Abstract
The bacterium Staphylococcus aureus is an important cause of the life-threatening condition toxic shock syndrome in humans. Bacterial toxins known as superantigens (SAgs) generate this illness by acting as broad activators of a substantial fraction of all T lymphocytes, bypassing the normally highly stringent T-cell receptor antigen specificity to cause a systemic inflammatory cytokine storm in the host. In a new study, Shaler et al. found that immune cells called mucosa-associated invariant T (MAIT) cells make an unexpectedly large contribution to the SAg response in a largely T-cell receptor–independent, cytokine-driven manner. Subsequent to such activation, the MAIT cells remain unresponsive to stimulation with bacterial antigen. Thus, S. aureus hijacks MAIT cells in the cytokine storm and leaves them functionally impaired. This work provides new insight into the role of MAIT cells in antibacterial immunity and opens new avenues of investigation to understand and possibly treat bacterial toxic shock and sepsis.
T cells are lymphocytes that play critical and multifaceted roles in the immune defense of the human host. Situations where the T-cell compartment is compromised and functions poorly or in a misguided fashion often lead to very serious conditions ranging from immunodeficiency to autoimmunity and immunopathology. The majority of T cells respond in an adaptive fashion to peptide antigens derived from pathogen proteins and this recognition is governed by major histocompatibility complex (MHC)-encoded antigen-presenting molecules. However, the T-cell compartment also includes several types of unconventional T-cell subsets that recognize antigens presented by nonclassical MHC-like molecules [1]. Mucosa-associated invariant T (MAIT) cells are a subset of such unconventional T cells that have recently gained considerable attention (Box 1). MAIT cells exist in expanded numbers in adult humans and have a mature phenotype ready to respond to antigens. The differentiation and maturation of MAIT cells can be observed already in fetal tissues [2], while the expansion to 1%–10% of T cells in peripheral blood occurs after birth [3].
Box 1. What are MAIT cells?
In humans, mucosa-associated invariant T (MAIT) cells are classically defined by their semi-invariant T-cell receptor (TCR) containing the invariant TCR α-chain variable region 1–2 (TRAV1-2 or commonly known as Vα7.2) coupled with TCR α-chain joining region 12, 20, or 33 (TRAJ12, 20, or 33 or Jα12, 20, 33), and a restricted TCR β-chain repertoire, namely, TCR β-chain variable region 6 (TRBV6 or Vβ13) and TRBV20 (Vβ2) [4–6]. The concomitant expression of their unique semi-invariant TCR with high levels of the C-type lectin receptor CD161 or the IL-18 receptor α subunit (IL-18Rα) identifies the MAIT cell population in blood and tissues [7,8]. The majority of human MAIT cells express the CD8 coreceptor, with a minor population expressing neither CD4 nor CD8 and only a minute population expressing CD4 [7,8]. MAIT cells express the tissue-homing and inflammatory-chemokine receptors, such as CCR5, CCR6, CCR9, CXCR3, CXCR4, and CXCR6 [3,9], allowing migration to both mucosal and other tissues as well as to sites of inflammation. Consequently, MAIT cells are found in high abundance in peripheral blood, the intestines, lungs, and liver of healthy humans [3,8]. Recent discoveries showed that MAIT cells recognize microbial riboflavin metabolites presented by the major histocompatibility complex (MHC) class-Ib related protein 1 (MR1) [10,11]. The critical and conserved nature of microbial riboflavin synthesis allows MAIT cells to respond to a wide range of microbes, including both pathogens and commensals alike [8] (see Box 2). Because MR1 also displays an extraordinary level of evolutionary conservation among placental and marsupial mammals [12,13], it is not surprising that MAIT cells have been found in a variety of mammals to date, including primates, rodents, and ruminants [12,14,15]. Interestingly, MHC-Ib-restricted T cells’ evolutionary conservation extends to the amphibian Xenopus spp., where invariant T cells are a critical component of early antiviral immunity [16]. MAIT cells develop in the thymus and follow several developmental stages under the control of multiple key factors, including the restriction element MR1, the transcription factor promyelocytic leukaemia zinc finger (PLZF, also known as zinc finger and BTB domain-containing protein 16 [ZBTB16]), and possibly microbial colonization [2,7,17]. The transcription factor PLZF is critical for MAIT cell maturation and functionality, a universal feature of the innate-like T-cell lineages [18]. Interestingly, while maturation of MAIT cells in mice requires the establishment of the gut microbiota [5,7], human fetal MAIT cells are already functionally mature in the mucosal tissues devoid of such established microbial colonization [2]. In summary, the abundance of MAIT cells, the highly conserved nature of MR1 across mammals, and equally conserved nature of the MAIT cell antigens across microbial species strongly suggest that this population of innate-like T cells plays important roles in protection of the host.
MAIT cells carry a T-cell receptor (TCR), which has conserved and invariant features. Using this TCR, they recognize antigens in complex with the MHC-Ib-related protein 1 (MR1) [5]. MR1 displays an extraordinary level of evolutionary conservation among placental and marsupial mammals [12,13], strongly supportive of the notion that MR1 and MAIT cells perform critical functions in the immune system. The nature of MR1-presented antigens was long elusive. In 2012, it was discovered that MAIT cells recognize microbial vitamin B2 (riboflavin) metabolites from a wide range of microbes presented by MR1 molecules [10] (Box 2) (Fig 1A). The conserved nature of this pathway underlies the ability of MAIT cells to respond to a diverse range of microbes, including Escherichia coli, Mycobacterium tuberculosis, Candida albicans, and S. aureus. Once activated by an antigen, MAIT cells rapidly produce pro-inflammatory cytokines, including interferon-gamma (IFNγ), tumor necrosis factor (TNF), and interleukin-17 (IL-17) [8,19]. This mixed cytokine profile most probably contributes to the reported role of MAIT cells in the protection against bacterial and mycobacterial lung infections in animal models as well as in human pulmonary tuberculosis [8,20–23].
Box 2. Antigenic vitamin B metabolites presented by major histocompatibility complex class-Ib related protein 1 (MR1)
Two recent seminal studies discovered that MR1 molecules bind and present vitamin B2 (riboflavin) and vitamin B9 (folic acid) metabolites [10,11]. Interestingly, the vast majority of MAIT cells are reactive only to metabolic intermediates and derivatives of microbial riboflavin biosynthesis. Consistent with these findings, a key distinction between the microbes that are stimulatory and nonstimulatory to MAIT cells is that the former synthesize riboflavin, whereas the latter do not [8,10]. Riboflavin is a critical component of the cofactors flavin adenine dinucleotide and flavin mononucleotide, key players in a wide variety of microbial cellular processes, including redox reactions, energy metabolism, and biosynthesis of many macromolecules. MAIT cell activation requires key genes encoding critical enzymes, including ribA and ribD, that form 5-amino-6-ribityl aminouracil (5-A-RU), an early aminopyrimidine intermediate in riboflavin biosynthesis [11]. 5-A-RU then gives rise to MAIT cell–activating antigens by the nonenzymatic condensation with host- or bacterial-derived small-molecule metabolic intermediates, such as glyoxal and methylglyoxal [11]. These otherwise unstable aminopyrimidine antigens are subsequently “captured” by MR1 and stabilized as the antigen-MR1 complex within the host cells’ endoplasmic reticulum before translocating to the plasma membrane, leading to presentation at the cell surface and recognition by MAIT cells [24].
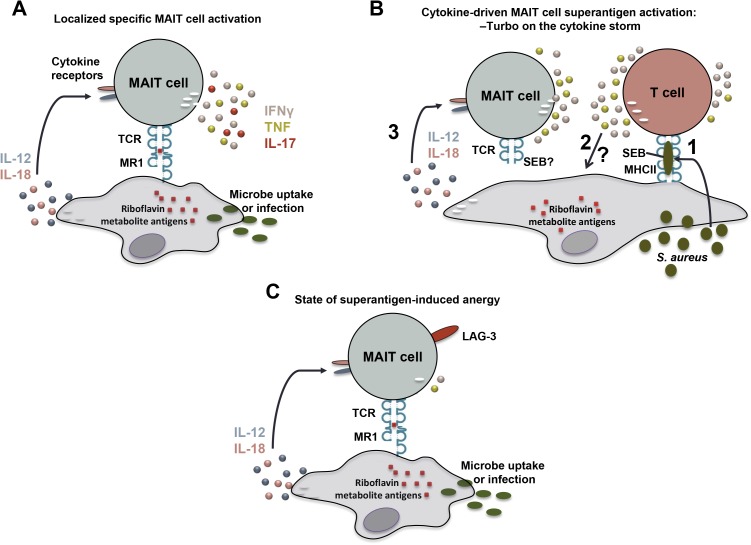
Fig 1. Deception and diversion of MAIT cell responses by bacterial superantigen.
(A) Mucosa-associated invariant T (MAIT) cells recognize bacterial riboflavin metabolites presented by major histocompatibility complex class-Ib related protein 1 (MR1) molecules. This response is enhanced by interleukin 12 (IL-12) and interleukin 18 (IL-18) produced by the antigen-presenting cell. (B) Staphylococcal enterotoxin B (SEB) activates a cytokine storm by conventional T cells, which in turn strongly activates MAIT cells. This MAIT-cell response enhances the already ongoing cytokine storm. There is also some contribution of direct SEB effects on a small fraction of MAIT cells via their T-cell receptor (TCR). (C) After the cytokine storm, MAIT cells become hyporesponsive and this anergic state depends at least partly on the inhibitory receptor lymphocyte-activation gene 3 (LAG-3).
In addition to their capacity for TCR-mediated recognition of microbial riboflavin metabolites, MAIT cells also express a range of receptors usually associated with so-called “innate” immune cells including natural killer (NK) cells [25]. This includes expression of receptors for ILs that can mediate “innate-like” activation of immune cells, including IL-18 and IL-12. Interestingly, Ussher et al. showed that MAIT cells as well as other T cells sharing the marker CD161 can be activated to produce IFNγ independently of the TCR by stimulation with IL-12 and IL-18 [26]. This finding broadened the potential role of MAIT cells in immune responses beyond infections caused by microbes carrying the riboflavin biosynthesis pathway to also include other situations where IL-12 and IL-18 might be produced. This cytokine mode of activation may be a useful tool for the immune system in the context of viral infection where IL-12 and IL-18 are induced, and MAIT cell production of IFNγ in the local milieu can have direct antiviral effects. Indeed, activation of MAIT cells was recently shown to occur in several different types of human viral infections [27]. In this viral context, MAIT cell activation was dependent on IL-18 in synergy with IL-12, IL-15, and interferons, and the IFNγ produced was able to inhibit hepatitis C virus replication in vitro, suggesting direct disease relevance. Thus, in the setting of acute viral infection, MAIT cells recruited to the site of infection may in this way act as amplifiers of the innate antiviral response.
Strong and largely nonspecific activation of an immune cell subset such as MAIT cells may, however, also pose severe danger to the host, in particular since MAIT cells represent up to 10% of T cells in blood and even up to 35% in liver and some mucosal sites [3,5,12,28]. In the June issue of PLOS Biology, Shaler et al. presents evidence that this mode of MAIT cell activation can be triggered by the superantigen (SAg) staphylococcal enterotoxin B (SEB) and that MAIT cells represent a substantial source of pro-inflammatory cytokines following SAg exposure, suggesting a role in staphylococcal immunopathology and immunosuppression [29]. SAgs are potent bacterial exotoxins secreted by bacteria including S. aureus and Streptococcus pyogenes, which cause local symptoms such as food poisoning as well as systemic life-threatening conditions such as toxic shock syndrome (TSS; Box 3) [30]. SAgs can do this because they act as oligoclonal activators of significant parts of the T-cell repertoire, where the fine MHC-peptide specificity of T cells is bypassed when the SAg cross-links MHC class II molecules on antigen-presenting cells and TCRs on T cells. This leads to TCR signalling and massive T-cell activation with concomitant release of inflammatory mediators. However, the interaction between SAgs and TCRs is not completely indiscriminate as only certain TCR β-chain variable (Vβ) segments can be engaged in this way, leaving only a minority of T cells susceptible to triggering by any given SAg. Shaler et al. dissected the human T-cell response to SEB, one of the main SAgs produced by S. aureus, and found that MAIT cells make an oversized contribution to the IFNγ response against SEB relative to their frequency among T cells. This MAIT cell response was dependent on MHC class II but not on MR1 and the magnitude of the response was surprising given that only a small minority of MAIT cells normally express TCR Vβ segments that would bind SEB. In subsequent experiments, Shaler et al. found that this strong MAIT cell activation and IFNγ production was instead primarily driven by IL-12 and IL-18 produced by other cells among peripheral blood mononuclear cells as a result of the SEB-mediated activation of polyclonal T cells (Fig 1B). Both IL-12 and IL-18 were required for full activation and inhibitors of components of the mitogen-activated protein (MAP) kinase pathway were able to block this MAIT cell response. The need for IL-12 and MHC class II in this cytokine-driven mode of activation indicates that so-called professional antigen-presenting cells, such as dendritic cells, initiate this MAIT cell response.
Box 3. Superantigens and their role during gram-positive bacterial infections
The gram-positive bacteria Staphylococcus aureus and Streptococcus pyogenes are important causes of life-threatening toxic shock syndrome (TSS), a fulminant systemic disease characterized by fever, hypotension, and multiorgan dysfunction. Key mediators of TSS are streptococcal and staphylococcal exotoxins that belong to the family of superantigens (SAgs) (reviewed in [31]). In S. pyogenes and S. aureus, 11 and 23 genetically distinct SAgs have been identified, respectively, and most clinical isolates secrete at least 1 SAg [32]. SAgs are defined by their ability to interact with and activate innate immune cells and T cells in an unconventional manner, bypassing normal rules for antigen processing and presentation. They bind directly to major histocompatibility complex class II molecules without prior cellular processing and outside the antigen-binding cleft as well as to specific Vβ regions of the T-cell receptor [30]. This leads to massive T-cell activation and proliferation and consequently a cytokine storm resulting in systemic toxicity and shock. Although the SAg contribution to disease pathophysiology is undeniable, the question that has puzzled the scientific field since their discovery is how they influence bacterial fitness and survival. Many studies have indicated that the SAg-triggered overwhelming cytokine storm results in an unbalanced and, hence, dysfunctional immune response (reviewed in [33]). More recently, it was also demonstrated that SAgs are critical for the bacterial life cycle. Kasper et al. demonstrated that SAgs supported bacterial colonization of nasal mucosa and thereby the initial stages of infection [34]. Furthermore, a role in bacterial survival could be linked to SAg-triggered neutrophil chemotaxis and consequent abscess formation in the liver [35]. This shows the versatility of SAgs influencing a broad range of immune cells and promoting bacterial infections during different stages of infections.
These findings may at first glance not make total sense. Why would an efficient colonizer and pathogen such as S. aureus keep a toxin which not only triggers a broad T-cell response but also generates a secondary cytokine-driven activation of MAIT cells? This is especially intriguing given that S. aureus has the riboflavin biosynthesis pathway and generates the MR1- presented antigens that MAIT cells can detect [8]. Here, the paper by Shaler et al. provides a possible answer with their finding that SEB-stimulated hyperactivation of MAIT cells renders them unresponsive to a subsequent encounter with an MR1-presented bacterial antigen (Fig 1C). This state of cellular unresponsiveness after activation is often referred to as “anergy.” In this system, anergy was associated with the induction of the inhibitory receptors known as T cell immunoglobulin and mucin-3 (TIM-3) and lymphocyte-activation gene 3 (LAG-3) on MAIT cells, and blocking of LAG-3 could restore their ability to respond to bacteria. Finally, the up-regulation of these inhibitory receptors on MAIT cells was also seen in vivo in humanized mice injected with SEB. Altogether, these findings suggest that these effects may be a deception and diversion strategy by S. aureus to avoid direct recognition by MAIT cells.
The IL-12– and IL-18–mediated activation of MAIT cells is likely to have desired consequences during, for example, a local virus infection, in which enhanced IFNγ production can help limit virus replication, as discussed above. In the context of S. aureus infection, this mechanism appears to have been hijacked as a diversion mechanism that anergizes the MAIT cells and makes them unable to respond to MR1-restricted antigen. This way, the bacterium may avoid direct recognition, with detrimental consequences for host antibacterial immunity, not only against S. aureus but also against many other microbial infections. A potentially even more serious consequence for the host may occur in the context of systemic spread of bacterial toxin SAgs, in which the MAIT-cell contribution to the cytokine storm may provide a turbo effect and increase the risk of shock and systemic toxicity. This is a life-threatening condition and the realization that MAIT cells contribute to the SAg-triggered cytokine storm may open new possible avenues for patient treatment.
The work by Shaler et al. provides significant new insight into the role of MAIT cells in the complex interaction between a bacterial pathogen and the host. Still, many open questions remain regarding the specific S. aureus SAg effects on MAIT cells and the downstream consequences, as well as broader questions concerning other SAg-triggered diseases and how MAIT cell contribution to antibacterial immunity may be affected. For example, the perspective of time may be very important here; what is the relative contribution of TCR Vβ-dependent and cytokine-dependent MAIT cell activation during various stages of activation and during bacterial encounter? To what extent will the induced anergy prevent MAIT cell recognition of different bacteria? Will the MAIT cell anergy be local or systemic, and how long will it last? Will these effects limit immune defenses to subsequent encounters of unrelated bacterial, fungal, or viral pathogens? Can pharmaceutical intervention be designed to limit MAIT cell activation in this setting to prevent the immunopathology and the downstream effects? The answers to questions such as these will be very important to help understand the complex relationship between MAIT cells and bacterial pathogens and help define the role that these cells play in host protection.
Abbreviations
- 5-A-RU
5-amino-6-ribityl aminouracil
- IFNγ
interferon-gamma
- IL
interleukin
- LAG-3
lymphocyte-activation gene 3
- MAIT
mucosa-associated invariant T
- MAP
mitogen-activated protein
- MHC
major histocompatibility complex
- MR1
MHC class-Ib related protein 1
- NK
natural killer
- PLZF
promyelocytic leukaemia zinc finger
- SAg
superantigen
- SEB
staphylococcal enterotoxin B
- TCR
T-cell receptor
- TIM-3
T cell immunoglobulin and mucin-3
- TNF
tumor necrosis factor
- TRAJ
TCR α-chain joining region
- TRAV
TCR α-chain variable region
- TRBV
TCR β-chain variable region
- TSS
toxic shock syndrome
- Vβ
β-chain variable
- ZBTB16
zinc finger and BTB domain-containing protein 16
Funding Statement
Swedish Research Council www.vr.se (grant number 2016-03052). Received by JKS. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. National Institute of Diabetes and Digestive and Kidney Diseases www.niddh.nih.gov (grant number R01DK108350). Received by JKS. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The Swedish Cancer Society www.cancerfonden.se (grant number CAN 2014/879). Received by JKS. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Swedish Research Council www.vr.se (grant number 2015-00174). Received by EL. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. European Union FP7 https://ec.europa.eu/research/fp7/index_en.cfm (grant number 305340). Received by ANT. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Swedish Research Council www.vr.se (grant number 2014-6722). Received by ANT. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Provenance: Commissioned; externally peer reviewed
References
- 1.Godfrey DI, Uldrich AP, McCluskey J, Rossjohn J, Moody DB. The burgeoning family of unconventional T cells. Nat Immunol. 2015;16(11):1114–23. doi: 10.1038/ni.3298 . [DOI] [PubMed] [Google Scholar]
- 2.Leeansyah E, Loh L, Nixon DF, Sandberg JK. Acquisition of innate-like microbial reactivity in mucosal tissues during human fetal MAIT-cell development. Nature communications. 2014;5:3143 doi: 10.1038/ncomms4143 ; PubMed Central PMCID: PMC3916833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dusseaux M, Martin E, Serriari N, Peguillet I, Premel V, Louis D, et al. Human MAIT cells are xenobiotic-resistant, tissue-targeted, CD161hi IL-17-secreting T cells. Blood. 2011;117(4):1250–9. Epub 2010/11/19. blood-2010-08-303339 [pii] doi: 10.1182/blood-2010-08-303339 . [DOI] [PubMed] [Google Scholar]
- 4.Porcelli S, Yockey CE, Brenner MB, Balk SP. Analysis of T cell antigen receptor (TCR) expression by human peripheral blood CD4-8- alpha/beta T cells demonstrates preferential use of several V beta genes and an invariant TCR alpha chain. J Exp Med. 1993;178(1):1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Treiner E, Duban L, Bahram S, Radosavljevic M, Wanner V, Tilloy F, et al. Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1 (vol 422, pg 164, 2003). Nature. 2003;423(6943):1018–. doi: 10.1038/Nature01700 ISI:000183753900057. [DOI] [PubMed] [Google Scholar]
- 6.Lepore M, Kalinichenko A, Colone A, Paleja B, Singhal A, Tschumi A, et al. Parallel T-cell cloning and deep sequencing of human MAIT cells reveal stable oligoclonal TCRbeta repertoire. Nature communications. 2014;5:3866 doi: 10.1038/ncomms4866 . [DOI] [PubMed] [Google Scholar]
- 7.Martin E, Treiner E, Duban L, Guerri L, Laude H, Toly C, et al. Stepwise development of MAIT cells in mouse and human. PLoS Biol. 2009;7(3):e54 Epub 2009/03/13. 08-PLBI-RA-3994 [pii] doi: 10.1371/journal.pbio.1000054 ; PubMed Central PMCID: PMC2653554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Le Bourhis L, Martin E, Peguillet I, Guihot A, Froux N, Core M, et al. Antimicrobial activity of mucosal-associated invariant T cells. Nat Immunol. 2010;11(8):701–8. Epub 2010/06/29. ni.1890 [pii] doi: 10.1038/ni.1890 . [DOI] [PubMed] [Google Scholar]
- 9.Walker LJ, Kang YH, Smith MO, Tharmalingham H, Ramamurthy N, Fleming VM, et al. Human MAIT and CD8alphaalpha cells develop from a pool of type-17 precommitted CD8+ T cells. Blood. 2012;119(2):422–33. Epub 2011/11/17. blood-2011-05-353789 [pii] doi: 10.1182/blood-2011-05-353789 ; PubMed Central PMCID: PMC3257008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kjer-Nielsen L, Patel O, Corbett AJ, Le Nours J, Meehan B, Liu L, et al. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature. 2012;491(7426):717–23. Epub 2012/10/12. nature11605 [pii] doi: 10.1038/nature11605 . [DOI] [PubMed] [Google Scholar]
- 11.Corbett AJ, Eckle SB, Birkinshaw RW, Liu L, Patel O, Mahony J, et al. T-cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature. 2014;509(7500):361–5. doi: 10.1038/nature13160 . [DOI] [PubMed] [Google Scholar]
- 12.Huang S, Martin E, Kim S, Yu L, Soudais C, Fremont DH, et al. MR1 antigen presentation to mucosal-associated invariant T cells was highly conserved in evolution. Proc Natl Acad Sci U S A. 2009;106(20):8290–5. Epub 2009/05/07. 0903196106 [pii] doi: 10.1073/pnas.0903196106 ; PubMed Central PMCID: PMC2688861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsukamoto K, Deakin JE, Graves JA, Hashimoto K. Exceptionally high conservation of the MHC class I-related gene, MR1, among mammals. Immunogenetics. 2013;65(2):115–24. Epub 2012/12/12. doi: 10.1007/s00251-012-0666-5 . [DOI] [PubMed] [Google Scholar]
- 14.Goldfinch N, Reinink P, Connelley T, Koets A, Morrison I, Van Rhijn I. Conservation of mucosal associated invariant T (MAIT) cells and the MR1 restriction element in ruminants, and abundance of MAIT cells in spleen. Veterinary research. 2010;41(5):62 doi: 10.1051/vetres/2010034 ; PubMed Central PMCID: PMC2896809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greene JM, Dash P, Roy S, McMurtrey C, Awad W, Reed JS, et al. MR1-restricted mucosal-associated invariant T (MAIT) cells respond to mycobacterial vaccination and infection in nonhuman primates. Mucosal Immunol. 2017;10(3):802–13. doi: 10.1038/mi.2016.91 ; PubMed Central PMCID: PMC5397382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edholm ES, Albertorio Saez LM, Gill AL, Gill SR, Grayfer L, Haynes N, et al. Nonclassical MHC class I-dependent invariant T cells are evolutionarily conserved and prominent from early development in amphibians. Proc Natl Acad Sci U S A. 2013;110(35):14342–7. doi: 10.1073/pnas.1309840110 ; PubMed Central PMCID: PMC3761575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koay HF, Gherardin NA, Enders A, Loh L, Mackay LK, Almeida CF, et al. A three-stage intrathymic development pathway for the mucosal-associated invariant T cell lineage. Nat Immunol. 2016;17(11):1300–11. doi: 10.1038/ni.3565 . [DOI] [PubMed] [Google Scholar]
- 18.Mao AP, Constantinides MG, Mathew R, Zuo Z, Chen X, Weirauch MT, et al. Multiple layers of transcriptional regulation by PLZF in NKT-cell development. Proc Natl Acad Sci U S A. 2016;113(27):7602–7. doi: 10.1073/pnas.1601504113 ; PubMed Central PMCID: PMC4941452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leeansyah E, Ganesh A, Quigley MF, Sonnerborg A, Andersson J, Hunt PW, et al. Activation, exhaustion, and persistent decline of the antimicrobial MR1-restricted MAIT-cell population in chronic HIV-1 infection. Blood. 2013;121(7):1124–35. Epub 2012/12/18. blood-2012-07-445429 [pii] doi: 10.1182/blood-2012-07-445429 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chua WJ, Truscott SM, Eickhoff CS, Blazevic A, Hoft DF, Hansen TH. Polyclonal mucosa-associated invariant T cells have unique innate functions in bacterial infection. Infect Immun. 2012;80(9):3256–67. Epub 2012/07/11. IAI.00279-12 [pii] doi: 10.1128/IAI.00279-12 ; PubMed Central PMCID: PMC3418730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gold MC, Cerri S, Smyk-Pearson S, Cansler ME, Vogt TM, Delepine J, et al. Human mucosal associated invariant T cells detect bacterially infected cells. PLoS Biol. 2010;8(6):e1000407 Epub 2010/07/09. doi: 10.1371/journal.pbio.1000407 ; PubMed Central PMCID: PMC2893946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Georgel P, Radosavljevic M, Macquin C, Bahram S. The non-conventional MHC class I MR1 molecule controls infection by Klebsiella pneumoniae in mice. Mol Immunol. 2011;48(5):769–75. Epub 2010/12/31. S0161-5890(10)00659-0 [pii] doi: 10.1016/j.molimm.2010.12.002 . [DOI] [PubMed] [Google Scholar]
- 23.Meierovics A, Yankelevich WJ, Cowley SC. MAIT cells are critical for optimal mucosal immune responses during in vivo pulmonary bacterial infection. Proc Natl Acad Sci U S A. 2013. doi: 10.1073/pnas.1302799110 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McWilliam HE, Eckle SB, Theodossis A, Liu L, Chen Z, Wubben JM, et al. The intracellular pathway for the presentation of vitamin B-related antigens by the antigen-presenting molecule MR1. Nat Immunol. 2016;17(5):531–7. doi: 10.1038/ni.3416 . [DOI] [PubMed] [Google Scholar]
- 25.Dias J, Leeansyah E, Sandberg JK. Multiple layers of heterogeneity and subset diversity in human MAIT cell responses to distinct microorganisms and to innate cytokines. Proceedings of the National Academy of Sciences. 2017. doi: 10.1073/pnas.1705759114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ussher JE, Bilton M, Attwod E, Shadwell J, Richardson R, de Lara C, et al. CD161++ CD8+ T cells, including the MAIT cell subset, are specifically activated by IL-12+IL-18 in a TCR-independent manner. Eur J Immunol. 2014;44(1):195–203. doi: 10.1002/eji.201343509 ; PubMed Central PMCID: PMC3947164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Wilgenburg B, Scherwitzl I, Hutchinson EC, Leng T, Kurioka A, Kulicke C, et al. MAIT cells are activated during human viral infections. Nature communications. 2016;7:11653 doi: 10.1038/ncomms11653 ; PubMed Central PMCID: PMC4931007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le Bourhis L, Mburu YK, Lantz O. MAIT cells, surveyors of a new class of antigen: development and functions. Curr Opin Immunol. 2013;25(2):174–80. Epub 2013/02/21. S0952-7915(13)00006-X [pii] doi: 10.1016/j.coi.2013.01.005 . [DOI] [PubMed] [Google Scholar]
- 29.Shaler CR, Choi J, Rudak PT, Memarnejadian A, Szabo PA, Tun-Abraham ME, et al. MAIT cells launch a rapid, robust and distinct hyperinflammatory response to bacterial superantigens and quickly acquire an anergic phenotype that impedes their cognate antimicrobial function: Defining a novel mechanism of superantigen-induced immunopathology and immunosuppression. PLoS Biol. 2017;15(6):e2001930 doi: 10.1371/journal.pbio.2001930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kotzin BL, Leung DY, Kappler J, Marrack P. Superantigens and their potential role in human disease. Adv Immunol. 1993;54:99–166. [DOI] [PubMed] [Google Scholar]
- 31.Lappin E, Ferguson AJ. Gram-positive toxic shock syndromes. Lancet Infect Dis. 2009;9(5):281–90. doi: 10.1016/S1473-3099(09)70066-0 . [DOI] [PubMed] [Google Scholar]
- 32.Spaulding AR, Salgado-Pabon W, Kohler PL, Horswill AR, Leung DY, Schlievert PM. Staphylococcal and streptococcal superantigen exotoxins. Clinical microbiology reviews. 2013;26(3):422–47. doi: 10.1128/CMR.00104-12 ; PubMed Central PMCID: PMC3719495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Llewelyn M, Cohen J. Superantigens: microbial agents that corrupt immunity. Lancet Infect Dis. 2002;2(3):156–62. . [DOI] [PubMed] [Google Scholar]
- 34.Kasper KJ, Zeppa JJ, Wakabayashi AT, Xu SX, Mazzuca DM, Welch I, et al. Bacterial superantigens promote acute nasopharyngeal infection by Streptococcus pyogenes in a human MHC Class II-dependent manner. PLoS Pathog. 2014;10(5):e1004155 doi: 10.1371/journal.ppat.1004155 ; PubMed Central PMCID: PMC4038607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu SX, Gilmore KJ, Szabo PA, Zeppa JJ, Baroja ML, Haeryfar SM, et al. Superantigens subvert the neutrophil response to promote abscess formation and enhance Staphylococcus aureus survival in vivo. Infect Immun. 2014;82(9):3588–98. doi: 10.1128/IAI.02110-14 ; PubMed Central PMCID: PMC4187807. [DOI] [PMC free article] [PubMed] [Google Scholar]