Abstract
Small subunit ribosomal DNA (SSU rDNA) is widely used for phylogenetic inference, barcoding and other taxonomy-based analyses. Recent studies indicate that SSU rDNA of ciliates may have a high level of sequence variation within a single cell, which impacts the interpretation of rDNA-based surveys. However, sequence variation can come from a variety of sources including experimental errors, especially the mutations generated by DNA polymerase in PCR. In the present study, we explore the impact of four DNA polymerases on sequence variation and find that low-fidelity polymerases exaggerate the estimates of single-cell sequence variation. Therefore, using a polymerase with high fidelity is essential for surveys of sequence variation. Another source of variation results from errors during amplification of SSU rDNA within the polyploidy somatic macronuclei of ciliates. To investigate further the impact of SSU rDNA copy number variation, we use a high-fidelity polymerase to examine the intra-individual SSU rDNA polymorphism in ciliates with varying levels of macronuclear amplification: Halteria grandinella, Blepharisma americanum and Strombidium stylifer. We estimate the rDNA copy numbers of these three species by single-cell quantitative PCR. The results indicate that: (i) sequence variation of SSU rDNA within a single cell is authentic in ciliates, but the level of intra-individual SSU rDNA polymorphism varies greatly among species; (ii) rDNA copy numbers vary greatly among species, even those within the same class; (iii) the average rDNA copy number of Halteria grandinella is about 567 893 (s.d. = 165 481), which is the highest record of rDNA copy number in ciliates to date; and (iv) based on our data and the records from previous studies, it is not always true in ciliates that rDNA copy numbers are positively correlated with cell or genome size.
Keywords: ciliophora, polymerase fidelity, rDNA polymorphism, rDNA copy number, quantitative polymerase chain reaction
1. Introduction
The nuclear ribosomal DNA (rDNA) locus, which includes the small subunit (SSU) rDNA, the large subunit (LSU) rDNA, the 5.8S rDNA and the internal transcribed spacers (ITS1 and ITS2), is a useful marker for comparisons of organisms from a range of taxonomic levels [1,2]. It has been widely used for phylogenetic inference and barcoding technology of eukaryotic microbes [3–9]. In particular, the SSU rDNA is a universal marker for phylogenetic analyses, as well as identifications and classifications of microbes [10–12]. Moreover, rDNA-based barcoding and high-throughput environmental sequencing have become the mainstream approaches to address fundamental questions of microbial diversity, ecology and biogeography [13–15].
The rDNA copy number of a broad range of eukaryotes is highly variable, and extrachromosomal copies are often generated in eukaryotic species [16]. In animals and plants, the rDNA copy number ranges are 39–19 300 and 150–26 048, respectively [17], while in fungi estimates are from 60 to 220 [18]. In eukaryotic microbes, rDNA copy numbers range from 61 to 36 896 in dinoflagellates, and 200 to 12 812 in diatoms [19,20]. Ciliates' extensive processing of the germline rDNA locus yields many extrachromosomal copies in somatic macronuclei [21]. In the class Spirotrichea, estimates are 100 000 rDNA copies per macronucleus in Oxytricha nova and 200 000 in Stylonychia lemnae [21,22]. Other estimates are approximately 316 000 rDNA copies in Vorticella sp. (CI: Oligohymenophorea) [23] and 59 000 to 80 000 in Chilodonella uncinata (CI: Phyllopharyngea) [24].
Numerous studies indicate both intra-specific (among the individuals of a given species) and intra-individual (among the copies of a given individual) variability in rDNA sequences [25–28]. Intra-specific variability of rDNA is documented in a range of organisms, including pinyon pine [29], dinoflagellates [30–32] and diatoms [33]. Intra-individual variation has been detected in some fungal species [34] and dinoflagellates [35] using PCR amplification, cloning and sequencing approaches. Ciliates have also been argued to have intra-specific and intra-individual rDNA variation [23,36–38]. For example, the intra-specific SSU rDNA variation can reach up to 1.6% in Gastrostyla pulchra (CI: Spirotrichea) between the marine and estuarine strains [37], and the intra-individual ITS variation is estimated to be as high as 0.96% in a Vorticella species [23]. However, sequence variation can be biological (i.e. generated through DNA amplification and replication during the life cycle of the organism) or experimental errors (i.e. mutations introduced by polymerase during PCR amplification).
To explore the sources of high intra-individual polymorphism in ciliates, we assess the impacts of four polymerases on the sequence variation. Based on these findings, we then use a high-fidelity polymerase to investigate the intra-individual polymorphism of three ciliate species: Blepharisma americanum (CI: Heterotrichea), Halteria grandinella and Strombidium stylifer (CI: Spirotrichea; figure 1). We also assess the rDNA copy number within a single cell of these species using quantitative PCR (qPCR) to examine the relationship between polymorphism and rDNA copy number.
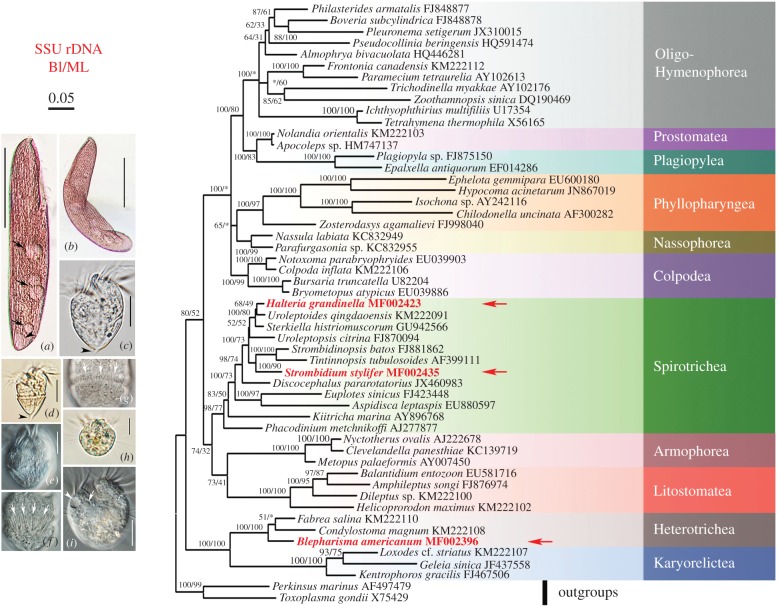
Figure 1.
Phylogeny and morphology of ciliates, highlighting target taxa Blepharisma americanum (a,b), Strombidium stylifer (c–g) and Halteria grandinella (h,i). The phylogenetic tree based on small subunit ribosomal RNA gene sequences shows the positions of the three focal taxa: B. americanum, S. stylifer and H. grandinella. Asterisk indicates the disagreement in topology of BI and ML trees. The scale bar corresponds to five substitutions per 100 nucleotide positions. (a) ventral view of a representative of B. americanum; arrows mark the food vacuoles and arrowhead points out the contractile vacuole. (b) Photograph of a bending B. americanum, to show the flexibility of the body. (c–g) Photographs of S. stylifer; arrowheads in (c) and (d) indicate the apparent tail, and arrows in (f) and (g) indicate the extrusomes of ventral and dorsal sides, respectively. (h,i) Pictures of H. grandinella; arrow and arrowhead in (i) mark the contractile vacuole and micronucleus, respectively. Scale bars, 70 µm (a,b); 15 µm (c–g); and 10 µm (h,i). (Online version in colour.)
2. Material and methods
(a). Ciliate culture and identification
In October 2014, we collected Halteria grandinella and Strombidium stylifer from a pond of Baihuayuan Park (36°04′ N, 120°22′ E) and from Golden Beach (35°58′ N, 120°15′ E) in Qingdao, China, respectively. We isolated Blepharisma americanum from Yangtze River in Chongqing, China (29°36′ N, 106°59′ E) in September 2014. All the three species were picked up with a micropipette from water samples and cultured at room temperature (25°C) in filtered and autoclaved water taken from each site, with rice grains added to enrich bacterial food. We determined species identity by observation of living morphology and protargol impregnation method [39].
(b). DNA extraction
We washed a mid-sized single cell with filtered and autoclaved water five times and then transferred it to a 1.5 ml Eppendorf tube with about 0.5 µl water. Genomic DNA was isolated using Extraction Solution, Tissue Preparation Solution, and Neutralization Solution B in REDExtract-N-Amp Tissue PCR Kit (Sigma, St. Louis, MO) following the manufacturer's protocol, which we modified by using only 1/10 of suggested volume for each solution. The final volume of the solution was about 23 µl. We sampled three cells for each morphospecies.
(c). Fidelity verification test of four DNA polymerases
We amplified the full length SSU rDNA of B. americanum with universal primers [40] using PfuTurbo DNA polymerase (Agilent Technologies, USA). PCR products were purified by EasyPure PCR Purification Kit (Transgen Biotech, China), and then cloned using pEASY-T1 Cloning Kit (Transgen Biotech, China). One clone was picked randomly and cultured in LB broth medium for 15 h to extract the plasmid using Sanprep Plasmid Miniprep Kit (Sangon Biotech, Shanghai). Afterwards, PfuTurbo DNA polymerase (Cat. #600250, Agilent Technologies, USA), Q5 Hot Start High-Fidelity DNA Polymerase (Cat. #M0493 L, New England Biolabs, USA), ExTaq DNA polymerase (Cat. #RR001A & #RR003A, TaKaRa, Japan) and Taq DNA Polymerase (Cat. #EP0402, Thermo Fisher Scientific, USA) were used to amplify the SSU rDNA in the plasmid with universal primers [40]. PCR and cloning were performed as described above. The SSU rDNA in the plasmid was sequenced bidirectionally both in GENEWIZ Incorporated Company (Beijing, China) and Shanghai Sunny Biotechnology Company (Shanghai, China) to reduce the impact of errors caused by sequencing. For each polymerase, we sequenced 20 clones at the Shanghai Sunny Biotechnology Company and also sequenced four of these at the GENEWIZ Incorporated Company. The sequencing data from the two companies are identical, which indicates that no error was introduced by sequencing.
(d). DNA polymorphism and nucleotide diversity
The full length SSU rDNA of B. americanum was amplified by Q5 and ExTaq DNA polymerases. The SSU rDNA of H. grandinella and S. stylifer was amplified using Q5 DNA polymerase. PCR and cloning were performed as described above. For each individual and polymerase, 20 to 25 clones were sequenced at the GENEWIZ Incorporated Company.
Contigs were assembled by SeqMan (DNAStar) and chromatograms were inspected individually to confirm that polymorphisms were indeed real. Sequences were aligned using BioEdit v. 7.0.1 to identify the polymorphic sites [41]. MEGA v. 6.06 was used to calculate pairwise distance [42]. We calculated the number of polymorphic sites, haplotype diversity (Hd) and nucleotide diversity (π) using DnaSP v. 5.10 [43]. Pearson's correlation analyses were calculated by SPSS v. 18.0 with default parameters [44].
(e). Quantitative real-time PCR assays
The plasmids containing the SSU rDNA of B. americanum, H. grandinella and S. stylifer were constructed according to the procedures described above and used as standards for qPCR assays, respectively. We used serial 10-fold dilutions (10−1 to 10−7) to obtain standard curves. The concentrations of plasmids were measured by Qubit 3.0 (Invitrogen, USA). In order to avoid contaminations, specific primers were designed in variable regions of SSU rDNA for each species (electronic supplementary material, table S2).
Reactions were performed using EvaGreen qPCR MasterMix–Low Rox (Applied Biological Materials Inc., Canada) in a final volume of 25 µl containing 12.5 µl 2 × qPCR mix, 0.5 µM of each primer, 1 µl of total genomic DNA and 6.5 µl of tri-distilled and autoclaved water. All reactions were performed in triplicate with an ABI 7500 Fast Real-Time PCR System (Applied Biosystems). The PCR programme started with an initial soaking step at 50°C for 2 min and 98°C for 2 min; followed by 40 cycles of denaturation at 98°C for 10 s, annealing at 55°C for 10 s, and extension at 68°C for 30 s; and finally a melting curve stage (preprogramed in system as following: 95°C for 15 s, 60°C for 1 min, 95°C for 30 s and 60°C for 15 s). The number of molecules in the standards was calculated using the website http://cels.uri.edu/gsc/cndna.html [45]. The efficiency of amplification (E) was calculated as E = (10−1/k − 1) × 100%, where k is the slope of standard curve. As we used only 1 µl out of 23 µl total genomic DNA in the qPCR, the final copy number for each individual was multiplied by 23.
(f). Phylogenetic analyses
Phylogenetic analyses include three most common SSU rDNA sequences of H. grandinella, B. americanum and S. stylifer, and 50 sequences downloaded from NCBI GenBank database (accession numbers as shown in figure 1). All sequences were aligned using the GUIDANCE2 Server [46] with default settings and further modified manually using BioEdit v. 7.0.1 [41]. Maximum-likelihood (ML) analyses were performed in CIPRES Science Gateway using RAxML-HPC2 on XSEDE v. 8.1.24 [47] with the model of GTRGAMMA + I. The reliability of internal branches was assessed using a non-parametric bootstrap method with 1000 replicates. Bayesian inference (BI) analysis was carried out using MrBayes on XSEDE v. 3.2.6 with the model GTR + I + G (selected by MrModeltest v. 2.0 [48]) in CIPRES Science Gateway. Markov chain Monte Carlo simulations were run with two sets of four chains for 6 000 000 generations with a frequency of 100 generations, and 25% were discarded as burn-in. MEGA v. 6.06 [42] was used to visualize tree topologies.
The SSU rRNA sequence of H. grandinella was selected as an example to predict the secondary structure following the previous model of Tetrahymena canadensis (M26359, http://rrna.uia.ac.be) using Mfold (http://unafold.rna.albany.edu/?q=mfold) with default parameters [49]. Rnaviz v. 2.0.0 was used for aesthetic adjustment [50].
3. Results
(a). The impact of varying polymerases on estimates of sequence variation
In order to test the impact of varying DNA polymerases on estimates of rDNA diversity, we used four DNA polymerases (PfuTurbo DNA polymerase, Q5 Hot Start High-Fidelity DNA polymerase, ExTaq DNA polymerase and Taq DNA polymerase) to amplify the plasmid containing the SSU rDNA sequence of B. americanum. We sequenced 20 clones for each polymerase and found substantial variation in experimental error rates as estimated by SSU rDNA sequence variation (table 1). None of the sequences generated by the Taq and ExTaq DNA polymerases is identical to the template DNA. Compared with the template DNA, as many as eight and five substitutions per sequence are generated by Taq and ExTaq polymerase, respectively (table 1). The sequences amplified by Taq polymerase have the highest average pairwise distance of 0.466% and the most polymorphic sites of 76 (4.52% of the full length). The sequences amplified by ExTaq polymerase have an average pairwise difference of 0.280% and 46 polymorphic sites among the 20 clones (2.73%; table 1). PfuTurbo polymerase has the highest fidelity as all 20 clones generated with this polymerase are identical to the template DNA (table 1). For the Q5 polymerase, there is only one polymorphic site in one clone that differs from the template DNA (table 1).
Table 1.
Genetic distances and polymorphic sites of SSU rDNA from 20 clones generated in PCRs using four DNA polymerases. π, nucleotide diversity; Hd, haplotype diversity; max and min, the maximum and minimum value of pairwise genetic distances; n, range of polymorphic sites per sequence compared with template; p, numbers of polymorphic sites in relation to SSU rDNA length in %; s.d., standard deviation.
| polymerase | pairwise genetic distance (×10−2) |
no. polymorphic sites (p) | no. haplotypes | Hd | π (×10−2) | n | |||
|---|---|---|---|---|---|---|---|---|---|
| mean | max | min | s.d. | ||||||
| ExTaq | 0.280 | 0.538 | 0.119 | 0.099 | 46 (2.73) | 20 | 1.000 | 0.279 | 1–5 |
| Taq | 0.466 | 0.962 | 0.119 | 0.198 | 76 (4.52) | 20 | 1.000 | 0.463 | 1–8 |
| Q5 | 0.006 | 0.059 | 0 | 0.018 | 1 (0.06) | 2 | 0.100 | 0.006 | 0–1 |
| PfuTurbo | 0 | 0 | 0 | 0 | 0 (0) | 1 | 0 | 0 | 0 |
We also used ExTaq and Q5 polymerases to amplify the SSU rDNA from the three individuals of B. americanum (electronic supplementary material, table S1). The results show a substantial difference between the two polymerases as the mean pairwise genetic distance of sequences amplified by ExTaq is two to eight times higher than that amplified by Q5 polymerase (electronic supplementary material, table S1). The comparison between ExTaq and Q5 further demonstrates that the low-fidelity DNA polymerase dramatically increases the level of sequence variation by generating errors during amplification.
A t-test for equality of means of pairwise genetic distance indicates that the differences between each pair of the four DNA polymerases except for the pair of Q5 and PfuTurbo are significant (table 2). Even though PfuTurbo has high fidelity, it is more expensive and has low efficiency in PCR amplifications. Given that the fidelity of Q5 is comparable with PfuTurbo, we selected Q5 to perform the subsequent research.
Table 2.
Analyses of significant difference about the four DNA polymerases. The numbers in lower left diagonal are p values. In upper right diagonal, * means a significant difference at 95% level; ** means an extremely significant difference at 99% level.
| p-value | PfuTurbo | Q5 | ExTaq | Taq |
|---|---|---|---|---|
| PfuTurbo | — | ** | ** | |
| Q5 | 0.330 | — | ** | ** |
| ExTaq | 6.0 × 10−8 | 2.33 × 10−8 | — | * |
| Taq | 9.18 × 10−7 | 1.29 × 10−6 | 0.02 | — |
(b). Sequence variation among the three individuals of each species
To assess variation among individuals, we amplified the SSU rDNA locus of H. grandinella, B. americanum and S. stylifer using Q5 Hot Start High-Fidelity DNA polymerase and sequenced at least 30 clones per individual (table 3). Among the clones of each individual, there is one most common version of the SSU rDNA sequence, which may represent the germline micronuclear template SSU rDNA. Compared with the most common sequence, 1–5 and 1–3 polymorphic sites are detected in H. grandinella and B. americanum, respectively. Pairwise sequence comparisons within individuals reveal 0–8 and 0–4 polymorphic sites in H. grandinella and B. americanum, respectively.
Table 3.
Genetic distances, polymorphic sites and copy numbers of SSU rDNA amplified from three different cells of three species. π, nucleotide diversity; Hd, haplotype diversity; max and min, the maximum and minimum value of pairwise genetic distances; n, ranges of polymorphic sites compared with the most common sequences of each individual; p, numbers of polymorphic sites in relation to SSU rDNA length in %; s.d., standard deviation. The rDNA copy number of each individual is the mean value of three estimates.
| DNA polymerase |
Q5 Hot Start High-Fidelity |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| species | B. americanum (1683 bp) | H. grandinella (1730 bp) | S. stylifer (1729 bp) | |||||||
| individual | 1 | 2 | 3 | 1 | 2 | 3 | 1 | 2 | 3 | |
| clones | 30 | 30 | 30 | 30 | 31 | 35 | 34 | 33 | 33 | |
| pairwise genetic distance (×10−2) | mean | 0.062 | 0.025 | 0.061 | 0.123 | 0.014 | 0.059 | 0.003 | 0 | 0 |
| max | 0.238 | 0.119 | 0.179 | 0.446 | 0.116 | 0.289 | 0.003 | 0 | 0 | |
| min | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| s.d. | 0.062 | 0.032 | 0.055 | 0.085 | 0.032 | 0.065 | 0.014 | 0 | 0 | |
| no. polymorphic sites (p) | 7 (0.42) | 2 (0.12) | 6 (0.30) | 29 (1.68) | 2 (0.12) | 10 (0.58) | 1 (0.06) | 0 (0) | 0 (0) | |
| no. of haplotypes | 8 | 3 | 6 | 21 | 3 | 7 | 2 | 1 | 1 | |
| Hd | 0.623 | 0.393 | 0.669 | 0.915 | 0.185 | 0.605 | 0.059 | 0 | 0 | |
| π (×10−2) | 0.062 | 0.025 | 0.061 | 0.129 | 0.014 | 0.059 | 0.003 | 0 | 0 | |
| n | 1–3 | 1 | 1–2 | 1–5 | 1–2 | 1–4 | 1 | 0 | 0 | |
| no. common sequences (%) | 18 (60.0) | 23 (76.7) | 15 (50.0) | 9 (30.0) | 28 (90.3) | 20 (57.1) | 33 (97.1) | 33 (100.0) | 33 (100.0) | |
| rDNA copy number | 134 852 | 105 313 | 9984 | 705 287 | 335 128 | 663 265 | 4596 | 1082 | 16 995 | |
| s.d. of rDNA copies | 23 042 | 6787 | 4884 | 35 116 | 14 089 | 20 920 | 334 | 31 | 267 | |
We also counted the number of polymorphic sites in our data to estimate variation within and among species. Only one polymorphic site is present among the three S. stylifer cells, while we observed 14 polymorphic sites in B. americanum and 41 polymorphic sites in H. grandinella. In the cell Hal-1, we found 29 polymorphic sites (1.68%), representing the highest level of polymorphism among all the examined individuals. The numbers of polymorphic sites vary among cells of H. grandinella, ranging from 2 (0.12%) to 29 (1.68%). For example, we find 21 unique sequences among 30 clones in Hal-1, with the haplotype diversity of 0.915; this number is about seven times higher than that in Hal-2, which has three unique sequences among 30 clones with the haplotype diversity of 0.185.
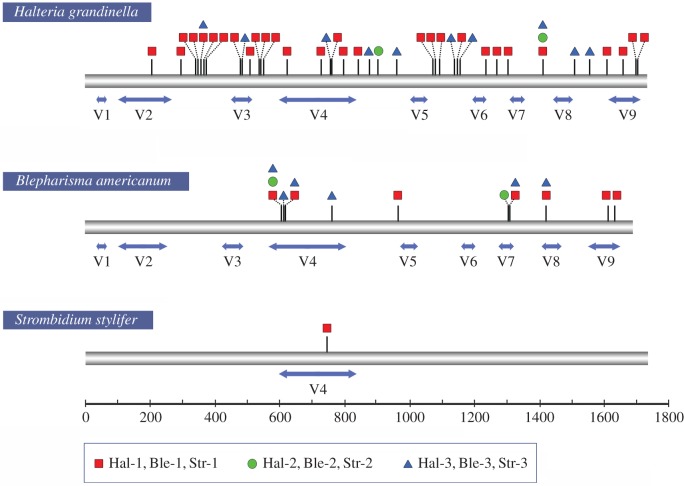
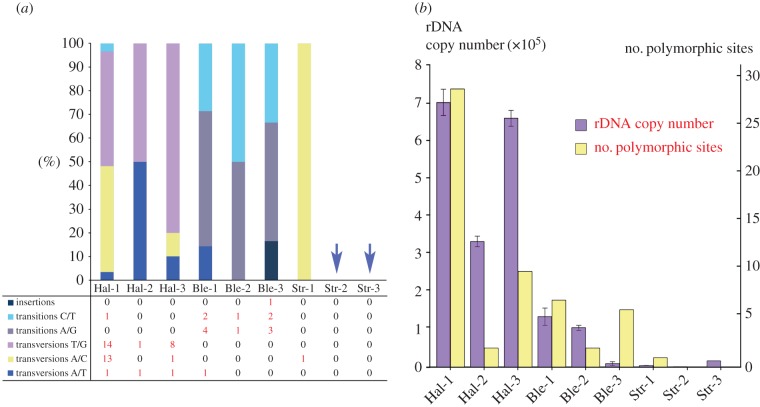
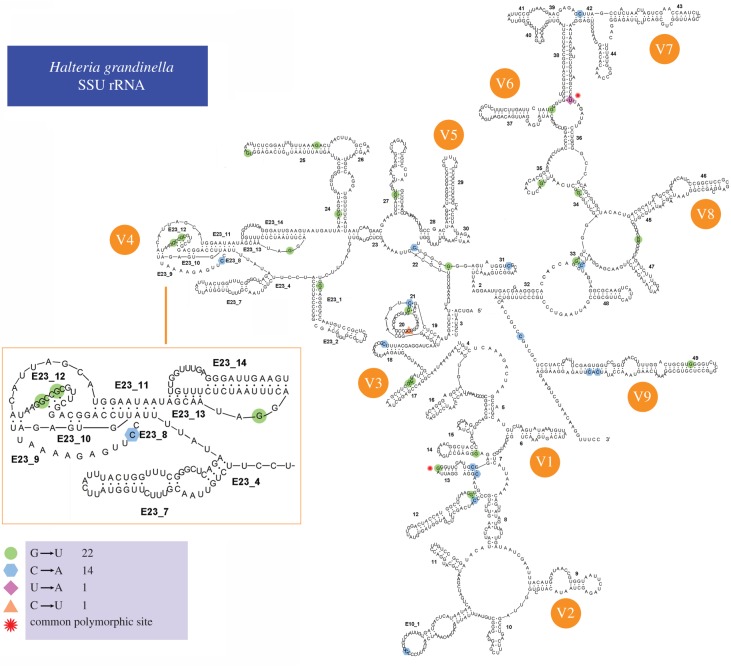
Variation among cloned rDNA sequences is primarily due to single nucleotide polymorphisms (SNPs) (figure 2). In total, 57 polymorphic sites are detected in all the nine individuals, caused by transitions, transversions or insertions (figure 3a). We detect a total of six common polymorphic sites in H. grandinella and B. americanum that are shared by more than one individual (two in H. grandinella and four in B. americanum). Taking H. grandinella as an example, polymorphic sites from the three individuals are mapped in the predicted secondary structure, which shows that polymorphisms are found in both stems and loops (figure 4).
Figure 2.
Distribution of polymorphic sites in the SSU rDNA of Halteria grandinella, Blepharisma americanum and Strombidium stylifer amplified using Q5 Hot Start High-Fidelity DNA polymerase. Polymorphic sites are indicated by short vertical lines. The squares, circles and triangles represent the three individuals of each species. The two-way arrows indicate the hypervariable regions of SSU rDNA and the lengths of sequences are to scale. Hal, Halteria grandinella; Ble, Blepharisma americanum; Str, Strombidium stylifer. (Online version in colour.)
Figure 3.
Polymorphic sites and rDNA copy numbers of Halteria grandinella, Blepharisma americanum and Strombidium stylifer. (a) Proportion of insertions, transitions and transversions of each individual. Transitions are further split into substitutions between C and T and between A and G. Transversions are split into substitutions between T and G, A and C, and A and T. (b) Graph of rDNA copy numbers and the number of polymorphic sites. The left vertical axis is copy number and the right is the number of polymorphic sites. The left columns represent copy numbers and the right columns indicate the number of polymorphic sites. Hal, Halteria grandinella; Ble, Blepharisma americanum; Str, Strombidium stylifer. (Online version in colour.)
Figure 4.
Position of polymorphic sites in secondary structure of SSU rRNA of Halteria grandinella. The polymorphic sites of the three individuals are shown in four different colours and shapes. The star indicate the positions of shared polymorphic sites among the individuals. (Online version in colour.)
(c). rDNA copy number per cell
We estimated rDNA copy number per cell using qPCR analyses of genomic DNA extracted from single cell. For the DNA extraction kit, there is no washing or filtering step involved in the procedure, so it can be assumed that there is no loss of genomic DNA. The linear relationships obtained between the cycle threshold and rDNA copy number are shown in electronic supplementary material, figure S1.
Based on the standard curves and the CT values of each single cell, we estimated the rDNA copies per cell and found that rDNA copies vary greatly among species (table 3). The rDNA copy number in H. grandinella is extremely high, with an average of 567 893 (s.d. = 165 482), while it is 83 383 (s.d. = 53 284) in B. americanum and only 7558 (s.d. = 6826) in S. stylifer. The highest copy number is found in Hal-1, with 705 287 ± 35 116 copies per cell, and the lowest is found in Str-2 (1082 ± 31). Within species, rDNA copy numbers vary among individuals. For example, the rDNA copies among the three individuals differ by approximately 13-fold in B. americanum and approximately 15-fold in S. stylifer.
(d). Correlations between rDNA copy number and SSU rDNA polymorphism
We used Pearson's correlation analysis to access the relationship between SSU rDNA sequence variation and copy number. The results indicate that both nucleotide diversity (r = 0.708, p = 0.033) and polymorphic site number (r = 0.799, p = 0.010) are positively correlated with rDNA copy number, and the correlations are significant (p < 0.05; figure 3b; electronic supplementary material, figure S2).
4. Discussion
(a). The fidelity of polymerase influences estimates of sequence variation
Understanding the sources of sequence variation is critical to biological research. For example, evolutionary relationships between different species are estimated in phylogenetic trees based on sequence variation [51], and variation is the basis for interpreting results of high-throughput environmental sequencing approaches used to estimate biodiversity [52]. However, sequence variation can be generated by both the biology of the system and experimental errors. Consistent with previous studies, we find that levels of sequence variation in PCR amplifications are impacted by the fidelity of DNA polymerase [53–55], with Taq polymerase leading to a higher level of variation than Q5 and PfuTurbo polymerases (table 1). The substantial differences in variation estimated from SSU rDNA sequences amplified by Q5 and ExTaq polymerases in B. americanum further confirm that the polymerase with high fidelity is essential in studies of genetic variation to avoid inaccurate taxon identification (barcoding) and phylogenetic reconstruction.
(b). Intra-individual rDNA polymorphism in ciliates
Intra-individual rDNA polymorphism exists in a wide range of organisms [34,35,56–59]. In oligotrichous (CI: Spirotrichea) and peritrichous ciliates (CI: Oligohymenophorea), high intra-individual polymorphisms of rDNA were reported with the highest pairwise genetic distance of 0.96% in the ITS region of a Vorticella species [23]. However, the polymerase used in their research is Taq polymerase, which could generate an amount of misreading sites in PCR and exaggerate the sequence variation. Using Q5 Hot Start High-Fidelity polymerase, our results reveal the existence of intra-individual SSU rDNA polymorphism in ciliates, but the level of SSU rDNA polymorphism is lower (table 3).
The level of intra-individual SSU rDNA polymorphism varies greatly among the three ciliate species we studied and is positively correlated with the rDNA copy number, which is consistent with previous studies [23]. The high rDNA copy number increases the probability of mutations as DNA is replicated during the life cycle of the organisms. The rDNA copy number of H. grandinella is about seven times higher than that of B. americanum and 75 times higher than that of S. stylifer, increasing the probability of mutation immensely. The varying levels of intra-individual polymorphism may also reflect the age of macronucleus. Following sexual conjugation, macronucleus rDNA are amplified from the zygotic template during macronuclear development [60,61]. In vegetative growth, the macronucleus divides through amitosis, which can allow the accumulation of mutations [21]. Therefore, more polymorphic sites may reflect the age, or time since conjugation, of a macronucleus.
(c). rDNA copy number in ciliates
The rDNA copy number can be extremely high in ciliates. Before this study, the highest record of rDNA copy number in ciliates is about 316 000 in Vorticella sp. [23]. In the present study, the average rDNA copy number of H. grandinella is 567 893 (n = 3, s.d. = 165 482), higher than any data reported previously in ciliates. However, rDNA copy numbers vary greatly within and between species, even when they fall within the same class (figure 1). For example, the peritrichs (e.g. Vorticella, Epistylis, Zoothamnium, etc.) have rDNA copies ranging from 3385 to 315 786, and the rDNA copy numbers in oligotrichs (CI: Spirotrichea) range from 30 247 to 172 889 [23]. In this study, both H. grandinella and S. stylifer are assigned as oligotrichs based on morphological data, but their rDNA copy numbers are 567 893 (s.d. = 165 482) and 7558 (s.d. = 6826), respectively.
The rDNA copy numbers vary greatly not only among species, but also among individuals within species. For example, the rDNA copy numbers among the three individuals of B. americanum differ over 13-fold, and they differ over 15-fold in S. stylifer. The high level of divergence may reflect that individuals are in different stages of growth or under different nutritional conditions, as reported in T. pyriformis [10,21]. Other explanations include the accumulation of mutations in ageing somatic macronuclei and the presence of unidentified cryptic species. The imprecise distribution of chromosomes following amitosis of macronuclei may also influence copy number [16,62]. The substantial variation of rDNA copy number among individuals of the same morphospecies may also reveal the presence of unidentified cryptic species. In C. uncinata, different cryptic species have 59 000 to 80 000 copies of rDNA [24].
Copy number of rDNA is suggested to be positively correlated with cell size and biovolume [20,63]. However, it is not always true in ciliates based on the data we generated combined with previously published estimates. For example, the rDNA copy number of H. grandinella is about seven times higher than that of B. americanum even though cells of B. americanum are much bigger than H. grandinella (180–260 × 60–130 µm versus 24–36 × 22–32 µm) [64,65]. Furthermore, even though the cell size and biovolume of S. stylifer (40–70 × 20–45 µm) are comparable with that of H. grandinella, the rDNA copy number of H. grandinella is about 80 times higher than that of S. stylifer (table 3) [66].
The rDNA copy number is positively correlated with genome size in eukaryotes but not in ciliates [17]. Ciliates have two distinct nuclei within each cell: the germline micronucleus and the highly processed (i.e. chromosomal fragmentation, DNA elimination, and DNA amplification) somatic macronucleus [21]. Therefore, the genome size of the micronucleus and macronucleus could be largely different, not only within but also among species [21,67–69]. As macronuclei are transcriptionally active, contributing virtually all expression during vegetative growth, one may argue that the macronucleus genome size should be counted. However, the macronucleus genome size of Tetrahymena thermophila is about 100 MB, with the rDNA copy number about 9000, while Oxytricha and Stylonychia have much higher rDNA copy numbers, but their macronucleus genome sizes are only about 50 MB [70–72]. Even though macronucleus genome sizes of ciliates range from 50 MB to 105 MB, which are smaller than most animals and plants [17,73,74], rDNA copy number in ciliates is much higher than that in animals and plants [23]. The extremely high copy number of rDNA in ciliates might be an advantage that allows rapid adaptation to changing environments through fast synthesis of proteins [23]. On the other hand, the gene copy number does not always correlate with its expression level, which means that DNA with high copy number may be expressed at low level [24,75,76]. Further studies of single cells isolated under varying conditions will allow these issues to be disentangled.
(d). Ecological implications
High-throughput sequencing has been applied to investigate the microbial diversity in a wide variety of systems, including deep marine waters, lakes, soils and marine sediments [77–79]. SSU rDNA is a universal marker for environmental surveys, especially the variable regions V4 and V9 [13,80]. However, the number of predicted operational taxonomic units (OTUs) can increase due to the existence of rDNA copy number variation, pseudogenes and intracellular polymorphisms, then resulting in an overestimate of the community complexity [63,81]. The high rDNA copy number and the considerable variation within and between ciliate species highlight the difficulty of using rDNA variation to estimate the abundance of microbial eukaryotes in environmental samples [81,82]. The highest level of interspecific polymorphism for the full-length SSU rDNA among the three ciliate species we studied is 8 base pairs (0.46%) in H. grandinella, whereas the lowest level is 0 in S. stylifer. The highest levels of interspecific polymorphism in V4 and V9 regions among the three species are 3 (1.34%) and 2 (2.25%) base pairs in B. americanum, respectively. The substantial variation in patterns between species creates difficulty in setting cut-off to account for intra-individual polymorphism. Considering the existing data, different levels of cut-off (1% for the full length SSU rDNA, 2% for V4 region and 3% for V9 region) should be used to exclude the interspecific sequence variation. However, it may be too strict for some groups and results in underestimate of the biodiversity. A relatively complete database of intra-individual polymorphism and copy number variation among ciliates would help in estimating diversity from high-throughput sequencing based environmental researches.
Supplementary Material
Acknowledgements
Many thanks are due to Mrs Ying Yan, Wen Song, Xiaotian Luo and Chunyu Lian, graduate students at OUC, for their help in species identification.
Data accessibility
All sequence information has been archived on NCBI/GenBank database under the accession numbers MF002385–MF002436. The datasets supporting this article have been uploaded as part of the electronic supplementary material.
Authors' contributions
F.G. conceived the study. C.W., T.Z. and Y.W. did the experiments and analysed the data. C.W. wrote the manuscript. F.G., L.A.K. and W.S. edited the manuscript.
Competing interests
We declare that we have no competing interests.
Funding
This work is supported by the Natural Science Foundation of China (grant numbers 31430077, 31522051, 41406135), a research grant by Qingdao government to W.S. (project no. 15-12-1-1-jch), an NIH grant to L.A.K. (1R15GM113177-01) and the Fundamental Research Funds for the Central Universities (201562029).
References
- 1.Hillis DM, Dixon MT. 1991. Ribosomal DNA: molecular evolution and phylogenetic inference. Q. Rev. Biol. 66, 411–453. ( 10.1086/417338) [DOI] [PubMed] [Google Scholar]
- 2.Wang P, Gao F, Huang J, Strüder-Kypke M, Yi Z. 2015. A case study to estimate the applicability of secondary structures of SSU–rRNA gene in taxonomy and phylogenetic analyses of ciliates. Zool. Scr. 44, 574–585. ( 10.1111/zsc.12122) [DOI] [Google Scholar]
- 3.Chen X, Ma H, Al-Rasheid KA, Miao M. 2015. Molecular data suggests the ciliate Mesodinium (Protista: Ciliophora) might represent an undescribed taxon at class level. Zool. Syst. 40, 31–40. ( 10.11865/zs.20150102) [DOI] [Google Scholar]
- 4.Elder JF Jr, Turner BJ. 1995. Concerted evolution of repetitive DNA sequences in eukaryotes. Q. Rev. Biol. 70, 297–320. ( 10.1086/419073) [DOI] [PubMed] [Google Scholar]
- 5.Hibbett DS, et al. 2007. A higher-level phylogenetic classification of the fungi. Mycol. Res. 111, 509–547. ( 10.1016/j.mycres.2007.03.004) [DOI] [PubMed] [Google Scholar]
- 6.Jiang J, Zhang Q, Warren A, Al-Rasheid KA, Song W. 2010. Morphology and SSU rRNA gene-based phylogeny of two marine Euplotes species E. orientalis spec. nov. and E. raikovi (Ciliophora, Euplotida). Eur. J. Protistol. 46, 121–132. ( 10.1016/j.ejop.2009.11.003) [DOI] [PubMed] [Google Scholar]
- 7.Saldarriaga JF, Taylor F, Keeling PJ, Cavalier-Smith T. 2001. Dinoflagellate nuclear SSU rRNA phylogeny suggests multiple plastid losses and replacements. J. Mol. Evol. 53, 204–213. ( 10.1007/s002390010210) [DOI] [PubMed] [Google Scholar]
- 8.Woese CR, Kandler O, Wheelis ML. 1990. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl Acad. Sci. USA 87, 4576–4579. ( 10.1073/pnas.87.12.4576) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wylezich C, Nies G, Mylnikov AP, Tautz D, Arndt H. 2010. An evaluation of the use of the LSU rRNA D1-D5 domain for DNA-based taxonomy of eukaryotic protists. Protist 161, 342–352. ( 10.1016/j.protis.2010.01.003) [DOI] [PubMed] [Google Scholar]
- 10.Engberg J, Pearlman RE. 1972. The amount of ribosomal RNA genes in Tetrahymena pyriformis in different physiological states. Eur. J. Biochem. 26, 393–400. ( 10.1111/j.1432-1033.1972.tb01779.x) [DOI] [PubMed] [Google Scholar]
- 11.Marie D, Zhu F, Balagué V., Ras J, Vaulot D. 2006. Eukaryotic picoplankton communities of the Mediterranean Sea in summer assessed by molecular approaches (DGGE, TTGE, QPCR). FEMS Microbiol. Ecol. 55, 403–415. ( 10.1111/j.1574-6941.2005.00058.x) [DOI] [PubMed] [Google Scholar]
- 12.Pace NR. 1997. A molecular view of microbial diversity and the biosphere. Science 276, 734–740. ( 10.1126/science.276.5313.734) [DOI] [PubMed] [Google Scholar]
- 13.Amaral-Zettler LA, McCliment EA, Ducklow HW, Huse SM. 2009. A method for studying protistan diversity using massively parallel sequencing of V9 hypervariable regions of small-subunit ribosomal RNA genes. PLoS ONE 4, e6372 ( 10.1371/annotation/50c43133-0df5-4b8b-8975-8cc37d4f2f26) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moreira D, López-García P. 2002. The molecular ecology of microbial eukaryotes unveils a hidden world. Trends Microbiol. 10, 31–38. ( 10.1016/S0966-842X(01)02257-0) [DOI] [PubMed] [Google Scholar]
- 15.Stoeck T, Behnke A, Christen R, Amaral-Zettler L, Rodriguez-Mora MJ, Chistoserdov A, Orsi W, Edgcomb VP. 2009. Massively parallel tag sequencing reveals the complexity of anaerobic marine protistan communities. BMC Biol. 7, 1 ( 10.1186/1741-7007-7-72) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parfrey LW, Lahr DJ, Katz LA. 2008. The dynamic nature of eukaryotic genomes. Mol. Biol. Evol. 25, 787–794. ( 10.1093/molbev/msn032) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prokopowich CD, Gregory TR, Crease TJ. 2003. The correlation between rDNA copy number and genome size in eukaryotes. Genome 46, 48–50. ( 10.1139/G02-103) [DOI] [PubMed] [Google Scholar]
- 18.Simon D, Moline J, Helms G, Friedl T, Bhattacharya D. 2005. Divergent histories of rDNA group I introns in the lichen family Physciaceae. J. Mol. Evol. 60, 434–446. ( 10.1007/s00239-004-0152-2) [DOI] [PubMed] [Google Scholar]
- 19.Galluzzi L, Penna A, Bertozzini E, Vila M, Garcés E, Magnani M. 2004. Development of a real-time PCR assay for rapid detection and quantification of Alexandrium minutum (a dinoflagellate). Appl. Environ. Microbiol. 70, 1199–1206. ( 10.1128/AEM.70.2.1199-1206.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Godhe A, Asplund ME, Härnström K, Saravanan V, Tyagi A, Karunasagar I. 2008. Quantification of diatom and dinoflagellate biomasses in coastal marine seawater samples by real-time PCR. Appl. Environ. Microbiol. 74, 7174–7182. ( 10.1128/AEM.01298-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prescott DM. 1994. The DNA of ciliated protozoa. Microbiol. Rev. 58, 233–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heyse G, Jönsson F, Chang W-J, Lipps HJ. 2010. RNA-dependent control of gene amplification. Proc. Natl Acad. Sci. USA 107, 22 134–22 139. ( 10.1073/pnas.1009284107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gong J, Dong J, Liu X, Massana R. 2013. Extremely high copy numbers and polymorphisms of the rDNA operon estimated from single cell analysis of oligotrich and peritrich ciliates. Protist 164, 369–379. ( 10.1016/j.protis.2012.11.006) [DOI] [PubMed] [Google Scholar]
- 24.Huang J, Katz LA. 2014. Nanochromosome copy number does not correlate with RNA levels though patterns are conserved between strains of the ciliate morphospecies Chilodonella uncinata. Protist 165, 445–451. ( 10.1016/j.protis.2014.04.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Appel DJ, Gordon TR. 1995. Intraspecific variation within populations of Fusarium oxysporum based on RFLP analysis of the intergenic spacer region of the rDNA. Exp. Mycol. 19, 120–128. ( 10.1006/emyc.1995.1014) [DOI] [PubMed] [Google Scholar]
- 26.DePriest PT. 1993. Small subunit rDNA variation in a population of lichen fungi due to optional group-I introns. Gene 134, 67–74. ( 10.1016/0378-1119(93)90175-3) [DOI] [PubMed] [Google Scholar]
- 27.Odorico D, Miller D. 1997. Variation in the ribosomal internal transcribed spacers and 5.8 S rDNA among five species of Acropora (Cnidaria; Scleractinia): patterns of variation consistent with reticulate evolution. Mol. Biol. Evol. 14, 465–473. ( 10.1093/oxfordjournals.molbev.a025783) [DOI] [PubMed] [Google Scholar]
- 28.Thornhill DJ, Lajeunesse TC, Santos SR. 2007. Measuring rDNA diversity in eukaryotic microbial systems: how intragenomic variation, pseudogenes, and PCR artifacts confound biodiversity estimates. Mol. Ecol. 16, 5326–5340. ( 10.1111/j.1365-294X.2007.03576.x) [DOI] [PubMed] [Google Scholar]
- 29.Gernandt DS, Liston A, Piñero D. 2001. Variation in the nrDNA ITS of Pinus subsection Cembroides: implications for molecular systematic studies of pine species complexes. Mol. Phylogenet. Evol. 21, 449–467. ( 10.1006/mpev.2001.1026) [DOI] [PubMed] [Google Scholar]
- 30.Rehnstam-Holm A.-S., Godhe A, Anderson DM. 2002. Molecular studies of Dinophysis (Dinophyceae) species from Sweden and North America. Phycologia 41, 348–357. ( 10.2216/i0031-8884-41-4-348.1) [DOI] [Google Scholar]
- 31.Santos SR, Kinzie RA, Sakai K, Coffroth MA. 2003. Molecular characterization of nuclear small subunit (18S) rDNA pseudogenes in a symbiotic dinoflagellate (Symbiodinium, Dinophyta). J. Eukaryot. Microbiol. 50, 417–421. ( 10.1111/j.1550-7408.2003.tb00264.x) [DOI] [PubMed] [Google Scholar]
- 32.Yeung P, Kong K, Wong F, Wong J. 1996. Sequence data for two large-subunit rRNA genes from an Asian strain of Alexandrium catenella. Appl. Environ. Microbiol. 62, 4199–4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alverson AJ, Kolnick L. 2005. Intragenomic nucleotide polymorphism among small subunit (18 s) rDNA paralogs in the diatom genus Skeletonema (Bacillariophyta) 1. J. Phycol. 41, 1248–1257. ( 10.1111/j.1529-8817.2005.00136.x) [DOI] [Google Scholar]
- 34.Simon UK, Weiß M. 2008. Intragenomic variation of fungal ribosomal genes is higher than previously thought. Mol. Biol. Evol. 25, 2251–2254. ( 10.1093/molbev/msn188) [DOI] [PubMed] [Google Scholar]
- 35.Gribble KE, Anderson DM. 2007. High intraindividual, intraspecific, and interspecific variability in large-subunit ribosomal DNA in the heterotrophic dinoflagellates Protoperidinium, Diplopsalis, and Preperidinium (Dinophyceae). Phycologia 46, 315–324. ( 10.2216/06-68.1) [DOI] [Google Scholar]
- 36.Coleman AW. 2005. Paramecium aurelia revisited. J. Eukaryot. Microbiol. 52, 68–77. ( 10.1111/j.1550-7408.2005.3327r.x) [DOI] [PubMed] [Google Scholar]
- 37.Gong J, Kim S-J, Kim S-Y, Min G-S, Roberts DM, Warren A, Choi J-K. 2007. Taxonomic redescriptions of two ciliates, Protogastrostyla pulchra ng, n. comb. and Hemigastrostyla enigmatica (Ciliophora: Spirotrichea, Stichotrichia), with phylogenetic analyses based on 18S and 28S rRNA gene sequences. J. Eukaryot. Microbiol. 54, 468–478. ( 10.1111/j.1550-7408.2007.00288.x) [DOI] [PubMed] [Google Scholar]
- 38.Miao W, Fen WS, He YY, Yuan ZX, Shwn YF. 2004. Phylogenetic relationships of the subclass Peritrichia (Oligohymenophorea, Ciliophora) inferred from small subunit rRNA gene sequences. J. Eukaryot. Microbiol. 51, 180–186. ( 10.1111/j.1550-7408.2004.tb00543.x) [DOI] [PubMed] [Google Scholar]
- 39.Wilbert N. 1975. Eine verbesserte technik der protargolimprägnation für ciliaten. Mikrokosmos 64, 171–179. [Google Scholar]
- 40.Medlin L, Elwood HJ, Stickel S, Sogin ML. 1988. The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene 71, 491–499. ( 10.1016/0378-1119(88)90066-2) [DOI] [PubMed] [Google Scholar]
- 41.Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98. [Google Scholar]
- 42.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. ( 10.1093/molbev/mst197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Librado P, Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. ( 10.1093/bioinformatics/btp187) [DOI] [PubMed] [Google Scholar]
- 44.Allen PJ, Bennett K. 2010. PASW statistics by SPSS: a practical guide: version 18.0. Melbourne, Australia: Cengage Learning Press. [Google Scholar]
- 45.Staroscik A.2016. Calculator for determining the number of copies of a template. See http://cels.uri.edu/gsc/cndna.html .
- 46.Sela I, Ashkenazy H, Katoh K, Pupko T. 2015. GUIDANCE2: accurate detection of unreliable alignment regions accounting for the uncertainty of multiple parameters. Nucleic Acids Res. 43, 7–14. ( 10.1093/nar/gkv318) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 57, 758–771. ( 10.1080/10635150802429642) [DOI] [PubMed] [Google Scholar]
- 48.Nylander J. 2004. Mrmodeltest v2: program distributed by the author, vol. 2 Uppsala, Sweden: Uppsala University Press. [Google Scholar]
- 49.Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31, 3406–3415. ( 10.1093/nar/gkg595) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.De Rijk P, De Wachter R. 1997. RnaViz, a program for the visualisation of RNA secondary structure. Nucleic Acids Res. 25, 4679–4684. ( 10.1093/nar/25.22.4679) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fitch WM, Margoliash E. 1967. Construction of phylogenetic trees. Science 155, 279–284. ( 10.1126/science.155.3760.279) [DOI] [PubMed] [Google Scholar]
- 52.Logares R, Haverkamp TH, Kumar S, Lanzén A, Nederbragt AJ, Quince C, Kauserud H. 2012. Environmental microbiology through the lens of high-throughput DNA sequencing: synopsis of current platforms and bioinformatics approaches. J. Microbiol. Methods 91, 106–113. ( 10.1016/j.mimet.2012.07.017) [DOI] [PubMed] [Google Scholar]
- 53.McInerney P, Adams P, Hadi MZ. 2014. Error rate comparison during polymerase chain reaction by DNA polymerase. Mol. Biol. Int. 2014, 8 ( 10.1155/2014/287430) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cline J, Braman JC, Hogrefe HH. 1996. PCR fidelity of Pfu DNA polymerase and other thermostable DNA polymerases. Nucleic Acids Res. 24, 3546–3551. ( 10.1093/nar/24.18.3546) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brandariz-Fontes C, Camacho-Sanchez M, Vilà C, Vega-Pla JL, Rico C, Leonard JA. 2015. Effect of the enzyme and PCR conditions on the quality of high-throughput DNA sequencing results. Sci. Rep. 5, 8056 ( 10.1038/srep08056) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Campbell CS, Wojciechowski MF, Baldwin BG, Alice LA, Donoghue MJ. 1997. Persistent nuclear ribosomal DNA sequence polymorphism in the Amelanchier agamic complex (Rosaceae). Mol. Biol. Evol. 14, 81–90. ( 10.1093/oxfordjournals.molbev.a025705) [DOI] [PubMed] [Google Scholar]
- 57.Hartmann S, Nason JD, Bhattacharya D. 2001. Extensive ribosomal DNA genic variation in the columnar cactus Lophocereus. J. Mol. Evol. 53, 124–134. ( 10.1007/s002390010200) [DOI] [PubMed] [Google Scholar]
- 58.Hugall A, Stanton J, Moritz C. 1999. Reticulate evolution and the origins of ribosomal internal transcribed spacer diversity in apomictic Meloidogyne. Mol. Biol. Evol. 16, 157–164. ( 10.1093/oxfordjournals.molbev.a026098) [DOI] [PubMed] [Google Scholar]
- 59.Reed KM, Hackett JD, Phillips RB. 2000. Comparative analysis of intra-individual and inter-species DNA sequence variation in salmonid ribosomal DNA cistrons. Gene 249, 115–125. ( 10.1016/S0378-1119(00)00156-6) [DOI] [PubMed] [Google Scholar]
- 60.Juranek SA, Lipps HJ. 2007. New insights into the macronuclear development in ciliates. Int. Rev. Cytol. 262, 219–251. ( 10.1016/S0074-7696(07)62005-1) [DOI] [PubMed] [Google Scholar]
- 61.McGrath C, Zufall R, Katz L. 2006. Genome evolution in ciliates. In Genomics and evolution of eukaryotic microbes (eds Katz LA, Bhattacharya D), pp. 64–77. New York, NY: Oxford University Press. [Google Scholar]
- 62.Robinson T, Katz LA. 2007. Non-mendelian inheritance of paralogs of 2 cytoskeletal genes in the ciliate Chilodonella uncinata. Mol. Biol. Evol. 24, 2495–2503. ( 10.1093/molbev/msm203) [DOI] [PubMed] [Google Scholar]
- 63.Zhu F, Massana R, Not F, Marie D, Vaulot D. 2005. Mapping of picoeucaryotes in marine ecosystems with quantitative PCR of the 18S rRNA gene. FEMS Microbiol. Ecol. 52, 79–92. ( 10.1016/j.femsec.2004.10.006) [DOI] [PubMed] [Google Scholar]
- 64.Foissner W, O'donoghue P. 1990. Morphology and infraciliature of some freshwater ciliates (Protozoa: Ciliophora) from Western and South Australia. Invertebr. Syst. 3, 661–696. ( 10.1071/IT9890661) [DOI] [Google Scholar]
- 65.Song W. 1993. Studies on the cortical morphogenesis during cell division in Halteria grandinella (Muller, 1773) (Ciliophora, Oligotrichida). Chin. J. Oceanol. Limnol. 11, 122–129. ( 10.1007/BF02850818) [DOI] [Google Scholar]
- 66.Song W, Li J, Liu W, Al-Rasheid KA, Hu X, Lin X. 2015. Taxonomy and molecular phylogeny of four Strombidium species, including description of S. pseudostylifer sp. nov. (Ciliophora, Oligotrichia). Syst. Biodivers. 13, 76–92. ( 10.1080/14772000.2014.970674) [DOI] [Google Scholar]
- 67.Prescott DM. 1992. Cutting, splicing, reordering, and elimination of DNA sequences in hypotrichous ciliates. Bioessays 14, 317–324. ( 10.1002/bies.950140505) [DOI] [PubMed] [Google Scholar]
- 68.Goldman AD, Stein EM, Bracht JR, Landweber LF. 2014. Programmed genome processing in ciliates. In Discrete and topological models in molecular biology (eds Jonoska N, Saito M), pp. 273–287. Berlin, Germany: Springer. [Google Scholar]
- 69.Jahn CL, Klobutcher LA. 2002. Genome remodeling in ciliated protozoa. Annu. Rev. Microbiol. 56, 489–520. ( 10.1146/annurev.micro.56.012302.160916) [DOI] [PubMed] [Google Scholar]
- 70.Eisen JA, et al. 2006. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 4, e286 ( 10.1371/journal.pbio.0040286) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Swart EC, et al. 2013. The Oxytricha trifallax macronuclear genome: a complex eukaryotic genome with 16 000 tiny chromosomes. PLoS Biol. 11, e1001473 ( 10.1371/journal.pbio.1001473) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Aeschlimann SH, Jönsson F, Postberg J, Stover NA, Petera RL, Lipps H.-J., Nowacki M, Swart EC. 2014. The draft assembly of the radically organized Stylonychia lemnae macronuclear genome. Genome Biol. Evol. 6, 1707–1723. ( 10.1093/gbe/evu139) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xiong J, Wang G, Cheng J, Tian M, Pan X, Warren A, Jiang C, Yuan D, Miao W. 2015. Genome of the facultative scuticociliatosis pathogen Pseudocohnilembus persalinus provides insight into its virulence through horizontal gene transfer. Sci. Rep. 5, 15470 ( 10.1038/srep15470) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Slabodnick MM, et al. 2017. The macronuclear genome of Stentor coeruleus reveals tiny introns in a giant cell. Curr. Biol. 20, 569–575. ( 10.1016/j.cub.2016.12.057) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.La Terza A, Miceli C, Luporini P. 1995. Differential amplification of pheromone genes of the ciliate Euplotes raikovi. Dev. Genet. 17, 272–279. ( 10.1002/dvg.1020170312) [DOI] [PubMed] [Google Scholar]
- 76.Xu K, Doak TG, Lipps HJ, Wang J, Swart EC, Chang W.-J. 2012. Copy number variations of 11 macronuclear chromosomes and their gene expression in Oxytricha trifallax. Gene 505, 75–80. ( 10.1016/j.gene.2012.05.045) [DOI] [PubMed] [Google Scholar]
- 77.Edgcomb V, et al. 2011. Protistan microbial observatory in the Cariaco Basin, Caribbean. I. Pyrosequencing vs Sanger insights into species richness. ISME J. 5, 1344–1356. ( 10.1038/ismej.2011.6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mangot JF, Domaizon I, Taib N, Marouni N, Duffaud E, Bronner G, Debroas D. 2013. Short-term dynamics of diversity patterns: evidence of continual reassembly within lacustrine small eukaryotes. Environ. Microbiol. 15, 1745–1758. ( 10.1111/1462-2920.12065) [DOI] [PubMed] [Google Scholar]
- 79.Bik HM, Sung W, De Ley P, Baldwin JG, Sharma J, Rocha-Olivares A, Thomas WK. 2012. Metagenetic community analysis of microbial eukaryotes illuminates biogeographic patterns in deep-sea and shallow water sediments. Mol. Ecol. 21, 1048–1059. ( 10.1111/j.1365-294X.2011.05297.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stoeck T, Bass D, Nebel M, Christen R, Jones MD, Breiner HW, Richards TA. 2010. Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water. Mol. Ecol. 19, 21–31. ( 10.1111/j.1365-294X.2009.04480.x) [DOI] [PubMed] [Google Scholar]
- 81.Medinger R, Nolte V, Pandey RV, Jost S, Ottenwaelder B, Schloetterer C, Boenigk J. 2010. Diversity in a hidden world: potential and limitation of next–generation sequencing for surveys of molecular diversity of eukaryotic microorganisms. Mol. Ecol. 19, 32–40. ( 10.1111/j.1365-294X.2009.04478.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Terrado R, Medrinal E, Dasilva C, Thaler M, Vincent WF, Lovejoy C. 2011. Protist community composition during spring in an Arctic flaw lead polynya. Polar Biol. 34, 1901–1914. ( 10.1007/s00300-011-1039-5) [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequence information has been archived on NCBI/GenBank database under the accession numbers MF002385–MF002436. The datasets supporting this article have been uploaded as part of the electronic supplementary material.