Abstract
Bilateral neovascular age-related macular degeneration (AMD) causes much more handicaps for patients than unilateral neovascular AMD. Although several AMD-susceptibility genes have been evaluated for their associations to bilaterality, genome-wide association study (GWAS) on bilaterality has been rarely reported. In the present study, we performed GWAS using neovascular AMD cases in East Asian. The discovery stage compared 581,252 single nucleotide polymorphisms (SNPs) between 803 unilateral and 321 bilateral Japanese cases but no SNP showed genome-wide significance, while SNPs at six regions showed P-value < 1.0 × 10−5, STON1-GTF2A1L/LHCGR/FSHR, PLXNA1, CTNNA3, ARMS2/HTRA1, LHFP, and FLJ38725. The first replication study for these six regions comparing 36 bilateral and 132 unilateral Japanese cases confirmed significant associations of rs4482537 (STON1-GTF2A1L/LHCGR/FSHR), rs2284665 (ARMS2/HTRA1), and rs8002574 (LHFP) to bilaterality. In the second replication study comparing 24 bilateral and 78 unilateral cases from Singapore, rs4482537 (STON1-GTF2A1L/LHCGR/FSHR) only showed significant association. Meta-analysis of discovery and replication studies confirmed genome-wide level significant association (P = 2.61 × 10−9) of rs4482537 (STON1-GTF2A1L/LHCGR/FSHR) and strong associations (P = 5.76 × 10−7 and 9.73 × 10−7, respectively) of rs2284665 (ARMS2/HTRA1) and rs8002574 (LHFP). Our GWAS for neovascular AMD bilaterality found new genetic loci STON1-GTF2A1L/LHCGR/FSHR and confirmed the previously reported association of ARMS2/HTRA1.
Introduction
Age-related macular degeneration (AMD) is one of the major causes of visual impairment in developed countries. Although early stage AMD does not affect visual function, late stage AMD induces severe visual loss. AMD is a complex disease caused by multiple environmental and genetic risk factors. Previous genome-wide association studies (GWASs) identified two major susceptibility loci for AMD; complement factor H (CFH) and age-related maculopathy susceptibility 2/high temperature requirement A1 (ARMS2/HTRA1). Recently, AMD Gene Consortium performed meta-analysis of GWASs and found 34 loci were associated with AMD development1. Furthermore, GWASs in East Asian populations revealed new loci for AMD and suggested ethnic differences in susceptibility to AMD2, 3.
Compared with patients with unilateral late AMD, patients with bilateral late AMD are more prone to visual handicaps. The prevalence of bilateral AMD was reported to be 40–50% in Caucasian and 10–20% in Asian4–10. Previous studies investigated the association between bilaterality of AMD and the known AMD susceptibility loci. Several studies reported that ARMS2/HTRA1 contributes to the bilaterality of late AMD10–16. In contrast, it is still controversial whether CFH increases the risk of AMD bilaterality10, 12–14, 16–19. To identify the genetic determinants associated with bilaterality of late AMD in East Asian, we conducted a GWAS comparing bilateral late AMD patients with unilateral late AMD patients. Since most late AMD is neovascular AMD (wet type) in East Asian and geographic atrophy (dry type) is rare, we focused on only neovascular AMD. After finding genes associated with the bilaterality of neovascular AMD, we confirmed their susceptibility to AMD occurrence by comparing all neovascular AMD cases including both bilateral cases and unilateral cases with controls of Japanese general populations.
Results
Japanese patients with neovascular AMD were recruited at the Center for Macular Diseases of Kyoto University Hospital (n = 821) and Fukushima Medical University (n = 333) for the discovery stage. From these 1154 cases, 10 cases were excluded due to lack of detailed fundus examination of the fellow eye and 20 cases were excluded because of the quality control for their genotype count analysis. Of the 1124 cases analyzed in the discovery stage, 803 had unilateral neovascular AMD and 321 had bilateral neovascular AMD. Subtypes of AMD were typical AMD in 507 patients and PCV in 617 patients. Clinical features of subjects are summarized in Table 1. Patients with bilateral neovascular AMD were older than patients with unilateral neovascular AMD (P < 0.001). While 70–80% of patients were male, the percentages were similar between bilateral and unilateral groups. The number of SNPs evaluated after quality control was 581,252. We plotted our genome-wide association findings on quantile-quantile (QQ) plots, and genomic control method revealed only a slight inflation of the test statistics (Inflation factor λ = 1.01).
Table 1.
Neovascular age-related macular degeneration samples used in the study
| Bilateral neovascular AMD | Unilateral neovascular AMD | P-value | ||||||
|---|---|---|---|---|---|---|---|---|
| n | Age | Sex (%, male) | n | Age | Sex (%, male) | Age | Sex | |
| Discovery stage | ||||||||
| Kyoto | 247 | 80.9 ± 7.8 | 70.5 | 546 | 76.4 ± 8.4 | 69.6 | <0.001 | 0.81 |
| Fukushima | 74 | 81.6 ± 6.3 | 83.8 | 257 | 77.6 ± 7.9 | 75.1 | <0.001 | 0.12 |
| Total | 321 | 81.1 ± 7.4 | 73.5 | 803 | 76.8 ± 8.3 | 71.4 | <0.001 | 0.47 |
| Replication stage | ||||||||
| Kobe | 36 | 83.4 ± 7.0 | 77.8 | 132 | 77.4 ± 8.0 | 71.2 | <0.001 | 0.43 |
| Singapore | 24 | 71.6 ± 5.9 | 70.8 | 78 | 66.0 ± 10.2 | 61.5 | 0.0013 | 0.56 |
Results of the genome-wide association analysis are shown in Fig. 1. Although no chromosomal loci showed genome-wide significance, fourteen SNPs showed P-value < 1.0 × 10−5; 7 SNPs in STON1-GTF2A1L/LHCGR/FSHR region, 1 SNP in PLXNA1, 2 SNPs in CTNNA3, 1 SNP in ARMS2/HTRA1, 1 SNP in LHFP, and 2 SNPs near FLJ38725 (Table 2). No SNPs in CFH showed significant association with bilaterality of neovascular AMD (P > 0.05). Supplementary Table 1 shows the association between the AMD bilaterality and the analyzed 440 SNPs within 10 genes for which associations with AMD were verified in Asian individuals. Although rs11963725 of C2/CFB (P = 0.0124), rs1054060 of C3 (P = 0.0454), rs17310296 of CETP (P = 0.00250), rs4714699 of VEGFA (P = 0.0368), rs6822976 of CFI (P = 0.00256), and rs12638651 of ADAMTS9 (P = 0.00742) showed nominally significant associations, these associations should be interpreted as negative results after permutation tests.
Figure 1.
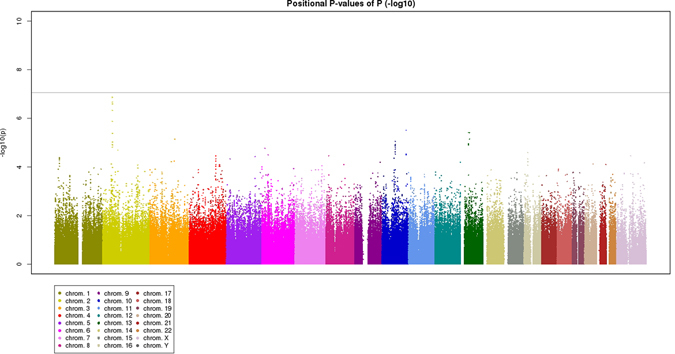
Minus log-transformed P-values are shown in a signal intensity (Manhattan) plot relative to their genomic position for bilaterality of age-related macular degeneration. P-values are adjusted for age and sex.
Table 2.
Results of discovery study for the 14 SNPs that showed P-value < 10−5, comparing bilateral neovascular age-related macular degeneration cases with unilateral those.
| SNP | Chr | Nearest Gene | Allele | Bilateral neovascular AMD | Unilateral neovascular AMD | P-value | OR (95% CI) | P-value* | OR (95% CI)* | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 11 | 12 | 22 | MAF | 11 | 12 | 22 | MAF | |||||||
| rs7589251 | 2 | STON1- GTF2A1L/LHCGR/FSHR | G | T | 52 | 133 | 136 | 0.37 | 58 | 321 | 423 | 0.27 | 6.22 × 10−6 | 1.56 (1.28–1.91) | 4.16 × 10−6 | 1.60 (1.31–1.96) |
| rs10208693 | 2 | A | G | 51 | 138 | 132 | 0.37 | 56 | 314 | 433 | 0.27 | 3.53 × 10−7 | 1.65 (1.35–2.01) | 4.74 × 10−7 | 1.68 (1.37–2.05) | |
| rs4538253 | 2 | G | A | 58 | 139 | 124 | 0.40 | 69 | 344 | 390 | 0.30 | 9.49 × 10−6 | 1.54 (1.26–1.88) | 9.55 × 10−6 | 1.56 (1.28–1.90) | |
| rs7603311 | 2 | T | C | 48 | 137 | 134 | 0.37 | 54 | 311 | 438 | 0.26 | 9.14 × 10−7 | 1.63 (1.33–2.00) | 1.35 × 10−6 | 1.65 (1.35–2.02) | |
| rs4482537 | 2 | C | T | 52 | 142 | 121 | 0.39 | 60 | 326 | 411 | 0.28 | 3.78 × 10−7 | 1.65 (1.34–2.02) | 1.36 × 10−7 | 1.73 (1.41–2.12) | |
| rs6545074 | 2 | G | T | 50 | 142 | 129 | 0.38 | 54 | 326 | 420 | 0.27 | 8.23 × 10−7 | 1.63 (1.32–2.00) | 2.54 × 10−7 | 1.71 (1.39–2.10) | |
| rs7037739 | 2 | G | A | 48 | 141 | 132 | 0.37 | 48 | 325 | 430 | 0.26 | 4.73 × 10−7 | 1.65 (1.34–2.02) | 2.14 × 10−7 | 1.73 (1.40–2.12) | |
| rs900429 | 3 | PLXNA1 | C | T | 8 | 98 | 214 | 0.18 | 45 | 329 | 428 | 0.26 | 2.96 × 10−5 | 0.61 (0.48–0.78) | 7.18 × 10−6 | 0.57 (0.45–0.73) |
| rs1925616 | 10 | CTNNA3 | G | A | 24 | 129 | 168 | 0.28 | 25 | 263 | 515 | 0.19 | 2.77 × 10−5 | 1.57 (1.25–1.97) | 8.79 × 10−6 | 1.67 (1.33–2.10) |
| rs10997482 | 10 | A | G | 24 | 129 | 168 | 0.28 | 25 | 263 | 515 | 0.19 | 2.77 × 10−5 | 1.57 (1.25–1.97) | 8.79 × 10−6 | 1.67 (1.33–2.10) | |
| rs2284665 | 10 | ARMS2/HTRA1 | G | T | 43 | 99 | 179 | 0.29 | 147 | 359 | 296 | 0.41 | 1.39 × 10−7 | 0.59 (0.48–0.71) | 3.08 × 10−6 | 0.63 (0.52–0.76) |
| rs8002574 | 13 | LHFP | T | C | 2 | 33 | 285 | 0.06 | 12 | 177 | 614 | 0.13 | 2.86 × 10−6 | 0.43 (0.29–0.63) | 3.84 × 10−6 | 0.41 (0.28–0.60) |
| rs9525873 | 13 | FLJ38725 | T | C | 49 | 172 | 100 | 0.42 | 97 | 327 | 378 | 0.32 | 1.77 × 10−5 | 1.51 (1.25–1.84) | 7.13 × 10−6 | 1.56 (1.29–1.90) |
| rs895266 | 13 | T | C | 74 | 168 | 79 | 0.49 | 135 | 362 | 305 | 0.39 | 2.07 × 10−5 | 1.49 (1.23–1.80) | 3.86 × 10−6 | 1.57 (1.29–1.90) | |
SNP, single nucleotide polymorphism; Chr, chromosome; MAF, minor allele frequency; OR, odds ratio; CI, confidence interval.
*Adjusted for age and sex.
One representative SNP from each of these six loci was examined in the replication studies (Table 3). For the first replication stage, Japanese patients with neovascular AMD were recruited at Kobe City Medical Center General Hospital (n = 170). Two cases were excluded after quality control for the genotype count analysis, and the genotypes of 36 bilateral AMD and 132 unilateral AMD were compared. The associations with neovascular AMD bilaterality were confirmed in rs4482537 of STON1-GTF2A1L/LHCGR/FSHR (P = 4.97 × 10−2), rs2284665 of ARMS2/HTRA1 (P = 3.83 × 10−2), and rs8002574 of LHFP (P = 3.66 × 10−2). For the second replication stage, Singaporean patients with neovascular AMD (n = 102) were recruited at Singapore National Eye Center. In the second replication study with 24 bilateral and 78 unilateral cases, only rs4482537 of STON1-GTF2A1L/LHCGR/FSHR showed significant association (P = 3.34 × 10−2), while rs2284665 of ARMS2/HTRA1 and rs8002574 of LHFP showed same association direction as in the discovery stage analysis and the first replication study with Japanese without statistically significant P-value. The meta-analysis of discovery and replication studies confirmed genome-wide significant association of STON1-GTF2A1L/LHCGR/FSHR (rs4482537; P = 2.61 × 10−9) with bilaterality. As for ARMS2/HTRA1 and LHFP, the effect direction was same among three cohorts and meta-analysis further confirmed their strong associations with bilaterality (P = 5.76 × 10−7 for rs2284665 of ARMS2/HTRA1 and P = 9.73 × 10−7 for rs8002574 of LHFP).
Table 3.
Results of two replication studies and meta-analysis for the six loci associated in discovery study, comparing bilateral neovascular age-related macular degeneration cases with unilateral those.
| SNP | Nearest Gene | Replication study | Discovery and replication study | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Kobe (n = 168) | Singapore (n = 102) | Combined meta-analysis | |||||||||||
| P value | OR (95% CI) | P value* | OR (95% CI) | P value | OR (95% CI) | P value* | OR (95% CI)* | P value | OR(95% CI) | P value* | OR (95% CI)* | ||
| rs4482537 | STON1- GTF2A1L/ LHCGR/ FSHR | 0.048 | 1.71 (1.01–2.90) | 0.050 | 1.78 (1.00–3.16) | 0.034 | 2.42 (1.07–5.49) | 0.033 | 2.53 (1.08–5.93) | 2.88 × 10−8 | 1.69 (1.40–2.03) | 2.61 × 10−9 | 1.76 (1.46–2.13) |
| rs2284665 | ARMS2/HTRA1 | 0.002 | 0.39 (0.21–0.72) | 0.038 | 0.52 (0.28–0.97) | 0.601 | 0.83 (0.42–1.66) | 0.711 | 0.88 (0.43–1.77) | 1.65 × 10−9 | 0.58 (0.49–0.69) | 5.76 × 10−7 | 0.63 (0.53–0.76) |
| rs8002574 | LHFP | 0.124 | 0.47 (0.16–1.36) | 0.037 | 0.32 (0.11–0.93) | 0.801 | 0.89 (0.38–2.13) | 0.584 | 0.78 (0.32–1.90) | 1.60 × 10−5 | 0.48 (0.35–0.67) | 9.73 × 10−7 | 0.44 (0.32–0.61) |
| rs895266 | FLJ38725 | 0.726 | 0.91 (0.53–1.56) | 0.552 | 0.85 (0.49–1.46) | 0.353 | 0.72 (0.38–1.43) | 0.364 | 0.72 (0.36–1.46) | 8.86 × 10−4 | 1.35 (1.13–1.61) | 1.40 × 10−4 | 1.40 (1.18–1.67) |
| rs900429 | PLXNA1 | 0.423 | 1.28 (0.67–2.43) | 0.367 | 1.34 (0.71–2.54) | 0.133 | 1.71 (0.85–3.44) | 0.235 | 1.57 (0.75–3.28) | 4.84 × 10−3 | 0.73 (0.59–0.91) | 8.46 × 10−4 | 0.69 (0.55–0.86) |
| rs1925616 | CTNNA3 | 0.822 | 0.93 (0.46–1.90) | 0.928 | 1.03 (0.51–2.10) | — | — | 0.709 | 0.85 (0.37–1.96) | — | — | 5.59 × 10−5 | 1.54 (1.25–1.90) |
SNP, single nucleotide polymorphism; OR, odds ratio; CI, confidence interval.
*Adjusted for age and sex.
To evaluate the associations of these three SNPs with the occurrence of AMD, genotypes of all neovascular AMD cases including both bilateral cases and unilateral cases were compared with the Nagahama cohort as Japanese general control (Table 4). The analysis showed that only ARMS2/HTRA1 was significantly associated with the occurrence of AMD (P = 1.78 × 10−81).
Table 4.
Results of the association study for three loci about the occurrence of AMD, comparing all neovascular age-related macular degeneration cases with Japanese general population cohort.
| SNP | Allele | Control (Nagahama cohort, n = 3265) | AMD cases (n = 1394) | P -value* (OR, 95%CI) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 11 | 12 | 22 | AF1 | 11 | 12 | 22 | MAF | |||
| rs4482537 (LHCGR/FSHR) | C | T | 280 | 1430 | 1555 | 0.30 | 134 | 582 | 666 | 0.31 | 0.79 (1.01, 0.92–1.12) | |
| rs2284665 (ARMS2/HTRA1) | G | T | 1165 | 1556 | 544 | 0.60 | 232 | 567 | 594 | 0.37 | 1.78 × 10−81 (0.40, 0.36–0.44) | |
| rs8002574 (LHFP) | T | C | 53 | 690 | 2522 | 0.12 | 21 | 265 | 1107 | 0.11 | 0.11 (0.89, 0.78–1.03) | |
SNP, single nucleotide polymorphism; AF, allele frequency; MAF, minor allele frequency; OR, odds ratio; CI, confidence interval, *Chi-trend.
Discussion
In this current study, we performed GWAS on bilaterality of neovascular AMD for the first time and identified the association of new genetic loci STON1-GTF2A1L/LHCGR/FSHR with neovascular AMD bilaterality. ARMS2/HTRA1 could be also associated with bilaterality of neovascular AMD and the association of LHFP should be further investigated.
Regional association plot (Supplementary Figure 1) for the studied SNPs within STON1-GTF2A1L/LHCGR/FSHR region did not elucidate which of these genes was correspond to the bilaterality of AMD. STON1 encodes stonin 1. Although stonin 2 controls synaptic transmission, the function of stonin 1 has not been elucidated20. GTF2A1L is expressed mainly in testis to form a counterpart of general transcription factor IIA subunit. In drosophila, transcription factor IIA regulates development of photoreceptor cells21. Polymorphisms in STON1-GTF2A1L might promote bilateral development of AMD through their effects on photoreceptor cells synaptic transmission.
LHCGR encodes luteinizing hormone/choriogonadotropin receptor, a receptor for luteinizing hormone (LH) and human chorionic gonadotropin (hCG), and FSHR encodes follicle-stimulating hormone receptor, a receptor for follicle-stimulating hormone (FSH). Both LH and FSH are released from the pituitary gland and stimulate estrogen secretion. Estrogen is associated with inhibition of AMD development22–24 and a previous study reported that polymorphisms of estrogen receptor gene were associated with AMD25. Aging decreases estrogen production, leading to increased LH/FSH secretion in the elderly via feedback mechanism.
Age-related decrease in estrogen production and increase in LH/FSH secretion are associated, partly, with increased risk of atherosclerosis and heart diseases by affecting lipoprotein/cholesterol metabolism26–28. Considering that several genes in lipoprotein/cholesterol metabolism are associated with AMD development, LHCGR/FSHR would affect the bilaterality of neovascular AMD in part by altering lipoprotein/cholesterol metabolism. Another possibility is that LHCGR/FSHR polymorphism would have localized effect, thereby affecting neovascular AMD bilaterality. Müller cells and retinal pigmented epithelial cells produce hCG and cone photoreceptor cells express its receptor LHCGR29. Further study on the roles of LHCGR of photoreceptor cells and the roles of LHCGR/FSHR-induced lipoprotein/cholesterol metabolism alteration would lead to prevention of neovascular AMD development in the fellow eye.
STON1-GTF2A1L/LHCGR/FSHR region also includes long intergenic noncoding RNA (lincRNA, RP11-460M2.1). LincRNA can control gene expression and some lincRNAs might be able to control ocular neovascularization30. LincRNA RP11-460M2.1 might have some ability to control bilaterality of neovascular AMD.
Although CFH is a major susceptibility gene for AMD, SNPs in CFH did not have any association with the bilaterality of neovascular AMD in our study. Previous studies also showed that CFH was not associated with the bilaterality of neovascular AMD4, 7, 10, 11. In contrast, rs4482537 in LHCGR/FSHR locus had significant association with the bilaterality of neovascular AMD, but not with the occurrence of AMD. On the other hand, previous studies and current study support that ARMS2/HTRA1 is associated with both occurrence and bilaterality of AMD. AMD-associated genes can be classified into three types, genes associated with both AMD occurrence and bilaterality, genes associated with only AMD occurrence, and genes associated with only AMD bilaterality. The second eye involvement in AMD might be regulated by a unique mechanism in addition to the factors associated with the first eye involvement.
LHFP is a HMGIC fusion partner gene in lipoma, one of the most common mesenchymal tumors31. LHFP is also associated with mesenchymal differentiation in gliosarcoma32. Recent studies suggest that epithelial–mesenchymal transition (EMT) has important roles in the development of AMD33, 34. LHFP might affect the bilaterality of neovascular AMD by affecting the EMT process. LHFP is also associated with Alzheimer’s disease35 that shares common clinical and pathological features with AMD; both Alzheimer’s disease and AMD are preceded by accumulation of amyloid beta36. LHFP might affect accumulation of amyloid beta and trigger the second eye involvement of AMD.
Although anti-VEGF treatment has improved treatment outcome of exudative AMD, the SEVEN-UP study reported that 51% of the patients in their study suffered from bilateral neovascular AMD during 7 years of follow-up37. The second eye involvement in patients with late AMD is a matter of concern because patients with bilateral late AMD are more prone to visual handicaps than patients with unilateral late AMD. The SEVEN-UP study also suggested that reduced frequency of treatments contributed to the decline of visual acuity. Increasing the frequency of injection might maintain visual acuity for an extended period. Therefore intensive treatment should be initiated in patients with unilateral neovascular AMD who are most likely to develop bilateral neovascular AMD in the future, to maintain good visual acuity. Prediction of the second eye involvement in neovascular AMD would be beneficial for patients with unilateral neovascular AMD. Elucidation of the mechanism to control the second eye involvement might lead to prevention of the second eye involvement.
The limitations of this study are its retrospective nature and relatively small sample size. Studies involving large size might successfully replicate the association of LHFP, and prospective study would further confirm the association of STON1-GTF2A1L/LHCGR/FSHR and ARMS2/HTRA1 to the bilaterality of AMD. Considering that CATT study could not detect any genetic associations between polymorphisms and the second eye involvement within 2 years of follow-up17, studies with longer follow-up period should be performed. The unilateral patients included in the current study were significantly younger than the bilateral patients. The unilateral patients may go on to develop bilateral disease with longer follow up, which would reduce the power of detecting genetic associations. Statistical adjustment for the time of diagnosis with unilateral neovascular AMD would also be helpful. Current study was performed only in Asians including both typical AMD and PCV. The prevalence of PCV is higher in Asian than Caucasian4–10, 38. Although the reported prevalence of bilateral involvement is similar between typical AMD and PCV, the ethnic difference cannot be ignored; 40–50% in Caucasian4, 5 and 10–20% in Asian6–10. Recent studies also suggested the role of ethnic differences in AMD genetic susceptibility3. Further study is needed to evaluate whether STON1-GTF2A1L/LHCGR/FSHR and LHFP are associated with neovascular AMD bilaterality in other races.
In conclusion, our GWAS for neovascular AMD bilaterality identified novel genetic loci STON1-GTF2A1L/LHCGR/FSHR and confirmed the association of ARMS2/HTRA1 with the bilaterality. LHFP might also be associated with AMD bilaterality. Prediction of the second eye involvement would be beneficial in determining the long-term management strategy for the first eye of AMD, and elucidation of the mechanisms for the second eye involvement would lead to prevention of the second eye involvement in AMD.
Methods
All procedures used in this study confirmed to the tenets of the Declaration of Helsinki. The Institutional Review Board and the Ethics Committee of each institution approved the experimental protocols; The Kyoto University Graduate School and Faculty of Medicine Ethics Committee, Fukushima Medical University Ethics Committee, Kobe City Medical Center General Hospital Ethics Committee, Singapore National Eye Center Ethics Committee, the Ad hoc Review Board of the Nagahama Cohort Project, and the Nagahama Municipal Review Board of Personal Information Protection. All the participants were fully informed of the purpose and procedures and a written consent was obtained from each.
Study subjects in GWAS for bilaterality
Japanese patients with neovascular AMD were recruited at the Center for Macular Diseases of Kyoto University Hospital (n = 821) and Fukushima Medical University (n = 333) for the discovery stage, and at Kobe City Medical Center General Hospital (n = 170) for the replication stage. Further, patients with neovascular AMD (n = 112) were recruited at Singapore National Eye Center for the second replication stage. All subjects underwent comprehensive ophthalmologic examinations, including dilated contact lens slit-lamp biomicroscopy, fundus photography, fluorescein and indocyanine green angiography (HRA2, Heidelberg Engineering, Heidelberg, Germany), and optical coherence tomography (Spectralis HRA + OCT, Heidelberg Engineering, Heidelberg, Germany).
Neovascular AMD was defined as the presence of exudative AMD as described in the international classification system for age-related maculopathy. Typical AMD involved classic choroidal neovascularization (CNV), occult CNV, or a combination of both. The diagnosis of polypoidal choroidal vasculopathy (PCV) was based on indocyanine green angiography, which showed a branching vascular network terminated in polypoidal lesions. Diagnosis and grading of AMD were performed in a masked manner by two ophthalmologists independently. In cases of disagreement, the third retinal specialist made the final decision. Bilaterality of neovascular AMD and subject age at final visit was used for analysis.
Genotyping
Genomic DNAs were extracted from peripheral blood leukocytes using QuickGene-610L DNA extraction kit (FUJIFILM Co., Tokyo, Japan). Genotyping was performed using Illumina BeadChip, both OmniExpress and HumanExome, HumanOmni2.5–8, or OmniExpress. The distortion of Hardy-Weinberg equilibrium (HWE) was not considered in this study because all samples comprised AMD cases. Stringent quality control, including minor allele frequency (MAF) ≥ 1% and genotype call rate ≥ 95% (per SNP and per individual), was performed using PLINK ver1.07 (http://pngu.mgh.harvard.edu/~purcell/plink/).
Statistical analysis
Association between genotypic distribution of each SNP and the bilaterality of neovascular AMD was examined using logistic regression analysis by adjusting for age and sex using Software R (R Foundation for Statistical Computing, Vienna, Austria). Inflation of the test statistics was assessed using the genomic-control method. SNPs with P-value < 1.0 × 10−5 were selected as candidates of replication stages. Among the candidate SNPs, one representative SNP was selected from each locus (r2 > 0.8 in the discovery stage samples) and these SNPs were tested for association in replication stages. The genotypic counts of the first and second stages were also evaluated in meta-analysis.
Furthermore, association between genotypic distribution of each candidate SNP of neovascular AMD bilaterality and the occurrence of neovascular AMD was examined by chi-square test for trend. All neovascular AMD cases were compared with Japanese general cohort, the Nagahama Study39, as control.
Control cohort for the AMD susceptibility test
A fixed dataset of 3,265 unrelated healthy Japanese subjects from the Nagahama prospective genome cohort for the Comprehensive Human Bioscience (The Nagahama Study) was used as a control group in the AMD susceptibility test. In detail, a total of 3,712 individuals from the Nagahama study were genotyped using HumanHap610K Quad Arrays, HumanOmni2.5 M Arrays, and/or HumanExome Arrays (Illumina Inc., CA, USA). To ensure high-quality genotype data, a series of quality control filters were applied to the data from each platform before imputation, including MAF cut-offs (>0.01), HWE (P > 1.0 × 10−6), genotypic success rate (>95%), individual call rate (>99%), and estimated relatedness (PI-HAT < 0.35). Quality controls were performed using PLINK (ver.1.07; http://pngu.mgh.harvard.edu/,purcell/plink/). For consistent genotyping data across each platform, we performed genomic imputation on available 1000 Genome Project data from East Asian subjects using MACH software (http://www.sph.umich.edu/csg/abecasis/MACH/tour/imputation.html). After imputation, we again performed quality control including MAF cut-offs (>0.01), HWE (P > 1.0 × 10−7), genotypic success rate (>90%), individual call rate (>90%), and imputation quality (R2 > 0.3).
Electronic supplementary material
Acknowledgements
Supported in part by grants-in-aid for scientific research (No. 24592624) from the Japan Society for the Promotion of Science, Tokyo, Japan, and the Japan National Society for the Prevention of Blindness, Tokyo, Japan. This research was also supported by the National Medical Research Council in Singapore (CSA/033/2012), Singapore.
Author Contributions
K.K.-K., K.Y., F.M. and N.Y. have designed the study. K.K.-K., K.Y., M.Y., M.M., G.C.-C.-M., Q.F., J.Y.-K., M.S., M.S.-K., M.O., Y.A.-K., I.N., H.N., N.G., A.O., H.T., S.O., A.T., Y.K., and T.S. acquired the data. K.K.-K., K.Y., M.Y., M.M., G.C.-C.-M., Q.F., J.Y.-K., F.M., C.-C.K., C.-Y.C., T.-Y.W. and N.Y. analyzed and interpreted data. F.M., C.-C.K., C.-Y.C., T.-Y.W. and N.Y. supervised the study. K.K.-K., K.Y., M.Y., and M.M. wrote the manuscript. All authors reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-07526-9
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Fritsche LG, et al. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat Genet. 2016;48:134–143. doi: 10.1038/ng.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang LZ, et al. Whole-exome sequencing implicates UBE3D in age-related macular degeneration in East Asian populations. Nat Commun. 2015;6:6687. doi: 10.1038/ncomms7687. [DOI] [PubMed] [Google Scholar]
- 3.Cheng CY, et al. New loci and coding variants confer risk for age-related macular degeneration in East Asians. Nat Commun. 2015;6:6063. doi: 10.1038/ncomms7063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yannuzzi LA, et al. Polypoidal choroidal vasculopathy and neovascularized age-related macular degeneration. Arch Ophthalmol. 1999;117:1503–1510. doi: 10.1001/archopht.117.11.1503. [DOI] [PubMed] [Google Scholar]
- 5.Ladas ID, et al. Polypoidal choroidal vasculopathy and exudative age-related macular degeneration in Greek population. Eye (Lond). 2004;18:455–459. doi: 10.1038/sj.eye.6700706. [DOI] [PubMed] [Google Scholar]
- 6.Sho K, et al. Polypoidal choroidal vasculopathy: incidence, demographic features, and clinical characteristics. Arch Ophthalmol. 2003;121:1392–1396. doi: 10.1001/archopht.121.10.1392. [DOI] [PubMed] [Google Scholar]
- 7.Maruko I, et al. Clinical characteristics of exudative age-related macular degeneration in Japanese patients. Am J Ophthalmol. 2007;144:15–22. doi: 10.1016/j.ajo.2007.03.047. [DOI] [PubMed] [Google Scholar]
- 8.Liu Y, et al. Subtype lesions of neovascular age-related macular degeneration in Chinese patients. Graefes Arch Clin Exp Ophthalmol. 2007;245:1441–1445. doi: 10.1007/s00417-007-0575-8. [DOI] [PubMed] [Google Scholar]
- 9.Hou J, et al. Clinical characteristics of polypoidal choroidal vasculopathy in Chinese patients. Graefes Arch Clin Exp Ophthalmol. 2011;249:975–979. doi: 10.1007/s00417-010-1575-7. [DOI] [PubMed] [Google Scholar]
- 10.Seddon JM, et al. Association of CFH Y402H and LOC387715 A69S with progression of age-related macular degeneration. JAMA. 2007;297:1793–1800. doi: 10.1001/jama.297.16.1793. [DOI] [PubMed] [Google Scholar]
- 11.Chen H, et al. Association of HTRA1 polymorphism and bilaterality in advanced age-related macular degeneration. Vision Res. 2008;48:690–694. doi: 10.1016/j.visres.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 12.Seddon JM, et al. Prediction model for prevalence and incidence of advanced age-related macular degeneration based on genetic, demographic, and environmental variables. Invest Ophthalmol Vis Sci. 2009;50:2044–2053. doi: 10.1167/iovs.08-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sakurada Y, et al. Role of complement factor H I62V and age-related maculopathy susceptibility 2 A69S variants in the clinical expression of polypoidal choroidal vasculopathy. Ophthalmology. 2011;118:1402–1407. doi: 10.1016/j.ophtha.2010.12.010. [DOI] [PubMed] [Google Scholar]
- 14.Chen Y, et al. Assessing susceptibility to age-related macular degeneration with genetic markers and environmental factors. Arch Ophthalmol. 2011;129:344–351. doi: 10.1001/archophthalmol.2011.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tamura H, et al. Association of ARMS2 genotype with bilateral involvement of exudative age-related macular degeneration. Am J Ophthalmol. 2012;154:542–548. doi: 10.1016/j.ajo.2012.03.042. [DOI] [PubMed] [Google Scholar]
- 16.Schwartz SG, et al. The ARMS2 A69S variant and bilateral advanced age-related macular degeneration. Retina. 2012;32:1486–1491. doi: 10.1097/IAE.0b013e318240a540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maguire MG, et al. Incidence of choroidal neovascularization in the fellow eye in the comparison of age-related macular degeneration treatments trials. Ophthalmology. 2013;120:2035–2041. doi: 10.1016/j.ophtha.2013.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tedeschi-Blok N, Buckley J, Varma R, Triche TJ, Hinton DR. Population-based study of early age-related macular degeneration: role of the complement factor H Y402H polymorphism in bilateral but not unilateral disease. Ophthalmology. 2007;114:99–103. doi: 10.1016/j.ophtha.2006.07.043. [DOI] [PubMed] [Google Scholar]
- 19.Pai AS, Mitchell P, Rochtchina E, Iyengar S, Wang JJ. Complement factor H and the bilaterality of age-related macular degeneration. Arch Ophthalmol. 2009;127:1339–1344. doi: 10.1001/archophthalmol.2009.239. [DOI] [PubMed] [Google Scholar]
- 20.Maritzen T, Podufall J, Haucke V. Stonins–specialized adaptors for synaptic vesicle recycling and beyond? Traffic. 2010;11:8–15. doi: 10.1111/j.1600-0854.2009.00971.x. [DOI] [PubMed] [Google Scholar]
- 21.Zeidler MP, Yokomori K, Tjian R, Mlodzik M. Drosophila TFIIA-S is up-regulated and required during Ras-mediated photoreceptor determination. Genes Dev. 1996;10:50–9. doi: 10.1101/gad.10.1.50. [DOI] [PubMed] [Google Scholar]
- 22.Feskanich D, Cho E, Schaumberg DA, Colditz GA, Hankinson SE. Menopausal and reproductive factors and risk of age-related macular degeneration. Arch Ophthalmol. 2008;126:519–524. doi: 10.1001/archopht.126.4.519. [DOI] [PubMed] [Google Scholar]
- 23.Fraser-Bell S, Wu J, Klein R, Azen SP, Varma R. Smoking, alcohol intake, estrogen use, and age-related macular degeneration in Latinos: the Los Angeles Latino Eye Study. Am J Ophthalmol. 2006;141:79–87. doi: 10.1016/j.ajo.2005.08.024. [DOI] [PubMed] [Google Scholar]
- 24.Freeman EE, Munoz B, Bressler SB, West SK. Hormone replacement therapy, reproductive factors, and age-related macular degeneration: the Salisbury Eye Evaluation Project. Ophthalmic Epidemiol. 2005;12:37–45. doi: 10.1080/09286580490907779. [DOI] [PubMed] [Google Scholar]
- 25.Boekhoorn SS, et al. Estrogen receptor alpha gene polymorphisms associated with incident aging macula disorder. Invest Ophthalmol Vis Sci. 2007;48:1012–1017. doi: 10.1167/iovs.06-0577. [DOI] [PubMed] [Google Scholar]
- 26.Bush TL, Fried LP, Barrett-Connor E. Cholesterol, lipoproteins, and coronary heart disease in women. Clin Chem. 1988;34:B60–70. [PubMed] [Google Scholar]
- 27.Stampfer MJ, Colditz GA. Estrogen replacement therapy and coronary heart disease: a quantitative assessment of the epidemiologic evidence. Prev Med. 1991;20:47–63. doi: 10.1016/0091-7435(91)90006-P. [DOI] [PubMed] [Google Scholar]
- 28.Kavanagh K, et al. Estrogen decreases atherosclerosis in part by reducing hepatic acyl-CoA:cholesterol acyltransferase 2 (ACAT2) in monkeys. Arterioscler Thromb Vasc Biol. 2009;29:1471–1477. doi: 10.1161/ATVBAHA.109.191825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dukic-Stefanovic S, et al. Chorionic gonadotropin and its receptor are both expressed in human retina, possible implications in normal and pathological conditions. PLoS One. 2012;7:e52567. doi: 10.1371/journal.pone.0052567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu XD, et al. Long non-coding RNAs: new players in ocular neovascularization. Mol Biol Rep. 2014;41:4493–505. doi: 10.1007/s11033-014-3320-5. [DOI] [PubMed] [Google Scholar]
- 31.Petit MM, et al. LHFP, a novel translocation partner gene of HMGIC in a lipoma, is a member of a new family of LHFP-like genes. Genomics. 1999;57:438–41. doi: 10.1006/geno.1999.5778. [DOI] [PubMed] [Google Scholar]
- 32.Nagaishi M, et al. Amplification of the STOML3, FREM2, and LHFP genes is associated with mesenchymal differentiation in gliosarcoma. Am J Pathol. 2012;180:1816–1823. doi: 10.1016/j.ajpath.2012.01.027. [DOI] [PubMed] [Google Scholar]
- 33.Hirasawa M, et al. Transcriptional factors associated with epithelial-mesenchymal transition in choroidal neovascularization. Mol Vis. 2011;17:1222–1230. [PMC free article] [PubMed] [Google Scholar]
- 34.Ishikawa, K., Kannan, R. & Hinton, D. R. Molecular mechanisms of subretinal fibrosis in age-related macular degeneration. Exp Eye Res. (2015). [DOI] [PMC free article] [PubMed]
- 35.Melville SA, et al. Multiple loci influencing hippocampal degeneration identified by genome scan. Ann Neurol. 2012;72:65–75. doi: 10.1002/ana.23644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ohno-Matsui K. Parallel findings in age-related macular degeneration and Alzheimer’s disease. Prog Retin Eye Res. 2011;30:217–238. doi: 10.1016/j.preteyeres.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 37.Rofagha S, Bhisitkul RB, Boyer DS, Sadda SR, Zhang K. Seven-year outcomes in ranibizumab-treated patients in ANCHOR, MARINA, and HORIZON: a multicenter cohort study (SEVEN-UP) Ophthalmology. 2013;120:2292–2299. doi: 10.1016/j.ophtha.2013.03.046. [DOI] [PubMed] [Google Scholar]
- 38.Coscas G, et al. Comparison of exudative age-related macular degeneration subtypes in Japanese and French Patients: multicenter diagnosis with multimodal imaging. Am J Ophthalmol. 2014;158:309–318. doi: 10.1016/j.ajo.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 39.Nakata I, et al. Calcium, ARMS2 genotype, and Chlamydia pneumoniae infection in early age-related macular degeneration: a multivariate analysis from the Nagahama study. Sci Rep. 2015;5:9345. doi: 10.1038/srep09345. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


