (S1 Fig and S1 Table). U-box-ARM protein AtPUB30 acts in salt stress tolerance as a negative factor independent of ABA during seed germination [13], and it is homologous to MtPUB38 (S1 Fig). In rice, the U-box-ARM E3 ligase SPL11 negatively regulates plant cell death and defense[14]. OsPUB15, another U-box-ARM protein in rice, helps reduce cellular oxidative stress during seedling establishment [15]. OsPUB15 is homologous to MtPUB29 in M. truncatula (S1 Fig).
Eight MtPUB proteins were found to have a kinase domain, indicating their potential involvement in signal transduction cascades via phosphorylation. The KAP (kinesin-associated protein) domain, found in four MtPUB proteins, is associated with motor function, consistent with the role of kinesins as intracellular multimeric transport motor proteins that move cellular cargo on microtubule tracks.
Two MtPUB proteins had WD40 domains. WD40 domain-containing proteins are made up of highly conserved repeating units approximately 40 amino acids long and usually ending with Trp-Asp (WD) [25]. They are found in all eukaryotes but not in prokaryotes, and they regulate numerous cellular functions, such as cell division, cell-fate determination, gene transcription, transmembrane signaling, mRNA modification, and vesicle fusion. The USP, Ufd2p, TPR, HEAT, and Pro isomerase domains were each present in only one MtPUB protein (Fig 1). WD40 and TPR domains are known to be involved in protein interactions [26,27]. Rice and Arabidopsis U-box proteins containing WD40 repeats are homologous to animal and human Prp19p proteins and are involved in preRNA splicing and other biological processes [7,28,29]. AtCHIP, the only TPR domain-containing U-box protein in Arabidopsis, is homologous to the mammalian CHIP (carboxyl terminus of Hsc70-interacting protein) and participates in abiotic stress response and the regulation of chloroplast protein turnover [30,31]. In humans and animals, CHIP interacts with molecular chaperones, such as Hsp70 and Hsp90, and acts as a partner in the cell to ensure protein stability. CHIP is involved in cell stress protection and several neurodegenerative diseases [32,33]. The homolog of AtCHIP in M. truncatula is MtPUB58 (S1 Fig).
Phylogenetic and evolutionary analysis of U-box proteins in M. truncatula
For the phylogenetic analysis of the identified U-box proteins, we used HMMER 3.0 software (http://hmmer.janelia.org/) to analyze the motif sequences of each U-box protein. All of the U-box proteins were found to contain only one U-box motif. Using the U-box motif sequence for the alignment, an unrooted phylogenetic tree of the entire dataset was created (Fig 2). The phylogenetic tree divided the 64 MtPUB proteins into six subfamilies according to the distribution of various branches, the length of each branch, and the phylogenetic relationship between MtPUB proteins.
Fig 2. Phylogenetic tree of the U-box protein family from Medicago truncatula.
The 70-amino-acid U-box motifs from the 64 putative U-box proteins were aligned by CLUSTAL X 2.0, and the unrooted NJ phylogenetic tree was constructed by MEGA 5.1, using the p-distance method and a bootstrap value of 1,000. The six groups identified from the phylogenetic analysis are marked on the outside. The bar represents the branch length equivalent to 0.05 amino acid changes per residue. Table 1 provides additional information for the corresponding genes.
The phylogenetic tree was color-coded according to the different functional domains (Fig 2). Most of the kinase domain-containing MtPUB proteins were in the G6 family. The ARM-containing MtPUB proteins generally localized in clades within the G1 family. This correlation further supports the phylogenetic relationships in the U-box tree and suggests a co-evolution of the U-box motif with other domains.
Locations of the U-box protein-encoding genes on M. truncatula chromosomes
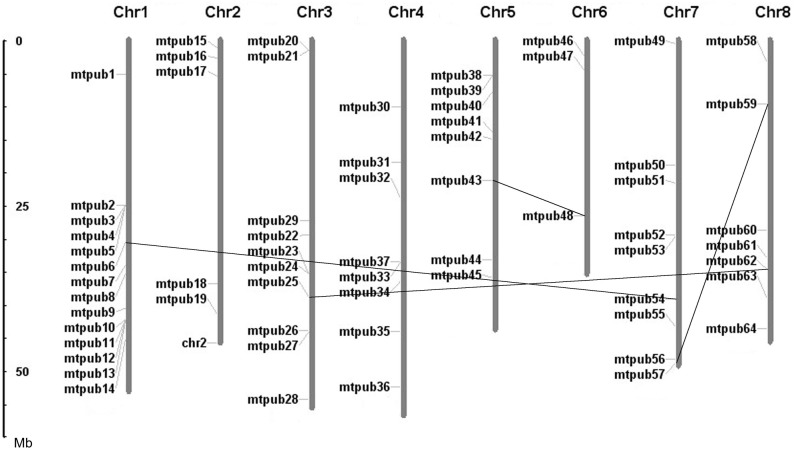
The U-box protein-encoding genes were distributed randomly on all eight M. truncatula chromosomes. To determine whether the gene family in M. truncatula evolved through duplication events, we obtained the chromosomal locations of the U-box protein-encoding genes from the M. truncatula genomic database and mapped the loci on the chromosomes (Fig 3). With 14 U-box genes, chromosome 1 had the largest number, whereas chromosome 6 had only three U-box genes. Some U-box genes were arranged in tandem repeats either in the same or inverse orientation, representing local gene duplications. As shown in Fig 3, there were four segmental duplication events between chromosomes, suggesting that tandem duplications of chromosomal regions played a major role in the expansion of this gene family.
Fig 3. Locations and duplications of Medicago truncatula U-box genes on chromosomes 1–8.
Genes lying on duplicated segments of genome have been joined by lines. The scale represents megabases (Mb). The chromosome numbers are indicated at the top of each bar.
Expression analysis of U-box protein-encoding genes in various tissues
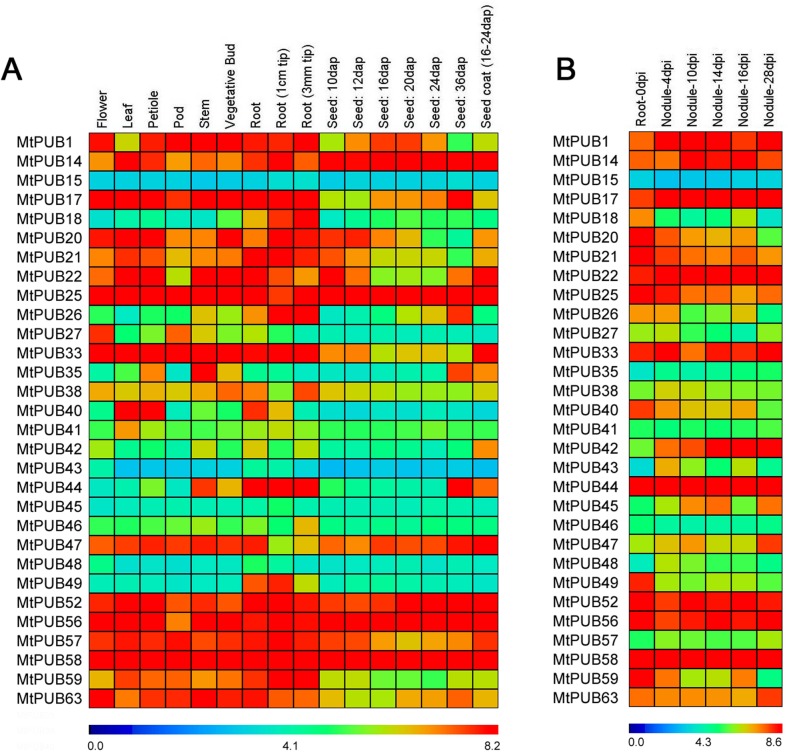
Using an existing database (http://mtgea.noble.org/v2/), we were able to survey the expression of many MtPUB genes in different tissues. A few MtPUBs were expressed only in certain tissues. For example, MtPUB18 and MtPUB49 were mainly expressed in roots; MtPUB40 expression was largely restricted to leaves and roots; MtPUB27 was expressed in flowers and pods; and MtPUB44 was expressed in roots and mature seeds (Fig 4A). Because legume root nodules plays an important role in symbiotic nitrogen fixation, we also identified MtPUBs that were differentially expressed in the nodule. Strong expression of MtPUB42 and MtPUB47 could be seen in root nodules, while the expression of MtPUB18, MtPUB40, and MtPUB49 in root nodules was low (Fig 4B).
Fig 4.
Expression profiles of Medicago truncatula U-box protein-encoding genes during panicle development (A) and root development (B). The average log signal values of U-box protein-encoding genes in various tissues/organs and developmental stages (mentioned at the top of each lane) are presented. The data comes from this site (http://mtgea.noble.org/v2/annotation_search_form.php#gid). The color scale (representing log signal values) is shown at the bottom. dap: days after pollination.
Identification of stress-responsive MtPUBs
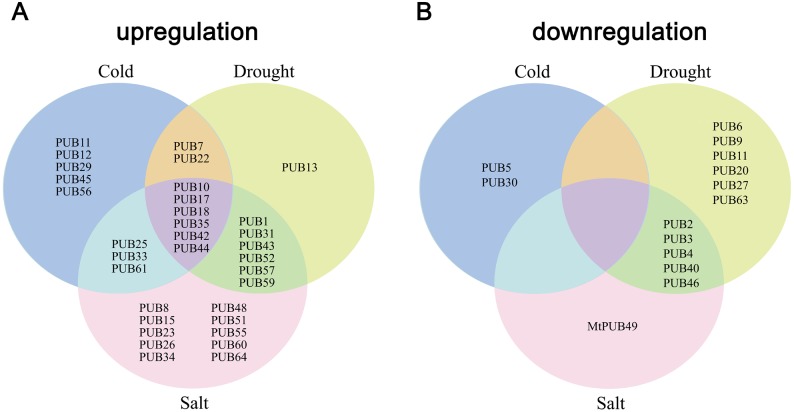
To study the expression of the U-box family genes under abiotic stress, 4-week-old M. truncatula seedlings were collected and treated with drought, salt, or cold stress for 0, 2, 6, and 12 h. Total RNA was extracted, and libraries were constructed for RNA-seq. In general, under drought, salt, and cold stress, there were more up-regulated genes than down-regulated genes, and the difference was most obvious at 2 h (S2 Fig). Salt stress had the strongest correlation with drought stress, and the R value was more than 0.95 at 2, 6, and 12 h (S3 Fig). The analysis showed that some of the 64 U-box family genes could be induced by salt, drought, or cold stress, but a few genes were down-regulated (Fig 5, Tables 2–4). After drought treatment, MtPUB1, MtPUB7, MtPUB10, MtPUB13, MtPUB17, MtPUB18, MtPUB22, MtPUB31, MtPUB35, MtPUB42, MtPUB43, MtPUB44, MtPUB52, MtPUB57, and MtPUB59 were up-regulated (Table 2). (A gene was considered up-regulated if its expression was increased at 2, 6, and 12 h and if the log2 fold change > 1 for at least one of these time points.) Using the same criteria, we found that MtPUB1, MtPUB8, MtPUB10, MtPUB15, MtPUB17, MtPUB18, MtPUB23, MtPUB25, MtPUB26, MtPUB31, MtPUB33, MtPUB34, MtPUB35, MtPUB42, MtPUB43, MtPUB44, MtPUB48, MtPUB51, MtPUB52, MtPUB55, MtPUB57, MtPUB59, MtPUB60, MtPUB61, and MtPUB64 were up-regulated under salt stress (Table 3). After cold treatment, MtPUB7, MtPUB10, MtPUB11, MtPUB12, MtPUB17, MtPUB18, MtPUB22, MtPUB25, MtPUB29, MtPUB33, MtPUB35, MtPUB42, MtPUB44, MtPUB45, MtPUB56, and MtPUB61 were up-regulated (Table 4).
Fig 5. Venn diagram showing common and unique differential MtPUB gene expression under three treatment conditions.
Among them, 25 high-salinity-, 15 drought-, and 16 cold- up regulated U-box genes were detected and 6 U-box genes were observed to respond remarkably to all three stresses. in contrast, 6 high-salinity-, 11 drought-, and 2 cold- down regulated U-box genes were detected.
Table 2. Read abundance of MtPUB genes in the drought-0, drought-2, drought-6, and drought-12 libraries.
| Gene_ID | Drought-0 | Drought-2 | Drought-6 | Drought-12 | log2 (Drought-2/ Drought-0) |
log2 (Drought-6/ Drought-0) |
log2 (Drought-12/ Drought-0) |
|---|---|---|---|---|---|---|---|
| MtPUB1 | 79 | 146 | 178 | 111 | 0.89 | 1.17* | 0.49 |
| MtPUB2 | 59 | 27 | 13 | 4 | -1.11* | -2.17* | -3.91* |
| MtPUB3 | 69 | 41 | 14 | 13 | -0.76 | -2.29* | -2.45* |
| MtPUB4 | 158 | 76 | 41 | 14 | -1.05* | -1.97* | -3.49* |
| MtPUB5 | 62 | 176 | 53 | 55 | 1.51* | -0.23 | -0.16 |
| MtPUB6 | 299 | 200 | 91 | 131 | -0.58 | -1.71* | -1.19* |
| MtPUB7 | 348 | 841 | 784 | 717 | 1.27* | 1.17* | 1.04* |
| MtPUB8 | 3 | 6 | 1 | 4 | 1.07* | -1.53* | 0.44 |
| MtPUB9 | 941 | 633 | 624 | 343 | -0.57 | -0.59 | -1.46* |
| MtPUB10 | 10 | 11 | 23 | 35 | 0.09 | 1.13* | 1.72* |
| MtPUB11 | 4 | 4 | 1 | 2 | -0.12 | -1.94* | -0.64 |
| MtPUB12 | 5 | 9 | 9 | 7 | 0.85 | 0.86 | 0.51 |
| MtPUB13 | 463 | 911 | 1106 | 901 | 0.98 | 1.26* | 0.96 |
| MtPUB14 | 393 | 446 | 539 | 442 | 0.18 | 0.46 | 0.17 |
| MtPUB15 | 2 | 3 | 2 | 2 | 0.47 | 0.11 | 0.34 |
| MtPUB16 | 1 | 1 | 1 | 1 | 0 | 0 | 0 |
| MtPUB17 | 520 | 730 | 1218 | 905 | 0.49 | 1.23* | 0.80 |
| MtPUB18 | 51 | 155 | 122 | 67 | 1.59* | 1.25* | 0.38 |
| MtPUB19 | 526 | 661 | 798 | 914 | 0.33 | 0.60 | 0.80 |
| MtPUB20 | 449 | 393 | 191 | 232 | -0.19 | -1.23* | -0.95 |
| MtPUB21 | 448 | 478 | 281 | 325 | 0.09 | -0.67 | -0.46 |
| MtPUB22 | 119 | 336 | 237 | 176 | 1.50* | 1.00* | 0.57 |
| MtPUB23 | 174 | 96 | 452 | 560 | -0.85 | 1.38* | 1.69* |
| MtPUB24 | 444 | 449 | 484 | 468 | 0.02 | 0.13 | 0.08 |
| MtPUB25 | 274 | 304 | 286 | 354 | 0.15 | 0.06 | 0.37 |
| MtPUB26 | 219 | 340 | 233 | 222 | 0.63 | 0.09 | 0.02 |
| MtPUB27 | 143 | 143 | 69 | 46 | 0.00 | -1.05* | -1.63* |
| MtPUB28 | 1009 | 1266 | 1309 | 1224 | 0.33 | 0.38 | 0.28 |
| MtPUB29 | 768 | 883 | 1182 | 953 | 0.20 | 0.62 | 0.31 |
| MtPUB30 | 361 | 308 | 313 | 283 | -0.23 | -0.21 | -0.35 |
| MtPUB31 | 26 | 362 | 60 | 36 | 3.82* | 1.23* | 0.49 |
| MtPUB32 | 1348 | 1690 | 1839 | 1330 | 0.33 | 0.45 | -0.02 |
| MtPUB33 | 159 | 328 | 150 | 165 | 1.04* | -0.08 | 0.05 |
| MtPUB34 | 238 | 209 | 254 | 251 | -0.19 | 0.09 | 0.07 |
| MtPUB35 | 47 | 1924 | 3780 | 3587 | 5.34* | 6.32* | 6.24* |
| MtPUB36 | 461 | 771 | 700 | 702 | 0.74 | 0.60 | 0.61 |
| MtPUB37 | 561 | 394 | 355 | 321 | -0.51 | -0.66 | -0.81 |
| MtPUB38 | 169 | 233 | 97 | 81 | 0.47 | -0.81 | -1.05* |
| MtPUB39 | 415 | 462 | 571 | 540 | 0.15 | 0.46 | 0.38 |
| MtPUB40 | 688 | 662 | 176 | 184 | -0.06 | -1.97* | -1.91* |
| MtPUB41 | 405 | 361 | 681 | 712 | -0.16 | 0.75 | 0.81 |
| MtPUB42 | 36 | 439 | 1544 | 912 | 3.61* | 5.42* | 4.66* |
| MtPUB43 | 10 | 70 | 10 | 27 | 2.87* | 0.04 | 1.52* |
| MtPUB44 | 226 | 1168 | 693 | 341 | 2.37* | 1.62* | 0.60 |
| MtPUB45 | 6 | 21 | 9 | 5 | 1.90* | 0.60 | -0.09 |
| MtPUB46 | 208 | 95 | 109 | 96 | -1.13* | -0.94 | -1.11* |
| MtPUB47 | 1422 | 1781 | 2647 | 2632 | 0.32 | 0.90 | 0.89 |
| MtPUB48 | 32 | 99 | 32 | 45 | 1.62* | -0.02 | 0.47 |
| MtPUB49 | 279 | 153 | 160 | 143 | -0.87 | -0.80 | -0.97 |
| MtPUB50 | 178 | 126 | 136 | 154 | -0.50 | -0.39 | -0.21 |
| MtPUB51 | 119 | 110 | 113 | 127 | -0.10 | -0.07 | 0.09 |
| MtPUB52 | 806 | 1059 | 1823 | 2081 | 0.39 | 1.18* | 1.37* |
| MtPUB53 | 1 | 1 | 1 | 1 | 0 | 0 | 0 |
| MtPUB54 | 222 | 156 | 237 | 157 | -0.51 | 0.10 | -0.50 |
| MtPUB55 | 96 | 436 | 89 | 29 | 2.19* | -0.11 | -1.74* |
| MtPUB56 | 261 | 284 | 156 | 220 | 0.12 | -0.74 | -0.24 |
| MtPUB57 | 832 | 1663 | 1887 | 1736 | 1.00* | 1.18* | 1.06* |
| MtPUB58 | 230 | 300 | 244 | 241 | 0.38 | 0.09 | 0.07 |
| MtPUB59 | 328 | 584 | 609 | 728 | 0.83 | 0.89 | 1.15* |
| MtPUB60 | 1 | 1 | 6 | 1 | 0 | 2.70* | 0 |
| MtPUB61 | 262 | 456 | 431 | 470 | 0.80 | 0.72 | 0.84 |
| MtPUB62 | 1125 | 1514 | 1961 | 1846 | 0.43 | 0.80 | 0.71 |
| MtPUB63 | 149 | 135 | 76 | 67 | -0.14 | -0.98 | -1.16* |
| MtPUB64 | 2 | 10 | 16 | 1 | 2.41* | 3.07* | -0.96 |
Values indicate number of reads.
* indicates a significant difference in expression compared to the 0 h time point (P < 0.01 and |log2N| ≥ 1). Drought-0, Drought-2, Drought-6, and Drought-12 indicate 0, 2, 6, and 12 h drought treatment, respectively.
Table 4. Read abundance of MtPUB genes in the cold-0, cold-2, cold-6, and cold-12 libraries.
| Gene_ID | Cold-0 | Cold-2 | Cold-6 | Cold-12 | log2 (Cold-2/Cold-0) |
log2 (Cold-6/Cold-0) |
log2 (Cold-12/Cold-0) |
|---|---|---|---|---|---|---|---|
| MtPUB1 | 83 | 154 | 43 | 50 | 0.90 | -0.94 | -0.72 |
| MtPUB2 | 57 | 146 | 63 | 30 | 1.37* | 0.14 | -0.90 |
| MtPUB3 | 68 | 149 | 71 | 54 | 1.13* | 0.05 | -0.33 |
| MtPUB4 | 162 | 380 | 205 | 121 | 1.23* | 0.34 | -0.42 |
| MtPUB5 | 60 | 48 | 28 | 21 | -0.31 | -1.11* | -1.54* |
| MtPUB6 | 283 | 290 | 400 | 446 | 0.04 | 0.50 | 0.66 |
| MtPUB7 | 340 | 1346 | 1024 | 771 | 1.99* | 1.59* | 1.18* |
| MtPUB8 | 1 | 1 | 2 | 1 | 0.74 | ||
| MtPUB9 | 969 | 743 | 1315 | 1567 | -0.38 | 0.44 | 0.69 |
| MtPUB10 | 7 | 17 | 20 | 15 | 1.26* | 1.50* | 1.06* |
| MtPUB11 | 3 | 13 | 12 | 9 | 2.13* | 2.01* | 1.62* |
| MtPUB12 | 4 | 5 | 11 | 8 | 0.25 | 1.43* | 0.96 |
| MtPUB13 | 459 | 558 | 555 | 627 | 0.28 | 0.27 | 0.45 |
| MtPUB14 | 370 | 401 | 396 | 446 | 0.12 | 0.10 | 0.27 |
| MtPUB15 | 1 | 1 | 2 | 1 | 0 | 0.74 | 0 |
| MtPUB16 | 1 | 1 | 1 | 1 | 0 | 0 | 0 |
| MtPUB17 | 505 | 1059 | 686 | 750 | 1.07* | 0.44 | 0.57 |
| MtPUB18 | 56 | 76 | 271 | 152 | 0.45 | 2.28* | 1.45* |
| MtPUB19 | 509 | 570 | 867 | 719 | 0.16 | 0.77 | 0.50 |
| MtPUB20 | 448 | 487 | 483 | 508 | 0.12 | 0.11 | 0.18 |
| MtPUB21 | 372 | 520 | 322 | 404 | 0.48 | -0.21 | 0.12 |
| MtPUB22 | 193 | 2724 | 268 | 295 | 3.82* | 0.47 | 0.61 |
| MtPUB23 | 188 | 72 | 450 | 278 | -1.38* | 1.26* | 0.57 |
| MtPUB24 | 420 | 438 | 355 | 439 | 0.06 | -0.24 | 0.07 |
| MtPUB25 | 296 | 330 | 619 | 797 | 0.16 | 1.07* | 1.43* |
| MtPUB26 | 254 | 168 | 163 | 229 | -0.60 | -0.64 | -0.15 |
| MtPUB27 | 125 | 89 | 123 | 102 | -0.49 | -0.02 | -0.30 |
| MtPUB28 | 960 | 963 | 1088 | 1170 | 0.00 | 0.18 | 0.29 |
| MtPUB29 | 689 | 1078 | 1388 | 1378 | 0.65 | 1.01* | 1.00* |
| MtPUB30 | 364 | 232 | 164 | 240 | -0.65 | -1.15* | -0.60 |
| MtPUB31 | 26 | 23 | 16 | 49 | -0.14 | -0.71 | 0.91 |
| MtPUB32 | 1314 | 1815 | 2248 | 1965 | 0.47 | 0.77 | 0.58 |
| MtPUB33 | 176 | 898 | 367 | 276 | 2.35* | 1.06* | 0.65 |
| MtPUB34 | 247 | 232 | 148 | 191 | -0.09 | -0.74 | -0.37 |
| MtPUB35 | 33 | 392 | 158 | 162 | 3.57* | 2.26* | 2.29* |
| MtPUB36 | 407 | 413 | 544 | 654 | 0.02 | 0.42 | 0.68 |
| MtPUB37 | 526 | 480 | 270 | 270 | -0.13 | -0.96 | -0.96 |
| MtPUB38 | 166 | 169 | 138 | 170 | 0.02 | -0.27 | 0.04 |
| MtPUB39 | 389 | 453 | 432 | 579 | 0.22 | 0.15 | 0.58 |
| MtPUB40 | 713 | 720 | 760 | 728 | 0.01 | 0.09 | 0.03 |
| MtPUB41 | 413 | 348 | 273 | 399 | -0.25 | -0.60 | -0.05 |
| MtPUB42 | 37 | 222 | 684 | 449 | 2.58* | 4.20* | 3.60* |
| MtPUB43 | 17 | 68 | 23 | 8 | 2.05* | 0.49 | -1.04* |
| MtPUB44 | 274 | 2081 | 506 | 532 | 2.93* | 0.89 | 0.96 |
| MtPUB45 | 3 | 17 | 7 | 5 | 2.49* | 1.20* | 0.76 |
| MtPUB46 | 212 | 215 | 218 | 229 | 0.02 | 0.04 | 0.11 |
| MtPUB47 | 1392 | 1537 | 1539 | 1790 | 0.14 | 0.14 | 0.36 |
| MtPUB48 | 62 | 148 | 50 | 25 | 1.26* | -0.31 | -1.32* |
| MtPUB49 | 350 | 269 | 237 | 525 | -0.38 | -0.56 | 0.58 |
| MtPUB50 | 174 | 194 | 174 | 148 | 0.16 | 0.00 | -0.23 |
| MtPUB51 | 121 | 136 | 143 | 152 | 0.17 | 0.24 | 0.33 |
| MtPUB52 | 792 | 937 | 775 | 958 | 0.24 | -0.03 | 0.27 |
| MtPUB53 | 1 | 1 | 1 | 1 | 0 | 0 | 0 |
| MtPUB54 | 205 | 226 | 195 | 219 | 0.14 | -0.07 | 0.10 |
| MtPUB55 | 95 | 949 | 227 | 77 | 3.32* | 1.25* | -0.31 |
| MtPUB56 | 298 | 626 | 925 | 467 | 1.07* | 1.64* | 0.65 |
| MtPUB57 | 874 | 1148 | 1370 | 1357 | 0.39 | 0.65 | 0.63 |
| MtPUB58 | 255 | 250 | 261 | 297 | -0.03 | 0.03 | 0.22 |
| MtPUB59 | 278 | 358 | 353 | 385 | 0.37 | 0.34 | 0.47 |
| MtPUB60 | 1 | 1 | 1 | 1 | 0 | 0 | 0 |
| MtPUB61 | 198 | 432 | 337 | 378 | 1.12* | 0.77 | 0.93 |
| MtPUB62 | 1146 | 891 | 2446 | 2168 | -0.36 | 1.09* | 0.92 |
| MtPUB63 | 160 | 188 | 170 | 100 | 0.23 | 0.08 | -0.67 |
| MtPUB64 | 4 | 4 | 2 | 8 | 0.00 | -0.81 | 0.96 |
Values indicate number of reads.
* indicates a significant difference in expression compared to the 0 h time point (P < 0.01 and |log2N| ≥ 1). Cold-0, Cold-2, Cold-6, and Cold-12 indicate 0, 2, 6, and 12 h cold treatment, respectively.
Table 3. Read abundance of MtPUB genes in the salt-0, salt-2, salt-6, and salt-12 libraries.
| Gene_ID | Salt-0 | Salt-2 | Salt-6 | Salt-12 | log2 (Salt-2/Salt-0) |
log2 (Salt-6/Salt-0) |
log2 (Salt-12/Salt-0) |
|---|---|---|---|---|---|---|---|
| MtPUB1 | 109 | 142 | 158 | 223 | 0.38 | 0.54 | 1.03* |
| MtPUB2 | 44 | 18 | 13 | 24 | -1.31* | -1.81* | -0.91 |
| MtPUB3 | 44 | 42 | 13 | 13 | -0.09 | -1.81* | -1.78* |
| MtPUB4 | 159 | 81 | 54 | 42 | -0.97 | -1.56* | -1.91* |
| MtPUB5 | 61 | 43 | 70 | 147 | -0.52 | 0.20 | 1.27* |
| MtPUB6 | 249 | 183 | 242 | 179 | -0.45 | -0.04 | -0.48 |
| MtPUB7 | 375 | 518 | 578 | 621 | 0.46 | 0.62 | 0.73 |
| MtPUB8 | 1 | 3 | 1 | 10 | 1.73* | 0 | 3.37* |
| MtPUB9 | 975 | 654 | 837 | 944 | -0.58 | -0.22 | -0.05 |
| MtPUB10 | 13 | 13 | 37 | 25 | 0.01 | 1.47* | 0.91 |
| MtPUB11 | 1 | 3 | 1 | 1 | 1.35* | 0 | 0 |
| MtPUB12 | 1 | 2 | 2 | 1 | 0.82 | 0.83 | 0 |
| MtPUB13 | 484 | 809 | 775 | 927 | 0.74 | 0.68 | 0.94 |
| MtPUB14 | 375 | 509 | 615 | 377 | 0.44 | 0.71 | 0.01 |
| MtPUB15 | 1 | 9 | 7 | 4 | 3.25* | 2.85* | 1.87* |
| MtPUB16 | 1 | 1 | 3 | 1 | 0 | 1.74* | 0 |
| MtPUB17 | 527 | 752 | 854 | 1116 | 0.51 | 0.70 | 1.08* |
| MtPUB18 | 34 | 117 | 103 | 171 | 1.76* | 1.58* | 2.32* |
| MtPUB19 | 587 | 525 | 601 | 526 | -0.16 | 0.04 | -0.16 |
| MtPUB20 | 534 | 351 | 428 | 483 | -0.61 | -0.32 | -0.15 |
| MtPUB21 | 464 | 383 | 408 | 667 | -0.28 | -0.18 | 0.52 |
| MtPUB22 | 131 | 242 | 223 | 240 | 0.88 | 0.76 | 0.87 |
| MtPUB23 | 191 | 217 | 348 | 428 | 0.18 | 0.86 | 1.16* |
| MtPUB24 | 393 | 458 | 450 | 585 | 0.22 | 0.19 | 0.57 |
| MtPUB25 | 220 | 411 | 397 | 526 | 0.90 | 0.85 | 1.26* |
| MtPUB26 | 237 | 281 | 395 | 797 | 0.24 | 0.74 | 1.75* |
| MtPUB27 | 110 | 63 | 104 | 58 | -0.80 | -0.08 | -0.92 |
| MtPUB28 | 929 | 1282 | 1146 | 1239 | 0.47 | 0.30 | 0.42 |
| MtPUB29 | 745 | 963 | 1003 | 1122 | 0.37 | 0.43 | 0.59 |
| MtPUB30 | 419 | 340 | 396 | 989 | -0.30 | -0.08 | 1.24* |
| MtPUB31 | 28 | 92 | 108 | 367 | 1.73* | 1.97* | 3.73* |
| MtPUB32 | 1387 | 1959 | 2172 | 2297 | 0.50 | 0.65 | 0.73 |
| MtPUB33 | 123 | 264 | 169 | 302 | 1.10* | 0.45 | 1.29* |
| MtPUB34 | 177 | 249 | 254 | 388 | 0.49 | 0.52 | 1.13* |
| MtPUB35 | 80 | 1755 | 1351 | 1129 | 4.45* | 4.08* | 3.82* |
| MtPUB36 | 513 | 703 | 683 | 589 | 0.45 | 0.41 | 0.20 |
| MtPUB37 | 483 | 491 | 490 | 550 | 0.02 | 0.02 | 0.19 |
| MtPUB38 | 158 | 106 | 127 | 151 | -0.58 | -0.32 | -0.06 |
| MtPUB39 | 458 | 508 | 509 | 674 | 0.15 | 0.16 | 0.56 |
| MtPUB40 | 807 | 220 | 240 | 153 | -1.88* | -1.75* | -2.40* |
| MtPUB41 | 421 | 314 | 520 | 514 | -0.42 | 0.31 | 0.29 |
| MtPUB42 | 25 | 1145 | 1127 | 1003 | 5.49* | 5.47* | 5.30* |
| MtPUB43 | 13 | 32 | 34 | 72 | 1.27* | 1.34* | 2.43* |
| MtPUB44 | 248 | 528 | 507 | 1083 | 1.09* | 1.03* | 2.12* |
| MtPUB45 | 5 | 17 | 4 | 1 | 1.66* | -0.41 | -2.45* |
| MtPUB46 | 274 | 104 | 142 | 119 | -1.40* | -0.94 | -1.20* |
| MtPUB47 | 1416 | 1755 | 2165 | 2306 | 0.31 | 0.61 | 0.70 |
| MtPUB48 | 47 | 86 | 83 | 131 | 0.88 | 0.84 | 1.49* |
| MtPUB49 | 272 | 176 | 187 | 94 | -0.63 | -0.54 | -1.53* |
| MtPUB50 | 207 | 208 | 240 | 137 | 0.01 | 0.21 | -0.60 |
| MtPUB51 | 110 | 127 | 168 | 251 | 0.20 | 0.61 | 1.19* |
| MtPUB52 | 908 | 1296 | 1477 | 1886 | 0.51 | 0.70 | 1.05* |
| MtPUB53 | 1 | 1 | 1 | 1 | 0 | 0 | 0 |
| MtPUB54 | 209 | 191 | 351 | 307 | -0.13 | 0.75 | 0.55 |
| MtPUB55 | 83 | 289 | 93 | 259 | 1.79* | 0.15 | 1.64* |
| MtPUB56 | 295 | 268 | 190 | 218 | -0.14 | -0.64 | -0.44 |
| MtPUB57 | 797 | 1701 | 1589 | 2026 | 1.09* | 1.00* | 1.35* |
| MtPUB58 | 236 | 298 | 291 | 302 | 0.34 | 0.30 | 0.35 |
| MtPUB59 | 286 | 585 | 647 | 694 | 1.03* | 1.18* | 1.28* |
| MtPUB60 | 1 | 9 | 5 | 2 | 3.12* | 2.29* | 1.22* |
| MtPUB61 | 204 | 319 | 465 | 638 | 0.65 | 1.19* | 1.65* |
| MtPUB62 | 1122 | 1812 | 1784 | 1790 | 0.69 | 0.67 | 0.67 |
| MtPUB63 | 134 | 84 | 90 | 98 | -0.68 | -0.57 | -0.44 |
| MtPUB64 | 1 | 10 | 12 | 18 | 3.36* | 3.57* | 4.19* |
Values indicate number of reads.
* indicates a significant difference in expression compared to the 0 h time point (P < 0.01 and |log2N| ≥ 1). Salt-0, Salt-2, Salt-6, and Salt-12 indicate 0, 2, 6, and 12 h salt treatment, respectively.
As indicated above, fewer MtPUB genes were down-regulated under the analyzed stress conditions. Under drought treatment, MtPUB2, MtPUB3, MtPUB4, MtPUB6, MtPUB9, MtPUB11, MtPUB20, MtPUB27, MtPUB40, MtPUB46, and MtPUB63 were down-regulated (Fig 5B, Table 2). (A gene was considered down-regulated if its expression was decreased at 2, 6, and 12 h and if the log2 fold change < -1 for at least one of these time points). Using the same criteria, MtPUB2, MtPUB3, MtPUB4, MtPUB40, MtPUB46, and MtPUB49 were down-regulated under salt stress (Table 3). After cold treatment, only MtPUB5 and MtPUB30 were down-regulated. We also identified MtPUB genes that were induced by more than one stress condition (Fig 5). For example, MtPUB10, MtPUB17, MtPUB18, MtPUB35, MtPUB42, and MtPUB44 were induced by salt, drought, and cold treatment. In addition, MtPUB2, MtPUB3, MtPUB4, MtPUB40, and MtPUB46 were down-regulated under salt stress and under drought stress (Fig 5).
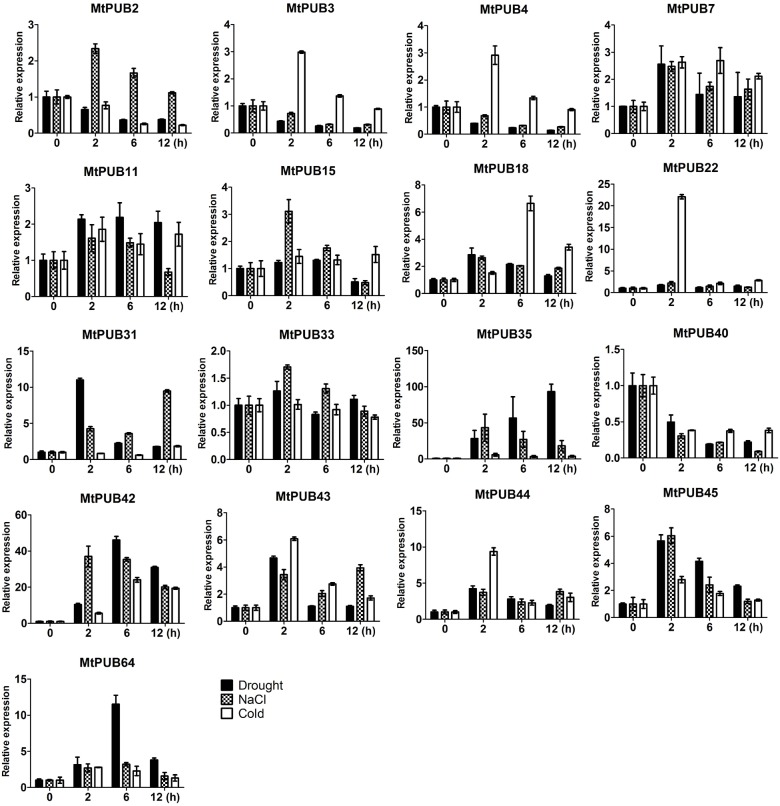
To verify the above data, we conducted qRT-PCR to examine the expression patterns of 17 MtPUB genes under the different stress conditions (Fig 6 and S3 Table). Under drought stress, the transcript levels of the following U-box protein-encoding genes increased: MtPUB7, MtPUB11, MtPUB18, MtPUB22, MtPUB31, MtPUB35, MtPUB42, MtPUB43, MtPUB44, MtPUB45, and MtPUB64. Among these, MtPUB31, MtPUB35, MtPUB42, MtPUB43, MtPUB44, MtPUB45, and MtPUB64 were strongly induced. MtPUB35, MtPUB42, MtPUB43, MtPUB44, and MtPUB45 were also strongly induced by salt stress treatment. Under cold stress, the transcript levels of MtPUB3, MtPUB4, MtPUB22, MtPUB35, MtPUB42, MtPUB43, MtPUB44, MtPUB45, and MtPUB64 increased, and among these, MtPUB18, MtPUB22, MtPUB35, MtPUB42, MtPUB43, and MtPUB44 were strongly induced. It is worth noting that the domain analysis identified MtPUB35 and MtPUB42 as U-box-ARM proteins and that U-box-ARM proteins in Arabidopsis and rice are known to have important roles in plant stress response [15].
Fig 6. The expression of U-box protein-encoding genes induced by drought, salt, and cold stress as determined by qRT-PCR.
Four-week-old seedlings were treated with drought (by transferring them to dry Whatman 3MM paper in a sterile petri dish), NaCl (300 mM), or cold (4°C) for 0, 2, 6, and 12 h.
A few MtPUBs were down-regulated under stress, including MtPUB40, which was down-regulated under all three abiotic stress conditions. MtPUB3 and MtPUB4 were down-regulated under drought and salt stress, and MtPUB2 was down-regulated under drought and cold stress. These data illustrate the consistency between the qRT-PCR and high-throughput sequencing analyses (Fig 5 and Fig 6, Tables 2–4). Some U-box protein-encoding genes were induced by all three stress conditions and may therefore have important roles in response to abiotic stress; however, further study is required to characterize the functions of these and other MtPUB genes.
Stress-associated cis-acting elements in MtPUB promoters
Cis-regulatory elements and trans-acting factors involved in stress-induced gene expression have been extensively analyzed [7]. To identify promoter elements at MtPUB loci, we analyzed the 1500 bp upstream promoter sequences of the 64 MtPUBs using the PlantCARE database (http://intra.psb.ugent.be:8080/PlantCARE) [34]. The elements listed in S2 Table include several known stress-related elements, including the MYB binding site involved in drought inducibility (MBS), anaerobic induction elements (AREs), heat-stress-responsive elements (HSEs), low-temperature-responsive elements (LTRs), ABA-responsive elements (ABREs), and stress-responsive elements (TC-rich repeats) and so on [35,36]. Among the 64 MtPUBs, 27 had ABREs, suggesting they might be involved in ABA-mediated stress response processes. Forty-five MtPUBs had AREs, elements involved in the response to hypoxic, low-temperature, and dehydration stresses [37]. The presence of ABREs and AREs in some MtPUBs suggests that they might be regulated by stress conditions. For example, we found more than two AREs and ABREs in the promoters of MtPUB13, MtPUB17, MtPUB42, MtPUB48, and MtPUB57. These findings from the analysis of stress-responsive cis elements provide auxiliary evidence that some MtPUBs are likely to be involved in the response to abiotic stresses.
Discussion
2.1 U-box family genes structure and evolution
The global identification of U-box genes should help improve the understanding of gene expression and regulatory mechanisms that underlie plant tolerance to abiotic stresses such as salinity, drought, and cold. This study identified 64 U-box genes from M. truncatula, which is similar to the number identified in Arabidopsis (61) (S1 Table) [38] and rice (77) (S1 Table) [5]. Compared to higher plants, there are far fewer U-box proteins in yeast (3) and human (20) [39], indicating an uneven distribution of U-box proteins among species of different kingdoms. Considering the percentage of U-box genes among total genes in the genome, the percentage in M. truncatula (0.134%) was lower than that in Arabidopsis (0.249%). Through the phylogenetic tree analysis, we found that multiple members in each class of U-box proteins raised the possibility of functional redundancy among the members, such as MtPUB10 and MtPUB11 (Fig 2). Such functional redundancy may represent a daunting challenge for the functional characterization of PUB genes.
In addition to the U-box domain, other important domains, including the ARM, kinase, KAP, and WD40 domains, were present in the identified proteins. The most highly represented was the ARM domain, an approximately 40-amino-acid long tandemly repeated sequence motif (Fig 1). This domain was first identified in the Drosophila melanogaster segment polarity protein Armadillo, which is involved in Wingless signal transduction [40]. Structural characteristics of the ARM motif suggest its involvement in protein-protein interaction, which has been demonstrated in several cases [41]. In a few cases, HEAT repeats were detected in proximity to the ARM repeats. In animals, the functions of ARM-repeat proteins are significant, including cytoskeletal regulation and intracellular signaling transduction.
We analyzed the chromosomal locations of the U-box protein-encoding genes on the M. truncatula genome (Fig 3). Profiling of the gene distribution on the eight M. truncatula chromosomes indicated that the gene family evolved in this species through a large number of duplication events. Gene duplication was defined according to the following criteria: (1) The length of the sequence alignment covered ≥80% of the longest gene, and (2) the similarity of the aligned gene regions was ≥70% [22,23]. The 64 U-box genes in M. truncatula were distributed on all eight chromosomes, but in some cases, the genes were concentrated in certain chromosomal regions, such as the bottom half of chromosome 1. In addition, we found some U-box genes were arranged in tandem repeats of two genes, representative of local gene duplications. This finding suggests that tandem duplications of chromosomal regions may have played an important role in the expansion of this gene family. On the other hand, we also found tandem U-box genes harboring different functional domains, indicative of diversification by domain shuffling after tandem duplication, which would promote functional diversity of the U-box genes.
2.2 U-box family genes tissue-differentially expression and function
The functions of U-box genes in M. truncatula remain poorly understood. Some of them were constitutively expressed, such as MtPUB25, MtPUB52, MtPUB56, and MtPUB58, whose high expression levels in tissues suggest they may be essential for M. truncatula growth and development (Fig 4). Other U-box genes, such as MtPUB42, had low expression levels in all tissues but were clearly induced by stress according to the RNA-seq data, indicating a potential role in abiotic stress. Finally, tissue-specific expression was also observed, such as the root-specific expression of MtPUB49, indicating that some U-box genes may have tissue-specific or organ-specific functions (Fig 4).
2.3 U-box family genes in response to various abiotic stresses
It remains unclear why plants have more U-box proteins than other organisms. One possibility is that U-box proteins significantly contribute to the ability of plants to respond to diverse environmental stresses, due to plant immobility and the lack of an animal-like immune system [39]. There has been increasing evidence supporting this hypothesis in recent years, which prompted us to investigate whether M. truncatula PUB proteins are induced by abiotic stress. The number of up-regulated U-box genes was 15, 25, and 16 under drought, salt, and cold stress, respectively. In contrast, the number of down-regulated U-box genes was 11, 6, and 3, respectively (Fig 5, Tables 2–4). Thus, abiotic stress mainly induces U-box gene expression. Many genes were induced by two or three stress conditions and may therefore play a role under various environmental stresses. Our results showed that, as in other species, the expression of many MtPUB genes, such as MtPUB10, MtPUB17, MtPUB18, MtPUB35, MtPUB42, and MtPUB44, could be induced by drought, salt, and cold stress (Fig 5, Tables 2–4).
In higher plants, U-box-ARM proteins have been implicated in the regulation of cell death and defense [9] and in reducing cellular oxidative stress during seedling establishment in rice [15]. MtPUB35 and MtPUB42 were found to encode ARM domain-containing proteins and were up-regulated more than 10-fold at different time points under all three stresses (Fig 5, Tables 2–4). In addition to their classification as U-box-ARM protein-encoding genes with markedly induced expression under abiotic stresses, the proteins encoded by MtPUB35 and MtPUB42 were grouped together in the G1 subfamily in the phylogenetic analysis. Analysis of cis sequences revealed 4 and 3 ABRE elements in MtPUB35 and MtPUB42, respectively, as well as 5 ARE elements in MtPUB42 (S2 Table), further indicating that the two U-Box-ARM genes are important for stress response. Further study of these genes is therefore warranted. In short, these results are consistent with the findings in other plants that U-box-ARM proteins have the potential to regulate plant responses to abiotic stresses. M. truncatula homologs of other characterized PUB genes were also identified in the present study. For example, the Arabidopsis genes AtPUB22 and AtPUB23 play a key role in drought stress response [10], so MtPUB18, the homologous gene in M. truncatula, may also be associated with drought stress. Similarly, MtPUB44 may be involved in disease resistance, as it is homologous to tobacco NtCMPG1, which has been shown to be essential for disease resistance [42]. Taken together, the present findings suggest that PUB proteins likely play critical roles in stress response in M. truncatula.
Supporting information
(PDF)
(PDF)
(PDF)
Detailed genomic information, including the gene name, gene ID, and protein sequence, is provided for each U-box gene.
(XLS)
(XLS)
(XLS)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by the National Natural Science Foundation of China, grant number 31560076 (https://isisn.nsfc.gov.cn/egrantindex/funcindex/prjsearch-list) to JIANBO SONG. The role of the sponsors was RNA sequencing and data analysis related fee. This study was also supported by the National Natural Science Foundation of China, grant number 91440105 (https://isisn.nsfc.gov.cn/egrantindex/funcindex/prjsearch-list) to XIAOWEI MO, LUMING YUE. The role of the sponsors was study design and RNA sequencing. This study was also supported by the National Natural Science Foundation of China, grant number 31571332 (https://isisn.nsfc.gov.cn/egrantindex/funcindex/prjsearch-list) to BEIXIN MO. The role of the sponsors was data collection and analysis. This study was also supported by the Guangdong Innovation Research Team Fund, grant number 2014ZT05S078 (http://cxtd.gdstc.gov.cn) to BEIXIN MO, JUN SONG. The role of the sponsors was decision to publish and data collection and analysis. This study was also supported by the China Postdoctoral Science Foundation, grant number 2016M592523 (http://jj.chinapostdoctor.org.cn/V1/Program3/Default.aspx) to JIANBO SONG, HAIQI YANG. The role of the sponsors was preparation of the manuscript.
References
- 1.Ciechanover A (1998) The ubiquitin–proteasome pathway: on protein death and cell life. The EMBO journal 17: 7151–7160. doi: 10.1093/emboj/17.24.7151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ciechanover A (2005) Intracellular protein degradation: from a vague idea thru the lysosome and the ubiquitin–proteasome system and onto human diseases and drug targeting. Cell Death & Differentiation 12: 1178–1190. [DOI] [PubMed] [Google Scholar]
- 3.Yee D, Goring DR (2009) The diversity of plant U-box E3 ubiquitin ligases: from upstream activators to downstream target substrates. Journal of Experimental Botany: ern369. [DOI] [PubMed] [Google Scholar]
- 4.Koegl M, Hoppe T, Schlenker S, Ulrich HD, Mayer TU, Jentsch S (1999) A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell 96: 635–644. [DOI] [PubMed] [Google Scholar]
- 5.Zeng L-R, Park CH, Venu R, Gough J, Wang G-L (2008) Classification, expression pattern, and E3 ligase activity assay of rice U-box-containing proteins. Molecular Plant 1: 800–815. doi: 10.1093/mp/ssn044 [DOI] [PubMed] [Google Scholar]
- 6.Hatakeyama S, Yada M, Matsumoto M, Ishida N, Nakayama K-I (2001) U box proteins as a new family of ubiquitin-protein ligases. Journal of Biological Chemistry 276: 33111–33120. doi: 10.1074/jbc.M102755200 [DOI] [PubMed] [Google Scholar]
- 7.Cho SK, Bae H, Ryu MY, Yang SW, Kim WT (2015) PUB22 and PUB23 U-BOX E3 ligases directly ubiquitinate RPN6, a 26S proteasome lid subunit, for subsequent degradation in Arabidopsis thaliana. Biochemical and biophysical research communications 464: 994–999. doi: 10.1016/j.bbrc.2015.07.030 [DOI] [PubMed] [Google Scholar]
- 8.Yan J, Wang J, Li Q, Hwang JR, Patterson C, Zhang H (2003) AtCHIP, a U-box-containing E3 ubiquitin ligase, plays a critical role in temperature stress tolerance in Arabidopsis. Plant Physiology 132: 861–869. doi: 10.1104/pp.103.020800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang C-W, González-Lamothe R, Ewan RA, Rowland O, Yoshioka H, Shenton M, et al. (2006) The E3 ubiquitin ligase activity of Arabidopsis PLANT U-BOX17 and its functional tobacco homolog ACRE276 are required for cell death and defense. The Plant Cell 18: 1084–1098. doi: 10.1105/tpc.105.039198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho SK, Ryu MY, Song C, Kwak JM, Kim WT (2008) Arabidopsis PUB22 and PUB23 are homologous U-Box E3 ubiquitin ligases that play combinatory roles in response to drought stress. The Plant Cell 20: 1899–1914. doi: 10.1105/tpc.108.060699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y-C, Wu Y-R, Huang X-H, Sun J, Xie Q (2011) AtPUB19, a U-box E3 ubiquitin ligase, negatively regulates abscisic acid and drought responses in Arabidopsis thaliana. Molecular plant 4: 938–946. doi: 10.1093/mp/ssr030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li W, Ahn I-P, Ning Y, Park C-H, Zeng L, Whitehill JG, et al. (2012) The U-Box/ARM E3 ligase PUB13 regulates cell death, defense, and flowering time in Arabidopsis. Plant physiology 159: 239–250. doi: 10.1104/pp.111.192617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hwang JH, Seo DH, Kang BG, Kwak JM, Kim WT (2015) Suppression of Arabidopsis AtPUB30 resulted in increased tolerance to salt stress during germination. Plant cell reports 34: 277–289. doi: 10.1007/s00299-014-1706-4 [DOI] [PubMed] [Google Scholar]
- 14.Zeng L-R, Qu S, Bordeos A, Yang C, Baraoidan M, Yan H, et al. (2004) Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. The Plant Cell 16: 2795–2808. doi: 10.1105/tpc.104.025171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park JJ, Yi J, Yoon J, Cho LH, Ping J, Jeong HJ, et al. (2011) OsPUB15, an E3 ubiquitin ligase, functions to reduce cellular oxidative stress during seedling establishment. The Plant Journal 65: 194–205. doi: 10.1111/j.1365-313X.2010.04416.x [DOI] [PubMed] [Google Scholar]
- 16.Wang J, Qu B, Dou S, Li L, Yin D, Pang Z, et al. (2015) The E3 ligase OsPUB15 interacts with the receptor-like kinase PID2 and regulates plant cell death and innate immunity. BMC plant biology 15: 1 doi: 10.1186/s12870-014-0410-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hur Y-J, Yi YB, Lee JH, Chung YS, Jung HW, Yun DJ, et al. (2012) Molecular cloning and characterization of OsUPS, a U-box containing E3 ligase gene that respond to phosphate starvation in rice (Oryza sativa). Molecular biology reports 39: 5883–5888. doi: 10.1007/s11033-011-1399-5 [DOI] [PubMed] [Google Scholar]
- 18.Cho SK, Chung HS, Ryu MY, Park MJ, Lee MM, Bahk Y-Y, et al. (2006) Heterologous expression and molecular and cellular characterization of CaPUB1 encoding a hot pepper U-Box E3 ubiquitin ligase homolog. Plant physiology 142: 1664–1682. doi: 10.1104/pp.106.087965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo Q, Li Y, Wang W, Fei X, Deng X (2015) Genome-wide survey and expression analysis of Chlamydomonas reinhardtii U-box E3 ubiquitin ligases (CrPUBs) reveal a functional lipid metabolism module. PloS one 10: e0122600 doi: 10.1371/journal.pone.0122600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wiborg J, O'Shea C, Skriver K (2008) Biochemical function of typical and variant Arabidopsis thaliana U-box E3 ubiquitin-protein ligases. Biochemical Journal 413: 447–457. doi: 10.1042/BJ20071568 [DOI] [PubMed] [Google Scholar]
- 21.Bae H, Kim WT (2014) Classification and interaction modes of 40 rice E2 ubiquitin-conjugating enzymes with 17 rice ARM-U-box E3 ubiquitin ligases. Biochemical and biophysical research communications 444: 575–580. doi: 10.1016/j.bbrc.2014.01.098 [DOI] [PubMed] [Google Scholar]
- 22.Yang X, Kalluri UC, Jawdy S, Gunter LE, Yin T, Tschaplinski TJ, et al. (2008) The F-box gene family is expanded in herbaceous annual plants relative to woody perennial plants. Plant physiology 148: 1189–1200. doi: 10.1104/pp.108.121921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gu Z, Cavalcanti A, Chen F-C, Bouman P, Li W-H (2002) Extent of gene duplication in the genomes of Drosophila, nematode, and yeast. Molecular biology and evolution 19: 256–262. [DOI] [PubMed] [Google Scholar]
- 24.Zhou ZS, Yang SN, Li H, Zhu CC, Liu ZP, Yang ZM (2013) Molecular dissection of mercury-responsive transcriptome and sense/antisense genes in Medicago truncatula. Journal of hazardous materials 252: 123–131. doi: 10.1016/j.jhazmat.2013.02.011 [DOI] [PubMed] [Google Scholar]
- 25.Smith TF, Gaitatzes C, Saxena K, Neer EJ (1999) The WD repeat: a common architecture for diverse functions. Trends in biochemical sciences 24: 181–185. [DOI] [PubMed] [Google Scholar]
- 26.Das AK, Cohen PT, Barford D (1998) The structure of the tetratricopeptide repeats of protein phosphatase 5: implications for TPR‐mediated protein–protein interactions. The EMBO journal 17: 1192–1199. doi: 10.1093/emboj/17.5.1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.LiD R (2001) WD-repeatproteins: Structurecharacteristics, biologicalfunction, andtheirinvolvementinhumandiseases. CellMolLifeSci 58: 2085–2097. [Google Scholar]
- 28.Löscher M, Fortschegger K, Ritter G, Wostry M, Voglauer R, Schmid JA, et al. (2005) Interaction of U-box E3 ligase SNEV with PSMB4, the β7 subunit of the 20 S proteasome. Biochemical Journal 388: 593–603. doi: 10.1042/BJ20041517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohi MD, Gould KL (2002) Characterization of interactions among the Cef1p-Prp19p-associated splicing complex. Rna 8: 798–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo J, Shen G, Yan J, He C, Zhang H (2006) AtCHIP functions as an E3 ubiquitin ligase of protein phosphatase 2A subunits and alters plant response to abscisic acid treatment. The Plant Journal 46: 649–657. doi: 10.1111/j.1365-313X.2006.02730.x [DOI] [PubMed] [Google Scholar]
- 31.Shen G, Adam Z, Zhang H (2007) The E3 ligase AtCHIP ubiquitylates FtsH1, a component of the chloroplast FtsH protease, and affects protein degradation in chloroplasts. The Plant Journal 52: 309–321. doi: 10.1111/j.1365-313X.2007.03239.x [DOI] [PubMed] [Google Scholar]
- 32.Rosser MF, Washburn E, Muchowski PJ, Patterson C, Cyr DM (2007) Chaperone functions of the E3 ubiquitin ligase CHIP. Journal of Biological Chemistry 282: 22267–22277. doi: 10.1074/jbc.M700513200 [DOI] [PubMed] [Google Scholar]
- 33.Sahara N, Murayama M, Mizoroki T, Urushitani M, Imai Y, Takahashi R, et al. (2005) In vivo evidence of CHIP up‐regulation attenuating tau aggregation. Journal of neurochemistry 94: 1254–1263. doi: 10.1111/j.1471-4159.2005.03272.x [DOI] [PubMed] [Google Scholar]
- 34.Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, et al. (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic acids research 30: 325–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mundy J, Yamaguchi-Shinozaki K, Chua N-H (1990) Nuclear proteins bind conserved elements in the abscisic acid-responsive promoter of a rice rab gene. Proceedings of the National Academy of Sciences 87: 1406–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu D, Duan X, Wang B, Hong B, Ho T-HD, Wu R (1996) Expression of a late embryogenesis abundant protein gene, HVA1, from barley confers tolerance to water deficit and salt stress in transgenic rice. Plant physiology 110: 249–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dolferus R, Jacobs M, Peacock WJ, Dennis ES (1994) Differential interactions of promoter elements in stress responses of the Arabidopsis Adh gene. Plant Physiology 105: 1075–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Azevedo C, Santos-Rosa MJ, Shirasu K (2001) The U-box protein family in plants. Trends in plant science 6: 354–358. [DOI] [PubMed] [Google Scholar]
- 39.Patterson C (2002) A new gun in town: the U box is a ubiquitin ligase domain. Science Signaling 2002: pe4–pe4. [DOI] [PubMed] [Google Scholar]
- 40.Riggleman B, Wieschaus E, Schedl P (1989) Molecular analysis of the armadillo locus: uniformly distributed transcripts and a protein with novel internal repeats are associated with a Drosophila segment polarity gene. Genes & development 3: 96–113. [DOI] [PubMed] [Google Scholar]
- 41.Huber AH, Nelson WJ, Weis WI (1997) Three-dimensional structure of the armadillo repeat region of β-catenin. Cell 90: 871–882. [DOI] [PubMed] [Google Scholar]
- 42.González-Lamothe R, Tsitsigiannis DI, Ludwig AA, Panicot M, Shirasu K, Jones JD (2006) The U-box protein CMPG1 is required for efficient activation of defense mechanisms triggered by multiple resistance genes in tobacco and tomato. The Plant Cell 18: 1067–1083. doi: 10.1105/tpc.106.040998 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
(PDF)
Detailed genomic information, including the gene name, gene ID, and protein sequence, is provided for each U-box gene.
(XLS)
(XLS)
(XLS)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.