Abstract
Purpose
Retinitis pigmentosa (RP) is a genetically heterogeneous inherited retinal dystrophy. To date, over 80 genes have been implicated in RP. However, the disease demonstrates significant locus and allelic heterogeneity not entirely captured by current testing platforms. The purpose of the present study was to characterize the underlying mutation in a patient with RP without a molecular diagnosis after initial genetic testing.
Methods
Whole-exome sequencing of the affected proband was performed. Candidate gene mutations were selected based on adherence to expected genetic inheritance pattern and predicted pathogenicity. Sanger sequencing of MERTK was completed on the patient’s unaffected mother, affected brother, and unaffected sister to determine genetic phase.
Results
Eight sequence variants were identified in the proband in known RP-associated genes. Sequence analysis revealed that the proband was a compound heterozygote with two independent mutations in MERTK, a novel nonsense mutation (c.2179C>T) and a previously reported missense variant (c.2530C>T). The proband’s affected brother also had both mutations. Predicted phase was confirmed in unaffected family members.
Conclusion
Our study identifies a novel nonsense mutation in MERTK in a family with RP and no prior molecular diagnosis. The present study also demonstrates the clinical value of exome sequencing in determining the genetic basis of Mendelian diseases when standard genetic testing is unsuccessful.
Keywords: Retina, genetics, retinitis pigmentosa, dystrophy
Introduction
Retinitis pigmentosa (RP) represents a genetically heterogeneous group of retinal dystrophies. The disease mainly affects rod photoreceptor cells and manifests progressively with night blindness, constriction of the visual field, and eventual blindness[1, 2]. Ophthalmoscopic findings include retinal vessel attenuation, optic disc pallor, and bone spicule pigmentary changes in the retina [3, 4]. To date, more than 80 genes have been implicated in non-syndromic cases of RP. The mutations in these genes are inherited in autosomal dominant, autosomal recessive, and X-linked patterns, accounting for approximately 20–25%, 15–20%, and 10–15% of non-syndromic cases, respectively, with the remaining attributable to sporadic cases[5, 6]. However, these mutations are only responsible for 60% of known RP cases, indicating 40% of patients do not have a molecular diagnosis [7–9]. The present study focuses on a proband with clinically established RP yet without a molecular diagnosis after commercial genetic testing. Identifying such novel mutations helps to elucidate the pathology of RP, confirm the inheritance pattern, identify potential carriers, shed insight into prognosis, and uncover novel therapeutic targets.
One of the genes implicated in the pathogenesis of RP, Mer tyrosine kinase (MERTK – OMIM#604705) is a receptor tyrosine kinase of the Tyro3/Axl/Mer family of tyrosine kinases[10]. This enzyme class shares a conserved sequence within the kinase domain and in the adhesion molecule-like extracellular domain[11]. The tyrosine kinase is involved in many cellular processes, among them cell proliferation, adhesion, and migration, regulation of inflammatory processes, and blood clotting[12, 13]. Within the retina, MERTK is involved in the retinal pigment epithelium’s (RPE) role in recycling photoreceptor outer segments. Photoreceptors continually renew the light-sensitive disks in their outer segments as they bear significant light stress. These shed disks are phagocytized by the RPE [14, 15]. Prior studies have demonstrated that initial photoreceptor outer segment binding to RPE cells is facilitated by the αvβ5 integrin while MERTK is necessary for phagocytosis of the outer segments [16–18].
Methods
Subjects
Four subjects were ascertained from a family affected by retinitis pigmentosa yet without an associated molecular diagnosis after commercial genetic testing. After IRB approval from the University of Illinois at Chicago was granted for the study, informed consent was obtained from the proband and three family members. Research was performed in concordance with the Declaration of Helsinki.
Proband Whole-Exome Sequencing
A 10mL whole blood sample was collected from the proband in an EDTA tube. A total of 3.5 µg of genomic DNA (gDNA) was extracted using the prepIT-L2P kit (Cat#PT-L2P-5, Genotek Inc., Ottawa, Ontario, Canada) according to manufacturer’s instructions. Prior to library construction, the concentration of the gDNA was determined with the Qubit dsDNA HS Assay Kit (Cat#Q32851, Life Technologies, Carslbad, CA, USA). gDNA integrity was assessed with DNA agarose gel electrophoresis (agarose Gel: 1%, voltage 150V, electrophoresis time: 40 min). Whole-exome sequencing was performed using the BGI Exome Capture Kit (59Mb) on the Complete Genomics platform using Combinatorial Probe-Anchor Ligation (cPAL) technology at 100X depth of coverage (BGI, Hong Kong, China). Quality control of whole-exome sequencing was provided by validation of 140 SNP loci with WaferGen variant validation.
Mapping and Variant Analysis
Initial mapping was performed using TeraMap. Raw read data was aligned to the NCBI human reference genome (NCBI, GRCh37)[19]. Regions found to differ from the reference were identified, and individual reads lying in these regions of interest were collected for local de novo assembly using mate alignment information. Based on the results from reference mapping and sequence assembly, a Bayesian model was applied to call variants from which only those with statistical significance—based on the best scoring hypotheses—were reported [20, 21]. Finally, ambiguous variants, found in duplicative regions or with high similarity, were rescored based on a correlation analysis. Complete Genomics in-house tools were used for the analyses (BGI, Hong Kong, China).
Candidate Gene Identification
To select candidate genes, identified single nucleotide polymorphisms (SNPs) were removed if present in the 1000 Genomes Project (www.1000genomes.org) or the Single Nucleotide Polymorphism Database (dbSNP) (NCBI). Once novel SNPs were identified, predicted pathogenicity was determined using SIFT, PolyPhen2, MutationAssesser, RadialSVM, and snpEff. Based on predicted pathogenicity and expected patterns of inheritance, two gene mutations in MERTK (NM_006343.2:p.Arg727*/c.2179C>T) and (NM_006343.2:p.Arg844Cys/c.2530C>T) were identified as the most likely candidate mutations. The ExAC Browser (The Broad Institute) and the NHLBI database were surveyed to search for previous reports of the two candidate gene mutations.
Sanger Sequencing
After candidate gene mutations were identified, saliva samples were obtained from the proband’s unaffected mother, unaffected sister, and affected brother using the Oragene DNA Self-Collection Kit (REF OG-300, Genotek Inc., Ottawa, Ontario, Canada). gDNA was extracted using the prepIT-L2P kit (Cat#PT-L2P-5, Genotek Inc., Ottawa, Ontario, Canada) according to manufacturer’s instructions. PCR amplification was performed using Green DreamTaq Master Mix (Cat#K1081, Life Technologies, Grand Island, NY) according to manufacturer’s protocols and pre-designed primer sets (Primer set IDs Hs00446022_CE and Hs00360031_CE, Life Technologies, Grand Island, NY, USA). PCR products were purified with AMPure beads (Cat #A63880, Beckman Coulter, Brea, CA, USA) and sequenced using 3730xl DNA Analyzer (Life Technologies, Grand Island, NY, USA).
Results
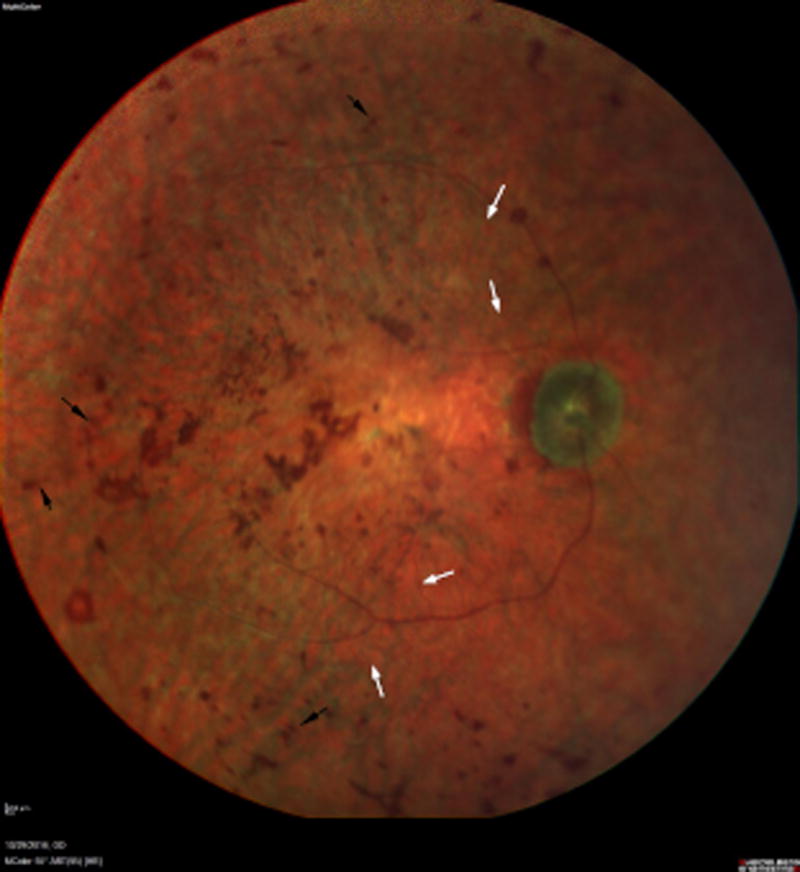
The proband had an established clinical diagnosis of RP. His visual acuity was light perception in both eyes. Ophthalmoscopy demonstrated retinal bone-spicule pigmentary changes in both eyes, retinal vessel attenuation, and diffuse macular atrophy (Figure 1). The proband’s brother also carried the diagnosis of RP. An unaffected sister did not have any retinal disease on examination. Lastly, the proband’s mother was also unaffected by RP though she had been diagnosed with age-related macular degeneration.
Fig 1.
Multicolor 55 Degree Image of the retina (right) reveals a pallorous optic nerve with severely attenuated retinal arterioles (white arrows). Central atrophy is evident with associated coarse, placoid pigmentation in the macula. The midperiphery reveals diffuse retinal pigment epithelial atrophy with associated bone spicule-like pigmentation (black arrows). Similar findings were present in the left eye.
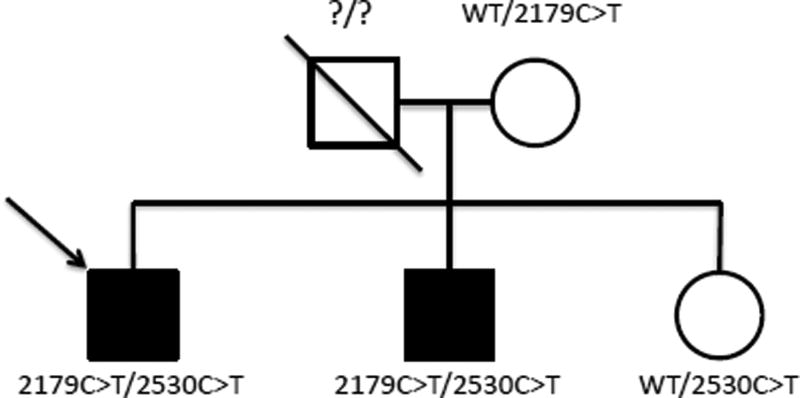
The proband had not demonstrated a previously reported RP gene mutation in genetic testing using the GeneDx RP panel (Cat# 368, GeneDx, Gaithersburg, MD, USA). Based on pedigree analysis, the predicted mode of inheritance was determined to most likely be autosomal recessive (Figure 2). Testing of X-linked RP genes was also performed as an X-linked pattern of inheritance could not be ruled out and given the significance of family planning for carriers (Cat# 326, GeneDx, Gaithersburg, MD, USA). No X-linked RP mutations were identified.
Fig 2.
Family Pedigree: The arrow identifies the proband. Shaded individuals carried established diagnoses of retinitis pigmentosa. The proband’s mother and sister were unaffected, though the mother had age-related macular degeneration. Carried MERTK variants are shown next to each individual (WT = wild type).
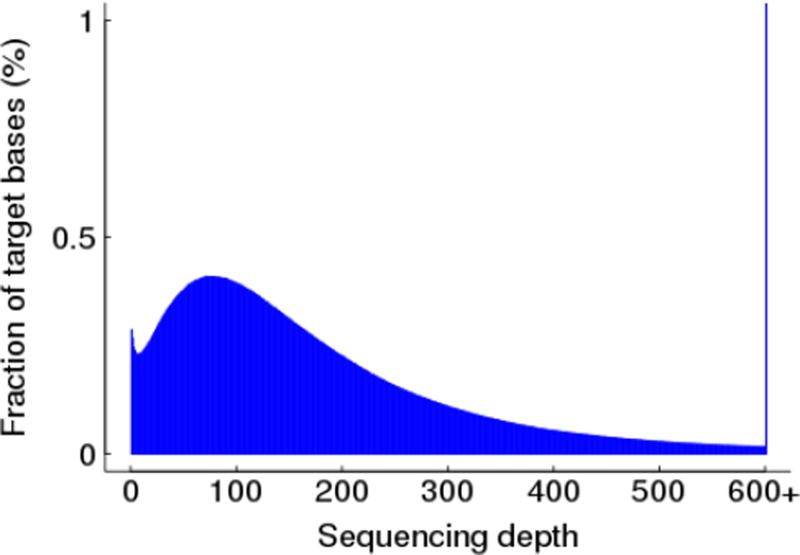
Whole-exome sequencing of the proband revealed 19,756 SNP variants (Figure 3). Of these, 265 SNPs were not reported in both the 1000 Genomes and dbSNP databases. Next, the candidate mutation pool was further narrowed by identifying known RP-associated genes. Eight of the 265 novel variants were located in genes previously associated with RP: 2 in MERTK4 in RP1L11 in CNGB3, and 1 in PRPF31 (Table 1). The 2 mutations in MERTK, a nonsense variant resulting in a premature stop (NM_006343.2:p.Arg727*/c.2179C>T) and a missense variant resulting in an arginine to cysteine change (NM_006343.2:p.Arg844Cys/c.2530C>T) were both heterozygous. The missense MERTK mutation was predicted to be damaging by SIFT, PolyPhen2, MutationAssessor, and RadialSVM. The nonsense mutation was analyzed by snpEff and found to likely cause loss of function and to be targeted by nonsense-mediated decay (SIFT, PolyPhen2, MutationAssessor, and RadialSVM are not well suited to predict pathogenicity in nonsense mutations).
Fig 3.
Whole-exome Sequencing Coverage: 97.3% of targets were covered with at least 10X coverage. 20X depth of coverage or more was achieved for 94.82% of targets. Average sequencing depth was 192X.
Table 1. Eight Candidate Gene Mutations.
Eight novel variants were identified in known RP-associated genes. Predicted pathogenicity was determined using four predictive tools (SIFT, PolyPhen2, MuationAssessor, and RadialSVM) for missense mutations. The nonsense mutation in MERTK was evaluated using snpEff.
| Gene | Mutation Site | Mutation Type |
SIFT Score | PolyPhen2 Score |
Mutation Assessor Score |
Radial SVM Score |
|---|---|---|---|---|---|---|
| MERTK | Arg727*/2179C>T | Nonsense | Above predictive tools are not well-suited to evaluate nonsense mutations | |||
| Arg844Cys/2530C>T | Missense | 0 (Damaging) | 1 (Probably Damaging) | 4.695 (High Functional Impact) | 0.8992 (Damaging) | |
| RP1L1 | Glu1379Ala/4136A>C | Missense | 0.04 (Damaging) | 0.636 (Possibly Damaging) | 0.345 (Neutral) | −0.9360 (Tolerated) |
| Glu1379Lys/4135G>A | Missense | 0.04 (Damaging) | 0.636 (Possibly Damaging) | 0.345 (Neutral) | −0.9613 (Tolerated) | |
| Gly1363Ala/4088G>C | Missense | 0.69 (Tolerated) | 0.007 (Benign) | 0 (Neutral) | −0.9248 (Tolerated) | |
| Gly1363Arg/4087G>A | Missense | 0.46 (Tolerated) | 0.075 (Benign) | 0 (Neutral) | −0.9015 (Tolerated) | |
| CNGB3 | Arg26Gln/77G>A | Missense | 0.54 (Tolerated) | 0.0 (Benign) | 0 (Neutral) | −0.9924 (Tolerated) |
| PRPF31 | Gly436Asp/1307G>A | Missense | 0 (Damaging) | 1.0 (Probably Damaging) | 2.85 (Medium Functional Impact) | 0.0704 (Damaging) |
The proband was also heterozygous for a missense mutation in CNGB3 (NM_019098.4:p.Arg26Gln/c.77G>A) and another in PRPF31 (NM_015629.3:p.Gly436Asp/c.1307G>A). The CNGB3 and PRPF31 mutations were unlikely candidates given that a single heterozygous mutation did not conform to the expected autosomal recessive disease model. Additionally, the CNGB3 mutation was not predicted to be pathogenic. The remaining four RP1L1 variants were all missense mutations, however, two of the RP1L1 mutations were not predicted to be damaging (NM_178857.5:p.Gly1363Arg/c.4087G>A and NM_178857.5:p.Gly1363Ala/c.4088G>C) and thus excluded from consideration as candidate mutations. The remaining two RP1LI mutations (NM_178857.5:p.Glu1379Ala/c.4136A>C and NM_178857.5:p.Glu1379Lys/c.4135G>A) were only predicted to be damaging by SIFT. Though the proband could have been a compound heterozygote for these latter RP1L1 mutations, given their lower predicted probability of pathogenicity, the MERTK mutations were investigated first.
Based on the predicted pathogenicity of the mutations as well as the expected mode of inheritance, the two mutations in MERTK were selected as the primary candidate mutations to further investigate. The proband was predicted to be a compound heterozygote with an independent mutation in each MERTK allele based on the pedigree. The ExAC database, which contains exome sequencing data from more than 60,000 individuals, was surveyed for previous reports of either mutation. The nonsense mutation that was identified in MERTK had not previously been reported. The MERTK missense mutation had been documented previously in just two European individuals in the ExAC database (of unknown phenotype) making it exceedingly rare. It has been previously reported in cases of RP in the scientific literature [22]. Neither variant was found in a search of the NHLBI database, which contains exome-sequencing data from 6,503 individuals recruited for common disease studies.
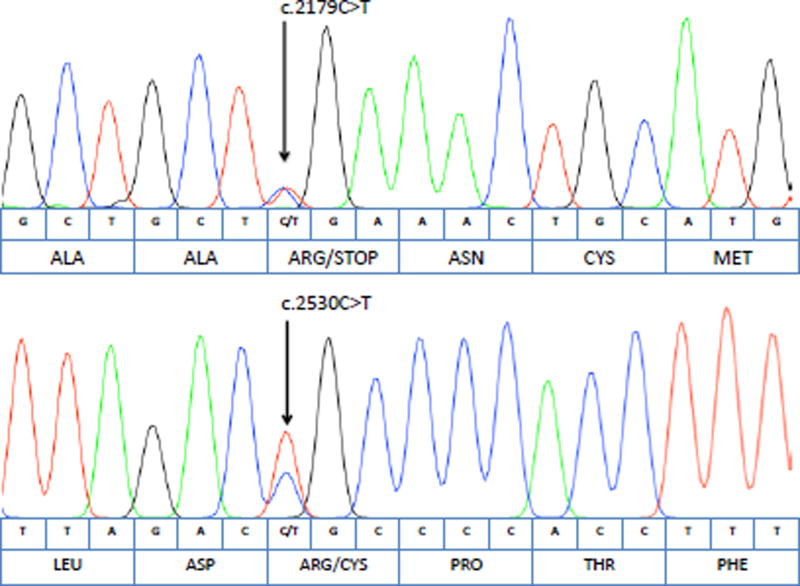
To establish phase and demonstrate that the MERTK mutations were on separate alleles as would be expected in a compound heterozygous model of inheritance, the proband’s unaffected mother, unaffected sister, and affected brother’s MERTK genes were sequenced. The proband’s affected brother was also found to be heterozygous at each mutation site as expected in a compound heterozygous model (Figure 4). Meanwhile, the unaffected mother carried the nonsense MERTK mutation but was wild type at the missense site. The unaffected sister carried a copy of the missense MERTK mutation and was wild type at the nonsense site. The sequencing results from the unaffected mother and sister suggest that the MERTK mutations are located on separate alleles, supporting the predicted compound heterozygous model of inheritance.
Fig 4.
Chromatograms of the Proband’s Affected Brother: Sanger sequencing revealed that the proband’s affected brother carried the same two heterozygous MERTK mutations as the proband. The nucleotide sequence and corresponding amino acid sequences are shown below each chromatogram.
Discussion
In this study, we present a family with retinitis pigmentosa with an autosomal recessive pattern of inheritance and no prior molecular diagnosis. To determine the likely causative gene mutation, whole-exome sequencing was performed on the proband. Among RP-associated genes, mutations were identified through whole-exome sequencing in MERTK, RP1L1, CNGB3, and PRPF31. Ultimately, the two mutations in MERTK, a novel nonsense mutation and a missense mutation, were selected as the primary candidate mutations given their predicted pathogenicity as well as their adherence to the observed inheritance pattern.
MERTK has previously been associated with RP. The initial work uncovering the role of MERTK mutations in the pathogenesis of RP evolved from the Royal College of Surgeons’s (RCS) inherited retinal degeneration rat line, which was the first animal model with inherited retinal degeneration[23]. Subsequent studies uncovered the causative mutation, a 1.5kb deletion in MERTK, in the RCS rat [24, 25]. Molecular studies demonstrated that while the RPE was able to bind to photoreceptor outer segments, it could not successfully engulf them[26, 27]. The outer segment debris accumulated in the subretinal space [28]. After these initial studies, multiple MERTK mutations were documented in patients with RP. These patients’ disease progression is thought to be more severe than typical RP cases, with early onset in the first decade of life and quickly worsening macular atrophy as seen in our family[15].
The present study also further demonstrates the clinical utility of targeted exome-sequencing in patients presenting without diagnoses on initial genetic panel testing. While many genetic panels for RP do include MERTK, the commercial panels used in the initial diagnosis of this patient did not (GeneDx). Targeted gene capture panels are advantageous in that they are cheaper and produce results more rapidly than WES. Therefore, targeted genetic panels are an appropriate first-line molecular diagnostic tool. However, if initial screening does not yield a molecular diagnosis, especially in genetically heterogeneous diseases like RP, WES should be considered. WES can lead to higher diagnostic yields and the discovery of novel gene-disease associations that would otherwise be missed on targeted panels at an increasingly cost-effective rate[29]. Uncovering such novel gene mutations furthers our understanding of Mendelian diseases and offers novel therapeutic targets. Understanding the role of these mutations in the function of the retina will aid in therapy design. Furthermore, as targeted gene therapy becomes available, the identification of specific genetic causes of retinal dystrophy will be critical to matching patients with appropriate therapies.
Acknowledgments
The authors extend our deep gratitude to the proband and his family. We also thank Vaiva Liakaite at the University of Illinois at Chicago’s DNA Services Facility and the sequencing center at BGI for assisting with the present study. Lastly, the authors thank Darshana Patel for assisting with the coordination of this study.
Funding:
This report was supported by funding from Search for Vision, R01EY023644, NEI core grant EY001792, and Research to Prevent Blindness (departmental support). The sponsor had no role in the design or conduct of this research.
Footnotes
Conflict of Interest:
All authors certify that they have no affiliations with or involvement in any organization or entity with any financial interest (such as honoraria; educational grants; participation in speakers' bureaus; membership, employment, consultancies, stock ownership, or other equity interest; and expert testimony or patent-licensing arrangements), or non-financial interest (such as personal or professional relationships, affiliations, knowledge or beliefs) in the subject matter or materials discussed in this manuscript.
Ethical Approval:
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent:
Informed consent was obtained from all individual participants included in the study.
References
- 1.Evans RJ, Schwarz N, Nagel-Wolfrum K, Wolfrum U, Hardcastle AJ, Cheetham ME. The retinitis pigmentosa protein RP2 links pericentriolar vesicle transport between the Golgi and the primary cilium. Hum Mol Gen. 2010;19:1358–1367. doi: 10.1093/hmg/ddq012. [DOI] [PubMed] [Google Scholar]
- 2.Busskamp V, Duebel J, Balya D, et al. Genetic reactivation of cone photoreceptors restores visual responses in retinitis pigmentosa. Science. 2010;329:413–417. doi: 10.1126/science.1190897. [DOI] [PubMed] [Google Scholar]
- 3.Hartong DT, Berson EL, Dryja TP. Retinitis pigmentosa. The Lancet. 2006;368:1795–1809. doi: 10.1016/S0140-6736(06)69740-7. [DOI] [PubMed] [Google Scholar]
- 4.Farrar G, Kenna PF, Humphries P. New EMBO member's review: On the genetics of retinitis pigmentosa and on mutation-independent approaches to therapeutic intervention. EMBO J. 2002;21:857–864. doi: 10.1093/emboj/21.5.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferrari S, Di Iorio E, Barbaro V, Ponzin D, Sorrentino FS, Parmeggiani F. Retinitis pigmentosa: genes and disease mechanisms. Curr Genomics. 2011;12:238–249. doi: 10.2174/138920211795860107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fernandez-San Jose P, Blanco-Kelly F, Corton M, Trujillo-Tiebas M-J, Gimenez A, Avila-Fernandez A, Garcia-Sandoval B, Lopez-Molina M-I, Hernan I, Carballo M, Riveiro-Alvarez R, Ayuso C. Prevalence of rhodopsin mutations in autosomal dominant retinitis pigmentosa in spain: clinical and analytical review in 200 families. Acta Ophthalmol. 2015;93:e38–e44. doi: 10.1111/aos.12486. [DOI] [PubMed] [Google Scholar]
- 7.Daiger SP, Sullivan LS, Bowne SJ. Genes and mutations causing retinitis pigmentosa. Clin Genet. 2013;84 doi: 10.1111/cge.12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang NH-H, Chen S-J, Yang C-F, Chen H-W, Chuang H-P, Lu Y-H, Chen C-H, Wu J-Y, Niu D-M, Chen Y-T. Homozygosity mapping and whole-genome sequencing links a missense mutation in POMGNT1 to autosomal recessive retinitis pigmentosa missense mutation in POMGNT1 and autosomal recessive RP. Invest Ophthalmol Vis Sci. 2016;57:3601–3609. doi: 10.1167/iovs.16-19463. [DOI] [PubMed] [Google Scholar]
- 9.Anasagasti A, Irigoyen C, Barandika O, López de Munain A, Ruiz-Ederra J. Current mutation discovery approaches in Retinitis Pigmentosa. Vision Res. 2012;75:117–129. doi: 10.1016/j.visres.2012.09.012. [DOI] [PubMed] [Google Scholar]
- 10.Boye SE, Boye SL, Lewin AS, Hauswirth WW. A comprehensive review of retinal gene therapy. Mol Ther. 2013;21:509–519. doi: 10.1038/mt.2012.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsou W-I, Nguyen K-QN, Calarese DA, Garforth SJ, Antes AL, Smirnov SV, Almo SC, Birge RB, Kotenko SV. Receptor tyrosine kinases, TYRO3, AXL, and MER, demonstrate distinct patterns and complex regulation of ligand-induced activation. J Biol Chem. 2014;289:25750–25763. doi: 10.1074/jbc.M114.569020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zagorska A, Traves PG, Lew ED, Dransfield I, Lemke G. Diversification of TAM receptor tyrosine kinase function. Nat Immunol. 2014;15:920–928. doi: 10.1038/ni.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen C, Li Q, Darrow AL, Wang Y, Derian CK, Yang J, de Garavilla L, Andrade-Gordon P, Damiano BP. Mer receptor tyrosine kinase signaling participates in platelet function. Arterioscler Thromb Vasc Biol. 2004;24:1118–1123. doi: 10.1161/01.ATV.0000130662.30537.08. [DOI] [PubMed] [Google Scholar]
- 14.Linger RMA, Keating AK, Earp HS, Graham DK. TAM receptor tyrosine kinases: biologic functions, signaling, and potential therapeutic targeting in human cancer. Adv Cancer Res. 2008;100:35–83. doi: 10.1016/S0065-230X(08)00002-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parinot C, Nandrot FE. A Comprehensive Review of Mutations in the MERTK Proto-Oncogene. In: Bowes Rickman C, LaVail MM, Anderson ER, Grimm C, Hollyfield J, Ash J, editors. Retinal Degenerative Diseases: Mechanisms and Experimental Therapy. Springer International Publishing, Cham; 2016. pp. 259–265. [DOI] [PubMed] [Google Scholar]
- 16.Nandrot EF, Kim Y, Brodie SE, Huang X, Sheppard D, Finnemann SC. Loss of synchronized retinal phagocytosis and age-related blindness in mice lacking αvβ5 Integrin. J Exp Med. 2004;200:1539–1545. doi: 10.1084/jem.20041447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nandrot EF, Anand M, Almeida D, Atabai K, Sheppard D, Finnemann SC. Essential role for MFG-E8 as ligand for αvβ5 integrin in diurnal retinal phagocytosis. Proc Natl Acad Scie U.S.A. 2007;104:12005–12010. doi: 10.1073/pnas.0704756104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qin S. Blockade of MerTK Activation by AMPK Inhibits RPE Cell Phagocytosis. In: Bowes Rickman C, LaVail MM, Anderson ER, Grimm C, Hollyfield J, Ash J, editors. Retinal Degenerative Diseases: Mechanisms and Experimental Therapy. Springer International Publishing, Cham; 2016. pp. 773–778. [DOI] [PubMed] [Google Scholar]
- 19.International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 20.Carnevali P, Baccash J, Halpern AL, Nazarenko I, Nilsen GB, Pant KP, Ebert JC, Brownley A, Morenzoni M, Karpinchyk V, Martin B, Ballinger DG, Drmanac R. Computational techniques for human genome resequencing using mated gapped reads. J Comput Biol. 2012;19:279–292. doi: 10.1089/cmb.2011.0201. [DOI] [PubMed] [Google Scholar]
- 21.Drmanac R, Sparks AB, Callow MJ, et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science. 2010;327:78–81. doi: 10.1126/science.1181498. [DOI] [PubMed] [Google Scholar]
- 22.McHenry CL, Liu Y, Feng W, Nair AR, Feathers KL, Ding X, Gal A, Vollrath D, Sieving PA, Thompson DA. MERTK Arginine-844-Cysteine in a patient with severe rod–cone dystrophy: loss of mutant protein function in transfected cells. Invest Ophthalmol Vis Sci. 2004;45:1456–1463. doi: 10.1167/iovs.03-0909. [DOI] [PubMed] [Google Scholar]
- 23.Bourne MC, Campbell DA, Tansley K. Hereditary degeneration of the rat retina. Br J Ophthalmol. 1938;22:613–623. doi: 10.1136/bjo.22.10.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.D’Cruz PM, Yasumura D, Weir J, Matthes MT, Abderrahim H, LaVail MM, Vollrath D. Mutation of the receptor tyrosine kinase gene Mertk in the retinal dystrophic RCS rat. Hum Mol Gen. 2000;9:645–651. doi: 10.1093/hmg/9.4.645. [DOI] [PubMed] [Google Scholar]
- 25.Nandrot E, Dufour EM, Provost AC, Péquignot MO, Bonnel S, Gogat Kn, Marchant D, Rouillac C, Sépulchre de Condé B, Bihoreau M-T, Shaver C, Dufier J-L, Marsac C, Lathrop M, Menasche M, Abitbol MM. Homozygous deletion in the coding sequence of the C-MER gene in RCS rats unravels general mechanisms of physiological cell adhesion and apoptosis. Neurobiol Dis. 2000;7:586–599. doi: 10.1006/nbdi.2000.0328. [DOI] [PubMed] [Google Scholar]
- 26.Bok D, Hall MO. The role of the pigment epithelium in the etiology of inherited retinal dystrophy in the rat. J Cell Biol. 1971;49:664–682. doi: 10.1083/jcb.49.3.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Finnemann SC, Nandrot EF. MERTK activation during rpe phagocytosis in vivo requires αvβ5 integrin. Adv Exp Med Biol. 2006;572:499–503. doi: 10.1007/0-387-32442-9_69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dowling JE, Sidman RL. Inherited retinal dystrophy in the rat. J Cell Biol. 1962;14:73–109. doi: 10.1083/jcb.14.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weisschuh N, Mayer AK, Strom TM, et al. Mutation detection in patients with retinal dystrophies using targeted next generation sequencing. PLoS ONE. 2016;11:e0145951. doi: 10.1371/journal.pone.0145951. [DOI] [PMC free article] [PubMed] [Google Scholar]