Abstract
Signal transducer and activator of transcription factor 3 (STAT-3) is known to be overexpressed in cancer stem cells. Poor solubility and variable drug absorption are linked to low bioavailability and decreased efficacy. Many of the drugs regulating STAT-3 expression lack aqueous solubility; hence hindering efficient bioavailability. A theranostics nanoplatform based on luminescent carbon particles decorated with cucurbit[6]uril is introduced for enhancing the solubility of niclosamide, a STAT-3 inhibitor. The host–guest chemistry between cucurbit[6]uril and niclosamide makes the delivery of the hydrophobic drug feasible while carbon nanoparticles enhance cellular internalization. Extensive physicochemical characterizations confirm successful synthesis. Subsequently, the host–guest chemistry of niclosamide and cucurbit[6]uril is studied experimentally and computationally. In vitro assessments in human breast cancer cells indicate approximately twofold enhancement in IC50 of drug. Fourier transform infrared and fluorescence imaging demonstrate efficient cellular internalization. Furthermore, the catalytic biodegradation of the nanoplatforms occur upon exposure to human myeloperoxidase in short time. In vivo studies on athymic mice with MCF-7 xenograft indicate the size of tumor in the treatment group is half of the controls after 40 d. Immunohistochemistry corroborates the downregulation of STAT-3 phosphorylation. Overall, the host–guest chemistry on nanocarbon acts as a novel arsenal for STAT-3 inhibition.
1. Introduction
Breast cancer is the most prevalent malignancy among women worldwide. In the United States alone, it is projected that 249 260 new cases of breast cancer will be diagnosed among both genders in 2016 with predicted 40 890 people succumbing to the disease.[1] Common methods of therapy such as primary tumor excision and chemotherapy are partially successful as they preferentially attack mature epithelial cells whilst sparing the small population of self-renewing, tumorigenic cancer stem cells (CSCs), which show resistance to treatment and cause tumor recurrence.[2] CSCs are resilient to anti neoplastic therapy through various defense mechanisms, including the activation of self-renewal pathways such as JAK/STAT, NOTCH, MAP-Kinase/ERK, and PI3K/AKT.[3] Instead of generalized chemotherapy, over the past years there has been a paradigm shift toward novel cancer treatments which specifically target cellular pathways for sustained and complete inhibition of cancer.[4,5] Janus-Kinase (JAK)/signal transducer and activator of transcription (STAT) is one of the common cytokine signaling pathways.[6] Of particular interest in the STAT protein family, STAT-3 has prominent role in regulating cellular functions.[7] Despite its essential role, the anomalous activity of STAT-3 protein has been observed in several cancers.[8] STAT-3 has been linked to CSC expansion, survival, and maintenance in glioblastoma, bone sarcomas, and breast cancer.[5,9] However, this pathway is considered to be “druggable” target at various stages of signaling and thus it is promising to engineer therapeutic approaches that specifically inhibit STAT-3 for comprehensively combating cancer.[6]
Repurposing a food and drug administration (FDA)-approved drug is exceptionally attractive in reducing time and cost burdens associated with the invention of a new formulation and its translation from bench to bedside.[10] Niclosamide (Nic) is an FDA-approved anthelminthic drug that has been acknowledged to demonstrate anti-neoplastic properties via the active inhibition of multiple pathways modulated in CSCs.[11,12] The potent inhibitory effect of Nic toward the STAT-3 pathway has been recently attributed to the disruption of transcriptional activities due to phosphorylation abrogation and hindering of nuclear translocation.[12,13] While the proven efficacy and biological understanding provide enthusiasm, the limited aqueous solubility of Nic (5–8 mg L−1 at 20 °C) sufficiently limits its bioavailability.[14] Therefore, the development of new delivery strategies is highly prized. The poor aqueous solubility of many drugs precludes their use for cancer therapy. Effective drug candidates are typically lipophilic as they exert their pharmacological action at or in biological membranes and/or membrane-linked proteins. Oral bioavailability of these lipophilic molecules often becomes insufficient and highly variable, and leads to the application of undesired surface-active excipients for stabilization. Hence, the development of carrier systems for the optimal oral and parenteral administration of these poorly soluble agents is of tremendous practical and broad interest which can be achieved by improving of the solubility of these agents.
Seeking to develop a generalizable approach to enhance the solubility of the hydrophobic drugs, such as Nic, in this work we propose a direct associating system based on specific molecular recognition of a guest by its corresponding host molecule presented at the nanoscale. Molecular containers such as dendrimers, cyclodextrins, and calixarene have been already utilized to enhance the solubility of Nic with some degree of success.[15] Cucurbit[n]urils (n = 5, 6, 7, 8, 10), a class of organic capsules, have shown utility as drug delivery vehicles. As a member of this family, CB6 possesses a hydrophobic cavity similar to α-CD; however, unlike its CD analogue, it has the symmetrical hydrophilic portals lined ureido carbonyl rims which enable the encapsulation of neutral molecules and positively charged species through ion–dipole, hydrophobic–hydrophobic, and hydrogen bonding interactions. The presence of two linkers holding subunits together endows the structure with improved rigidity, high degree of preorganization and superior binding selectivity.[16] In addition, the investigation of the biological response of cucurbituril (CB) in vivo and in vitro, revealed no cytotoxicity or acute toxicity in mice.[17] These merits make CB6 a favorable candidate for the next generation drug delivery systems. We envision that a direct, reversible, and noncovalent interaction will enhance the solubility of Nic, enabling complex formation between CB and the drug molecule.
In the past few years, extensive research has been directed to exploit the luminescence properties of carbon nanoparticles (CNPs) for in vitro and in vivo fluorescence, Raman, and photoacoustic imaging.[18] CNPs can also serve as a vehicle for controlled drug and gene delivery due to their high surface area to volume ratio, excellent biocompatibility, small size for being taken up by solid tumors and most importantly, being rich in surface functional moieties appropriate for the incorporation of various compounds as homing agents.[19] These features integrated with their remarkable optical properties can put forth the concept of “find, fight, and follow” in a single platform.[20]
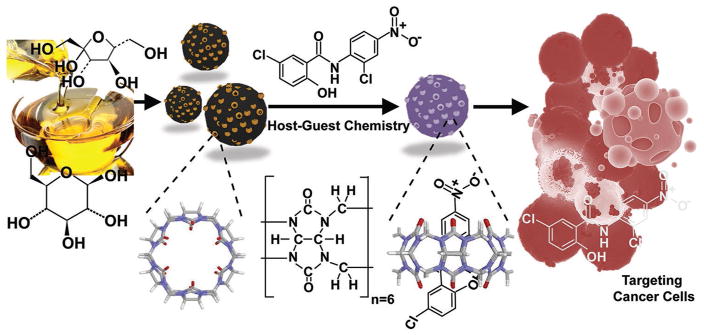
We hypothesize that a defined host–guest chemistry would enhance the solubility of a candidate hydrophobic drug while nano-enabled delivery through carbon nanoparticles would result in facilitated cellular internalization owing to the pronounced surface effect. Moreover, the utilization of multiple signature functionalities and unique optical luminescent properties of carbon nanoparticles would make the delivery of the cargo trackable by vibrational and fluorescence imaging. Furthermore, we demonstrated the feasibility of engineered nanoparticles for in vitro and in vivo drug delivery toward effective control of a xenograft breast cancer model through intratumoral (IT) administration route. Hence, this highly versatile multifunctional host–guest nano-system is a stride toward synchronous imaging, enhanced target specific therapy, and expedited cellular uptake which can be exploited as an effective strategy against STAT-3 modulation in tumor bearing mice (Figure 1). To the best of our knowledge, a molecular guest–host chemistry at the nanoscale has not been explored in the past to enhance the solubility of a poorly soluble therapeutic agent.
Figure 1.
Schematic representation of preparation, loading mechanism, and treatment of cancer cells. The CNP coated with CB6 was loaded with STAT-3 inhibitor (niclosamide) to effectively treat cancer cells via apoptosis.
2. Results and Discussion
2.1. Host–Guest Interaction Studies
The host–guest interaction of CB and Nic was first studied analytically and computationally to pave the path for applications. A classical molecular dynamics (MD) simulation using ReaxFF was performed to determine which CB6 or CB7 variants would result in lower formation energy and in turn higher stability. Contrasting with previous numerical simulations on host–guest interactions, which employed nonreactive force fields such as AMBER,[21] CHARMM,[22] and GROMOS,[23] ReaxFF is capable of capturing bond formation and bond dissociation within molecular statics/dynamics (MS/D) simulations. ReaxFF is able to describe a smooth transition between the nonbonded states and single, double or triple bonded systems by employing a bond order/distance based empirical formalism in a highly transferable fashion.[24] The total interaction energy in the ReaxFF potential can be divided into several energy contributions given by Equation (1) as follows
| (1) |
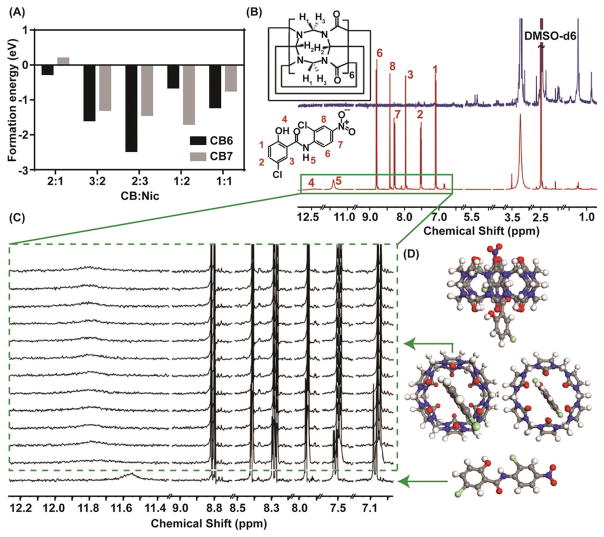
The energy terms in Equation (1) comprise the bond energy (Ebond), valance-angle (Eval), torsion-angle energy (Etor), lone pair (Elp), over-coordinate (Eover), and under coordinate (Eunder) energy penalty terms as well as nonbonded interactions, i.e., van der Waals (Evdw) and coulombic energy (Ecoulombic). All the connectivity dependent energy terms (e.g., valance and torsion-angles) are bond- order dependent and are updated at every MD or energy minimization step. Thus, the energy and forces pertinent to these components approach zero upon bond-dissociation. Furthermore, nonbonded interactions such as van der Waals and coulombic terms were calculated for the entire system between each pair of atoms. All the simulations in this study were performed using ReaxFF parameters set as it was implemented in large-scale atomic/molecular massively parallel simulator (LAMMPS).[25] The implementation of ReaxFF in LAMMPS consisted of a charge equilibration (QEq) approach[26] to account for charge distribution based on the electronegativity and geometry changes of atoms. Another advantage of using ReaxFF compared to nonreactive force fields is the capability to model direct interactions between ionic residues (H+ and Cl−). According to the binding enthalpies reported by Figure 2A, it is immediately noticeable that except the case of CB:Nic = 2:1, the formation of Nic guest is enthalpically more favored when it is inserted into the circular structures of CB6 compared to CB7. Higher energies and thus less stability of 2:1 case might be attributed to the poorer steric fit of Nic into the CB6 host compared with the same ratio in CB7. Thus, the simulation results were utilized to conclude that CB6 would offer a better candidate for the delivery of Nic.
Figure 2.
A) The formation energy comparison obtained from computational studies revealed that in most of the given ratios, CB6 yields more stable complex compared to CB7; hence, justifying the application of CB6 for the experiments. B) 1H NMR titration studies to determine the feasibility of the interaction between host and guest. The Red graph is the 1H NMR spectrum of niclosamide and the navy blue one is the CB6 spectrum. All the measurements were done in DMSO- d6. C) The magnified box shows the region of interest in the titration studies. The green box indicates how the peak positions shift upon the addition of CB6 while maintaining niclosamide concentration. D) The ball and stick graphs indicate the energy minimized configuration obtained from ReaxFF simulation seen from various directions. The gray, white, blue, red, green correspond to C, H, N, O and Cl atoms, respectively.
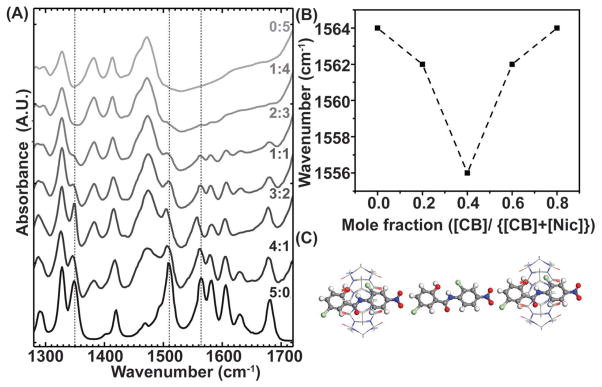
Furthermore, the host (H)–guest (G) complexation of CB6 and Nic was experimentally verified by titration studies utilizing multiple spectroscopic techniques, including UV–vis, fluorescence and 1H NMR. Titration studies with UV–vis and fluorescence spectroscopy were hindered in the aqueous media due to the strong scattering from CB6 (Figure S1, Supporting Information), whereas 1H NMR spectroscopy yielded accurate results.[27] In a typical titration investigation, the concentration of Nic was kept constant while the concentration of CB6 was gradually varied. An expected outcome of the host–guest chemistry would be a change in the magnetic field experienced by guest protons located within the host cavity. Thus, changes in chemical shift of the drug resonances, together with changes in line width, provide information on the location and the orientation of the bound drug to CB6. The assigned peaks of Nic in the 1H NMR spectrum (Figure 2B,C) demonstrated pronounced splitting and broadening after the addition of 0.028 × 10−3 M of CB6. This is likely due to the presence of mixed magnetic species of Nic when complexed within CB6 cavity. In other words, the species interacting with CB6 experience a different magnetic environment as a result of interaction while the uncomplexed species remain unaffected at lower concentrations. This trend virtually disappeared upon further addition of Nic, suggesting the magnetic homogenization of the species. The titration revealed that the amide peak moved downfield which is not simply justified based on the shielding/deshielding effect. One explanation for this observation might be the keto-enol tautomerisation where proton from the N–H can move to carbonyl unit, generating more electronegative OH. Migration of this proton to the ketonic moiety reduces the strength of the amide II mode, resulting in its red-shift of its vibrational frequency as shown in Figure 3A. Therefore, the corresponding peak of N–H disappears and originates an O–H peak on slightly downfield region thus leading to Nic stabilization upon encapsulation. Protons # 1, 2, 3, 7, and 8 were all shifted upfield, suggesting encapsulation by CB6, whereas proton #6 shifted downfield, likely due to being excluded from CB6 cavity. The peak at 3.33 ppm, which is attributed to water, was shifted downfield (Figure S3, Supporting Information). Since water is released from the CB cavity upon complexation, the corresponding peak moved downfield to the deshielded region. In fact, the first guest molecule that was proven to be encapsulated in hydrophobic CB6 cavity was water.[28] Our suggested molecular orientation was confirmed by atomistic simulation through the energy minimized studies. The ball and stick model of the energy minimized configuration, as predicted in ReaxFF simulation, is shown in Figure 2D. Consistent with the experimentally observed results ReaxFF simulations demonstrate the amide encapsulation is within the CB6 cavity. To determine the stoichiometry of host–guest chemistry, the method of continuous variation was utilized where the molar fraction of one component was varied between zero and one and a parameter dependent on the concentration of [HG] complex was plotted versus mole fraction. Since wavenumber is directly proportional to the energy in IR spectra (Figure 3A), the minimum of the Job’s plot was used to determine reaction stoichiometry (Figure 3B). While most of the vibrational modes corresponding to both Nic and CB6 remained constant (Figure 3A), the amide II modes (C–N) shifted to lower energies at a 2:3 concentration ratio of CB6 to Nic. We hypothesized that this is due to the favorable interaction between the Nic molecules inside the cavity of CB6. To ascertain these interactions, ReaxFF calculations were carried out with four possible binding scenarios (Figure S4, Supporting Information). Out of the these cases, the ratio of CB6:Nic 2:3 (Figure 3C) yielded the lowest energy where the negative binding energy (Gibbs free energy) signifies that the insertion of the Nic molecule into the host molecule is energetically favorable. This observation is also compatible with the ratio obtained for CB6 as has been already presented in Figure 2A. The formation energy values obtained from each scenario are summarized in Table 1.
Figure 3.
The stoichiometry of the interaction was concluded from both experimental and in silico studies. A) The FT-IR spectra showing decreasing level of niclosamide: CB6 ratio from bottom to the top. B) The derived Job’s plot obtained from Amide I peak. The minimum of the graph corresponds to the most energetically favorable configuration. C) 2:3 configuration after energy minimization.
Table 1.
The possible scenarios of binding and the corresponding host-guest complex formation energy at T = 0 K obtained from ReaxFF simulations normalized per CB6. The lowest formation energy is the most favorable state.
| #Nic | #CB6 | KCal mol−1 | eV/#CB6 | |||
|---|---|---|---|---|---|---|
|
|
|
|||||
| N | M | E(mCB6 + nNic) | E(CB6) | E(Nic) | Ef(mCB6 + nNic) | Ef(mCB6 + nNic) |
| 1 | 2 | −27782.195 | −11984.983 | −3799.826 | −6.202 | −0.267 |
| 2 | 2 | −31643.590 | −11984.983 | −3799.826 | −36.986 | −1.590 |
| 3 | 2 | −35484.391 | −11984.983 | −3799.826 | −57.473 | −2.471 |
| 2 | 1 | −19599.787 | −11984.983 | −3799.826 | −15.152 | −0.652 |
| 1 | 1 | −15813.103 | −11984.983 | −3799.826 | −28.294 | −1.217 |
| 0 | 1 | −11984.983 | −11984.983 | −3799.826 | – | – |
| 1 | 0 | −3799.826 | −11984.983 | −3799.826 | – | – |
2.2. Synthesis and Physicochemical Characterization of the Cucurbit[6]uril Passivated Carbon Nanoparticles
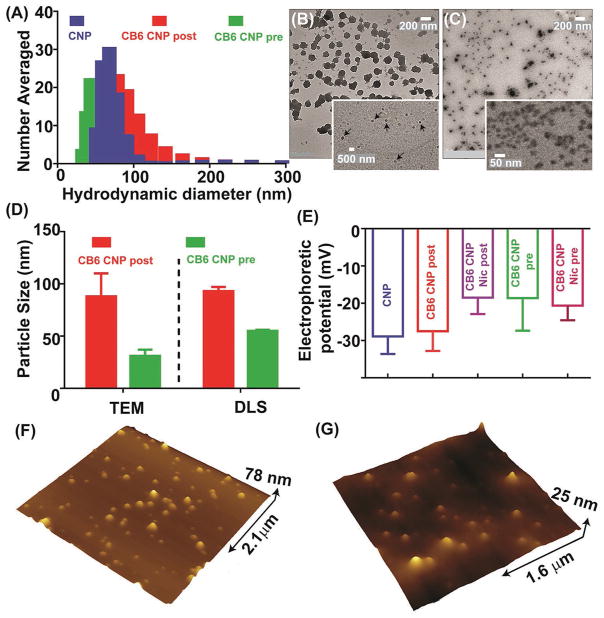
Translating nanoparticles for clinical use requires characterization of efficiency, biocompatibility, reproducibility, and real time monitoring of the drug release.[29] In this work, we adopted a facile green route of synthesis for the preparation of Nic loaded carbon nanoparticles. The synthesis involved the hydrothermal treatment of the food grade agave nectar which is a rich carbohydrate source (batch composition; 47%–56% fructose and 16%–20% glucose, rest is a mixture of other sugars and water) using a commercial hot plate followed by probe sonication, dialysis, and filtration. The introduction of CB6 on CNP surfaces was then achieved by either pre- or post-synthesis functionalization. Prefunctionalization was carried out in the presence of CB6 in one pot whereas postfunctionalization involved coating the preformed CNPs with CB6 via noncovalent interactions. Niclosamide was introduced to the system through a controlled host–guest interaction. Extensive physicochemical characterizations of the nanoparticles were subsequently carried out (Figure 4) to confirm the integrity of the structure, morphology and robustness of the system. The hydrodynamic size of the prepared carbon nanoparticles decorated with CB6 was initially optimized by making various dilutions of CB6 and a concentration of ×10−6 M was found to be most suitable for this purpose as it yielded the optimum hydrodynamic size (Figure S5, Supporting Information). Therefore, this concentration was adopted for the rest of the studies. The mean number-averaged hydrodynamic size for CNP as well as CB6 CNP postfunctionalized and prefunctionalized particles was determined to be 76 ± 13, 93 ± 04, and 55 ± 01 nm, respectively (Figure 4A). It is important to determine the hydrodynamic size of the nanoparticles for systemic applications as it directly influences their accumulation and clearance profile from a subject body.[30] The average anhydrous diameter was determined using transmission electron microscopy (TEM) images of 100 particles (Figure 4B,C and Figure S6, Supporting Information) to be 88 ± 22 and 31 ± 06 nm for post- and prefunctionalized samples, respectively. As shown in Figure 4D, the discrepancy between hydrodynamic and anhydrous diameter was marginal suggesting the semi-rigid nature of these particles. The smaller size of prefunctionalized particles compared to postfunctionalized ones could be attributed to the more controlled nucleation of CNPs in the presence of CB6 as a result of the nanoscale confinement of CB6 exerting on CNP growth. Nanoparticle agglomeration in high-ionic strength solutions such as biological media as a result of pronounced effect of particle functionalities is a known issue in biomedicine.[31] Therefore, we investigated the colloidal stability of nano-particles over an extended period to avoid potential issues such as blockage of blood capillaries in vivo, opsonization, systemic degradation, and elimination as foreign entities by the reticuloendothelial system (RES).[31] Shelf life of the nanoparticles was examined in various physiologically relevant media over the period of four weeks (Figure S7, Supporting Information). Hydrodynamic size measurements revealed that the nano-particles were remarkably stable, without any noticeable aggregation in aqueous media at neutral and pH of 4.5 at 37 °C. Nanoparticles incubated in aqueous media containing 10% FBS, a widely used serum in cell culture and biological fluids, also demonstrated a stable profile (Figure S7C, Supporting Information). The stability of particles in different conditions can also be correlated with the negative electrophoretic potential values which result in particle repulsion and prevent aggregation or clumping. The electrophoretic potential of the pristine CNP and CB6 CNP, in the absence and the presence of Nic, were determined by zeta potential measurements (Figure 4E). The negative zeta potential on the surface of CNP (−29 ± 05 mV) represented the abundance of carboxyl, carbonyl, and hydroxyl groups. In the presence of CB6, zeta potential values became marginally less negative both for postfunctionalized (−27 ± 05 mV) and prefunctionalized samples (−19 ± 09 mV) which is consistent with the surface charge on the outer surface of CB cavity carrying slightly positive charge.[32] The addition of Nic further contributed to the less negative zeta potential for drug-loaded postfunctionalized (−18 ± 5 mV) and prefunctionalized (−20 ± 4 mV) samples. This reduction is likely due to electrostatic interaction of CB6 and Nic and complexation, which masks negatively charged groups on Nic.
Figure 4.
Materials characterization tests on the nanoplatforms. A) Hydrodynamic size distribution of the pristine CNPs and pre- and postfunctionalized CB6 coated CNPs. Transmission electron images of B) post- and C) prefunctionalized CB6 coated CNPs. The arrows in (B) indicate CB6 coating. (scale bar on (B) and (C) =200 nm; inset (B) =500 nm and inset (C) =50 nm). D) Comparison between the hydrodynamic and anhydrous sizes of post- and prefunctionalized samples showing the smaller size of prefunctionalized compared to the postfunctionalized samples. E) Electrophoretic potential measurements. The negative zeta potential on the nanoparticles effectively prevented their agglomeration on the extended period. AFM height profiles of F) post- and G) prefunctionalized samples.
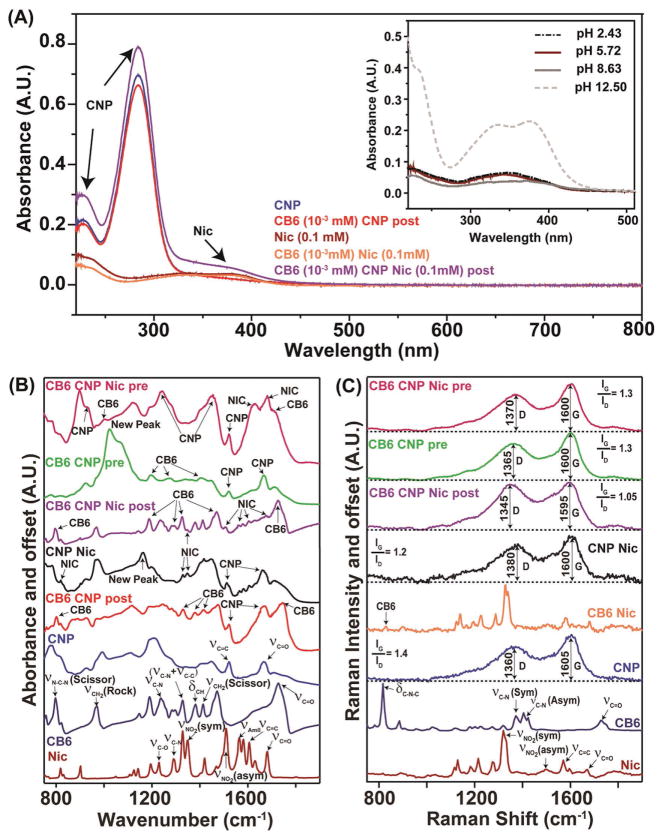
The nanoparticles’ morphology and size distribution are shown in Figure 4B,C. A bright halo around postfunctionalized samples indicates the formation of a uniform coating around the bare CNP (Figure 4B inset). The particles prepared in the presence of CB6 showed network formation into several clusters (Figure 4C inset), likely due to CB6 assisted self-assembly during dehydration.[33] Atomic force microscopy (AFM) measurements (Figure 4F, G) provided average heights of post- and prefunctionalized particles to be 25 ± 5 and 20 ± 6 nm, respectively. The UV–vis spectra of CNP, Nic, CB6 CNP (pre), and CB6 CNP (post) in the absence and the presence of drug are shown in Figure 5A. The absorption band for CNP was seen to be monotonically increasing and is devoid of any band gap transition that is typical of amorphous carbon substrates. The peak around 280 nm can be attributed to the n–π* transition of the C=O band and the π–π* transition of the conjugated C=C band.[34] CB6 did not show any UV–vis signature peak due to the lack of chromophoric units. Niclosamide, demonstrated a pH-dependent absorption spectrum as shown in Figure 5A (inset). Niclosamide was barely soluble in water at low pH but solubility was greatly enhanced by increasing the pH. Nic indicated four distinct peaks under basic pH conditions whereas in acidic or neutral conditions only two bands appeared (Figure 5A inset), which could be explained based on the resonance effects as well as the dehydration of phenolic –OH group to phenolate in aqueous base.[35] Importantly, a lack of aqueous solubility at physiologic pH (7.4) is evident. The introduction of CB6 diminished Nic characteristic peaks, confirming complexation (Figure 5A). CB6 CNP postfunctionalized samples followed the same trend. UV–vis studies on drug loaded particles demonstrate the characteristic absorption band of Nic at 377 nm in drug loaded CB6 CNP particles despite being largely masked otherwise by the CNP spectrum.
Figure 5.
Spectroscopic characterization of formulations. Panel A) demonstrates vis absorption efficiency associated with each of the constituents and their interaction. The continuous absorption profile of CNPs with characteristic broad peak starting in the UV region and extending to the visible range is discernable. Also, the Nic peak signature is readily detectable in the CB6 CNP Nic suggesting the presence of Nic. The inset investigates the effect of pH on the UV–vis spectrum of Nic from which the pattern of solubility of the drug can be concluded. B,C) Not only show the FT-IR and Raman spectra of the individual chemical components (Nic, CB6, and CNPs), but also the CNPs loaded with them using different synthetic protocols described in the text.
To identify functional groups and their molecular interactions, Fourier transform infrared (FT-IR) and Raman spectroscopy were utilized. Figure 5B,C shows the IR and Raman spectra of the constituents and all combinations of nanoparticles. Characteristic features of both Nic and CB6 could be identified in both types of spectra. For the IR characterization (Figure 5B), the νC=O (1680 cm−1), ν AmideI (1550 cm−1), νC=C, νNO2 (symmetric and asymmetric) stretch for the Nic molecule were used as the characteristic signatures. The characteristic band assignment of CB6 was made with bands at 1732 cm−1 C=O,[36] 3485 cm−1 N–H,[37] 1473 cm−1 C–N stretching vibration[37] and 2930 cm−1 –CH2 stretching vibration.[38] CNPs displayed a variety of IR peaks, including νC=O and C=C vibrations. Upon CB6 and/or Nic introduction on the CNPs surface, features corresponding to both Nic and CB6 can be identified, demonstrating successful incorporation of the molecules on CNPs. To compare the preparation routes of functionalized CNPs with CB6 and Nic, the IR spectra of the products from the pre- and postfunctionalization synthesis were investigated. While in the postfunctionalized samples, CB6 and the Nic peaks can be easily identified, the CNP features were less distinct. On the other hand, in the prefunctionalized samples, the CNP features overwhelm the vibrational peaks from Nic and CB6. While the vibrational features from each of the components (Nic and CB6) were distinct in the IR, intense Raman scattering from CNPs characterized by the D (sp3) and G (sp2) characteristic bands (Figure 5C) masked the contributions from the other decorating molecules. The extent and type of surface decoration through the ratio of the G and D bands showed a change with varying in the nature of the functionalization. The change could be attributed to the collective addition of all the molecular vibrational features in the 1200–1400 cm−1 region which affected the intensity ratio. From IR and Raman data, it seems that the postfunctionalized samples were more reproducibly synthesized and provided better functionalization control, hence they were adopted for the following experiments.
Analytical studies confirmed that under the physiologic condition the interaction between host (CB6) and guest (Nic) was simply of the physical type and no apparent evidence of chemical transformation of the drug itself was noticed. This result is conceptually expected since only noncovalent interaction between cucurbituril and the guest molecules are known to encounter followed by the release of the payload under specific circumstances.
2.3. In Vitro Cellular Studies
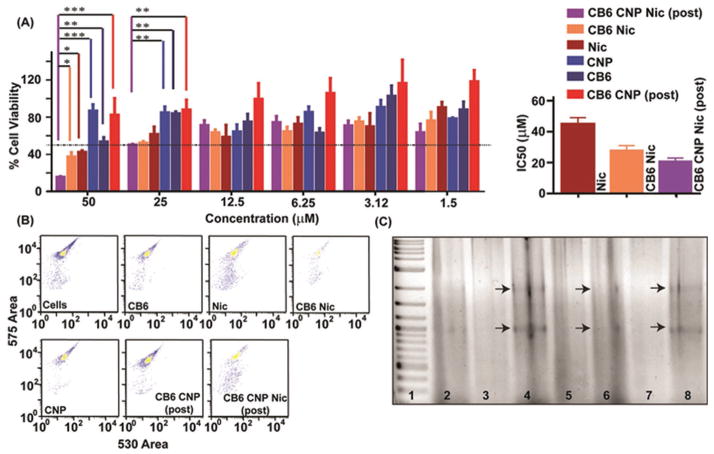
To evaluate the growth inhibitory effects of nanoparticles, MTT assay was performed on an estrogen receptor positive (ER(+)) human breast cancer cell line, MCF-7 (Figure 6A). Bright field imaging of treated MCF-7 cells revealed the loss of cell growth density and cellular morphology selectively in populations treated with Nic, CB6 Nic, CB6 CNP Nic postfunctionalized samples (Figure S9, Supporting Information). On the other hand, CB6, CNP, CB6 CNP (postfunctionalized) did not alter growth density or morphology significantly compared to control MCF-7 cells (Figure S9, Supporting Information). The MTT results showed that after 48h incubation, CNP, CB6, and CB6 CNP (postfunctionalized) did not induce any significant cytotoxic effect (Figure 6A) for a range of applied concentrations (1.5 × 10−6 to 50 × 10−6 M) which is a crucial factor as a drug delivery platform and biological applications. The half maximal inhibitory concentrations (IC50) of Nic, CB6 Nic, and CB6 CNP Nic (postfunctionalized) were determined to be (45 ± 04) × 10−6 M, (28 ± 03) × 10−6 M, and (21 ± 02) × 10−6 M, respectively (Figure 6A right panel). The effectiveness of Nic obtained in this study was found to be quite comparable to some of the previously reported literature. Bai et al.[39] reported the fabrication of micro and nanoparticles for the encapsulation of Nic using electrospraying. MTS assay conducted in CP70 and SKOV-3 could not achieve IC50 at least in SKOV-3 cells till concentration of 12 × 10−6 M. Dubey and Gopinath[40] developed an electrospun system based on PCL- PEO for the codelivery of Ag and Nic. The MTT assay revealed that IC50 for Nic was improved in A549 cells and MCF-7 cells merely by ≈1.2-fold although IC50 decreased from 1.45 × 10−6 to 1.21 × 10−6 M.
Figure 6.
A) Cell toxicity assay in MCF-7 cells and the corresponding IC50 values. A two-way ANOVA test with Bonferroni posttests was performed between % cell viability from treatment of each formulation to acquire significance level of *, **, and *** for p-values <0.05, 0.01, and 0.001, respectively. MTT assay was performed at Nic concentration ranging from 1.5 × 10−6 to 50 × 10−6 M in various formulation form including CB6 Nic, Nic, and CB6 CNP Nic (postfunctionalized) while CB6, CNP, and CB6 CNP were used as positive control. B) FACS pattern of PI stained MCF-7 cells after incubation with CB6, Nic, CB6 Nic, CNP, CB6 CNP, (post) and CB6 CNP Nic (post); C) Gel electrophoresis of extracted genomic DNA after incubating MCF-7 cells with Lane 2: Cells alone; Lane 3: CNP; Lane 4: Nic; lane 5: CB6; Lane 6: CB6 Nic; Lane 7: CB6 CNP and CNP CB6 Nic (post) Lane 8. Lane 1 represents ladder from 1 Kb DNA ladder.
The enhancement in the inhibitory effect of Nic delivered through host–guest interaction emphasizes the improved stability and suspendability of Nic in the presence of CB6 compared to the free drug. Furthermore, the nanoparticles’ decoration with CB6 further elevated the potency of the drug which can be attributed to the facile internalization and translocation inside the cell, sustained release of the drug and enhanced bioavailability of the drug due to its solubilization in CB6 CNP Nic sample.[41] By far, solubility is regarded as the most important parameter to achieve desired concentration of drug at the site of the action for intended pharmacological response. The inadequate dissolution rate of the drug is also the limiting factor in the bioavailability of poorly water soluble compounds. We have demonstrated through multiple studies that this defined host–guest chemistry helps to “enclose” the poorly soluble candidate drug, Nic, at the nanoscale of CNP, resulting in high potency as compared to the equimolar drug by itself. The enhanced solubility is clearly evident in spectroscopic titration studies with 1H NMR (Figure 2 and Figures S2 and S3, Supporting Information) and FT-IR (Figure 3). The entrapment of the multiple small molecular Nic to the CB6 moieties is influencing the down-field shift of water peak in 1H NMR titration in the presence of increasing CB6 concentration indicating the release of water molecule upon Nic encapsulation (Figure S2, Supporting Information). The gradual complexation of Nic inside the cavity of CB6 is observed in Figure 2 and Figure S2 of the Supporting Information indicating a marked increase in its solubility in the aqueous suspension of CB6 Nic. Furthermore, the dissolution study revealed a sustained release of the drug molecule from the nanoformulation over 5 d (Figure S8, Supporting Information), suggesting a highly solubilized drug-carrier system.
Mechanistic studies of cell regression after incubation with free Nic and its nanoformulation were performed to investigate genomic DNA degradation after probable induction of apoptosis. A turned on suicidal switch of apoptosis leads to the degradation of genomic DNA, decreasing the intercalation of propidium iodide (PI) and also generating DNA fragments.[42] Calculated cell population with fragmented genomic DNA with low PI intercalation were found ≈28% for the treatment of free Nic which increased to 37, 46% for CB6 Nic and CNP CB6 Nic while CNP and CB6 showed a nominal population of ≈5% and 11%, respectively (Figure 6B and Table 2). Gel electrophoresis of extracted genomic DNA after incubating MCF-7 cells was analyzed for all cases—Lane 2: Cells alone; Lane 3: CNP; Lane 4: Nic; lane 5: CB6; Lane 6: CB6 Nic; Lane 7: CB6 CNP and CB6 CNP Nic (postfunctionalized) Lane 8. Results demonstrate the generation of ladders only in cases of Nic in free (Lane 4) or form of host–guest complex (Lane 6) or nano-formulation (Lane 8), thus establishing induced apoptosis with genomic DNA fragmentation assays (Figure 6D). The biological properties of the free drug, complex of drug with CB6 and nanoparticles loaded drug are summarized in Table 2 for comparison.
Table 2.
In vitro biological assessment of the free drug, drug CB6 complex, and drug loaded nanoparticles for MCF-7 cell line. Here, CNP and CB6 were used as negative control.
| Formulation | IC50 [×10−6 M] | % Apoptotic cells |
|---|---|---|
| CNP | – | 5 |
| CB6 | – | 11 |
| Nic | 45 ± 4 | 28 |
| CB6 Nic | 28 ± 3 | 37 |
| CB6 CNP Nic (postfunctionalized) | 21 ± 2 | 46 |
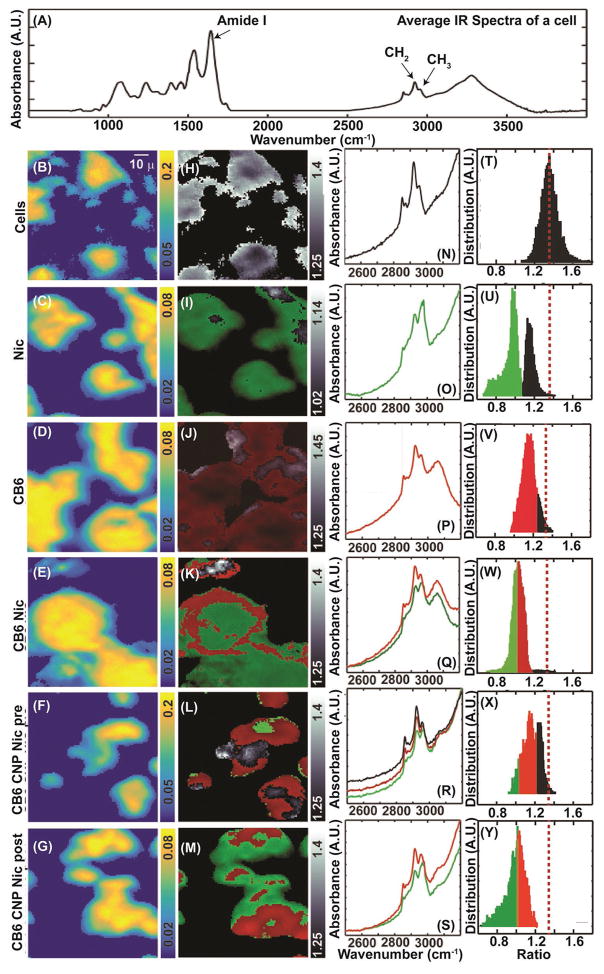
We examined the distribution of CNPs and their payload inside the cells using FT-IR, Raman and fluorescence imaging techniques. Although CNPs have no significant infrared signatures, the components of CB6 and Nic have prominent IR characteristics that can be measured. Figure 7A shows the average IR spectra of the untreated cells with a chemical image of a typical untreated MCF-7 cell line (Figure 7B). The intensity of each pixel in Figure 7B–G corresponds to the Amide I characteristic peak found as part of the cellular features. The neighboring black and white images indicate the C–H intensity ratio between the CH2 and the CH3 bands’ absorbance (Figure 7H–M), which is ≈1.34 for a typical cell (Figure 7N,T). Incubation of the cells with either free Nic or CB6 resulted in dramatic changes in the average IR spectra (Figure 7O,P). Niclosamide (shown as green) and CB6 (shown as red) changed the mean CH2:CH3 ratio to 0.94 and 1.15, respectively (Figure 7U,V). The exclusive internalization of the CB6 and Nic can be concluded from the intensity ratio map of these molecules in the cells. The histogram of the intensity distribution of Nic (Figure 7U) follows a bimodal distribution whereas for CB6 a normal distribution is seen (Figure 7V). For cells incubated with Nic, regions with a ratio less than 1.03 (one standard deviation away from the mean) were associated with the presence of Nic (spectra shown in green). A ratio between 1.03 and 1.23 (shown in red: within one standard deviation for CB6) was attributed to the presence of CB6. For the cells incubated with CB6 Nic, the characteristic vibrational features of both were observed (Figure 7E,K). While Nic appeared to spread throughout all cellular regions, CB6 accumulated mostly near the edges when using CB6 Nic formulation. When CB6 CNP Nic (post- and prefunctionalized) samples were used, the histogram of the absorbance ratio shows a broad distribution. Using the intensity ratios image, regions rich in Nic (<1.03), CB6 (between 1.03 and 1.22), and cellular matrix (>1.22) were identified. A representative image of cells incubated with CB6 CNP Nic prefunctionalized (Figure 7F,L) exhibits a relatively large unaffected cellular domain compared with the postfunctionalized sample (Figure 7G,M). Although the difference in surface loading for pre- and postfunctionalized samples cannot be ruled out, the above results can potentially lead to the conclusion that postfunctionalized samples can make a better candidate.
Figure 7.
Infrared imaging and intracellular analysis for presence of CB6 and Nic. A) The average infrared spectra of the cell. Panels B–G) show the amide I intensities from the cellular regions. A typical infrared absorption spectra in the C–H stretching region is shown in (N) and the intensity ratio (ICH2/ICH3) distribution centers at 1.23 is shown in (T). H) plots the intensity ratio in grayscale over the same region as shown in (B). Panels O–S) exhibit the C–H spectral region and the intensity ratio distribution corresponding to the conditions displayed in the extreme left. Intensity distributions below 0.8 (green) and 1.15 (red) were attributed to the presence of Nic and CB6, respectively. Panels I–M) plots the regions corresponding to (C–H) with the cellular regions (grayscale), Nic (green), and CB6 (red). Panels U–Y) indicate the distribution intensity of the probed signatures.
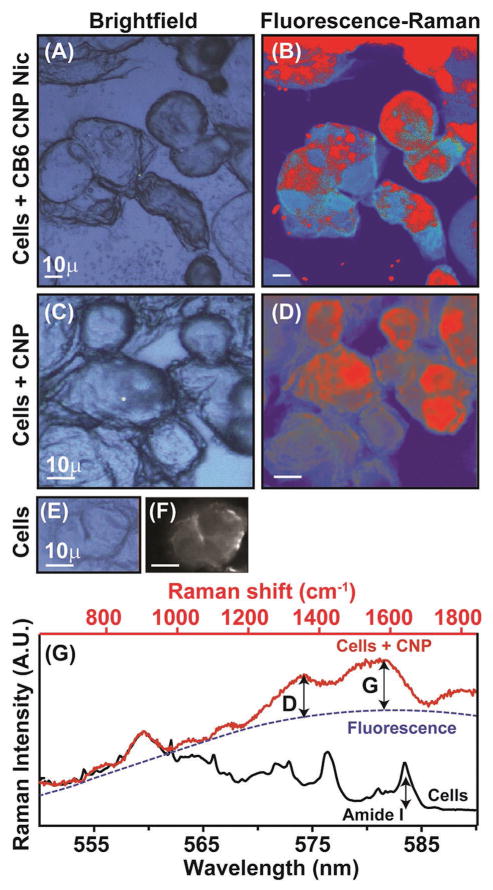
While the lack of a strong IR label does not permit detecting CNPs in cells, strong fluorescence and Raman signatures allow visualization after incubation. Shown in Figure 8A,C,E are the bright field images of cells incubated with CB6 CNP Nic, CNP, and control cells. Figure 8B,D displays the fluorescence images (in blue) corresponding to Figure 8A,C, respectively, with average fluorescence spectra shown in Figure 8G (red). The fluorescence contribution was extracted by fitting spectra to a 3rd order polynomial (dashed blue) and integrated. The integrated value (blue) in Figure 8B,D depicts the spread of the CNPs across the cellular matrix. The intensity of the G-band for the CNPs, extracted by subtracting the fluorescence, is plotted above the fluorescence images (colored in red). It is apparent that the G-band of the Raman spectra is scattered with respect to the fluorescence distribution, which can be explained by the adherence of CNPs in those regions shown in red, thus resulting in an intense G or D band. In contrast, the Fluorescence merged Raman spectrum of the cells under control conditions are shown in Figure 8G (black). Autofluorescence from the cells is markedly reduced compared to cells incubated with CNPs. The intensities of the Amide I (Figure 8F) represent a typical Raman image of the control cells. Together, the three imaging modalities demonstrate efficient cellular internalization of the formulations in the cells, their distribution and likely activity.
Figure 8.
Label free Fluorescence merged Raman images overlaid with bright field images of the MCF-7 cells treated with CNP containing formulations. A), C), and E) are the bright field images while B), D), and F) represent the Fluorescence merged Raman images. The red region in the latter corresponds to the CNPs distribution in the cells signifying the fluorescent signature and D and G bands originating from CNPs. Panel G) shows the average fluorescence and Raman spectra obtained from the cells.
2.4. In Vivo Studies and Tumor Growth Inhibition
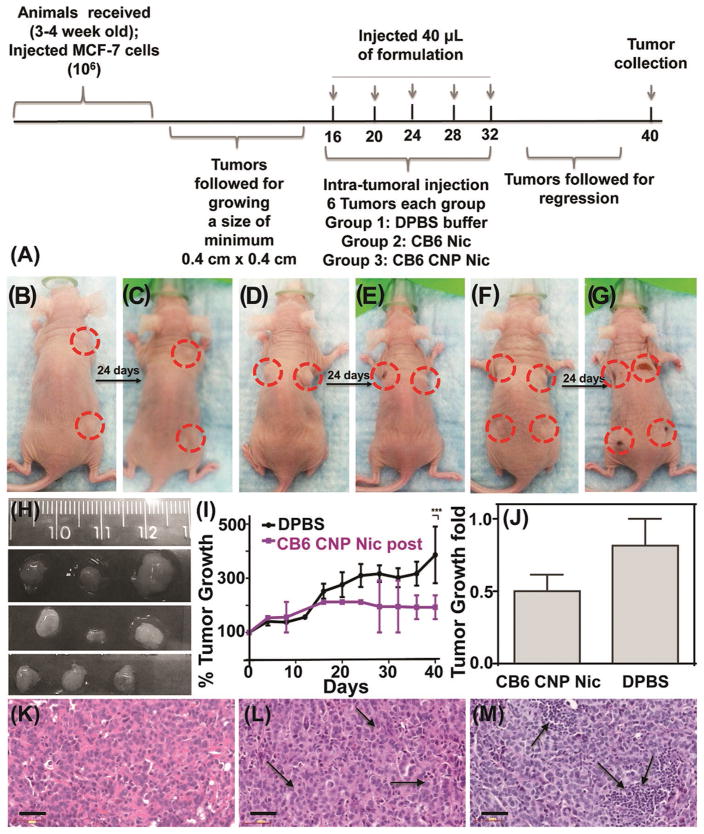
The anti-tumor capability of Nic was verified in vivo in a xenograft generated from MCF-7 cells in athymic nude mice. Tumors grown to a minimum size of 0.4 × 0.4 cm2 (Figure 9A) were intratumorally injected with phosphate buffer saline (DPBS) (Figure 9B); CB6 Nic (Figure 9D); and CB6 CNP Nic (Figure 9F) in 40 μL volume on day 16, 20, 24, 28, and 32 d following the first measurement after cell implantation. The growth was followed for 40 d before sacrificing and excising the tumor (Figure 9I and Figure S10, Supporting Information). During this period, no mice suffered from adverse clinical signs, distress or weight loss, an observation compatible with the in vitro studies. The bodies’ normal functions were not impaired compared to control group and were maintained at similar level indicating no in vivo toxicity. It was observed that the tumor growth could significantly be harnessed using CB6 Nic and CB6 CNP Nic, compared to tumors treated with buffer in same order (Figure 9H,I and Figure S10, Supporting Information). The buffer treated tumors continued to grow up to ≈400% compared to those inhibited by CB6 Nic and CB6 CNP Nic treated samples. After dose withdrawal of CB6 Nic, an upsurge in tumor growth was noticed compared to the constant growth inhibition in CB6 CNP Nic treated animals (Figure S10, Supporting Information). In vivo results were corroborated with histologic analyses. Hematoxylin/eosin (H&E)[43] stained sections of tumors revealed nuclear fragmentation and retracted cytoplasm, indicating the occurrence of apoptotic features in tumor sections treated with CB6 Nic and CB6 CNP Nic (Figure 9K–M).
Figure 9.
In vivo results on xenograft mouse model. A) Timeline of experiment; representative animals with tumors before and after treatment with B,C) DPBS; D,E) CB6 Nic and F,G) CB6 CNP Nic. H) Tumors collected after sacrificing the animals treated with DPBS (Top line); CB6 Nic (Middle line) and CB6 CNP Nic (Bottom line). I) Tumor growth curves with time and J) fold change. H&E images of tumor sections for treatments with K) DPBS; L) CB6 Nic and M) CB6 CNP Nic. An unpaired t test was performed between % tumor growth values in animals with DPBS treatment and CB6 CNP Nic to reach a two tailed p-value < 0.001, represented as ***. Scale bar is 50 μm.
The systemic administration (i.e., IV) of the agent was deliberately avoided to minimize circulatory biological barriers. In order to confirm the efficient sustained release of the agent in a tumor environment and its ability to inhibit the intended path of action, i.e., STAT-3, an intratumoral (IT) administration route was envisioned. Administration through IT would limit the diffusion of the nanoparticles to the tumor boundaries and accumulation will tend to occur more effectively due to the poor lymphatic drainage of tumor vessels.[44] The results indicated that our designed system was highly capable of delivering the poorly soluble agent in an efficacious manner and the intended action of the drug was retained at the molecular level. We realize that an IV route would provide information about the systemic biodistribution and other pharmacokinetic properties assessments of the agent. However, this extended biological study was beyond the scope of this introductory investigation. An ongoing study in our lab revealed that the similar particles can actively be targeted to stem cell surface marker and release Nic for highly efficacious inhibition of STAT-3 when administered using IV route. This also points to the intended future application of the system for targeting stem-like cancer cells.
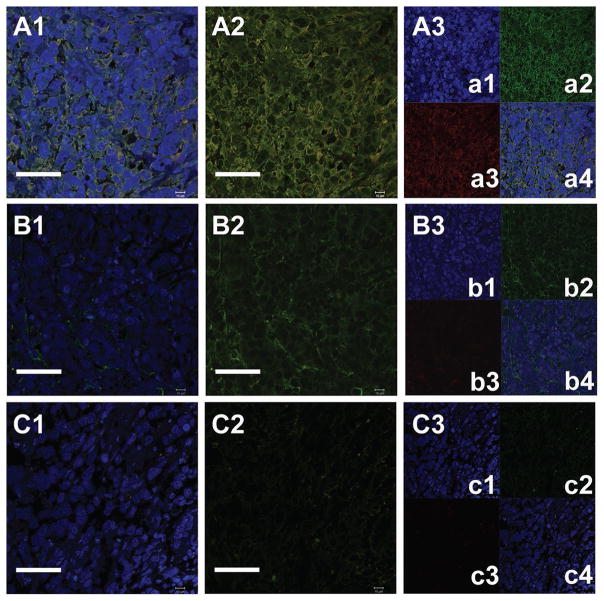
STAT-3 inhibition has been reported to play a crucial role, especially in that the phosphorylation of STAT-3 causes active participation in cancer cell survival.[45] Thus, a loss in the level of STAT-3 phosphorylation could be used to establish cancer growth regression via STAT-3 inhibition. pSTAT-3 protein expression was analyzed by immunostaining sections of tumors treated with buffer, CB6 Nic, and CB6 CNP Nic using standard protocol. β-actin was used as background protein for all sections.[46] Representative immune-labeled cross sections of tumors treated with phosphate buffer saline (Figure 10A1–A3 and Figure S11A1–A3, Supporting Information), CB6 Nic (Figure 10B1–B3 and Figure S11B1–B3, Supporting Information) and CB6 CNP Nic (Figure 10C1–C3 and Figure S11C1–C3, Supporting Information) revealed the downregulation of pSTAT-3 activity in increasing order of CB6 Nic and CB6 CNP Nic. Sections were stained with pSTAT-3 antibody (Red) against antibody for common unaffected protein β-actin (Green) around cell nuclei. Cellular nuclei were stained with DAPI (Blue). A higher level of pSTAT-3 in phosphate buffer saline treated tumors was seen compared to tumors treated with Nic formulations. Sections were also stained with DAPI and only secondary antibodies for pSTAT-3 and β-actin without incubation with primary antibodies to avoid any false positive staining (Figure S12, Supporting Information).
Figure 10.
Representative immunohistochemical analysis of xenograft sections treated with (A1–A3) DPBS, (B1–B3) CB6 Nic, and (C1–C3) CB6 CNP Nic observed in animal #7, 5, and 16, respectively. A1, B1, and C1 represent the β-actin stained cells with DAPI stained nucleus; A2, B2, C2 β-actin stained cells without nuclear staining and A3, B3, and C3 for individual staining of nucleus (a1, b1, c1); β-actin (a2, b2, c2); pSTAT3 (a3, b3, c3) and overlapped images (a4, b4, c4, and d4). It represents decrease in pSTAT3 staining (red) for CB6 Nic and CB6 CNP Nic treated tumor sections. The scale bar corresponds to 50 μm.
2.5. Enzyme Mediated Biodegradation of Carbon Nanoparticles
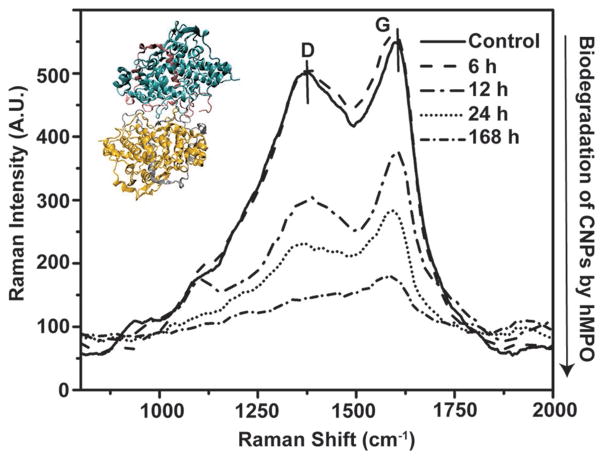
The detrimental health effects and bioresistance of certain nanomaterials, e.g., carbon nanotubes and quantum dots, have raised uncertainties about eventual clearance, fate, and biodegradability in general.[47] It is largely believed that the nanometer sized agents should either be cleared from the body following the drug delivery and/or imaging action, or be biodigested internally. It is also known that particles with hydrodynamic radii below 10 nm are likely to be cleared through renal pathway. For larger particles, therefore, it is ideal to design particles that can be degraded in the physiologic temperatures via naturally abundant enzymes expressed in vivo within a relatively short time frame. Human myeloperoxidase (hMPO)[48] was utilized for the first time in this study as a model human enzyme to ensure the catalytic in vitro degradation of the carbon nanoparticles as a result of generated enzymatic byproducts. CB6 CNP Nic postfunctionalized sample was incubated with hMPO and H2O2 at 37 °C and subsequently the activity of enzyme was maintained by replenishing H2O2. The activity of the enzyme was quenched at specific time points by transferring the samples to 4 °C. Raman spectra were used to monitor degradation of the samples. Analysis of the D and G bands revealed the progressive suppression of both characteristic peaks of CNPs, suggesting the success of the catalytic degradation of the CNPs by the oxidative pathway (Figure 11). Analysis of the spectra of CB6 CNP Nic postfunctionalized samples incubated with hMPO in the presence of H2O2, demonstrated almost complete disappearance of the D band after a week. The intensity ratio of G to D band, as the measure of the integrity of the CNPs, yielded no particular trend. This can be attributed to the particle to particle variation in terms of graphitic content during our hydrothermal synthesis. In this primary study, to indicate the safe application of CB6 CNPs, we demonstrated that these particles can successfully be degraded in the presence of a human enzyme. However, more mechanistic study to understand the interaction of hMPO with CNP is outside the scope of this work but would be an interesting topic for upcoming research from our laboratories.
Figure 11.
Raman spectra of the CB6 CNP Nic (post) and (hMPO and H2O2)-treated sample indicating the weakening of the D and G band overtime implying the degradation via enzymatically catalyzed reactions. The inset shows hMPO structure which has been obtained from protein data bank (1MHL).[49]
3. Conclusions
We have designed and synthesized a nanoscale supramolecular system to improve the delivery and solubility of a poorly soluble drug through host–guest, noncovalent interaction resulting in an enhanced control of cancer. The common approach to fabricate supramolecular networks from guest–host interactions is the modification of polymers with guest and host groups either as end groups or pendant modification; our route offers an alternative and facile method using simply tuned interactions. Our systematic development first analyzed interaction between CB and Nic through experimental and computational methods. After establishing the complexation, cucurbit[6]uril tethered carbon nanoparticles were derived by either prefunctionalization or postfunctionalization routes. Extensive physiochemical tests were conducted, which robustly indicated the success of synthesis. Subsequently, biological assays on the in vitro breast cancer model demonstrated the promoted inhibitory effect of the nanoformulation compared with free drug. Chemical and microscopic imaging modalities demonstrated the feasibility of the screening for cargo internalization in cells. In vivo studies also revealed the controlled growth of tumors treated with our approach compared to controls while a suppression of STAT-3 pathway was observed in the treatment group via immunohistochemical assessments. Enzymatic degradation of the nanoplatform was achieved upon exposure to human myeloperoxidase. Overall, these results imply a high potential of our nanoplatform for the enhanced inhibition of STAT-3 pathway. We envision that this methodology can be exploited in the future for the delivery of other poorly soluble agents. Specifically, our particles can be utilized against the cancer cell enriched with stem-like properties. Moreover, although this study was focused on the breast cancer models, it can be tuned toward the downregulation of STAT-3 as a common pathway modulated in other types of cancer. The smart nanocarriers equipped with defined functionality for tailored interaction through “host–guest chemistry”, and biodegradable properties can ferry poorly soluble agents to selectively target cancer cells with stem-like properties and controllably release their payload in response to environment, thus resulting in an improved antitumor activity in vivo.
4. Experimental Section
Materials
Food grade agave nectar (HoneyTree’s Organic Agave Nectar, Onsted, MI) was purchased from local store. Cucurbit[6]uril (CB[6]) hydrate (CAS Number 80262-44-8) and niclosamide, 98% (HPLC) (2′,5-Dichloro-4′-nitrosalicylanilide, CAS Number 50-65-7) were supplied from Sigma-Aldrich (St. Louis, MO, USA) and AKA Scientific Inc. (Union City, CA, USA), respectively. Nanopure water (0.2 × 10−6 M, 18 MΩ cm) was used throughout the experiment. Human breast cancer MCF-7 cell line was obtained from American Type Culture Collection (ATCC). High Glucose Dulbecco’s Modified Eagle’s Medium (DMEM; Sigma) was supplemented with 10% fetal bovine serum (FBS (Seradigm, US)) and 1% Penicillin Streptomycin (PenStrep) (Lonza). Trypsin (EDTA 0.02%, dextrose 0.05%, and trypsin 0.1%) in Dulbecco’s phosphate buffer saline (DPBS, Gibco) was prepared. Tetrazolium salt 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide was obtained from Sigma. 1H NMR measurements were carried out in DMSO-d6 (Cambridge Isotope Laboratories, Inc., MA, USA). Other reagents were obtained from Aldrich Chemical Co. (St. Louis, MO, USA). Athymic mice were purchased from Charles River Laboratories International, Inc., USA. All the antibodies were purchased from Molecular probes, Thermo fisher scientific, USA.
Host–Guest Complex Formation between CB6 and Nic: Computational Method
The starting geometries of the studied CB6 Nic complexes were generated by the manual insertion of the guest molecule (Nic) into the desired position of the host molecule (CB). Prior to the molecular dynamics simulations, the initial configurations of CB6, 7, and Nic, borrowed from protein data bank (PDB), were minimized using ReaxFF. In order to determine the stability of Nic CB6 complexes, the binding energy of nNic:mCB6 complexes was calculated based on Equation (2)
| (2) |
The negative sign of binding energies signifies that the insertion of the Nic molecules into the host molecule is energetically favorable.
Each solvated complex was subjected to energy minimization using the conjugate gradient algorithm prior to starting the MD simulation. The minimized complex was then equilibrated at 300 K for 60 ps, followed by a 500 ps equilibration step at 300 K at 1 atm. To account for the energetics of Nic release in water, the bond-restraint sampling was repeated through dragging of the Nic molecules away from CB6 host. Subsequently, the production runs were carried out for 1 ns at 300 K and 1.0 atm with a 0.1 fs time step under NVT ensemble (Brendsen thermostat with Tdamp = 100.0 fs). The Materials Studio-8 program was used to visualize the structures.
In addition, we computed the enthalpy of binding as the difference of the energy of complex and the individual molecules, where the initial states were obtained from series of energy minimization steps consisting of a conjugate gradient (CG) geometry optimization using ReaxFF with a convergence criterion of 0.5 kcal mol−1 between subsequent CG steps.
Experimental Studies on Host–Guest Interaction
For the determination of the stoichiometry of the complexation, Job’s plot was constructed by plotting the amide I peak wavenumber versus the mole fraction. The total molar concentration was preserved at 0.5 × 10−3 M. The minimum of the plot was taken as the Stoichiometric ratio. 1H NMR titration measurements were carried out on Varian VXR 500 (Varian, Inc., Palo Alto, CA) spectrometer operating at 500 MHz equipped with 5 mm Nalorac QUAD probe at 37 °C to achieve accurate shimming.[50] The starting volume Nic was set to 700 μL (2 × 10−3 M) and 10 μL of titrant containing CB6 (30 × 10−3 M) and Nic (2 × 10−3 M) was added at each titration step. A total of 128 acquisitions were made and averaged out. The data was analyzed with MestRenova 8.1 software (Mestrelab Research SL; Santiago de Compostela, Spain).
Nanoparticles Synthesis and Characterization: Carbon Nano-particles Decorated with Cucurbit[6]uril Synthesis
Pristine carbon nanoparticles (CNP) were carbonized by facile hydrothermal method using commercially available food grade agave nectar as the carbohydrate source (batch composition; 47%–56% fructose and 16%–20% glucose, rest is the mixture of other sugars and water). Agave nectar dispersed in water (0.2 mg mL−1) was slowly heated on a hotplate at 300 °C (Corning) so as to evaporate water and the product visually changed color from amber to dark brown. Subsequently, the burnt residue was resuspended in water and probe sonicated (Q700, Qsonica Sonicators, Newtown, CT) in the ice bath for 5 min at Pulsed Amp, 1; on, 5 s; off, 3 s. The samples were dialyzed for prolonged period and the content underwent two steps of additional filtration (Acrodisc syringe) with mesh size of 0.45 and 0.22 μm, respectively.
Two methods of synthesis were applied for CNPs decorated with CB6 preparation. In the first method, CB6 was introduced on the surface of CNP via vigorous stirring of the respective concentrations of CB6 for 5 min using vortex machine. These particles were labeled CB6 CNP postfunctionalized henceforth. In the second method, one pot nucleation of CB6 CNP was carried out. Briefly, the mixture of agave nectar and CB6 suspended in water at the desired concentration was heated on the hot plate and similar steps as for CNP preparation was followed. These particles were denoted as CB6 CNP prefunctionalized. For the preparation of CB6 CNP Nic, a stock solution of Nic was prepared (2 × 10−3 M) and then diluted with water to reach the desired concentration depending on the experiment. The drug was loaded on the CB6 CNP (pre-/postfunctionalized) using simple mixing method for 5 min.
Physicochemical Characterization of the Nanoparticles
The hydrodynamic size and the distribution of the nanoparticles were determined through dynamic light scattering measurements on Malvern Zetasizer ZS90 instrument (Malvern Instruments Ltd, United Kingdom) at fixed angle of 90°. TEM images were acquired on the Cu drop- deposited grid (200 mesh) using JEOL 2100 Cryo TEM machine (Tokyo, Japan) equipped with Gatan UltraScan 2k × 2k CCD camera. Anhydrous diameter was determined using image J software (NIH, Bethesda, MD). AFM images were obtained in the tapping mode on Asylum Cypher (Asylum Research, Santa Barbara, CA) device using freshly cleaved mica attached to the stainless steel discs substrate. Zeta potential ζ values were determined using a Malvern Zetasizer (Malvern Instruments Ltd, United Kingdom) of nanoseries. The experiment was performed at 25 °C and at pH = 7 at the light scattering mode.[51] Absorption spectra from different samples were recorded by GENESYS 10 UV–vis spectrometer (Thermo Scientific, MA, USA). The IR spectra was recorded by PerkinElmer Spotlight 400 (PerkinElmer, Waltham, MA, USA) equipped with a thermal source, and a raster-scanning linear array detector. Spectra were collected using a 1 cm s−1 mirror speed for acquisition, and zero padding was not used. Background scans were taken at 8 cm−1 spectral resolution; 150 × 150 μm images were collected at 8 cm−1 spectral resolutions with 8 scans per pixel and a 6.25 × 6.25 μm pixel size. Data were atmosphere corrected on the spotlight and further processing was done using Matlab R2013 software. In addition, Raman spectra were acquired in reflection mode on (LabRAM Horiba) machine coupled with Andor Newton back-illuminated EMCCD camera (LAB Ram, Horiba) and setting the excitation wavelength at 532 nm and the power at 25 mW with a 10 s acquisition time (spectral resolution: 8 cm−1, NA: 0.8).
In Vitro Biological Studies: Cancer Cell Culture
Cell toxicity assay was performed on human breast cancer ER (+) MCF-7 (ATCC) cells. Cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum (FBS) incubated at 37 °C in a 99% humidified atmosphere containing 5% CO2. Cells were regularly passaged by trypsinization with 0.1% trypsin (EDTA 0.02%, dextrose 0.05%, and trypsin 0.1%) in DPBS (pH 7.4).
Raman and Fluorescence Imaging of the Cells
A high resolution research grade Horiba LabRAM HR Raman imaging system coupled to an Andor Newton back-illuminated EMCCD camera DU970P was used to measure the Raman and the fluorescence images. (λexcitation = 532nm, grating = 150 g mm−1, slit aperture = 100 μm, confocal aperture = 50 μm, laser power = 25 mW, acquisition time = 1 s).
Cell Toxicity Assay
The cell toxicity assay measures the potency of the drug in inhibiting the cell growth and compared by calculating IC50 (inhibitory concentration for 50% cell population). MTT assay was performed on MCF-7 cells (104 cells per well) in 96-well plate using the previously established method.[52] Briefly, 24 h after plating the cells, they were treated with the samples at concentrations ranging from 1.5 × 10−6 M to 50 × 10−6 M. After 44 h, 20 μL of MTT solution (5 mg mL−1) was added to the growth medium and the cells were incubated for an additional 4 h. Subsequently, the solution was removed and replaced by 200 μL of dimethyl sulfoxide (DMSO) to solubilize formazan blue crystals. Absorption was read on multiwell plate reader machine (BioTek Synergy HT, US) using the standard wavelengths of 592 nm. An average of the triplicate wells was calculated and the data were analyzed using Graph pad prism 6.0 software.
Cell Mechanistic Studies
Cells were treated with Nic formulations followed by flow assisted cell sorting (FACS) analysis on propidium iodide (PI) stained treated and untreated cells. Results were analyzed to calculate the apoptotic cell population. MCF-7 cells (0.4 × 106 per well) were plated in 6-well plates and grown for ≈24 h. Cells were treated with 50 × 10−6 M of free Nic or loaded with CB6 and CB6 CNP. At the end of 72 h time point, cells were trypsinized and collected in 100 μL of reconstituted medium (10% FBS in DMEM). Cell pellets were fixed in chilled ethanol while vortexing. Fixed cells were stored at −20 °C for >12 h. At the end of the incubation, cells were washed with DPBS and incubated with RNase A (1 μg mL−1) at 37 °C for >12 h. Cells were incubated with PI (2 μg mL−1) for 30 min before scanning on FACS machine. Cells were treated in triplicate and mixed well before acquiring the FACS data.
Laddering Assay for Fragmented Genomic DNA Post Apoptosis
DNA fragmentation assay was performed on MCF-7 cells to further probe the cell apoptosis as described earlier.[53] Briefly, ≈80% confluent MCF-7 cells were treated for 72 h with Nic formulations. At the end of the incubation period, cells were washed, spun and collected in 400 μL of lysis buffer. DNA extraction was performed using Thermo Scientific DNA extraction kit. Extracted genomic DNA was washed with 70% EtOH and dissolved in water after air drying. Collected genomic DNA was used to load in 2% agarose gel and ran at 100 mV for 40 min before imaging on Gel doc (Bio-Rad Laboratories, Inc., Hercules, CA).
Cell Fixing for Imaging and Spectroscopy
MCF-7 cells were grown in a 6-well plate to reach ≈70% confluency. The cells were then trypisinized and seeded onto sterilized low E-slides (for infrared imaging) and coverslips (for Raman and confocal measurements). The cultured cells on the mentioned substrates were incubated with various formulations for 4 h at 37 °C. Subsequently, they were washed with DPBS and fixed using 4% formaldehyde.
In Vivo Experiments
Advanced 3D cell culture techniques can only mimic some of the aspects of the in vivo tumor environment; hence, in vivo studies make better models. To evaluate the efficacy of adopted strategy, animal experiments were performed in athymic mice. All experiments were designed to minimize the use of animals. We generated four tumors per animal including three mice per group. Animals were housed in Carl R. Woese Institute for Genomic Biology, University of Illinois at Urbana-Champaign and were allowed a week for acclimation.
Administration of MCF-7 (ER(+) Human Breast Cancer Cells) in Flanks of Athymic Mice
Approximately 5 × 106 MCF-7 human breast cancer cells suspended in 40 μL of Matrigel (50 %, v/v) were subcutaneously injected on the flanks of mice. No significant discomfort was noticed in the tumor bearing mice in the time frame of completing the experiment. Mice body weights were measured every week. Criteria for interventions were set as tumor increase to 17 mm × 17 mm. Tumor size was determined by measuring the length and width of the tumor using caliper and then calculating the tumor volume via formulae
Administration of Treatments
Animals were followed till tumors grew to a minimum of 4 mm × 4 mm in size before starting the treatment. CB6 Nic and CB6 CNP Nic were prepared at 1.5 × 10−3 M concentration of Nic and a total of 40 μL formulations were injected to each tumor every 4 d including 16th, 20th, 24th, 28th, and 32nd and followed till 40th day for measuring tumor growth and regression.
Animal Dissection, Tumor Collection, Processing, Embedding, and Sectioning
At the end of the experiment, animals were euthanized with CO2 influx. Animals were dissected to collect tumors and stored in tissue cassettes dipped in 10% formalin before performing the tissue fixation protocol in Leica ASP300 tissue processor following IGB facility protocol. Embedded tumor blocks were clamped in microtome (Leica) and sectioned at 6 μm thickness.
Hematoxylin and Eosin (H&E) Staining
Paraffin-embedded sections (7 μm) were subjected to hematoxylin and eosin (H&E) staining following the standard protocol supplied at core facility IGB, UIUC. Staining was performed in Mayer hematoxylin solution for 3 min followed by counterstaining in eosin-phloxine B solution (or eosin Y solution) for 3 min. The morphological changes of H&E-stained tissue with each fixation were analyzed at magnification ×40.
Immunolabeling of Tissue Sections for pSTAT3 and β-actin
To image the extent of pSTAT3 protein expression in treated and untreated tumors, immunolabeling was performed on paraffin-embedded sections (7 μm). immunolabeling was performed by following standard protocol supplied by core facility IGB, UIUC. Sections were further incubated with primary antibody for >12 h at RT in dark. It was further incubated for 2 h at RT in dark with secondary antibody of β-actin (FITC conjugated) and phosphoSTAT3 (Cy3 conjugated) with dilution of 1:200 each in 1x FX medium. Tumor sections only incubated with primary anti-body were considered as negative controls. At the end of incubations, sections were washed with 1x PBS (three times) before mounting them under coverslips using mounting medium containing DAPI. Samples were bound with nail polish before performing confocal microscopy.
Enzymatic degradation of CB6 CNP Nic
Five suspensions of CB6 CNP Nic (carbon content: 10 mg mL−1) were prepared in Dulbecco’s phosphate buffered solution and 200 × 10−6 M of hydrogen peroxide. Subsequently, 100 μg of human myeloperoxidase (hMPO) enzyme (activity > 50 units mg−1 protein) was added to four of the five samples and statically incubated at 37 °C in the dark. Hydrogen peroxide was replenished every 5 h to sustain the activity of enzyme. To quench the enzyme activity, samples were transferred to 4 °C after 6, 12, 24, and 168 h. The collected samples were drop cast on glass slides for Raman analysis (120 to 2700 nm at 2% laser power for 20 s). All Raman measurements were taken on Nanophoton RAMAN 11 laser Raman microscope (Osaka, Japan) with a 532 nm wavelength laser for 20 s at 2% laser power using a 20x objective. For each spectrum a grating (600 l mm−1) scan was taken over the range 120–2700 cm−1. An average of 20 spectra were recorded and averaged for each measurement sample.
Statistical Analysis
The results were expressed as mean ± standard deviation and the data were analyzed using Graph pad prism 6.0 software. The p-value < 0.05 (95% confidence interval) was considered to be significantly different.
Supplementary Material
Acknowledgments
F.O. and S.K.M. contributed equally to this work. P.M., and A.O. co-second authored the paper. D.P. conceived the idea, F.O., S.K.M., and P.M. designed the experiments. F.O. performed syntheses, characterization, host–guest study, stability experiments, and cell experiments. S.K.M. performed in vitro and in vivo studies. A.O. performed and analyzed the computational studies. P.M., S.T., S.M., and M.G. performed FT-IR and Raman studies. E.D. performed the enzyme degradation study. D.P., F.O., R.B., P.M., and S.K.M. analyzed data and wrote the paper. All authors have given approval to the final version of the manuscript. The authors wish to thank Frederick Seitz Materials Research Laboratory staffs, Roger Adams NMR Lab director (D. Olson), Illinois sustainability center (J. Scott), Rodríguez-López Lab members (M. Burgess, B. Simpson), J. F. Serrano and L. Luu for the experimental and technical support. The authors are also grateful for the funding support from the University of Illinois at Urbana-Champaign and Children’s Discovery Institute. TEM, AFM, and zeta potential measurements were carried out at Frederick Seitz Materials Research Laboratory, UIUC. NMR studies were performed at Roger Adams Lab, UIUC. The study was also partly supported by the National Institutes of Health via Award No. 5R01EB009745.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Fatemeh Ostadhossein, Department of Bioengineering, University of Illinois at Urbana-Champaign, 1304 W. Springfield Ave., Urbana, IL 61801, USA.
Dr. Santosh K. Misra, Department of Bioengineering, University of Illinois at Urbana-Champaign, 1304 W. Springfield Ave., Urbana, IL 61801, USA
Dr. Prabuddha Mukherjee, Department of Bioengineering, University of Illinois at Urbana-Champaign, 1304 W. Springfield Ave., Urbana, IL 61801, USA
Dr. Alireza Ostadhossein, Department of Engineering Science and Mechanics, Pennsylvania State University, University Park, PA 16802, USA.
Enrique Daza, Department of Bioengineering, University of Illinois at Urbana-Champaign, 1304 W. Springfield Ave., Urbana, IL 61801, USA.
Saumya Tiwari, Department of Bioengineering, University of Illinois at Urbana-Champaign, 1304 W. Springfield Ave., Urbana, IL 61801, USA.
Shachi Mittal, Department of Bioengineering, University of Illinois at Urbana-Champaign, 1304 W. Springfield Ave., Urbana, IL 61801, USA.
Mark C. Gryka, Department of Bioengineering, University of Illinois at Urbana-Champaign, 1304 W. Springfield Ave., Urbana, IL 61801, USA
Prof. Rohit Bhargava, Departments of Bioengineering, Electrical and Computer Engineering, Chemical and Biomolecular Engineering, Chemistry, and Mechanical Science and Engineering, Beckman Institute for Advanced Science and Technology, University of Illinois at Urbana-Champaign, 1304 W. Springfield Ave., Urbana, IL 61801, USA
Prof. Dipanjan Pan, Carle Foundation Hospital, 502 N. Busey St., Urbana, IL 61801, USA. Departments of Bioengineering and Materials Science and Engineering, Beckman Institute for Advanced Science and Technology, Institute for Sustainability in Energy and Environment, 502 N. Busey St., Urbana, IL 61801, USA
References
- 1.Siegel R, Miller KD, Jemal AL. Ca-Cancer J Clin. 2016;66:7. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Abdullah LN, Chow EKH. Clin Transl Med. 2013;2:3. doi: 10.1186/2001-1326-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shen S, Xia JX, Wang J. Biomaterials. 2016;74:1. doi: 10.1016/j.biomaterials.2015.09.037. [DOI] [PubMed] [Google Scholar]
- 4.a) Takebe N, Harris PJ, Warren RQ, Ivy SP. Nat Rev Clin Oncol. 2011;8:97. doi: 10.1038/nrclinonc.2010.196. [DOI] [PubMed] [Google Scholar]; b) Dubrovska A, Kim S, Salamone RJ, Walker JR, Maira S-M, García-Echeverría C, Schultz PG, Reddy VA. Proc Natl Acad Sci USA. 2009;106:268. doi: 10.1073/pnas.0810956106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou J, Wulfkuhle J, Zhang H, Gu P, Yang Y, Deng J, Margolick JB, Liotta LA, Petricoin E, Zhang Y. Proc Natl Acad Sci USA. 2007;104:16158. doi: 10.1073/pnas.0702596104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alison MR, Lin WR, Lim SM, Nicholson LJ. Cancer Treat Rev. 2012;38:589. doi: 10.1016/j.ctrv.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 7.Bromberg J, Darnell JE., Jr Oncogene. 2000;19:2468. doi: 10.1038/sj.onc.1203476. [DOI] [PubMed] [Google Scholar]
- 8.a) Looyenga BD, Hutchings D, Cherni I, Kingsley C, Weiss GJ, MacKeigan JP. PloS One. 2012;7:e30820. doi: 10.1371/journal.pone.0030820. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Lin L, Fuchs J, Li C, Olson V, Bekaii-Saab T, Lin J. Biochem Biophys Res Commun. 2011;416:246. doi: 10.1016/j.bbrc.2011.10.112. [DOI] [PubMed] [Google Scholar]; c) Zhang Y, Du XL, Wang CJ, Lin DC, Ruan X, Feng YB, Huo YQ, Peng H, Cui JL, Zhang TT. Gastroenterology. 2012;142:521. doi: 10.1053/j.gastro.2011.11.023. [DOI] [PubMed] [Google Scholar]; d) Chen RJ, Ho YS, Guo HR, Wang YJ. Toxicol Sci. 2008;104:283. doi: 10.1093/toxsci/kfn086. [DOI] [PubMed] [Google Scholar]
- 9.a) Sherry MM, Reeves A, Wu JK, Cochran BH. Stem Cells. 2009;27:2383. doi: 10.1002/stem.185. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Gibbs CP, Kukekov VG, Reith JD, Tchigrinova O, Suslov ON, Scott EW, Ghivizzani SC, Ignatova TN, Steindler DA. Neoplasia. 2005;7:967. doi: 10.1593/neo.05394. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Yu H, Lee H, Herrmann A, Buettner R, Jove R. Nat Rev Cancer. 2014;14:736. doi: 10.1038/nrc3818. [DOI] [PubMed] [Google Scholar]
- 10.Gupta S, Sung B, Prasad S, Webb LJ, Aggarwal BBC. Trends Pharmacol Sci. 2013;34:508. doi: 10.1016/j.tips.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 11.a) Jin Y, Lu Z, Ding K, Li J, Du X, Chen C, Sun X, Wu Y, Zhou J, Pan J. Cancer Res. 2010;70:2516. doi: 10.1158/0008-5472.CAN-09-3950. [DOI] [PubMed] [Google Scholar]; b) Wang AM, Ku HH, Liang YC, Chen YC, Hwu YM, Yeh TS. J Cell Biochem. 2009;106:682. doi: 10.1002/jcb.22065. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Balgi AD, Fonseca BD, Donohue E, Tsang T, Lajoie P, Proud CG, Nabi IR, Roberge M. PLoS One. 2009;4:e7124. doi: 10.1371/journal.pone.0007124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ren X, Duan L, He Q, Zhang Z, Zhou Y, Wu D, Pan J, Pei D, Ding K. ACS Med Chem Lett. 2010;1:454. doi: 10.1021/ml100146z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.a) Li R, Hu Z, Sun SY, Chen ZG, Owonikoko TK, Sica GL, Ramalingam SS, Curran WJ, Khuri FR, Deng X. Mol Cancer Ther. 2013;12:2200. doi: 10.1158/1535-7163.MCT-13-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) You S, Li R, Park D, Xie M, Sica GL, Cao Y, Xiao ZQ, Deng X. Mol Cancer Ther. 2014;13:606. doi: 10.1158/1535-7163.MCT-13-0608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Misra SK, Jensen TW, Pan D. Nanoscale. 2015;7:7127. doi: 10.1039/c5nr00403a. [DOI] [PubMed] [Google Scholar]
- 15.a) Devarakonda B, Hill RA, Liebenberg W, Brits M, de Villiers MM. Int J Pharm. 2005;304:193. doi: 10.1016/j.ijpharm.2005.07.023. [DOI] [PubMed] [Google Scholar]; b) Yang W, de Villiers MM. AAPS J. 2005;7:E241. doi: 10.1208/aapsj070123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Assaf KI, Nau WM. Chem Soc Rev. 2015;44:394. doi: 10.1039/c4cs00273c. [DOI] [PubMed] [Google Scholar]
- 17.Uzunova VD, Cullinane C, Brix K, Nau WM, Day AI. Org Biomol Chem. 2010;8:2037. doi: 10.1039/b925555a. [DOI] [PubMed] [Google Scholar]
- 18.a) Yang ST, Cao L, Luo PG, Lu F, Wang X, Wang H, Meziani MJ, Liu Y, Qi G, Sun YP. J Am Chem Soc. 2009;131:11308. doi: 10.1021/ja904843x. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Mukherjee P, Misra SK, Gryka MC, Chang HH, Tiwari S, Wilson WL, Scott JW, Bhargava R, Pan D. Small. 2015;11:4691. doi: 10.1002/smll.201500728. [DOI] [PubMed] [Google Scholar]; c) Wu L, Cai X, Nelson K, Xing W, Xia J, Zhang R, Stacy AJ, Luderer M, Lanza GM, Wang LV. Nano Res. 2013;6:312. doi: 10.1007/s12274-013-0308-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Zhu S, Zhang J, Tang S, Qiao C, Wang L, Wang H, Liu X, Li B, Li Y, Yu W. Adv Funct Mater. 2012;22:4732. [Google Scholar]
- 19.a) Huang P, Lin J, Wang X, Wang Z, Zhang C, He M, Wang K, Chen F, Li Z, Shen G. Adv Funct Mater. 2012;24:5104. [Google Scholar]; b) Misra SK, Ohoka A, Kolmodin NJ, Pan D. Mol Pharmaceutics. 2015;12:375. doi: 10.1021/mp500742y. [DOI] [PubMed] [Google Scholar]; c) Tang J, Kong B, Wu H, Xu M, Wang Y, Wang Y, Zhao D, Zheng G. Adv Mater. 2013;25:6569. doi: 10.1002/adma.201303124. [DOI] [PubMed] [Google Scholar]; d) Xie Z, Wang F, Liu Cy. Adv Mater. 2012;24:1716. doi: 10.1002/adma.201104962. [DOI] [PubMed] [Google Scholar]
- 20.Choi Y, Kim S, Choi MH, Ryoo SR, Park J, Min DH, Kim BS. Adv Funct Mater. 2014;24:5781. [Google Scholar]
- 21.Fenley AT, Henriksen NM, Muddana HS, Gilson MK. J Chem Theory Comput. 2014;10:4069. doi: 10.1021/ct5004109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Banerjee R, Chakraborty H, Sarkar M. Biopolymers. 2004;75:355. doi: 10.1002/bip.20147. [DOI] [PubMed] [Google Scholar]
- 23.Biedermann F, Uzunova VD, Scherman OA, Nau WM, De Simone A. J Am Chem Soc. 2012;134:15318. doi: 10.1021/ja303309e. [DOI] [PubMed] [Google Scholar]
- 24.a) Yoon K, Ostadhossein A, van Duin AC. Carbon. 2016;99:58. [Google Scholar]; b) Islam MM, Ostadhossein A, Borodin O, Yeates AT, Tipton WW, Hennig RG, Kumar N, van Duin AC. Phys Chem Chem Phys. 2015;17:3383. doi: 10.1039/c4cp04532g. [DOI] [PubMed] [Google Scholar]; c) Misra SK, Ray T, Ostadhossein F, Kim B, Ray PS, Pan D. Bioconjugate Chem. 2015;27:594. doi: 10.1021/acs.bioconjchem.5b00601. [DOI] [PubMed] [Google Scholar]
- 25.Plimpton S. J Comput Phys. 1995;117:1. [Google Scholar]
- 26.Rappe AK, Goddard WA., III J Phys Chem. 1991;95:3358. [Google Scholar]
- 27.Barrow SJ, Kasera S, Rowland MJ, del Barrio J, Scherman OA. Chem Rev. 2015;115:12320. doi: 10.1021/acs.chemrev.5b00341. [DOI] [PubMed] [Google Scholar]
- 28.Kim J, Jung IS, Kim SY, Lee E, Kang JK, Sakamoto S, Yamaguchi K, Kim K. J Am Chem Soc. 2000;122:540. [Google Scholar]
- 29.Kumar R, Shin WS, Sunwoo K, Kim WY, Koo S, Bhuniya S, Kim JS. Chem Soc Rev. 2015;44:6670. doi: 10.1039/c5cs00224a. [DOI] [PubMed] [Google Scholar]
- 30.Choi HS, Liu W, Misra P, Tanaka E, Zimmer JP, Ipe BI, Bawendi MG, Frangioni JV. Nat Biotechnol. 2007;25:1165. doi: 10.1038/nbt1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wiogo HT, Lim M, Bulmus V, Yun J, Amal R. Langmuir. 2010;27:843. doi: 10.1021/la104278m. [DOI] [PubMed] [Google Scholar]
- 32.Lee JW, Samal S, Selvapalam N, Kim HJ, Kim K. Acc Chem Res. 2003;36:621. doi: 10.1021/ar020254k. [DOI] [PubMed] [Google Scholar]
- 33.Taylor RW, Lee TC, Scherman OA, Esteban R, Aizpurua J, Huang FM, Baumberg JJ, Mahajan S. ACS Nano. 2011;5:3878. doi: 10.1021/nn200250v. [DOI] [PubMed] [Google Scholar]
- 34.Wu L, Luderer M, Yang X, Swain C, Zhang H, Nelson K, Stacy AJ, Shen B, Lanza GM, Pan D. Theranostics. 2013;3:677. doi: 10.7150/thno.6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Al-Hadiya BM. Profiles Drug Subst, Excipients, Relat Methodol. 2005;32:67. doi: 10.1016/S0099-5428(05)32002-8. [DOI] [PubMed] [Google Scholar]
- 36.Hou Z-S, Tan Y-B, Kim K, Zhou Q-F. Polymer. 2006;47:742. [Google Scholar]
- 37.Munteanu M, Choi S, Ritter H. Macromolecules. 2009;42:3887. [Google Scholar]
- 38.Yang Q, Jiang Y, Li X, Yang Y, Hu L. Bull Mater Sci. 2014;37:1167. [Google Scholar]
- 39.Bai MY, Yang HC. Colloids Surf, A. 2013;419:248. [Google Scholar]
- 40.Dubey P, Gopinath P. J Mater Chem B. 2016;4:726. doi: 10.1039/c5tb02351c. [DOI] [PubMed] [Google Scholar]
- 41.Ostadhossein F, Daza EA, Frankowski D, Goatz D, Imgruet M, Kus J, Lake R, Modak M, Olsen N, Schwartz-Duval A, Zimmer A, Kolmodin N, Pan D. In: Personalized Medicine with a Nanochemistry Twist: Nanomedicine. Pan D, editor. Springer International Publishing; Cham, Switzerland: 2016. p. 105. [Google Scholar]
- 42.Misra SK, Ye M, Ostadhossein F, Pan D. Sci Rep. 2016;6:28196. doi: 10.1038/srep28196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Graf N, Bielenberg DR, Kolishetti N, Muus C, Banyard J, Farokhzad OC, Lippard SJ. ACS Nano. 2012;6:4530. doi: 10.1021/nn301148e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.a) Niu Q, Wang W, Li Q, Li Y, Ruden DM, He B. Transl Oncol. 2014;7:303. doi: 10.1016/j.tranon.2014.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Lammers T, Peschke P, Kühnlein R, Subr V, Ulbrich K, Huber P, Hennink W, Storm G. Neoplasia. 2006;8:788. doi: 10.1593/neo.06436. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Dickerson EB, Dreaden EC, Huang X, El-Sayed IH, Chu H, Pushpanketh S, McDonald JF, El-Sayed MA. Cancer Lett. 2008;269:57. doi: 10.1016/j.canlet.2008.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu H, Jove R. Nat Rev Cancer. 2004;4:97. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 46.Roqueta-Rivera M, Abbott TL, Sivaguru M, Hess RA, Nakamura MT. Biol Reprod. 2011;85:721. doi: 10.1095/biolreprod.110.089524. [DOI] [PubMed] [Google Scholar]
- 47.Liu Y, Zhao Y, Sun B, Chen C. Acc Chem Res. 2012;46:702. doi: 10.1021/ar300028m. [DOI] [PubMed] [Google Scholar]
- 48.Kagan VE, Konduru NV, Feng W, Allen BL, Conroy J, Volkov Y, Vlasova II, Belikova NA, Yanamala N, Kapralov A. Nat Nanotechnol. 2010;5:354. doi: 10.1038/nnano.2010.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fenna R, Zeng J, Davey C. Arch Biochem Biophys. 1995;316:653. doi: 10.1006/abbi.1995.1086. [DOI] [PubMed] [Google Scholar]
- 50.Misra SK, Ray T, Ostadhossein F, Kim B, Ray PS, Pan D. Bioconjugate Chem. 2016;27:594. doi: 10.1021/acs.bioconjchem.5b00601. [DOI] [PubMed] [Google Scholar]
- 51.Vulugundam G, Misra SK, Ostadhossein F, Schwartz-Duval AS, Daza E, Pan D. Chem Commun. 2016;52:7513. doi: 10.1039/c6cc02525k. [DOI] [PubMed] [Google Scholar]
- 52.Ostadhossein F, Mahmoudi N, Morales-Cid G, Tamjid E, Navas-Martos FJ, Soriano-Cuadrado B, Paniza JML, Simchi A. Materials. 2015;8:6401. doi: 10.3390/ma8095309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Misra SK, Ghoshal G, Gartia MR, Wu Z, De AK, Ye M, Bromfield CR, Williams EM, Singh K, Tangella KV. ACS Nano. 2015;9:10695. doi: 10.1021/acsnano.5b05974. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.