Graphical Abstract
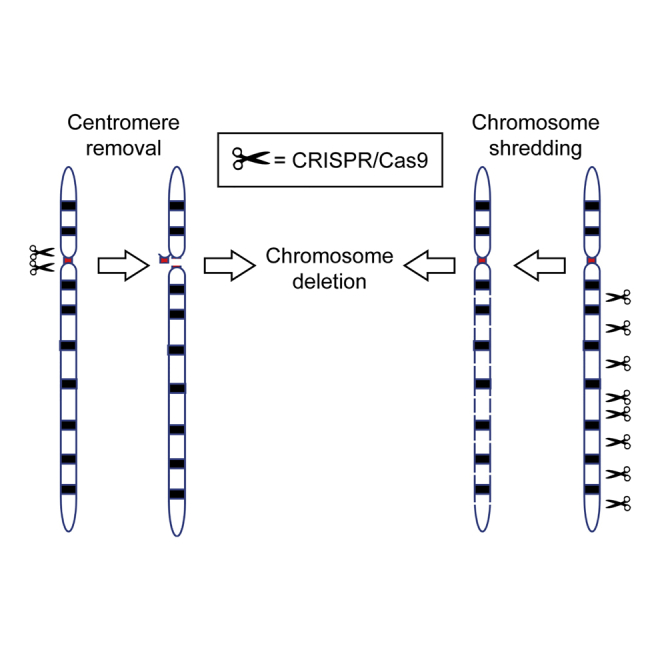
CRISPR/Cas9 genome editing can facilitate efficient deletion of genomic region, but it has not been used to delete an entire chromosome. Here, Adikusuma et al. show proof-of-concept for efficient CRISPR-mediated selective chromosome deletion by removing the centromere or shredding the chromosome arm in mouse embryonic stem cells and zygotes.
Main Text
The recent emergence of gene editing technologies, in particular CRISPR/Cas, has enabled rapid generation of disease models and provides a novel approach for the treatment of monogenic disorders through correction of disease-causing mutations.1, 2 In contrast, the therapeutic potential of CRIPSR/Cas technology for aneuploidies, such as Down syndrome (Trisomy 21), remains unexplored. Indeed, disorders that are caused by supernumerary chromosomes represent a significant challenge, because genetic correction requires targeted ablation of an entire chromosome, which, to our knowledge, has not been demonstrated using genome editing technology.1
To assess the potential of CRISPR/Cas technology to effect chromosomal loss, we investigated the hypothesis that simultaneous generation of multiple DNA double-strand breaks (DSBs) at targeted chromosomal locations can induce directed chromosomal deletion.3 We selected the 90 Mb acrocentric mouse Y chromosome for deletion because loss of this chromosome does not overtly impact cell/mouse viability and it is only present in one copy in male cells, thus facilitating screening.4
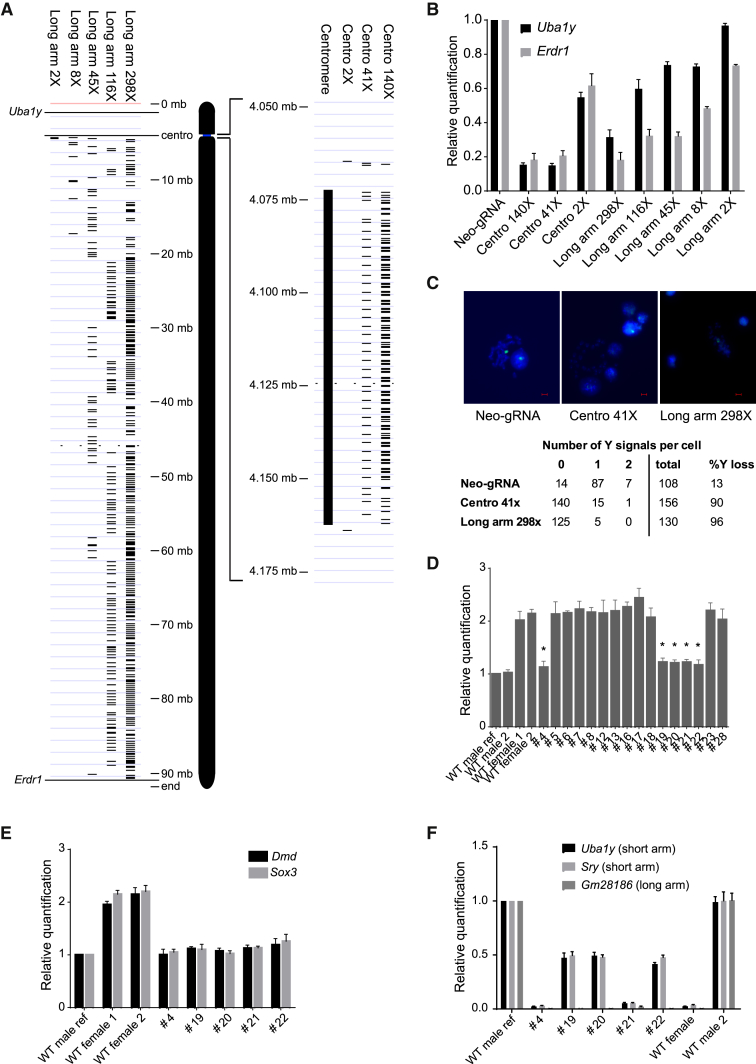
Our first strategy used CRISPR/Cas to fragment the centromere, which is indispensable for chromosome segregation during mitosis.5 We screened the 90 kb Y centromere for guide RNA (gRNA) sequences in repetitive elements that would enable targeted cleavage at multiple sites. We identified two gRNA candidates that target the centromere 140 or 41 times (centro 140X and centro 41X, respectively; Figure 1A). For comparison, we also tested a gRNA pair targeting two unique sequences immediately flanking the centromere (centro 2X; Figure 1A). Cas9 and single-guide RNA (sgRNA) were expressed in R1 XY mouse embryonic stem cells (ESCs) using plasmid PX459 V.2, followed by transient puromycin selection, to ensure only transfectants were harvested.6 Quantification of Y chromosome dosage was performed by genomic qPCR amplification of Uba1y and Erdr1, genes located at the end of the Y chromosome short and long arm, respectively (Figure 1A). Strikingly, Uba1y and Erdr1 qPCR signal was reduced by 80%–85% for both centro 140X and centro 41X compared with the sgRNA-expressing negative control (Neo-gRNA; Figure 1B). Further, a reduction of ∼40% was achieved using the centro 2X gRNA (Figure 1B). To confirm that the reduction of qPCR signal was caused by Y chromosome loss, we performed fluorescence in situ hybridization (FISH) using Y chromosome paint on centro 41X-treated samples. Consistent with the qPCR data, the Y chromosome was not detected in 90% of centro 41X cells compared to 13% of control cells (Figure 1C). We also noted that 6% of control cells had two Y chromosomes, and this was reduced to less than 1% in centro 41X-treated cells. These findings confirm that CRISPR/Cas-mediated centromere cleavage leads to Y chromosome loss at high efficiency.
Figure 1.
Deletion of Y Chromosome Using CRISPR/Cas9 in Mouse ESCs and In Vivo Mouse Zygote Injection
(A) Schematic showing the position of gRNA target sites in the long arm and centromere of the Y chromosome. (B) qPCR of genomic DNA to quantify Y chromosome dosage. Sox1 qPCR was used as the internal reference control. Data are presented as mean ± SEM from n ≥ 3 biological replicates. Statistical analysis using two-way ANOVA is presented in Table S3. (C) FISH analysis detection of Y chromosome loss. Y chromosome and DAPI staining was indicated by green and blue signals, respectively. Scale bar, 5 μm. (D) Xist genomic qPCR of phenotypically female mice generated through zygote injection of centro 41X gRNA. Asterisks indicate female candidates with single X. (E) Dmd and Sox3 genomic qPCR confirming single X chromosome in female XO candidates. (F) Genomic qPCR quantifying dosage of Y short and long arms. Sox1 qPCR was used as the internal reference control. Results are presented as mean ± SD from n ≥ 3 replicates.
Next, we tested an alternative strategy for chromosome deletion in which the long arm is targeted for fragmentation by cleavage at multiple sites. As this approach does not target the centromere, it has potential for application in both dividing and non-dividing cells. We again identified gRNAs that targeted repetitive sequences in the Y chromosome (Figure 1A). However, the selected gRNAs sequences were specific to the long arm to ensure the centromere was left intact. Expression of sgRNAs that targeted the long arm 298X, 116X, 45X, 8X, and 2X resulted in Uba1y qPCR signal loss of 69%, 40%, 26%, 27%, and 3%, respectively, and Erdr1 qPCR signal loss of 82%, 68%, 68%, 52%, and 27%, respectively (Figure 1B). These data indicate that targeted fragmentation of a chromosomal arm can induce chromosome deletion and the frequency of deletion is proportional to the number of cuts. Notably, apart from long arm 298X Edr1, all long gRNAs resulted in significantly higher Uba1y and Edr1 signals than the centro 41X and 140X gRNAs (Table S3). Given that Uba1y qPCR signal was significantly higher than Erdr1 for all long gRNAs (Table S3), we speculate that fragmentation of the long arm occasionally results in chromosome truncation or translocation, with retention of the Y short arm sequence containing Uba1y. FISH Y painting analysis in 298X-treated samples revealed 95% of cells contained no Y chromosome signal, confirming that the long arm fragmentation strategy was indeed effective (Figure 1C).
Notably, the degree of Y chromosome depletion induced by 8X and 45X are similar. This is significant, because targeted deletion of potentially any chromosome could be achieved relatively easily by transfection of a single vector expressing eight unique gRNAs.7 We were also impressed with the activity of the long arm 2X gRNA. Although this gRNA induced negligible loss of short arm signal (3%), it appears to truncate the Y long arm relatively efficiently based on an Erdr1 qPCR signal loss of 27%.
Having successfully deleted an entire chromosome in vitro, we next tested our centromere deletion strategy in vivo in mouse zygotes with the expectation that successful Y chromosome deletion in male zygotes would result in an XO female phenotype.4 We selected gRNA centro 41X due to its high efficiency in vitro and low off-target prediction (Table S2).8
After zygote injection of centro 41X gRNA and Cas9 mRNA, we collected 27 E15.5 embryos, of which 11 were phenotypically male and 16 were female based on gonadal assessment (Figure S1). We then screened the female embryos for X chromosome dosage and identified five embryos with only one X chromosome (Figures 1D and 1E). No evidence of XO karyotype was detected in control females injected with autosomal targeted gRNAs (Figure S2).
To directly assess Y chromosome loss in the five single X females, we performed Y chromosome genomic qPCR. Y short and long arm signals were undetectable in two of these embryos, indicating an XO karyotype. The remaining three embryos contained approximately 50% Y short arm signal and no long arm signal, suggesting that these mice were mosaic, with half of the cells containing translocated/truncated Y short arm and the other half containing no detectable Y (Figure 1F).
Given mosaic outcomes are common following CRISPR/Cas zygote injection,9 we extended our screening to look for phenotypic males that were mosaic for Y chromosome loss. We identified 3 of 11 males with 10%–20% reduction of Y dosage (Figures S3A and S3B). Testis development in these embryos is unsurprising given this level of XY cells.10 In summary, from 27 embryos, we identified 11 XX females, 8 XY males, 2 XO females, 3 mosaic XO females, and 3 mosaic XO males (Table S1). These results provide proof of concept for efficient chromosome deletion in vivo.
This study shows that targeted chromosome deletion is achievable and relatively efficient both in vitro and in vivo using CRISPR/Cas genome editing. This approach should be applicable for other chromosomes and could be utilized in a variety of cellular contexts and species. Accordingly, we envisage that this strategy will be applied to modeling of aneuploidy syndromes and therapeutic intervention by targeting parental-specific polymorphisms.
Author Contributions
F.A., J.H., and P.T. conceived the study. F.A. performed all of the experiments apart from the FISH analysis, which was performed by N.W. and F.G. F.A., J.H., and P.T. drafted the manuscript, which was reviewed and edited by all authors.
Conflicts of Interest
The authors declare that they have no competing financial interests.
Acknowledgments
We acknowledge Sandra Piltz for performing zygote injections and Daniel Pederick for assistance with statistical analysis. F.A. was supported by a scholarship from Beasiswa Unggulan DIKTI (Directorate General of Higher Education, Indonesian Government).
Footnotes
Supplemental Information includes Supplemental Materials and Methods, three figures, and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.ymthe.2017.05.021.
Supplemental Information
References
- 1.Barrangou R., Doudna J.A. Applications of CRISPR technologies in research and beyond. Nat. Biotechnol. 2016;34:933–941. doi: 10.1038/nbt.3659. [DOI] [PubMed] [Google Scholar]
- 2.Mali P., Esvelt K.M., Church G.M. Cas9 as a versatile tool for engineering biology. Nat. Methods. 2013;10:957–963. doi: 10.1038/nmeth.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dumanski J.P., Rasi C., Lönn M., Davies H., Ingelsson M., Giedraitis V., Lannfelt L., Magnusson P.K., Lindgren C.M., Morris A.P. Mutagenesis. Smoking is associated with mosaic loss of chromosome Y. Science. 2015;347:81–83. doi: 10.1126/science.1262092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Probst F.J., Cooper M.L., Cheung S.W., Justice M.J. Genotype, phenotype, and karyotype correlation in the XO mouse model of Turner Syndrome. J. Hered. 2008;99:512–517. doi: 10.1093/jhered/esn027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Westhorpe F.G., Straight A.F. Functions of the centromere and kinetochore in chromosome segregation. Curr. Opin. Cell Biol. 2013;25:334–340. doi: 10.1016/j.ceb.2013.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sakuma T., Nishikawa A., Kume S., Chayama K., Yamamoto T. Multiplex genome engineering in human cells using all-in-one CRISPR/Cas9 vector system. Sci. Rep. 2014;4:5400. doi: 10.1038/srep05400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stemmer M., Thumberger T., Del Sol Keyer M., Wittbrodt J., Mateo J.L. CCTop: An Intuitive, Flexible and Reliable CRISPR/Cas9 Target Prediction Tool. PLoS ONE. 2015;10:e0124633. doi: 10.1371/journal.pone.0124633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang H., Wang H., Jaenisch R. Generating genetically modified mice using CRISPR/Cas-mediated genome engineering. Nat. Protoc. 2014;9:1956–1968. doi: 10.1038/nprot.2014.134. [DOI] [PubMed] [Google Scholar]
- 10.Chang H.J., Clark R.D., Bachman H. The phenotype of 45,X/46,XY mosaicism: an analysis of 92 prenatally diagnosed cases. Am. J. Hum. Genet. 1990;46:156–167. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.