Abstract
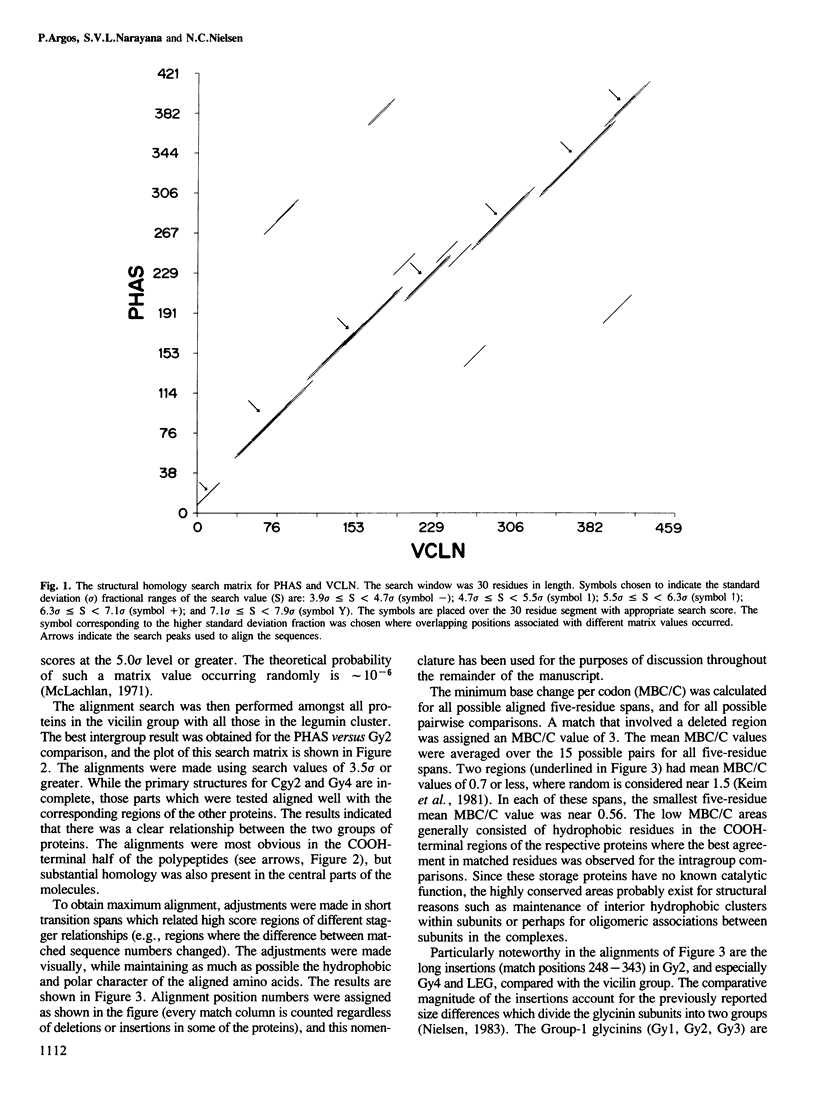
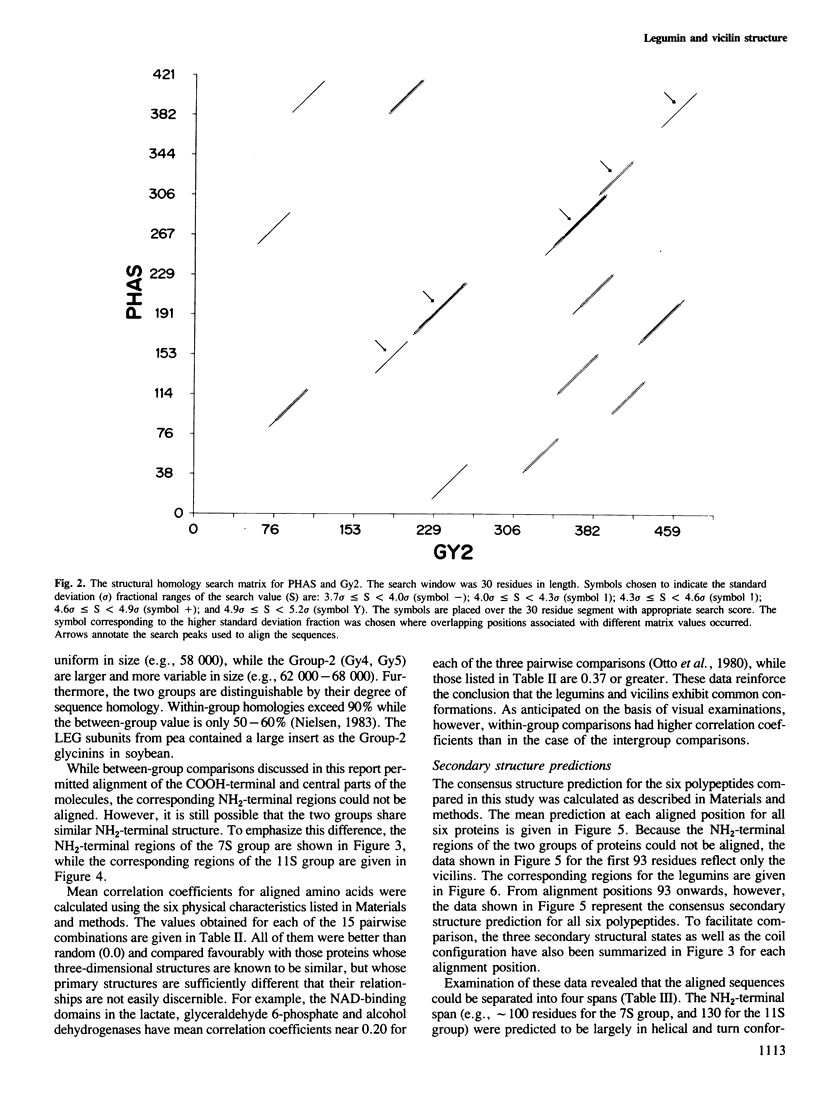
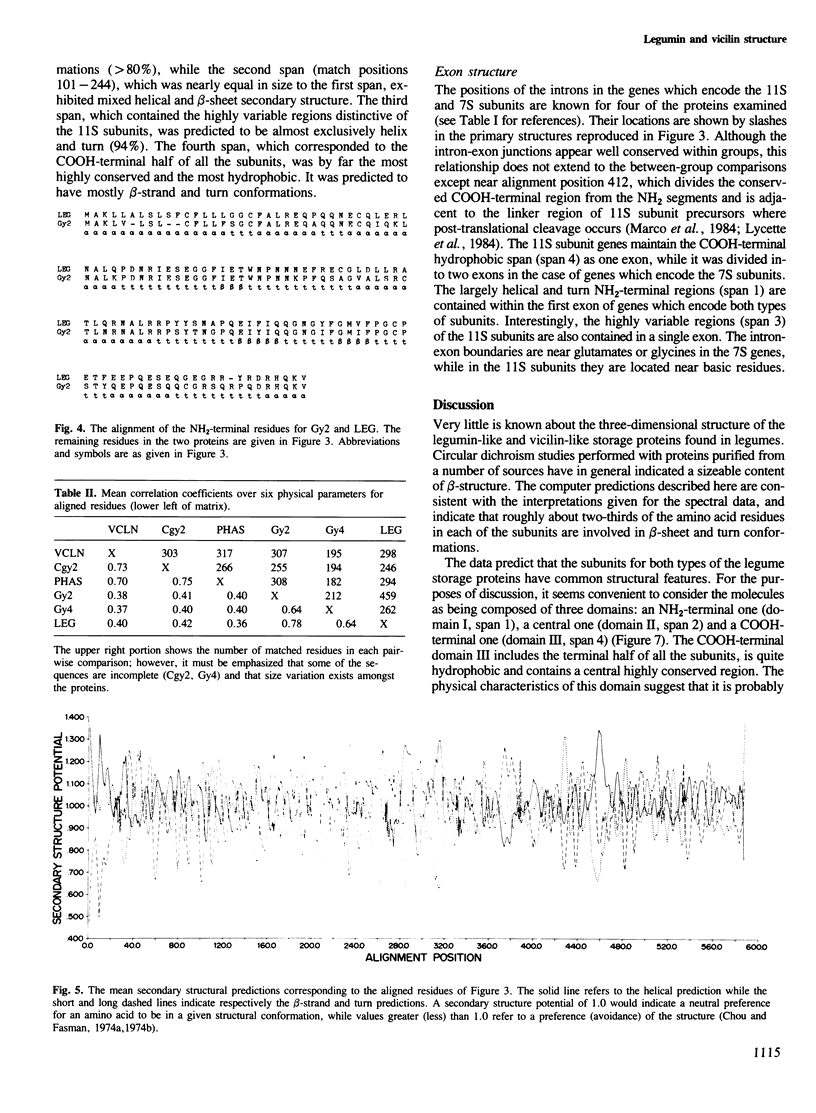
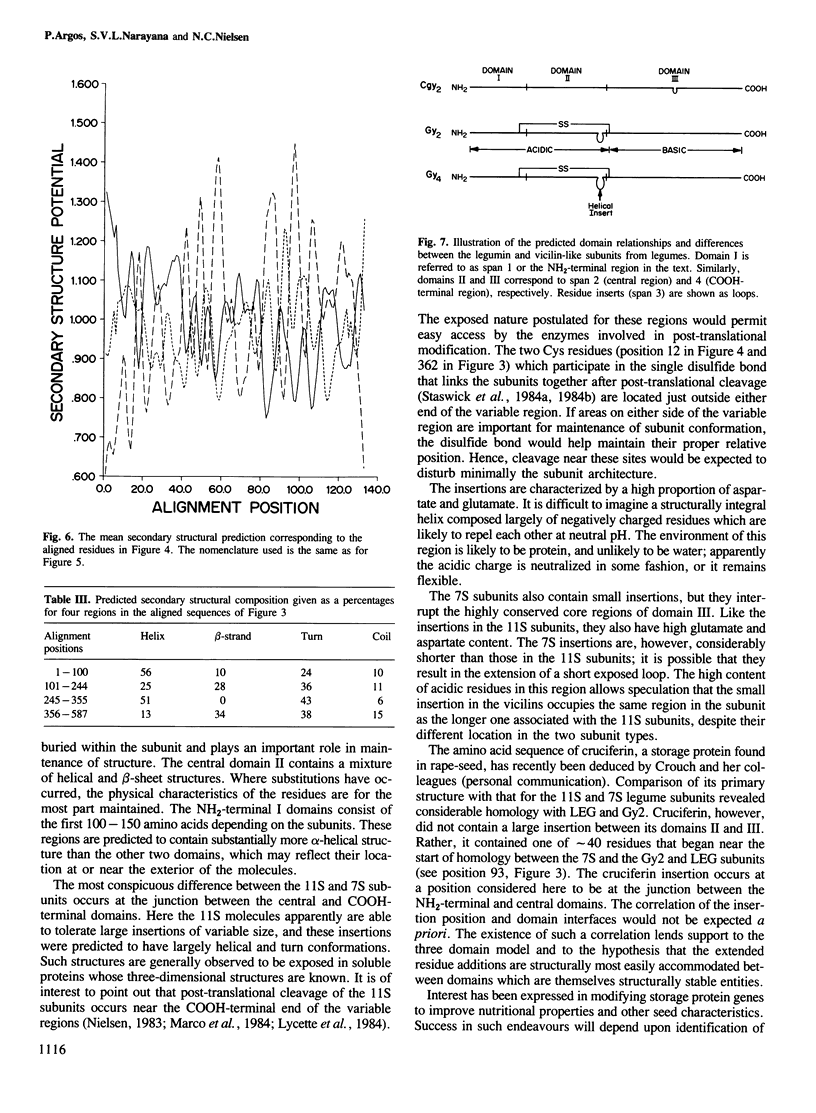
The primary structures for several members of both the vicilin and legumin families of storage proteins were examined using a computer routine based on amino acid physical characteristics. The comparison algorithm revealed that sequences from the two families could be aligned and share a number of predicted secondary structural features. The COOH-terminal half of the subunits in both families displayed a highly conserved core region that was largely hydrophobic and in which a high proportion of the residues were predicted to be in beta-sheet conformations. The central region of the molecules which contained mixed areas of predicted helical and sheet conformations showed more variability in residue selection than the COOH-terminal regions. The NH2-terminal segments of subunits from the two different families could not be aligned though they characteristically had a high proportion of residues predicted to be in helical conformations. The feature which most clearly distinguished subunits between the two families was an inserted span in the legumin group with a high proportion of acidic amino acids located between the central and COOH-terminal domains. Residues in this insertion were predicted to exist mainly in helical conformation. Since considerable size variation occurs in this area amongst the legumin subunits, alterations in this region may have a minimal detrimental effect on the structure of the proteins.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Argos P., Hanei M., Wilson J. M., Kelley W. N. A possible nucleotide-binding domain in the tertiary fold of phosphoribosyltransferases. J Biol Chem. 1983 May 25;258(10):6450–6457. [PubMed] [Google Scholar]
- Argos P., Palau J. Amino acid distribution in protein secondary structures. Int J Pept Protein Res. 1982 Apr;19(4):380–393. doi: 10.1111/j.1399-3011.1982.tb02619.x. [DOI] [PubMed] [Google Scholar]
- Argos P., Schwarz J., Schwarz J. An assessment of protein secondary structure prediction methods based on amino acid sequence. Biochim Biophys Acta. 1976 Aug 9;439(2):261–273. doi: 10.1016/0005-2795(76)90062-3. [DOI] [PubMed] [Google Scholar]
- Argos P., Siezen R. J. Structural homology of lens crystallins. A method to detect protein structural homology from primary sequences. Eur J Biochem. 1983 Mar 1;131(1):143–148. doi: 10.1111/j.1432-1033.1983.tb07241.x. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Conformational parameters for amino acids in helical, beta-sheet, and random coil regions calculated from proteins. Biochemistry. 1974 Jan 15;13(2):211–222. doi: 10.1021/bi00699a001. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Prediction of protein conformation. Biochemistry. 1974 Jan 15;13(2):222–245. doi: 10.1021/bi00699a002. [DOI] [PubMed] [Google Scholar]
- Creighton T. E. Experimental studies of protein folding and unfolding. Prog Biophys Mol Biol. 1978;33(3):231–297. doi: 10.1016/0079-6107(79)90030-0. [DOI] [PubMed] [Google Scholar]
- Keim P., Heinrikson R. L., Fitch W. M. An examination of the expected degree of sequence similarity that might arise in proteins that have converged to similar conformational states. The impact of such expectations on the search for homology between the structurally similar domains of rhodanese. J Mol Biol. 1981 Sep 5;151(1):179–197. doi: 10.1016/0022-2836(81)90227-8. [DOI] [PubMed] [Google Scholar]
- Kubota Y., Nishikawa K., Takahashi S., Ooi T. Correspondence of homologies in amino acid sequence and tertiary structure of protein molecules. Biochim Biophys Acta. 1982 Feb 18;701(2):242–252. doi: 10.1016/0167-4838(82)90120-0. [DOI] [PubMed] [Google Scholar]
- Kubota Y., Takahashi S., Nishikawa K., Ooi T. Homology in protein sequences expressed by correlation coefficients. J Theor Biol. 1981 Jul 21;91(2):347–361. doi: 10.1016/0022-5193(81)90237-x. [DOI] [PubMed] [Google Scholar]
- Lycett G. W., Croy R. R., Shirsat A. H., Boulter D. The complete nucleotide sequence of a legumin gene from pea (Pisum sativum L.). Nucleic Acids Res. 1984 Jun 11;12(11):4493–4506. doi: 10.1093/nar/12.11.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lycett G. W., Delauney A. J., Gatehouse J. A., Gilroy J., Croy R. R., Boulter D. The vicilin gene family of pea (Pisum sativum L.): a complete cDNA coding sequence for preprovicilin. Nucleic Acids Res. 1983 Apr 25;11(8):2367–2380. doi: 10.1093/nar/11.8.2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marco Y. A., Thanh V. H., Tumer N. E., Scallon B. J., Nielsen N. C. Cloning and structural analysis of DNA encoding an A2B1a subunit of glycinin. J Biol Chem. 1984 Nov 10;259(21):13436–13441. [PubMed] [Google Scholar]
- McLachlan A. D. Tests for comparing related amino-acid sequences. Cytochrome c and cytochrome c 551 . J Mol Biol. 1971 Oct 28;61(2):409–424. doi: 10.1016/0022-2836(71)90390-1. [DOI] [PubMed] [Google Scholar]
- Otto J., Argos P., Rossmann M. G. Prediction of secondary structural elements in glycerol-3-phosphate dehydrogenase by comparison with other dehydrogenases. Eur J Biochem. 1980 Aug;109(2):325–330. doi: 10.1111/j.1432-1033.1980.tb04798.x. [DOI] [PubMed] [Google Scholar]
- Schuler M. A., Ladin B. F., Pollaco J. C., Freyer G., Beachy R. N. Structural sequences are conserved in the genes coding for the alpha, alpha' and beta-subunits of the soybean 7S seed storage protein. Nucleic Acids Res. 1982 Dec 20;10(24):8245–8261. doi: 10.1093/nar/10.24.8245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slightom J. L., Sun S. M., Hall T. C. Complete nucleotide sequence of a French bean storage protein gene: Phaseolin. Proc Natl Acad Sci U S A. 1983 Apr;80(7):1897–1901. doi: 10.1073/pnas.80.7.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. An interactive graphics program for comparing and aligning nucleic acid and amino acid sequences. Nucleic Acids Res. 1982 May 11;10(9):2951–2961. doi: 10.1093/nar/10.9.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. An interactive graphics program for comparing and aligning nucleic acid and amino acid sequences. Nucleic Acids Res. 1982 May 11;10(9):2951–2961. doi: 10.1093/nar/10.9.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick P. E., Hermodson M. A., Nielsen N. C. Identification of the cystines which link the acidic and basic components of the glycinin subunits. J Biol Chem. 1984 Nov 10;259(21):13431–13435. [PubMed] [Google Scholar]
- Staswick P. E., Hermodson M. A., Nielsen N. C. The amino acid sequence of the A2B1a subunit of glycinin. J Biol Chem. 1984 Nov 10;259(21):13424–13430. [PubMed] [Google Scholar]


