Abstract
In a range of taxa, the relatedness between mates influences both pre- and post-mating processes of sexual selection. However, relatively little is known about the genetic loci facilitating such a bias, with the exception of the major histocompatibility complex. Here, we performed tightly controlled replicated in vitro fertilization trials to explore the impact of relatedness on two possible mechanisms of cryptic female choice (CFC) in Chinook salmon (Oncorhynchus tshawytscha). We tested (i) whether relatedness of mates, assessed using 682 single nucleotide polymorphisms (SNPs) on 29 SNP-linkage groups (LGs), biases a male's sperm velocity in ovarian fluid (a parameter previously shown to predict male fertilization success), and (ii) whether relatedness of mates governs fertilization success via other mechanisms, probably via sperm–egg interactions. We found that relatedness on three LGs explained the variation in sperm velocity, and relatedness on two LGs explained fertilization success, which might indicate the presence of genes important in sperm–ovarian fluid and sperm–egg interactions in these genomic regions. Mapping of the SNPs on these LGs to the rainbow trout genome revealed two genes that affect fertility in humans and represent candidate genes for further studies. Our results thereby provide a novel contribution to the understanding of the mechanism of CFC.
Keywords: Chinook salmon, fish, sexual selection, cryptic female choice, SNPs, genetic compatibility
1. Introduction
It is now widely recognized that post-mating processes of sexual selection, through both sperm competition and cryptic female choice (CFC), can each have profound effects on fertilization outcomes [1]. While the former has received significant attention [2,3], CFC is less well studied and poorly understood at a mechanistic level, except in a few systems [4–6]. A growing appreciation of the importance of female choice during or after mating [7,8] led to a variety of studies that demonstrated the presence of processes that bias fertilization outcomes towards certain males in order to increase female reproductive success [3,7,8]. The majority of studies focused on internally fertilizing species, particularly insects [9,10], and many of these studies have revealed that CFC biases fertilization outcomes towards unrelated males, including Drosophila spp. [9], field crickets [10] and sand lizards [11]. However, the study of CFC in internal fertilizers is complicated by male competitive effects that may confound the effect of CFC [3]. By contrast, external fertilizers offer the advantage that male effects, such as the density of spermatozoa, can be well controlled in vitro to tease apart male and female effects on fertilization outcomes, and those that result from the interaction between both systems [12].
In externally fertilizing species, CFC may act via (i) the impact of ovarian fluid on sperm traits [13,14] and/or (ii) sperm–egg interactions once the sperm have reached the micropyle of the ova [15]. There is evidence in several externally fertilizing fishes that ovarian fluid, a viscous substance surrounding the spawned eggs, enhances sperm velocity compared with water [13,16], whereby a given female's ovarian fluid differentially enhances the sperm velocity of different males. As sperm velocity strongly predicts male fertilization success, it is likely that the varying degree of velocity enhancement is a mechanism of CFC [14,17–19]. Sperm–egg interactions, as observed in external fertilizers [5,6], may represent another mechanism of CFC. Here, CFC manifests as non-random gamete fusions mediated by proteins on the egg and sperm surfaces. Possibly a similar mechanism exists in externally fertilizing teleost fish, where sperm might interact with the inner walls of the micropyle [15,20].
Prior work suggests a genetic basis for CFC mechanisms. In vitro fertilization experiments in the Arctic charr [21] and the Peron's tree frog [22] revealed that fertilization success was higher for mates genetically more similar. In lake trout, sperm velocity measured in a female's ovarian fluid was positively correlated with mate relatedness [23]. However, relatedness was derived either from pedigree information [23] or from small panels of microsatellites [21,22], which does not allow the identification of genes that may play a role in CFC.
Here, we used tightly controlled in vitro fertilization trials and employed single nucleotide polymorphism (SNP)-based relatedness of mates to explore the role of relatedness in two mechanisms of CFC in Chinook salmon (Oncorhynchus tshawytscha). Chinook salmon are ideal to study CFC for genetic compatibility, because they exhibit a non-resource-based mating system (i.e. no nuptial gifts or other direct benefits are received by the female [24]). Species with this mating system, in particular, are expected to base mate selection on genetic benefits [25]. Further, Chinook salmon have a polyandrous mating system [26], allowing sexual selection to occur post mating.
We tested whether relatedness biases males' sperm velocity in the ovarian fluid, which predicts fertilization success in Chinook salmon [14,27]. Further, we explored the impact of mate relatedness on the variation of fertilization success that could not be explained by differences in sperm velocity, which might indicate the presence of sperm–egg interactions. Finally, we mapped the SNPs of LGs that explained the variation in sperm velocity and fertilization success to the rainbow trout [28] and Atlantic salmon genomes [29], to identify genes in these genomic regions that might play a role in CFC.
2. Material and methods
(a). Fish sampling
Chinook salmon were caught in a trap at the Kaiapoi River, Canterbury, New Zealand, during their spawning run in April–May in the years 2010 and 2011. Sexually mature 2–3-year-old ‘hooknose’ males and 3-year-old females were individually tagged and maintained using standard husbandry procedures at Salmon Smolt NZ, Canterbury, New Zealand [12–14,30]. The animals were handled and maintained using protocols approved by the Animal Ethics Committee for the University of Otago, New Zealand.
(b). Experimental overview
We set up a series of in vitro paired-male fertilization trials using equal amounts of sperm from two males. We measured the sperm velocity of males in the ovarian fluid of the females they were crossed to in our fertilization trials. We performed two replicate tests for each sperm velocity measurement and each in vitro fertilization. We used ten females (four in 2010 and six in 2011) and generated 90 different fertilization trials and 60 female–male combinations. We used 27 males, of which some were used in more than one trial (10 males were used in only one trial, 6 in two, 9 in three and 2 in four trials).
(c). Measurement of sperm velocity in ovarian fluid
The sperm velocity of randomly collected males was assessed in the ovarian fluid of a randomly sampled female using a CEROS sperm tracker (v. 12, Hamilton Thorne Research, Beverly, MA). Sperm velocity was recorded as previously described [12–14]. Briefly, milt was pipetted onto a 20 µl Leja slide (Leja Products B.V., Nieuw-Vennep, The Netherlands) and activated with ovarian fluid of the focal female. We recorded the average path velocity (VAP in μm s−1) 10 s post activation in ovarian fluid.
Ovarian fluid is a viscous substance surrounding the spawned eggs and the concentration is probably very high near the egg surface, reducing with increase in distance from the egg mass [20,30]. Possibly, there is no one concentration for in vitro experiments that best mimics natural spawning conditions, but we expect the sperm to swim up a gradient of ovarian fluid experiencing different concentrations while approaching the egg. We examined sperm velocity in 50% of ovarian fluid in 2010 and in 100% in 2011, and controlled for that difference in our statistical analysis.
(d). In vitro fertilization trials
(i). Paired-male trials
We performed trials with two randomly chosen males and one female. A batch of approximately 100 eggs (approx. 50 ml of the eggs including 25 ml of ovarian fluid) was placed in a plastic beaker and fertilized with equal amounts of milt from both males—milt was added simultaneously with 250 ml of river water to imitate natural spawning. Thus, the ovarian fluid concentration present during fertilization was 10%, which is a concentration previously used to document male–female interactions [12,14]. We determined sperm densities using an improved Neubauer haemocytometer and added respective volumes of milt from each male containing 1 × 108 spermatozoa. Each fertilized egg batch was reared separately under conditions imitating natural conditions (i.e. in the dark with a constant water flow and a water temperature of 12–12.5°C [31]).
(ii). Single-male control trials
We performed control trials to ensure that no infertile male and/or prematurely activated sperm would confound our results. To ensure that the number of fertilized eggs represented the proportion of functional sperm, we used a low sperm number (1 × 105 spermatozoa). Males had fertile sperm (fertilization rate: 57 ± 4% s.e., 60 control trials, approx. 100 eggs per trial) except for two males with fertilization rates of 1.5%, which were removed from further analyses. We therefore analysed 84 paired-male trials (each male contributed to three trials) and 58 female–male combinations.
(e). DNA extraction
DNA was extracted from 60 mg of dorsal fin tissue using a Proteinase K digestion as described by Gemmell & Akiyama [32], except that chloroform was substituted with NaCl (100 mM).
(f). Microsatellite-based paternity assignment
To determine male fertilization success in paired-male trials, we randomly collected 48 offspring from each family (24 offspring from each replicate) at the eyed stage of embryonic development (five weeks post fertilization). For all 84 fertilization trials, a total of 4032 offspring were genotyped using nine microsatellite markers (Ocl-1 [33], Omy-325, Ssa-85 [34] Ots-101, Ots-104, Ots-107, Ots-2, Ots-3 [35], Ssa197 [36] and assigned to a sire using the maximum likelihood approach in the program Cervus v. 3.0 [37].
(g). Relatedness of mates
All 37 parental fish were genotyped with a 6000 SNP array established for Chinook salmon (see the electronic supplementary material) using the Illumina Infinium II BeadChip Technology and the Genome Studio software v. 2010.3/v. 1.8 (Illumina) for analysis. Genome-wide relatedness and LG-specific relatedness estimates between mates were based on 682 SNPs and 583 SNPs, respectively. LG-specific relatedness was calculated for a total of 29 LGs with at least 8 SNPs (see electronic supplementary material, table S1). See the electronic supplementary material for details on the SNP discovery and the selection of informative SNPs. Relatedness between mates was determined using the triadic likelihood estimator [38] implemented in the program Coancestry v. 1.0.1.2 [39].
(h). Statistical analysis
Statistical analyses were carried out in the R v. 3.0.1 statistics program [40] using the MCMCglmm package [41] which implements Markov chain Monte Carlo (MCMC) routines to fit multi-response generalized linear mixed models. The individual fish, females and males, and the female–male interactions were modelled as random effects. The genome-wide relatedness and relatedness on each LG were modelled as fixed effects in separate models. See the electronic supplementary material for more details.
(i). Analysis of the variation in sperm velocity
The two values of sperm velocity recorded for each male in a focal female's ovarian fluid were used as the response variable with a Gaussian error with the identity link. To control for a possible impact of the year of experiment and the sperm velocity measured in 50% ovarian fluid in season 2010 and 100% in season 2011, the season was modelled as a fixed effect. This allowed us to distinguish how much of the variation in sperm velocity was attributed to the season (i.e. year of experiment and difference in ovarian fluid concentration) and how much was explained by other variance components (e.g. female–male interaction).
(ii). Analysis of the variation in fertilization success
The relative fertilization success of a male in a paired-male trial was set as the response variable (i.e. the number of eggs fertilized by a male was used as count data and the total number of eggs fertilized by both males was modelled as a fixed effect). MCMCglmms were modelled with a Poisson error distribution with the logit link. All trials were performed twice and the two replicates (first trial denoted ‘replicate 1’, second trial ‘replicate 2’) were analysed separately. The sperm velocity of a male relative to the male he was paired with was modelled as a fixed effect (i.e. we used a binary predictor by labelling the faster male ‘F’ and the slower male ‘S’ in order to control for the difference in sperm velocity measured in 50% and 100% ovarian fluid in season 2010 and 2011, respectively). ‘SF’ was also modelled as the random slope for each pair of males (denoted by ‘Pair’), because some males were used in more than one female and, thus, the designation of S and F can change depending on the sperm velocity of the male he was paired with. Further, we modelled the z-transformed sperm velocity data as a fixed effect to adjust for the difference in velocity recordings between seasons. As we explored the effect of relatedness at 29 LGs on sperm velocity and fertilization success, we corrected for type I errors using the false discovery rate [42].
(i). Genes on SNP-linkage groups correlated with sperm velocity and/or fertilization success
To identify genes with a potential role in CFC mechanisms, we mapped all SNPs on LGs correlated with sperm velocity and/or fertilization success to the rainbow trout genome [28] using BLAST [43]. We additionally mapped SNPs to the Atlantic salmon genome [29]. We extracted genes and sequence 1 kb upstream and downstream using the gff2fast.pl script modified for our purposes (see the electronic supplementary material). We report genes when SNPs mapped in a gene, or to positions within 1 kb upstream of or downstream from a gene, as prior work suggests that SNPs in these regions exhibit a stronger association with phenotypes than SNPs beyond a 1 kb distance from a gene [44]. Atlantic salmon annotations were retrieved from SalmoBase (http://salmobase.org). The peptide sequence of rainbow trout genes were accessed via the Trout Genome Browser (https://www.genoscope.cns.fr/trout/) and searched against the Uniprot v. 2017_03 protein database using BLASTP v. 2.6.0 [45] to find highly similar sequences identified in other species. All BLAST searches were used with an E-value cut-off of less than 10−50 and sequence similarities above 90%.
3. Results
(a). Relatedness and sperm velocity in ovarian fluid
Mean genome-wide relatedness of mates, estimated using 682 SNPs, was low (r = 0.007, min = 0, max = 0.08 across mates; see electronic supplementary material, figure S1). The relatedness of mates varied across LGs (r = 0.06–0.23; see electronic supplementary material, figure S1). The mean sperm velocity in ovarian fluid was 61 µm s−1 ± 1.86 (s.e.) in season 2010 (n = 22) and 79 µm s−1 ± 2.29 (s.e.) in season 2011 (n = 36). The difference between spawning seasons (i.e. year and ovarian fluid concentrations) explained a significant amount, but less than 20%, of the variation in sperm velocity (pMCMC = 0.004–0.042 across models; table 1). The amount of variation not explained by the difference in seasons was significantly explained by mate relatedness on LG10, LG12 and LG24 (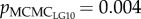 ,
, 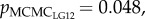
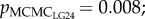 table 1). Males less closely related to the female on these LGs had a higher sperm velocity in the ovarian fluid than males more related to the female. Further, the female–male interaction explained on average 67% of the variation in sperm velocity, indicating that relatedness on the given LG did not fully characterize the female–male compatibility. Relatedness on the other 26 LGs (pMCMC = 0.14–0.92) and genome-wide relatedness of mates (pMCMC = 0.22; see electronic supplementary material, table S2) were not significantly correlated with sperm velocity. When corrected for multiple testing, pMCMC values for LG10, LG12 and LG24 did not remain significant (see electronic supplementary material, table S2).
table 1). Males less closely related to the female on these LGs had a higher sperm velocity in the ovarian fluid than males more related to the female. Further, the female–male interaction explained on average 67% of the variation in sperm velocity, indicating that relatedness on the given LG did not fully characterize the female–male compatibility. Relatedness on the other 26 LGs (pMCMC = 0.14–0.92) and genome-wide relatedness of mates (pMCMC = 0.22; see electronic supplementary material, table S2) were not significantly correlated with sperm velocity. When corrected for multiple testing, pMCMC values for LG10, LG12 and LG24 did not remain significant (see electronic supplementary material, table S2).
Table 1.
Relatedness of mates on SNP-linkage groups LG10, LG12 and LG24 was negatively correlated with sperm velocity in the ovarian fluid in Chinook salmon. Significant values of fixed effects of the MCMCglmm analyses are in italics. The variance (%) explained by each random effect refers to the total variance by all random effects (female identity (n = 10), male identity (n = 27), female–male interaction (n = 58)) and the residual variance. For each model, the amount of variation explained by the fixed effects (marginal R2) and by the whole model (conditional R2) are reported.
| estimate | variance (%) | lower 95% CI | upper 95% CI | pMCMC | |
|---|---|---|---|---|---|
| LG10 | |||||
| fixed effects | |||||
| intercept | 67.88 | 56.019 | 78.496 | ||
| LG10 | −61.06 | −102.330 | −25.586 | 0.004 | |
| season | 16.45 | 2.915 | 28.788 | 0.016 | |
| random effects | |||||
| female (10) | 21.76 | 4.77 | <0.001 | 122.300 | |
| male (27) | 2.44 | 53.00 | <0.001 | 10.870 | |
| female–male (58) | 291.50 | 63.86 | 153.600 | 458.700 | |
| residual | 140.80 | 30.84 | 97.880 | 183.600 | |
| summary statistics | |||||
| marginal R2 | 0.21 | ||||
| conditional R2 | 0.75 | ||||
| LG12 | |||||
| fixed effects | |||||
| intercept | 66.95 | 55.052 | 77.683 | ||
| LG12 | −31.94 | −61.688 | 0.806 | 0.048 | |
| season | 17.56 | 4.922 | 31.650 | 0.022 | |
| random effects | |||||
| female (10) | 11.71 | 2.41 | <0.001 | 80.440 | |
| male (27) | 3.13 | 0.64 | <0.001 | 19.460 | |
| female–male (58) | 328.70 | 67.64 | 187.800 | 486.900 | |
| residual | 142.40 | 29.30 | 99.670 | 189.800 | |
| summary statistics | |||||
| marginal R2 | 0.15 | ||||
| conditional R2 | 0.76 | ||||
| LG24 | |||||
| fixed effects | |||||
| intercept | 66.08 | 56.465 | 77.047 | ||
| LG24 | −40.00 | −66.410 | −12.392 | 0.008 | |
| season | 20.54 | 9.147 | 34.599 | 0.004 | |
| random effects | |||||
| female (10) | 6.99 | 1.52 | <0.001 | 42.020 | |
| male (27) | 2.67 | 0.58 | <0.001 | 7.773 | |
| female–male (58) | 309.20 | 67.09 | 183.700 | 467.800 | |
| residual | 142.00 | 30.81 | 102.900 | 188.600 | |
| summary statistics | |||||
| marginal R2 | 0.20 | ||||
| conditional R2 | 0.75 | ||||
Female identity and male identity explained on average only 3.36% and 0.75% of the variation in sperm velocity, respectively (see electronic supplementary material, table S2), and thus there were no females in which males had a clearly elevated sperm velocity compared with other females' ovarian fluid. Similarly, there were no males whose sperm velocity was overall faster compared with other males.
(b). Relatedness and fertilization success
Each paired-male fertilization trial was performed twice (replicate 1 and 2) and 24 offspring were randomly sampled from each replicate. On average, 47 offspring were successfully assigned to one of the paired males. The male fertilization success was the proportion of the 24 offspring sired by a given male and ranged from 0% to 100% in both replicates. The male with the higher sperm velocity in the focal female's ovarian fluid fertilized significantly more eggs than the male with the lower sperm velocity (pSF = less than 0.001–0.002 across models; table 2 for LG12 and LG23). Relatedness of mates on LG12 and LG23 were negatively correlated with fertilization success, whereby some pMCMC values were marginally significant (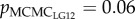 and 0.036;
and 0.036; 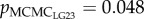 and 0.054 in replicate 1 and 2, respectively; table 2). Relatedness of mates on the other LGs and genome-wide did not explain the variation in fertilization success (see electronic supplementary material, table S3). The results for LG12 and LG23 were confirmed when the z-transformed velocity data was modelled (
and 0.054 in replicate 1 and 2, respectively; table 2). Relatedness of mates on the other LGs and genome-wide did not explain the variation in fertilization success (see electronic supplementary material, table S3). The results for LG12 and LG23 were confirmed when the z-transformed velocity data was modelled (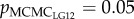 and 0.012; pMCMCLG23 = 0.06 and 0.075 in replicates 1 and 2, respectively). The pMCMC values, corrected for multiple testing, did not remain (marginally) significant for LG12 and 23 (see electronic supplementary material, table S3).
and 0.012; pMCMCLG23 = 0.06 and 0.075 in replicates 1 and 2, respectively). The pMCMC values, corrected for multiple testing, did not remain (marginally) significant for LG12 and 23 (see electronic supplementary material, table S3).
Table 2.
Relatedness of mates on SNP-linkage groups LG12 and LG23 explained the variation in fertilization success in Chinook salmon. The MCMCglmm results for each replicated fertilization trial (replicate 1 and 2) are reported. Significant and marginally significant values of fixed effects are in italics.
| estimate |
variance (%) |
lower 95% CI |
upper 95% CI |
pMCMC |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| replicate 1 | replicate 2 | replicate 1 | replicate 2 | replicate 1 | replicate 2 | replicate 1 | replicate 2 | replicate 1 | replicate 2 | |
| LG12 | ||||||||||
| fixed effects | ||||||||||
| intercept | 1.217 | 1.171 | 0.320 | −0.100 | 2.112 | 2.678 | ||||
| LG12 | −0.445 | −0.562 | −0.916 | −1.057 | −0.018 | −0.051 | 0.050 | 0.036 | ||
| SF | 0.508 | 0.377 | 0.281 | 0.147 | 0.751 | 0.608 | <0.001 | 0.002 | ||
| random effects | ||||||||||
| female (10) | 0.003 | 0.003 | 0.83 | 0.30 | <0.001 | <0.001 | 0.009 | 0.009 | ||
| male (27) | 0.006 | 0.005 | 1.57 | 0.49 | <0.001 | <0.001 | 0.022 | 0.016 | ||
| male–male pair (84) | 0.371 | 1.000 | 96.87 | 98.91 | 0.208 | 0.178 | 0.539 | 0.475 | ||
| SF (random slope) | 1.049 | 0.991 | 0.685 | 0.579 | 1.481 | 1.362 | ||||
| additive overdispersion | 0.003 | 0.003 | 0.78 | 0.30 | <0.001 | <0.001 | 0.009 | 0.009 | ||
| summary statistics | ||||||||||
| marginal R2 | 0.11 | 0.10 | ||||||||
| conditional R2 | 0.42 | 0.37 | ||||||||
| LG23 | ||||||||||
| fixed effects | ||||||||||
| intercept | 1.270 | 1.009 | 0.348 | −0.361 | 2.148 | 2.391 | ||||
| LG23 | −0.281 | −0.273 | −0.542 | −0.569 | 0.011 | <0.001 | 0.048 | 0.054 | ||
| SF | 0.527 | 0.387 | 0.300 | 0.159 | 0.784 | 0.662 | <0.001 | 0.002 | ||
| random effects | ||||||||||
| female (10) | 0.004 | 0.004 | 1.03 | 1.10 | <0.001 | <0.001 | 0.011 | 0.013 | ||
| male (27) | 0.007 | 0.007 | 1.81 | 1.93 | <0.001 | <0.001 | 0.024 | 0.024 | ||
| male–male pair (84) | 0.373 | 0.349 | 96.38 | 96.14 | 0.231 | 0.208 | 0.553 | 0.526 | ||
| SF (random slope) | 1.041 | 1.015 | 0.674 | 0.629 | 1.463 | 1.431 | ||||
| additive overdispersion | 0.003 | 0.030 | 0.78 | 0.83 | <0.001 | <0.001 | 0.008 | 0.008 | ||
| summary statistics | ||||||||||
| marginal R2 | 0.14 | 0.09 | ||||||||
| conditional R2 | 0.46 | 0.48 | ||||||||
The female identity, male identity and the identity of the male competitor (denoted Pair), explained on average 0.83% (0.7–1.05% across models), 2% (1.5–2.4% across models) and 96% (89–98% across models) of the variation in fertilization success, respectively.
(c). Genes on SNP-linkage groups correlated with sperm velocity and/or fertilization success
LG10, LG12 and LG24 were correlated with sperm velocity and LG12 and LG23 with fertilization success before correcting for multiple testing, and thus SNPs mapping within or 1 kb upstream of and downstream from genes in the rainbow trout genome are reported (see electronic supplementary material, table S4). Across all five LGs, we identified 10 SNPs with intragenic locations and 3 SNPs in regions within of a gene 1 kb upstream or downstream of a gene. Twenty-four of a total of 65 SNPs could not be successfully mapped to the rainbow trout genome. Additional mapping against the Atlantic salmon genome revealed another 19 SNPs with intragenic locations and 7 SNPs in the 1kb upstream or downstream regions (see electronic supplementary material, table S4).
The genes identified have a variety of roles (e.g. in fatty acid biosynthesis, cell-division cycle, bone formation and sucrose transport) and include genes whose function is yet to be characterized (see electronic supplementary material, table S4).
4. Discussion
In a variety of species, CFC is determined by the relatedness of females and males [9–11]. A first step in understanding the underlying mechanisms is to identify genomic regions of importance in the processes of CFC. In this study, we investigated the role of mate relatedness (genome-wide and on 29 SNP-LGs) on sperm velocity in ovarian fluid and on fertilization success.
(a). Genome-wide relatedness and cryptic female choice
Our study shows that the variation in sperm velocity in ovarian fluid and in fertilization success was random with respect to genome-wide relatedness of mates, assessed using 682 SNPs. The strong natal homing and the distinct timing of spawning runs in salmonids result in smaller subpopulations with higher probabilities of inbreeding that might be important for preserving local adaptation [46]. Inbreeding could be facilitated by post-mating mechanisms for more closely related mates. However, our data indicate that CFC was independent of genome-wide relatedness. By contrast, in a hatchery population of lake trout, sperm velocity was higher for full-sibling mates compared with unrelated mates [23]. The low variation in genome-wide relatedness between our wild-caught individuals might better represent natural conditions. On the other hand, the low variation might hamper the detection of relatedness-dependent CFC.
Differences in life cycles and environments of lake trout and Chinook salmon might also explain the different findings. Lake trout live in closed-system freshwater lakes and appear to exhibit a stronger degree of inbreeding, to which they might have become adapted [23]. By contrast, Chinook salmon migrate to sea and return to their natal freshwater stream to spawn. Chinook salmon might prefer mates similar in some genomic regions to preserve local adaptations and dissimilar in other regions to cope with a variety of challenges (e.g. exposure to a set of pathogens with low predictability) at sea. In that case CFC can be random with respect to genome-wide relatedness because positive and negative correlations across the genome may be equalized.
(b). Relatedness on specific SNP-linkage groups determined sperm velocity in ovarian fluid
We found that the majority of the variation in sperm velocity in ovarian fluid was explained by the female–male interaction; a given male's sperm velocity varied depending on the female source of the ovarian fluid. This finding confirms previous work in Chinook salmon [13,14,27] and suggests the presence of a female–male compatibility that determines sperm velocity.
While genome-wide relatedness varied little between mates because higher and lower relatedness were equalized, relatedness on specific LGs varied. We found that males less closely related to the female on LG10, LG12 and LG24 had a higher sperm velocity (i.e. a significant amount of the female–male compatibility was characterized by relatedness on these LGs), suggesting the presence of genes that might determine sperm velocity in ovarian fluid. We identified several predicted genes on these LGs by mapping the Chinook salmon SNPs to the genomes of the rainbow trout [28] and Atlantic salmon [29]. These genes span a variety of predicted or known functions including fatty acid biosynthesis, bone formation, cell cycle mechanisms, cell metabolism and the regulation of chromosome structure. Some genes with functions in, for example, bone formation are unlikely candidates for roles in CFC. Other genes, like Elovl7, which contributes to the biosynthesis of fatty acids [47], might be candidates, because another elongase, Elovl2, has been associated with sperm function and fertility in mice [47]. In humans, Elovl7 is expressed in sperm and implicated in male fertility [48]. There is evidence for several elongase enzymes in fish [49], although a possible importance for gametes has not been studied. SLC45A3, located on LG12, which was correlated with sperm velocity and fertilization success, might be another candidate gene. SLC45A3 codes for the sucrose transporter solute carrier family 45 member A3 and is linked to male fertility in humans [48].
Related research might provide ideas for roles of the candidate genes. In sea urchins, oocyte-associated peptides bind to receptors on the sperm plasma membrane and stimulate respiration, motility and chemoattraction of spermatozoa to the oocyte [50]. In salmonids, egg-associated factors stimulate sperm motility and guide the sperm to the micropyle [20]. It remains yet unknown whether these factors have a selective nature that could explain the varying compatibility among mates.
(c). Relatedness on specific SNP-linkage groups determined fertilization success
We found that a male's sperm velocity relative to his rival significantly influenced his fertilization success. As sperm velocity was mainly driven by female–male compatibility, it confirms earlier suggestions that CFC mechanisms determining sperm velocity are a way to effectively bias fertilization outcomes towards certain males [13].
The variation in fertilization success not explained by sperm velocity was explained by the relatedness of mates on LG12 and LG23. Males less closely related to the female on these LGs had a significantly higher fertilization success, hinting at genes mediating non-random gamete fusions. These correlations were found in both replicates of our trials, although it should be noted that some pMCMC values were only marginally significant. Two of the genes found on LG12 and LG23 have neuronal functions and appear unlikely to be involved in CFC. However, thrombospondin 3b ensuring cell adhesion and calcium ion binding might be a candidate. In other external fertilizers, such as sea urchins [5] and tunicates [6], peptides expressed on the gamete surface facilitate a non-random gamete fusion, and oocytes preferably fuse with sperm with a similar [5] or dissimilar peptide [6] to their own. One possibility is that genes in our candidate regions code for peptides with a similar function in salmon.
We found that sperm velocity and fertilization success were higher for mates less closely related to each other at specific LGs but not genome-wide. Thus, CFC mechanisms in Chinook salmon do not appear to avoid overall inbreeding. Instead, the large number of markers enabled us to identify genomic regions that may hold genes involved in post-mating processes that might ensure genetic compatibility at linked loci important in fitness. Prior work documenting a fitness benefit from this post-mating selection in Chinook salmon [14] supports this concept.
However, the possibility that the CFC might be linked to decreasing the effects of inbreeding cannot be completely dismissed. Salmonids show local adaptation to their breeding grounds [51], and low levels of inbreeding can be beneficial to preserve this adaptation [52]. However, mechanisms to balance the benefits and the costs of inbreeding are thought to exist. For example, alternative mating tactics by sneaker males are thought to counteract the costs of body-size-dependent assortative mating [53]. Possibly, post-mating mechanisms are another option to counteract inbreeding on specific genomic regions. Unfortunately, the lack of knowledge on the genetic loci involved in post-mating processes makes this speculative until further studies have provided empirical evidence.
We controlled for many specific female and male effects such as sperm number and ovarian fluid volume, so we might focus on the effects of the female–male interactions previously documented [13,14,27]. Nevertheless, we modelled the female and male identities to account for all individual characteristics (e.g. age or milt and ovarian fluid traits, which could not be explicitly measured or controlled for in this study). Although female and male effects might be more influential in natural spawning, they explained only minor amounts of variation in our data. We detected variation in fertilization success that was neither caused by differences in sperm velocity nor by female or male effects. Thus, our data indicate that other factors, possibly sperm–egg interactions as found in other external fertilizers [5,6], might explain the non-random gamete fusion.
Relatedness on specific LGs did not fully explain the variation in sperm velocity and fertilization success. Possibly, a higher marker density would be more powerful to detect genomic regions of importance that we might have missed. Further, the composition and protein levels in the ovarian fluid [30,54] could potentially explain some of the remaining variance in female–male compatibility.
We confirmed our results in experimental replicates for all trials, but correcting for multiple testing suggested that some of the results should be considered with some caution. However, the necessity of adjusting for multiple testing is questioned by some researchers [55]. Moreover, LG12 was linked with both sperm velocity and differences in fertilization success that were not explained by sperm velocity. This increases the likelihood that LG12 contains genes important to CFC.
In conclusion, we detected that female–male relatedness on four SNP-LGs predicted sperm velocity in ovarian fluid and fertilization success. As sperm velocity in ovarian fluid determines male fertilization success in Chinook salmon ([14] and this study), our data suggest the presence of genes on these LGs that influence CFC outcomes. Further, we found that mate relatedness on two LGs explained the variation in fertilization success that was not explained by sperm velocity, indicating that these LGs might contain genes mediating non-random gamete fusion. Possibly, these genes code for peptides expressed on the gamete surfaces similar to peptides in marine invertebrates [5,6]. On two LGs, we detected genes with known roles in fertility in other species, thus representing candidate genes for further studies of a possible role in CFC.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We express our gratitude to New Zealand King Salmon for access to the pedigree information and Chinook salmon family samples to develop the SNP chip and linkage map. We are especially thankful to Dr Patrice Rosengrave for her efforts at field work. We are grateful to Dr Shinichi Nakagawa, Dianne Hyndmann, Ken Dodds, Janine Wing, Joanne Gillum and Dr Monika Zavodna for their help with data analysis, field work, Sanger sequencing and for general advice. We are also thankful to the staff at the Silverstream hatchery of Salmon Smolt NZ.
Ethics
All experiments were performed according to protocols approved by the Animal Ethics Committee of the University of Otago.
Data accessibility
This article has no additional data.
Authors' contribution
N.J.G. developed the original idea and the experimental design. S.L.J., N.J.G. and C.G. contributed to field work. C.G. conducted the laboratory work and analysed the data with the help of P.F. and K.R. C.G. wrote the manuscript. All authors contributed to refining the manuscript. J.S., P.F. and S.C. developed the Chinook salmon SNP resources.
Competing interests
The authors declare that there is no conflict of interest.
Funding
This research was supported via a Royal Society of New Zealand Marsden Fund grant UOO-0913 to N.J.G. and a University of Otago Doctoral Scholarship to C.G. Funding for the development of the 6000 SNP chip was received through the National Institute of Water and Atmospheric Research Breeding and Genetic Technologies Programme 3 (2010/2011, 2011/12 SCI).
References
- 1.Birkhead TR, Pizzari T. 2002. Postcopulatory sexual selection. Nat. Rev. Genet. 3, 262–273. ( 10.1038/nrg774) [DOI] [PubMed] [Google Scholar]
- 2.Parker GA. 1970. Sperm competition and its evolutionary consequences in the insects. Biol. Rev. 45, 525–567. ( 10.1111/j.1469-185X.1970.tb01176.x) [DOI] [Google Scholar]
- 3.Birkhead TR, Møller AP. 1998. Sperm competition and sexual selection. New York, NY: Academic Press. [Google Scholar]
- 4.Ward PI. 2007. Postcopulatory selection in the yellow dung fly Scathophaga stercoraria (L.) and the mate-now-choose-later mechanism of cryptic female choice. Adv. Study Behav. 37, 343–369. ( 10.1016/S0065-3454(07)37007-1) [DOI] [Google Scholar]
- 5.Palumbi SR. 1999. All males are not created equal: fertility differences depend on gamete recognition polymorphisms in sea urchins. Proc. Natl Acad. Sci. USA 96, 12 632–12 637. ( 10.1073/pnas.96.22.12632) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scofield VL, Schlumpberger JM, West LA, Weissman I. 1982. Protochordate allorecognition is controlled by a MHC-like gene system. Nature 295, 499–502. ( 10.1038/295499a0) [DOI] [PubMed] [Google Scholar]
- 7.Eberhard WG. 1996. Female control: sexual selection by cryptic female choice. Princeton, NJ: Princeton University Press. [Google Scholar]
- 8.Thornhill R, Alcock J. 1983. The evolution of insect mating systems. Cambridge, MA: Harvard University Press. [Google Scholar]
- 9.Markow TA. 1997. Assortative fertilization in Drosophila. Proc. Natl Acad. Sci. USA 94, 7756–7760. ( 10.1073/pnas.94.15.7756) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stockley P. 1999. Sperm selection and genetic incompatibility: does relatedness of mates affect male success in sperm competition? Proc. R. Soc. Lond. B 266, 1663–1669. ( 10.1098/rspb.1999.0829) [DOI] [Google Scholar]
- 11.Olsson M, Shine R, Madsen T, Gullberg A, Tegelstrom H. 1996. Sperm selection by females. Nature 383, 585 ( 10.1038/383585a0) [DOI] [Google Scholar]
- 12.Evans JP, Rosengrave P, Gasparini C, Gemmell NJ. 2013. Delineating the roles of males and females in sperm competition. Proc. R. Soc. B 280, 20132047 ( 10.1098/rspb.2013.2047) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosengrave P, Gemmell NJ, Metcalf V, McBride K, Montgomerie R. 2008. A mechanism for cryptic female choice in Chinook salmon. Behav. Ecol. 19, 1179–1185. ( 10.1093/beheco/arn089) [DOI] [Google Scholar]
- 14.Rosengrave P, Montgomerie R, Gemmell N. 2016. Cryptic female choice enhances fertilization success and embryo survival in Chinook salmon. Proc. R. Soc. B 283, 20160001 ( 10.1098/rspb.2016.0001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hart N. 1990. of Sperm-Egg Interactions. Int. Rev. Cytol. 121, 1–66. ( 10.1016/S0074-7696(08)60658-0) [DOI] [PubMed] [Google Scholar]
- 16.Turner E, Montgomerie R. 2002. Ovarian fluid enhances sperm movement in Arctic charr. J. Fish Biol. 60, 1570–1579. ( 10.1111/j.1095-8649.2002.tb02449.x) [DOI] [Google Scholar]
- 17.Boschetto C, Gasparini C, Pilastro A. 2011. Sperm number and velocity affect sperm competition success in the guppy (Poecilia reticulata). Behav. Ecol. Sociobiol. 65, 813–821. ( 10.1007/s00265-010-1085-y) [DOI] [Google Scholar]
- 18.Gage MJ, Macfarlane CP, Yeates S, Ward RG, Searle JB, Parker GA. 2004. Spermatozoal traits and sperm competition in Atlantic salmon: relative sperm velocity is the primary determinant of fertilization success. Curr. Biol. 14, 44–47. ( 10.1007/s00265-010-1085-y) [DOI] [PubMed] [Google Scholar]
- 19.Alonzo SH, Stiver KA, Marsh-Rollo SE. 2016. Ovarian fluid allows directional cryptic female choice despite external fertilization. Nat. Commun. 7, 12452 ( 10.1038/ncomms12452) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yanagimachi R, et al. 2013. Sperm attractant in the micropyle region of fish and insect eggs. Biol. Reprod. 88, 47 ( 10.1095/biolreprod.112.105072) [DOI] [PubMed] [Google Scholar]
- 21.Liljedal S, Rudolfsen G, Folstad I. 2008. Factors predicting male fertilization success in an external fertilizer. Behav. Ecol. Sociobiol. 62, 1805–1811. ( 10.1007/s00265-008-0609-1) [DOI] [Google Scholar]
- 22.Sherman C, Wapstra E, Uller T, Olsson M. 2008. Males with high genetic similarity to females sire more offspring in sperm competition in Peron's tree frog Litoria peronii. Proc. R. Soc. B 275, 971–978. ( 10.1098/rspb.2007.1626) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Butts IA, Johnson K, Wilson CC, Pitcher TE. 2012. Ovarian fluid enhances sperm velocity based on relatedness in lake trout, Salvelinus namaycush. Theriogenology 78, e2101 ( 10.1016/j.theriogenology.2012.06.031) [DOI] [PubMed] [Google Scholar]
- 24.Foote CJ. 1989. Female mate preference in Pacific salmon. Anim. Behav. 38, 721–723. ( 10.1016/S0003-3472(89)80022-3) [DOI] [Google Scholar]
- 25.Searcy WA. 1982. The evolutionary effects of mate selection. Annu. Rev. Ecol. Syst. 13, 57–85. ( 10.1146/annurev.es.13.110182.000421) [DOI] [Google Scholar]
- 26.Gross MR. 1984. Sunfish, salmon, and the evolution of alternative reproductive strategies and tactics in fishes. In Fish reproduction: strategies and tactics (eds Potts GW, Wootton RJ), pp. 55–75. London, UK: Academic Press. [Google Scholar]
- 27.Gessner C, Nakagawa S, Zavodna M, Gemmell N. 2017. Sexual selection for genetic compatibility: the role of the major histocompatibility complex on cryptic female choice in Chinook salmon (Oncorhynchus tshawytscha). Heredity 118, 442–452. ( 10.1038/hdy.2016.116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berthelot C, et al. 2014. The rainbow trout genome provides novel insights into evolution after whole-genome duplication in vertebrates. Nature Commun. 5, 946 ( 10.1038/ncomms4657) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lien S, et al. 2016. The Atlantic salmon genome provides insights into rediploidization. Nature 533, 200–205. ( 10.1038/nature17164) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson SL, Villarroel M, Rosengrave P, Carne A, Kleffmann T, Lokman PM, Gemmell NJ. 2014. Proteomic analysis of Chinook salmon (Oncorhynchus tshawytscha) ovarian fluid. PLoS ONE 9, e104155 ( 10.1371/journal.pone.0104155) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Billard R, Jensen JJO. 1996. Gamete removal, fertilization and incubation. Dev. Aquacult. Fish. Sci. 29, 291–364. ( 10.1016/S0167-9309(96)80008-1) [DOI] [Google Scholar]
- 32.Gemmell NJ, Akiyama S. 1996. An efficient method for the extraction of DNA from vertebrate tissues. Trends Genet. 12, 338–339. ( 10.1016/S0168-9525(96)80005-9) [DOI] [PubMed] [Google Scholar]
- 33.Condrey M, Bentzen P. 1998. Characterization of coastal cutthroat trout (Oncorhynchus clarki clarki) microsatellites and their conservation in other salmonids. Mol. Ecol. (UK) 7, 787–789. [PubMed] [Google Scholar]
- 34.Heath DD, Bryden CA, Shrimpton JM, Iwama GK, Kelly J, Heath JW. 2002. Relationships between heterozygosity, allelic distance (d 2), and reproductive traits in Chinook salmon, Oncorhynchus tshawytscha. Can. J. Fish. Aquat. Sci. 59, 77–84. ( 10.1139/f01-192) [DOI] [Google Scholar]
- 35.Beacham TD, Candy JR, Le KD, Wetklo M. 2009. Population structure of chum salmon (Oncorhynchus keta) across the Pacific Rim, determined from microsatellite analysis. Fish. Bull. 107, 244–260. [Google Scholar]
- 36.Banks M, Blouin M, Baldwin B, Rashbrook V, Fitzgerald H, Blankenship S, Hedgecock D. 1999. Isolation and inheritance of novel microsatellites in Chinook salmon (Oncorhynchus tschawytscha). J. Hered. 90, 281–288. ( 10.1093/jhered/90.2.281) [DOI] [Google Scholar]
- 37.Kalinowski ST, Taper ML, Marshall TC. 2007. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 16, 1099–1106. ( 10.1111/j.1365-294X.2007.03089.x) [DOI] [PubMed] [Google Scholar]
- 38.Wang J. 2007. Triadic IBD coefficients and applications to estimating pairwise relatedness. Genet. Res. 89, 135–153. ( 10.1017/S0016672307008798) [DOI] [PubMed] [Google Scholar]
- 39.Wang J. 2011. COANCESTRY: a program for simulating, estimating and analysing relatedness and inbreeding coefficients. Mol. Ecol. Res. 11, 141–145. ( 10.1111/j.1755-0998.2010.02885.x) [DOI] [PubMed] [Google Scholar]
- 40.R Core Team 2013. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 41.Hadfield JD. 2010. MCMC methods for multi-response generalized linear mixed models: the MCMCglmm R package. J. Stat. Soft. 33, 1–22. ( 10.18637/jss.v033.i02) [DOI] [Google Scholar]
- 42.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser B (Methodological), 57, 289–300. [Google Scholar]
- 43.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. ( 10.1093/nar/25.17.3389) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schork AJ, et al. 2013. All SNPs are not created equal: genome-wide association studies reveal a consistent pattern of enrichment among functionally annotated SNPs. PLoS Genet. 9, e1003449 ( 10.1371/journal.pgen.1003449) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Altschul SF, Wootton JC, Gertz EM, Agarwala R, Morgulis A, Schäffer AA, Yu YK. 2005. Protein database searches using compositionally adjusted substitution matrices. Febs J. 272, 5101–5109. ( 10.1111/j.1742-4658.2005.04945.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Allendorf FW, Waples RS. 1996. Conservation and genetics of salmonid fishes. In Conservation genetics: case histories from nature (eds Avise JC, Hamrick JL), pp. 238–280. New York, NY: Chapman & Hall. [Google Scholar]
- 47.Zadravec D. 2010. Metabolic significance of fatty acid elongation. See https://core.ac.uk/display/36177013. [Google Scholar]
- 48.García-Herrero S, Meseguer M, Martínez-Conejero JA, Remohí J, Pellicer A, Garrido N. 2010. The transcriptome of spermatozoa used in homologous intrauterine insemination varies considerably between samples that achieve pregnancy and those that do not. Fertil. Steril. 94, 1360–1373. ( 10.1016/j.fertnstert.2009.07.1671) [DOI] [PubMed] [Google Scholar]
- 49.Monroig Ó, Webb K, Ibarra-Castro L, Holt GJ, Tocher DR. 2011. Biosynthesis of long-chain polyunsaturated fatty acids in marine fish: Characterization of an Elovl4-like elongase from cobia Rachycentron canadum and activation of the pathway during early life stages. Aquaculture 312, 145–153. ( 10.1016/j.aquaculture.2010.12.024) [DOI] [Google Scholar]
- 50.Hansbrough J, Garbers D. 1981. Speract. Purification and characterization of a peptide associated with eggs that activates spermatozoa. J. Biol. Chem. 256, 1447–1452. [PubMed] [Google Scholar]
- 51.Taylor EB. 1991. A review of local adaptation in Salmonidac, with particular reference to Pacific and Atlantic salmon. Aquaculture 98, 185–207. ( 10.1016/0044-8486(91)90383-I) [DOI] [Google Scholar]
- 52.Wang S, Hard JJ, Utter F. 2002. Salmonid inbreeding: a review. Rev. Fish Biol. Fish. 11, 301–319. ( 10.1023/A:1021330500365) [DOI] [Google Scholar]
- 53.Foote CJ, Brown GS, Wood CC. 1997. Spawning success of males using alternative mating tactics in sockeye salmon, Oncorhynchus nerka. Can. J. Fish. Aquat. Sci. 54, 1785–1795. ( 10.1139/f97-080) [DOI] [Google Scholar]
- 54.Rosengrave P, Montgomerie R, Metcalf V, McBride K, Gemmell N. 2009. Sperm traits in Chinook salmon depend upon activation medium: implications for studies of sperm competition in fishes. Can. J. Zool. 87, 920–927. ( 10.1139/Z09-081) [DOI] [Google Scholar]
- 55.Gelman A, Hill J, Yajima M. 2012. Why we (usually) don't have to worry about multiple comparisons. J. Res. Educ. Effectiveness 5, 189–211. ( 10.1080/19345747.2011.618213) [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This article has no additional data.


